Abstract
Benign Adult Familial Myoclonic Epilepsy (BAFME) is an autosomal dominant disorder characterized by adult-onset cortical tremor or action myoclonus predominantly in the upper limbs, and generalized seizures. We investigated a Thai BAFME family. Clinical and electrophysiological studies revealed that 13 were affected with BAFME. There were a total of 24 individuals studied. Genetic analysis by genome-wide linkage study (GWLS) was performed using 400 microsatellite markers and excluded linkage of the previous BAFME loci, 8q23.3-q24.1, and 2p11.1-q12.2. GWLS showed that the disease-associated region in our Thai family was linked to a newly identified locus on chromosome 3q26.32-3q28. This locus represents the fourth chromosomal region for BAFME.
Keywords: benign adult familial myoclonic epilepsy, BAFME, genome-wide linkage study
Introduction
Benign Adult Familial Myoclonic Epilepsy (BAFME) is an autosomal dominant disorder characterized by adult-onset cortical tremor or action myoclonus predominantly in the upper limbs, and generalized seizures.1 It is also known as familial cortical myoclonic tremor with epilepsy. BAFME diagnosis is based on clinical and electrophysiological criteria. An electrophysiological study is essential to confirm the cortical origin of myoclonus. It consists of features favoring cortical reflex myoclonus (enhanced long latency reflexes at rest, giant cortical somatosensory evoked potentials (SEPs), premyoclonus cortical spikes detected by the jerk-locked back averaging (JLA) method), and a good response to antiepileptic drugs.2 BAFME was first reported in 1990 in the Japanese population.3 Until now about 50 Japanese, 10 European families, and a French family were reported.4
Previous studies revealed that BAFME was mapped to three chromosomal regions on chromosome 8,5, 6 chromosome 2,7, 8 and chromosome 54 in the Japanese, European and French populations, respectively. Mori et al9 remapped the BAFME locus in a large Japanese pedigree and they reconfirmed positive linkage on chromosome 8q. However, a study of one BAFME family in China showed the absence of linkage to the first two loci.10 This study provided evidence for the fourth locus causing BAFME.
Here, we studied the first BAFME pedigree in Thailand consisting of 13 affected members. Genome-wide linkage analysis identified a new locus spanning 10.0 Mb on chromosome 3.
Materials and methods
Clinical descriptions
We identified a BAFME family in Thailand (Figure 1). General information including age, gender, age of onset of cortical tremor and generalized seizures, frequency of seizures, severity of tremors, family history, and medical history was collected (Table 1). Of 24 individuals, 13 were affected. All had cortical tremor while seven had generalized seizures. Only one patient, III-16 (Table 1), had seizures 2 years before the onset of tremor, while the others had hand tremor before the onset of seizures. The average age of onset was 19.5 (range 10–33) years for tremor and 25 (range 19–33) years for seizures. None had cognitive impairment. After adequate treatment with either valproic acid (VPA), levetiracetam or clonazepam (CZP), all except one patient with seizures remained seizure free (Table 1).
Figure 1.
(a) Pedigree of a Thai family with BAFME. Genotypes for the six markers of the linked loci are shown. The fourth BAFME locus is indicated in the rectangle. (b) Genetic map of the 11 markers for fine mapping (left) and their two-point LOD scores (right).
Table 1. Clinical and neurophysiological findings in 13 patients with benign adult familial myoclonic epilepsy.
