Abstract
Gene duplicates generated via retroposition were long thought to be pseudogenized and consequently decayed. However, a significant number of these genes escaped their evolutionary destiny and evolved into functional genes. Despite multiple studies, the number of functional retrogenes in human and other genomes remains unclear. We performed a comparative analysis of human, chicken, and worm genomes to identify “orphan” retrogenes, that is, retrogenes that have replaced their progenitors. We located 25 such candidates in the human genome. All of these genes were previously known, and the majority has been intensively studied. Despite this, they have never been recognized as retrogenes. Analysis revealed that the phenomenon of replacing parental genes with their retrocopies has been taking place over the entire span of animal evolution. This process was often species specific and contributed to interspecies differences. Surprisingly, these retrogenes, which should evolve in a more relaxed mode, are subject to a very strong purifying selection, which is, on average, two and a half times stronger than other human genes. Also, for retrogenes, they do not show a typical overall tendency for a testis-specific expression. Notably, seven of them are associated with human diseases. Recognizing them as “orphan” retrocopies, which have different regulatory machinery than their parents, is important for any disease studies in model organisms, especially when discoveries made in one species are transferred to humans.
Keywords: retrogene, gene duplication, gene expression, human genetic disease
Introduction
Despite advances in molecular biology and plethora of genomic and transcriptomic data, understanding genetic basis of diseases and turning basic science discoveries into therapies remains challenging. Animal experiments have contributed a lot to decoding the mechanisms of diseases. However, the value of animal studies in predicting the effectiveness of treatment is often controversial (Hackam 2007; Perel et al. 2007; van der Worp et al. 2010). Inconsistency between animal models and clinical trials may be explained by inadequate animal data or simply because animal models do not reflect disease in humans in a satisfactory way.
The key in deciphering this disparity is in understanding interspecies differences and translating genomes into phenotypes. Phenotypic diversity, beside environmental factors, is generated through changes in the genomic sequence. Without knowing which genomic features result in phenotypic differences between species, we will not be able to predict functional consequences of transferring model organism research results to medical treatment of humans. One of the fundamental factors in the evolution of lineage-specific and species-specific traits is the birth of new genes. Gene duplication is the major process contributing to the origin of these genes. There are two mechanisms for gene duplication: DNA-based creating copies with genetic features similar to their parental genes and RNA based. In RNA-based duplication, mRNA is reverse-transcribed into cDNA and reintegrated into a new location in the genome (Vanin 1984; Weiner et al. 1986; Brosius 1991). Although the mechanism of this process has not been widely studied, there is experimental evidence that in humans the machinery of long interspersed repeats is used (Esnault et al. 2000). In this type of duplication, multi-exon genes give birth to single-exon copies which, in most cases, lack regulatory elements and are commonly believed to be pseudogenes (Mighell et al. 2000). However, many of them are known to produce new, very often lineage-specific genes (Betran, Wang, et al. 2002; Marques et al. 2005; Svensson et al. 2006). They can also lead to new protein domains through fusion with other genes (Vinckenbosch et al. 2006; Baertsch et al. 2008), regulatory RNAs (Yano et al. 2004; Devor 2006), or other regulatory elements (Nozawa et al. 2005).
Soares et al. (1985) discovered for the first time a functional retrosequence in the rodent genome in 1985. They found that the rat insulin I gene is a functional retrocopy of the insulin II gene. This finding was followed by the number of discoveries of functional retrogenes in mammalian genomes (McCarrey and Thomas 1987; Ashworth et al. 1990) (for review see Brosius 1999) as well as in the fruit fly (Long and Langley 1993; Betran, Thornton, et al. 2002). Although several genome-wide surveys have been performed over the last decade, it is still unknown how many retrogenes are actually transcribed in human and other genomes. It is estimated that the human genome contains approximately 8,000 retrogenes (Zhang et al. 2003). Harrison et al. (2005) found that some 4–6% of them are abundantly expressed. Utilizing in silico assays Vinckenbosh et al. (2006) identified over 1,000 transcribed retrogenes, out of which 120 evolved into bona fide genes. Other investigators reported that only 2–3% of processed pseudogenes are transcribed in the human genome (Yano et al. 2004; Yu et al. 2007) and an even lower number of functional retrogenes in the human genome come from the studies of Sakai et al. (2007). Only 79 of retrogenes studied by them had evidence for transcription and they estimated that 1.08% of all processed pseudogenes are transcribed. In the most recent studies, Pan and Zhang (2009) identified 163 functional human retrogenes.
Retrogenes, for a long time considered being “dead on arrival” copies of parental genes, are nowadays often called “seeds of evolution” (Brosius 1991) because they made a significant contribution to molecular evolution. As duplicates of their parental genes, these retrocopies evolve fast because duplication events allow a relaxed purifying selection, so that these genes may acquire novel functions. They are important source of functional innovations and species-specific traits. For example, retrogene fgf4 is responsible for the dogs’ chondrodisplasia. All breeds with short legs are carriers of the fgf4 retrogene (Parker et al. 2009). Another example of retrogenes contribution in shaping interspecies differences is retrogene RNF113B, which gained an intron in primates and has two splicing forms with distinct expression patterns while in other mammals it has only one single-exon form (Szczesniak et al. 2011).
Retrogenes are also known to be involved in many diseases. A good example is the RHOB gene, a tumor suppressor of the Rho GTPases family (Prendergast 2001), which arose by retroposition in the early stage of vertebrate evolution (Sakai et al. 2007). Mutation in another retrogene, TACSTD2 (tumor-associated calcium signal transducer 2) causes gelatinous drop-like corneal dystrophy leading to blindness (Tsujikawa et al. 1999).
Although several efforts have been made to detect functional retrogenes, their number remains unclear. A genome-wide study showed that 20% of mammalian protein encoding genes lack introns in their coding sequence (Sakharkar et al. 2002). Therefore, it is conceivable that many genes lacking introns arose by retroposition. In published studies, the identification of retrogenes was always based on the assumption that both, the parental gene and its retrocopy, are present in the genome. Therefore, only genomic sequence loci that were homologous to multi-exon genes were considered and single-exon genes without close paralogs were automatically eliminated from the set of putative retrogenes. However, we cannot exclude the possibility that the parental gene was lost or pseudogenized after the duplication and the retrogene, which took over its function, does not have any multi-exon homologs. Here, we present a comparative analysis of human, chicken, and worm genes leading to the identification of 25 “orphan” retrogenes, which likely replaced their progenitors, in the human genome. All of them are functional and although most were studied more intensively, none of them were ever recognized as a retrogene.
Materials and Methods
Identification of “Orphan” Retrogenes
The sequence collection used in this study consisted of 5,342 human transcripts encoded by single exon genes, and 60,922 human and 4,613 chicken mRNAs encoded by multi-exon genes as annotated in the UCSC Genome Browser database (Fujita et al. 2011), assemblies hg18 and galGal3, respectively. We deliberately used all human transcripts encoded by single-exon genes to avoid the exclusion of transcribed retrogenes annotated as noncoding due to the frameshift, premature stop codons, missing 3′- or 5′-end of coding sequence, and annotation errors. In addition to human and chicken genes, sequences of 4,649 human–worm orthologs were downloaded from the InParanoid database (Ostlund et al. 2010).
“Orphan” retrogenes in the human genome, that is, retrocopies without their parental genes present in the genome, were identified using three approaches. The first two were based on the analysis of sequence similarity between human and chicken genes. Furthermore, in the second approach, the genomic location was taken into consideration. The third approach relied on the gene structure analysis of already predefined human and Caenorhabditis elegans orthologs.
Method I
mRNA sequences from single-exon and multi-exon human genes and chicken multi-exon genes were downloaded using the UCSC Table Browser. The set of human single-exon genes was next filtered to exclude out histone sequences, which are known to be intronless in all vertebrates, as well as all sequences equal or shorter than 200 bp to eliminate putative small RNAs. In this step, we removed 79 and 2006 sequences, respectively. The remaining 3,257 sequences were used as a query in translated similarity searches, using TBLASTX (Altschul et al. 1997), against mRNAs of multi-exon chicken genes and against mRNAs of human multi-exon genes. Following the similarity searches, results were filtered based on three criteria: 1) identity percentage, 2) score in the BLAST searches, and 3) query coverage in the alignment with chicken mRNAs. Approved for further analysis were single-exon human genes that showed a higher alignment score and a higher similarity to chicken multi-exon genes than to human multi-exon gene and with an alignment covering at least 35% of the chicken mRNA sequence. After filtering, the resulting set of sequences was manually checked and all cases with an uncertain status were removed.
