Abstract
Cancer associated glycoconjugates are important biomarkers, as exemplified by globo-H, CA125, CA15.3 and CA27.29. However, the exact chemical structures of many such biomarkers remain unknown because of technological limitations. In this article, we propose the “immunologic mapping” of cancer glycomes based on specific immune recognition of glycan structures, which can be hypothesized theoretically, produced chemically, and examined biologically by immuno-assays. Immunologic mapping of glycans not only provides a unique perspective on cancer glycomes, but also may lead to the invention of powerful reagents for diagnosis and therapy.
Keywords: Glycomics, Mass spectrometry, Cancer, Immunopathogenesis, Immunotherapy, Review
2. INTRODUCTION
The immune system identifies pathogenic microbes and tumor cells by recognizing pathogenic epitopes that originate from microorganisms and cancers (1, 2) and are not expressed by healthy tissues and cells. Because of the complex and dynamic nature of microbial and cancerous epitopes, multiple mechanisms have evolved to recognize and counteract the pathogens and cancers from which they arise. Defects in recognizing pathogenic immune epitopes result in lethal infectious diseases and cancer. On the other hand, immune rejection toward epitopes expressed by normal self tissues and cells can lead to autoimmune diseases such as allergies, asthma, rheumatoid arthritis, diabetes mellitus type 1, and lupus erythematosus. Identifying the chemical nature of pathogenic immune epitopes facilitates both vaccine development for cancer and pathogens, and invention of therapeutic approaches to prevent and treat autoimmune diseases.
Immune epitopes are expressed by diseased self tissues and cells. To improve the efficacy of recognizing immune epitopes, professional antigen presenting cells called dendritic cells (3) have evolved to process and present immune epitopes. Meanwhile, pathogens and cancer cells mutate to evade the attacks by cytotoxic immune components such as complement and cytotoxic T cells. Furthermore, pathogens alter their expression of epitopes at different stages in their lives. Thus identifying immune epitopes can be a very challenging task. In addition, biochemical identification of clinically relevant immune epitopes is often hindered by a lack of materials. It is not unusual that receptors for a cancerous or infectious epitope are identified in immunobiological studies, but the exact chemical nature of the epitope itself is only identified years after.
2.1. The role of immune receptors as signal sensors in both innate and adaptive immunity
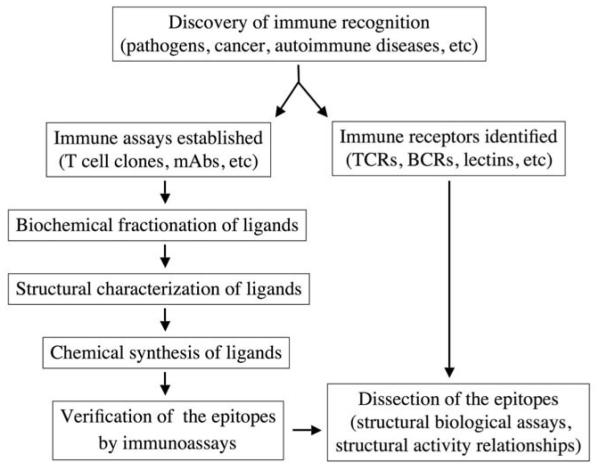
Immune system receptors have evolved in both the innate and adaptive arms of immunity. Receptors of innate immunity (pattern recognition molecules) recognize structures of microbial and self origin, and transduce signals in a highly sensitive mode to activate adaptive immune responses. Recent progress in biochemical and structural biological studies has shed light on the mechanisms of interaction between innate receptors and their ligands (4). Receptors of adaptive immunity include B cell receptors and T cell receptors. B cell receptors recognize antigens and transduce activation signals that drive the differentiation of B cells and the production of opsonization antibodies. In identifying a ligand or epitope recognized by B cells, peptide fragments of candidate proteins may be purified and analyzed by mass spectrometry (MS), and candidate peptides can be chemically synthesized to test specific recognition using antibody binding assays (Figure 1). T cell receptors recognize antigens presented by major histocompatibility complex (MHC) molecules, which means they are restricted to self MHC molecules; the receptors’ fine specificity is regulated by the recognition of antigens loaded on the surface of MHC molecules. T cell epitopes can be identified by purifying peptides bound to MHC molecules, and the identified candidate peptides can be examined by testing their recognition by T cell clones expressing specific T cell receptor genes (5, 6).
Figure 1.
Molecular identification of an immune epitope. mAb, monoclonal antibody; TCR, T cell receptor; BCR, B cell receptor.
2.2. The impact of immune receptor signaling on the biology of immune cells
Receptors expressed on immune cells mediate important signaling functions that drive the cells’ development and function.
2.2.1. T cells
T cell precursors originate from bone marrow and migrate to the thymus, where they undergo several stages of development. A critical step in the development of T cells is the expression of the T cell receptor on the cell surface, which allows them to receive positive selection signals mediated by self MHC molecules and putative self, low-affinity TCR ligands (7). After they mature in the thymus, T cells migrate to peripheral lymphoid organs, where they are activated by foreign (pathogen-derived) antigens. For conventional T cells, the self ligands that mediate positive selection are never the same as the foreign ligands they recognize in the peripheral organs. However, some non-conventional T cells, such as natural killer T (NKT) cells, show evidence that when they are in peripheral organs they can be activated by ligands expressed in the thymus. Such auto-reactivity (8) may be responsible for the important immune regulation function of NKT cells.
2.2.2. B cells
It is not clear whether the B cells require ligand recognition during their development. When mature B cells enter lymphoid organs and encounter pathogens there, B cell receptors receive antigen stimulation, and turn on a series of signal transduction pathways responsible for their differentiation, class switching, and immunoglobulin secretion.
2.2.3. Dendritic cells and macrophages
Dendritic cells and macrophages express surface and cytosolic receptors that transduce microbial signals. The chemical nature of the microbial ligands determines the pro-inflammatory or anti-inflammatory cytokine profiles, as well as organelle functions and the fate of dendritic cells and macrophages. For example, the mycobacterial cell wall component lipoarabinomannan (LAM) inhibits phagosome maturation and autophagosome formation in macrophages (9). LAM-regulated proteins responsible for such changes have been studied using a proteomics approach.
3. IMMUNOBIOLOGY OF RECEPTORS RECOGNIZING GLYCOCONJUGATES
In the past several decades, many immune receptors specific for complex glycoconjugates have been molecularly identified and characterized, including C-type and other animal lectins, B cell receptors, and T cell receptors.