| Age of onset (years) | Electrophysiological study | ||||||||||
|
Patient ID |
Gender |
Age at follow-up (years) |
Cortical tremor |
Generalized seizures |
EEG |
Photic |
Giant SEPs |
C-reflex |
JLA |
Medication |
Seizure frequency in the past year |
| II-1 | M | 63 | 33 | 33 | Multifocal PSW | − | N/A | N/A | N/A | Rivotril, PB | GTC once a year, Myoclonus once a month |
| II-4 | F | 62 | 20 | None | Multifocal PSW | + | N/A | N/A | N/A | None | None |
| II-5 | F | 62 | 28 | None | Generalized sharp wave | − | + | + | N/A | None | None |
| II-9 | M | 59 | 12 | 26 | Multifocal PSW | + | − | + | N/A | VPA 400, CZP 0.5 | None |
| II-11 | M | 57 | 10 | None | N/A | N/A | + | + | N/A | None | None |
| III-2 | M | 31 | 21 | 25 | N/A | − | − | + | N/A | CZP 0.5, PPN 20 | None |
| III-3 | M | 30 | 17 | 20 | Multifocal PSW | − | − | + | N/A | VPA 500, CZP 2 | None |
| III-4 | M | 34 | 20 | None | Normal | − | − | + | N/A | PPN 80 | None |
| III-7 | F | 31 | 25 | 30 | N/A | N/A | + | + | N/A | None | None |
| III-10 | F | 32 | 10 | None | Multifocal PSW | + | + | + | N/A | None | None |
| III-14 | F | 22 | 17 | None | Multifocal PSW | + | + | + | N/A | None | None |
| III-15 | M | 28 | 19 | 24 | Multifocal PSW | − | + | + | + | LVT 4000, CZP 4 | None |
| III-16 | F | F, 27 | 21 | 19 | Generalized sharp wave | + | + | + | + | CZP 2, LVT 1500 | None |
Abbreviations: CZP, clonazepam; EEG, electroencephalographic; GTC, generalized tonic-clonic convulsion; JLA, jerk-locked back averaging; LVT, levetiracetam; N/A, not available; PB, Phenobarbital; PPN, propanolol; PSW, polyspikes and wave; SEPs, somatosensory evoked potentials; VPA, valproic acid; −, absence; +, presence.
Electrophysiological findings
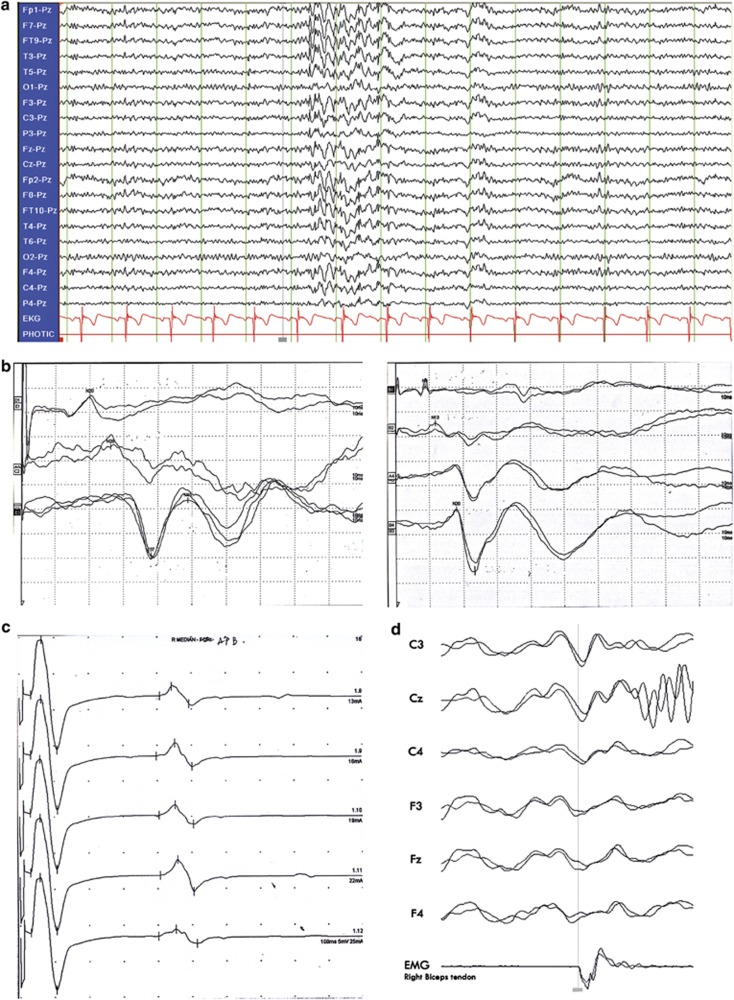
Of the 10 patients available for electroencephalographic studies, 9 had spike and wave or polyspike and wave discharges (Figure 2a). Of these nine, seven had multifocal discharges corresponding with contralateral jerky movement and photosensitivity (photoparoxysmal response, photomyoclonic response). SEPs and long latency reflex (C-reflex) were studied in 11 patients (Table 1). Seven patients had increased amplitudes of the cortical SEPs in the median SEPs study (Figure 2b) and four patients in the tibial SEPs study, while all had the C-reflex (Figure 2c and Table 1). We could demonstrate the cortical discharges by JLA (Figure 2d), which indicated cortical myoclonus, in two available patients. Clinical and neurophysiological studies are summarized in Table 1.
Figure 2.