The manual checking included BLASTX searches against human and other genomes, synteny analysis of a retrogene and the parental gene orthologs, analysis of annotations in several resources such ENSEMBL, UCSC Genome Browser, NCBI genomic maps, as well as alignment analysis to confirm that alignment of retrogene and its parental gene ortholog covers more than two exons. The main reasons for rejecting candidates were incorrect annotations in the chicken genome, gaps in the sequence creating artificial introns, and the alignment spanning only one exon of the parental gene ortholog. In few cases, the candidate was discarded due to the presence of parental gene paralogs and uncertainty, which of the gene was a progenitor of a given retrogene.
Method II
In the second approach, filtered transcripts from human intronless genes were used for a BLAST search against chicken multi-exon genes. Sequences with no hits to the chicken mRNAs and those with alignments to chicken transcripts shorter than 100 bp were removed from the set. The remaining pairs, a human single-exon gene and its matching chicken multi-exon gene, were analyzed in regard to their chromosomal localization and surrounding genomic sequence. We compared, by BLAST searches, genes in the nearest vicinity of candidate retrogene in the human genome and in the region near the multi-exon gene in the chicken genome. Based on the assumption that a retroposed gene will have different neighbors than its parental gene, all pairs that have as neighbors orthologous genes at one or both sides were eliminated from the data set. All gene pairs that passed this filtering were manually examined and, similarly to method I, all cases with an uncertain status were removed.
Method III
In the last approach, identifiers of human and C. elegans proteins coded by orthologous genes were downloaded from the InParanoid database (version 7.0) (Ostlund et al. 2010). All proteins identifiers were converted into nucleotide accession numbers using Galaxy (Goecks et al. 2010) and for each gene the exon number was obtained using the UCSC Table Browser (Karolchik et al. 2004). All pairs where a human gene had only one exon and the matching C. elegans gene had two or more exons were selected and manually inspected.
In the search for “orphan” retrogenes, we intentionally did not use a standard practice applied in the retrogenes identification studies, which is mapping all multi-exon genes to the genomic sequence. This approach, although very efficient in identifying retrocopies, would return a lot of pseudoretrogenes, which were beyond our interests.
Identification of Orthologous Genes in Other Species of Animals
To determine the evolutionary history of identified human “orphan” retrogenes, we looked for their orthologs and/or orthologs of their parental genes in seven vertebrate species: Mus musculus (house mouse), Bos Taurus (cattle), Monodelphis domestica (opossum), Ornithorhyncus anatinus (platypus), Gallus gallus (chicken), Xenopus tropicalis (western clawed frog), and Danio rerio (zebrafish) as well as in one insect species: Drosophila melanogaster (fruit fly). Orthology relations between genes were established based on the annotations in the NCBI Gene database (Maglott et al. 2011) and the Ensembl database (Flicek et al. 2011) as well as BLAST (Sayers et al. 2011) similarity searches.
Gene Expression Analysis
Expression of identified “orphan” retrogenes was analyzed in MTC Multiple Tissue cDNA Panels, Human I and Human II, from Clontech. The selected panels represented together cDNA libraries from 16 human tissues and organs: heart, brain, placenta, lung, liver, skeletal muscle, kidney, pancreas, spleen, thymus, prostate, testis, ovary, small intestine w/o mucosal lining, colon, and peripheral leucocytes. As a positive and a negative control, GAPHD and GYS2, respectively, were used as recommended by the cDNA libraries provider.
Forward and reverse primers for all genes were designed using Primer-BLAST (Sayers et al. 2011) with the following parameters: product length 120–160 bp; primers melting temperature (Tm) 58–62°C; GC content between 40% and 60%.
The expression of analyzed genes was determined by a real-time polymerase chain reaction (PCR) method (Kubista et al. 2006) performed in Applied Biosystems 7900HT System with Power SYBR Green PCR Master Mix (Applied Biosystems) and the results were interpreted using SDS Software 2.3. The cut-off value for CT (cycle threshold) was established as 32 based on the optimal cut-off for real-time PCR experiments obtained in other studies. Results were visualized through the construction of a heatmap in the R software environment (version 2.11.1).
Identification of MicroRNA Target Sites and TFBS Analysis
Information about microRNA target sites was obtained from TargetScan Release 5.1, a database of target site predictions (Friedman et al. 2009). Identification of potential binding sites for transcription factors in DNA sequences was performed using Match™ – 1.0 Public (Alamanova et al. 2010). We analyzed 1,000 nt upstream sequence for each gene and looked for transcription factor binding sites with the highest two most important parameters: the matrix similarity score and the core similarity score. Identification was limited to vertebrate-specific weight matrices.
Calculation of KA/KS Ratio
The KA/KS ratio for human retrogenes and their orthologs in mice was calculated using the KAKS_Calculator, which uses the MYN method (modified version of the Yang–Nielsen method) (Zhang et al. 2006).
Results
Identification of Retrogenes without Parents
As proposed in several papers by Nei and coworkers (Ota and Nei 1994; Nei et al. 2000; Nikolaidis et al. 2005) gene families may evolve by the “birth-and-death process.” Therefore, after the speciation event, the divergence between two resultant species may be shaped by the gradual accumulation of gene gains and losses. Retroposition provides a wealth of gene duplicates. These so-called processed pseudogenes are considered to have little evolutionary significance as they are “dead on arrival” and represent disabled copies of functional parental gene (Li et al. 1981; Lynch and Conery 2000). However, some of them gain a function and become functional paralogs (Soares et al. 1985; McCarrey and Thomas 1987; Ashworth et al. 1990; Long and Langley 1993; Brosius 1999). Thus, according to the “birth-and-death evolution,” we may expect that after divergence in one lineage both copies may be retained, in another the retrocopy may be lost, and yet in another the parental gene will lose its function and the retrogene will be left as the only functional copy.
Zhang et al. (2010) described what they called unitary pseudogenes in the primate lineage. They identified 87 unprocessed pseudogenes without functioning counterparts. These genes, although well established in the vertebrate lineage, are extinct in humans and/or other primates. In this study, we also looked for well-established genes that were lost, for example, due to deletion, or pseudogenized in the human genome. However, the function of these genes was undertaken by their duplicates—retrocopies. These presumed “orphan” retrogenes were identified based on the comparative analysis of human, chicken, and worm genes using three different approaches as described in the Materials and Methods section. In the first one, putative orphan retrogenes were selected based on similarity searches, in which human single-exon genes were run against human and chicken multi-exon gene transcripts. The results of both BLAST searches were compared and sequences showing higher similarity to chicken genes than to human genes were selected. Seventeen single-exon human genes met these rigorous filtering criteria. However, after manual checking only four pairs of human retrogenes and chicken orthologs of their parental genes remained.
In the second approach, the results of a similarity search for human single-exon genes versus chicken multi-exon genes were filtered and pairs of human–chicken sequences with at least 100 bp alignments were selected for further studies. Only 915 pairs met this criterion. For further data processing, considering the mechanism of retroposition, we made a rather obvious assumption that a retrogene and its parental gene, or in this case the ortholog of parental gene, should have different genomic locations. Based on this deduction, we analyzed sequences surrounding genes from each human–chicken pair and removed those that had orthologous genes at one or both sides. This analysis returned 260 potential pairs of “orphan” retrogenes in the human genome and orthologs of its parental gene in the chicken genome. Nevertheless, only nine pairs were confirmed after manual examination, out of which four were identified in the previous approach.
It is noticeable that the ratio of false-positives in methods I and II was relatively high. This may imply inaccuracy in the methodology. However, majority of false positives come from incorrect annotations of the chicken genome. In addition, gaps in the chicken genomic sequence were generating artificial introns and often single-exon chicken genes would appear, according to annotations, as multi-exon.
The third strategy relied on the orthology relationships established in the InParanoid database (Ostlund et al. 2010). 4649 human–Caenorhabditis elegans orthologous groups were identified in the database. After filtering followed by an exon number comparison, as described in Material and Methods, 58 pairs were selected. Twenty pairs passed manual verification and four of them were already identified by methods I and II. This gave 16 new “orphan” retrogenes. Therefore, overall we identified 25 unique retrogenes, which do not have their parental gene in the human genome. All of these genes are listed in table 1. Interestingly, only for one retrogene, CHMP1B, we were able to find traces of the parental gene in the human genome. In other cases, the region where the parental gene was located was either deleted or mutated to the degree in which no similarity can be found.
Table 1.
