Abstract
Protein interaction networks play central roles in biological systems, from simple metabolic pathways through complex programs permitting the development of organisms. Multicellularity could only have arisen from a careful orchestration of cellular and molecular roles and responsibilities, all properly controlled and regulated. Disease reflects a breakdown of this organismal homeostasis. To better understand the evolution of interactions whose dysfunction may be contributing factors to disease, we derived the human protein coevolution network using our MatrixMatchMaker algorithm and using the Orthologous MAtrix project (OMA) database as a source for protein orthologs from 103 eukaryotic genomes. We annotated the coevolution network using protein–protein interaction data, many functional data sources, and we explored the evolutionary rates and dates of emergence of the proteins in our data set. Strikingly, clustering based only on the topology of the coevolution network partitions it into two subnetworks, one generally representing ancient eukaryotic functions and the other functions more recently acquired during animal evolution. That latter subnetwork is enriched for proteins with roles in cell–cell communication, the control of cell division, and related multicellular functions. Further annotation using data from genetic disease databases and cancer genome sequences strongly implicates these proteins in both ciliopathies and cancer. The enrichment for such disease markers in the animal network suggests a functional link between these coevolving proteins. Genetic validation corroborates the recruitment of ancient cilia in the evolution of multicellularity.
Keywords: coevolution, multicellularity, protein–protein interactions, cilia, cancer
Introduction
At the heart of elucidating the complexity of cellular cooperation in metazoans is determining the protein–protein interactions (PPI) that enable cell–cell communication and the dynamic functioning of cellular pathways that respond to internal and environmental signals. Evolution of multicellularity would necessitate the formation of new interactions between new genes while accommodating and adapting proteins from the precursor unicellular network. New systems of cellular communication and cell division control must have evolved early on in the evolution of multicellularity. Breakdown of such systems can lead to disease, particularly cancer, because cancer arises from the loss of a cell’s normal compliance with the ground rules of multicellular phenotypic organization. Most recently, cancer-related “gatekeeper” genes, which are involved in cellular signaling and growth processes, were phylogenetically mapped to the emergence of multicellularity in metazoa (Domazet-Loso and Tautz 2010), and the rise of cancer together with multicellularity was further highlighted in the context of sequencing the Amphimedon queenslandica (sponge) genome (Srivastava et al. 2010).
Networks of PPI have evolved under natural selection over millions of years, leaving their mark in the molecular sequences of proteins, which we can now study thanks to the abundance of sequence data provided by advances in sequencing technology. Interactions between proteins can be predicted through their correlated evolutionary rates (Pazos and Valencia 2001; Juan et al. 2008). Detecting molecular coevolution can thus help to elucidate functional interactions between molecules within and between cells to gain insight into biological processes, pathways, and the networks of interactions important for cellular function.
Our recently developed method MatrixMatchMaker (MMM) implements an efficient computational strategy to detect sequence coevolution (Tillier and Charlebois 2009; Rodionov et al. 2011). It was shown to be more accurate than previous coevolutionary methods, more accurate than coabundance predictions, and most accurate in predicting protein complexes (Clark et al. 2011). Using this tool on recently available data, we are now able to resolve two subnetworks of human protein coevolution: one involving ancestral eukaryotic proteins and the other proteins specific to animals or having been recruited in animals to play roles in multicellular communication and control. That latter subnetwork exposes associations among genes required for multicellularity, providing opportunities to study interactions among systems contributing to organismal homeostasis. More specifically, we here describe a human protein coevolution network that contains many proteins previously uncharacterized in terms of protein interactions and expression patterns. We, however, find strong evidence for genetic interaction among these proteins when considering human disease data, particularly links among genes involved in ciliopathies and cancer. The nonmotile primary cilia play important roles in chemo-, mechano-, and thermosensation in vertebrates and coordinate signaling with motility or with cell division and differentiation (Satir and Christensen 2008). Proteins associated with centrioles, centrosomes, and cilia have been implicated in both ciliopathies and cancer in human (Bettencourt-Dias et al. 2011).
Materials and Methods
Input Data for MMM
For the construction of the human coevolution network, we used the version of the OMA Browser (Altenhoff et al. 2011) dated November 2010, to obtain the 20,804 OMA groups containing a human protein. Only eukaryotic orthologs (from 103 genomes) were considered in these groups, having at least two sequences. We also considered an alternative clustering scheme to that used in OMA. For this, all eukaryotic proteins reciprocally best matching with human proteins (1:1 orthologs) were added to the human sequence to form a group, though we did not require all pairwise 1:1 orthologs to hold over all species. This alternative clustering yielded 20,225 groups.
MAFFT (Katoh and Toh 2008) was used to create multiple sequence alignments for each group; evolutionary distance matrices were obtained by using protdist (from PHYLIP 3.69 [Felsenstein 1989], modified to allow selenocysteine and pyrolysine amino acids and for identical sequences to have a distance of 0.0 [protdist sets these to 0.00001]).
MMM searches for pairs of evolutionary distance submatrices that are similar within a specified tolerance. Such matching submatrices represent similar phylogenetic subtrees, indicating coevolution over the evolutionary history of the subset of taxa included in the submatrices. The number of taxa found in the matching submatrices is the MMM score; higher scores provide stronger evidence for covariation of the two protein families. MMM is scale independent and thus allows for the detection of correlated distances, not just identical distances, so that protein coevolution may be detected despite the proteins evolving at different rates. We used MMMvII (Rodionov et al. 2011), an efficient and exact algorithm for identifying the submatrices. All-by-all pairwise combinations (216,850,725 for the OMA groups and another 204,535,425 for the 1:1 groups) were analyzed, only allowing proteins belonging to the same species to be included in possible MMM solutions (parameter: u, so that only distances between the species in common in the two alignments were considered) and with a match tolerance (parameter: a) set to 0.1 (10%), which has been previously shown (Tillier and Charlebois 2009; Clark et al. 2011) to provide adequate sensitivity and specificity.
An average matrix was obtained by averaging the matrix entries of each protein family for each pairwise species comparison, over all of the OMA groups’ matrices. The MMM score for each matrix to the average matrix was used to calculate the so-called G score and the sum of MMM plus G scores (MMMpG), which is used to correct MMM scores between groups for the part of their coevolution that is due to the species phylogeny (see Clark et al. 2011 for details).
Evolutionary Age of Orthologous Groups
We also used the average matrix described earlier to compute the relative rate of an OMA group’s evolution, as the ratio of its rate (average distance to the human ortholog) over the average matrix’s rate (same subset of species). Neighbor Joining (Felsenstein 1989) and Minimum Evolution (ME) trees (Rzhetsky and Nei 1992) were obtained from the average matrix, showing generally good agreement with the eukaryotic species phylogeny but with some artifacts arising due to long-branch effects. We chose to use the ME tree (supplementary fig. S1, Supplementary Material online). For each OMA group, its species distribution was used to determine the position in the ME tree for the last common ancestor of all the sequences in the group. The age of the protein is simply then the evolutionary distance from the human sequence to this internal node. We chose the edge between the branch node leading to the unicellular Monosiga brevicollis (at a distance of 0.48) and the node leading to early multicellular animals (at a distance of 0.45) as the delimiter for “new” groups because all species with a smaller distance to human are multicellular animals. The “old” OMA groups include human orthologous proteins from nonanimals and animals.
Map Equation Clusters
Map equation (Rosvall and Bergstrom 2008) was used to cluster the network with MMM and MMMpG scores 12 and higher according to its topology. A total of 145 clusters were obtained for the OMA groups’ network and 97 for the 1:1 network. The age of each cluster was taken as the average age of the nodes included in the cluster, and each cluster was then labeled either “NEW” or “OLD” according to their average distance being <0.46 or >0.46, respectively. Because there are some “old” nodes in the “NEW” clusters, and conversely, “new” nodes in the “OLD” clusters, we use capitalization to differentiate the age of clusters and genes.
Orthologous Groups
The division of the MMM network into two distinct groups of NEW and OLD clusters is partly an artifact of the methodology used in defining sets of orthologs, because it is more difficult to identify distant orthologs for rapidly evolving proteins (making old proteins appear newer). The ages of the proteins are only gross estimates, because missing sequences from unfinished genomes, Basic Local Alignment Search Tool (BLAST) parameters, and clustering approaches will all affect the composition of the orthologous clusters. We attempted to add potentially more distant human orthologs by creating protein families that considered only the 1:1 reciprocal best BLAST hits to the human proteins, which is not nearly as strict a criterion for orthology as that used by OMA, which does not permit paralogous proteins to be included in their groups (Altenhoff et al. 2011). We think this more relaxed approach would be less likely to underestimate the age of proteins. This indeed resulted in larger matrices on average and older ages (supplementary data set S2, Supplementary Material online). The resulting 1:1 MMM network was quite similar to the one obtained from the OMA groups, however, and does not alter our conclusions. We therefore chose to present the network from the better established orthologous clusters assembled by OMA, which are most likely to represent single-protein functions and also to yield better quality alignments and distance matrices.