(a) EEG showed generalized paroxysmal sharp and slow complex wave (mainly anterior head regions). (b) SEP of right tibial (left panel) and left median nerve (right panel) showed giant cortical potential in Patient III-14. (c) C-reflex was found during submaximal stimulation of the right median nerve of Patient III-15. (d) Jerk-locked averaging analysis showing a positive–negative potential over the contralateral centroparietal electrodes, preceding myoclonus about 22 ms (right biceps brachii; 2 averaging, 200 each).
Genotyping genetic markers and linkage analysis
After informed consent, genomic DNA was extracted from peripheral blood leukocytes of 24 family members using ArchivePure DNA Blood Kit (5 Prime Inc., Gaithersburg, MD, USA). This study was approved by the institutional review board of the Faculty of Medicine of Chulalongkorn University. We first performed linkage analysis with the first two known loci on chromosome 8q23.3-q24.1 and 2p11.1-q12.2. The third locus of BAFME was published after we finished the GWLS.4 Using seven microsatellite markers (D8S1830, D8S555, D8S588, D8S1112, D8S1826, D8S572-18, and D8S1799) on chromosome 8 and three markers (D2S388, D2S2175, D2S2264) on chromosome 2, we were able to exclude linkage to these loci. The details of the primers were obtained from Marshfield map (http://research.marshfieldclinic.org/ genetics/home/index.asp). We typed all fluorescently labeled primers on an ABI Prism 3100 genetic analyzer (Applied Biosystems, Foster City, CA, USA) with GeneMapper software (Applied Biosystems). MLINK program (available from http://linkage.rockefeller.edu/) was used to calculate the two-point linkage analysis with the following model: autosomal dominant inheritance with high penetrance, set at 0.01 and 0.99.
Genome-wide linkage study (GWLS)
GWLS was performed using ABI Prism Linkage Mapping Sets-MD10 Version 2.5 (Applied Biosystems). This set consists of 400 fluorescently labeled primer pairs selected to amplify dinucleotide repeats that define an average 10 centimorgan (cM) resolution human index map.
Narrowing down the critical region
After the linked locus was identified, we selected 11 additional markers (D3S2421, D3S3676, D3S2427, D3S3037, D3S3730, D3S1571, D3S3609, D3S3592, D3S1602, D3S3686, and D3S3651) in the region on chromosome 3 for fine mapping.
Results
Absence of linkage to 8q23.3-q24.1 and 2p11.1-q12.2
Two-point linkage analysis generated negative LOD scores in every microsatellite marker at all the recombination values from 0.00 to 0.50. The LOD scores were equal to minus infinity at recombination rate of 0.00, indicating no linkage to any of the selected markers (data not shown). As a result, linkage to the two chromosome regions 8q23.3-q24.1 and 2p11.1-q12.2 was excluded.
Our results from GWLS also showed the absence of linkage to chromosome 5p15.31-p15, the third locus for BAFME. The LOD scores were equal to minus infinity with the markers D5S630 and D5S416, which were located at the linked region on chromosome 5p15.31-p15 (data not shown).
GWLS and narrowing down the critical region
We detected preliminary evidence for linkage at D3S1262 that gave the maximum two-point LOD score of 5.419 at θ=0.00. The critical region was 15 Mb in size and located on chromosome 3q26.31-3q28. This prompted us to select 11 additional markers on chromosome 3q (Figure 1) to refine the critical region to 10 Mb between D3S3730 and D3S1580 on 3q26.32-3q28. The results of haplotype analysis for these markers and two-point LOD scores between disease phenotype and each marker locus are shown in Figure 1, respectively.
Discussion
Clinical and electrophysiological features of our patients (Table 1) confirmed the diagnosis of BAFME. Of the 13 affected members, one (7.7%) developed epileptic seizure before tremor, originally observed in 16% of cases.2
The patients' phenotypes were similar to those previously reported families in other populations particularly the Japanese patients.5, 9 Electrophysiologic studies revealed the cortical hyperexcitability with cortical origin of tremor. Our patients had a non-progressive clinical course. Myoclonic tremor and seizure responded well to VPA, CZP, or levetiracetam. No cognitive deficit was found in our cases. BAFME has previously been mapped to three chromosomal regions. We performed genome-wide linkage analysis with 400 microsatellite markers. The D3S1262 on chromosome 3q26.32-3q28 was the only marker with the LOD scores greater than 3 (5.419). Using 11 additional markers, we successfully narrowed down the critical region to 10 Mb, between the markers D3S3730 and D3S1580. This locus represents the fourth chromosomal region for BAFME. Genetic heterogeneity is not uncommon in human diseases including neurological disorders. A striking example is spinocerebellar ataxia, which has at least 33 underlying genes.11
Discovery of the fourth BAFME chromosomal region will facilitate the identification of the responsible gene. There are possible candidates considering their expression pattern and protein function. These include CLCN2 and KCNMB2. Both are expressed in brain tissue and function as ion channel proteins. Additional studies to elucidate the role of these genes on disease etiology are warranted. This will provide further understanding into the molecular basis of epilepsy and better insight into the disease mechanism leading to more effective treatment of this disorder.