3.1. C-type lectins, Siglecs, galectins, and other animal lectins
Multiple animal lectins (including C-type lectins, Siglecs, galectins, and other newly discovered lectins) are expressed by immune cells or by tissues that interact with immune cells bearing lectin ligands. For example, C-type lectins are calcium-dependent carbohydrate-binding animal proteins (10). The role of C-type lectins as immune receptors was established after lectins expressed by dendritic cells were found to sense the environmental carbohydrate structures and regulate the signaling events of dendritic cells. In addition to serving as signaling receptors of dendritic cells and other antigen presenting cells such as macrophages, C-type lectins are also expressed by natural killer cells, T cells, and plasmacytoid dendritic cells, and their functions are currently being studied closely. Interested readers are urged to consult other extensive and excellent reviews about this field (10, 11). Siglecs are another family of lectins widely expressed on immune cells and exert signaling functions upon encountering their ligands (12-14). Some lectins (such as galectins) are mainly expressed by tissues that interact with immune cells bearing glycan ligands (15). Galectin expression in the microenvironment of cancer and infectious diseases may regulate the function of immune cells (16-19).
The development of glycan-array technologies by the Consortium of Functional Glycomics has played an essential role in identifying the physiological ligands for lectins. In many cases such arrays also generate inconclusive results. It is well-known that lectin binding to glycans is dependent on multivalent interactions of ligands. Technologies are being developed to measure these multivalent interactions. For example, Kurmyshkina et al. designed “glycoprobes” in which fluorescence-labeled polyacrylamide was conjugated to multivalent glycan ligands (20). Such glycoprobes were used to determine the expression of galectins in tumor cells by immunohistochemical staining. However, whether the multivalent glycans displayed by polyacrylamide are similar to those displayed by native glycoproteins remains to be studied. In another study, Zhang et al. displayed glycan ligands on a carrier protein, bovine serum albumin (21). Such “neoglycoproteins” were used as tools to evaluate binding to more than 150 antibodies and lectins. However, it is unclear whether the density and 3-dimensional conformation of multivalent glycans displayed in “neoglycoproteins” are similar to those of native glycoproteins (21). Thus there is an urgent need to develop technologies to determine the density and 3-dimensional conformation of displayed glycans.
3.2. B cell receptors
B cell receptors and monoclonal antibodies have been identified for a series of glycoconjugates: 1) receptors only specific for carbohydrate moieties, such as the blood group antibodies (A and B), which recognize blood group A or B antigens present in glycoproteins or glycosphingolipids (GSLs), and the xeno-reactive antibodies toward the Galα1,3Gal expressed on both glycoproteins and GSLs (22); and 2) receptors specific for a carbohydrate moiety plus an additional structural motif, such as a peptide. This second type of B cell receptor is exemplified by antibodies that require both the sugar part and the peptide part for recognition (23).
3.3. Special concerns for glycoconjugate-specific antibodies
Many antibodies “specific” for a glycan are known for their cross-reactivity with structurally related glycoconjugates. Whether staining by such an antibody, like immunohistochemical staining of tissue sections, should be recognized as a standard for biochemical relevance remains controversial (24, 25). Several questions remain unanswered: 1) whether such antibodies bind to yet-unknown glycan structures; 2) whether such antibodies bind to other glycans with much stronger affinity than to the target glycan structure; 3) what the sensitivity of such antibodies is in immunohistochemical staining or flow cytometry analysis.
3.4. T cell receptors for glycans
T cell receptors have been identified for glycopeptides, GSLs and non-GSL glycolipids.
3.4.1. Glycopeptides
T cell epitopes that recognize both the sugar and peptide parts of a glycopeptide have been reported, such as Galα1,3 Gal-HEL (hen egg lysosome, 26). Glycopeptide epitopes have also been found in pathogens, such as bacteria-derived glycopeptides recognized by CD4-positive T cells restricted to mouse MHC class II molecules (27).
3.4.2. GSLs
GSL epitopes of T cells were first discovered for CD1-restricted T cells. A marine sponge-derived GSL, α-galactosylceramide, was found to be a superagonist antigen for invariant NKT cells (28). Similar structure of GSLs were later discovered in Sphingomonas (29-31), a bacterium related to an autoimmune disease (primary liver cirrhosis), in which the bacterial GSL α-glucuronic acid ceramide is recognized by the immune system simultaneously with bacterial mitochondria proteins that share homology with human hepatocyte membrane proteins. Thus, the bacterial GSL-induced NKT activation elicits autoimmune antibodies toward hepatocytes (32). Recognition of self GSLs by CD1-restricted T cells was first discovered by the De Libero group in multiple sclerosis patients (33, 34). Another self GSL, isoglobotriaosylceramide (iGb3), although reported by several groups to be a stimulatory antigen for invariant NKT cells with characteristics of natural ligands mediating NKT cell development, has unclear physiological functions (35-39).
3.4.3. Non-GSL lipids
Non-GSL lipid epitopes for CD1-restricted T cells were first discovered in mycobacteria (40, 41). Furthermore, mycobacteria-derived lipopeptides were reported as T cell epitopes as well (42). Their physiological relevance to disease progression is unclear. Mycobacterial lipids, which induce CD1-restricted adaptive T cell responses in animal models, have been proposed as vaccine candidates for Mycobacterium tuberculosis (43).
α-Galactosyl diacylglycerol, a non-GSL structure expressed by the pathogenic bacteria Borrelia burgdorferi which causes Lyme disease (44, 45), is another bacterial glycolipid antigen for CD1d-restricted invariant NKT cells. These glycolipids constitute toll-like receptor-independent activation of innate immunity and may play important roles in the human immune defense against these bacteria.
4. GENETICS AND BIOCHEMISTRY FOR GLYCAN IMMUNE EPITOPES
4.1. Glycoproteins and glycolipids are metabolically unique and structurally challenging
Complex carbohydrate structures bear an important function of information storage. Therefore, it is not surprising that these chemical structures are epitopes recognized by the immune system. The most well-known of this type of epitope is that defining the blood group ABO system, which was discovered in 1900 (46). However, it was not until one century later that the system’s chemical and genetic basis was elucidated. The technical difficulties of carbohydrate biochemistry are obvious and are due to the unique feature of glycosidic linkage: one typical hexose sugar may have five hydroxyl (OH) groups in different positions with which another sugar can form glycosidic linkages. Furthermore, sugars have anomers and form branches (Figure 2). The identification of a complex carbohydrate structure from biomaterials thus is often hampered by two layers of barriers: 1) the need to separate the multiple isomers into individual homogenous fractions; and 2) the limited material available to analyze the sugar identities, sequence, and linkages of the glycan structure.
Figure 2.
