Abstract
Alcohol is oxidized to acetaldehyde, which in turn is oxidized to acetate. The aldehyde dehydrogenase 2 gene (ALDH2) is the most important gene responsible for acetaldehyde metabolism. Individuals heterozygous or homozygous for the lys (A or *2) allele at the single nucleotide polymorphism (SNP) glu504lys (rs671) of ALDH2 have greatly reduced ability to metabolize acetaldehyde, which greatly decreases their risk for alcohol dependence (AD). Case-control studies have shown association between this SNP and alcohol dependence as well as alcohol-induced liver disease. However, some studies have produced insignificant results. Using cumulative data from the past 20 years predominately from Asian populations (from both English and Chinese publications), this meta-analysis sought to examine and update whether the aggregate data provide new evidence of statistical significance for the proposed association. Our results (9,678 cases and 7,331 controls from 53 studies) support a strong association of alcohol abuse and dependence, with allelic P value of 3×10−56 and OR of 0.23 (0.2, 0.28) under the random effects model. The dominant model (lys-lys + lys-glu vs. glu-glu) also showed strong association with P value of 1×10−44 and OR of 0.22 (0.18, 0.27). When stricter criteria and various sub-group analyses were applied, the association remained strong (for example, OR = 0.23 (0.18, 0.3) and P = 2×10−28 for the alcoholic patients with alcoholic liver disease, cirrhosis, or pancreatitis). These findings provide confirmation of the involvement of the human ALDH2 gene in the pathogenesis of AD as well as alcohol-induced medical illnesses in East-Asians.
Keywords: Meta-analysis, Association, Linkage Disequilibrium, Ethanol Metabolism
Introduction
Alcohol dependence, which constitutes major public health problems, is a chronic relapsing disorder characterized by compulsive seeking, abuse, tolerance, and physical dependence on alcohol. It is a multifactorial disorder caused by complex interaction of genetic and environmental factors (Gelernter and Kranzler 2009; Kendler et al. 2007). The alcohol dehydrogenases and aldehyde dehydrogenases metabolize alcohol into acetaldehyde and then into acetate. There are two major aldehyde dehydrogenase (ALDH) liver isoforms, cytosolic and mitochondrial, encoded by the aldehyde dehydrogenase 1 gene (ALDH1) and aldehyde dehydrogenase 2 gene (ALDH2), respectively. The ALDH2 gene was hypothesized to alter genetic susceptibility to alcohol dependence (AD) and alcohol-induced liver diseases.
The ALDH2 gene maps to chromosome 12q24.2. A single nucleotide polymorphism (SNP) at exon 12 predicts lysine at residue 504 instead of glutamic acid. The common form of the SNP (rs671) (504glu) encodes the glu (G) allele and was previously referred to as the ALDH2 *1 allele; the 504lys (A, formerly ALDH2 *2 and 487lys) allele produces a catalytically inactive isozyme (Yoshida et al. 1991), which has greatly reduced or no ability to metabolize acetaldehyde (Yoshida et al. 1984). Individuals homozygous for the lys allele (lower frequency in alcoholic subjects) therefore have greatly reduced capacity to clear acetaldehyde and typically show facial flushing and nausea after alcohol consumption, whereas heterozygotes exhibit similar but less severe reactions (Mizoi et al. 1994). This variant can partially determine blood acetaldehyde concentrations after drinking: heterozygotes or homozygotes for the 504lys allele showed peak blood acetaldehyde concentrations after alcohol consumption 6- and 19-fold higher than homozygous common-allele individuals, respectively (Mizoi et al. 1994). Due to delayed oxidation in individuals with the 504lys allele, these individuals have high blood acetaldehyde concentrations, which can cause adverse reactions sufficient to deter drinking. Some individuals with a high daily intake of alcohol develop alcohol-induced medical diseases, e.g., alcoholic cirrhosis, which occurs in around 10% and hepatitis in 10–35% (Grant et al. 1988) of AD individuals. It was hypothesized that the 504lys allele could decrease the risk of AD and consequently influence risk for alcohol-induced medical illnesses, including alcohol liver disease, cirrhosis, and pancreatitis (Thomasson et al. 1991; Yoshida et al. 1991). In addition, genome-wide association studies showed that rs671 was associated with mean corpuscular hemoglobin concentration (Kamatani et al. 2010) and esophageal squamous cell carcinoma (Cui et al. 2009). Other ALDH2 variants were found to be associated with blood pressure (Kato et al. 2011) or upper aerodigestive tract cancers (McKay et al. 2011).
Several issues prompted us to carry out the present meta-analysis. First, it is difficult to determine the effect of the minor allele without using a large sample, which can most readily be obtained by meta-analysis. Second, rates of AD differ across population subgroups, even within Asian populations, and this is further complicated because of the consequences of differing sampling strategies. Third, many new studies have been reported from various populations in recent years since the previous meta-analyses (Luczak et al. 2006; Zintzaras et al. 2006). Therefore, based on these factors together with the well known critical role of the gene product in alcohol and acetaldehyde metabolism and the fact that the SNP is known to alter protein activity, we performed a comprehensive meta-analysis of the ALDH2 gene with AD as well as alcohol-induced diseases including alcohol liver disease, cirrhosis, and pancreatitis, based on all identifiable published studies in both English and Chinese publications, to update the association and to compare the results with those previously described.
Methods
Literature Search
The publications included in the analysis were selected from both PubMed and the database of Chinese Academic Journals with keywords 'aldehyde dehydrogenase', ‘ALDH’, 'association', 'associated', 'drug’, 'substance', 'alcoholism’, 'alcohol’, 'alcoholics', 'heroin’, 'opiate', and 'opioid'. Both English and Chinese language keywords were used in searching the Chinese academic journals (the first author, who performed the search, is proficient in Chinese). All references cited in these studies and in published reviews were examined in order to identify additional works not indexed by the databases. The analyzed data cover all identified English and Chinese publications up to August 2010.
Inclusion Criteria
Eligible studies had to meet the following criteria: they (i) were published in peer-reviewed journals and contained original data; (ii) presented sufficient data to calculate the odds ratio (OR) with confidence interval (CI) and P value; (iii) were association studies investigating the specific SNP discussed here (rs671); (iv) described or referenced appropriate genotyping methods or protocols; (v) investigated alcohol or drug dependence (or abuse) diagnosed by valid and published criteria. For the studies investigating alcohol-induced liver disease, cirrhosis, or chronic pancreatitis, the cases were considered as alcoholics with the diseases attributable to alcoholism. Cirrhosis was diagnosed by histological, clinical, radiological, and (or) endoscopic findings; (vi) had no description of other major psychiatric disorders for the patients except anxiety or depressive disorder (patients of AD are often comorbid with anxiety disorder or depressive disorder, or the status was unknown in some studies); and (vii) used unrelated individuals in case-control studies and healthy individuals (or random population) as controls. Authors were contacted in cases where we determined it would be useful to have additional information regarding their studies.
Statistical Analyses
Studies were classified according to inclusion criteria, as “alcoholics with alcoholic liver disease”; “alcoholics with cirrhosis or pancreatitis”; “alcoholics not known to have alcoholic liver disease, cirrhosis, or pancreatitis (or without medical comorbidity specified)”; and “other groups”, e.g., heroin dependence (abuse). A study that contained data from more than one population groups (e.g., Asian and Mexican American) was considered effectively as multiple independent studies. Data from the included studies were summarized by two-by-two tables. From each table a log-odds ratio and its sampling variance were calculated. The Cochran’s χ2-based Q statistic test was performed in order to assess heterogeneity (Zintzaras and Ioannidis 2005) to ensure that each group of studies was suitable for meta-analysis. Where heterogeneity was found, the random effects model, which yields a wider CI, was adopted; otherwise, both the fixed and random effects models were used. Heterogeneity Q tests were also performed for differences in OR between studies using the World Health Organization’s International Statistical Classification of Diseases and Related Health Problems (ICD) (World Health Organization) or the American Psychiatric Association’s Diagnostic and Statistical Manual of Mental Disorders (DSM) (American Psychiatric Association) system and others; between the English-language publications and Chinese-language publications; between studies originating in mainland China and others (identified by institutions and addresses of the authors); and between studies originating in the United States and others.
A test for funnel plot asymmetry, described by Egger et al. (Egger et al. 1997), was applied to assess evidence for publication bias. The test used a linear regression approach to measure funnel plot asymmetry on the natural logarithm of the OR. Degree of funnel plot asymmetry was measured by the intercept from regression of standard normal deviates against precision. The larger the deviation of each study from the funnel curve, the more pronounced the asymmetry. Results from small studies are expected to scatter widely at the bottom of the graph, with the spread narrowing among larger studies. The significance of the intercept was evaluated using the T test. The classic fail-safe analysis (Rosenthal 1979) was also applied to address potential publication bias. If publication bias is found, the “Duval and Tweedie's Trim and Fill” procedure (Duval and Tweedie 2000) is used to impute the number of potentially-missing studies. The Trim and Fill procedure imputes these missing studies, adds them to the analysis, and then re-computes the adjusted overall effect size.
ORs were pooled using the method of DerSimonian and Laird (DerSimonian and Laird 1986), and 95% CIs were constructed using Woolf’s method (Woolf 1955). The significance of the overall OR was determined using the Z test. To measure sensitivity of our analysis results, each study was removed in turn from the total, and the remainder then reanalyzed. This procedure was used to ensure that no individual study was entirely responsible for the combined results. Both allelic and genotypic analyses (e.g., dominant model) were applied, and different combinations of the alcohol-related medical conditions (e.g., alcohol liver disease, cirrhosis, and pancreatitis) were also analyzed. Retrospective analysis was performed to better understand the potential effect of year of publication upon the results. The type I error rate was set at 0.05. The tests were two-tailed.
