Abstract
Cells deficient in the recQ-like helicase BLM are characterized by chromosome changes that suggest the disruption of normal mechanisms needed to resolve recombination intermediates and to maintain chromosome stability. Human BLM and topoisomerase IIα interact directly via amino acids 489-587 of BLM and co-localize predominantly in late G2- and M-phases of the cell cycle. Deletion of this region does not affect the inherent in vitro helicase activity of BLM but inhibits the topoisomerase IIα-dependent enhancement of its activity, based on analysis of specific DNA substrates that represent some recombination intermediates. Deletion of the interaction domain from BLM fails to correct the elevated chromosome breakage of transfected BLM-deficient cells. Our results demonstrate that the BLM-topoisomerase IIα interaction is important for preventing chromosome breakage and elucidate a DNA repair mechanism that is critical to maintain chromosome stability in cells and prevent tumor formation.
Introduction
The BLM helicase is a recQ-like, structure-specific helicase with 3’-5’ directionality (1) mutated in the autosomal recessive chromosome breakage disorder Bloom syndrome (BS) (2). Those affected are small in stature, sun-sensitive, immunodeficient, and predisposed to multiple cancers (3). Cytogenetically, BS cells demonstrate increased sister chromatid exchanges (4), quadriradial structures (4), telomere associations (5) and chromosome breaks (6). These chromosomal abnormalities resulting from BLM loss first suggested a role for BLM in processes such as homologous recombination (HR) and double strand break (DSB) repair
BLM is a component of at least three unique protein complexes, including the BRCA1-associated complex (7), a telomere-specific complex in telomerase-negative immortalized cells (8), and the BTB multienzyme complex (9–11). BLM is one of the first proteins to localize to DSBs, recruit other repair and checkpoint proteins, and ensure proper DSB processing (12–15). BLM localizes to promyelocytic leukemia protein (PML) bodies and the nucleolus (16), as well as to a newly defined class of ultrafine anaphase bridges (UFBs) (17), a large number of which contain fragile site DNA and where it may function in sister chromatid decatenation (18). BS cells display excessive numbers of anaphase bridges and lagging chromosomes, structures that represent incompletely segregated chromosomes most likely caused by unresolved recombination events that then give rise to chromosome breaks generated by mitotic spindle forces (19). Indeed, chromosome breakage was the first distinguishing cytological characteristic observed in BS cells (6).
The elevated chromosome breakage of BS cells is highly reminiscent of that observed in cells where topoisomerase IIα activity is inhibited (20). Topoisomerase IIα, a type II topoisomerase that functions as a homodimer, cleaves both strands of a DNA duplex to allow passage of another DNA duplex, unknotting or decatenating DNA (21). The BLM ortholog Sgs1 in the budding yeast Saccharomyces cerevisiae binds a type II topoisomerase (22); both are required for proper chromosome segregation (23). Recent work on telomeric protein complexes in mammalian cells using the alternative lengthening of telomeres (ALT) mechanisms also demonstrated a direct interaction of BLM and topoisomerase IIα (8). These observations suggest that the chromosome breakage of BS cells could result from the improper resolution of tangled DNA intermediates residual to impaired DNA repair or recombination earlier in the cell cycle. In this study, we tested the idea that an interaction of BLM with topoisomerase IIα might regulate these events.
Our results demonstrate that BLM and topoisomerase IIα associate in human cells in a cell cycle-dependent manner via amino acids 489-587 within BLM. Equimolar topoisomerase IIα enhances BLM helicase activity in vitro using substrates that resemble early HR intermediates, but not those resembling late intermediates. A BLM deletion mutant lacking the topoisomerase IIα binding domain has comparable in vitro helicase activity to BLM but is not stimulated by topoisomerase IIα and cannot correct elevated micronuclei formation in transfections of BS cells. In total, these data show that BLM and topoisomerase IIα interaction is required to prevent chromosome breakage and elucidate a DNA repair mechanism that is critical to maintain chromosome stability in cells.
Materials and Methods
Cell lines and synchronization
All cell lines were obtained directly from ATCC and/or Coriell Cell Repository and used within 6 months of receipt. The cell lines were authenticated using routine cell biology programs by these institutions which included certification that each cell line was negative for mycoplasma, bacteria, fungi contamination; confirmation of species identity and detection of possible cellular contamination or misidentification using COI for interspecies identification and STR analysis (DNA profiling) for intra-species identification; conducting additional test methods, such as cytogenetic analysis (G-banding, FISH), flow cytometry and immunocytochemistry as well as consistent refinement of cell growth conditions as well as documentation systems, ensuring traceability. 293T, HeLa, MCF7 and HCT116 are from ATCC, GM08505 (BLM−/−) from Coriell Cell Repositories, and cultured using standard techniques.
Synchronization used double-thymidine block and harvest post-release to obtain cells at S- or G2/M-phases. Cells were treated with 200 ng/ml nocodazole for 16 hrs and released to harvested G1- or M-phase cells.
Generation of BLM protein segments and mutants
BLM segments were generated by in vitro transcription and translation (IVTT) (Supplemental Experimental Procedures; 24). pEGFP-BLMΔ489-587 was generated by mutation of codons for amino acids 488-489 and 587-588 to BspE1 sites in pEGFP-BLM using QuikChange PCR (Stratagene), BspE1 digestion, vector re-ligation, correction of codons for amino acids 488 and 588 (Supplemental Experimental Procedures).
Immunoprecipitations, western assays and immunofluorescence
Immunoprecipitations used nuclear lysates (25) and a BLM-specific antibody (A300-110A, Bethyl Laboratories). For some immunoprecipitations, DNase I (10 µg/mL) was added prior to immunoprecipitation. Western blotting used antibodies for BLM, topoisomerase IIα (Ab-1, Calbiochem), or topoisomerase IIIα (N-20, Santa Cruz Biotechnology). BLM segment immunoprecipitations used 400 ng of topoisomerase IIα (USB) and 3 µg of topoisomerase IIα-specific antibody (ab2987, AbCam) (24). Immunoprecipitations of EGFP-tagged proteins used an anti-GFP antibody (T-19, Santa Cruz Biotechnologies). Immunofluorescence assays used antibodies for topoisomerase IIα (Ab-1, Lab Vision), PML (PG-M3, Santa Cruz Biotechnology) or NPM23 (FC82291, Abcam) and viewed using an Axiovert 200M Zeiss microscope and Axio Vision 4.5 software.
Protein purification and helicase assays
6xHis-tagged BLM proteins were over-expressed in S. cerevisiae lysed using a french press at 20K psi, cleared by ultracentrifugation and purified by FPLC on nickel resin, Ni-NTA (Qiagen) and a Q-sepharose column (Sigma). Bubble, X-junction and 3’ overhang substrates were generated as described (24, 26; Supplemental Experimental Procedures). Helicase assays used the indicated amounts of BLM and topoisomerase IIα (USB) with 2 fmoles of substrate (24).
Micronuclei assays
GM08505 cells (BLM−/−) were transfected with Effectene (Qiagen) and treated at 30 hrs post-transfection with 5 µg/mL cytochalasin B. Cells were fixed with 10% formalin and stained with anti-β-tubulin (Novus) at 48 hrs and DAPI. Binucleate cells were viewed using an Axiovert 200M Zeiss microscope and Axio Vision 4.5 software (27).
