Abstract
Intellectual disability (ID) with autosomal recessive (AR) inheritance is believed to be common; however, very little is known about causative genes and genotype–phenotype correlations. The broad genetic heterogeneity of AR-ID, and its usually nonsyndromic nature make it difficult to pool multiple pedigrees with the same underlying genetic defect to achieve consistent nosology. Nearly all autosomal genes responsible for recessive cognitive disorders have been identified in large consanguineous families from the Middle East, and nonsense mutations in TRAPPC9 have been reported in a total of 5. Although several recurrent phenotypic abnormalities are described in some of these patients, the associated phenotype is usually referred to as nonsyndromic. By means of single-nucleotide polymorphism-array first and then by exome sequencing, we identified a new pathogenic mutation in TRAPPC9 in two Italian sisters born to healthy and apparently nonconsanguineous parents. It consists of a homozygous splice site mutation causing exon skipping with frameshift and premature termination, as confirmed by mRNA sequencing. By detailed phenotypic analysis of our patients, and by critical literature review, we found that homozygous TRAPPC9 loss-of-function mutations cause a distinctive phenotype, characterized by peculiar facial appearance, obesity, hypotonia (all signs resembling a Prader–Willi-like phenotype), moderate-to-severe ID, and consistent brain abnormalities.
Keywords: autosomal recessive intellectual disability, TRAPPC9 , exome sequencing
Introduction
Autosomal recessive (AR) gene defects are thought to be responsible for about a quarter of idiopathic nonsyndromic intellectual disability (NSID) cases.1, 2, 3
Few autosomal recessive (AR) genes and several other loci have been identified by means of linkage analysis and genome-wide homozigosity mapping in large consanguineous families followed by resenquencing of candidate genes. These include PRSS12 (MIM 606709),4 CRBN (MIM 609262),5 CC2D1A (MIM 610055),6 GRIK2 (MIM 138244),7 TUSC3 (MIM 601385),8, 9 and TRAPPC9 (MIM 611966).10, 11, 12, 13
The introduction of next generation sequencing techniques dramatically changed this scenario, leading to the discovery of a rapidly increasing number of AR-NSID causative genes. In addition to mutations in TECR (MIM 610057),14 MAN1B1 (MIM 604346),15 and ST3GAL3 (MIM 606494),16 32 putative novel genes causative of AR-NSID have been recently reported by Najmabadi et al.17
Of note, the latter studies used an exome sequencing approach targeted to chromosome regions identified by means of linkage analysis, limiting the search of causative genes to families with high imbreeding coefficient, rarely found in western countries.
Mutations in the above mentioned genes were detected in single kindreds, with the exception of variants in TUSC3, TRAPPC9, MAN1B1, ST3GAL3, ELP2, and ZNF526, that were reported in two to five pedigrees.
Genetic heterogeneity of AR-ID, (intellectual disability) its usual nonsyndromic nature and the few reported cases make it difficult to achieve consistent genotype–phenotype correlations.
Here, we describe two sisters with ID and phenotypic abnormalities, in whom a homozygous splice site mutation of TRAPPC9 was identified by means of exome sequencing analysis. Homozygous mutations of TRAPPC9 have so far been considered to cause a nonsyndromic form of ID. However, on the basis of deep phenotyping of our patients and critical literature review, we found that definition of a distinctive phenotype is possible.
Materials and methods
Patients
Two sisters were referred to our clinical genetics service for severe developmental delay. They are the only children of healthy parents, originating from the South of Italy. As referred, parents are not related.
The older sister was born at 38 weeks of gestation by cesarean section because of placenta previa. Birth-weight was 3000, g (50th centile) and length 50 cm (75th centile). Hypotonia was diagnosed soon after birth. At 14 months, following the diagnosis of subclinical hypothyroidism, opotherapy was started successfully. She walked unsupported at 18 months. Frequent sleep awakenings were referred and at the age of 19 and 26 months, respectively, she suffered from two generalized myoclonic seizures under hyperpyrexia. Mild focal epileptic alterations were recorded by asleep EEG. Since the last seizure she has been successfully treated with sodium valproate. On examination at 5 years, her height was 117 cm (50th centile), weight 31 kg (97th centile) and head circumference 48.5 cm (10th centile). She presented with a peculiar craniofacial appearance, including brachycephaly, bitemporal narrowing, round face, hypoplastic supraorbital ridges, hypertelorism, straight eyebrows, synophrys, wide nasal bridge, fleshy ears, and thin upper lip with downturned mouth (Figures 1a and b). She also had tapering fingers and clinodactyly of the fifth finger, bilaterally.