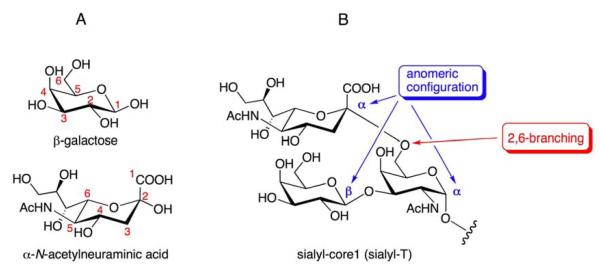
Structural basis of diverse glycosidic linkages. A. Numbering of representative hexose sugars (galactose and N-acetylneuraminic acid). Hydroxyl groups at different positions (OH-2, OH-3, OH-4, and OH-6) of a typical hexose acceptor may be involved in glycosidic linkages. B. Two possible anomeric configurations (α versus β) and branching are the basis for further structural diversity. In this example, the T antigen (Galβ1,3GalNAc) is branched with an α-N-acetylneuraminic acid moiety at position 6 of N-acetylgalactosamine.
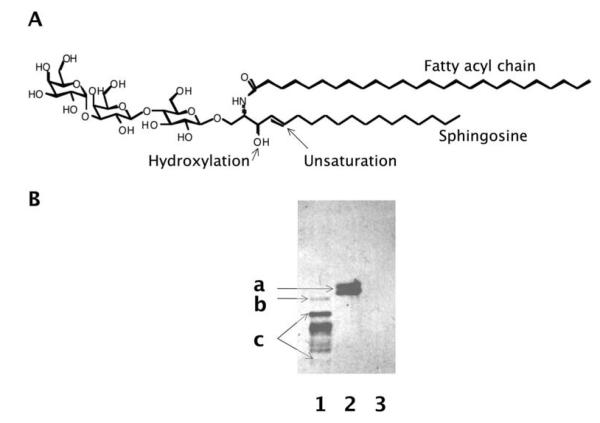
Complex lipid structures include GSLs and non-GSL structures such as phospholipids, the major components of the bilayers of the plasma membrane. The heterogeneous nature of GSLs (47, 48) is also caused by: 1) variations in the length of their fatty acid components (typically from C16 N-fatty acyl to C26 N-fatty acyl); 2) unsaturation of the N-fatty acyl chain; and 3) hydroxyl modification of the N-fatty acyl chain or sphingosine chain (Figure 3). Taking the trisaccharide-ceramide GSL iGb3 as an example, the ceramide part of a chemically synthesized iGb3 is d18:1/C26:0, while iGb3 in a leukemia cell line (RBL) has a ceramide core mixed with d18:1/C24:0 and d18:1/C24:1. When iGb3s with different forms of ceramide are separated in thin layer chromatography, they appear as different bands and are often misunderstood as different glycans by non-experts.
Figure 3.
GSLs are heterogeneous because of variations in their ceramide parts. A The ceramide part of GSLs includes a sphingosine and an N-fatty acyl chain. Both the sphingosine and N-fatty acyl chains may be modified by hydroxyl groups and unsaturation. B. Immunostaining of GSLs separated by thin layer chromatography (TLC). Lane 1, leukemia cell line RBL, which expresses iGb3 and other α1,3Gal-terminated GSLs, as stained by a monoclonal antibody specific for Galα1,3Gal; Lane 2, a chemically synthesized iGb3 from Alexis Biochemicals, CA; Lane 3, a chemically synthesized Gb3 from Alexis Biochemicals, CA. The retention factor of GSLs on thin layer chromatography is dependent on its sugar moiety (hydrophilic) and ceramide moiety (hydrophobic). The hydroxylation and unsaturation of a GSL also influences its retention factor. a, a chemically synthesized iGb3 (h18:1/C26:0, which means it contains one hydroxylation and one unsaturation in the sphingosine chain, and no unsaturation in N-fatty acyl chain); b, iGb3 expressed in a leukemia cell line, which has a different ceramide part (mixed d18:1/C24:0 and d18:1/C24:1); c, multiple bands representing other α1,3Gal-terminated GSLs expressed by RBL cells.
4.2. Gene chips and microarrays cannot be used to detect glycan epitopes
Unlikely polynucleotides and proteins, glycans are synthesized by glycosyltransferases and glycosidases according to a different central dogma, characterized as an “assembly line” by Rosemarie and Stuart Kornfeld (49). Glycan structures are not copied or translated from any existing template. Instead, each individual glycan is assembled by glycosyltransferases and glycosidases sequentially (Figure 4). Therefore, the expression level of glycosyltransferase or glycosidase genes is not predictive of the presence or absence of related glycan structures because they are subjected to further modification by other enzymes.
Figure 4.
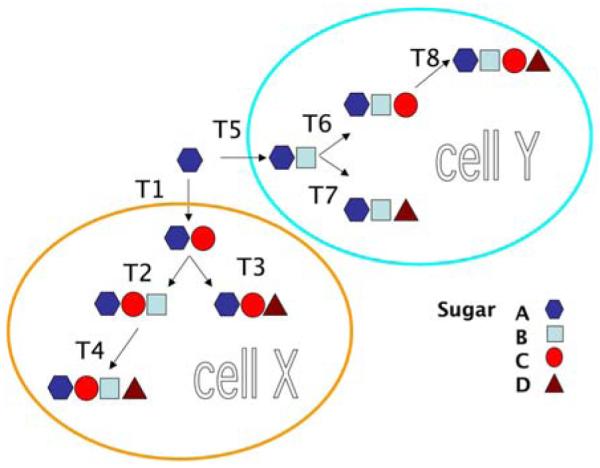
The assembly line of glyco-enzymes. Glycans are assembled in a stepwise manner catalyzed by glycosyltransferases, instead of from DNA or RNA templates. In different cells (here represented as X and Y), different glycan structures are formed by different glycosyltransferases (T1-T4 in cell X, and T5-T8 in cell Y). The presence of specific glycan structures cannot be predicted by gene expressions, because each glycan can be converted to another structure by downstream glyco-enzyme modification.
4.3. Assembly of glycan immune epitopes in unique cellular compartments
Immune epitopes can be generated from both exogenous phagocytosed pathogens and endogenously assembled structures. Particular pathogens, such as bacteria, are typically phagocytosed and digested in the endolysosomal pathways of professional antigen presenting cells, resulting in peptide epitopes presented to CD4 T cells (through the MHC class II antigen presenting pathway) and B cells. Viruses hijack the host’s protein synthesis pathway and assemble viral proteins in the endoplasmic reticulum compartment, which is surveyed by the MHC class I antigen presenting pathway, and trigger CD8 responses.