Mapping of OMA Proteins and MMM Networks to Known Interactions, Coexpression Data, Gene Ontology Functional Annotation, Pathways, and Diseases
The OMA proteins and the MMM networks were annotated with many resources, and these annotations can be found in supplementary data sets S2 (protein annotations) and S1 (MMM network annotation), Supplementary Material online. The descriptions and the references for these are given in supplementary text S1, Supplementary Material online (supplementary methods and results, Supplementary Material online).
Coexpression Data
Data from the E-MTAB-62 data set, a meta analysis of gene expression data from ∼5,400 human samples representing 369 different cell and tissue types, disease states, and cell lines, all on the same platform of the Affymetrix GeneChip Human Genome HG-U133A (G-U133A) obtained from GEO and ArrayExpress (Lukk et al. 2010), were obtained from ArrayExpress (http://www.ebi.ac.uk/arrayexpress/, last accessed 2012 September 23). The processed data were mapped to the OMA human proteins, and all expression values for a gene were averaged. The Pearson correlation coefficient was then calculated for all pairwise comparisons of genes over all the samples. Correlation values of expression data were also obtained from COXPRESdb (Obayashi and Kinoshita 2011).
Ciliopathies
“Cilia” and “ciliopathy” were both used as search terms in Gene Ontology (GO), Online Mendelian Inheritance in Man (OMIM), and National Center for Biotechnology Information (NCBI) Gene and by searching through reviews of ciliopathies in the literature (Gerdes et al. 2009; Tobin and Beales 2009); the ciliome database (http://ciliome.com, last accessed 2012 September 23) (Inglis et al. 2006) that had at least three studies linking a gene to cilia; and from the http://ciliaproteome.org (last accessed 2012 September 23) database (Gherman et al. 2006). We found 323 human genes involved in cilia or ciliopathies that we could map to the OMA groups.
Cancer Genomes
Data from cancer genome sequencing were obtained from the International Cancer Genome Consortium Data Portal on 27 May 2011. Ensembl IDs for the mutations were mapped to the OMA group human proteins, and the number of samples in which the gene was (nonsilently) mutated was counted. Genes mutated in fewer than three samples were ignored. Ovarian cancer data were updated on 29 June 2011 from the supplementary table in The Cancer Genome Atlas Research Network (2011). The OMA protein annotation file (supplementary data set S2, Supplementary Material online) shows the total number of samples over all cancer genomes.
Results
The MMM Coevolution Network Splits According to Evolutionary Age
We have previously shown that MMM scores have predictive value for known PPI (Tillier and Charlebois 2009; Clark et al. 2011). Here, we obtained MMM scores in an all-by-all analysis of distance matrices from orthologous groups containing human proteins (supplementary data set S2, Supplementary Material online), retaining scores of at least 12 in what we refer to below as the MMM12+ network (supplementary data set S1, Supplementary Material online). MMM-D, a database of scores from the MMM12+ network (containing 6,422 pairs from 1,608 protein families) and all known as well as orthologous protein interactions (323,702 interactions between 11,836 protein families), is available at http://tillier.uhnres.utoronto.ca/MMMD.php (last accessed 2012 September 23).
Clustering of the network, strictly based on topology using the Map equation (Rosvall and Bergstrom 2008), revealed two large subnetworks separated in their evolutionary age: one on average younger than the origin of animal multicellularity (MMM12+NEW) and one predating that origin (MMM12+OLD) (see Materials and Methods and supplementary fig. S1, Supplementary Material online). Connecting the two subnetworks are MMM12+NEW/OLD edges. For clarity, we display the smaller MMM13+ network in figure 1A.
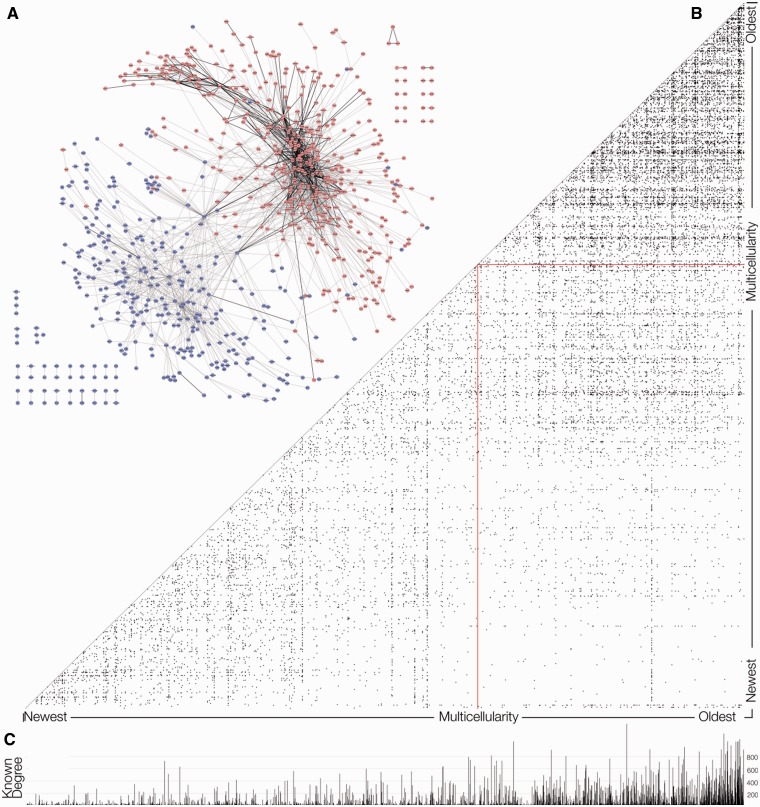
Fig. 1.
The MMM13+ network. (A) This display of the MMM13+ network was produced using Cytoscape v.8.0’s spring-embedded layout (Shannon et al. 2003). Clustering of the network according to its network topology was separately done using the Map equation algorithm (see Materials and Methods). The pink nodes indicate that they are found in evolutionarily OLD clusters, that is, Map equation clusters of the MMM12+ network with an average distance dating to before the origin of animals. The blue nodes are in clusters that have an average age no older than the origin of animals, although some individual nodes within those clusters are older. (B) The MMM12+ network, represented as a heat map (Tarassov and Michnick 2005), also shows fewer old-to-new edges than new-to-new or old-to-old edges. (C) The degree for nodes in the known interaction network is higher for older nodes, indicating that more interactions are known among the older proteins.
The random expectations for old–old, new–old, and new–new frequencies in the network are simply derived from the binomial expansion. A χ2 test (supplementary table S1, Supplementary Material online) revealed a highly significant overrepresentation of new–new and old–old node connections within the MMM12+ network, whether using OMA groups (P < 1.1E – 311) (as is shown in fig. 1B) or MAP clusters (P < 1.1E – 311). This remained true when we analyzed the network of all known protein interactions (P < 1.1E – 311), such that on average, old proteins interact with old proteins and new with new. Figure 1C shows that proteins in the known interaction network also have more connections when they are old.
Agreement with Known Interactions
Overall, higher MMM scores have predictive value for known PPI. Figure 2A shows that higher scores give greater precision (the frequency of predictions that were previously known interactions), which is approximately 10% at MMM ≥ 12 and climbs to over 30% at MMM ≥ 16. These values are comparable to biochemical high-throughput methods applied to identify protein interactions in human, such as yeast-2-hybrid and immunoprecipitation/mass spectrometry, whose predicted interactomes overlap with literature-curated PPI networks between 2–8% and 6–11%, respectively, and that have mutual overlap of 7.9% (Rual et al. 2005; Ewing et al. 2007). We thus set a threshold of 12 for our network, which we call MMM12+. As seen in figure 2A, the highest coevolution scores are not found in the Human Protein Reference Database (HPRD) database, the gold standard for human PPI, but are validated by other data sets, particularly the Comprehensive Resource of Mammalian Protein Complexes (CORUM) (see supplementary methods, Supplementary Material online, for a list of databases used for known interactions). We also considered the scores from the Drosophila interactome (Guruharsha et al. 2011) and found that these rapidly increased with higher MMM scores (giving 36.3% precision for PPI prediction with MMM12+), also indicating agreement with this orthologous network (fig. 2B).