“Orphan” Retrogenes in the Human Genome.
| Gene Symbol | Gene Name | Chromosomal Localization | Ka | Ks | Ka/Ks | |
|---|---|---|---|---|---|---|
| 1 | MAB21L1 | Mab-21-like 1 | 13 | 0 | 0.74 | 0 |
| 2 | MAB21L2 | Mab-21-like 2 | 4 | 0.001 | 0.806 | 0.001 |
| 3 | PURA | Purine-rich element binding protein A | 5 | 0.001 | 0.29 | 0.004 |
| 4 | ADRA2Aa | Adrenergic, alpha-2A-, receptor | 10 | 0.036 | 2,112 | 0.017 |
| 5 | CHMP1Ba | Chromatin modifying protein 1B | 18 | 0.009 | 0.398 | 0.022 |
| 6 | IMP3a | U3 small nucleolar ribonucleoprotein | 15 | 0.017 | 0.681 | 0.024 |
| 7 | EXOC8 | Exocyst complex component 8 | 1 | 0.03 | 1.214 | 0.024 |
| 8 | B3GALT6 | UDP-Gal:betaGal beta 1,3-galactosyltransferase polypeptide 6 | 1 | 0.073 | 1.79 | 0.041 |
| 9 | RRS1a | RRS1 ribosome biogenesis regulator | 8 | 0.042 | 0.963 | 0.043 |
| 10 | TTC30B | Tetratricopeptide repeat domain 30B | 2 | 0.037 | 0.594 | 0.063 |
| 11 | PIGMa | Phosphatidylinositol glycan anchor biosynthesis, class M | 1 | 0.051 | 0.698 | 0.073 |
| 12 | MOCS3 | Molybdenum cofactor synthesis 3 | 20 | 0.117 | 1.391 | 0.084 |
| 13 | TBCC | Tubulin folding cofactor C | 6 | 0.126 | 1.489 | 0.085 |
| 14 | CH25H | Cholesterol 25-hydroxylase | 10 | 0.11 | 1.151 | 0.095 |
| 15 | CEBPB | CCAAT/enhancer binding protein (C/EBP), beta | 20 | 0.068 | 0.687 | 0.099 |
| 16 | ADRA2B | Adrenergic, alpha-2B-, receptor | 2 | 0.079 | 0.769 | 0.103 |
| 17 | MARS2 | Methionyl-tRNA synthetase 2 | 2 | 0.073 | 0.697 | 0.105 |
| 18 | UTP3 | Small subunit (SSU) processome component | 4 | 0.063 | 0.589 | 0.108 |
| 19 | KTI12 | KTI12 homolog, chromatin associated | 1 | 0.129 | 1.165 | 0.111 |
| 20 | MGAT2a | Mannosyl (alpha-1,6-)-glycoprotein beta-1,2-N-acetylglucosaminyltransferase | 14 | 0.058 | 0.407 | 0.144 |
| 21 | RNF113A | Ring finger protein 113A | X | 0.066 | 0.423 | 0.156 |
| 22 | SFT2D3 | SFT2 domain containing 3 | 2 | 0.129 | 0.822 | 0.157 |
| 23 | ZNF830 | Zinc finger protein 830 | 17 | 0.09 | 0.459 | 0.197 |
| 24 | TRMT12a | tRNA methyltransferase 12 homolog | 8 | 0.107 | 0.515 | 0.208 |
| 25 | LCMT2 | Leucine carboxyl methyltransferase 2 | 15 | 0.131 | 0.54 | 0.242 |
aGene associated with human disease.
Zhang et al. (2011) pointed out that partial DNA-level duplications of intron containing genes can make a significant contribution to the existence of intronless genes. Therefore, even relatively long alignments between single-exon genes and intron-containing parents may not be sufficient to define a new copy as retrogene. Keeping this in mind, in the process of manual evaluation, we looked not only at the alignment length but also checked whether the alignment covers exon–exon junctions of putative parental gene ortholog. The graphical representation of this comparison is shown in supplementary figure S1, Supplementary Material online. It is visible that in all identified by us retrogene–parental ortholog pairs alignments cover all or majority of introns located in the coding region.
Retroposition and Loss of Parental Gene
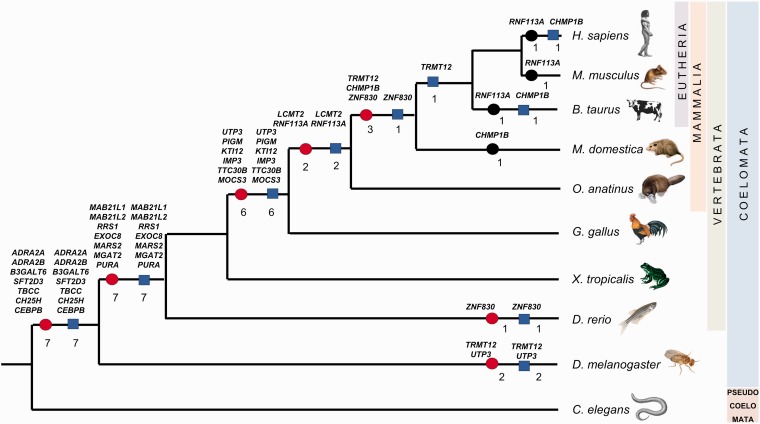
Each pair of genes, either human–chicken or human–C. elegans, was further examined in selected animal species: house mouse, cattle, opossum, platypus, zebrafish, frog, and fruit fly. In addition, genes identified in method III were investigated in the chicken genome. Using genome annotations and similarity searches, we looked for orthologs of retrogenes as well as orthologs of multi-exonic parental genes. The main goal of this analysis was to estimate the time when the retroposition took place and when the parental gene was lost or pseudogenized. We were able to identify the time of these events for all genes. Interestingly, the loss of the parental gene occurred, in most cases, almost simultaneously with retroposition, before the next major phylogenetic split (fig. 1). The exceptions are genes CHMP1B and TRMT12 in the mammalian lineage. The first of these, retrogene CHMP1B, arose in a common ancestor of placental mammals but the parental gene is still functioning in some mammals, for example, in rodents. In other species, such as humans and cattle, the parental gene was pseudogenized. This loss of function in the human and cow genomes occurred independently. TRMT12 was also retroposed in the genome of the placental mammals' ancestor but the parental gene was lost after the divergence of Metatheria and Eutheria (fig. 1).
Fig. 1.
Phylogenetic tree showing points of retroposition and parental gene loss for each retrocopy. Red circle represents retroposition; blue square, parental gene loss; black circle, retrogene duplication or retroposition.
We cannot exclude that in some cases, the parental gene is not observed in the genomic sequence due to the sequencing gaps. However, this is not very likely in the case of the human genome and genomes of model organisms such as mouse, fruit fly, and C. elegans, which were sequenced with high coverage and are well annotated. For other genomes used in the analysis, we cannot completely rule out the possibility that the parental gene exists but was not sequenced.
It is known that retroposition has a remarkably high rate in placental mammals (Moran et al. 1996; Ostlund et al. 2010), and therefore we expected that the turnover between the parental gene and its retrocopy will be especially intensive in this taxonomic group. Surprisingly, the highest rate of parental gene loss subsequent to the retroposition was before the divergence of vertebrates. Seven genes were retroposed and eventually lost right after the divergence of Pseudocelomata and Celomata and also seven retrogenes replaced their parental genes in the common ancestor of vertebrates (fig. 1). The next wave of the birth of “orphan” retrogenes started in the genome of the warm-blooded animals’ predecessor. Six retrogenes substituted parental genes at this point of the evolution and two parental genes were lost in the genome of the mammalian ancestor. Only three retrogenes took the place of their progenitors in placental mammals, out of which two in Eutheria.
Our analyses also revealed that four parental genes, which are lost in the human genome, independently vanished in other species (fig. 1). It was already mentioned in this article that the progenitor of the CHMP1B retrogene was pseudogenized in the human as well as in the cattle genome. In addition ZNF830 was replaced by its retrocopy in Danio rerio. Two retrogenes, TRMT12 and UTP3, took the place of their parents in the D. melanogaster genome.
Disease Association
As we have already mentioned, retrogenes can be involved in human diseases (Tsujikawa et al. 1999; Prendergast 2001; Zemojtel et al. 2010). Identified by us “orphan” retrogenes are not the exception in this matter. However, in all previously described cases both genes, a retrocopy and its parent, were present. Here, we identified disease-associated retrogenes, which functionally replaced their parental genes. These genes, although coding for the same protein as the pseudogenized parent, have different regulatory machinery, as promoter regions are not inherited in the process of retrotransposition. There is an evidence for functional evolution of retrogenes and differences in the expression scheme between the parental gene and its functional retrocopy (Zhang et al. 2002; Marques et al. 2005; Vinckenbosch et al. 2006; Zemojtel et al. 2010). Therefore, we may anticipate that “orphan” retrogenes are not necessarily regulated in the same way as their parents were. This should be kept in mind in any disease studies in model organisms, where discoveries made in one species are transferred to humans, especially when one organism has functional parental gene and the other only its retrocopy.