The generation of glycan immune epitopes may involve multiple cellular compartments. For endogenous antigens, glycans can be synthesized in the endoplasmic reticulum and Golgi compartments and subsequently processed in the lysosomal compartment (50). For external antigens, bacterial glycan and lipid antigens can be processed in the endolysosomal compartment, similar to protein antigens (51). In contrast to peptide antigens which are generated by proteases, lipid and glycan epitopes are generated by glycosidases and lipidases (50, 51). For CD1-restricted T cell glycolipid antigens, chaperone proteins, such as saposins, are required for antigen loading (50, 52-53). These chaperone proteins are responsible for removing lipids loaded to CD1 when it is synthesized and secreted to the cell surface, and facilitate the loading of self and foreign glycolipid antigens in the lysosome.
4.4. Conventional biochemical purification approach for immune epitope discovery
Successful identification of a biologically active structure from biological samples is dependent on the establishment of a detection system, the availability of sufficient initial samples for detection and characterization, effective structural analysis methods, and the ability to chemically synthesize a structure for verifying the natural structure’s biological activity.
4.4.1. Detection system
An absolute prerequisite for identifying a biologically active epitope is a reliable and robust detection system, which must have 1) sufficient sensitivity to detect the biologically active epitope, thereby allowing further downstream enrichment; 2) a specificity that discriminates the biologically active epitope from the vast array of non-active epitopes; and 3) a reproducibility reliable enough for repeated fractionation and enrichment of the immune epitope.
For B cell epitopes, the availability of a specific and sensitive monoclonal antibody itself will allow the detection of the antigen by technologies such as enzyme-linked immunosorbent assays (ELISA). For T cell epitopes, it is much more difficult to directly use T cell receptor protein to measure the binding to an antigen presented by MHC molecules or MHC-like molecules such as CD1. T cell clones must be generated based on their specific recognition of certain antigen epitopes. Such T cell clones produce cytokines or exert cytotoxicity when recognizing antigen presenting cells that present the epitope of interest. Pippa Marrack invented T cell hybridoma technology, which uses cell fusion between fresh T cells and a myeloma tumor cell line. T cell hybridomas can be repeatedly sub-cloned to select those producing high amounts of cytokines upon stimulation by a T cell antigen, and thus can have a sensitivity 2 to 3 orders of magnitude higher than that of non-fused T cell clones.
4.4.2. Sufficient initial samples for detection and characterization
To successfully identify a biologically active epitope, one must acquire a considerable amount of biological material that contains the structurally unknown immune epitopes. The concentration of a desired structure in samples can be estimated quantitatively before large-scale fractionation. Furthermore, a desired structure may suffer considerable loss in every step of fractionation; thus the percentage of recovery in every purification step should be estimated quantitatively as well. For example, according to our previous studies, we can typically recover 160 μg of acidic GSLs and 80 μg of neutral GSLs from each thymus of 4-week-old C57B6 strain of mice (54). From 2000 mouse thymuses, we expect to purify 320 mg of acidic GSLs and 160 mg of neutral GSLs. Assuming 5 μg of a natural ligand is required for biological assays, a ligand must have an abundance among total GSLs of higher than 0.001% to be detectable, assuming its 100% recovery. Using a conservative estimation of recovery for a GSL ligand of 50%, we would need to start with 4000 mouse thymuses.
4.4.3. Analytic method to prove the chemical nature of the fraction of interest
A bottleneck in immune epitope discovery lies in the lack of analytical technologies available for analyzing the epitopes’ chemical compositions and structures. A highly sensitive and specific technology would not only precisely determine the chemical nature of an immune epitope, but would also significantly reduce the amount of purified epitope required for structural determination, and the amount of starting material initially needed for fractionation and purification. In general, structural analysis requires a highly purified fraction that contains only the structure representing the immune epitope; contaminants or mixtures are not allowed. This is a particularly challenging task for identification of carbohydrate-containing epitopes because of heterogeneity in both the O-glycans and N-glycans of naturally occurring glycoconjugates.
4.4.4. Synthetic method to verify the chemical nature of an immune epitope
After a candidate structure of an immune epitope is established, larger quantities of such a potential epitope can be synthesized by chemical or biochemical methods, and tested for their immunogenicity. Such synthesis may also provide the basis for the manufacturing of vaccines and diagnosis reagents. Purity of a desired synthetic immune epitope is essential for biological assays. It is well-known that less than 1% of contaminants introduced during chemical synthesis can be recognized by other immune receptors, and trigger potent undesired immune responses.
4.5. Limitations of the conventional biochemical purification approach
Conventional biochemical purification may be hampered by the nature of the immune epitope, the resolution of separating technologies, and the power of analytic technologies.
4.5.1. Weak antigenicity or affinity
Biochemical isolation will be difficult if an immune epitope has weak affinity. C-type lectins expressed by dendritic cells are known to bind to sugar ligands with low affinity (with Kd values in the micromolar range). A natural ligand for invariant NKT cells, iGb3, also binds to T cell receptors with Kd values in the micromolar range and serves as a weak agonist.
4.5.2. Low abundance
A good example of low-abundance epitopes is the natural ligands for NKT cells (40). Although such natural ligands exist in the mouse and human thymus, lipidomic and glycosphingolipidomic studies have found no natural ligands among the known lipids (including GSLs) other than iGb3. We have estimated that the ratio of iGb3 to Gb3 in mouse thymus is 0.4% (54). If each mouse thymocyte expresses about 20,000 copies of Gb3 (38), 80 copies of iGb3 will be present. If iGb3 is not the ligand responsible for NKT cell development, it is likely that the abundance of the major ligand would be above 80 copies per thymocyte.
4.5.3. Resolution of separation technologies
Most technologies used to separate an immune epitope from others are based on chromatography. These include size exclusion chromatography, hydrophobic interaction chromatography, hydrophilic interaction chromatography, and affinity chromatography. Currently, peptide immune epitopes can be separated in proteomics core facilities in most academic or industrial institutions. However, the separation of glycoconjugate and lipid immune epitopes remains challenging because of the presence of multiple glycoconjugate isomers, which are similar in size and polarity, and the difficulty in separating lipid species with subtle structural variations.
4.5.4. Power of analytic technologies
It is generally accepted that mass spectrometry (MS) is the standard technology for immune epitope identification. Although high-performance liquid chromatography (HPLC) is still occasionally used for identifying immune epitopes, it cannot provide exact structural information, nor can it reveal possible contaminants that are invisible to the conventional detectors used in HPLC, such as ultraviolet and fluorescence detectors. Current U.S. Food and Drug Administration regulations related to peptide vaccines require MS data, which may disclose low levels of contaminant peptide even though the HPLC profile may show a single “pure” peak. Modern liquid chromatography (LC)-MS instruments combine separation and mass spectrometric analysis of mixtures and provide information about the retention times as well as the masses of individual compounds if the peaks are base-line resolved.