Results
Interactions of BLM and topoisomerase IIα are cell cycle-dependent and most prominent in late G2- and M-phases
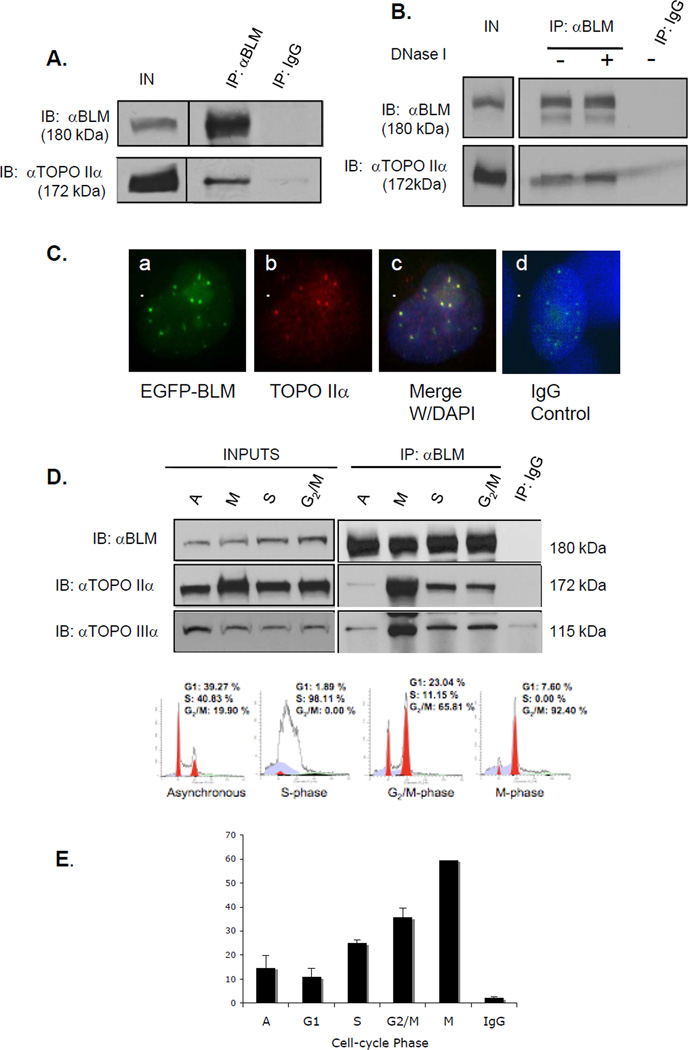
In S. cerevisiae, the BLM ortholog Sgs1 interacts with topoisomerase II, an interaction necessary for proper chromosome segregation (22). To determine whether BLM and topoisomerase IIα associate in human cells, co-immunoprecipitation experiments were carried out using extracts prepared from the embryonic kidney cell line 293T (Figure 1A) and HeLa cells (data not shown). An anti-BLM antibody co-immunoprecipitated BLM and topoisomerase IIα as demonstrated by subsequent western analysis with an antibody for topoisomerase IIα (Figure 1A). Similar immunoprecipitation and western analyses using extracts from BS cells did not identify topoisomerase IIα (Supplementary Figure S1). The reverse co-immunoprecipitation did not yield detectable levels of BLM protein by western blotting (data not shown), perhaps the result of the BLM and topoisomerase IIα antibodies used. DNA does not mediate this interaction as the association persists following DNase I treatment of lysates (Figure 1B).
Figure 1. BLM and topoisomerase IIα associate and co-localize in human cells.
(A) BLM was immunoprecipitated from 293T nuclear extracts with a rabbit anti-BLM antibody. Rabbit IgG was used as a control. Proteins were separated using SDS-PAGE and analyzed by western blotting with anti-BLM or anti-topoisomerase IIα antibody. (B) 293T cells were transfected with pEGFP-BLM and BLM immunoprecipitated as in A, with or without DNase I. (C) MCF7 cells were transfected with pEGFP-BLM, fixed and stained with an anti-topoisomerase IIα antibody and DAPI for nuclear staining. Panel a. shows EGFP-BLM-positive foci; panel b. shows topoisomerase IIα staining; panel c. shows merged images and DAPI staining; panel d. shows staining with rhodamine-labeled mαIgG and DAPI staining. (D) Nuclear extracts from synchronized 293T cells were used in immunoprecipitations. Western blotting with an anti-topoisomerase IIIα antibody was used as a positive control. Inputs are shown in the left panel; immunoprecipitations in the right panels. The bottom panel shows FACS analysis of representative cell populations used for immunoprecipitations. (E) The percentage of BLM foci that co-localize with topoisomerase IIα foci during cell cycle phases in HCT 116 cells is shown graphically.
Co-immunofluorescence studies determined that BLM and topoisomerase IIα co-localize within the nucleus. The breast cancer cell line MCF7 was chosen for its transfection efficiency using a vector expressing EGFP-tagged BLM. GFP fluorescence demonstrated BLM expression (Figure 1C, panel a), while an antibody for topoisomerase IIα detected topoisomerase IIα (Figure 1C, panel b). Merged images for both proteins and DAPI nuclear staining show co-localization of EGFP-BLM and topoisomerase IIα (Figure 1C, panel c); staining with a rhodamine-labeled IgG control antibody does not overlap with EGFP expression (Figure 1C, panel d). These results are consistent with the previously reported association of BLM and topoisomerase IIα (8).
The independent expression patterns of BLM and topoisomerase IIα are cell cycle-regulated, with low expression of both proteins in G1-phase followed by increases in S-phase and peaks in G2/M-phase (28, 29). To test whether the association of BLM and topoisomerase IIα also varies during the cell cycle, 293T cells were synchronized by double thymidine block and collected 1-hour post-release to obtain cells in S-phase or 6-hours post-release to obtain cells in G2/M-phases. Cells were arrested with nocodazole and harvested 16-hours post-release to obtain M-phase cells. FACS analysis confirmed cell synchronization (Figure 1D, lower panel). Extracts from asynchronous and synchronized cells were used for BLM immunoprecipitation (Figure 1D, upper left panel). Western blotting demonstrated the association of BLM and topoisomerase IIα in asynchronous cells (Figure 1D, upper left panel). The association of the two proteins was observed in S- and G2/M-phases and most predominantly in M-phase (Figure 1D, upper right panel). The slight mobility shift of the topoisomerase IIα protein most likely reflects its modification by M-phase phosphorylation (30, 31). The association of topoisomerase IIIα, a known protein partner of BLM required for resolution of Holliday structures (32), increased in a similar pattern. Co-immunofluorescence of BLM and topoisomerase IIα in another cell cycle-synchronized cell line, HCT116, demonstrated the percentage of BLM and topoisomerase IIα co-localization (Figure 1E). Co-localization and immunoprecipitation data show increases in the association of BLM and topoisomerase IIα in S- and G2/M-phases and its most prominent association in mitosis.