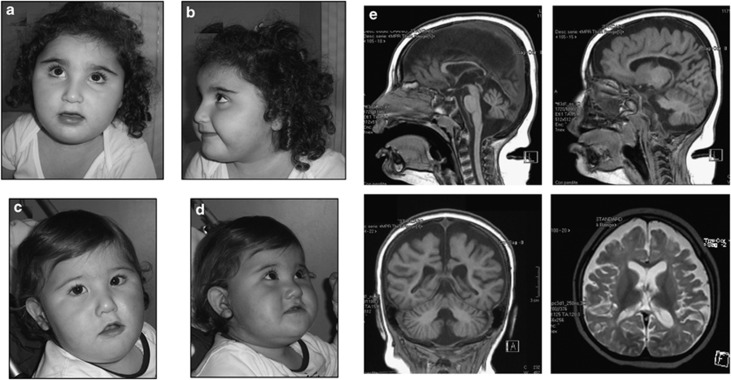
Figure 1.
Frontal (a) and lateral (b) views of older sister and frontal (c) and lateral (d) views of younger sister. Please note the common facial features: bitemporal narrowing, round face, hypoplastic supraorbital ridges, hypertelorism, straight eyebrows, synophrys, wide nasal bridge, and thin upper lip. (e) Brain MRI of the oldest sister at the age of 33/12 years showing: atrophy/hypoplasia of cerebellum and corpus callosum; multiple confluent areas of cerebral white matter signal alteration on T2-weighted sequences; atrophy/hypoplasia of the supratentorial white and gray matter.
She was severely intellectual disabled and exhibited a friendly disposition and could ask for a toy by pointing it out, but was not able to utter single words and understood only simple situational orders. The Composite Score at the Bayley Scales was 55 (corresponding to a mental age of 11 months) and Vineland Adaptive Behavior Scales detected a global maladaptive behavior corresponding to an equivalent age lower than 18 months in all domains. Brain MRI revealed diminished cerebral white matter volume, with marked sulcal enlargement, thinning of the corpus callosum, and reduced cerebellar volume. In addition, several areas of T2 hyperintensity were present in the subcortical white matter (Figure 1e). Heart and kidney ultrasound examinations were normal.
The second sister was born at 36 weeks of gestation by cesarean section. Birth-weight was 2550, g (25th centile) and length 50 cm (90th centile). Hypotonia was noted from early on. Her psychomotor milestones were delayed: she shuffled until the age of 36 months when she was able to walk unsupported, but she never was able to stand up or utter single words. On examination at 36/12 years, her height was 102 cm (75th centile), weight 18 kg (90th centile) and head circumference 46 cm (<2nd centile). She presented with a peculiar craniofacial appearance, with brachycephaly, round face, hypertelorism, epicantus, straight eyebrows, synophrys, wide and depressed nasal bridge, prominent auricles with uplifted lobe, and downslanting corners of the mouth (Figure 1c-d). She was severely intellectually disabled and suffered from sporadic temper tantrums and frequent awakenings, which were not associated with seizures or epileptic EEG abnormalities. Brain MRI showed alterations similar to but less severe than those detected in the sister.
Methods
We first performed array-CGH (Agilent 44 k, 100 kb resolution), methylation-sensitive MLPA of the PWS/AS region and UBE3A sequencing analysis, with normal results.
