Abstract
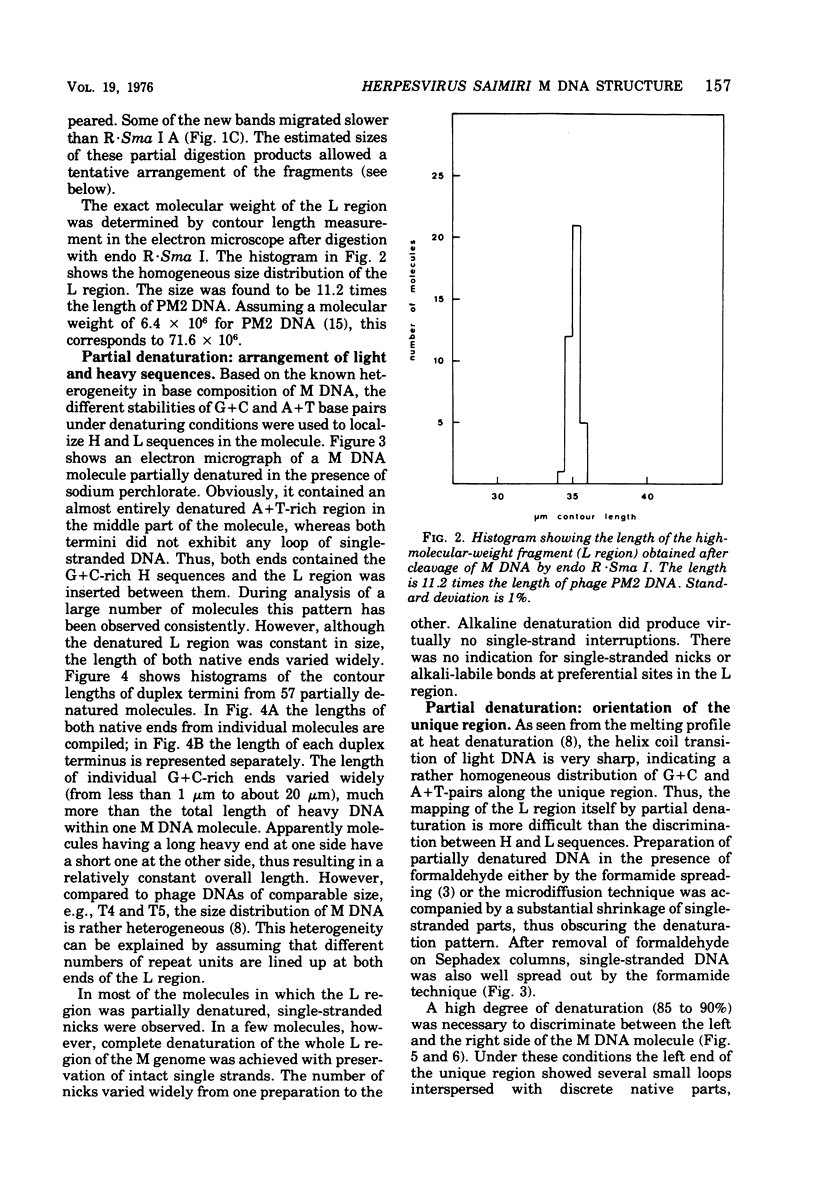
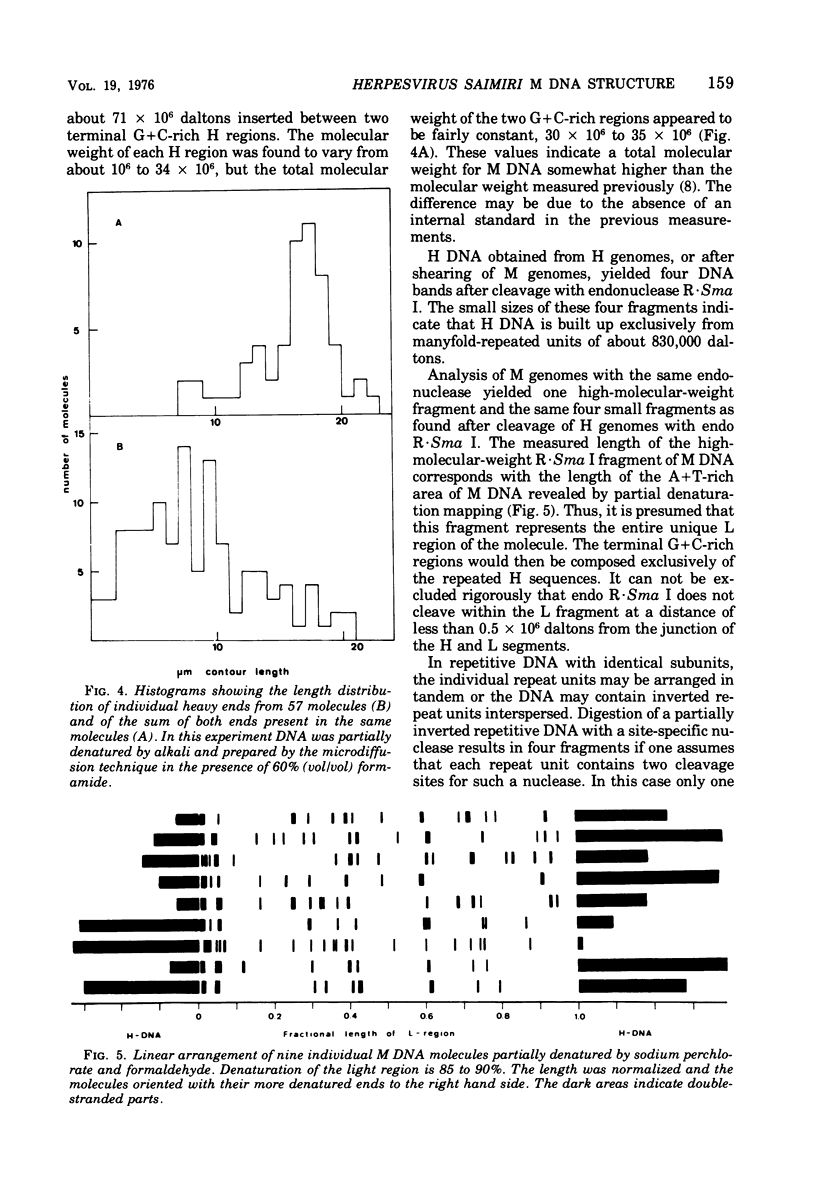
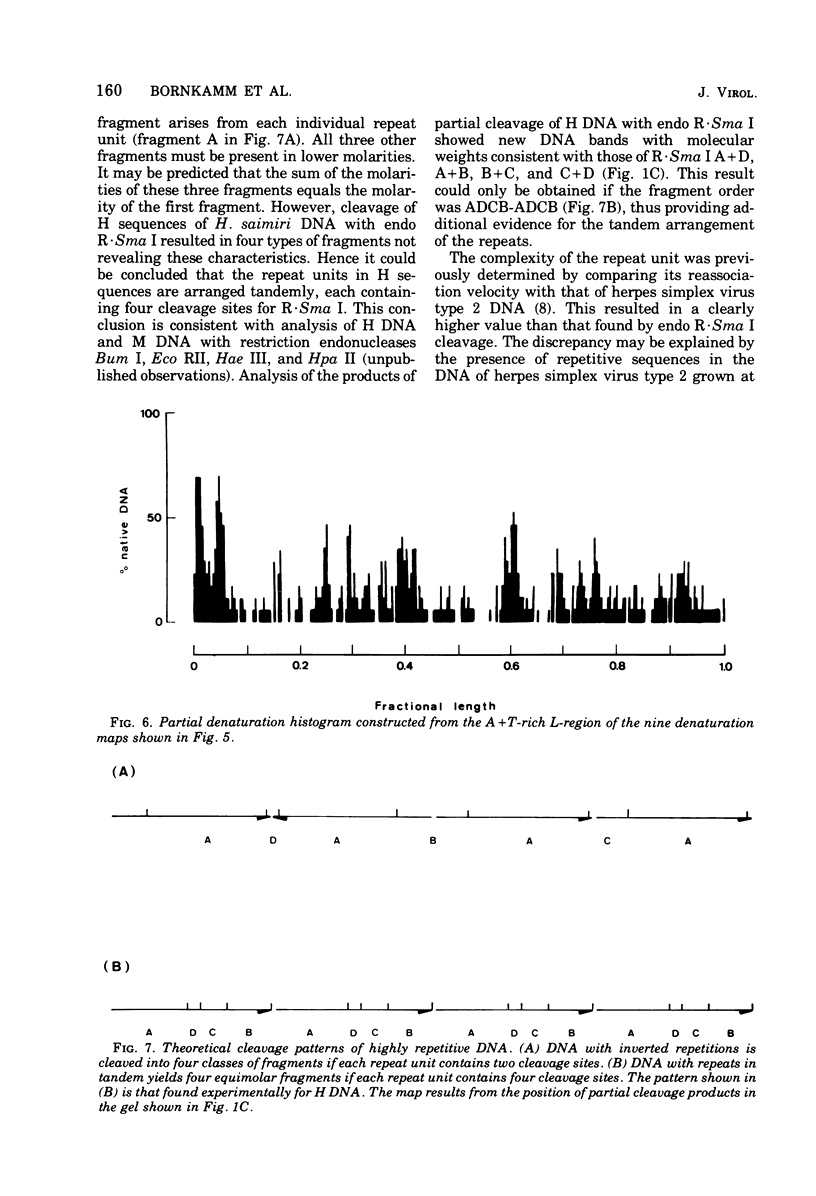
Herpesvirus saimiri contains two species of DNA molecules. (i) The M genome is composed of 70% light (L) DNA (36% cytosine plus guanine; density in CsCl, 1.695 g/ml), which consists of unique sequences, and 30% heavy (H) DNA (71% cytosine plus guanine; density, 1.729 g/ml). (ii) The H genome contains heavy sequences exclusively. H sequences in M and H genomes cross-hybridize completely and are cleaved identically by restriction endonuclease R-Sma I into four classes of fragments with molecular weights of about 360,000, 300,000, 130,000 and 40,000, respectively. H sequences are chains of identical repeat units in tandem arrangement. The molecular weight of each repeat unit is about 830,000. L sequences have no cleavage site for endo R-Sma I H sequences are terminally arranged at both ends of the M genome, as seen by electron microscopy after partial denaturation. The length of the individual heavy ends varies between 21 mum and less than 1 mum, whereas the light region is uniform in size (35.3+/-0.35 mum). As a rule, molecules with a long heavy end at one side have a short heavy end at the other side, thus giving rise to a limited size heterogeneity. Orientation of M DNA molecules by the denaturation map of the light region shows that the longer heavy end may be located at the left or at the right side of the M genome.