Topoisomerase IIα enhances in vitro BLM unwinding activity of DNA substrates representing early homologous recombination intermediates
The interaction of BLM and topoisomerase IIα suggests that topoisomerase IIα modulates BLM helicase activity or that BLM modulates topoisomerase IIα decatenation activity. His-tagged BLM protein was generated in S. cerevisiae and purified by fast protein liquid chromatography (FPLC) purification through nickel and Q-sepharose resins. Figure 2A shows a Coomassie blue-stained gel depicting protein purity; western blotting with anti-BLM and anti-His antibodies confirmed the protein as BLM (data not shown).
Figure 2. BLM helicase activity is enhanced by topoisomerase IIα using 3’ overhang and bubble, but not X-junction, substrates.
(A) Coomassie blue-stained polyacrylamide gel of purified recombinant 6xHis-tagged BLM (100 ng) used in helicase assays. (B) Graphical representation of percent unwinding of 3’ overhang substrate by indicated BLM concentrations. BLM was incubated with 2 fmoles of 3’ overhang substrate and resulting DNA products resolved by non-denaturing PAGE. Heat-denatured controls (HD) show migration of released radio-labeled strand (grey). (C) Autoradiograph of a time-course helicase assay with indicated amounts of BLM and topoisomerase IIα with 2 fmoles of 3’ overhang substrate. (D) Quantitation of percent unwinding of 3’ overhang substrate. Dotted line represents unwinding by 1 nM BLM. The solid line represents unwinding by 1 nM BLM with equimolar topoisomerase IIα. (E) This bar graph shows the percent unwinding by BLM alone or BLM with equimolar topoisomerase IIα of 3’ overhang, bubble and X-junction substrates. The time point is 12 minutes.
Helicase assays with increasing BLM concentrations show that purified BLM effectively processes a 3’ overhang substrate (Figure 2B), as 2 fmoles of substrate are almost completely unwound by 7.5 nM BLM within 15 minutes. Lower concentrations of BLM fully unwind bubble substrate (1.5 nM) and X-junction substrate (2.5 nM) (data not shown), repeating previously published findings that bubble and X-junction substrates are more attractive substrates for BLM than 3’ overhangs (26).
The effects of topoisomerase IIα on BLM activity were then tested. BLM concentrations that generated approximately 10% unwinding of each substrate were used in helicase assays with or without topoisomerase IIα, and with 2 fmoles of each substrate. BLM helicase activity (fmol·min−1·nM−1) was calculated and compared. Recombinant topoisomerase IIα (USB) possesses no significant unwinding ability with any of the substrates evaluated in this study (data not shown). One nM BLM led to 16.6% unwinding of the 3’ overhang substrate in 12 minutes (Figure 2C, left panel and Figure 2D, dotted line). In the presence of equimolar topoisomerase IIα, unwinding increased to 52.7%, an approximately 3-fold increase in activity from 0.02768 to 0.08787 fmol·min−1·nM−1 (Figure 3C, right panel and Figure 3D, solid line). With a bubble substrate, addition of BLM led to 13.1% unwinding in 12 minutes. This increased to 60.7% unwinding with equimolar topoisomerase IIα (an approximately 4.6-fold increase in activity from 0.04374 to 0.20245 fmol·min−1·nM−1; Figure 2E and Supplementary Figure S2). With the X-junction substrate, 0.75 nM BLM led to 8% unwinding (Figure 2E; Supplementary Figure S3). The presence of topoisomerase IIα resulted in a reduction of unwinding to 3.2%, a 2.4-fold reduction in activity from 0.01774 to 0.00725 fmol·min−1·nM−1 (Figures 2E; Supplementary Figure S3). These data suggest that topoisomerase IIα enhances BLM helicase activity with substrates resembling DNA structures predicted to be early HR intermediates (bubble and 3’ overhang), but not those resembling structures in later stages (X-junctions). Stimulation of unwinding using a 3’ overhang or a bubble substrate was verified using a fixed concentration of BLM (1 nM) at varying concentrations of topoisomerase IIα (Supplementary Figure S4). Finally, topoisomerase IIα does not stimulate a helicase-dead mutant BLM, BLMD795A, confirming that BLM helicase activity is required for topoisomerase IIα–dependent stimulation of unwinding (Supplementary Figure S5).
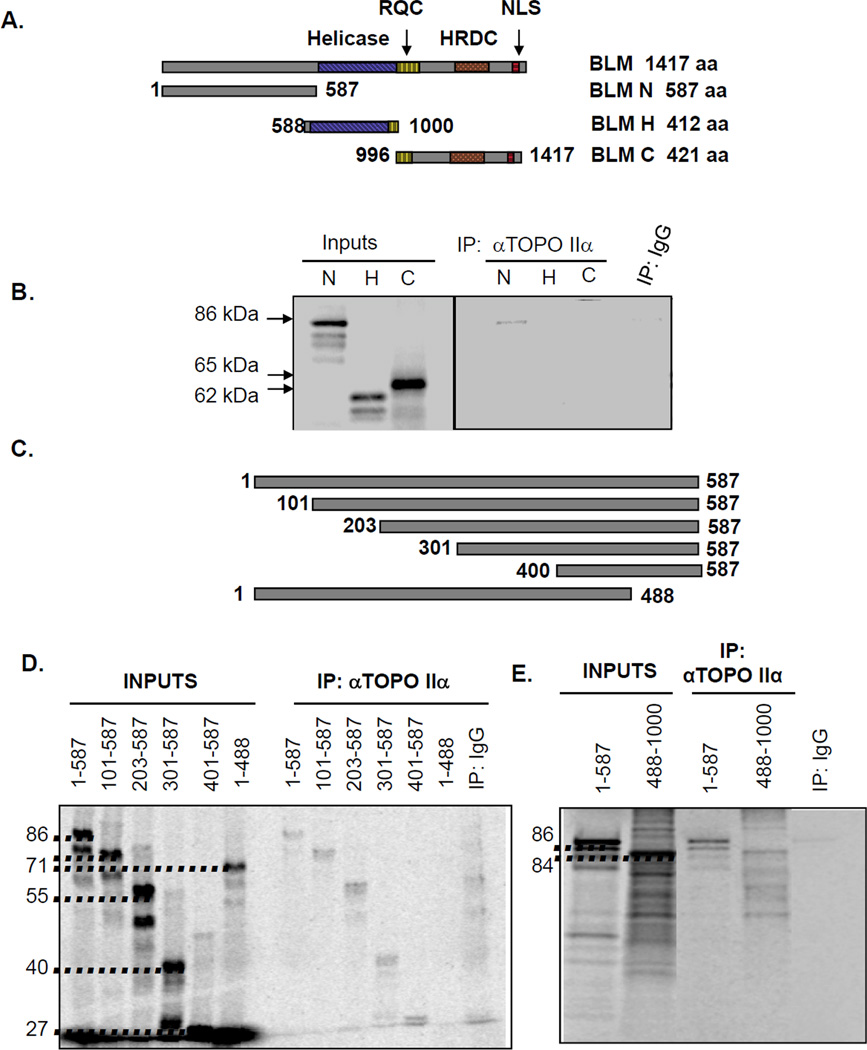
Figure 3. BLM interaction with topoisomerase IIα occurs within the BLM amino-terminus.