Among 25 “orphan” retrogenes identified by us, seven are involved in human diseases, which corresponds to 28% of all identified genes. Two of these genes are linked to cancer. The IMP3 gene is expressed in tumors and its expression level is associated with metastasis in renal cell carcinomas and patient’s survival rate (Jiang, Chu, et al. 2008; Jiang, Lohse, et al. 2008). Overexpression of another “orphan” retrogene, TRMT12, may lead to translation errors in breast tumor cells (Rodriguez et al. 2007). A high expression level of ADRA2A can increase type 2 diabetes risk (Rosengren et al. 2010). The same gene is also involved in attention-deficit/hyperactivity disorder (Roman et al. 2006). Other examples include MGAT2 responsible for defective brain development (Tan et al. 1996), mutation of ADRB1 is associated with congestive heart failure and beta-blocker response (Mason et al. 1999), RRS1 is involved in endoplasmic reticulum stress response in Huntington’s disease (Carnemolla et al. 2009), and PIGM is linked to glycosylphosphatidylinositol deficiency (Almeida et al. 2006).
It is expected that molecular evolution of retrogenes is selectively neutral and therefore these genes evolve relatively quickly, although there is evidence for retrogenes under strong purifying selection (Vinckenbosch et al. 2006; Yu et al. 2007). The degree and type of selection can be measured by the ratio of nonsynonymous substitutions (KA) to synonymous substitutions (KS). Under neutral evolution KA = KS, deviation of KA from KS may be due to positive selection when the KA/KS is >1, or purifying selection when KA/KS < 1. Nevertheless, genes are considered to be under strong purifying selection when KA/KS ratio is ≪1 (Hurst 2002). We calculated the KA/KS ratio for all “orphan” human retrogenes and their orthologs in mouse (table 1). As the results show, none of these genes are evolving neutrally and the KA/KS ratio is <0.25 for all of them, strongly indicating that retrogenes, which replaced their parents, are under purifying selection. The average ratio for all 25 genes is 0.088 and it is much lower than the average for human–mouse genes, which was estimated as 0.180 (Makalowski and Boguski 1998). An even stronger purifying selection is observed in the case of seven disease-associated “orphan” retrogenes. The average ratio for this group is 0.076. Interestingly, this value is lower than previously published. Tu et al. (2006) analyzed the evolutionary rate for human disease genes and obtained, for human–mouse orthologs, average KA/KS ratio 0.12. Another group (Thomas et al. 2003) analyzed 121 human genes implicated in cancer and calculated the average ratio to be 0.079, which is close to the value obtained by us. It is intriguing that the retrogenes studied by us, disease related or not, are under a similarly strong pressure as cancer-related genes.
Although we did not apply any minimum similarity filtering, it is possible that methods used by us led to the enrichment of slow evolving genes in our set. On the other hand, these genes represent single-copy or two-copy genes, which are known to be slowly evolving (Waterhouse et al. 2011).
A Study Case of CHMP1B Gene
An interesting case represents CHMP1B, a retrogene associated with hereditary spastic paraplegia (Reid et al. 2005). This gene was retroposed before the divergence of Theria. The retrogene was then either tandemly duplicated or retroposed in Metatheria as opossum has two single-exon genes and one multi-exon gene. In the Eutherian lineage, the retrogene and its parent coexist in the majority of the taxa. However, in the human and cattle genomes the parental genes do not function anymore. Pseudogenization of the CHMP1B parental gene was independent in both lineages since in mice and rats, which like humans belong to Euarchontoglires, the parental gene is intact and expressed in various tissues. In the primate lineage, the CHMP1B parent was pseudogenized in the genome of the ancestor of Old World and New World monkeys because this gene is fragmentary in all available primate genomes: marmoset, macaque, orangutan, chimpanzee, and human.
Proteins coded by the CHMP1B retrogene and its functional parents are highly conserved (fig. 2), which may indicate that retrogene gained its function shortly after the retroposition and immediately became subjected to purifying selection. The strong pressure to conserve protein sequences confirms the KA/KS ratio, which is 0.012 for the mouse retrogene and its parent and 0.022 for human and mouse retrogenes. This is an order of magnitude lower than average KA/KS ratio (0.18) for human–mouse coding sequences (Makalowski and Boguski 1998). The human parental gene, although pseudogenized, does get expressed; there is one mRNA sequence, CR627394, and two EST sequences deposited in the GenBank. Nevertheless, from the very low number of ESTs, we may conclude that the expression level of this gene is very low. Also, this gene is significantly different from its ortholog in mice. It contains only parts of exons coding for the prototype protein: fragment of exon 2 and most of exons 3 and 5 (fig. 2). In addition, there is a frameshift since a fragment of exon 2 is in frame +1 and the other two exons are in frame +3. Interestingly, nearly all the coding exons present in the mouse gene can be detected in the human genomic sequence but they are not used in any transcript.
Fig. 2.

Alignment of proteins coded by human and mouse CHMP1B retrogenes and their parental genes (functional gene in mouse and pseudogene in human genome).
Retroposed genes need to recruit regulatory elements to become transcribed and usually, as a consequence of hiring transcription regulation factors different from their parent, acquire a new function. We performed analysis of 1,000 bp upstream sequences of human and mouse CHMP1B retrogenes and the mouse parental gene. Indeed, regulatory elements present in upstream sequences of retrogenes differ from elements observed in parental gene’s regulatory region. Three transcription factor binding sites (TFBS): CREB, CRE-BP1, and E2F are specific for human and mouse retrogenes and are not found in the regulatory region of the mouse parental gene. On the other hand, the mouse parental gene has two unique TFBS: HNF-1 and Evi-1. There is no single TFBS shared between all three genes (fig. 3). However, the transcript level is not regulated exclusively by the transcription factors. Short RNA molecules like microRNA may bind to the complementary sequence on target transcripts leading to translational repression and gene silencing (Ambros 2004). MicroRNA target sites are located in 3′-UTR sequences and therefore, unlike transcription factor binding sites, are inherited by retrogenes. It is known that the conservation of 3′-UTRs is much lower than conservation of coding sequence (Makalowski and Boguski 1998). Nevertheless, most microRNA targets are well conserved in mammalian mRNAs (Friedman et al. 2009). Employing TargetScan (Friedman et al. 2009), we identified microRNA target sites in CHMP1B retrogenes and their parental genes, functional or pseudogenized, in several mammalian species. The TargetScan identified only one microRNA target site, site for miR-743ab/743b-3p, conserved in all functional parental genes. The target sequence for this microRNA, present in rodent, horse, and elephant genes, was clearly deleted in human and chimpanzee where the gene was pseudogenised (fig. 4A). None of the other target sites recognized by the program were conserved in all functional genes. For example, sites for miR-155 and miR-669f are conserved in rodent and elephant functional genes but not in horse genes. On the other hand, the target site for miR-9 is conserved in mouse, rat, and horse but not in elephant. All these four target sites are conserved in the human pseudogene and three of them in the chimpanzee pseudogene.
Fig. 3.
Upstream regions of human and mouse CHMP1B retrogenes and mouse parental gene with annotated positions of identified transcription factor binding sites. TFBS which are shared by retrogenes but not present in upstream sequence of parental gene have darker background.
Fig. 4.
microRNA target sites in 3′-UTR sequences of CHMP1B mammalian retrogenes (A) and available functional or pseudogenized parental genes (B).
CHMP1B retrogenes have two highly conserved microRNA target sites, miR-9 and miR-182, which are present in all available transcripts from placental mammals (fig. 4B). Interestingly, only one of them, target site for miR-9, is also present in some but not all functional parental genes. In addition, this site has a different location in parental genes and in retrogenes and the microRNA–mRNA pairing type is also different. Although in retrogenes the site for miR-9 is 7mer-1A type, in parental genes it is type 7mer-m8 (Friedman et al. 2009).
It is quite interesting that retrogenes, which are expected to evolve under a more relaxed selective pressure, have conserved microRNA target sites to a greater extent than that of parental genes. However, considering the pseudogenization of parental gene in some genomes, the lack of high conservation of microRNA target sites in the remaining functional genes may indicate that retrogenes took over the function in all genomes and the parental gene is an “unnecessary copy,” which eventually may also lose its function in other mammalian genomes.