The invention of matrix-assisted-laser-desorption/ionization time-of-flight MS (MALDI-TOF MS) and electro-spray ionization mass spectrometry (ESI-MS) has revolutionized the field of immune epitope discovery. The ionization methods in these technologies are highly efficient and relatively mild, which allows for the detection of molecular ion peaks of unfragmented species. For more detailed characterization of biomacromolecules, MS instruments have been invented for selecting individual molecular ion species and breaking them into smaller fragments (product ions) that provide structural information about the parent ions. This fragmentation process is normally done by collision-induced dissociation, which involves activating the biomacromolecules (ionized as parent ions) through collision with inert gas molecules. The increased energy induces bond breakage, and systematic analysis of the resulting fragments provides information about the molecular structure of the biomacromolecules. Often, “signature” fragments that are diagnostic for particular structural features can be generated, in addition to the molecular mass of the original parent biomacromolecules. These fragments can sometimes be sufficient for identifying biomacromolecules, particularly if standards, often chemically synthesized, are available. Multiple rounds of low-energy fragmentation, which can be effectively carried out in an ion-trap mass spectrometer (IT-MS), further expand the information yield from mass spectral analysis (55). With four or five rounds of fragmentation (which can only be done usng IT-MS, but not other MS instruments), it is possible to determine the structure of an immune epitope.
Two other analytical methods useful for the accurate structural determination of immune epitopes are nuclear magnetic resonance (NMR) spectrometry and X-ray crystallography. These methods allow for the distinction of constitutional isomers and stereoisomers, which can be a difficult task using MS, even with sophisticated MS capacities, particularly when no reference material is available. A drawback of immune epitope determination by NMR spectrometry and X-ray crystallography is that larger sample quantities (micrograms to milligrams) are required.
5. PROPOSING, TESTING AND VERIFYING HYPOTHETICAL GLYCAN IMMUNE EPITOPES
5.1. Why hypothetical epitopes
Because of the limitations of conventional approaches to biochemical purification, it may be impossible to identify those immune epitopes with weak immunogenicity or low abundance. When conventional biochemical purification approaches do not lead to unambiguous structural information of an immune epitope composed of sugar units and/or lipids, we propose that such epitopes may be identified through a “reverse” approach. In this process, the existing glycan structures from biological samples may be hypothesized based on limited structural information and candidate immune epitopes synthesized by methods of bio-organic chemistry. Chemically synthesized candidate epitopes would then be examined for their immunogenicity, revealing novel immune epitopes. For example, in our recent studies, we identified a candidate novel glycolipid structure from mouse thymus by MS analysis, but the abundance of this structure does not allow for extensive analysis. Based on partial structural information, we hypothesized it to be Galα1,3(Fuc α1,2)Galβ1,4Glcβ1,1Cer (fucosylated iGb3). Chemically synthesized fucosylated iGb3 was found to be stimulatory to mouse NKT cell clones (unpublished data). Thus fucosylated iGb3 serves as a candidate natural ligand for NKT cells.
5.2. Interdisciplinary nature of immunologic glycomics: synergized efforts by immunobiology, analytical chemistry, and synthetic chemistry
The success of the immunologic glycomics approach is based on advances in the fields of immunobiology, analytical chemistry, and synthetic chemistry. Sensitive immunoassays, such as those that use reporter genes fused to immune signaling molecules, may allow large-scale screening of candidate immune epitopes. In addition, chemo-enzymatic synthesis of glycans, glycolipids, and glycopeptides provides candidate immune epitopes for immunoassays in quantities often impossible to purify from natural materials.
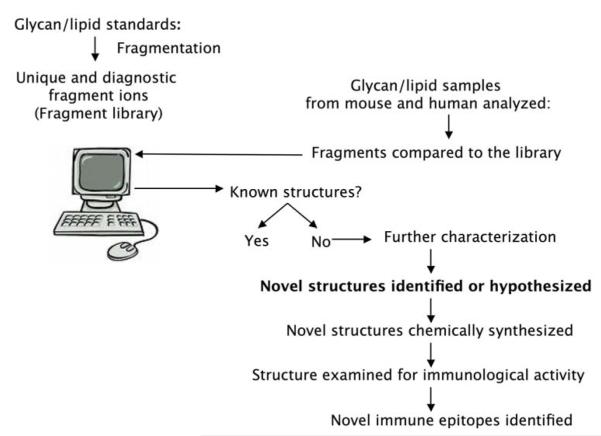
The essential driving force for this synergized effort is the development of specific, sensitive, and generally applicable analytical technologies. A complete picture of a cellular glycome is possible only after known structures are viewed (Figure 5). For identifying candidate unknown structures (the hypothesis-generating step), powerful analytical technologies allow finding of more structures that may not be visible by insensitive methods like HPLC. More importantly, such analytical technologies are essential to verify the structures and purities of chemically synthesized immune epitopes. Chemical synthesis will unavoidably introduce other byproducts, which may not be visible by HPLC but may interfere with immunoassays even at very low abundance.
Figure 5.
Generation of hypotheses in immunologic mapping of the glycome. Glycan structures are collected from immune cells and compared to a database of known glycans. Candidate novel glycans are isolated and their structures hypothesized. Novel structures are then chemically synthesized and examined for immunological activity.
6. ESSENTIAL ROLE OF ANALYTICAL METHODS AND TOOLS IN THE HYPOTHESIS-GENERATING PHASE
6.1. IT-MS as a unique tool in the study of glycomics and lipidomics
The presence of multiple isomers sharing the same molecular masses is a longstanding impediment to their structural characterization. Tandem MS (MS-MS) technology has significantly helped in solving part of the problem in that it permits identifying characteristic fragment ions generated by instruments with tandem MS capacity, such as MALDI-TOF-TOF and triple quadruple/time-of-flight (Q-TOF) instruments. However, tandem MS analysis often does not reveal the complex linkage information of glycans. The Reinhold group first demonstrated that multiple rounds of low-energy fragmentation, which can be effectively carried out in IT-MS, greatly improves the information yield from glycan mass spectral analysis (55).
6.1.1. Glycolipid analysis
Levery and coworkers combined all the potential advantages of IT-MS methodology, including the detection of isomeric structures using signature diagnostic ions that are observable only in MS4 and MS5 spectra (which involve three and four rounds of fragmentation, respectively), for the highly sensitive identification and quantitation of GSLs present in the form of multiple isobaric mixtures (56-57). In theory, this method may be applied to identify every existing glycolipid structure, pending the availability of standard glycolipids, which can be chemically synthesized.
6.1.2. N-glycans, O-glycans, and O-glycopeptides
The power of the IT-MSn method has also been demonstrated for N-glycan and O-glycan analysis (58-60), as well as the analysis of glycopeptides modified by O-glycosylation (60). The powerful multi-stage fragmentation (up to MS5 or MS6) function of ion trap instruments allows investigators to diagnose highly complex sugar linkages, which they can not do using other mass spectrometers.