(A) Schematic representation of BLM segments tested for topoisomerase IIα binding. RQC identifies the RecQ C-terminal domain, HRDC represents the helicase and Rnase D C-terminal domain, and NLS the nuclear localization sequence. (B) [35S]-methionine-labeled IVTT proteins representing the amino-terminal (N), helicase (H) and carboxy-terminal (C) segments were incubated with topoisomerase IIα and immunoprecipitated (IP) with topoisomerase IIα-specific antibody. Input lanes represent 2% of each IVTT reaction. Control reactions used mαIgG immunoprecipitations. (C) Schematic representation of N-terminal segments. (D) [35S]-methionine-labeled N-terminal segments were generated via IVTT and tested for binding. Input lanes represent 20% of each IVTT reaction. Control reactions used mαIgG immunoprecipitations. (E) [35S]-methionine-labeled IVTT proteins representing the amino-terminal (N) and binding domain tagged to the flanking helicase domain (488-1000) were tested for binding. Input lanes represent 10% of each IVTT reaction. Control reactions used mαIgG immunoprecipitations.
To determine whether binding of BLM to topoisomerase IIα alters the decatenation activity of topoisomerase IIα, a concentration range of purified BLM was tested with purified human topoisomerase IIα (USB) and catenated DNA. Addition of BLM or BLMD795A ranging from 0.1 nM to 100 nM did not alter the average percent decatenation of 0.1U topoisomerase IIα (Supplementary Figure S6). Neither BLM protein affected catenated DNA in the absence of topoisomerase IIα (Supplementary Figure S6).
Amino acids 489-587 of BLM are dispensable for helicase activity but required for topoisomerase IIα binding, enhancing BLM helicase activity and correcting elevated chromosome breakage
Bhattacharyya et al. demonstrated that BLM and topoisomerase IIα interact directly (8). To determine the region of BLM that mediates this interaction, three segments of BLM were in vitro transcribed and translated (IVTT): BLM-N (aa 1-587), BLM-H (aa 588-1000), and BLM-C (aa 996-1417) (Figure 3A). These three [35S]-labeled BLM segments were used in immunoprecipitations with human topoisomerase IIα. Topoisomerase IIα immunoprecipitated with the N-terminus of BLM (amino acids 1-587) but not with the helicase domain (BLM-H) or the C-terminus (BLM-C) (Figure 3B). The topoisomerase IIα-interacting region was then refined using six smaller segments of BLM-N (Figures 3C–D). Topoisomerase IIα was immunoprecipitated with five BLM segments containing the last 98 amino-terminal amino acids (489-587). The sixth segment lacked this region and did not immunoprecipitate with topoisomerase IIα, suggesting that amino acids 489-587 of BLM mediate the interaction (Figure 3D). This region is similar to that within Sgs1 necessary for interaction with yeast Top2 (22). Finally, to demonstrate that this region confers the ability of BLM to bind topoisomerase IIα, the last 98 amino acids were added to the central domain of BLM (amino acids 488-1000) using IVTT to immunoprecipitate topoisomerase IIα (Figure 3E).
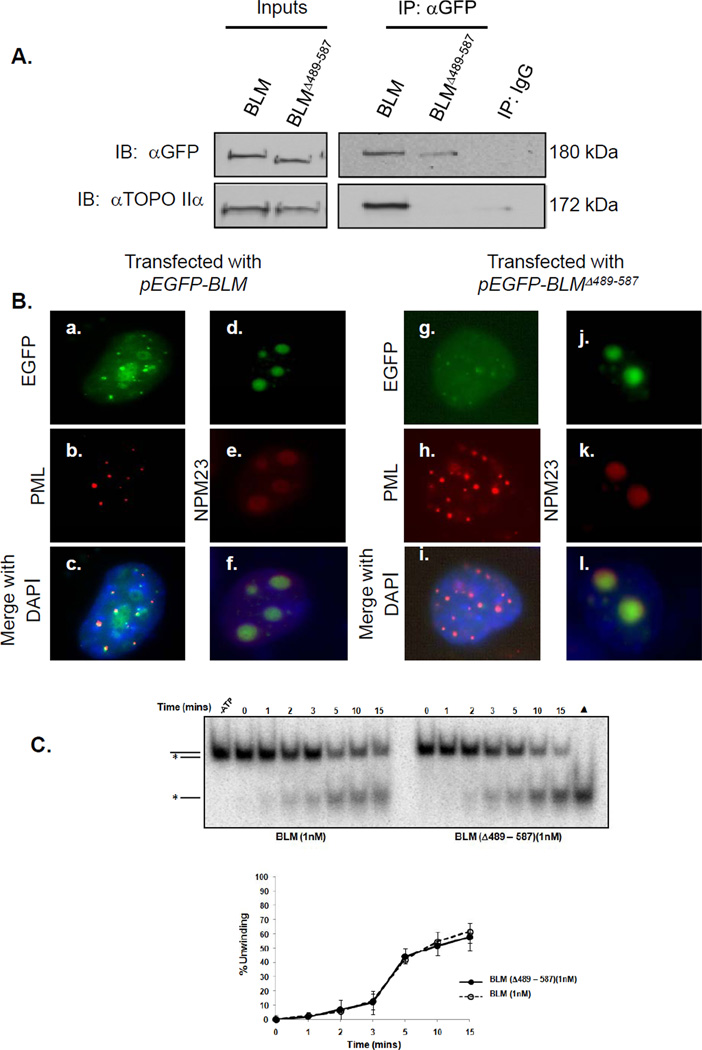
A deletion construct of pEGFP-BLM was generated without the topoisomerase IIα-interaction domain (pEGFP-BLMΔ489-587) to test the requirement for BLM and topoisomerase IIα interaction in preventing chromosome breakage. 293T cells were transfected with pEGFP-BLM or pEGFP-BLMΔ489-587; co-immunoprecipitations determined whether GFP-BLMΔ489-587 binds topoisomerase IIα. Western blot analysis of nuclear extracts from pEGFP-BLMΔ489-587-transfected cells with an anti-GFP antibody identifies a protein with the predicted shift in electrophoretic mobility (Figure 4A). Immunoprecipitations with anti-GFP antibodies using lysates from pEGFP-BLM- or pEGFP-BLMΔ489-587-transfected cells show a marked decrease in co-immunoprecipitated topoisomerase IIα in cells transfected with the mutant versus wild-type BLM (Figure 4A).
Figure 4. Amino acids 489-587 in BLM are necessary for interaction with topoisomerase IIα but are not required for localization or helicase activity.
(A) 293T cells were transfected with pEGFP-BLM or pEGFP-BLMΔ489-587. Post-transfection, wild-type and mutant BLM proteins were immunoprecipitated from nuclear extracts with mouse anti-GFP antibody or mouse IgG control, separated using SDS-PAGE and analyzed by western blotting with anti-GFP or anti-topoisomerase IIα antibody. (B) GM08505 BS cells were transfected with pEGFP-BLM or pEGFP-BLMΔ489-587. Cells were stained with anti-PML antibodies and anti-NPM23 antibodies to recognize PML bodies and the nucleolus, and DAPI for nuclear staining. EGFP-BLM or EGFP-BLMΔ489-587 co-localization with PML and NPM23 is shown. (C) Helicase activities of BLM and BLMΔ489-587 using 3’ DNA overhang substrates are equivalent. BLM proteins were incubated with 2 fmoles of substrate; resulting products were resolved by PAGE and visualized by autoradiography (top). A heat-denatured control (▲) shows migration of the released radio-labeled strand (grey). The broken line represents unwinding by 1 nM BLM. The solid line represents unwinding by 1 nM BLMΔ489-587.