Further genetic studies included: (1) single-nucleotide polymorphism (SNP) array analysis in propositae and parents, using Illumina Infinitum Platform (Illumina Inc., San Diego, CA, USA) with the HumanCytoSNP-12 BeadChip, according to the manufacturer's instructions (acquired data was visualized and analyzed with Genome Studio Data Analysis Software v.2008); (2) familial segregation analysis of markers D8S1836, D8S1717, D8S1837, and D8S537 mapping on 8q24; (3) whole exome sequencing, that was commissioned to an external service (IGA Technology Service, Udine, Italy) on DNAs enriched for exonic regions with SureSelect 38Mbp All exon kit v. 2.0 (Agilent Technologies, Santa Clara, CA, USA), prepared according to manufacturer protocols and 75 × 2 bp paired-end sequenced on HiSeq2000 (Illumina Inc.), with a 50–80x mean coverage. Reads were aligned to reference genome NCBI build 37/UCSC hg19 with CASAVA software (Illumina Inc.) and variant calling (SNPs, single-nucleotide polymorphisms, and DIPs, deletion insertion polymorphisms) was performed with CLC Bio Genomics Workbench software (CLC bio, Aarhus, Denmark). Finally, variant filtering was performed by using ANNOVAR software;18 (4) direct sequencing of PCR products of exon 18 of TRAPPC9 (forward primer: 5′-GCCTCCTGGTGGATCAAACT-3′ reverse primer: 5′-ACGGGATGCATGACAAAAAC-3′), and cDNA sequencing of exons 17 to 19 of TRAPPC9 (forward primer: 5′-CT TTTCGGCAGGTCGTTCGGC-3′ reverse primer: 5′-GGCGTGCAGGATGAGTGCCT-3′); 5) semi-quantitative real-time PCR on cDNA by using TRAPPC9-specific TaqMan Gene Expression Assay Hs00230278_m1 (Applied Biosystems, Foster City, CA, USA). The comparative threshold cycle method was used to analyze the expression of TRAPPC9 in affected individuals, in their parents and in control subjects.
Results
SNP-array analysis showed a single long continuous stretch of homozigosity of about 13 Mb on 8q24 in both sisters (Figure 2a). Of note, both parents shared a common haplotype in this region. On segregation analysis of the informative markers 1837 and 1717, we found that the single allele present in both sisters was present in heterozygous state in both parents, leading us to establish that the same haplotype was inherited from each parent. As more than 110 protein coding genes are included in the 8q homozygous region, we decided to perform exome sequencing analysis. In total, 550 variants within the 8q homozygous chromosome region were disclosed. Of these, 494 were non-coding, 31 were synomymous, and 25 were non-synonymous variants already reported as SNPs in the dbSNP 132 (and with allele frequencies higher than 13% in 1000genomes database). Among the non-coding variants there was a single novel acceptor splice site variant before exon 18 of TRAPPC9, c.2851-2A>C, that seems to be the only interesting variant in the region.
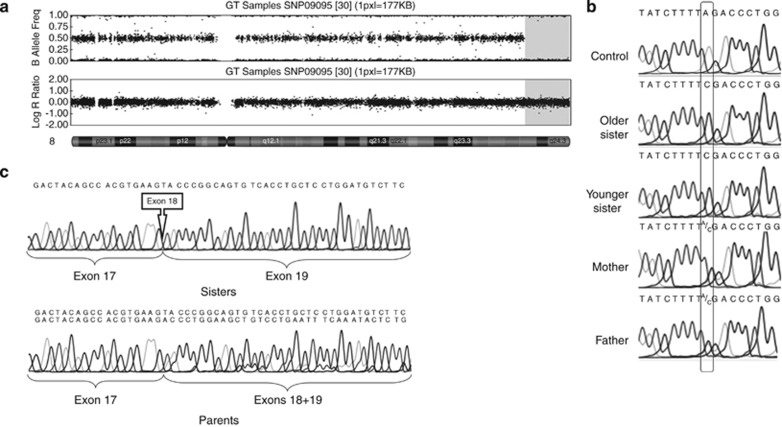
Figure 2.