Although IT-MS instruments can dissect the structure of complex sugars, for analyzing a glycopeptide, they may be limited in their ability to determine where the sugar modification is because the fragmentation induced by collision with high-energy gas cleaves the weakest bond in the molecule first, and this bond is often the glycosidic bond linking the glycan to the peptide backbone. This problem can be overcome by an alternative fragmentation method, electron transfer dissociation (ETD), which can cleave the peptide backbone stepwise without damaging glycosylated amino acids. Using this powerful technique, two independent groups recently generated a complete glycoprofile for alpha-dystroglycan (α-DG), an O-glycosylated protein related to congenital muscular dystrophies (61, 62).
6.2. Sample preparation and separation technology
Sample preparation and separation in glycomics studies critically influence the quality, reproducibility, and reliability of data. The combination of MS and liquid chromatography is now routinely practiced for proteomics studies. Efforts in combining these technologies for glycomics and glycolipidomics have also been made.
Separating glycans by HPLC after cleaving them from glycoproteins or glycolipids has proved to be an effective method to generate glycan profiles (48). Recently, native glycans have also been separated by means of hydrophilic interaction without labeling with fluorescence groups (63, 64). Several groups of investigators have reported separating certain categories of lipid structures by reverse-phase HPLC in combination with MS (65).
6.3. Controversies on the detection of immune epitopes in low abundance
Immune epitopes, such as T cell antigens, may function at extremely low abundance. For example, α-galactosylceramide, formulated by Kirin Pharma Co. as KRN7000 using a patented solvent technology, may stimulate mouse NKT cells in vivo at a serum concentration of 10 pg/mL, whereas current available MS technology can only measure α-galactosylceramide at a serum concentration of 12.5 ng/mL (66). Controversies may arise regarding which standard should be recognized as evidence of a chemical structure, if such a structure could not be detected by any chemical measurement such as HPLC or MS. So far, biological assays remain the most sensitive assays for detecting T cell epitopes.
The super-agonistic activity of some immune epitopes, such as α-galactosylceramide as mentioned above, also raise the question of which standard should be used to conclude that an immune epitope is absent. For example, in our own experience, we have not been able to develop an assay based on MS to detect less than 1% contamination of α-galactosylceramide in the presence of its β-isomer. Thus, when we prepare such β-glycosylceramides, the only reliable method we can use to exclude contamination by α-isomers is bioassays.
7. EARLY INSIGHT INTO CLINICAL APPLICATIONS
7.1. Demands and markets
Demand for lipidomics and glycomics in clinical research endeavors is emerging. Glycans and glycolipids are recognized as targets for immunodiagnosis and immunotherapy in multiple diseases; examples include 1) Tn antigens, a family of under-glycosylated mucin proteins expressed in more than 80% of cancer patients, which are being used both as cancer markers (CA125, CA15.3 and CA2.29) and vaccine candidates (67); 2) GM2, GD2, GD3, and other gangliosides, which are being evaluated as vaccine candidates in melanoma and neuroblastoma clinics (68, 69); and 3) GbH, which is being evaluated as a biomarker and vaccine candidate in breast cancer clinics (70). Important clinical applications have already been developed in several areas: 1) cancer vaccines, such as Tn glycans and GM2; 2) tumoricidal antibodies or toxins, such as anti-GD2 and anti-MUC1; and 3) diagnostic reagents for pathogens, such as the monoclonal antibodies for anthrax spore glycans (71, 72) and Chaga’s disease (73).
7.2. Future directions
Research trends in the field of immunologic glycomics are likely to include the following: 1) more precise, accurate, and complete structural determination of epitopes will be demanded: for example, not only should the sugar moiety of a glycoprotein or glycolipid be characterized, but also the sequence of the protein and the lipid moieties. 2) More sensitive analytical methods suitable for clinical samples will be needed: for instance, MS technology has been recently applied for detecting Gb3 from the urine of patients with Fabry disease (74). 3) Assays will be highly focused on detecting specific structures, while the extensive profiling of glycans and lipids in small samples may continue to encounter technical difficulties and provide little value for studying disease mechanisms or in developing products for clinical use. 4) Advances in synthetic bioorganic chemistry may improve the commercial availability and accessibility of homogeneous standards, which will help definitively unravel mechanisms in biological processes.
7.3. Five-year view
The last few years have seen much progress in our understanding of glycan and glycolipid functions. The next five years are likely to continue to reveal functional aspects of this type of biomarcromolecules. Synergistic efforts in immunology, biochemistry, and synthetic chemistry will be routine practice for future discoveries. Question-driven studies on important diseases such as cancer, allergies, and asthma may provide clues for ground-breaking discoveries. Furthermore, hypothesis-generating studies may also lead to interesting findings. Major goals for the next five years will be to complete the picture of the spectra of functionally specialized glycan and lipid structures, to improve our understanding of related genetic pathways responsible for synthesizing of functional glycan and lipid structures, and to use such information to develop preventive and therapeutic approaches for personalized therapy.
ACKNOWLEDGEMENTS
DZ is supported by MD Anderson Cancer Center and NIH grants AI079232, AI078898, a developmental award and a supplemental award from P30-AI36211. MD Anderson Cancer Center is supported in part by NIH grant CA16672. We thank the Consortium of Functional Glycomics (CFG), National Institute of General Medical Sciences, for providing glycan standards and glycan arrays and other CFG investigators for sharing unpublished data, Paul Savage, Albert Bendelac, Luc Teyton, Zachary Bohannan and Kathryn Carnes for helpful discussion. The main goal of this article is to emphasize the need for depth of interdisciplinary collaboration. Readers are referred to other extensive and outstanding reviews regarding specific topics of glycobiology and glycochemistry. The authors apologize for their inability to cite many excellent articles.
Footnotes
9. FINANCIAL DISCLOSURE DZ is a consultant for BioTex, Houston, TX, and an inventor involved in patents related to technologies mentioned in this article, issued or in application.