Expression Pattern
Gene retroposition, together with segmental duplication, belongs to the central mechanisms responsible for the creation of species-specific traits (Brosius 1991, 1999; Marques et al. 2005). Duplication of chromosomal segments tends to produce daughter copies that inherit features of their parental genes. Therefore, these copies show not only the same protein functions but also similar expression patterns. On the contrary, the retroposed cDNA is generally expected to lack regulatory elements and duplicated genes are considered to be “dead on arrival.” However, as a number of studies shows, many of them do acquire new functions (Burki and Kaessmann 2004; Krasnov et al. 2005; Sakai et al. 2007; Kaessmann et al. 2009). These new functions, usually different from the functions of parental genes, may come from the gain of new spatiotemporal expression patterns, imposed by the content of the genomic sequence surrounding inserted cDNA. Numerous studies revealed a tendency of retrogenes to be expressed in the testis (Marques et al. 2005; Vinckenbosch et al. 2006; Potrzebowski et al. 2008) and a significant excess of autosomal testis-expressed retrogenes were identified as duplicates of X-linked parental genes (Betran, Thornton, et al. 2002). This specific transcription of retrocopies may be resulting from the hypertranscription state observed in meiotic and postmeiotic spermatogenic cells (Kleene 2001). An alternative explanation may come from the hypothesis that retrocopies are preferentially inserted into actively transcribed, and therefore open chromatin (Fontanillas et al. 2007). As the retroposition occurs in the germ line, retrocopies may primarily be inserted into, or nearby genes expressed in the germ line. This could enable and/or enhance their expression in testis. Yet another hypothesis, based on the fact that there is an excess of retrogenes originated from the X chromosome, links this testis-specific expression with an escape from the male meiotic sex chromosome inactivation (Emerson et al. 2004; Wang 2004).
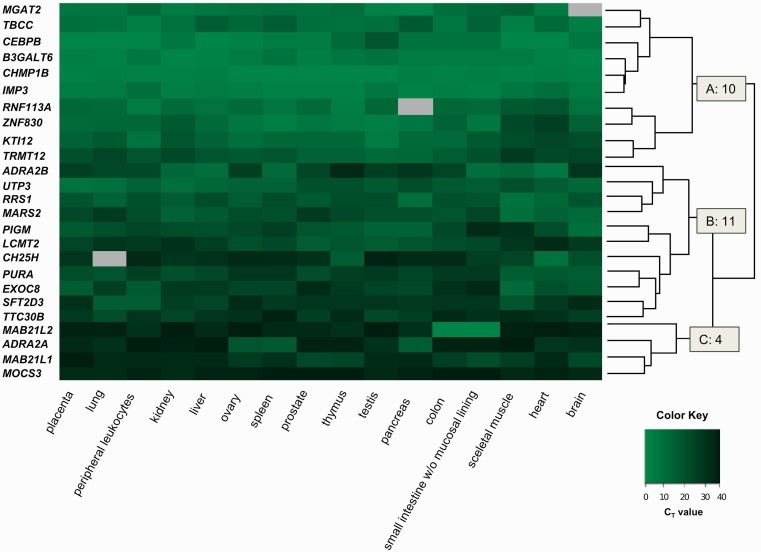
Preferential expression of retrogenes in testis was previously reported for retrocopies for which functional parent genes prevail in a given genome (Brosius 1991, 1999; Marques et al. 2005). To test if this specific pattern is also observable in “orphan” retrogenes we performed a real-time PCR for all 25 retrogenes in 16 human cDNA libraries including a cDNA library from testis. Real-time PCR CT values referring to the number of cycles during reaction in which product (dsDNA) appeared, with cut-off CT 32, were used to construct a heat map of expression profiles with a dendrogram (fig. 5). A majority of investigated retrogenes, 19 out of 25, was detected in all libraries. Five genes were expressed in 15 libraries and 1 in 14. No single retrogene revealed a testis-specific expression, including those that originated from genes located on chromosome X, like CHMP1B or TRMT12; both of them are ubiquitously expressed.
Fig. 5.
Heat map representing expression pattern of all identified human “orphan” retrogenes. Gray color indicates undetermined CT values.
Dai et al. (2006) found that new genes seem to be expressed in fewer tissues or organs in comparison with parental genes. From the presented data, obviously we cannot make any conclusions as for the change in the expression pattern in comparison with these genes progenitors because parental genes are not present in the human genome and comparison with other species would be questionable. However, we made one interesting observation. The expression pattern of studied retrogenes is related to their age. Younger retrocopies tend to be expressed in all tissues and have a higher expression level. Cluster A represents retrogenes with the strongest and broadest expression. Out of 10 genes in this cluster, six were retroposed in the ancestor of warmblooded animals or later. Clusters B (moderate expression) and C (lowest expression) are build in majority from genes retroposed before vertebrates. This is quite intriguing since, according to a previous study (Wolf et al. 2009), we should rather expect that retrogenes slowly gain functions as they get older and their regulatory regions “mature.” Apparently, it seems to be the opposite in the case of “orphan” retrogenes where younger copies have, on average, a broader and higher expression.
Discussion
Gene duplicates generated via retroposition were long thought to be pseudogenized and consequently decayed. However, a significant number of these genes escaped their evolutionary destiny and evolved into functional genes. The function of the retrogenes was usually discussed in the aspects of neofunctionalization and/or subfunctionalization (Kaessmann et al. 2009). Here, we presented the first genome wide analysis aimed at the identification of retrogenes which replaced their progenitors and took over their functions. We identified 25 functional retrogenes, for which parental genes do not exist or do not function anymore in the human genome. None of these genes were considered earlier as retrogenes. One of the most surprising discoveries was the fact that many of these genes have ancient origins dating back even more than 900 million years and are common for all Coelomata. Obviously, we cannot exclude that these intronless copies originated via other than retroposition mechanism of intron loss; however, retroposition is the most parsimonious and most plausible in the case where all introns from a given gene have disappeared. Unexpectedly, despite a very intensive retroposition in placental mammals (Moran et al. 1996), a relatively low number of retrogenes replaced their parent in the mammalian lineage. One explanation could be that they just need a long time to do so but the data does not verify this. The replacement of the parental gene, in the majority of cases, was in the same lineage, before the next major divergence.
It is postulated that molecular evolution of retrocopies is selectively neutral, whereas their parental genes are subject to purifying selection. Indeed, Yu et al. (2007) found that the majority of retrogenes are in the state of a “relaxed” selection. Nonetheless, they also discovered that some human retrogenes are undergoing a nonneutral evolution. Retrogenes under a strong purifying selection were also identified by Vinckenbosch et al. (2006). Apparently, all the identified here “orphan” retrogenes are under a strong purifying selection. We showed that the CHMP1B protein is highly conserved between mouse parental genes and retrogenes as well as between human and mouse retrogenes. This strong conservation and low KA/KS values are characteristic for all analyzed by us genes. As shown in table 1, the ratio of nonsynonymous to synonymous substitution for all but three genes is below the average value estimated for human–mouse genes, which is 0.18 (Makalowski and Boguski 1998) and the average for all “orphan” retrogenes is about two times lower: 0.088. Therefore, this particular group of retrogenes is not only, without any exception, under a strong purifying selection but also evolves at a lower than average rate. This rate is even lower for disease associated “orphan” retrogenes: 0.076. The high conservation level is in concordance with the observation that these genes replaced their parents soon after the retroposition. Consequently, they became the only functional copy of the gene and their evolution was immediately constrained by a purifying selection.
Large-scale analyses of retrogenes in mammals and fruit flies revealed the overall tendency to testis-specific expression (Marques et al. 2005; Vinckenbosch et al. 2006; Potrzebowski et al. 2008). This trend was observed independently of the parental gene expression pattern. Shiao et al. (2007) showed that mouse retrogenes are expressed at more restrictive pattern than parental paralogs and all of them were expressed predominantly in testis. Similar observation was made by Dai et al. (2006) based on the Drosophila retrogenes study. Our study does not confirm this bias. The majority of “orphan” retrogenes was expressed in all examined 16 tissues/organs. Not a single gene showed a testis-specific expression pattern. The simple explanation of this disparity may be in the fact that analyzed by us retrogenes naturally mimic the parental expression pattern and therefore, have much broader expression than expected. It was also suggested that the propensity to be expressed in testis observed in other studies might be related to the fact that in meiotic and postmeiotic spermatogenic cells chromosomes are in the state of hypertranscription. This state enables transcription of DNA that is usually not transcribed and therefore facilitates the transcription of retrocopies (Kleene 2001). Subsequently, these retrocopies could evolve into bona fide genes, enhance their regulatory elements, and broaden the range of tissues they get expressed in. If this would be a scenario for “orphan” retrogenes evolution we would see a limited expression in younger retrogenes and a wider expression in older copies. Evidently the picture is quite the opposite, younger genes from our set tend to be ubiquitously expressed at relatively high level and the older ones have more limited expression. These results are in disagreement with the studies of Wolf et al. (2009) who found that among human genes those that are eukaryote specific, “old” ones, are expressed at a higher level than younger, mammalian-specific genes.