To rule out a change in cellular localization that prevents co-immunoprecipitation of topoisomerase IIα and the EGFP-BLMΔ489-587 mutant, cells were transfected with pEGFP-BLMΔ489-587 or pEGFP-BLM and evaluated for subcellular localization. GFP fluorescence demonstrates EGFP-BLMΔ489-587 in small nuclear foci and within the nucleolus similar to wild-type BLM (Figure 4B) (16, 33, 34). EGFP-BLMΔ489-587 co-localizes with PML and the nucleolar marker NPM23, demonstrating similar localization of the deletion mutant and wild-type BLM (Figure 4B). There were no significant differences in helicase activity between the two BLM proteins using a 3’ overhang substrate (Figure 4C).
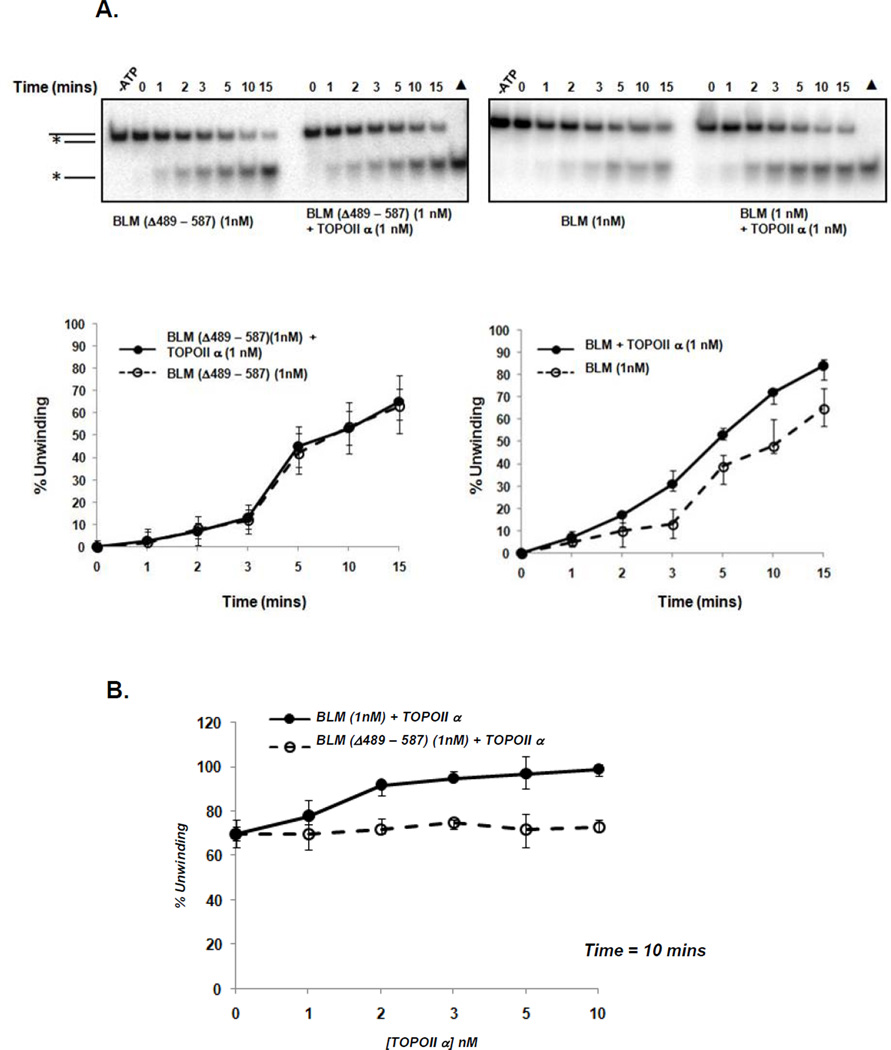
Topoisomerase IIα stimulates the helicase activity of BLM and amino acids 489-587 of BLM are required for the interaction of the two proteins. We then asked whether topoisomerase IIα and BLM binding is required for topoisomerase IIα to stimulate BLM. BLMΔ489-587 was expressed in S. cerevisiae, purified as previously described and used in 3’ overhang time-dependent helicase assays with BLM as a control. Equimolar topoisomerase IIα stimulates BLM helicase activity as shown previously but does not stimulate BLMΔ489-587 helicase activity (Figure 5A). Increasing concentrations of topoisomerase IIα do not alter the unwinding capability of BLMΔ489-587 (Figure 5B) and demonstrate that the BLM topoisomerase IIα interaction domain is required for topoisomerase IIα stimulation of BLM unwinding activity.
Figure 5. Amino acids 489-587 in BLM are necessary for enhancing BLM unwinding by topoisomerase IIα.
(A) Autoradiograph of time-course helicase assays with 2 fmoles of 3’ overhang substrate and indicated amounts of BLM or BLMΔ489-587 (left panel) or wild-type BLM (right panel) with and without topoisomerase IIα. Quantification of percent unwinding by BLMΔ489-587 or wild-type BLM with equimolar topoisomerase IIα is shown. Only wild-type BLM is stimulated by topoisomerase IIα. Minus ATP (-ATP) control indicates ATP-dependent unwinding of substrate and (▲) depicts completely unwound heat-denatured substrate. (B) Increasing concentrations of topoisomerase IIα (0 – 10 nM) do not stimulate BLMΔ489-587 but stimulate wild-type BLM. Helicase assays were carried out with different concentrations of topoisomerase IIα and a fixed concentration of BLM (1 nM) for 10 mins.