10. REFERENCES
- 1.Matzinger P. The danger model: a renewed sense of self. Science. 2002;296(5566):301–5. doi: 10.1126/science.1071059. [DOI] [PubMed] [Google Scholar]
- 2.Medzhitov R, Janeway CA., Jr Decoding the patterns of self and nonself by the innate immune system. Science. 2002;296(5566):298–300. doi: 10.1126/science.1068883. [DOI] [PubMed] [Google Scholar]
- 3.Mellman I, Turley SJ, Steinman RM. Antigen processing for amateurs and professionals. Trends Cell Biol. 1998;8(6):231–7. doi: 10.1016/s0962-8924(98)01276-8. [DOI] [PubMed] [Google Scholar]
- 4.Carpenter S, O’Neill LA. Recent insights into the structure of Toll-like receptors and post-translational modifications of their associated signalling proteins. Biochem J. 2009;422(1):1–10. doi: 10.1042/BJ20090616. [DOI] [PubMed] [Google Scholar]
- 5.Rudensky AY. Endogenous peptides associated with MHC class II and selection of CD4 T cells. Semin Immunol. 1995;7(6):399–409. doi: 10.1006/smim.1995.0044. [DOI] [PubMed] [Google Scholar]
- 6.Joyce S, Nathenson SG. Methods to study peptides associated with MHC class I molecules. Curr Opin Immunol. 1994;6(1):24–31. doi: 10.1016/0952-7915(94)90029-9. [DOI] [PubMed] [Google Scholar]; Curr Opin Immunol. 1994;6(2):334. Erratum in: [Google Scholar]
- 7.Germain RN. Ligand-dependent regulation of T cell development and activation. Immunol Res. 2003;27(2-3):277–86. doi: 10.1385/IR:27:2-3:277. [DOI] [PubMed] [Google Scholar]
- 8.Bendelac A. Nondeletional pathways for the development of autoreactive thymocytes. Nat Immunol. 2004;5(6):557–8. doi: 10.1038/ni0604-557. [DOI] [PubMed] [Google Scholar]
- 9.Shui W, Petzold CJ, Redding A, Liu J, Pitcher A, Sheu L, Hsieh TY, Keasling JD, Bertozzi CR. Organelle membrane proteomics reveals differential influence of mycobacterial lipoglycans on macrophage phagosome maturation and autophagosome accumulation. J Proteome Res. 2011;10(1):339–48. doi: 10.1021/pr100688h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cambi A, Figdor CG. Levels of complexity in pathogen recognition by C-type lectins. Curr Opin Immunol. 2005;17(4):345–51. doi: 10.1016/j.coi.2005.05.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.van Kooyk Y. C-type lectins on dendritic cells: key modulators for the induction of immune responses. Biochem Soc Trans. 2008;36(Pt 6):1478–81. doi: 10.1042/BST0361478. [DOI] [PubMed] [Google Scholar]
- 12.O’Reilly MK, Paulson JC. Siglecs as targets for therapy in immune-cell-mediated disease. Trends Pharmacol Sci. 2009;30(5):240–8. doi: 10.1016/j.tips.2009.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bochner BS, Alvarez RA, Mehta P, Bovin NV, Blixt O, White JR, Schnaar RL. Glycan array screening reveals a candidate ligand for Siglec-8. Sialoside analogue arrays for rapid identification of high affinity siglec ligands. J Biol Chem. 2005;280(6):4307–12. doi: 10.1074/jbc.M412378200. [DOI] [PubMed] [Google Scholar]
- 14.Blixt O, Han S, Liao L, Zeng Y, Hoffmann J, Futakawa S, Paulson JC. Sialoside analogue arrays for rapid identification of high affinity siglec ligands. J Am Chem Soc. 2008;130(21):6680–1. doi: 10.1021/ja801052g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Barondes SH, Castronovo V, Cooper DN, Cummings RD, Drickamer K, Feizi T, Gitt MA, Hirabayashi J, Hughes C, Kasai K, et al. Galectins: a family of animal beta-galactoside-binding lectins. Cell. 1994;76(4):597–8. doi: 10.1016/0092-8674(94)90498-7. [DOI] [PubMed] [Google Scholar]
- 16.Cerliani JP, Stowell SR, Mascanfroni ID, Arthur CM, Cummings RD, Rabinovich GA. Expanding the universe of cytokines and pattern recognition receptors: Galectins and glycans in innate immunity. J Clin Immunol. 2010 doi: 10.1007/s10875-010-9494-2. [Epub ahead of print] [DOI] [PubMed] [Google Scholar]
- 17.Dam TK, Brewer CF. Lectins as pattern recognition molecules: the effects of epitope density in innate immunity. Glycobiology. 2010;20(3):270–9. doi: 10.1093/glycob/cwp186. [DOI] [PubMed] [Google Scholar]
- 18.Vasta GR. Roles of galectins in infection. Nat Rev Microbiol. 2009;7(6):424–38. doi: 10.1038/nrmicro2146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dhirapong A, Lleo A, Leung P, Gershwin ME, Liu FT. The immunological potential of galectin-1 and -3. Autoimmun Rev. 2009;8(5):360–3. doi: 10.1016/j.autrev.2008.11.009. [DOI] [PubMed] [Google Scholar]
- 20.Kurmyshkina O, Rapoport E, Moiseeva E, Korchagina E, Ovchinnikova T, Pazynina G, Belyanchikov I, Bovin N. Glycoprobes as a tool for the study of lectins expressed on tumor cells. Acta Histochem. 2010;112(2):118–26. doi: 10.1016/j.acthis.2009.01.004. [DOI] [PubMed] [Google Scholar]
- 21.Zhang Y, Campbell C, Li Q, Gildersleeve JC. Multidimensional glycan arrays for enhanced antibody profiling. Mol Biosyst. 6(9):1583–91. doi: 10.1039/c002259d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Macher BA, Galili U. The Galalpha1,3Galbeta1,4GlcNAc-R (alpha-Gal) epitope: a carbohydrate of unique evolution and clinical relevance. Biochim Biophys Acta. 2008;1780(2):75–88. doi: 10.1016/j.bbagen.2007.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Schietinger A, Philip M, Yoshida BA, Azadi P, Liu H, Meredith SC, Schreiber H. A mutant chaperone converts a wild-type protein into a tumor-specific antigen. Science. 2006;314(5797):304–8. doi: 10.1126/science.1129200. [DOI] [PubMed] [Google Scholar]
- 24.Zhou D, Levery SB. Response to Milland et al.: Carbohydrate residues downstream of the terminal Galalpha(1,3)Gal epitope modulate the specificity of xenoreactive antibodies. Immunol Cell Biol. 2008;86(8):631–2. doi: 10.1038/icb.2008.65. author reply 633-4. [DOI] [PubMed] [Google Scholar]