Acknowledgments
We thank the family members for participating in this study and Dr Chaichon Locharernkul for providing excellent patient care. This study was supported by the Royal Golden Jubilee Ph.D. Program to PY (Grant No. PHD/0022/2550), the 90th Anniversary of the Chulalongkorn University Fund, the Thailand Research Fund, the National Science and Technology Development Agency and the Higher Education Research Promotion and National Research University Project of Thailand, Office of the Higher Education Commission (HR1163A).
The authors declare no conflict of interest.
References
- Striano P, de Falco FA, Minetti C, Zara F. Familial benign nonprogressive myoclonic epilepsies. Epilepsia. 2009;50 (Suppl 5:37–40. doi: 10.1111/j.1528-1167.2009.02118.x. [DOI] [PubMed] [Google Scholar]
- Regragui W, Gerdelat-Mas A, Simonetta-Moreau M. Cortical tremor (FCMTE: familial cortical myoclonic tremor with epilepsy) Neurophysiol Clin. 2006;36:345–349. doi: 10.1016/j.neucli.2006.12.005. [DOI] [PubMed] [Google Scholar]
- Ikeda A, Kakigi R, Funai N, Neshige R, Kuroda Y, Shibasaki H. Cortical tremor: a variant of cortical reflex myoclonus. Neurology. 1990;40:1561–1565. doi: 10.1212/wnl.40.10.1561. [DOI] [PubMed] [Google Scholar]
- Depienne C, Magnin E, Bouteiller D, et al. Familial cortical myoclonic tremor with epilepsy: the third locus (FCMTE3) maps to 5p. Neurology. 2010;74:2000–2003. doi: 10.1212/WNL.0b013e3181e396a8. [DOI] [PubMed] [Google Scholar]
- Mikami M, Yasuda T, Terao A, et al. Localization of a gene for benign adult familial myoclonic epilepsy to chromosome 8q23.3-q24.1. Am J Hum Genet. 1999;65:745–751. doi: 10.1086/302535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plaster NM, Uyama E, Uchino M, et al. Genetic localization of the familial adult myoclonic epilepsy (FAME) gene to chromosome 8q24. Neurology. 1999;53:1180–1183. doi: 10.1212/wnl.53.6.1180. [DOI] [PubMed] [Google Scholar]
- Guerrini R, Bonanni P, Patrignani A, et al. Autosomal dominant cortical myoclonus and epilepsy (ADCME) with complex partial and generalized seizures: a newly recognized epilepsy syndrome with linkage to chromosome 2p11.1-q12.2. Brain. 2001;124:2459–2475. doi: 10.1093/brain/124.12.2459. [DOI] [PubMed] [Google Scholar]
- de Falco FA, Striano P, de Falco A, et al. Benign adult familial myoclonic epilepsy: genetic heterogeneity and allelism with ADCME. Neurology. 2003;60:1381–1385. doi: 10.1212/01.wnl.0000055874.24000.4a. [DOI] [PubMed] [Google Scholar]
- Mori S, Nakamura M, Yasuda T, Ueno S, Kaneko S, Sano A. Remapping and mutation analysis of benign adult familial myoclonic epilepsy in a Japanese pedigree. J Hum Genet. 56:742–747. doi: 10.1038/jhg.2011.93. [DOI] [PubMed] [Google Scholar]
- Deng FY, Gong J, Zhang YC, et al. Absence of linkage to 8q23.3-q24.1 and 2p11.1-q12.2 in a new BAFME pedigree in China: indication of a third locus for BAFME. Epilepsy Res. 2005;65:147–152. doi: 10.1016/j.eplepsyres.2005.05.006. [DOI] [PubMed] [Google Scholar]
- Teive HA. Spinocerebellar ataxias. Arq Neuropsiquiatr. 2009;67:1133–1142. [PubMed] [Google Scholar]