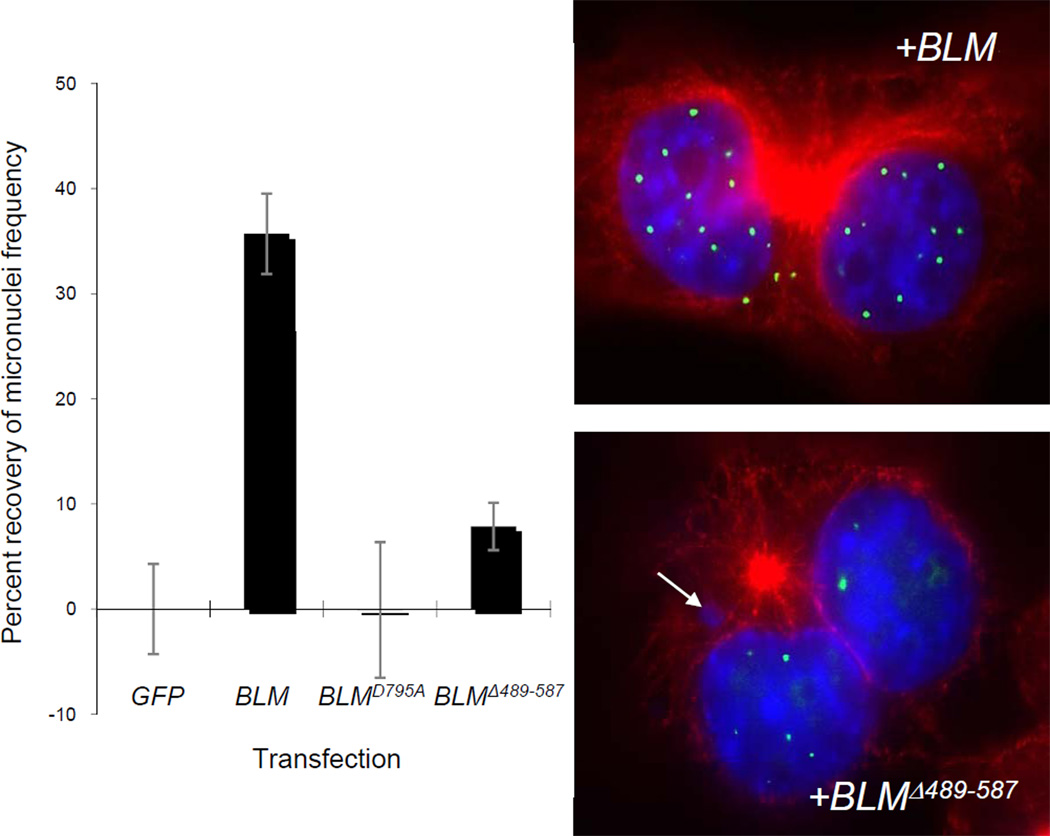
Finally, we asked whether the loss of the BLM-topoisomerase IIα interaction affects chromosome breakage as evaluated by the correction of elevated micronuclei formation in BS cells counted at M-phase. BS cell line GM08505 was transfected with pEGFPpEGFP-BLM, pEGFP-BLMD795A a helicase-dead BLM mutant) or pEGFP-BLMΔ489-587. Mock-transfected BS cells or cells transfected with vector have similarly high numbers of micronuclei. Transfection of pEGFP-BLM into BS cells partially corrects this damage (34.3% recovery) (Figure 6). In contrast, transfection of pEGFP-BLMD795A or pEGFP-BLMΔ489-587 does not significantly correct micronuclei in comparison to the wild-type BLM-transfected controls.
Figure 6. Amino acids 489-587 in BLM are necessary for correcting chromosome breakage in BS cells.
GM08505 cells (BLM−/−) were mock-transfected or transfected with a pEGFP-vector, pEGFP-BLM, pEGFP-BLMD795A or pEGFP-BLMΔ489-587 vector. Post-transfection, cells were treated with cytochalasin B to arrest binucleate cells in mitosis. Eighteen hours post-cytochalasin B block, cells were fixed and stained with anti-β-tubulin and DAPI for nuclear staining. Percentages represent the average percent recovery of micronuclei frequency over empty vector control. Averages reflect at least 300 cells from three independent blinded experiments. Only BLM transfection significantly reduced micronuclei frequency in BS cells (*). The panels on the right show BS cells with BLM foci in green within DAPI-stained nuclei and red β-tubulin. Top panel is representative of BLM transfections; lower panel is representative of BLMΔ489-587 transfections. Arrow identifies a micronucleus.
Discussion
Our experiments demonstrate an interaction between BLM and topoisomerase IIα that occurs in a cell cycle-dependent manner in S-, G2/M- and most prominently in M-phase. This interaction is mediated by residues 489-587 of BLM, a domain that falls within a larger region in the S. cerevisiae recQ-like helicase Sgs1 that interacts with Top2 (22). Equimolar topoisomerase IIα modulates BLM helicase activity in vitro using some recombination intermediate substrates. Deletion of the BLM topoisomerase IIα interaction region prevents BLM from interacting directly with topoisomerase IIα and extinguishes the ability of topoisomerase IIα to enhance BLM unwinding of substrates without altering innate helicase activity. Finally, the deletion mutant is unable to correct chromosome breakage as measured by micronuclei formation in BS cells.
Our previous work identified topoisomerase IIα and BLM in a telomere-specific complex in cells using ALT mechanisms. This interaction, however, does not enhance in vitro BLM activity with telomeric 3’ overhangs and telomeric D-loops, although it occurs in late-S and G2 phases of the cell cycle when ALT occurs (8). Unwinding of these substrates demonstrates an initial but unsustained inhibitory effect at high equimolar BLM and topoisomerase IIα concentrations (30 nM) (8). Our experiments support the functional specificity of BLM-topoisomerase IIα interactions and their role in mediating the resolution of unique substrates within the cell.
siRNA-mediated knock-down of either BLMTOPO2A or both increases DNA damage similarly suggesting common pathways for these proteins to repair DSBs and prevent chromosome breakage (Figure S7). More specifically, micronuclei studies show that the regulation of chromosome breakage is dependent upon the direct interaction of BLM and topoisomerase IIα, as a BLM mutant lacking the topoisomerase IIα binding domain, EGFP-BLMΔ489-587, cannot correct the elevated chromosome breakage of BS cells to the same levels as wild-type BLM after transfection (Figure 6). BLM helicase activity is also necessary for breakage correction. Immunolocalization data show that the inability of the mutant to revert the increased breakage phenotype of BS cells is not attributable to inappropriate compartmentalization but rather to the absence of the interaction (Figure 4A and B). Under normal circumstances, BLM facilitates efficient repair of DSB via HR, but commits cells to such a pathway. When DSBs occur in cells without BLM, cells can repair damage using other pathways such as non-homologous end joining or single-strand annealing pathways (35, 36). HR can also occur, albeit inefficiently, leading to the elevated basal levels of breakage in BS cells. When BLM is unable to bind topoisomerase IIα, repair is incomplete and chromosome breakage ensues. This work demonstrates that BLM and topoisomerase IIα cooperate to prevent chromosome breakage, perhaps via the resolution of early HR DNA intermediates accumulated from residual DSBs occurring earlier in the cell cycle.
More specific biochemical data suggest that BLM and topoisomerase IIα process early HR intermediates but not those that occur later (Figures 2, S3 and S4). Topoisomerase IIα increases BLM specific activity with 3’ overhang more than 3-fold and with bubble substrate almost 5-fold, but inhibits the specific activity of BLM with X-junction substrate. These findings suggest that BLM and topoisomerase IIα may enhance the generation of the 3’ overhang needed for strand invasion or process ends of a D-loop structure (same as one end of bubble or forked DNA structure) to facilitate melting and allow replication from the invading strand.
The main role of topoisomerase IIα is the decatenation of chromosomes before anaphase. Failure to fully decatenate chromosomes prior to or in mitosis leads to DNA breakage (20). The mitotic decatenation checkpoint, distinct from the G2/M DNA damage checkpoint in which topoisomerase IIα senses the catenation or tangling of sister chromatids following DNA replication (37), delays the onset of mitosis until chromosomes have been fully decatenated by topoisomerase IIα (38). One molecular mechanism controlling this checkpoint relies on ATR-mediated PLK1 inhibition (39) and cyclin B1 nuclear exclusion (38); BRCA1 and WRN, another recQ-like helicase, also maintain this checkpoint (38, 40). Interactions of BLM with topoisomerase IIα, BRCA1, WRN, PLK1 and ATR have been reported previously (7, 41, 42) and by association implicate BLM in this decatenation checkpoint. Although the most prominent association of BLM and topoisomerase IIα occurs in mitosis, our in vitro decatenation assays demonstrate no effect of BLM on topoisomerase IIα decatenation. This does not, however, eliminate a role for BLM and topoisomerase IIα in decatenation activities as other proteins or modifications may be required to facilitate decatenation and were not tested here.