(a) Graphical overview of SNP-array (HumanCytoSNP-12 BeadChip) results regarding chromosome 8, obtained with Genome Studio Data Analysis Software v.2008 (Illumina Inc., San Diego, CA, USA). Upper panel showing B-allele frequency demonstrates a long region of homozigosity in the terminal region of the long arm of chromosome 8. The second panel reports the Log R Ratio, indicating that no deletion is present in the same chromosome 8 region. (b) Electropherograms of sense strand sequence of TRAPPC9 exon 18 and flanking region. Nucleotide positions from 2851–7 to 2855 are shown. Rectangles indicate the mutated nucleotide of the acceptor splice site (c.2851-2A>C). (c) Electropherograms of TRAPPC9 cDNA direct sequencing. Upper panel: patients' TRAPPC9 cDNA sequences, showing the complete absence of exon 18 from TRAPPC9 mature mRNA. Lower panel: parents' TRAPPC9 cDNA sequences, demonstrating the simultaneous presence of both TRAPPC9 mature transcripts containing and missing exon 18, respectively (peaks corresponding to the longer transcript including exon 8 are lower than peaks from the shorter transcripts probably due to PCR artifacts).
By direct sequencing, this variant was confirmed to be present at a homozygous state in both sisters, while both parents were heterozygous carriers (Figure 2b). On cDNA sequencing, complete absence of exon 18 from the TRAPPC9 transcript was observed in both patients, while parents were heterozygous for both the normal and the shortened form of the transcript (Figure 2c).
As a result, the mutation causes skipping of exon 18 from the mature transcript (NM_031466.5), shift of the coding frame and introduction of a premature stop codon at amino-acid position 967 (p.T951Y fsX17).
Semi-quantitative real-time PCR analysis did not show reduction of TRAPPC9 transcript in both sisters.
Discussion
To date, five different families have been reported with AR-NSID caused by homozygous TRAPPC9 mutations.10, 11, 12, 13 TRAPPC9 variants consisted of the same nonsense mutation (p.R475X) in three families (a Pakistani, a Syrian and an Israeli-Arab family, respectively), suggesting the derivation from a common ancestor. The remaining two variants were a nonsense mutation (p.R570X) and an out-of-frame deletion of four nucleotides (c.2311-2314 delTGTT) resulting in the introduction of a premature stop codon (p.L772W fsX7) in a Tunisian and an Iranian family, respectively. Confirming that all the TRAPPC9 pathogenic variants reported so far are loss-of-function mutations, Mir et al10 and Philippe et al11 demonstrated a significant degree of nonsense-mediated mRNA decay in homozygous patients by means of quantitative real-time PCR, and Mochida et al12 and Philippe et al11 observed the almost complete absence of TRAPPC9 protein by western blot analysis.
The previously unreported TRAPPC9 variant found in our patients consisted of a splice site mutation causing skipping of exon 18 with frameshift and introduction of a premature stop codon at amino-acid position 967 (p.T951Y fsX17). A nonfunctional truncated protein can be reasonably expected, as no mRNA decay seemed to occur.
TRAPPC9 protein, also known as NIBP (NIK- and IKK-b-binding protein), has been shown to enhance in vitro TNFα-induced NF-kB activation and to be involved in nerve growth factor-induced neuronal differentiation.19 NF-kB signaling is involved in a wide range of biological processes, including inflammation, immunity, and synaptic plasticity, and it has also been suggested to regulate adult neurogenesis.20
Clinically, most literature patients with TRAPPC9 mutations are described as affected by a nonsyndromic form of ID, although recurrent brain abnormalities and obesity were described in some of them. We made an extensive phenotypic analysis of our patients, that included fine evaluation of both the physical and the brain phenotype, and a 2-year-period of follow-up. The more relevant clinical signs were a distinctive craniofacial appearance, true or relative microcephaly, obesity, early onset hypotonia, and peculiar brain abnormalities, consisting of shrunken cerebrum and cerebellum, thin corpus callosum and supratentorial white matter multifocal abnormalities. All the alterations so far reported in different cases occur in our patients.10, 11, 12, 13 Even though these features would suggest cortical atrophy with secondary degeneration of white matter, serial examinations are required to prove the progressive volume loss. Postnatal microcephaly as well as the less severe brain involvement in the younger sister suggest a progressive neuropathological disorder, at least during the first years of life.