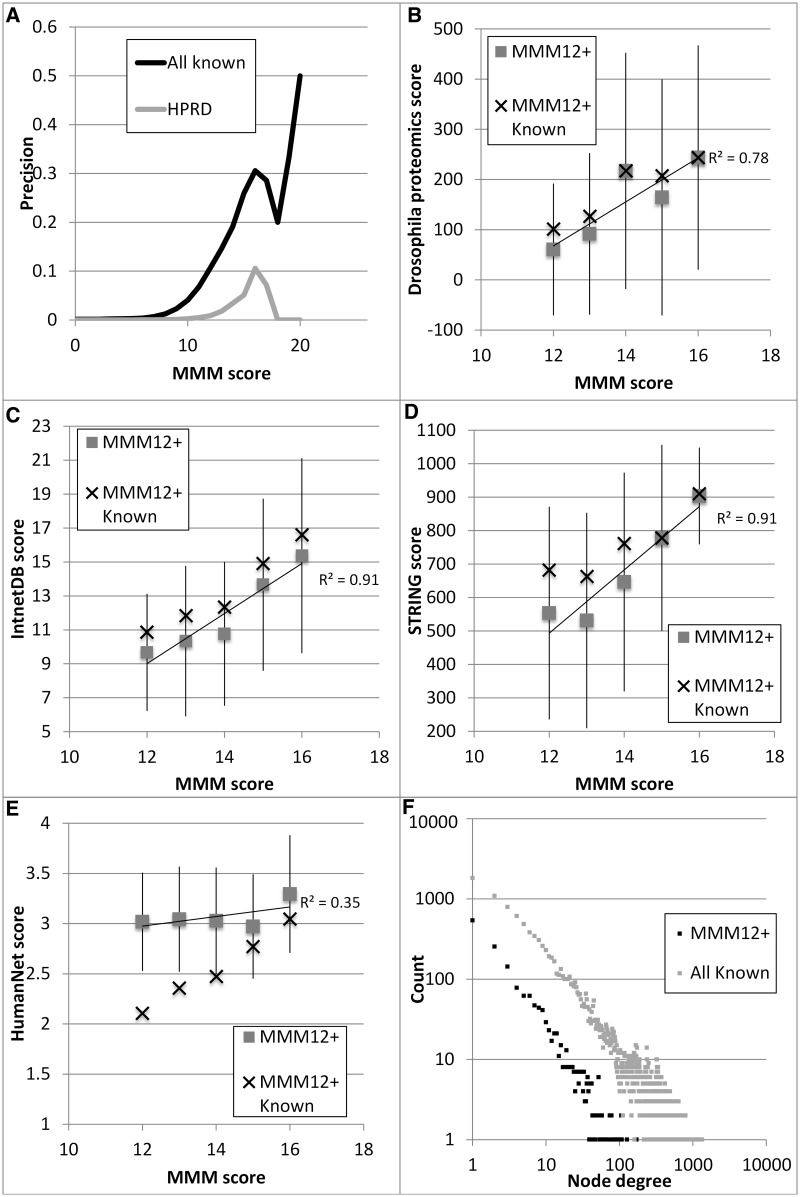
Fig. 2.
Accuracy of MMM. (A) Precision of MMM predictions (the frequency of coevolving pairs that are known interactions from PPI databases) increases with higher MMM score thresholds (x axis). Considering HPRD only, it contains far fewer interactions, and none with very high MMM scores. (B) MMM scores correlate with the average interaction scores from Drosophila based on mass spectrometry analyses. (C), (D), and (E) show a high correlation of MMM scores with average scores from other PPI prediction methods (IntnetDB, STRING, and HumanNet, respectively, each with their own scoring scale); this is true for all MMM predictions and for the subset that are known interactions. The correlation coefficient is shown for the MMM predictions, and error bars indicate one standard deviation over all MMM pairs. (F) Node degree frequency for proteins in the known interaction network and in the MMM12+ network shows that they are both scale free.
Our data also show agreement with data from other databases that include predicted interactions (fig. 2C–E). Although STRING (fig. 2D) uses phylogenetic profiles as part of its prediction score, which should be correlated with MMM scores, this approach is only used for their prediction of bacterial protein interactions. HumanNet (Lee et al. 2011) includes phylogenetic profiling as part of their prediction approach, and as expected, those scores are higher for gene pairs in the MMM12+ network (fig. 2E).
The structure of the network also agrees well with known interactions: figure 2F shows the degree distribution for the known interaction network of all 20,839 proteins in our data set along with the degree distribution, which is consistent with the power law and is typical for PPI networks.
Because interaction prediction data sets make use of the GO database, we also considered similarity of functional annotation using the Parent–Child algorithm (Alexa et al. 2006; Grossmann et al. 2007), which considers the GO hierarchy and thus assigns lower P values to shared more-specific GO terms (supplementary text S1, Supplementary Material online). MMM proteins in interactions are more commonly found to share the same GO annotation compared with random pairs. For cellular component, MMM12+ pairs are found to have the same annotation more often than expected (P = 1.1E − 124, Pearson’s χ2 with Yates’ continuity correction) and with significantly lower P values (P = 8.26E − 16, Mann–Whitney U test). Similarly, for biological process, MMM12+ pairs share the same annotation more often than expected (P = 9.2E − 281, Pearson’s χ2 with Yates’ continuity correction), also with significantly lower P values (P = 1.49E − 63, Mann–Whitney U test). This shows that MMM predictions are enriched for functionally interacting pairs of proteins. In supplementary figure S2, Supplementary Material online, we show a strong correlation between MMM scores and the P values indicating that higher MMM scores predict functionally related protein pairs.
The majority of known and interologous interactions were concentrated between the nodes within the ancient MMM12+OLD network (fig. 1). MMM12+NEW thus revealed a subnetwork of coevolving genes that are more evolutionarily recent and that are not generally known to have interaction partners.
The NEW network contained many proteins whose interactions remain uncharacterized. For example, of the 263 nodes included in the MMM12+ network with no known interactions in any of the databases interrogated, 235 (89%) were new nodes (219 of these, or 83% of the 263, are in NEW clusters). Figure 3A shows the frequency of NEW nodes in the MMM12+ network decreasing as their degree in the known interaction network increases, indicating that new nodes tend to have few known interactions.
Fig. 3.
New nodes and NEW network. (A) Frequency of NEW nodes decreases with increasing degree. (B) Frequency of node age for all proteins analyzed and for the known and MMM12+ networks. (C) The relative evolutionary rate (rates ratio to average matrix), as a function of the age of proteins is plotted for all proteins analyzed and for those in the MMM12+ network. The range was much reduced for proteins in the network, indicating that these neither evolve extremely quickly nor slowly. (D) The difference between the rates ratio of two proteins interacting (MMM12+ Known) or coevolving (MMM12+) decreases with MMM score, indicating that coevolving proteins also have similar rates of evolution.
We considered that this observation could be due to a bias in the known protein interaction network for proteins that are conserved from yeast to human, because the yeast interactome is better characterized, and we did use interologs in our assignment of interacting proteins. We thus compared the frequency distribution for the age of all proteins (20,839), of proteins in the known interaction network (11,581), and in our MMM12+ network (1,608), and did not see this bias (fig. 3B). MMM12+ was less biased toward new nodes (59%); however, both networks were similarly extremely biased against interactions with very new proteins.
Proteins with identified human orthologs in the Drosophila proteomics network (1,359) of protein complexes are predominantly old (889), and the interactions (2,152) found between these genes are statistically significantly biased (P < 6.6E − 117; supplementary table S1, Supplementary Material online) toward old–old interactions (1,456) and against new–new interactions (143). Overall, we found that known interactions favor new proteins but that proteins interacting in complexes and coevolving proteins tend to be old proteins with higher degree (fig. 1C).
Evolutionary Rates
Protein coevolution can only be recognized if the proteins in question have undergone a sufficient amount of evolutionary change. Therefore, for recently arisen proteins, the rate of their evolution must have been sufficiently high to detect their coevolution among a smaller set of species. We considered the relative rate of evolution of each OMA group to the average rate over all proteins. A ratio above 1 indicates a fast rate and a ratio below 1 a slow rate (see Materials and Methods). Figure 3C shows the rate ratio for all OMA groups, and for the subset in the MMM12+ network, plotted against the evolutionary age of the protein family. Very high rate ratios were not found in our network. We believe this is because the majority of such rapidly evolving proteins would evolve too quickly for enough orthologs to be detectable by BLAST or have emerged too recently to produce OMA groups big enough to yield high MMM scores. Conversely, slowly evolving proteins need to be old enough to have accumulated enough substitutions.
When we considered the nodes only present in our network, the range of evolutionary rate ratios was much reduced. Additionally, when looking at the difference in the rate ratio between the pairs of connected proteins (fig. 3D), we saw a decrease with MMM score, indicating that the most strongly coevolving proteins also tend to evolve at similar rates (not just correlated rates). This remained true in the MMM12+ subnetwork of known interactions (fig. 3D), which had on average even more highly similar rates (although the variance is quite high). In supplementary figure S3, Supplementary Material online, we show the frequency distribution of the difference in the rate ratio in the MMM12+ network and the known PPI network. In both of these networks, protein pairs evolve at more similar rates than do protein pairs drawn from a randomized network.