In order to evaluate whether there are other polymorphisms in strong linkage disequilibrium (LD) with this SNP, haplotype construction, counting, and LD block defining over a broader genomic region that includes ALDH2 were performed separately based on the published genotype data of 90 Asian (Chinese and Japanese) samples and 30 CEPH trios (Utah residents) from HapMap release 23a (http://ftp.hapmap.org/genotypes/2008-03/rs_strand/non-redundant). The multiallelic D’ was computed by performing a series of pairwise D’ calculations using each haplotype in turn as an allele, with all other haplotypes at the locus serving as the other allele. This was then repeated for each haplotype at each locus and averaged by haplotype frequency. Maximum likelihood haplotype blocks were calculated using the EM algorithm (Excoffier and Slatkin 1995). In addition, the genotype data from the 1000 Genomes Project were also used to calculate and build the LD block (Johnson et al. 2008).
Results
The search yielded 470 references. After discarding overlapping references and those which clearly did not meet the criteria, 70 studies remained. These studies were then filtered to ensure conformity with our inclusion criteria. One study (Higuchi et al. 1995) was excluded because subjects in this study were included in another study (Higuchi et al. 1996b); three studies (Choi et al. 2006; Harada et al. 1999; Yang et al. 2007) were excluded because they were investigating other polymorphisms, such as 1951G/A, rather than the SNP considered here; two (Shimosegawa et al. 2008; Tsuchihashi-Makaya et al. 2009) because the criteria to define “alcoholic” patients were not eligible (i.e., drinking and smoking behavior was assessed on the basis of the lifestyle questionnaire self-administered by the subjects (Tsuchihashi-Makaya et al. 2009); and patients with alcohol consumption of more than 80 g/day for at least two years were classified as alcoholic chronic pancreatitis (Shimosegawa et al. 2008)); two (Hendershot et al. 2009; Segado Soriano et al. 2005) because the patients were described nonspecifically as “drinkers”, rather than “dependent” or “abusers”; one (Nanakorn et al. 1999) because the patients had comorbid psychiatric disorders; one (Wei et al. 1999) because the cases and controls were related; one (Macgregor et al. 2009) because it was investigating twin pairs; four (Higuchi et al. 1996a; Kimura et al. 2009; Muramatsu et al. 1996; Vaswani et al. 2009) because no control subjects were recruited or control data were not available; one (Kuo et al. 2008) due to very low observed minor allele frequency (0.001) in their samples; and one (Shafe et al. 2009) because there was no genotype data available. In the end, 53 case-control studies met our criteria for inclusion. The studies included 52 studies (Chai et al. 2005; Chao et al. 1994; Chao et al. 2000; Chao et al. 2003; Chao et al. 1997; Chen et al. 1999; Chen et al. 1996; Chen et al. 1997; Fan et al. 1998; Guo et al. 2010; Hasegawa et al. 2002; Higuchi et al. 1996b; Huang et al. 2004; Iwahashi et al. 1995; Jing et al. 2009; Kim et al. 2008; Kim et al. 2004; Lee et al. 2001; Lee et al. 1999; Lee et al. 2009; Lee et al. 2010; Leng et al. 2008; Maezawa et al. 1995; Matsushita et al. 2001; Miyatake et al. 1995; Muramatsu et al. 1995; Nakamura et al. 1996; Nakamura et al. 1999; Nishizawa et al. 2006; Paik et al. 2000; Park et al. 2001; Sasabe et al. 2007; Shen et al. 1997a; Shen et al. 1997b; Shin et al. 2010; Tan et al. 2010; Tanaka et al. 1996; Thomasson et al. 1994; Thomasson et al. 1991; Xu et al. 2002; Yamauchi et al. 1995; Yan et al. 2003; Yu et al. 2002) of Asian populations (including Chinese aboriginal populations) and one study (Konishi et al. 2004) of Mexican Americans. One study (Xu et al. 2002) investigated heroin dependence and abuse and 52 studies investigated AD and alcohol abuse or AD only. Among these studies of alcoholics 48 studies (Chai et al. 2005; Chen et al. 1999; Chen et al. 1996; Chen et al. 1997; Fan et al. 1998; Guo et al. 2010; Hasegawa et al. 2002; Higuchi et al. 1996b; Huang et al. 2004; Iwahashi et al. 1995; Jing et al. 2009; Kim et al. 2008; Kim et al. 2004; Konishi et al. 2004; Lee et al. 2001; Lee et al. 1999; Lee et al. 2009; Lee et al. 2010; Leng et al. 2008; Maezawa et al. 1995; Matsushita et al. 2001; Miyatake et al. 1995; Muramatsu et al. 1995; Nakamura et al. 1996; Nakamura et al. 1999; Nishizawa et al. 2006; Paik et al. 2000; Park et al. 2001; Sasabe et al. 2007; Shen et al. 1997a; Shen et al. 1997b; Shin et al. 2010; Tan et al. 2010; Tanaka et al. 1996; Thomasson et al. 1994; Thomasson et al. 1991; Yamauchi et al. 1995; Yan et al. 2003; Yu et al. 2002) investigated alcoholics without (or without specifying) alcoholic liver disease, cirrhosis, or pancreatitis; seven studies (Chao et al. 1994; Chao et al. 2000; Chao et al. 2003; Chao et al. 1997; Lee et al. 2001; Tanaka et al. 1996; Yamauchi et al. 1995) investigated the alcoholics with alcoholic liver disease, cirrhosis, or pancreatitis; and three studies (Guo et al. 2010; Shin et al. 2010; Thomasson et al. 1994) included the mixed patients of AD and alcohol abuse. For the study by Lee et al. (Lee et al. 2009), some additional data, unavailable in the published paper, were kindly provided by the authors. These studies included 9,678 cases and 7,331 controls (supplementary Table 1). The results are detailed below.
Based on all these samples, the frequency of the protective 504lys allele was 14% (overall population estimate) but varied widely across different populations and disease statuses: high in the Asian normal populations 23% (1% – 53%), lower in affected subjects, 7% (0 – 23%); very low in Mexican Americans (1%); and undetected in the European-ancestry populations. Of the 53 studies included, 52 studies showed lower 504lys allele frequency in cases than in controls. Drug dependence as a primary diagnosis and non-Asian samples were considered as extra information for comparison (i.e., allele frequency calculation), and thus excluded from the following meta-analysis.
For the allelic analysis, the combined studies of alcohol abuse and dependence produced strong evidence of association with an overall P value of 3×10−56, the ln (OR) being −1.45 (−1.63, −1.27) with an OR of 0.23 (0.2, 0.28) under the random effects model (due to evidence of significant heterogeneity between studies (P(Q) = 3×10−14)). The subgroup of Asian populations also showed strong association with P value of 7×10−56 (Table 1).
Table 1.
Results of the overall and sub-grouped studies
| Groups | N* | LnOR (95% CI) | OR (95% CI) | P(Z) | P(Q) |
|---|---|---|---|---|---|
| Allelic Analysis (lys vs. glu) | |||||
| Alcoholics (alcohol dependence and abuse) | 50 | −1.45 (−1.63,−1.27) | 0.23 (0.2,0.28) | 3×10−56 | 3×10−14 |
| Alcoholics (Asian) | 49 | −1.45 (−1.63,−1.27) | 0.23 (0.2,0.28) | 7×10−56 | 2×10−14 |
| Alcohol-induced diseasesa | 7 | −1.45 (−1.71,−1.19) | 0.23 (0.18,0.3) | 2×10−28 | 0.48 |
| Alcoholic cirrhosis | 5 | −1.37 (−1.68,−1.07) | 0.25 (0.19,0.34) | 6×10−19 | 0.24 |
| Alcoholics without induced diseasesb | 46 | −1.46 (−1.67,−1.26) | 0.23 (0.19,0.28) | 7×10−46 | 7×10−15 |
| Alcoholics without induced diseasesb (Asian) | 45 | −1.46 (−1.67,−1.26) | 0.23 (0.19,0.28) | 1×10−45 | 4×10−15 |
| AD without induced diseasesc | 43 | −1.48 (−1.69,−1.27) | 0.23 (0.18,0.28) | 2×10−43 | 1×10−14 |
| AD without induced diseasesc (Asian) | 42 | −1.48 (−1.69,−1.27) | 0.23 (0.19,0.28) | 5×10−43 | 8×10−15 |
| Alcoholics and heroin addiction | 51 | −1.43 (−1.63,−1.22) | 0.24 (0.2,0.29) | 6×10−43 | 1×10−26 |
| Alcoholics and heroin addiction (Asian) | 50 | −1.43 (−1.63,−1.22) | 0.24 (0.2,0.29) | 1×10−42 | 6×10−27 |
| Dominant Model (lys-lys + lys-glu) vs. glu-glu | |||||
| Alcoholics | 52 | −1.51 (−1.72,−1.3) | 0.22 (0.18,0.27) | 1×10−44 | 2×10−21 |
| Alcoholics (Asian) | 51 | −1.51 (−1.72,−1.3) | 0.22 (0.18,0.27) | 2×10−44 | 8×10−22 |
| Alcohol-induced diseasesa | 7 | −1.54 (−1.83,−1.25) | 0.21 (0.16,0.29) | 8×10−26 | 0.27 |
| Alcoholic cirrhosis | 5 | −1.44 (−1.78,−1.1) | 0.24 (0.17,0.33) | 9×10−17 | 0.17 |
| Alcoholics without induced diseasesb | 48 | −1.53 (−1.76,−1.29) | 0.22 (0.17,0.27) | 1×10−37 | 3×10−22 |
| Alcoholics without induced diseasesb (Asian) | 47 | −1.52 (−1.76,−1.29) | 0.22 (0.17,0.28) | 2×10−37 | 2×10−22 |
| AD without induced diseasesc | 45 | −1.54 (−1.78,−1.3) | 0.21 (0.17,0.27) | 5×10−35 | 3×10−22 |
| AD without induced diseasesc (Asian) | 44 | −1.54 (−1.79,−1.3) | 0.21 (0.17,0.27) | 4×10−35 | 1×10−22 |
number of studies included in the analyses.
alcoholic patients with alcoholic liver disease, cirrhosis, or pancreatitis.
alcoholic patients without alcoholic liver disease, cirrhosis, or pancreatitis.
alcohol dependent patients without alcoholic liver disease, cirrhosis, or pancreatitis and those without liver disease status described.