In addition, both sisters presented with severe ID, absent speech and friendly behavior. One sister suffered from epilepsy, usually with fever, in the first 3 years of life. In contrast, stereotypic movements, hyperactivity and autism are no part of behavioral phenotype of our patients.
Of note, some of the above mentioned features are also reported in some of the literature patients, even though the whole clinical phenotype is sparsely described. We tentatively consider that the clinical phenotype associated with homozygous loss-of-function mutations of TRAPPC9 is defined by the association of typical facial appearance, obesity, early onset hypotonia (all signs resembling a Prader–Willi-like phenotype), moderate-to-severe ID and highly specific brain abnormalities (Table 1).
Table 1. : summary of the clinical signs associated with TRAPPC9-related AR-ID.
| Clinical sign | Literature patients (22/5 kindreeds) | Patient 1 | Patient 2 | Total | % |
|---|---|---|---|---|---|
| Microcephaly | 17/18 | + | + | 19/20 | 95 |
| Obesity | 4/6 | + | + | 6/8 | 75 |
| Brain abnormalities | 7/7 | + | + | 9/9 | 100 |
| Thin corpus callosum | 5 | + | + | 7/9 | |
| Cerebellar hypoplasia | 5 | + | + | 7/9 | |
| Cerebral white/gray matter hypoplasia | 5 | + | + | 7/9 | |
| Multifocal white matter lesions | 5 | + | + | 7/9 | |
| Peculiar facial appearancea | 9/18? | + | + | ||
| Epilepsy | 2/18 | + | − | 3/20 | 15 |
Abbreviations: Ar, autosomal recessive; ID, intellectual disability.
Insufficient literature data.
Our patients share a large 8q chromosome region, suggesting that different homozygous variants in this region can affect the final phenotype. However, by comparing the genetic profile with clinical manifestations, and also in considering their high frequencies in the general population, none of the 25 missense variants were considered to be functionally significant. Our family is the first Caucasian one described so far. Thus, a different genetic background could be hypothesized to affect the final phenotype. However, clinical manifestations of literature patients, although limited in some reports, are well consistent with our observations (Table 1).
The described cases represent the first report of TRAPPC9-related AR-ID in a western European family.
In addition to confirm the leading role that next generation sequencing techniques will has in the near future in the identification of the underlying causes of idiopathic ID, the present report provides evidence that a clinical nosology of AR forms of ID can be tentatively made by combining detailed phenotyping evaluation of patients with deep genotyping.
The authors declare no conflict of interest.
Footnotes
Web Resources
Online Mendelian Inheritance in Man (OMIM), http://www.ncbi.nlm.nih.gov/Omim/.
ANNOVAR website, http://www.openbioinformatics.org/annovar/.
The Single-Nucleotide Polymorphism database (dbSNP), www.ncbi.nlm.nih.gov/projects/SNP/.
1000genomes, http://www.1000genomes.org/.
Note: Patient consent was obtained for conducting the study.