Coexpression Data
To help elucidate the nature of the coevolution between the more recently evolved proteins and whether coevolution could possibly be entirely attributed to their coexpression (Hakes et al. 2007), we considered expression data using the Pearson correlation of genes from COXPRESdb and from E-MTAB-62, a more recent compilation of expression values across a wide range of conditions and tissues in human (Lukk et al. 2010).
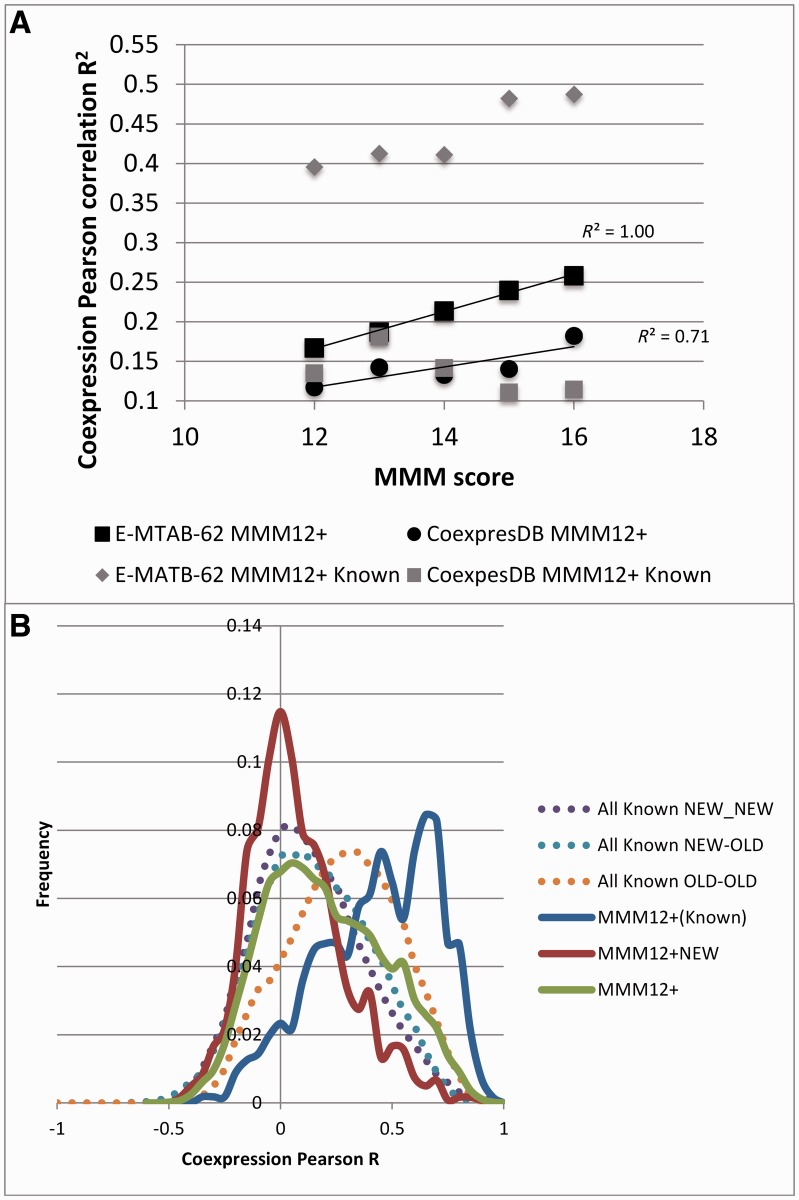
We found that coexpression values (as measured by Pearson correlation) do correlate with the MMM score, and as expected, known PPIs had high correlation values (fig. 4A). The E-MTAB-62 set showed these effects much more strongly than did the COXPRESdb data set, but only 66% of the edges in the MMM12+ network had both genes present in the E-MTAB-62 data set and only 8% of them could be mapped onto COXPRESdb data. There was no correlation between the two databases (Pearson R2 = 0.001).
Fig. 4.
MMM coevolution and coexpression. (A) The average Pearson correlation (R2) measuring the coexpression of gene pairs in the MMM12+ and its subset of known interactions (MMM12+ Known) increases with MMM score. (B) Frequency distribution of the Pearson correlation (R) of coexpression over the E-MTAB-62 data of gene pairs in the All Known and MMM12+ networks. For the known interactions found in MMM12+ (blue solid line), the frequency distribution was found to be skewed toward higher correlations. The overall MMM12+ network distribution had fewer high correlation values (green solid line), particularly when only the subnetwork of NEW clusters was considered (red solid line). When considering all the known interactions, we found higher correlation values when both genes were old (orange dotted line) but significantly less than if the genes were also coevolving (i.e., in MMM12+; blue solid line). Newly interacting genes are less likely to be coexpressed. The distribution of known interactions when one of the genes was “new” (blue and purple dotted lines) was similar to the distribution from the MMM12+NEW network (red solid line).
Although we could obtain the correlation of expression data for over 80% of the edges for the known interactions, only 56% of the edges in the NEW network had coexpression information in at least one of the two databases we considered. This suggests that many of the MMM12+NEW genes in our network were not on the expression arrays and were otherwise poorly characterized (data not shown).
The frequency distribution of Pearson correlations in the E-MTAB-62 data set (fig. 4B) was found to be skewed toward higher values when considering only the known interactions (blue solid line). The overall MMM12+ network distribution had fewer high correlation values (green solid line), particularly when only the subnetwork of NEW clusters was considered (red solid line). Of the MMM12+ known interactions, 76% had correlation values greater than 0.2 in either expression data set, whereas only 21% of the coevolving proteins in the NEW network reached that threshold.
Comparing these distributions in the network of all known interactions, we also found higher correlation values when both genes in the interaction were old (orange dotted line) but significantly less than if the genes were also coevolving (blue solid line). The distribution of known interactions when one of the genes was “new” (blue and purple dotted lines) was similar to the distribution in the MMM12+NEW network (red solid line).
Functional Annotation
To better annotate the MMM12+NEW coevolution network, we made use of GO and several pathway databases (annotations are found in supplementary data sets S1 and S2, Supplementary Material online). Statistically overrepresented GO terms are provided in supplementary data set S3, Supplementary Material online. The MMM12+NEW network was found to be enriched for extracellular proteins, cell anchoring and adhesion proteins, the cytoskeleton, and cilia. We also saw a link between these proteins and cell division, control of the cell cycle, and multicellular development.
Human Diseases
We used data from Online Mendelian Inheritance in Man (OMIM), Catalogue of Somatic Mutations in Cancer (COSMIC), and Gene Signature Database (GeneSigDB) to assess the relationship between protein age and disease (table 1). (Although also considered, the disease classifications in the Genetic Association Database did not yield any statistically significant results.) When considering all proteins mapped to OMA (not just in the MMM12+), we found that old genes were more likely to be implicated in OMIM diseases. This was especially true for proteins involved in enzyme or protein deficiencies and in ciliopathies. From COSMIC, however, we found that proteins involved in cancer were more likely to be new genes. The overrepresentation of genes involved in cancer among this set had been noticed previously (Domazet-Loso and Tautz 2010).
Table 1.
Disease Functional Annotation.
| Membership | New vs. Olda |
NEW vs. OLDb |
||||
|---|---|---|---|---|---|---|
| N | Overrepresented | P | N | Overrepresented | P | |
| OMIM | 2,476 | old | 1.74E-07 | 315 | — | — |
| OMIM "Deficiency" | 322 | old | 5.55E-14 | 55 | — | — |
| Cilia/ciliopathy | 323 | old | 5.8E-11 | 42 | NEW | 6.54E-03 |
| COSMIC mutation | 4,555 | new | 8.43E-06 | 558 | NEW | 1.92E-18 |
| COSMIC census | 399 | new | 9.40E-03 | 42 | NEW | 2.80E-03 |
| In signature | 10,189 | old | 3.25E-36 | 1,608 | OLD | 3.70E-03 |
| STEM signature | 5,836 | old | 3.49E-63 | 384 | OLD | 3.03E-07 |
| CD marker | 311 | new | 1.04E-16 | 26 | NEW | 1.37E-06 |
anew/old comparisons considered all genes mapped to OMA.
bNEW/OLD comparisons considered only the clusters in MMM12+.
The data from GeneSigDB were difficult to interpret because genes in a signature can be up or down for the disease considered, but interestingly, genes in signatures were mostly old (possibly due to the bias of expression studies). This is especially true of genes with signatures for stem cells, whereas cell differentiation markers were instead overrepresented in new genes.
Many of these observations still applied when considering the age of the protein clusters in the MMM12+ network. We saw a higher representation of NEW clusters involving genes of differentiated cells, and mutated in cancer, and we also saw an overrepresentation of ciliopathy genes in the NEW clusters.