P(Z): Z test used to determine significance of the overall OR. The P values < 0.05 are indicated in boldfaces.
P(Q): Cochran’s X2-based Q statistic test used to assess heterogeneity.
P(T): T test used to evaluate significance of publication bias (not shown). P (1-tailed) > 0.05.
For medical conditions related to alcoholism, the combined studies of alcoholics with alcoholic liver disease, cirrhosis (specifically), and (or) pancreatitis produced a strong association with P value of 2×10−28 (OR = 0.23 (0.18, 0.3)) with no evidence of significant heterogeneity (P = 0.5). The studies of alcoholic cirrhosis also showed strong association with P value of 6×10−19.
Strong association was also detected when the alcoholics with known (and reported) alcoholic liver disease, cirrhosis, or pancreatitis were excluded (P = 7×10−46 and OR = 0.23 (0.19, 0.28) under the random effects model). Furthermore, the association was still strong after all the three studies containing alcohol abuse were excluded (P = 2×10−43) or when the Mexican American subjects were excluded (P = 1×10−45); or both (P = 5×10−43). Additionally, when the study of heroin dependence and abuse was included, the between-studies heterogeneity was more significant but the association was still strong (P = 6×10−43).
For the dominant model (lys-lys + lys-glu vs. glu-glu) in genotypic analyses, overall the studies of alcohol abuse and dependence showed an overall P value of 1×10−44 with the ln (OR) of −1.51 (−1.72, −1.3) and OR of 0.22 (0.18, 0.27) under the random effects model (Table 1). The strong association did not change after the Mexican American subjects were excluded from the analysis (P = 2×10−44). Strong association was also found with alcoholic liver disease and alcoholics with cirrhosis and (or) pancreatitis with P value of 8×10−26 and OR of 0.21 (0.16, 0.29). No evidence of significant heterogeneity was found between studies. The combined studies of alcoholics without alcoholic liver disease, cirrhosis, or pancreatitis also produced strong association with P value of 1×10−37 and OR of 0.22 (0.17, 0.27). Furthermore, the consistently strong association was identified after the subjects of alcohol abuse or after the subjects of Mexican American were excluded with the P values of 5×10−35 and 2×10−37, respectively (Table 1).
The demography of the combined studies is shown in supplementary Table 1. The results of overall and subgroup meta-analyses are shown for both allelic and genotypic analyses in Table 1. The forest plots of the allelic and genotypic analyses are shown in Figures 1 and 2, respectively. The funnel plots of allelic and genotypic analyses are shown in Figure 3 and supplementary Figure 1.
Figure 1.
Forest plots of ln (OR) with 95% CI for the allelic analysis. Black squares indicate the ln (OR) (ln (OR) can be better fitted than OR), with the size of the square inversely proportional to its variance, and horizontal lines represent the 95% CIs. The pooled results are indicated by the unshaded black diamond. Six datasets including Chen 1997 (Atayal) (Chen et al. 1997), Chen 1997 (Bunun) (Chen et al. 1997), Chen 1997 (Paiwan) (Chen et al. 1997), Shen 1997 (b) (Elunchun) (Shen et al. 1997b), Fan 1998 (Elunchun) (Fan et al. 1998), and Konishi 2004(Konishi et al. 2004), are not shown on the forest plots because the wide CIs can not fit into the current version of the plots.
Figure 2.
Forest plots of ln (OR) with 95% CI for the dominant model of (lys-lys + lys-glu) vs. glu-glu. Six datasets are not shown on the forest plots as described above.
Figure 3.

Egger’s funnel plots of publication bias analysis for the allelic analysis. A larger deviation from the funnel curve of each study means more pronounced asymmetry. Results from small studies will scatter widely at the bottom of the graph, with the spread narrowing among larger studies.
Publication Bias and Fail-safe Analyses
In the present study, no evidence of significant publication bias was found in either allelic or genotypic analysis (P values > 0.05 in both Egger's regression and Begg’s rank correlation (Kendall's tau) (Begg and Mazumdar 1994) tests).
The classic fail-safe analysis showed that for the allelic analysis about 10,386 assumed non-significant studies would be required to bring the P(Z) value to > 0.05; and for the dominant model about 10,120 non-significant studies would be required to bring the P(Z) value to > 0.05. These results further supported the strong associations detected in the meta-analysis.
Other Heterogeneity Analyses
There was no evidence of significant heterogeneity regarding different diagnosis criteria, publication languages, or study countries (or regions) using either allelic or genotypic analysis. The results of AD and alcohol abuse are shown in supplementary Table 2 (P(Q) > 0.1).
Sensitivity and Retrospective Analyses
Sensitivity analysis showed that no individual group biased the findings to the extent that it could account for the overall strong association. For instance, the results of AD and alcohol abuse were strongly consistent, regardless of the data set removed, with the P values never > 6×10−51 among the 50 studies in the allelic analysis; under the dominant model the results also showed strong consistency, regardless of the data set removed, with the P value never > 3×10−41 among the 52 studies. The results are shown for the allelic and genotypic analyses in supplementary Tables 3 and 4, respectively.
The asymptote lines of the analyses in retrospect based on the 20 publication years showed that the cumulative synthesis of the SNP has tended to be stable, in line with the overall results of our meta-analysis. The results of the allelic and genotypic analyses are shown in Figure 4 and supplementary Figure 2 and the P values are shown in supplementary Tables 5 and 6, respectively.
Figure 4.
Retrospective analysis of the alleles. Analysis in retrospect was based on publication year since 1991.
LD and Haplotype Structure Analyses
In the Asian populations, the ALDH2 gene was located within a large region spanning 13 haplotype blocks with strong LD, and more than ten genes were found within this LD structure (Figure 5). Compared with the Asian populations, the European-ancestry populations showed stronger LD in this large structure, which was composed of three big haplotype blocks (supplementary Figure 3). Two other genes (BRAP and ACAD10) share the same haplotype block with ALDH2. Although it is unlikely that the association of this SNP was due to linkage disequilibrium with a different closely-mapped polymorphism at ALDH2 or another gene considering both the strength of the association and the compelling biological rationale, it is still desirable to investigate other variants within the large structure in subsequent studies. For example, one SNP rs4646776 was in strong LD with rs671 based on the 1000 Genomes Project (11,747 base pair; r2 = 0.81; and recombination rate = 0.99) (supplementary Figure 4).
Figure 5.
Graphical representation of the LD structure of the ALDH2 gene for the Asian populations. The LD structure, spanning 902kb, was constructed using the Asian genotype data of 351 SNPs. Vertical tick marks above the name indicate the relative genomic position of each SNP. The LD structure represents the pairwise calculation of D’ for each possible combination of SNPs. D’ < 0.5 is shown in white, D’ = 1.0 in dark red, with increasing shades of red between representing increasing D’ between the SNPs. The genes from left to right are: ATXN2, BRAP, ACAD10, ALDH2, MAPKAPK5, TMEM116, ERP29, C12orf30, TRAFD1, C12orf51. The ALDH2 gene and the SNP rs671 are shown in red; and rs1062136 (Val/Glu), which is a nonsynonymous SNP, is shown in blue. The other genes are in black.