It has been shown that many retrogenes, also those that are functional, are species-specific and contribute to interspecies differences. Some of these differences are of a high importance in medical research and may be responsible for the fact that results from animal studies cannot be transferred to humans. For example, the functional mouse retrogene Rps23r1 reduces Alzheimer’s beta-amyloid levels and tau phosphorylation (Zhang et al. 2009). However, results of this study cannot be applied to humans because this particular retrogene is rodent specific and does not exist in the human genome. Recognizing which retrogene is species specific, which replaced its parental gene, and which coexists with its progenitor is of high importance. In each of these scenarios genes would behave differently. If parental genes and retrogenes function as a single copy (i.e., parental only or retrogene only), they would code for the same protein but their expression regulation would be different. Therefore, it would be crucial to check if genes that seem to be very similar from the protein comparison level are truly orthologous before transferring animal studies to humans. If both copies exist, we may expect that there will be either subfunctionalization and functions previously carried out by parental genes will be divided between these two copies or alternatively a retrocopy could develop completely new functions. In the described example of the CHMP1B gene, the human retrogene was associated with hereditary spastic paraplegia (Reid et al. 2005). Mice are the most likely species of choice when one would like to study this gene in a model organism. However, mice have both a functional retrogene and its parent, coding for almost identical protein. In the human genome, the parental gene got pseudogenized and does not code for a functional protein anymore. Although the parental gene could compensate mutation in the CHMP1B retrogene in mice, in humans it could not. Therefore, studies on the CHMP1B gene in mice may not be, by any means, comparable with what is taking place in humans.
Here, we presumed that analyzed retrogenes functionally replaced pseudogenized parental genes. To consider these evolutionary events as perfect “replacement,” the retrogene would need to have the same regulatory sequences as parental gene and exhibit identical expression pattern. Because retrogenes, in most cases, do not inherit regulatory regions (the exception is the case when parental gene has alternative regulatory motifs in the 3′-UTR region), they need to acquire new regulatory machinery. This could happen either by mutations and positive selection leading to the origination of appropriate regulatory elements or by the “hitchhiking” of the existing elements regulating nearby gene. Without assurance that newly developed or adopted elements are the same as possessed by parental gene we cannot, in unquestionable way, determine whether the events described by us illustrate “replacement” or neofunctionalization. Because for the majority of retrogenes, there is no detectable trace of their parents in the human genome we cannot perform any considerable comparative studies. However, it would be interesting to see how evolutionary processes change the genomic sequence into the regulatory elements and to what degree these sequences mimic sequences of parental genes. To comprehend these processes a large-scale comparative analysis of functional retrogenes and their progenitors are required and such studies were recently launched in our laboratory.
Before the final conclusions, it is necessary to point out that the number of 25 “orphan” retrogenes in the human genome may seem to be low and not very appealing. At this point, it is impossible to form the opinion whether the number of such genes simply is so low or maybe the methodology needs to be worked out for better results as there are no studies to compare with. However, identifying retrogenes that lost their progenitors is very challenging due to the fact that many genes underwent multiple, and sometimes partial, duplications followed by significant changes in the gene structure, which often are difficult to trace. In addition, poorly annotated genomes likely produce false positives. Moreover, many retrogenes are known to gain exons and introns and in this particular study, we focused only on single exon genes. Nevertheless, we are currently conducting analyses concentrated on functional retrocopies, which acquired new exons and/or gain introns. It is quite conceivable that this study will reveal additional examples of human “orphan” retrogenes.
In summary, we may say that “orphan” retrogenes represent a very specific group of genes. They not only replaced their parental gene but also “behave” in unexpected ways. Although previous studies suggested that retrogenes evolve neutrally or under a relaxed functional constraint, they are actually more conserved than the average gene. They also seem to have a reversed expression pattern, that is, younger genes have higher expression and older ones are more limited. In addition, many of them are involved in serious human diseases. Altogether, these facts make this class of genes extremely interesting.
Supplementary Material
Supplementary figure S1 is available at Molecular Biology and Evolution online (http:www.mbe.oxfordjournals.org/).
Acknowledgments
The authors thank Jakub Dolata for technical support during real-time PCR experiments. This work was supported by Ministry of Science and Higher Education grant no. N303 320 437 (to I.M.), National Science Centre grant no. 2011/01/N/NZ2/01701 (to J.C.), and Seventh Frame Work Programme of the European Union, International Research Staff Exchange Scheme grant no. PIRSES-GA-2009-247633 (to I.M, W.M., and J.C).
References
- Alamanova D, Stegmaier P, Kel A. Creating PWMs of transcription factors using 3D structure-based computation of protein-DNA free binding energies. BMC Bioinformatics. 2010;11:225. doi: 10.1186/1471-2105-11-225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Almeida AM, Murakami Y, Layton DM, et al. (17 co-authors) Hypomorphic promoter mutation in PIGM causes inherited glycosylphosphatidylinositol deficiency. Nat Med. 2006;12:846–851. doi: 10.1038/nm1410. [DOI] [PubMed] [Google Scholar]
- Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ambros V. The functions of animal microRNAs. Nature. 2004;431:350–355. doi: 10.1038/nature02871. [DOI] [PubMed] [Google Scholar]
- Ashworth A, Skene B, Swift S, Lovell-Badge R. Zfa is an expressed retroposon derived from an alternative transcript of the Zfx gene. EMBO J. 1990;9:1529–1534. doi: 10.1002/j.1460-2075.1990.tb08271.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baertsch R, Diekhans M, Kent WJ, Haussler D, Brosius J. Retrocopy contributions to the evolution of the human genome. BMC Genomics. 2008;9:466. doi: 10.1186/1471-2164-9-466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Betran E, Thornton K, Long M. Retroposed new genes out of the X in Drosophila. Genome Res. 2002;12:1854–1859. doi: 10.1101/gr.604902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Betran E, Wang W, Jin L, Long M. Evolution of the phosphoglycerate mutase processed gene in human and chimpanzee revealing the origin of a new primate gene. Mol Biol Evol. 2002;19:654–663. doi: 10.1093/oxfordjournals.molbev.a004124. [DOI] [PubMed] [Google Scholar]
- Brosius J. Retroposons—seeds of evolution. Science. 1991;251:753. doi: 10.1126/science.1990437. [DOI] [PubMed] [Google Scholar]
- Brosius J. RNAs from all categories generate retrosequences that may be exapted as novel genes or regulatory elements. Gene. 1999;238:115–134. doi: 10.1016/s0378-1119(99)00227-9. [DOI] [PubMed] [Google Scholar]
- Burki F, Kaessmann H. Birth and adaptive evolution of a hominoid gene that supports high neurotransmitter flux. Nat Genet. 2004;36:1061–1063. doi: 10.1038/ng1431. [DOI] [PubMed] [Google Scholar]
- Carnemolla A, Fossale E, Agostoni E, Michelazzi S, Calligaris R, De Maso L, Del Sal G, MacDonald ME, Persichetti F. Rrs1 is involved in endoplasmic reticulum stress response in Huntington disease. J Biol Chem. 2009;284:18167–18173. doi: 10.1074/jbc.M109.018325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dai H, Yoshimatsu TF, Long M. Retrogene movement within- and between-chromosomes in the evolution of Drosophila genomes. Gene. 2006;385:96–102. doi: 10.1016/j.gene.2006.04.033. [DOI] [PubMed] [Google Scholar]