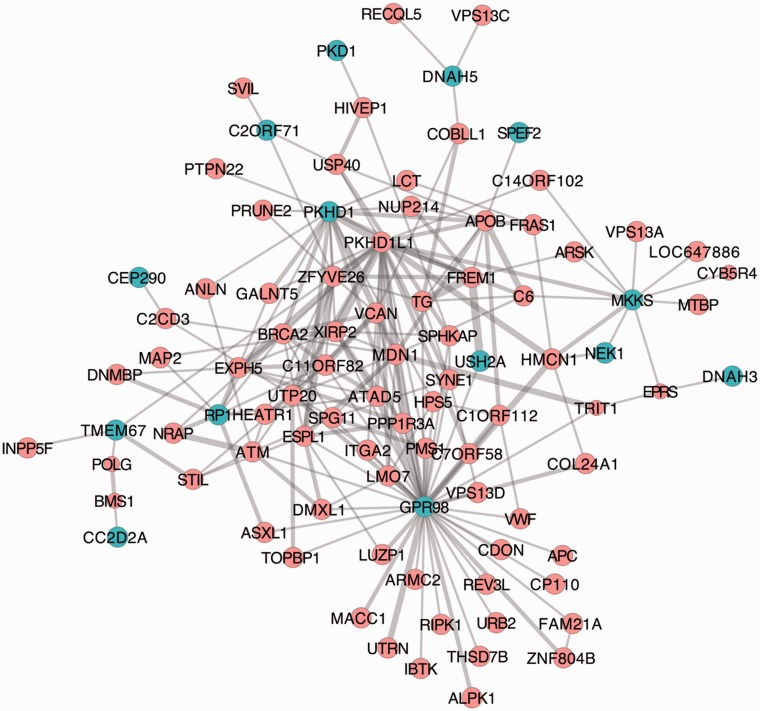
Although the ciliopathy genes are mostly old, many are clustering within the NEW network and are seen coevolving with newer animal-specific genes. USH2A and GPR98 have been found to be mutated in Usher syndrome, a ciliopathy leading to deafness and blindness, and found to potentially interact in the extracellular gap (Maerker et al. 2008). PKHD1 is involved in polycystic and hepatic disease. We also saw several axonemal dynein proteins, involved in ciliary dyskinesia. In figure 5, we show the MMM13+ network of only the ciliopathy genes and their first neighbors, indicating strong coevolution of genes particularly with GPR98, USH2A, PKHD1, RP1, and MKKS.
Fig. 5.
MMM13+ network of cilia/ciliopathy genes. Subnetwork of cilia/ciliopathy genes and their first neighbors in the MMM13+ network. Teal nodes indicate the cilia/ciliopathy genes. The thickness of the lines to their first neighbors is proportional to the MMM score.
Cancer Genomes
Mutation Data.
With the advent of cancer genome sequencing, we sought to determine whether genes mutated in cancer could also be found in our MMM12+ network, as the data from COSMIC indicated, and as tight control of multicellular homeostasis should warrant.
We considered the data from the recent sequencing of ovarian serous cystadenocarcinoma (OSC) tumors (The Cancer Genome Atlas Research Network 2011) because mutation of the BRCA2 gene is a risk factor for this cancer (Ford et al. 1998), which we found to be coevolving with cilia proteins in the NEW network. The TP53 gene is the most frequently mutated gene in this cancer, representing about half of all mutations, so the observed number of other mutated genes is small. Nevertheless, we saw an overrepresentation of genes in the MMM12+ network as mutated in a relatively high number of patients (fig. 6 and table 2), particularly USH2A, GPR98, PKHD1, and several dynein proteins.
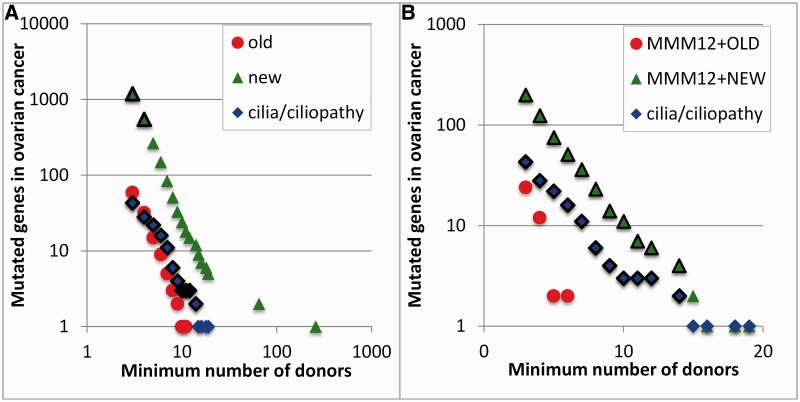
Fig. 6.
Frequency of mutated genes in ovarian serous cystadenocarcinoma tumors. The count of mutated genes (y axis) found in at least the number of tumor donor samples on the x axis is shown. Genes were annotated as being involved in cilia or ciliopathies (blue diamonds) or grouped by their evolutionary age (red circles for old and green triangles for new) and are black outlined when statistically significant (P < 0.05). (A) The results are for all genes. (B) We only considered genes in the MMM12+ network. The MMM12+OLD genes were never overrepresented in the samples, but the cilia genes were highly overrepresented in the samples, as were the MMM12+NEW genes.
Table 2.
Mutations in Ovarian Serous Cystadenocarcinoma from the TCGA.
| Gene Symbol | Donors | MMM12+ | Gene Age | Cilia | Gene Symbol | Donors | MMM12+ | Gene Age | Cilia |
|---|---|---|---|---|---|---|---|---|---|
| TP53 | 258 | NEW | PKHD1 | 9 | NEW | NEW | + | ||
| TTN | 65 | NEW | SI | 9 | NEW | NEW | |||
| CSMD3 | 19 | NEW | DNAH11 | 8 | NEW | NEW | + | ||
| FAT3 | 19 | NEW | GPR98 | 8 | NEW | NEW | + | ||
| USH2A | 19 | NEW | NEW | + | ANK2 | 8 | NEW | NEW | |
| MUC16 | 18 | NEW | APC | 8 | NEW | NEW | |||
| RYR2 | 16 | NEW | COL22A1 | 8 | NEW | NEW | |||
| DST | 15 | NEW | COL6A3 | 8 | NEW | NEW | |||
| HMCN1 | 15 | NEW | NEW | PKHD1L1 | 8 | NEW | NEW | ||
| DNAH5 | 14 | NEW | NEW | + | PPP1R3A | 8 | NEW | NEW | |
| LRP1B | 14 | NEW | TG | 8 | NEW | NEW | |||
| LRP2 | 14 | NEW | NEW | CRB1 | 7 | NEW | + | ||
| AHNAK | 12 | GLI2 | 7 | NEW | + | ||||
| APOB | 12 | NEW | NEW | RP1L1 | 7 | NEW | + | ||
| DNAH3 | 12 | NEW | NEW | + | FREM2 | 7 | NEW | NEW | |
| NF1 | 12 | NEW | IGSF10 | 7 | NEW | NEW | |||
| AHNAK2 | 11 | NEW | LAMA2 | 7 | NEW | NEW | |||
| FAT1 | 11 | NEW | NEW | LAMA3 | 7 | NEW | NEW | ||
| HYDIN | 11 | + | MAP2 | 7 | NEW | NEW | |||
| MUC17 | 11 | OLD | PRKDC | 7 | NEW | NEW | |||
| ODZ1 | 11 | NEW | PTPRZ1 | 7 | NEW | NEW | |||
| BRCA1 | 10 | NEW | NEW | STAB2 | 7 | NEW | NEW | ||
| BRCA2 | 10 | NEW | NEW | SYNE2 | 7 | NEW | NEW | ||
| LRRK2 | 10 | NEW | NEW | VPS13B | 7 | NEW | NEW | ||
| MACF1 | 10 | NEW | XIRP2 | 7 | NEW | NEW | |||
| RYR1 | 10 | NEW | YSK4 | 7 | NEW | NEW | |||
| SYNE1 | 10 | NEW | NEW | ZFYVE26 | 7 | NEW | NEW |
Note.—The genes most frequently mutated are listed (removing silent, in frame, untranslated regions, flanking, noncoding, nonstop, intron, and splice sites/region mutations). In between seven and nine donors (on the right), only genes involved in cilia/ciliopathies and/or in the MMM12+ network are listed. Underlined genes were highlighted in TCGA’s article (The Cancer Genome Atlas Research Network 2011).
Figure 6A plots the relationship between the total number of genes mutated against the cumulative number of donors with that number of mutations. Most of the relevant genes to cancer will thus appear in the most number of tumors. New genes appear most likely to be mutated (but they are also more frequent, so their overrepresentation in the set of cancer genes is not significant). In figure 6B, we see that cilia genes and MMM12+NEW genes are highly mutated in this cancer, and this is statistically significant (outlined markers indicate statistically significant P values < 0.05 for the number of mutations found in the cancer samples).