Discussion
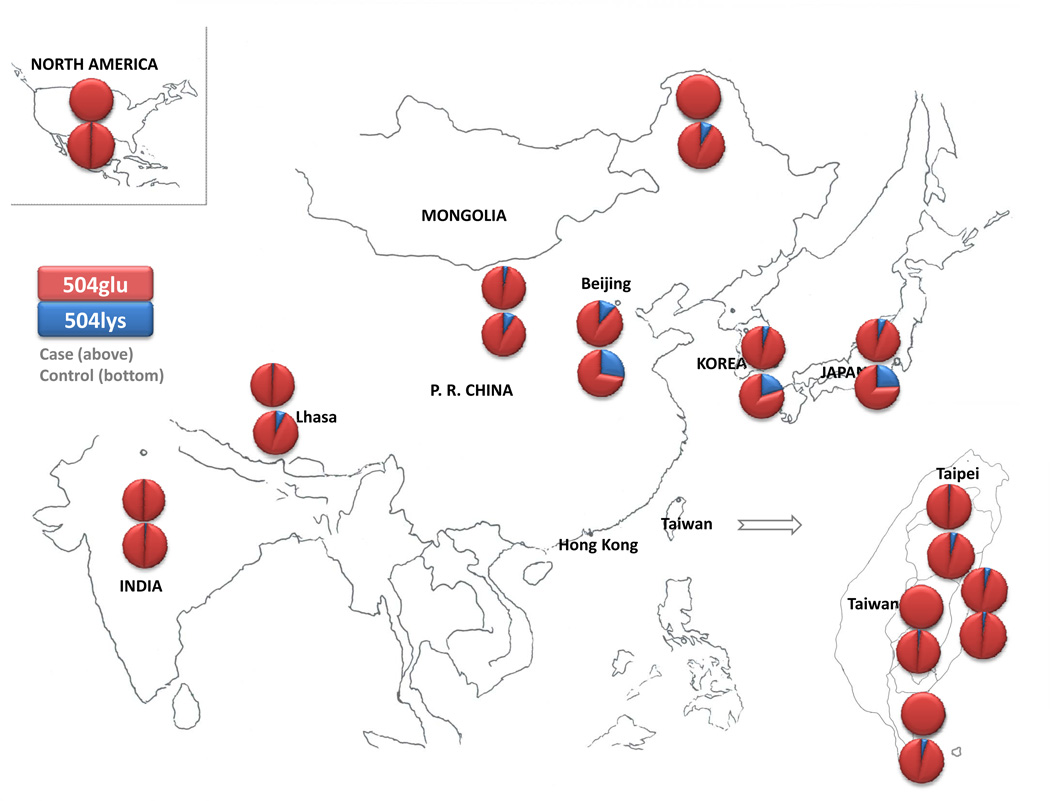
These findings support that the ALDH2 504lys allele can greatly lower the risk for AD as well as alcohol-induced diseases in the Asian populations. The reduction in ALDH2 activity in the 504lys/504glu heterozygotes is more than 100-fold compared with the ALDH2 activity of the 504glu/504glu homozygotes (Brooks et al. 2009; Crabb et al. 2004). Studies on liver extracts demonstrated that the ALDH2 504lys allele was nearly dominant, and the heterozygous and 504lys/504lys homozygous has almost no detectable ALDH2 activity in the liver (Edenberg 2007). The inactive ALDH2 504lys allele occurred mainly in some Asian populations (with the nearly complete absence of homozygous lys504lys alcoholics), but its prevalence varied across these samples. For instance, the 504lys allele was less commonly found in some aboriginal Chinese populations (e.g., Ami, Atayal, Bunun, Elunchan, Mongolian, and Paiwan) compared with Han Chinese. Figure 6 and supplementary Table 7 show the 504lys allele frequencies of by location. Most alcoholics with inactive ALDH2 alleles were heterozygous; homozygous lys-lys was rarely found in the alcoholic subjects, although it was often observed in controls. As expected, the recessive model based on the lys-lys genotype could not be applied in the present analysis due to insufficient power. Regarding the discrepant results in the individual association studies included in this meta-analysis, there are some possible explanations besides ethnic heterogeneity (e.g., one study (Chen et al. 1997) reported different direction of odd ratio in Ami): first, in most studies, subjects with AD or alcohol abuse were diagnosed according to the ICD (World Health Organization) or DSM (American Psychiatric Association) system. ICD-10 criteria for alcohol and drug dependence are more stringent than DSM-IV criteria, which in turn are more stringent than DSM-III-R (Schuckit et al. 1994). Two studies (Yan et al. 2003; Yu et al. 2002) applied local criteria and six studies had no specified criteria. Since these classification criteria differ, certain subjects would be eligible for inclusion as “affected” in some studies but not in others. Less stringent diagnostic criteria could differentiate the patient and control groups to a smaller extent, potentially altering observed effect sizes for ALDH2 504lys. Second, studies using mixed-sex samples may yield different results than studies using only male or female subjects, especially if the sexes are mixed in only patient or control group. Third, in terms of recruitment strategy, the allele differences between patients and controls may be greater in the samples recruited from clinical treatment than in the samples from general population due to well-recognized ascertainment bias. Fourth, the genetic effects of ALDH2 504lys on AD may change longitudinally over the course of illness. Fifth, the samples were not in Hardy-Weinberg equilibrium (HWE) (Zintzaras 2008) in several studies. Of the control samples that were not in HWE, either the samples were small or the disequilibrium was not highly significant. Disequilibrium in patients supports that the gene may be related to AD because the genotype distributions observed differ from those expected by chance; however, disequilibrium in controls is more likely to reflect genotyping error and thus distort the analysis in some degree (Luo et al. 2008). The sixth reason is different statistical power and type II error due to sample size as well as different genotyping methods. In addition, cultural differences may interact with the genetic components and result in differentially significance levels of protection of ALDH2 504lys against AD across different ethnic groups.
Figure 6.
504lys allele frequencies among different populations. Blue and red represent 504lys and 504glu, respectively. Upper graphs are based on the cases, lower graphs on controls. The geographical borders (Miyazaki et al. 1993) of Taiwan aboriginals were from a previous study.
The present comprehensive meta-analysis identified stronger association (P = 3×10−56) than the previous studies (Luczak et al. 2006; Zintzaras et al. 2006), which can be explained by several differences besides different research purposes and statistical methods (Minelli et al. 2005; Trikalinos et al. 2008; Zintzaras 2008 ; Zintzaras and Lau 2008; Zintzaras et al. 2009; Zintzaras et al. 2008) from ours (Li and He 2007a, b, c, 2008): the meta-analysis by Zintzaras et al.(Zintzaras et al. 2006) a) included 23 studies published in or before 2004 (compared to 53 studies in our meta-analysis); b) only included English literature; c) provided OR without specific P values; and d) found no association with liver disease using allelic analysis (the result was only positive under the dominant model). Another meta-analysis by Luczak et al.(Luczak et al. 2006), in which the latest dataset was also published in 2004, a) included 22 Asian studies including 1,980 cases and 2,550 controls (we included 9,678 cases and 7,331 controls); b) performed genotypic analysis but not allelic analysis; c) provided no specific P values; d) reported negative results (under the model of lys-lys vs. lys-glu); and e) found a difference between possessing one and two ALDH2 504lys alleles that approached but did not reach statistical significance. Compared with each published study, our meta-analysis a) included the largest sample size from 45 English and seven Chinese publications (it was important to include Chinese-language publications). Our analysis showed that there was no publication bias, thus, the Chinses literatutre did not introduce overestiamets of effect sizes in our meta-analysis (supplementary Table 2); b) performed both allelic and genotypic analyses using the random effects model; c) used both strict and extended criteria as well as carrying out subgroup analyses to detect the effects; d) applied comprehensive analysis methods, as shown in the results, to study additional questions not answered in those previous meta-analyses; and e) found strong evidence of association with both alcohol dependence and the alcohol-induced medical diseases. In addition, the procedure of ‘extended-quality score’ suggested in our previous study (Li et al. 2006) was also applied to assist the assessment of quality of the association studies. However, the limitations were that the majority of studies included were predominately from Asian populations, because of the protective 504lys allele was barely detected in the European-ancestry populations. The studies of both English and Chinese languages were included in this meta-analysis, and those of other Asian languages were not considered. In addition, it is possible that some negative results tend to be not published, which is hard to measure.
Findings of association studies depend on inclusion criteria and quality of the case and control subjects. Approximately 10% of Japanese alcoholics (Higuchi et al. 1994) develop their disease despite having the inactive (protective) ALDH2 encoded by either heterozygous lys-glu or homozygous lys-lys. Compared with the entire alcoholic population, the heterogeneity of alcoholism is considered to be relatively low in this genetically defined subgroup. Such alcoholics are considered to be advantageous for the identification of variations that produce minor effects; otherwise, the potential association may be masked by genetic variants with high effect.
In future studies, more precise analyses could be performed if information including sex, age, and age at onset was provided in most individual studies (sex differences and haplotypes (Liu et al. 2011) were not evaluated in this meta-analysis because the information was not available). The genetic risk effects of other factors can also be measured (see Dr. Zintzaras et al.’s papers (Trikalinos et al. 2008; Zintzaras ; Zintzaras and Lau 2008; Zintzaras et al. 2009; Zintzaras et al. 2008)), and other genetic models can be explored, as suggested in Minelli et al. study (Minelli et al. 2005). Additionally, full haplotypes as well as potential gene-gene and gene-environment interactions should be considered because these interactions have been found to alter the protective effect size. For example, Higuchi et al.(Higuchi et al. 1994) reported that rates of Japanese ALDH2 lys-glu individuals seeking treatment for AD increased from 2.5% in 1979 to 8% in 1986 then to 13% in 1992 in Japan while the rates of alcohol consumption also increased. Thus, the secular trend of increased cultural acceptance of alcohol consumption might have reduced the protective effect of the 504lys allele. Third, a previous longitudinal cohort study confirmed the significant roles of anxiety disorders and of the ADH1B Arg allele as antecedents of alcoholism among aboriginal Taiwanese (Cheng et al. 2004). Such studies will allow for determination of how the effects change over the course of lifetime alcohol use. Fourth, future studies should also try to use control samples that include older subjects which are more conservative regarding statistical analysis (as they will have passed through more of the age of risk for AD). Fifth, it is necessary to investigate the potential treatments for alcoholism based on the findings, for example, the alcohol aversion caused by this polymorphism compared to the effects of disulfiram.