References
- Wright SW, Tarjan G, Eyer L. Investigation of families with two or more mentally defective siblings: clinical observations. Am J Dis Child. 1959;97:445–456. doi: 10.1001/archpedi.1959.02070010447010. [DOI] [PubMed] [Google Scholar]
- Priest JH, Thuline HC, Laveck GD, Jarvis DB. An approach to genetic factors in mental retardation. Studies of families containing at least two siblings admitted to a state institution for the retarded. Am J Ment Defic. 1961;66:42–50. [PubMed] [Google Scholar]
- Bartley JA, Hall BD. Mental retardation and multiple congenital anomalies of unknown etiology: frequency of occurrence in similarly affected sibs of the proband. Birth Defects Defects Orig Artic Ser. 1978;14:127–137. [PubMed] [Google Scholar]
- Molinari F, Rio M, Meskenaite V, et al. Truncating neurotrypsin mutation in autosomal recessive nonsyndromic mental retardation. Science. 2002;298:1779–1781. doi: 10.1126/science.1076521. [DOI] [PubMed] [Google Scholar]
- Higgins JJ, Pucilowska J, Lombardi RQ, Rooney JP. A mutation in a novel ATP-dependent Lon protease gene in a kindred with mild mental retardation. Neurology. 2004;63:1927–1931. doi: 10.1212/01.wnl.0000146196.01316.a2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Basel-Vanagaite L, Attia R, Yahav M, et al. The CC2D1A, a member of a new gene family with C2 domains, is involved in autosomal recessive non-syndromic mental retardation. J Med Genet. 2006;43:203–210. doi: 10.1136/jmg.2005.035709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Motazacker MM, Rost BR, Hucho T, et al. A defect in the ionotropic glutamate receptor 6 gene (GRIK2) is associated with autosomal recessive mental retardation. Am J Hum Genet. 2007;81:792–798. doi: 10.1086/521275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garshasbi M, Hadavi V, Habibi H, et al. A defect in the TUSC3 gene is associated with autosomal recessive mental retardation. Am J Hum Genet. 2008;82:1158–1164. doi: 10.1016/j.ajhg.2008.03.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molinari F, Foulquier F, Tarpey PS, et al. Oligosaccharyltransferase-subunit mutations in nonsyndromic mental retardation. Am J Hum Genet. 2008;82:1150–1157. doi: 10.1016/j.ajhg.2008.03.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mir A, Kaufman L, Noor A, et al. Identification of mutations in TRAPPC9, which encodes the NIK- and IKK-beta-binding protein, in nonsyndromic autosomal-recessive mental retardation. Am J Hum Genet. 2009;85:909–915. doi: 10.1016/j.ajhg.2009.11.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Philippe O, Rio M, Carioux A, et al. Combination of linkage mapping and microarray-expression analysis identifies NF-kappaB signaling defect as a cause of autosomal-recessive mental retardation. Am J Hum Genet. 2009;85:903–908. doi: 10.1016/j.ajhg.2009.11.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mochida GH, Mahajnah M, Hill AD, et al. A truncating mutation of TRAPPC9 is associated with autosomal-recessive intellectual disability and postnatal microcephaly. Am J Hum Genet. 2009;85:897–902. doi: 10.1016/j.ajhg.2009.10.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abou Jamra R, Wohlfart S, Zweier M, et al. Homozygosity mapping in 64 Syrian consanguineous families with non-specific intellectual disability reveals 11 novel loci and high heterogeneity. Eur J Hum Genet. 2011;19:1161–1166. doi: 10.1038/ejhg.2011.98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Çalışkan M, Chong JX, Uricchio L, et al. Exome sequencing reveals a novel mutation for autosomal recessive non-syndromic mental retardation in the TECR gene on chromosome 19p13. Hum Mol Genet. 2011;20:1285–1289. doi: 10.1093/hmg/ddq569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rafiq MA, Kuss AW, Puettmann L, et al. Mutations in the alpha 1,2-mannosidase gene, MAN1B1, cause autosomal-recessive intellectual disability. Am J Hum Genet. 2011;89:176–182. doi: 10.1016/j.ajhg.2011.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu H, Eggers K, Chen W, et al. ST3GAL3 Mutations Impair the Development of Higher Cognitive Functions. Am J Hum Genet. 2011;89:407–414. doi: 10.1016/j.ajhg.2011.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Najmabadi H, Hu H, Garshasbi M, et al. Deep sequencing reveals 50 novel genes for recessive cognitive disorders. Nature. 2011;478:57–63. doi: 10.1038/nature10423. [DOI] [PubMed] [Google Scholar]
- Wang K, Li M, Hakonarson H. ANNOVAR: Functional annotation of genetic variants from next-generation sequencing data. Nucleic Acids Res. 2010;38:e164. doi: 10.1093/nar/gkq603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu WH, Pendergast JS, Mo XM, et al. NIBP, a novel NIK and IKK(beta)-binding protein that enhances NF-(kappa)B activation. J Biol Chem. 2005;280:29233–29241. doi: 10.1074/jbc.M501670200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denis-Donini S, Dellarole A, Crociara P, et al. Impaired adult neurogenesis associated with short-term memory defects in NF-kappaB p50-deficient mice. J Neurosci. 2008;28:3911–3919. doi: 10.1523/JNEUROSCI.0148-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]