Expression Data.
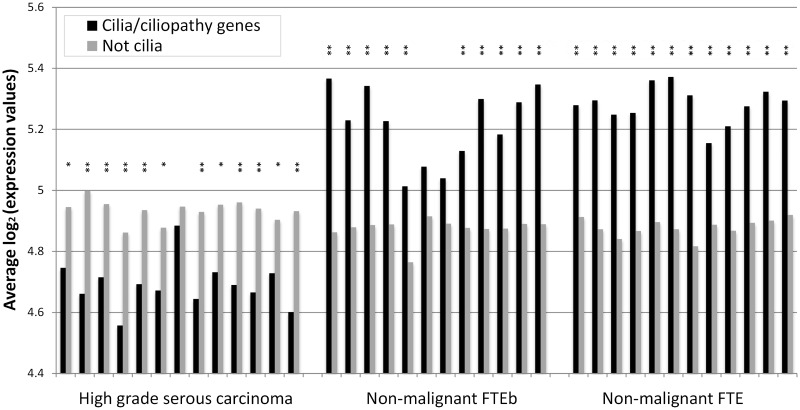
The analysis of the Cancer Genome Atlas (TCGA) ovarian cancer data from both Agilent and Affymetrix arrays similarly showed a significant underexpression of cilia genes in ovarian cancer samples (supplementary results and supplementary fig. S4, Supplementary Material online). Because these data contained few normal samples to compare to the tumor samples, to confirm, we analyzed a smaller sample of microdissected tumors of high-grade serous carcinoma from fallopian tube samples, against their nonmalignant counterparts (Tone et al. 2008). We considered the average difference in the log of expression values between the tumor samples and the normal samples and found significant underexpression of the new versus old genes over all the probes for the 17,271 genes mapped to OMA (P = 2.1E − 29) or in MMM12+(P = 3.6E − 18). Cilia/ciliopathy genes were strikingly underexpressed in these tumor samples (P = 6.6E − 105 in OMA and P = 2.8E − 31 in MMM12+). In figure 7, we show the average expression values for the probes to cilia/ciliopathy genes and to the rest of the genes in OMA. Although the expression of noncilia genes did not vary considerably between tumors and normal samples, cilia genes had lower expression in tumor samples and higher expression in normal samples.
Fig. 7.
Expression of cilia-related genes in high-grade serous ovarian carcinomas. The average log expression values for the cilia/ciliopathy genes and those not annotated as such (supplementary data set S2, Supplementary Material online) are shown for cancer samples (first 13 samples on the left), normal fallopian epithelial (FTE) cells from BRCA1-mutated donors (middle 12 samples), and normal fallopian epithelial cells from non-BRCA1 donors (rightmost 12 samples; see Tone et al. [2008] for details). Asterisks indicate statistical significance of a two-tailed t-test at *P < 0.05 or **P < 0.01, for significant over- or underexpression of cilia/ciliopathy genes.
There were a few exceptional examples of annotated cilia genes that we found overexpressed in the ovarian tumors (several kinesins and aurora kinase A [AURKA]; supplementary fig. S4, Supplementary Material online). However, such overexpression could indicate the misannotation of these as cilia genes or possibly be attributed to alternative functions such as mitotic activity, as these genes are also important for mitosis. Interestingly, AURKA promotes the destabilization of the axoneme and cilium (Pugacheva et al. 2007) and interacts with prometastatic protein NEDD9 (a member of the MMM12+NEW network). In ovarian cancers, AURKA is overexpressed and NEDD9 is amplified genetically (The Cancer Genome Atlas Research Network 2011) agreeing with the downregulation of cilia being associated with this disease.
Discussion
In this work, we have analyzed the coevolution network of human proteins found with MMM and considered the coevolution of interactions deposited in many PPI databases. To do this, we required an accurate set of orthologous identifications to the human proteins. Deducing orthology is difficult due to the prevalence of gene and domain duplications. To address potential problems with orthologous identification and the misdating of proteins in the network, we applied two strategies for the clustering of orthologous sequences, and although there were differences in detail, our conclusions remained the same: the MMM coevolution network separates into two loosely linked subnetworks (fig. 1A). Evolutionary dating of the two subnetworks, given the limitations of species distribution in our data set and the resolution of the phylogenetic tree, points to the separation around the emergence of multicellularity in animals. We thus investigated the functional differences between the proteins in these two subnetworks within this context.
MMM analysis revealed a network of coevolving genes that are more evolutionarily recent and not generally known to interact nor to be coexpressed. However, many of these MMM12+NEW network genes were absent from expression array surveys and are otherwise poorly characterized compared with genes that are conserved throughout the eukaryotes (fig. 4). Moreover, the MMM12+NEW network could be showing coevolution between genes that are potentially coexpressed and interacting only at specific stages of development and in specific tissues, as is likely to be the case for more newly evolved genes specialized for differentiated, multicellular animals.
The OLD network, similar to what we had found for highly conserved proteins (Tillier and Charlebois 2009) and for proteins with yeast orthologs (Clark et al. 2011), showed higher levels of overall protein expression and thus appears to reflect ancient and fundamental interactions important for basic cellular function. In support of this, we found an overrepresentation of cell differentiation markers in the MMM12+NEW network, whereas the OLD network showed more genes involving markers of stem cells (table 1).
Our previous study (Clark et al. 2011) showed that MMM predictions of coevolution most agreed with the (gold standard) interactions in the yeast interaction network coming from mass spectrometry studies. The recent study of protein complexes in Drosophila (Guruharsha et al. 2011) confirms this finding; however, that network is also extremely biased toward interactions between old proteins. The human network described here, especially when considering the NEW interactions, is thus more difficult to validate owing to the paucity of solid experimental data.
Many of the proteins in the MMM12+NEW network are extracellular, cytoskeletal, or nonsoluble membrane proteins, whose interactions are poorly characterized (supplementary data set S3, Supplementary Material online), so it is unclear whether coevolution predicts actual physical protein–protein contacts in this network. The predicted interaction between GPR98 and USH2A in mice was putatively confirmed as a physical interaction in Maerker et al. (2008), indicating that the known PPI databases are, unsurprisingly, incomplete. Despite the accumulation of literature-curated protein interaction data for model organisms such as yeast, allowing for the higher overlap of 20–30% for high-throughput PPI detection methods (Reguly et al. 2006), high-quality human PPI maps are still incomplete (Ewing et al. 2007; Havugimana et al. 2012). In addition, even though there has been significant progress in detecting PPI in multicellular organisms such as fly (Guruharsha et al. 2011), these data would not necessarily apply well to the human proteins, because 60% of the human protein complexes are thought to have originated in vertebrates (Havugimana et al. 2012). The significant under-representation of old–new pairs found in the MMM12+ network is also found in the known PPI databases, such that on average, old proteins interact with old proteins and new with new (supplementary table S1, Supplementary Material online).
An in silico approach has the strong advantage of not suffering from the same technical limitations and biases of laboratory experimentation, though our coevolutionary approach is itself limited in that it can only detect coevolution for proteins within a limited range of evolutionary rates (fig. 3). Sequence coevolution can only be detected in genes that evolve sufficiently quickly to provide a signal but not so quickly as to saturate that signal. We found the correlated evolutionary rates in the NEW network to be increased relative to the OLD network. The fact that these proteins have just the right evolutionary rates to be detected by coevolution may not be coincidental, because an adaptation to multicellularity should coordinately increase the rate of evolution of those proteins involved.
We found a network of coevolving genes involved in cell–cell communication, the cytoskeleton, and the cell cycle, appearing crucial to the evolution and maintenance of multicellularity. Defects in their control would be expected to perturb multicellular homeostasis, and we indeed found a significant overrepresentation in this network of genes involved in ciliopathies and in cancer. This MMM12+NEW network still contains several old proteins, many related to the cilium. The cilium is an ancient appendage, dating to the origin of the eukaryotic cell (Hartman and Smith 2009), but it is still incompletely defined. (Because many of the experimental protocols investigating human protein interactions opt to employ dividing cell lines that are transformed—which in itself is an abnormal phenotype—and lack cilia, it is not surprising that the interactions involving ciliary proteins may be systemically overlooked.) To assemble a working set of cilia/ciliopathy genes, we mostly used annotations from GO and OMIM to find genes involved in human ciliopathies, supplemented with genes commonly found in the ciliome and ciliaproteome databases. These two databases of high-throughput gene expression (Inglis et al. 2006) and proteomics and transcriptomics (Marshall 2008) data have little overlap with one another nor with human disease information and to some extent extrapolate from studies on ciliated or flagellated protists. Efforts to confirm these results in ciliated mammalian cell cultures have only just begun (Lai et al. 2011).