In conclusion, using the cumulative data of the past 20 years from both English and Chinese publications, predominately from East-Asian populations (but barely detected in the European populations), this meta-analysis found highly strong evidence of association using both allelic (P = 3×10−56) and genotypic (P = 1×10−44) analyses under the random effects model. When strict criteria and sub-group analyses were applied, the strong associations remained. Our meta-analysis provides strong evidence for the involvement of the human ALDH2 gene in the pathogenesis of AD and alcohol abuse as well as alcohol-induced medical diseases in East-Asians.
Supplementary Material
Acknowledgements
This work was supported by the research grants DA12849, DA12690, AA017535, AA12870, and AA11330 from the National Institutes of Health, USA. We also thank Dr. Xingguang Luo for very helpful comments on the manuscript.
Footnotes
Conflict of Interest
None
Electronic-database information
- GenBank, http://www.ncbi.nlm.nih.gov/Genbank/ for genomic structure of ALDH2;
- Online Mendelian Inheritance in Man (OMIM), http://www.ncbi.nlm.nih.gov/Omim for ALDH2;
- Genotype data, http://www.hapmap.org/ for ALDH2;
- Genome data, http://genome.ucsc.edu/ for ALDH2.
References
- American Psychiatric Association. Diagnostic and Statistical Manual of Mental Disorders (DSM) Washington, DC: American Psychiatric Press; [Google Scholar]
- Begg CB, Mazumdar M. Operating characteristics of a rank correlation test for publication bias. Biometrics. 1994;50:1088–1101. [PubMed] [Google Scholar]
- Brooks PJ, Enoch MA, Goldman D, Li TK, Yokoyama A. The alcohol flushing response: an unrecognized risk factor for esophageal cancer from alcohol consumption. PLoS Med. 2009;6:e50. doi: 10.1371/journal.pmed.1000050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chai YG, Oh DY, Chung EK, Kim GS, Kim L, Lee YS, Choi IG. Alcohol and aldehyde dehydrogenase polymorphisms in men with type I and Type II alcoholism. Am J Psychiatry. 2005;162:1003–1005. doi: 10.1176/appi.ajp.162.5.1003. [DOI] [PubMed] [Google Scholar]
- Chao YC, Liou SR, Chung YY, Tang HS, Hsu CT, Li TK, Yin SJ. Polymorphism of alcohol and aldehyde dehydrogenase genes and alcoholic cirrhosis in Chinese patients. Hepatology. 1994;19:360–366. [PubMed] [Google Scholar]
- Chao YC, Wang LS, Hsieh TY, Chu CW, Chang FY, Chu HC. Chinese alcoholic patients with esophageal cancer are genetically different from alcoholics with acute pancreatitis and liver cirrhosis. Am J Gastroenterol. 2000;95:2958–2964. doi: 10.1111/j.1572-0241.2000.02328.x. [DOI] [PubMed] [Google Scholar]
- Chao YC, Wang SJ, Chu HC, Chang WK, Hsieh TY. Investigation of alcohol metabolizing enzyme genes in Chinese alcoholics with avascular necrosis of hip joint, pancreatitis and cirrhosis of the liver. Alcohol Alcohol. 2003;38:431–436. doi: 10.1093/alcalc/agg106. [DOI] [PubMed] [Google Scholar]
- Chao YC, Young TH, Tang HS, Hsu CT. Alcoholism and alcoholic organ damage and genetic polymorphisms of alcohol metabolizing enzymes in Chinese patients. Hepatology. 1997;25:112–117. doi: 10.1002/hep.510250121. [DOI] [PubMed] [Google Scholar]
- Chen CC, Lu RB, Chen YC, Wang MF, Chang YC, Li TK, Yin SJ. Interaction between the functional polymorphisms of the alcohol-metabolism genes in protection against alcoholism. Am J Hum Genet. 1999;65:795–807. doi: 10.1086/302540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen WJ, Loh EW, Hsu YP, Chen CC, Yu JM, Cheng AT. Alcohol-metabolising genes and alcoholism among Taiwanese Han men: independent effect of ADH2, ADH3 and ALDH2. Br J Psychiatry. 1996;168:762–767. doi: 10.1192/bjp.168.6.762. [DOI] [PubMed] [Google Scholar]
- Chen WJ, Loh EW, Hsu YP, Cheng AT. Alcohol dehydrogenase and aldehyde dehydrogenase genotypes and alcoholism among Taiwanese aborigines. Biol Psychiatry. 1997;41:703–709. doi: 10.1016/S0006-3223(96)00072-8. [DOI] [PubMed] [Google Scholar]
- Cheng AT, Gau SF, Chen TH, Chang JC, Chang YT. A 4-year longitudinal study on risk factors for alcoholism. Arch Gen Psychiatry. 2004;61:184–191. doi: 10.1001/archpsyc.61.2.184. [DOI] [PubMed] [Google Scholar]
- Choi IG, Kee BS, Son HG, Ham BJ, Yang BH, Kim SH, Lee JS, Son BK, Lee BY, Lee SY, Chai YG, Shin HD. Genetic polymorphisms of alcohol and aldehyde dehydrogenase, dopamine and serotonin transporters in familial and non-familial alcoholism. Eur Neuropsychopharmacol. 2006;16:123–128. doi: 10.1016/j.euroneuro.2005.07.006. [DOI] [PubMed] [Google Scholar]
- Crabb DW, Matsumoto M, Chang D, You M. Overview of the role of alcohol dehydrogenase and aldehyde dehydrogenase and their variants in the genesis of alcohol-related pathology. Proc Nutr Soc. 2004;63:49–63. doi: 10.1079/pns2003327. [DOI] [PubMed] [Google Scholar]
- Cui R, Kamatani Y, Takahashi A, Usami M, Hosono N, Kawaguchi T, Tsunoda T, Kamatani N, Kubo M, Nakamura Y, Matsuda K. Functional variants in ADH1B and ALDH2 coupled with alcohol and smoking synergistically enhance esophageal cancer risk. Gastroenterology. 2009;137:1768–1775. doi: 10.1053/j.gastro.2009.07.070. [DOI] [PubMed] [Google Scholar]
- DerSimonian R, Laird N. Meta-analysis in clinical trials. Control Clin Trials. 1986;7:177–188. doi: 10.1016/0197-2456(86)90046-2. [DOI] [PubMed] [Google Scholar]
- Duval S, Tweedie R. Trim and fill: A simple funnel-plot-based method of testing and adjusting for publication bias in meta-analysis. Biometrics. 2000;56:455–463. doi: 10.1111/j.0006-341x.2000.00455.x. [DOI] [PubMed] [Google Scholar]
- Edenberg HJ. The genetics of alcohol metabolism: role of alcohol dehydrogenase and aldehyde dehydrogenase variants. Alcohol Res Health. 2007;30:5–13. [PMC free article] [PubMed] [Google Scholar]
- Egger M, Davey Smith G, Schneider M, Minder C. Bias in meta-analysis detected by a simple, graphical test. Bmj. 1997;315:629–634. doi: 10.1136/bmj.315.7109.629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Excoffier L, Slatkin M. Maximum-likelihood estimation of molecular haplotype frequencies in a diploid population. Mol Biol Evol. 1995;12:921–927. doi: 10.1093/oxfordjournals.molbev.a040269. [DOI] [PubMed] [Google Scholar]
- Fan J, Shen Y, Cui Y, Tian C, Zhou R, Zhou C, Wang J, Dan S, Kang Z, Teng X, Xia G, Yao W, Wei X, Zhou M. ADH and ALDH genes among Korea and Elunchun ethnic groups in China. Chin J Psychiatry. 1998;31:209–212. [Google Scholar]
- Gelernter J, Kranzler HR. Genetics of alcohol dependence. Hum Genet. 2009;126:91–99. doi: 10.1007/s00439-009-0701-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grant BF, Dufour MC, Harford TC. Epidemiology of alcoholic liver disease. Semin Liver Dis. 1988;8:12–25. doi: 10.1055/s-2008-1040525. [DOI] [PubMed] [Google Scholar]
- Guo W, Wang Q, Lanzi G, Luobu O, Ma X, Wang Y, Zhen P, Ji Y, Wei G, Wang Z, Deng W, Zhuoma B, Shi X, Yan C, He C, Liu X, Wu Y, Luo H, Collier DA, Ball D, Li T, Hu X. Interaction among genes influencing ethanol metabolism and sex is association with alcohol use disorders in a Tibet population. Am J Med Genet B Neuropsychiatr Genet. 2010;153B:561–569. doi: 10.1002/ajmg.b.31020. [DOI] [PubMed] [Google Scholar]
- Harada S, Okubo T, Nakamura T, Fujii C, Nomura F, Higuchi S, Tsutsumi M. A novel polymorphism (-357 G/A) of the ALDH2 gene: linkage disequilibrium and an association with alcoholism. Alcohol Clin Exp Res. 1999;23:958–962. [PubMed] [Google Scholar]
- Hasegawa Y, Higuchi S, Matsushita S, Miyaoka H. Association of a polymorphism of the serotonin 1B receptor gene and alcohol dependence with inactive aldehyde dehydrogenase-2. J Neural Transm. 2002;109:513–521. doi: 10.1007/s007020200042. [DOI] [PubMed] [Google Scholar]