- Devor EJ. Primate microRNAs miR-220 and miR-492 lie within processed pseudogenes. J Hered. 2006;97:186–190. doi: 10.1093/jhered/esj022. [DOI] [PubMed] [Google Scholar]
- Emerson JJ, Kaessmann H, Betran E, Long M. Extensive gene traffic on the mammalian X chromosome. Science. 2004;303:537–540. doi: 10.1126/science.1090042. [DOI] [PubMed] [Google Scholar]
- Esnault C, Maestre J, Heidmann T. Human LINE retrotransposons generate processed pseudogenes. Nat Genet. 2000;24:363–367. doi: 10.1038/74184. [DOI] [PubMed] [Google Scholar]
- Flicek P, Amode MR, Barrell D, et al. (52 co-authors) Ensembl 2011. Nucleic Acids Res. 2011;39:D800–D806. doi: 10.1093/nar/gkq1064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fontanillas P, Hartl DL, Reuter M. Genome organization and gene expression shape the transposable element distribution in the Drosophila melanogaster euchromatin. PLoS Genet. 2007;3:e210. doi: 10.1371/journal.pgen.0030210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedman RC, Farh KK, Burge CB, Bartel DP. Most mammalian mRNAs are conserved targets of microRNAs. Genome Res. 2009;19:92–105. doi: 10.1101/gr.082701.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujita PA, Rhead B, Zweig AS. The UCSC genome browser database: update 2011. Nucleic Acids Res. 2011;39:D876–D882. doi: 10.1093/nar/gkq963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goecks J, Nekrutenko A, Taylor J. Galaxy: a comprehensive approach for supporting accessible, reproducible, and transparent computational research in the life sciences. Genome Biol. 2010;11:R86. doi: 10.1186/gb-2010-11-8-r86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hackam DG. Translating animal research into clinical benefit. BMJ. 2007;334:163–164. doi: 10.1136/bmj.39104.362951.80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrison PM, Zheng D, Zhang Z, Carriero N, Gerstein M. Transcribed processed pseudogenes in the human genome: an intermediate form of expressed retrosequence lacking protein-coding ability. Nucleic Acids Res. 2005;33:2374–2383. doi: 10.1093/nar/gki531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hurst LD. The Ka/Ks ratio: diagnosing the form of sequence evolution. Trends Genet. 2002;18:486–487. doi: 10.1016/s0168-9525(02)02722-1. [DOI] [PubMed] [Google Scholar]
- Jiang Z, Chu PG, Woda BA, Liu Q, Balaji KC, Rock KL, Wu CL. Combination of quantitative IMP3 and tumor stage: a new system to predict metastasis for patients with localized renal cell carcinomas. Clin Cancer Res. 2008;14:5579–5584. doi: 10.1158/1078-0432.CCR-08-0504. [DOI] [PubMed] [Google Scholar]
- Jiang Z, Lohse CM, Chu PG, Wu CL, Woda BA, Rock KL, Kwon ED. Oncofetal protein IMP3: a novel molecular marker that predicts metastasis of papillary and chromophobe renal cell carcinomas. Cancer. 2008;112:2676–2682. doi: 10.1002/cncr.23484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaessmann H, Vinckenbosch N, Long M. RNA-based gene duplication: mechanistic and evolutionary insights. Nat Rev Genet. 2009;10:19–31. doi: 10.1038/nrg2487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karolchik D, Hinrichs AS, Furey TS, Roskin KM, Sugnet CW, Haussler D, Kent WJ. The UCSC table browser data retrieval tool. Nucleic Acids Res. 2004;32:D493–D496. doi: 10.1093/nar/gkh103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleene KC. A possible meiotic function of the peculiar patterns of gene expression in mammalian spermatogenic cells. Mech Dev. 2001;106:3–23. doi: 10.1016/s0925-4773(01)00413-0. [DOI] [PubMed] [Google Scholar]
- Krasnov AN, Kurshakova MM, Ramensky VE, Mardanov PV, Nabirochkina EN, Georgieva SG. A retrocopy of a gene can functionally displace the source gene in evolution. Nucleic Acids Res. 2005;33:6654–6661. doi: 10.1093/nar/gki969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kubista M, Andrade JM, Bengtsson M, et al. (12 co-authors) The real-time polymerase chain reaction. Mol Aspects Med. 2006;27:95–125. doi: 10.1016/j.mam.2005.12.007. [DOI] [PubMed] [Google Scholar]
- Li WH, Gojobori T, Nei M. Pseudogenes as a paradigm of neutral evolution. Nature. 1981;292:237–239. doi: 10.1038/292237a0. [DOI] [PubMed] [Google Scholar]
- Long M, Langley CH. Natural selection and the origin of jingwei, a chimeric processed functional gene in Drosophila. Science. 1993;260:91–95. doi: 10.1126/science.7682012. [DOI] [PubMed] [Google Scholar]
- Lynch M, Conery JS. The evolutionary fate and consequences of duplicate genes. Science. 2000;290:1151–1155. doi: 10.1126/science.290.5494.1151. [DOI] [PubMed] [Google Scholar]
- Maglott D, Ostell J, Pruitt KD, Tatusova T. Entrez gene: gene-centered information at NCBI. Nucleic Acids Res. 2011;39:D52–D57. doi: 10.1093/nar/gkq1237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makalowski W, Boguski MS. Evolutionary parameters of the transcribed mammalian genome: an analysis of 2,820 orthologous rodent and human sequences. Proc Natl Acad Sci U S A. 1998;95:9407–9412. doi: 10.1073/pnas.95.16.9407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marques AC, Dupanloup I, Vinckenbosch N, Reymond A, Kaessmann H. Emergence of young human genes after a burst of retroposition in primates. PLoS Biol. 2005;3:e357. doi: 10.1371/journal.pbio.0030357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mason DA, Moore JD, Green SA, Liggett SB. A gain-of-function polymorphism in a G-protein coupling domain of the human beta1-adrenergic receptor. J Biol Chem. 1999;274:12670–12674. doi: 10.1074/jbc.274.18.12670. [DOI] [PubMed] [Google Scholar]
- McCarrey JR, Thomas K. Human testis-specific PGK gene lacks introns and possesses characteristics of a processed gene. Nature. 1987;326:501–505. doi: 10.1038/326501a0. [DOI] [PubMed] [Google Scholar]
- Mighell AJ, Smith NR, Robinson PA, Markham AF. Vertebrate pseudogenes. FEBS Lett. 2000;468:109–114. doi: 10.1016/s0014-5793(00)01199-6. [DOI] [PubMed] [Google Scholar]
- Moran JV, Holmes SE, Naas TP, DeBerardinis RJ, Boeke JD, Kazazian HH., Jr High frequency retrotransposition in cultured mammalian cells. Cell. 1996;87:917–927. doi: 10.1016/s0092-8674(00)81998-4. [DOI] [PubMed] [Google Scholar]
- Nei M, Rogozin IB, Piontkivska H. Purifying selection and birth-and-death evolution in the ubiquitin gene family. Proc Natl Acad Sci U S A. 2000;97:10866–10871. doi: 10.1073/pnas.97.20.10866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nikolaidis N, Makalowska I, Chalkia D, Makalowski W, Klein J, Nei M. Origin and evolution of the chicken leukocyte receptor complex. Proc Natl Acad Sci U S A. 2005;102:4057–4062. doi: 10.1073/pnas.0501040102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nozawa M, Aotsuka T, Tamura K. A novel chimeric gene, siren, with retroposed promoter sequence in the Drosophila bipectinata complex. Genetics. 2005;171:1719–1727. doi: 10.1534/genetics.105.041699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ostlund G, Schmitt T, Forslund K, Kostler T, Messina DN, Roopra S, Frings O, Sonnhammer EL. InParanoid 7: new algorithms and tools for eukaryotic orthology analysis. Nucleic Acids Res. 2010;38:D196–D203. doi: 10.1093/nar/gkp931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ota T, Nei M. Divergent evolution and evolution by the birth-and-death process in the immunoglobulin VH gene family. Mol Biol Evol. 1994;11:469–482. doi: 10.1093/oxfordjournals.molbev.a040127. [DOI] [PubMed] [Google Scholar]
- Pan D, Zhang L. Burst of young retrogenes and independent retrogene formation in mammals. PLoS One. 2009;4:e5040. doi: 10.1371/journal.pone.0005040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parker HG, VonHoldt BM, Quignon P, et al. (17 co-authors) An expressed fgf4 retrogene is associated with breed-defining chondrodysplasia in domestic dogs. Science. 2009;325:995–998. doi: 10.1126/science.1173275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perel P, Roberts I, Sena E, Wheble P, Briscoe C, Sandercock P, Macleod M, Mignini LE, Jayaram P, Khan KS. Comparison of treatment effects between animal experiments and clinical trials: systematic review. BMJ. 2007;334:197. doi: 10.1136/bmj.39048.407928.BE. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Potrzebowski L, Vinckenbosch N, Marques AC, Chalmel F, Jegou B, Kaessmann H. Chromosomal gene movements reflect the recent origin and biology of therian sex chromosomes. PLoS Biol. 2008;6:e80. doi: 10.1371/journal.pbio.0060080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prendergast GC. Actin' up: RhoB in cancer and apoptosis. Nat Rev Cancer. 2001;1:162–168. doi: 10.1038/35101096. [DOI] [PubMed] [Google Scholar]