With the caveat that our set of human ciliome genes is probably incomplete, our coevolution network shows connections of several of these ancient proteins with the more recently arisen animal proteins. We also observed links to the centriole, implicating the centrosome and the control of cell division. Interestingly, this could indicate an adaptation of the originally motile cilia to so-called primary cilia, requiring new wiring to adapt the organelle to multicellularity. We did not observe coevolution with genes also involved in cytokinesis that localize at the basal body complex in vertebrate ciliated epithelial cells (Smith et al. 2011). Primary cilia play crucial roles in animals, particularly in vertebrate development (Goetz and Anderson 2010), and loss of their function results in human genetic diseases, both ciliopathies (Gerdes et al. 2009; Tobin and Beales 2009) and cancer.
Some cancers are thought to be linked to ciliopathies due to the importance of cilia and centrosomes in the control of the cell cycle (Tucker et al. 1979; Plotnikova et al. 2008) and in aberrant activation or suppression of the Hedgehog (Hh) and Wnt pathways (Nielsen et al. 2008; Han et al. 2009; Wong et al. 2009), PDGFα (Schneider et al. 2005), and other signaling pathways (Michaud and Yoder 2006). Loss of cilia has been observed in clear cell renal cell carcinoma (Schraml et al. 2008), medulloblastoma (Han et al. 2009), pancreatic cancer (Seeley et al. 2009), astrocytoma/glioblastoma (Moser et al. 2009), and breast cancer (Yuan et al. 2010).
Considering the data from the TCGA, we found the strongest evidence for mutated cilia genes in ovarian cancer and most particularly involving the subset of cilia-linked genes found to be coevolving in the MMM12+NEW network. Additional evidence from expression studies of these tumors also shows high-grade OSC tumors to have lower relative expression levels for many of the cilia genes compared with normal cells (supplementary results, Supplementary Material online). Investigation of tumors from microdissected fallopian epithelium samples confirmed these results. These cilia genes are at the center of the NEW network, which also includes BRCA1 and BRCA2. BRCA2 itself has been shown to localize to the centrosome and to nuclei; the dysfunction of BRCA2 in the centrosome causes abnormalities in cell division (Nakanishi et al. 2007). The dynein proteins DNAH3 and DNAH5 also seem particularly important in ovarian cancer and in our network. There is recent evidence that dynein-binding proteins may regulate the G1-S transition through an effect on cilia (Jackson 2011; Kim et al. 2011; Li et al. 2011), such that ciliated cells require the loss of their cilia to enter the cell cycle.
Mutations in genes in the OLD network would most likely have the most severe phenotypes (depending on heterozygosity and penetrance). The OLD network did show an overrepresentation of disease genes, linked to an overrepresentation of enzyme deficiencies in metabolism (table 1). The NEW network, on the other hand, was found to be enriched for proteins involved in cancer and ciliopathies, thus proteins involved in cell communication, control of cell division, and cell differentiation, providing community homeostasis in a multicellular organism. The enrichment for such disease markers in the NEW network indicates a functional link between these coevolving proteins and provides genetic validation for this network.
With a better understanding of the relationships between ciliary function, important in cell control and communication, and cancer, reflecting the loss of such control and communication, the human coevolution network can help elucidate the processes most relevant to these classes of disease and more generally to the control and maintenance of a cooperative multicellular phenotype. Concerted evolutionary changes implicating the recruitment of the ancient cilium to these new roles should provide a fruitful impetus for further study.
Supplementary Material
Supplementary text S1, table S1, figures S1–S4, and data sets S1–S3 are available at Molecular Biology and Evolution online (http://www.mbe.oxfordjournals.org/).
Acknowledgments
The authors thank Sophia George and Patricia Shaw for sharing their analyses of expression levels in the OSC tumors and for very interesting discussions. This work was supported in part by the Ontario Ministry of Health and Long Term Care and also by The Princess Margaret Hospital Foundation Graduate Fellowships in Cancer Research to A.B. and an NSERC discovery grant to E.R.M.T. The views expressed do not necessarily reflect those of the Ministry.
References
- Alexa A, Rahnenführer J, Lengauer T. Improved scoring of functional groups from gene expression data by decorrelating GO graph structure. Bioinformatics. 2006;22:1600–1607. doi: 10.1093/bioinformatics/btl140. [DOI] [PubMed] [Google Scholar]
- Altenhoff AM, Schneider A, Gonnet GH, Dessimoz C. OMA 2011: orthology inference among 1000 complete genomes. Nucleic Acids Res. 2011;39:D289–D294. doi: 10.1093/nar/gkq1238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bettencourt-Dias M, Hildebrandt F, Pellman D, Woods G, Godinho SA. Centrosomes and cilia in human disease. Trends Genet. 2011;27:307–315. doi: 10.1016/j.tig.2011.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- The Cancer Genome Atlas Research Network. Integrated genomic analyses of ovarian carcinoma. Nature. 2011;474:609–615. doi: 10.1038/nature10166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark GW, Dar V, Bezginov A, Yang JM, Charlebois RL, Tillier ERM. Using coevolution to predict protein-protein interactions. Methods Mol Biol. 2011;781:237–256. doi: 10.1007/978-1-61779-276-2_11. [DOI] [PubMed] [Google Scholar]
- Domazet-Loso T, Tautz D. Phylostratigraphic tracking of cancer genes suggests a link to the emergence of multicellularity in metazoa. BMC Biol. 2010;8:66. doi: 10.1186/1741-7007-8-66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ewing RM, Chu P, Elisma F, et al. (36 co-authors) Large-scale mapping of human protein-protein interactions by mass spectrometry. Mol Syst Biol. 2007;3:89. doi: 10.1038/msb4100134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Felsenstein J. PHYLIP (phylogeny inference package) Cladistics. 1989;5:164–166. [Google Scholar]
- Ford D, Easton DF, Stratton M, et al. (25 co-authors) Genetic heterogeneity and penetrance analysis of the BRCA1 and BRCA2 genes in breast cancer families. The Breast Cancer Linkage Consortium. Am J Hum Genet. 1998;62:676–689. doi: 10.1086/301749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerdes JM, Davis EE, Katsanis N. The vertebrate primary cilium in development, homeostasis, and disease. Cell. 2009;137:32–45. doi: 10.1016/j.cell.2009.03.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gherman A, Davis EE, Katsanis N. The ciliary proteome database: an integrated community resource for the genetic and functional dissection of cilia. Nat Genet. 2006;38:961–962. doi: 10.1038/ng0906-961. [DOI] [PubMed] [Google Scholar]
- Goetz SC, Anderson KV. The primary cilium: a signalling centre during vertebrate development. Nat Rev Genet. 2010;11:331–344. doi: 10.1038/nrg2774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grossmann S, Bauer S, Robinson PN, Vingron M. Improved detection of overrepresentation of Gene-Ontology annotations with parent child analysis. Bioinformatics. 2007;23:3024–3031. doi: 10.1093/bioinformatics/btm440. [DOI] [PubMed] [Google Scholar]
- Guruharsha KG, Rual JF, Zhai B, et al. (24 co-authors) A protein complex network of Drosophila melanogaster. Cell. 2011;147:690–703. doi: 10.1016/j.cell.2011.08.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hakes L, Lovell SC, Oliver SG, Robertson DL. Specificity in protein interactions and its relationship with sequence diversity and coevolution. Proc Natl Acad Sci U S A. 2007;104:7999–8004. doi: 10.1073/pnas.0609962104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han YG, Kim HJ, Dlugosz AA, Ellison DW, Gilbertson RJ, Alvarez-Buylla A. Dual and opposing roles of primary cilia in medulloblastoma development. Nat Med. 2009;15:1062–1065. doi: 10.1038/nm.2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartman H, Smith TF. The evolution of the cilium and the eukaryotic cell. Cell Motil Cytoskeleton. 2009;66:215–219. doi: 10.1002/cm.20344. [DOI] [PubMed] [Google Scholar]
- Havugimana PC, Hart GT, Nepusz T, et al. (24 co-authors) A census of human soluble protein complexes. Cell. 2012;150:1068–1081. doi: 10.1016/j.cell.2012.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inglis PN, Boroevich KA, Leroux MR. Piecing together a ciliome. Trends Genet. 2006;22:491–500. doi: 10.1016/j.tig.2006.07.006. [DOI] [PubMed] [Google Scholar]