- Hendershot CS, Collins SE, George WH, Wall TL, McCarthy DM, Liang T, Larimer ME. Associations of ALDH2 and ADH1B genotypes with alcohol-related phenotypes in Asian young adults. Alcohol Clin Exp Res. 2009;33:839–847. doi: 10.1111/j.1530-0277.2009.00903.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higuchi S, Matsushita S, Imazeki H, Kinoshita T, Takagi S, Kono H. Aldehyde dehydrogenase genotypes in Japanese alcoholics. Lancet. 1994;343:741–742. doi: 10.1016/s0140-6736(94)91629-2. [DOI] [PubMed] [Google Scholar]
- Higuchi S, Matsushita S, Murayama M, Takagi S, Hayashida M. Alcohol and aldehyde dehydrogenase polymorphisms and the risk for alcoholism. Am J Psychiatry. 1995;152:1219–1221. doi: 10.1176/ajp.152.8.1219. [DOI] [PubMed] [Google Scholar]
- Higuchi S, Muramatsu T, Matsushita S, Murayama M. No evidence of association between structural polymorphism at the dopamine D3 receptor locus and alcoholism in the Japanese. Am J Med Genet. 1996a;67:412–414. doi: 10.1002/(SICI)1096-8628(19960726)67:4<412::AID-AJMG17>3.0.CO;2-M. [DOI] [PubMed] [Google Scholar]
- Higuchi S, Muramatsu T, Matsushita S, Murayama M, Hayashida M. Polymorphisms of ethanol-oxidizing enzymes in alcoholics with inactive ALDH2. Hum Genet. 1996b;97:431–434. doi: 10.1007/BF02267061. [DOI] [PubMed] [Google Scholar]
- Huang SY, Lin WW, Ko HC, Lee JF, Wang TJ, Chou YH, Yin SJ, Lu RB. Possible interaction of alcohol dehydrogenase and aldehyde dehydrogenase genes with the dopamine D2 receptor gene in anxiety-depressive alcohol dependence. Alcohol Clin Exp Res. 2004;28:374–384. doi: 10.1097/01.alc.0000117832.62901.61. [DOI] [PubMed] [Google Scholar]
- Iwahashi K, Matsuo Y, Suwaki H, Nakamura K, Ichikawa Y. CYP2E1 and ALDH2 genotypes and alcohol dependence in Japanese. Alcohol Clin Exp Res. 1995;19:564–566. doi: 10.1111/j.1530-0277.1995.tb01549.x. [DOI] [PubMed] [Google Scholar]
- Jing Q, Zhong S, Gao L, Wang X, Dou S, He G, Ran Y, Bao J. Association analyses of ADH2, ALDH2 AND CYP4502E1 genetic polymorphisms with alcohol dependence syndrome in Yunnan Han population. Chin J Drug Depend. 2009;18:341–370. [Google Scholar]
- Johnson AD, Handsaker RE, Pulit SL, Nizzari MM, O'Donnell CJ, de Bakker PI. SNAP: a web-based tool for identification and annotation of proxy SNPs using HapMap. Bioinformatics. 2008;24:2938–2939. doi: 10.1093/bioinformatics/btn564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamatani Y, Matsuda K, Okada Y, Kubo M, Hosono N, Daigo Y, Nakamura Y, Kamatani N. Genome-wide association study of hematological and biochemical traits in a Japanese population. Nat Genet. 2010;42:210–215. doi: 10.1038/ng.531. [DOI] [PubMed] [Google Scholar]
- Kato N, Takeuchi F, Tabara Y, Kelly TN, Go MJ, Sim X, Tay WT, Chen CH, Zhang Y, Yamamoto K, Katsuya T, Yokota M, Kim YJ, Ong RT, Nabika T, Gu D, Chang LC, Kokubo Y, Huang W, Ohnaka K, Yamori Y, Nakashima E, Jaquish CE, Lee JY, Seielstad M, Isono M, Hixson JE, Chen YT, Miki T, Zhou X, Sugiyama T, Jeon JP, Liu JJ, Takayanagi R, Kim SS, Aung T, Sung YJ, Zhang X, Wong TY, Han BG, Kobayashi S, Ogihara T, Zhu D, Iwai N, Wu JY, Teo YY, Tai ES, Cho YS, He J. Meta-analysis of genome-wide association studies identifies common variants associated with blood pressure variation in east Asians. Nat Genet. 2011;43:531–538. doi: 10.1038/ng.834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kendler KS, Myers J, Prescott CA. Specificity of genetic and environmental risk factors for symptoms of cannabis, cocaine, alcohol, caffeine, and nicotine dependence. Arch Gen Psychiatry. 2007;64:1313–1320. doi: 10.1001/archpsyc.64.11.1313. [DOI] [PubMed] [Google Scholar]
- Kim DJ, Choi IG, Park BL, Lee BC, Ham BJ, Yoon S, Bae JS, Cheong HS, Shin HD. Major genetic components underlying alcoholism in Korean population. Hum Mol Genet. 2008;17:854–858. doi: 10.1093/hmg/ddm357. [DOI] [PubMed] [Google Scholar]
- Kim SA, Kim JW, Song JY, Park S, Lee HJ, Chung JH. Association of polymorphisms in nicotinic acetylcholine receptor alpha 4 subunit gene (CHRNA4), mu-opioid receptor gene (OPRM1), and ethanol-metabolizing enzyme genes with alcoholism in Korean patients. Alcohol. 2004;34:115–120. doi: 10.1016/j.alcohol.2004.06.004. [DOI] [PubMed] [Google Scholar]
- Kimura M, Sawayama T, Matsushita S, Higuchi S, Kashima H. Association between personality traits and ALDH2 polymorphism in Japanese male alcoholics. Alcohol Clin Exp Res. 2009;33:799–803. doi: 10.1111/j.1530-0277.2009.00898.x. [DOI] [PubMed] [Google Scholar]
- Konishi T, Luo HR, Calvillo M, Mayo MS, Lin KM, Wan YJ. ADH1B*1, ADH1C*2, DRD2 (-141C Ins), and 5-HTTLPR are associated with alcoholism in Mexican American men living in Los Angeles. Alcohol Clin Exp Res. 2004;28:1145–1152. doi: 10.1097/01.alc.0000134231.48395.42. [DOI] [PubMed] [Google Scholar]
- Kuo PH, Kalsi G, Prescott CA, Hodgkinson CA, Goldman D, van den Oord EJ, Alexander J, Jiang C, Sullivan PF, Patterson DG, Walsh D, Kendler KS, Riley BP. Association of ADH and ALDH genes with alcohol dependence in the Irish Affected Sib Pair Study of alcohol dependence (IASPSAD) sample. Alcohol Clin Exp Res. 2008;32:785–795. doi: 10.1111/j.1530-0277.2008.00642.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee HC, Lee HS, Jung SH, Yi SY, Jung HK, Yoon JH, Kim CY. Association between polymorphisms of ethanol-metabolizing enzymes and susceptibility to alcoholic cirrhosis in a Korean male population. J Korean Med Sci. 2001;16:745–750. doi: 10.3346/jkms.2001.16.6.745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee JF, Lu RB, Ko HC, Chang FM, Yin SJ, Pakstis AJ, Kidd KK. No association between DRD2 locus and alcoholism after controlling the ADH and ALDH genotypes in Chinese Han population. Alcohol Clin Exp Res. 1999;23:592–599. [PubMed] [Google Scholar]
- Lee SY, Hahn CY, Lee JF, Chen SL, Chen SH, Yeh TL, Kuo PH, Lee IH, Yang YK, Huang SY, Ko HC, Lu RB. MAOA-uVNTR polymorphism may modify the protective effect of ALDH2 gene against alcohol dependence in antisocial personality disorder. Alcohol Clin Exp Res. 2009;33:985–990. doi: 10.1111/j.1530-0277.2009.00919.x. [DOI] [PubMed] [Google Scholar]
- Lee SY, Hahn CY, Lee JF, Huang SY, Chen SL, Kuo PH, Lee IH, Yeh TL, Yang YK, Chen SH, Ko HC, Lu RB. MAOA interacts with the ALDH2 gene in anxiety-depression alcohol dependence. Alcohol Clin Exp Res. 2010;34:1212–1218. doi: 10.1111/j.1530-0277.2010.01198.x. [DOI] [PubMed] [Google Scholar]
- Leng A, Wang D, Yuan W. Research on the relationship between ALDH2 and CYP2E1 gene polymorphism and alcoholic liver disease in Han people. J Chongqing Med Univ. 2008;133:1108–1110. [Google Scholar]
- Li D, Collier DA, He L. Meta-analysis shows strong positive association of the neuregulin 1 (NRG1) gene with schizophrenia. Hum Mol Genet. 2006;15:1995–2002. doi: 10.1093/hmg/ddl122. [DOI] [PubMed] [Google Scholar]
- Li D, He L. Association study between the NMDA receptor 2B subunit gene (GRIN2B) and schizophrenia: a HuGE review and meta-analysis. Genet Med. 2007a;9:4–8. doi: 10.1097/01.gim.0000250507.96760.4b. [DOI] [PubMed] [Google Scholar]
- Li D, He L. G72/G30 genes and schizophrenia: a systematic meta-analysis of association studies. Genetics. 2007b;175:917–922. doi: 10.1534/genetics.106.061796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li D, He L. Meta-analysis supports association between serotonin transporter (5-HTT) and suicidal behavior. Mol Psychiatry. 2007c;12:47–54. doi: 10.1038/sj.mp.4001890. [DOI] [PubMed] [Google Scholar]