These experiments suggest that BLM and topoisomerase IIα, each independently required within the cell, are also required to interact physically to accommodate the resolution of chromosome breakage. The facilitation of BLM unwinding by topoisomerase IIα may permit the efficient unwinding of structures found at anaphase bridges where BLM localizes. Multiple roles for BLM in the homologous recombination (HR) repair pathway have been previously described, including early functions in the stimulation of the resection of DNA double-strand break ends or in the displacement of invading strands within DNA loops, and late functions in the dissolution of double Holliday junctions (43). Another BLM function described here is facilitated by interactions with topoisomerase IIα and is required for the correction of chromosome breaks as measured by micronuclei formation in BS cells. There is also some residual activity of the deleted BLM mutant in the correction of micronuclei as comparisons to the helicase-dead BLM mutant show some recovery in transfected cells.
The discovery of genetically determined chromosome breakage was made almost 50 years ago following the cytogenetic analysis of cells from a child with BS (6). These results were published in 1965 and demonstrated for the first time, in any species, constitutional chromosome breakage, that is breakage not caused by environmental mutagenic agents (6). The characterization of the BS disease gene, BLM, and the functions of the BLM helicase in nucleic acid transactions have suggested that through its disruption, or through the disruption of its interaction with topoisomerase IIα, chromosome breakage can be manipulated in ways that may be beneficial in therapeutic settings. Certainly, topoisomerase inhibitors in cancer treatment regimens are common although toxic, and can be improved by nuanced understandings of how protein interactions are regulated to escalate chromosome breakage and cell death more subtly.
Supplementary Material
Acknowledgments
We thank Cathy Ebert, Kyle Osterbrock and the OSU CCC Nucleic Acid Shared Resource for technical assistance.
Grant Support
The Bloom’s Syndrome Foundation (JG), NIH awards CA-117898 (JG) and CA-009338 (EP) and OSU Up on the Roof Program (SB) for financial support.
Footnotes
Supplementary data are appended.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest are disclosed.
References
- 1.Karow JK, Chakraverty RK, Hickson ID. The Bloom's syndrome gene product is a 3'–5' DNA helicase. J Biol Chem. 1997;272:30611–30614. doi: 10.1074/jbc.272.49.30611. [DOI] [PubMed] [Google Scholar]
- 2.Ellis NA, Groden J, Ye T-Z, Straughen J, Lennon DJ, Ciocci S, Proytcheva M, German J. The Bloom's syndrome gene product is homologous to RecQ helicases. Cell. 1995;83:655–666. doi: 10.1016/0092-8674(95)90105-1. [DOI] [PubMed] [Google Scholar]
- 3.German J. Bloom syndrome: A Mendelian prototype of somatic mutational disease. Medicine (Baltimore) 1993;72:393–406. [PubMed] [Google Scholar]
- 4.Chaganti RS, Schonberg S, German J. A manyfold increase in sister chromatid exchanges in Bloom's syndrome lymphocytes. Proc Natl Acad Sci USA. 1974;71:4508–4512. doi: 10.1073/pnas.71.11.4508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.German J, Crippa LP. Chromosomal breakage in diploid cell lines from Bloom's syndrome and Fanconi's anemia. Annales de Genetique. 1966;9:143–153. [Google Scholar]
- 6.German J, Archibald R, Bloom D. Chromosomal breakage in a rare and probably genetically determined syndrome of man. Science. 1965;148:506–507. doi: 10.1126/science.148.3669.506. [DOI] [PubMed] [Google Scholar]
- 7.Wang Y, Cortez D, Yazdi P, Neff N, Elledge SJ, Qin J. BASC, a super complex of BRCA1-associated protein involved in the recognition and repair of aberrant DNA structures. Genes Dev. 2000;14:927–939. [PMC free article] [PubMed] [Google Scholar]
- 8.Bhattacharyya S, Keirsey J, Russell B, Kavecansky J, Lillard-Wetherell K, Tahmaseb K, Turchi JJ, Groden J. Telomerase-associated protein 1, HSP90, and topoisomerase II{alpha} associate directly with the BLM helicase in immortalized cells using ALT and modulate its helicase activity using telomeric DNA substrates. J Biol Chem. 2009;284:14966–14977. doi: 10.1074/jbc.M900195200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Raynard S, Bussen W, Sung P. A double Holliday junction dissolvasome comprising BLM, topoisomerase IIIalpha, and BLAP75. J Biol Chem. 2006;281:13861–13864. doi: 10.1074/jbc.C600051200. [DOI] [PubMed] [Google Scholar]
- 10.Bussen W, Raynard S, Busygina V, Singh AK, Sung P. Holliday junction processing activity of the BLM-topo IIIalpha-BLAP75 complex. J Biol Chem. 2007;282:31484–31492. doi: 10.1074/jbc.M706116200. [DOI] [PubMed] [Google Scholar]
- 11.Singh TR, Ali AM, Busygina V, Raynard S, Fan Q, Du CH, Andreassen PR, Sung P, Meetei AR. BLAP18/RMI2, a novel OB-fold-containing protein, is an essential component of the Bloom helicase-double Holliday junction dissolvasome. Genes Dev. 2008;22:2856–2868. doi: 10.1101/gad.1725108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Davalos AR, Campisi J. Bloom syndrome cells undergo p53-dependent apoptosis and delayed assembly of BRCA1 and NBS1 repair complexes at stalled replication forks. J Cell Biol. 2003;162:1197–1209. doi: 10.1083/jcb.200304016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Franchitto A, Pichierri P. Bloom's syndrome protein is required for correct relocalization of RAD50/MRE11/NBS1 complex after replication fork arrest. J Cell Biol. 2002;157:19–30. doi: 10.1083/jcb.200110009. (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sengupta S, Robles AI, Linke SP, Sinogeeva NI, Zhang R, Pedeux R, Ward IM, Celeste A, Nussenzweig A, Chen J, et al. Functional interaction between BLM helicase and 53BP1 in a Chk1-mediated pathway during S-phase arrest. J Cell Biol. 2004;166:801–813. doi: 10.1083/jcb.200405128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tripathi V, Kaur S, Sengupta S. Phosphorylation-dependent interactions of BLM and 53BP1 are required for their anti-recombinogenic roles during homologous recombination. Carcinogenesis. 2008;29:52–61. doi: 10.1093/carcin/bgm238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sanz MM, Proytcheva M, Ellis NA, Holloman WK, German J. BLM, the Bloom's syndrome protein, varies during the cell cycle in its amount, distribution, and co-localization with other nuclear proteins. Cytogenet Cell Genet. 2000;91:217–223. doi: 10.1159/000056848. [DOI] [PubMed] [Google Scholar]