Full text
PDF

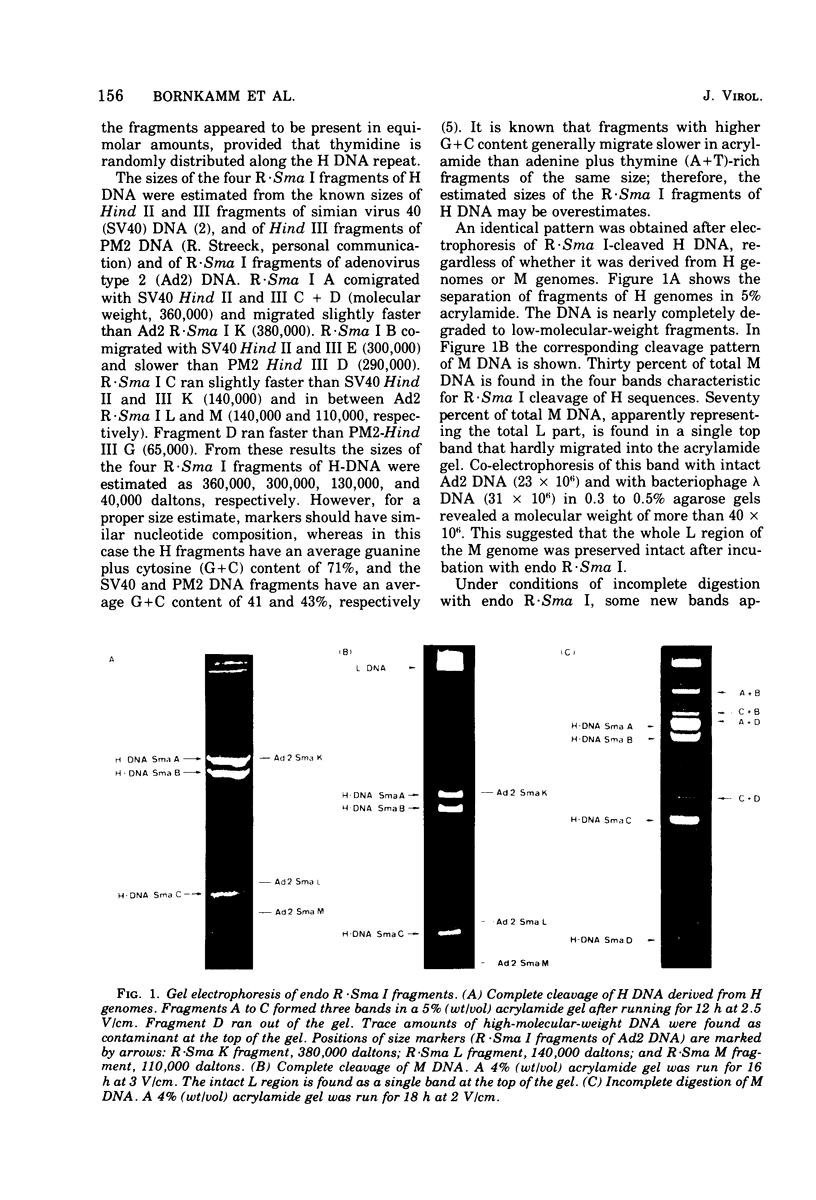


Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Adams A., Lindahl T. Epstein-Barr virus genomes with properties of circular DNA molecules in carrier cells. Proc Natl Acad Sci U S A. 1975 Apr;72(4):1477–1481. doi: 10.1073/pnas.72.4.1477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Danna K. J., Sack G. H., Jr, Nathans D. Studies of simian virus 40 DNA. VII. A cleavage map of the SV40 genome. J Mol Biol. 1973 Aug 5;78(2):363–376. doi: 10.1016/0022-2836(73)90122-8. [DOI] [PubMed] [Google Scholar]
- Espejo R. T., Canelo E. S. Properties of bacteriophage PM2: a lipid-containing bacterial virus. Virology. 1968 Apr;34(4):738–747. doi: 10.1016/0042-6822(68)90094-9. [DOI] [PubMed] [Google Scholar]
- Falk L. A., Wolfe L. G., Deinhardt F. Isolation of Herpesvirus saimiri from blood of squirrel monkeys (Saimiri sciureus). J Natl Cancer Inst. 1972 May;48(5):1499–1505. [PubMed] [Google Scholar]
- Fleckenstein B., Bornkamm G. W., Ludwig H. Repetitive sequences in complete and defective genomes of Herpesvirus saimiri. J Virol. 1975 Feb;15(2):398–406. doi: 10.1128/jvi.15.2.398-406.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fleckenstein B., Wolf H. Purification and properties of Herpesvirus saimiri DNA. Virology. 1974 Mar;58(1):55–64. doi: 10.1016/0042-6822(74)90140-8. [DOI] [PubMed] [Google Scholar]
- Gómez B., Lang D. Denaturation map of bacteriophage T7 DNA and direction of DNA transcription. J Mol Biol. 1972 Sep 28;70(2):239–251. doi: 10.1016/0022-2836(72)90536-0. [DOI] [PubMed] [Google Scholar]
- Hayward G. S., Jacob R. J., Wadsworth S. C., Roizman B. Anatomy of herpes simplex virus DNA: evidence for four populations of molecules that differ in the relative orientations of their long and short components. Proc Natl Acad Sci U S A. 1975 Nov;72(11):4243–4247. doi: 10.1073/pnas.72.11.4243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaschka-Dierich C., Adams A., Lindahl T., Bornkamm G. W., Bjursell G., Klein G., Giovanella B. C., Singh S. Intracellular forms of Epstein-Barr virus DNA in human tumour cells in vivo. Nature. 1976 Mar 25;260(5549):302–306. doi: 10.1038/260302a0. [DOI] [PubMed] [Google Scholar]