- Jackson PK. Do cilia put brakes on the cell cycle? Nat Cell Biol. 2011;13:340. doi: 10.1038/ncb0411-340. [DOI] [PubMed] [Google Scholar]
- Juan D, Pazos F, Valencia A. High-confidence prediction of global interactomes based on genome-wide coevolutionary networks. Proc Natl Acad Sci U S A. 2008;105:934–939. doi: 10.1073/pnas.0709671105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katoh K, Toh H. Recent developments in the MAFFT multiple sequence alignment program. Brief Bioinform. 2008;9:286–298. doi: 10.1093/bib/bbn013. [DOI] [PubMed] [Google Scholar]
- Kim S, Zaghloul NA, Bubenshchikova E, Oh EC, Rankin S, Katsanis N, Obara T, Tsiokas L. Nde1-mediated inhibition of ciliogenesis affects cell cycle re-entry. Nat Cell Biol. 2011;13:351–360. doi: 10.1038/ncb2183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai CK, Gupta N, Wen X, Rangell L, Chih B, Peterson AS, Bazan JF, Li L, Scales SJ. Functional characterization of putative cilia genes by high-content analysis. Mol Biol Cell. 2011;22:1104–1119. doi: 10.1091/mbc.E10-07-0596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee K, Battini L, Gusella GL. Cilium, centrosome and cell cycle regulation in polycystic kidneys disease. Biochim Biophys Acta. 2011;1812:1263–1271. doi: 10.1016/j.bbadis.2011.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li A, Saito M, Chuang JZ, Tseng YY, Dedesma C, Tomizawa K, Kaitsuka T, Sung CH. Ciliary transition zone activation of phosphorylated Tctex-1 controls ciliary resorption, S-phase entry and fate of neural progenitors. Nat Cell Biol. 2011;13:402–411. doi: 10.1038/ncb2218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lukk M, Kapushesky M, Nikkilä J, Parkinson H, Goncalves A, Huber W, Ukkonen E, Brazma A. A global map of human gene expression. Nat Biotechnol. 2010;28:322–324. doi: 10.1038/nbt0410-322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maerker T, van Wijk E, Overlack N, et al. (11 co-authors) A novel Usher protein network at the periciliary reloading point between molecular transport machineries in vertebrate photoreceptor cells. Hum Mol Genet. 2008;17:71–86. doi: 10.1093/hmg/ddm285. [DOI] [PubMed] [Google Scholar]
- Marshall WF. Use of transcriptomic data to support organelle proteomic analysis. Methods Mol Biol. 2008;432:403–414. doi: 10.1007/978-1-59745-028-7_27. [DOI] [PubMed] [Google Scholar]
- Michaud EJ, Yoder BK. The primary cilium in cell signaling and cancer. Cancer Res. 2006;66:6463–6467. doi: 10.1158/0008-5472.CAN-06-0462. [DOI] [PubMed] [Google Scholar]
- Moser JJ, Fritzler MJ, Rattner JB. Primary ciliogenesis defects are associated with human astrocytoma/glioblastoma cells. BMC Cancer. 2009;9:448. doi: 10.1186/1471-2407-9-448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakanishi A, Han X, Saito H, Taguchi K, Ohta Y, Imajoh-Ohmi S, Miki Y. Interference with BRCA2, which localizes to the centrosome during S and early M phase, leads to abnormal nuclear division. Biochem Biophys Res Commun. 2007;355:34–40. doi: 10.1016/j.bbrc.2007.01.100. [DOI] [PubMed] [Google Scholar]
- Nielsen SK, Møllgaard K, Clement CA, Veland IR, Awan A, Yoder BK, Novak I, Christensen ST. Characterization of primary cilia and Hedgehog signaling during development of the human pancreas and in human pancreatic duct cancer cell lines. Dev Dynamics. 2008;237:2039–2052. doi: 10.1002/dvdy.21610. [DOI] [PubMed] [Google Scholar]
- Obayashi T, Kinoshita K. COXPRESdb: a database to compare gene coexpression in seven model animals. Nucleic Acids Res. 2011;39:D1016–D1022. doi: 10.1093/nar/gkq1147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pazos F, Valencia A. Similarity of phylogenetic trees as indicator of protein-protein interaction. Protein Eng. 2001;14:609–614. doi: 10.1093/protein/14.9.609. [DOI] [PubMed] [Google Scholar]
- Plotnikova OV, Golemis EA, Pugacheva EN. Cell cycle-dependent ciliogenesis and cancer. Cancer Res. 2008;68:2058–2061. doi: 10.1158/0008-5472.CAN-07-5838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pugacheva EN, Jablonski SA, Hartman TR, Henske EP, Golemis EA. HEF1-dependent Aurora A activation induces disassembly of the primary cilium. Cell. 2007;129:1351–1363. doi: 10.1016/j.cell.2007.04.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reguly T, Breitkreutz A, Boucher L, et al. (20 co-authors) Comprehensive curation and analysis of global interaction networks in Saccharomyces cerevisiae. J Biol. 2006;5:11. doi: 10.1186/jbiol36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodionov A, Bezginov A, Rose J, Tillier ERM. A new, fast algorithm for detecting protein coevolution using maximum compatible cliques. Algorithms Mol Biol. 2011;6:17. doi: 10.1186/1748-7188-6-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosvall M, Bergstrom CT. Maps of random walks on complex networks reveal community structure. Proc Natl Acad Sci U S A. 2008;105:1118–1123. doi: 10.1073/pnas.0706851105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rual JF, Venkatesan K, Hao T, et al. (38 co-authors) Towards a proteome-scale map of the human protein-protein interaction network. Nature. 2005;437:1173–1178. doi: 10.1038/nature04209. [DOI] [PubMed] [Google Scholar]
- Rzhetsky A, Nei M. A simple method for estimating and testing minimum-evolution trees. Mol Biol Evol. 1992; 9:945–967. [Google Scholar]
- Satir P, Christensen ST. Structure and function of mammalian cilia. Histochem. Cell Biol. 2008;129:687–693. doi: 10.1007/s00418-008-0416-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider L, Clement CA, Teilmann SC, Pazour GJ, Hoffmann EK, Satir P, Christensen ST. PDGFR αα signaling is regulated through the primary cilium in fibroblasts. Curr Biol. 2005;15:1861–1866. doi: 10.1016/j.cub.2005.09.012. [DOI] [PubMed] [Google Scholar]
- Schraml P, Frew IJ, Thoma CR, Boysen G, Struckmann K, Krek W, Moch H. Sporadic clear cell renal cell carcinoma but not the papillary type is characterized by severely reduced frequency of primary cilia. Modern Pathol. 2008;22:31–36. doi: 10.1038/modpathol.2008.132. [DOI] [PubMed] [Google Scholar]
- Seeley ES, Carrière C, Goetze T, Longnecker DS, Korc M. Pancreatic cancer and precursor pancreatic intraepithelial neoplasia lesions are devoid of primary cilia. Cancer Res. 2009;69:422. doi: 10.1158/0008-5472.CAN-08-1290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shannon P, Markiel A, Ozier O, Baliga NS, Wang JT, Ramage D, Amin N, Schwikowski B, Ideker T. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 2003;13:2498–2504. doi: 10.1101/gr.1239303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith KR, Kieserman EK, Wang PI, Basten SG, Giles RH, Marcotte EM, Wallingford JB. A role for central spindle proteins in cilia structure and function. Cytoskeleton. 2011;68:112–124. doi: 10.1002/cm.20498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srivastava M, Simakov O, Chapman J, et al. (33 co-authors) The Amphimedon queenslandica genome and the evolution of animal complexity. Nature. 2010;466:720–726. doi: 10.1038/nature09201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tarassov K, Michnick SW. iVici: interrelational visualization and correlation interface. Genome Biol. 2005;6:R115. doi: 10.1186/gb-2005-6-13-r115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tillier ERM, Charlebois RL. The human protein coevolution network. Genome Res. 2009;19:1861–1871. doi: 10.1101/gr.092452.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tobin JL, Beales PL. The nonmotile ciliopathies. Genet Med. 2009;11:386–402. doi: 10.1097/GIM.0b013e3181a02882. [DOI] [PubMed] [Google Scholar]
- Tone AA, Begley H, Sharma M, Murphy J, Rosen B, Brown TJ, Shaw PA. Gene expression profiles of luteal phase fallopian tube epithelium from BRCA mutation carriers resemble high-grade serous carcinoma. Clin Cancer Res. 2008;14:4067–4078. doi: 10.1158/1078-0432.CCR-07-4959. [DOI] [PubMed] [Google Scholar]
- Tucker RW, Pardee AB, Fujiwara K. Centriole ciliation is related to quiescence and DNA synthesis in 3T3 cells. Cell. 1979;17:527–535. doi: 10.1016/0092-8674(79)90261-7. [DOI] [PubMed] [Google Scholar]
- Wong SY, Seol AD, So PL, Ermilov AN, Bichakjian CK, Epstein EH, Dlugosz AA, Reiter JF. Primary cilia can both mediate and suppress Hedgehog pathway-dependent tumorigenesis. Nat Med. 2009;15:1055–1061. doi: 10.1038/nm.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan K, Frolova N, Xie Y, Wang D, Cook L, Kwon YJ, Steg AD, Serra R, Frost AR. Primary cilia are decreased in breast cancer: analysis of a collection of human breast cancer cell lines and tissues. J Histochem Cytochem. 2010;58:857–870. doi: 10.1369/jhc.2010.955856. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.