- Reid E, Connell J, Edwards TL, Duley S, Brown SE, Sanderson CM. The hereditary spastic paraplegia protein spastin interacts with the ESCRT-III complex-associated endosomal protein CHMP1B. Hum Mol Genet. 2005;14:19–38. doi: 10.1093/hmg/ddi003. [DOI] [PubMed] [Google Scholar]
- Rodriguez V, Chen Y, Elkahloun A, Dutra A, Pak E, Chandrasekharappa S. Chromosome 8 BAC array comparative genomic hybridization and expression analysis identify amplification and overexpression of TRMT12 in breast cancer. Genes Chromosomes Cancer. 2007;46:694–707. doi: 10.1002/gcc.20454. [DOI] [PubMed] [Google Scholar]
- Roman T, Polanczyk GV, Zeni C, Genro JP, Rohde LA, Hutz MH. Further evidence of the involvement of alpha-2A-adrenergic receptor gene (ADRA2A) in inattentive dimensional scores of attention-deficit/hyperactivity disorder. Mol Psychiatry. 2006;11:8–10. doi: 10.1038/sj.mp.4001743. [DOI] [PubMed] [Google Scholar]
- Rosengren AH, Jokubka R, Tojjar D, et al. (16 co-authors) Overexpression of alpha2A-adrenergic receptors contributes to type 2 diabetes. Science. 2010;327:217–220. doi: 10.1126/science.1176827. [DOI] [PubMed] [Google Scholar]
- Sakai H, Koyanagi KO, Imanishi T, Itoh T, Gojobori T. Frequent emergence and functional resurrection of processed pseudogenes in the human and mouse genomes. Gene. 2007;389:196–203. doi: 10.1016/j.gene.2006.11.007. [DOI] [PubMed] [Google Scholar]
- Sakharkar MK, Kangueane P, Petrov DA, Kolaskar AS, Subbiah S. SEGE: a database on “intron less/single exonic” genes from eukaryotes. Bioinformatics. 2002;18:1266–1267. doi: 10.1093/bioinformatics/18.9.1266. [DOI] [PubMed] [Google Scholar]
- Sayers EW, Barrett T, Benson DA, et al. (42 co-authors) Database resources of the National Center for Biotechnology Information. Nucleic Acids Res. 2011;39:D38–D51. doi: 10.1093/nar/gkq1172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shiao MS, Khil P, Camerini-Otero RD, Shiroishi T, Moriwaki K, Yu HT, Long M. Origins of new male germ-line functions from X-derived autosomal retrogenes in the mouse. Mol Biol Evol. 2007;24:2242–2253. doi: 10.1093/molbev/msm153. [DOI] [PubMed] [Google Scholar]
- Soares MB, Schon E, Henderson A, Karathanasis SK, Cate R, Zeitlin S, Chirgwin J, Efstratiadis A. RNA-mediated gene duplication: the rat preproinsulin I gene is a functional retroposon. Mol Cell Biol. 1985;5:2090–2103. doi: 10.1128/mcb.5.8.2090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Svensson O, Arvestad L, Lagergren J. Genome-wide survey for biologically functional pseudogenes. PLoS Comput Biol. 2006;2:e46. doi: 10.1371/journal.pcbi.0020046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szczesniak MW, Ciomborowska J, Nowak W, Rogozin IB, Makalowska I. Primate and rodent specific intron gains and the origin of retrogenes with splice variants. Mol Biol Evol. 2011;28:33–37. doi: 10.1093/molbev/msq260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan J, Dunn J, Jaeken J, Schachter H. Mutations in the MGAT2 gene controlling complex N-glycan synthesis cause carbohydrate-deficient glycoprotein syndrome type II, an autosomal recessive disease with defective brain development. Am J Hum Genet. 1996;59:810–817. [PMC free article] [PubMed] [Google Scholar]
- Thomas MA, Weston B, Joseph M, Wu W, Nekrutenko A, Tonellato PJ. Evolutionary dynamics of oncogenes and tumor suppressor genes: higher intensities of purifying selection than other genes. Mol Biol Evol. 2003;20:964–968. doi: 10.1093/molbev/msg110. [DOI] [PubMed] [Google Scholar]
- Tsujikawa M, Kurahashi H, Tanaka T, Nishida K, Shimomura Y, Tano Y, Nakamura Y. Identification of the gene responsible for gelatinous drop-like corneal dystrophy. Nat Genet. 1999;21:420–423. doi: 10.1038/7759. [DOI] [PubMed] [Google Scholar]
- Tu Z, Wang L, Xu M, Zhou X, Chen T, Sun F. Further understanding human disease genes by comparing with housekeeping genes and other genes. BMC Genomics. 2006;7:31. doi: 10.1186/1471-2164-7-31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Worp HB, Howells DW, Sena ES, Porritt MJ, Rewell S, O'Collins V, Macleod MR. Can animal models of disease reliably inform human studies? PLoS Med. 2010;7:e1000245. doi: 10.1371/journal.pmed.1000245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanin EF. Processed pseudogenes: characteristics and evolution. Biochim Biophys Acta. 1984;782:231–241. doi: 10.1016/0167-4781(84)90057-5. [DOI] [PubMed] [Google Scholar]
- Vinckenbosch N, Dupanloup I, Kaessmann H. Evolutionary fate of retroposed gene copies in the human genome. Proc Natl Acad Sci U S A. 2006;103:3220–3225. doi: 10.1073/pnas.0511307103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang PJ. X chromosomes, retrogenes, and their role in male reproduction. Trends Endocrinol Metab. 2004;15:79–83. doi: 10.1016/j.tem.2004.01.007. [DOI] [PubMed] [Google Scholar]
- Waterhouse RM, Zdobnov EM, Kriventseva EV. Correlating traits of gene retention, sequence divergence, duplicability, and essentiality in vertebrates, arthropods, and fungi. Genome Biol Evol. 2011;3:75–86. doi: 10.1093/gbe/evq083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiner AM, Deininger PL, Efstratiadis A. Nonviral retroposons: genes, pseudogenes, and transposable elements generated by the reverse flow of genetic information. Annu Rev Biochem. 1986;55:631–661. doi: 10.1146/annurev.bi.55.070186.003215. [DOI] [PubMed] [Google Scholar]
- Wolf YI, Novichkov PS, Karev GP, Koonin EV, Lipman DJ. The universal distribution of evolutionary rates of genes and distinct characteristics of eukaryotic genes of different apparent ages. Proc Natl Acad Sci U S A. 2009;106:7273–7280. doi: 10.1073/pnas.0901808106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yano Y, Saito R, Yoshida N, Yoshiki A, Wynshaw-Boris A, Tomita M, Hirotsune S. A new role for expressed pseudogenes as ncRNA: regulation of mRNA stability of its homologous coding gene. J Mol Med. 2004;82:414–422. doi: 10.1007/s00109-004-0550-3. [DOI] [PubMed] [Google Scholar]
- Yu Z, Morais D, Ivanga M, Harrison PM. Analysis of the role of retrotransposition in gene evolution in vertebrates. BMC Bioinformatics. 2007;8:308. doi: 10.1186/1471-2105-8-308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zemojtel T, Duchniewicz M, Zhang Z, Paluch T, Luz H, Penzkofer T, Scheele JS, Zwartkruis FJ. Retrotransposition and mutation events yield Rap1 GTPases with differential signalling capacity. BMC Evol Biol. 2010;10:55. doi: 10.1186/1471-2148-10-55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J, Zhang YP, Rosenberg HF. Adaptive evolution of a duplicated pancreatic ribonuclease gene in a leaf-eating monkey. Nat Genet. 2002;30:411–415. doi: 10.1038/ng852. [DOI] [PubMed] [Google Scholar]
- Zhang YE, Vibranovski MD, Krinsky BH, Long M. A cautionary note for retrocopy identification: DNA-based duplication of intron-containing genes significantly contributes to the origination of single exon genes. Bioinformatics. 2011;27:1749–1753. doi: 10.1093/bioinformatics/btr280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang YW, Liu S, Zhang X, et al. (17 co-authors) A functional mouse retroposed gene Rps23r1 reduces Alzheimer's beta-amyloid levels and tau phosphorylation. Neuron. 2009;64:328–340. doi: 10.1016/j.neuron.2009.08.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z, Harrison PM, Liu Y, Gerstein M. Millions of years of evolution preserved: a comprehensive catalog of the processed pseudogenes in the human genome. Genome Res. 2003;13:2541–2558. doi: 10.1101/gr.1429003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z, Li J, Zhao XQ, Wang J, Wong GK, Yu J. KaKs_Calculator: calculating Ka and Ks through model selection and model averaging. Genomics Proteomics Bioinformatics. 2006;4:259–263. doi: 10.1016/S1672-0229(07)60007-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang ZD, Frankish A, Hunt T, Harrow J, Gerstein M. Identification and analysis of unitary pseudogenes: historic and contemporary gene losses in humans and other primates. Genome Biol. 2010;11:R26. doi: 10.1186/gb-2010-11-3-r26. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.