- 17.Chan KL, North PS, Hickson ID. BLM is required for faithful chromosome segregation and its localization defines a class of ultrafine anaphase bridges. EMBO J. 2007;26:3397–3409. doi: 10.1038/sj.emboj.7601777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chan KL, Palmai-Pallag T, Ying S, Hickson ID. Replication stress induces sister-chromatid bridging at fragile site loci in mitosis. Nat Cell Biol. 2009;11:753–760. doi: 10.1038/ncb1882. [DOI] [PubMed] [Google Scholar]
- 19.Payne M, Hickson ID. Genomic instability and cancer: Lessons from analysis of Bloom's syndrome. Biochem Soc Trans. 2009;37:553–559. doi: 10.1042/BST0370553. [DOI] [PubMed] [Google Scholar]
- 20.Holm C, Stearns T, Botstein D. DNA topoisomerase II must act at mitosis to prevent nondisjunction and chromosome breakage. Mol Cell Biol. 1989;9:159–168. doi: 10.1128/mcb.9.1.159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Watt PM, Hickson ID. Structure and function of type II DNA topoisomerases. Biochem J. 1994;303:681–695. doi: 10.1042/bj3030681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Watt PM, Louis EJ, Borts RH, Hickson ID. Sgs1: a eukaryotic homolog of E. coli RecQ that interacts with topoisomerase II in vivo and is required for faithful chromosome segregation. Cell. 1995;81:253–260. doi: 10.1016/0092-8674(95)90335-6. [DOI] [PubMed] [Google Scholar]
- 23.Watt PM, Hickson ID, Borts RH, Louis EJ. Sgs1, a homologue of the Bloom's and Werner's syndrome genes, is required for maintenance of genome stability in Saccharomyces cerevisiae. Genetics. 1996;144:935–945. doi: 10.1093/genetics/144.3.935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lillard-Wetherell K, Machwe A, Langland GT, Combs KA, Behbehani GK, Schonberg SA, German J, Turchi JJ, Orren DK, Groden J. Association and regulation of the BLM helicase by the telomere proteins TRF1 and TRF2. Hum Mol Genet. 2004;13:1919–1932. doi: 10.1093/hmg/ddh193. (2004). [DOI] [PubMed] [Google Scholar]
- 25.Jiang K, Pereira E, Maxfield M, Russell B, Goudelock DM, Sanchez Y. Regulation of Chk1 includes chromatin association and 14-3-3 binding following phosphorylation on Ser-345. J Biol Chem. 2003;278:25207–25217. doi: 10.1074/jbc.M300070200. [DOI] [PubMed] [Google Scholar]
- 26.Mohaghegh P, Karow JK, Brosh RM, Jr, Bohr VA, Hickson ID. The Bloom's and Werner's syndrome proteins are DNA structure-specific helicases. Nucl Acids Res. 2001;29:2843–2849. doi: 10.1093/nar/29.13.2843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Fenech M. In vitro micronucleus technique. Mutat Res. 2000;455:81–95. doi: 10.1016/s0027-5107(00)00065-8. [DOI] [PubMed] [Google Scholar]
- 28.Dutertre S, Ababou M, Onclercq R, Delic J, Chatton B, Jaulin C, Amor-Gueret M. Cell cycle regulation of the endogenous wild type Bloom's syndrome DNA helicase. Oncogene. 2000;19:2731–2738. doi: 10.1038/sj.onc.1203595. [DOI] [PubMed] [Google Scholar]
- 29.Heck MM, Hittelman WN, Earnshaw WC. Differential expression of DNA topoisomerase I and II during the eukaryotic cell cycle. Proc Natl Acad Sci USA. 1988;85:1086–1090. doi: 10.1073/pnas.85.4.1086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wells NJ, Fry AM, Guano F, Norbury C, Hickson ID. Cell cycle phase-specific phosphorylation of human topoisomerase II alpha. Evidence of a role for protein kinase C. J Biol Chem. 1995;270:28357–28363. doi: 10.1074/jbc.270.47.28357. [DOI] [PubMed] [Google Scholar]
- 31.Kimura K, Nozaki N, Enomoto T, Tanaka M, Kikuchi A. Analysis of M phase-specific phosphorylation of DNA topoisomerase II. J Biol Chem. 1996;271:21439–21445. doi: 10.1074/jbc.271.35.21439. [DOI] [PubMed] [Google Scholar]
- 32.Wu L, Hickson ID. The Bloom's syndrome helicase suppresses crossing over during homologous recombination. Nature. 2003;426:870–874. doi: 10.1038/nature02253. [DOI] [PubMed] [Google Scholar]
- 33.Bischof O, Kim SH, Irving J, Beresten S, Ellis NA, Campisi J. Regulation and localization of the Bloom syndrome protein in response to DNA damage. J Cell Biol. 2001;153:367–380. doi: 10.1083/jcb.153.2.367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yankiwski V, Marciniak RA, Guarente L, Neff NF. Nuclear structure in normal and Bloom syndrome cells. Proc Natl Acad Sci USA. 2000;97:5214–5219. doi: 10.1073/pnas.090525897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Adams MD, McVey M, Sekelsky JJ. Drosophila BLM in double-strand break repair by synthesis-dependent strand annealing. Science. 2003;299:265–267. doi: 10.1126/science.1077198. [DOI] [PubMed] [Google Scholar]
- 36.Iijima K, Ohara M, Seki R, Tauchi H. Dancing on damaged chromatin: Functions of ATM and the RAD50/MRE11/NBS1 complex in cellular responses to DNA damage. J Radiat Res. 2008;49:451–464. doi: 10.1269/jrr.08065. [DOI] [PubMed] [Google Scholar]
- 37.Kaufmann WK, Kies PE. DNA signals for G2 checkpoint response in diploid human fibroblasts. Mutat Res. 1998;400:153–167. doi: 10.1016/s0027-5107(98)00041-4. [DOI] [PubMed] [Google Scholar]
- 38.Deming PB, Cistulli CA, Zhao H, Graves PR, Piwnica-Worms H, Paules RS, Downes CS, Kaufmann WK. The human decatenation checkpoint. Proc Natl Acad Sci USA. 2001;98:12044–12049. doi: 10.1073/pnas.221430898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Deming PB, Flores KG, Downes CS, Paules RS, Kaufmann WK. ATR enforces the topoisomerase II-dependent G2 checkpoint through inhibition of PLK1 kinase. J Biol Chem. 2002;277:36832–36838. doi: 10.1074/jbc.M206109200. [DOI] [PubMed] [Google Scholar]
- 40.Franchitto A, Oshima J, Pichierri P. The G2-phase decatenation checkpoint is defective in Werner syndrome cells. Cancer Res. 2003;63:3289–3295. [PubMed] [Google Scholar]
- 41.Davies SL, North PS, Dart A, Lakin ND, Hickson ID. Phosphorylation of the Bloom's syndrome helicase and its role in recovery from S-phase arrest. Mol Cell Biol. 2004;24:1279–1291. doi: 10.1128/MCB.24.3.1279-1291.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.von Kobbe C, Karmakar P, Dawut L, Opresko P, Zeng X, Brosh RM, Jr, Hickson ID, Bohr VA. Colocalization, physical, and functional interaction between Werner and Bloom syndrome proteins. J Biol Chem. 2002;277:22035–22044. doi: 10.1074/jbc.M200914200. [DOI] [PubMed] [Google Scholar]
- 43.Chu WK, Hanada K, Kanaar R, Hickson ID. BLM has early and late functions in homologous recombination repair in mouse embryonic stem cells. Oncogene. 2010 doi: 10.1038/onc.2010.214. Epub ahead of print. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.