- Li D, He L. Meta-study on association between the monoamine oxidase A gene (MAOA) and schizophrenia. Am J Med Genet B Neuropsychiatr Genet. 2008;147B:174–178. doi: 10.1002/ajmg.b.30570. [DOI] [PubMed] [Google Scholar]
- Li Y. Issues on classification and diagnostic criteria of alcoholic liver disease. Chin J. Dig. 2002;22:38–39. [Google Scholar]
- Liu J, Zhou Z, Hodgkinson CA, Yuan Q, Shen PH, Mulligan CJ, Wang A, Gray RR, Roy A, Virkkunen M, Goldman D, Enoch MA. Haplotype-based study of the association of alcohol-metabolizing genes with alcohol dependence in four independent populations. Alcohol Clin Exp Res. 2011;35:304–316. doi: 10.1111/j.1530-0277.2010.01346.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luczak SE, Glatt SJ, Wall TL. Meta-analyses of ALDH2 and ADH1B with alcohol dependence in Asians. Psychol Bull. 2006;132:607–621. doi: 10.1037/0033-2909.132.4.607. [DOI] [PubMed] [Google Scholar]
- Luo X, Zuo L, Kranzler HR, Wang S, Anton RF, Gelernter J. Recessive genetic mode of an ADH4 variant in substance dependence in African-Americans: A model of utility of the HWD test. Behav Brain Funct. 2008;4:42. doi: 10.1186/1744-9081-4-42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macgregor S, Lind PA, Bucholz KK, Hansell NK, Madden PA, Richter MM, Montgomery GW, Martin NG, Heath AC, Whitfield JB. Associations of ADH and ALDH2 gene variation with self report alcohol reactions, consumption and dependence: an integrated analysis. Hum Mol Genet. 2009;18:580–593. doi: 10.1093/hmg/ddn372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maezawa Y, Yamauchi M, Toda G, Suzuki H, Sakurai S. Alcohol-metabolizing enzyme polymorphisms and alcoholism in Japan. Alcohol Clin Exp Res. 1995;19:951–954. doi: 10.1111/j.1530-0277.1995.tb00972.x. [DOI] [PubMed] [Google Scholar]
- Matsushita S, Muramatsu T, Murayama M, Nakane J, Higuchi S. Alcoholism, ALDH2*2 allele and the A1 allele of the dopamine D2 receptor gene: an association study. Psychiatry Res. 2001;104:19–26. doi: 10.1016/s0165-1781(01)00290-6. [DOI] [PubMed] [Google Scholar]
- McKay JD, Truong T, Gaborieau V, Chabrier A, Chuang SC, Byrnes G, Zaridze D, Shangina O, Szeszenia-Dabrowska N, Lissowska J, Rudnai P, Fabianova E, Bucur A, Bencko V, Holcatova I, Janout V, Foretova L, Lagiou P, Trichopoulos D, Benhamou S, Bouchardy C, Ahrens W, Merletti F, Richiardi L, Talamini R, Barzan L, Kjaerheim K, Macfarlane GJ, Macfarlane TV, Simonato L, Canova C, Agudo A, Castellsague X, Lowry R, Conway DI, McKinney PA, Healy CM, Toner ME, Znaor A, Curado MP, Koifman S, Menezes A, Wunsch-Filho V, Neto JE, Garrote LF, Boccia S, Cadoni G, Arzani D, Olshan AF, Weissler MC, Funkhouser WK, Luo J, Lubinski J, Trubicka J, Lener M, Oszutowska D, Schwartz SM, Chen C, Fish S, Doody DR, Muscat JE, Lazarus P, Gallagher CJ, Chang SC, Zhang ZF, Wei Q, Sturgis EM, Wang LE, Franceschi S, Herrero R, Kelsey KT, McClean MD, Marsit CJ, Nelson HH, Romkes M, Buch S, Nukui T, Zhong S, Lacko M, Manni JJ, Peters WH, Hung RJ, McLaughlin J, Vatten L, Njolstad I, Goodman GE, Field JK, Liloglou T, Vineis P, Clavel-Chapelon F, Palli D, Tumino R, Krogh V, Panico S, Gonzalez CA, Quiros JR, Martinez C, Navarro C, Ardanaz E, Larranaga N, et al. A genome-wide association study of upper aerodigestive tract cancers conducted within the INHANCE consortium. PLoS Genet. 2011;7:e1001333. doi: 10.1371/journal.pgen.1001333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minelli C, Thompson JR, Abrams KR, Thakkinstian A, Attia J. The choice of a genetic model in the meta-analysis of molecular association studies. Int J Epidemiol. 2005;34:1319–1328. doi: 10.1093/ije/dyi169. [DOI] [PubMed] [Google Scholar]
- Miyatake R, Suwaki H, Nakamura K, Matsuo Y, Iwahashi K. CYP2E1 genotypes and serum LAP in Japanese alcoholics. Life Sci. 1995;56:1121–1126. doi: 10.1016/0024-3205(95)00049-c. [DOI] [PubMed] [Google Scholar]
- Miyazaki H, Yamaguchi Y, Takehara T. Dental arch and palate in Taiwan aboriginals--Ami, Bunun, Paiwan and Rukai tribes. Arch Oral Biol. 1993;38:729–735. doi: 10.1016/0003-9969(93)90067-v. [DOI] [PubMed] [Google Scholar]
- Mizoi Y, Yamamoto K, Ueno Y, Fukunaga T, Harada S. Involvement of genetic polymorphism of alcohol and aldehyde dehydrogenases in individual variation of alcohol metabolism. Alcohol Alcohol. 1994;29:707–710. [PubMed] [Google Scholar]
- Muramatsu T, Higuchi S, Murayama M, Matsushita S, Hayashida M. Association between alcoholism and the dopamine D4 receptor gene. J Med Genet. 1996;33:113–115. doi: 10.1136/jmg.33.2.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muramatsu T, Wang ZC, Fang YR, Hu KB, Yan H, Yamada K, Higuchi S, Harada S, Kono H. Alcohol and aldehyde dehydrogenase genotypes and drinking behavior of Chinese living in Shanghai. Hum Genet. 1995;96:151–154. doi: 10.1007/BF00207371. [DOI] [PubMed] [Google Scholar]
- Nakamura K, Iwahashi K, Matsuo Y, Miyatake R, Ichikawa Y, Suwaki H. Characteristics of Japanese alcoholics with the atypical aldehyde dehydrogenase 2*2. I. A comparison of the genotypes of ALDH2, ADH2, ADH3, and cytochrome P-4502E1 between alcoholics and nonalcoholics. Alcohol Clin Exp Res. 1996;20:52–55. doi: 10.1111/j.1530-0277.1996.tb01043.x. [DOI] [PubMed] [Google Scholar]
- Nakamura T, Matsushita S, Nishiguchi N, Kimura M, Yoshino A, Higuchi S. Association of a polymorphism of the 5HT2A receptor gene promoter region with alcohol dependence. Mol Psychiatry. 1999;4:85–88. doi: 10.1038/sj.mp.4000474. [DOI] [PubMed] [Google Scholar]
- Nanakorn S, Fukuda K, Nishiyori A, Shibata A, Nakamura J. Aldehyde dehydrogenase genotypes and male alcohol use disorders: a case-control study in Khon Kaen, north-east Thailand. Psychiatry Clin Neurosci. 1999;53:397–405. doi: 10.1046/j.1440-1819.1999.00563.x. [DOI] [PubMed] [Google Scholar]
- Nishizawa D, Han W, Hasegawa J, Ishida T, Numata Y, Sato T, Kawai A, Ikeda K. Association of mu-opioid receptor gene polymorphism A118G with alcohol dependence in a Japanese population. Neuropsychobiology. 2006;53:137–141. doi: 10.1159/000093099. [DOI] [PubMed] [Google Scholar]
- Paik YK, Choi Y, Lee CG, Kim IK. Differences in Genetic Variation of ADH2 and ALDH2 between Alcoholics and Healthy Persons in Korea. Korean J. Genetics. 2000;22:117–126. [Google Scholar]
- Park KS, Mok JW, Chung TH. Genetic aspects and relative risk factors in alcoholism among Koreans. Korean J. Genetics. 2001;23:143–150. [Google Scholar]
- Rosenthal R. The file drawer problem and tolerance for null results. Psychol Bull. 1979;86:638–641. [Google Scholar]
- Sasabe T, Furukawa A, Matsusita S, Higuchi S, Ishiura S. Association analysis of the dopamine receptor D2 (DRD2) SNP rs1076560 in alcoholic patients. Neurosci Lett. 2007;412:139–142. doi: 10.1016/j.neulet.2006.10.064. [DOI] [PubMed] [Google Scholar]
- Saunders JB, Aasland OG, Babor TF, de la Fuente JR, Grant M. Development of the Alcohol Use Disorders Identification Test (AUDIT): WHO Collaborative Project on Early Detection of Persons with Harmful Alcohol Consumption--II. Addiction. 1993;88:791–804. doi: 10.1111/j.1360-0443.1993.tb02093.x. [DOI] [PubMed] [Google Scholar]
- Schuckit MA, Hesselbrock V, Tipp J, Anthenelli R, Bucholz K, Radziminski S. A comparison of DSM-III-R, DSM-IV and ICD-10 substance use disorders diagnoses in 1922 men and women subjects in the COGA study. Collaborative Study on the Genetics of Alcoholism. Addiction. 1994;89:1629–1638. doi: 10.1111/j.1360-0443.1994.tb03764.x. [DOI] [PubMed] [Google Scholar]