- Lang D., Mitani M. Simplified quantitative electron microscopy of biopolymers. Biopolymers. 1970;9(3):373–379. doi: 10.1002/bip.1970.360090310. [DOI] [PubMed] [Google Scholar]
- Laufs R., Fleckenstein B. Susceptibility to Herpesvirus saimiri and antibody development in old and new world monkeys. Med Microbiol Immunol. 1973 Mar 8;158(3):227–236. doi: 10.1007/BF02120558. [DOI] [PubMed] [Google Scholar]
- Lindahl T., Adams A., Bjursell G., Bornkamm G. W., Kaschka-Dierich C., Jehn U. Covalently closed circular duplex DNA of Epstein-Barr virus in a human lymphoid cell line. J Mol Biol. 1976 Apr 15;102(3):511–530. doi: 10.1016/0022-2836(76)90331-4. [DOI] [PubMed] [Google Scholar]
- Meléndez L. V., Hunt R. D., Daniel M. D., García F. G., Fraser C. E. Herpesvirus saimiri. II. Experimentally induced malignant lymphoma in primates. Lab Anim Care. 1969 Jun;19(3):378–386. [PubMed] [Google Scholar]
- Pettersson U., Mulder C., Deluis H., Sharp P. A. Cleavage of adenovirus type 2 DNA into six unique fragments by endonuclease R-RI. Proc Natl Acad Sci U S A. 1973 Jan;70(1):200–204. doi: 10.1073/pnas.70.1.200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schnös M., Inman R. B. Position of branch points in replicating lambda DNA. J Mol Biol. 1970 Jul 14;51(1):61–73. doi: 10.1016/0022-2836(70)90270-6. [DOI] [PubMed] [Google Scholar]
- Sheldrick P., Berthelot N. Inverted repetitions in the chromosome of herpes simplex virus. Cold Spring Harb Symp Quant Biol. 1975;39(Pt 2):667–678. doi: 10.1101/sqb.1974.039.01.080. [DOI] [PubMed] [Google Scholar]
- Wadsworth S., Jacob R. J., Roizman B. Anatomy of herpes simplex virus DNA. II. Size, composition, and arrangement of inverted terminal repetitions. J Virol. 1975 Jun;15(6):1487–1497. doi: 10.1128/jvi.15.6.1487-1497.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]