- Segado Soriano A, Santiago Dorrego C, Banares Canizares R, Alvarez Fernandez E, Bandres Moya F, Gomez-Gallego F. Genetic susceptibility to the development of acute alcoholic hepatitis: role of genetic mutations in dehydrogenase alcohol, aldehyde dehydrogenase and cytochrome P450 2E1. Rev Clin Esp. 2005;205:528–532. doi: 10.1016/s0014-2565(05)72632-1. [DOI] [PubMed] [Google Scholar]
- Shafe S, Gilder DA, Montane-Jaime LK, Joseph R, Moore S, Crooks H, Ramcharan C, Ehlers CL. Co-morbidity of Alcohol Dependence and Select Affective and Anxiety Disorders among Individuals of East Indian and African Ancestry in Trinidad and Tobago. West Indian Med. J. 2009;58:164–172. [PMC free article] [PubMed] [Google Scholar]
- Shen Y, Fan J, Cui Y, Zhou R, Wang Y, Tian C, Zhou C, Li TK, Edenberg HJ, Wang J, Zhao Z, Bai Y, Dan S, Kang Z, Teng X, Zhang X, Fan Z, Liu J, Zhang Z. A study of the correlation between alcohol dependence and polymorphism of alcoholdehydrogenase genes and aldehyde-dehydrogenase genes among Mongolian and Han ethnic groups in China. Chin J Psychiatry. 1997a;30:3–6. [Google Scholar]
- Shen YC, Fan JH, Edenberg HJ, Li TK, Cui YH, Wang YF, Tian CH, Zhou CF, Zhou RL, Wang J, Zhao ZL, Xia GY. Polymorphism of ADH and ALDH genes among four ethnic groups in China and effects upon the risk for alcoholism. Alcohol Clin Exp Res. 1997b;21:1272–1277. [PubMed] [Google Scholar]
- Shimosegawa T, Kume K, Masamune A. SPINK1, ADH2, and ALDH2 gene variants and alcoholic chronic pancreatitis in Japan. J Gastroenterol Hepatol. 2008;23(Suppl 1):S82–S86. doi: 10.1111/j.1440-1746.2007.05291.x. [DOI] [PubMed] [Google Scholar]
- Shin S, Stewart R, Ferri CP, Kim JM, Shin IS, Kim SW, Yang SJ, Yoon JS. An investigation of associations between alcohol use disorder and polymorphisms on ALDH2, BDNF, 5-HTTLPR, and MTHFR genes in older Korean men. Int J Geriatr Psychiatry. 2010;25:441–448. doi: 10.1002/gps.2358. [DOI] [PubMed] [Google Scholar]
- Tan EC, Lim L, Leong JY, Lim JY, Lee A, Yang J, Tan CH, Winslow M. Alcohol and aldehyde dehydrogenase polymorphisms in Chinese and Indian populations. Subst Use Misuse. 2010;45:1–14. doi: 10.3109/10826080802490584. [DOI] [PubMed] [Google Scholar]
- Tanaka F, Shiratori Y, Yokosuka O, Imazeki F, Tsukada Y, Omata M. High incidence of ADH2*1/ALDH2*1 genes among Japanese alcohol dependents and patients with alcoholic liver disease. Hepatology. 1996;23:234–239. doi: 10.1053/jhep.1996.v23.pm0008591846. [DOI] [PubMed] [Google Scholar]
- Thomasson HR, Crabb DW, Edenberg HJ, Li TK, Hwu HG, Chen CC, Yeh EK, Yin SJ. Low frequency of the ADH2*2 allele among Atayal natives of Taiwan with alcohol use disorders. Alcohol Clin Exp Res. 1994;18:640–643. doi: 10.1111/j.1530-0277.1994.tb00923.x. [DOI] [PubMed] [Google Scholar]
- Thomasson HR, Edenberg HJ, Crabb DW, Mai XL, Jerome RE, Li TK, Wang SP, Lin YT, Lu RB, Yin SJ. Alcohol and aldehyde dehydrogenase genotypes and alcoholism in Chinese men. Am J Hum Genet. 1991;48:677–681. [PMC free article] [PubMed] [Google Scholar]
- Trikalinos TA, Salanti G, Zintzaras E, Ioannidis JP. Meta-analysis methods. Adv Genet. 2008;60:311–334. doi: 10.1016/S0065-2660(07)00413-0. [DOI] [PubMed] [Google Scholar]
- Tsuchihashi-Makaya M, Serizawa M, Yanai K, Katsuya T, Takeuchi F, Fujioka A, Yamori Y, Ogihara T, Kato N. Gene-environmental interaction regarding alcohol-metabolizing enzymes in the Japanese general population. Hypertens Res. 2009;32:207–213. doi: 10.1038/hr.2009.3. [DOI] [PubMed] [Google Scholar]
- Vaswani M, Prasad P, Kapur S. Association of ADH1B and ALDH2 gene polymorphisms with alcohol dependence: a pilot study from India. Hum Genomics. 2009;3:213–220. doi: 10.1186/1479-7364-3-3-213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei F, Fan J, Shen Y, Tian C, Zhou R, Zheng X, Peng H, Zhao H, Li Y, Zhou C, Zhang W. A control study on the polymorphism of the ADH and ALDH genes in high risk alcoholic families of Han ethnic population. Chin J Psychiatry. 1999;32:164–166. [Google Scholar]
- Woolf B. On estimating the relation between blood group and disease. Ann Hum Genet. 1955;19:251–253. doi: 10.1111/j.1469-1809.1955.tb01348.x. [DOI] [PubMed] [Google Scholar]
- World Health Organization. Geneva: WHO; World Health Organization’s International Statistical Classification of Diseases and Related Health Problems (ICD) [Google Scholar]
- Xu K, Liu XH, Nagarajan S, Gu XY, Goldman D. Relationship of the delta-opioid receptor gene to heroin abuse in a large Chinese case/control sample. Am J Med Genet. 2002;110:45–50. doi: 10.1002/ajmg.10374. [DOI] [PubMed] [Google Scholar]
- Yamauchi M, Maezawa Y, Toda G, Suzuki H, Sakurai S. Association of a restriction fragment length polymorphism in the alcohol dehydrogenase 2 gene with Japanese alcoholic liver cirrhosis. J Hepatol. 1995;23:519–523. doi: 10.1016/0168-8278(95)80056-5. [DOI] [PubMed] [Google Scholar]
- Yan M, Zhu KX, Meng FL, Wang HJ, Wu ML. Relationship between ALDH gene polymorphism and alcoholic liver diseases. Zhonghua Gan Zang Bing Za Zhi. 2003;11:654–656. [PubMed] [Google Scholar]
- Yang J, Li N, Li T, Cao M, Shao L, Y M. Aldehyde Dehydrogenase Gene G1951A Polymorphism and Drinking Behavior in Males. Sci Tech Engng. 2007;7:5493–5497. [Google Scholar]
- Yoshida A, Hsu LC, Yasunami M. Genetics of human alcohol-metabolizing enzymes. Prog Nucleic Acid Res Mol Biol. 1991;40:255–287. doi: 10.1016/s0079-6603(08)60844-2. [DOI] [PubMed] [Google Scholar]
- Yoshida A, Huang IY, Ikawa M. Molecular abnormality of an inactive aldehyde dehydrogenase variant commonly found in Orientals. Proc Natl Acad Sci U S A. 1984;81:258–261. doi: 10.1073/pnas.81.1.258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu C, Li Y, Chen W, Yue M. Genotype of ethanol metabolizing enzyme genes by oligonucleotide microarray in alcoholic liver disease in Chinese people. Chin Med J (Engl) 2002;115:1085–1087. [PubMed] [Google Scholar]
- Zintzaras E. The generalized odds ratio as a measure of genetic risk effect in the analysis and meta-analysis of association studies. Stat Appl Genet Mol Biol. 9 doi: 10.2202/1544-6115.1542. Article21. [DOI] [PubMed] [Google Scholar]
- Zintzaras E. Variance estimation of allele-based odds ratio in the absence of Hardy-Weinberg equilibrium. Eur J Epidemiol. 2008;23:323–326. doi: 10.1007/s10654-008-9242-6. [DOI] [PubMed] [Google Scholar]
- Zintzaras E, Ioannidis JP. Heterogeneity testing in meta-analysis of genome searches. Genet Epidemiol. 2005;28:123–137. doi: 10.1002/gepi.20048. [DOI] [PubMed] [Google Scholar]
- Zintzaras E, Lau J. Synthesis of genetic association studies for pertinent gene-disease associations requires appropriate methodological and statistical approaches. J Clin Epidemiol. 2008;61:634–645. doi: 10.1016/j.jclinepi.2007.12.011. [DOI] [PubMed] [Google Scholar]
- Zintzaras E, Papathanasiou AA, Stefanidis I. Endothelial nitric oxide synthase gene polymorphisms and diabetic nephropathy: a HuGE review and meta-analysis. Genet Med. 2009;11:695–706. doi: 10.1097/GIM.0b013e3181b2046b. [DOI] [PubMed] [Google Scholar]
- Zintzaras E, Raman G, Kitsios G, Lau J. Angiotensin-converting enzyme insertion/deletion gene polymorphic variant as a marker of coronary artery disease: a meta-analysis. Arch Intern Med. 2008;168:1077–1089. doi: 10.1001/archinte.168.10.1077. [DOI] [PubMed] [Google Scholar]
- Zintzaras E, Stefanidis I, Santos M, Vidal F. Do alcohol-metabolizing enzyme gene polymorphisms increase the risk of alcoholism and alcoholic liver disease? Hepatology. 2006;43:352–361. doi: 10.1002/hep.21023. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.