- 25.Milland J, Christiansen D, Lazarus BD, Taylor SG, Xing PX, Sandrin MS. The molecular basis for galalpha(1,3)gal expression in animals with a deletion of the alpha1,3galactosyltransferase gene. J Immunol. 2006;176(4):2448–54. doi: 10.4049/jimmunol.176.4.2448. [DOI] [PubMed] [Google Scholar]
- 26.Harding CV, Kihlberg J, Elofsson M, Magnusson G, Unanue ER. Glycopeptides bind MHC molecules and elicit specific T cell responses. J Immunol. 1993;151(5):2419–25. [PubMed] [Google Scholar]
- 27.Cobb BA, Kasper DL. Coming of age: carbohydrates and immunity. Eur J Immunol. 2005 Feb;35(2):352–6. doi: 10.1002/eji.200425889. 2005. [DOI] [PubMed] [Google Scholar]
- 28.Taniguchi M, Nakayama T. Recognition and function of Valpha14 NKT cells. Semin Immunol. 2000;12(6):543–50. doi: 10.1006/smim.2000.0270. [DOI] [PubMed] [Google Scholar]
- 29.Mattner J, Debord KL, Ismail N, Goff RD, Cantu C, 3rd, Zhou D, Saint-Mezard P, Wang V, Gao Y, Yin N, Hoebe K, Schneewind O, Walker D, Beutler B, Teyton L, Savage PB, Bendelac A. Exogenous and endogenous glycolipid antigens activate NKT cells during microbial infections. Nature. 2005;434(7032):525–9. doi: 10.1038/nature03408. [DOI] [PubMed] [Google Scholar]
- 30.Kinjo Y, Wu D, Kim G, Xing GW, Poles MA, Ho DD, Tsuji M, Kawahara K, Wong CH, Kronenberg M. Recognition of bacterial glycosphingolipids by natural killer T cells. Nature. 2005;434(7032):520–5. doi: 10.1038/nature03407. [DOI] [PubMed] [Google Scholar]
- 31.Sriram V, Du W, Gervay-Hague J, Brutkiewicz RR. Cell wall glycosphingolipids of Sphingomonas paucimobilis are CD1d-specific ligands for NKT cells. Eur J Immunol. 2005;35(6):1692–701. doi: 10.1002/eji.200526157. [DOI] [PubMed] [Google Scholar]
- 32.Mattner J, Savage PB, Leung P, Oertelt SS, Wang V, Trivedi O, Scanlon ST, Pendem K, Teyton L, Hart J, Ridgway WM, Wicker LS, Gershwin ME, Bendelac A. Liver autoimmunity triggered by microbial activation of natural killer T cells. Cell Host Microbe. 2008;3(5):304–15. doi: 10.1016/j.chom.2008.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Shamshiev A, Gober HJ, Donda A, Mazorra Z, Mori L, De Libero G. Presentation of the same glycolipid by different CD1 molecules. J Exp Med. 2002;195(8):1013–21. doi: 10.1084/jem.20011963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.De Libero G, Mori L. Self glycosphingolipids: new antigens recognized by autoreactive T lymphocytes. News Physiol Sci. 2003;18:71–6. doi: 10.1152/nips.01418.2002. [DOI] [PubMed] [Google Scholar]
- 35.Dias BR, Rodrigues EG, Nimrichter L, Nakayasu ES, Almeida IC, Travassos LR. Identification of iGb3 and iGb4 in melanoma B16F10-Nex2 cells and the iNKT cell-mediated antitumor effect of dendritic cells primed with iGb3. Mol Cancer. 2009;8:116. doi: 10.1186/1476-4598-8-116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Christiansen D, Milland J, Mouhtouris E, Vaughan H, Pellicci DG, McConville MJ, Godfrey DI, Sandrin MS. Humans lack iGb3 due to the absence of functional iGb3-synthase: implications for NKT cell development and transplantation. PLoS Biol. 2008;6(7):e172. doi: 10.1371/journal.pbio.0060172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Xia C, Schümann J, Emmanuel R, Zhang Y, Chen W, Zhang W, De Libero G, Wang PG. Modification of the ceramide moiety of isoglobotrihexosylceramide on its agonist activity in stimulation of invariant natural killer T cells. J Med Chem. 2007;50(15):3489–96. doi: 10.1021/jm0701066. [DOI] [PubMed] [Google Scholar]
- 38.Speak AO, Salio M, Neville DC, Fontaine J, Priestman DA, Platt N, Heare T, Butters TD, Dwek RA, Trottein F, Exley MA, Cerundolo V, Platt FM. Implications for invariant natural killer T cell ligands due to the restricted presence of isoglobotrihexosylceramide in mammals. Proc Natl Acad Sci USA. 2007;104(14):5971–6. doi: 10.1073/pnas.0607285104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zhou D, Mattner J, Cantu C, 3rd, Schrantz N, Yin N, Gao Y, Sagiv Y, Hudspeth K, Wu YP, Yamashita T, Teneberg S, Wang D, Proia RL, Levery SB, Savage PB, Teyton L, Bendelac A. Lysosomal glycosphingolipid recognition by NKT cells. Science. 2004;306(5702):1786–9. doi: 10.1126/science.1103440. [DOI] [PubMed] [Google Scholar]
- 40.Behar SM, Porcelli SA. CD1-restricted T cells in host defense to infectious diseases. Curr Top Microbiol Immunol. 2007;314:215–50. doi: 10.1007/978-3-540-69511-0_9. [DOI] [PubMed] [Google Scholar]
- 41.Gilleron M, Stenger S, Mazorra Z, Wittke F, Mariotti S, Böhmer G, Prandi J, Mori L, Puzo G, De Libero G. Diacylated sulfoglycolipids are novel mycobacterial antigens stimulating CD1-restricted T cells during infection with Mycobacterium tuberculosis. J Exp Med. 2004;199(5):649–59. doi: 10.1084/jem.20031097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Moody DB, Young DC, Cheng TY, Rosat JP, Roura-Mir C, O’Connor PB, Zajonc DM, Walz A, Miller MJ, Levery SB, Wilson IA, Costello CE, Brenner MB. T cell activation by lipopeptide antigens. Science. 303(5657):527–31. doi: 10.1126/science.1089353. [DOI] [PubMed] [Google Scholar]; Science. 2004;304(5668):211. doi: 10.1126/science.304.5668.187a. Erratum in: [DOI] [PubMed] [Google Scholar]
- 43.Felio K, Nguyen H, Dascher CC, Choi HJ, Li S, Zimmer MI, Colmone A, Moody DB, Brenner MB, Wang CR. CD1-restricted adaptive immune responses to Mycobacteria in human group 1 CD1 transgenic mice. J Exp Med. 2009;206(11):2497–509. doi: 10.1084/jem.20090898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kinjo Y, Tupin E, Wu D, Fujio M, Garcia-Navarro R, Benhnia MR, Zajonc DM, Ben-Menachem G, Ainge GD, Painter GF, Khurana A, Hoebe K, Behar SM, Beutler B, Wilson IA, Tsuji M, Sellati TJ, Wong CH, Kronenberg M. Natural killer T cells recognize diacylglycerol antigens from pathogenic bacteria. Nat Immunol. 2006;7(9):978–86. doi: 10.1038/ni1380. [DOI] [PubMed] [Google Scholar]
- 45.Wang J, Li Y, Kinjo Y, Mac TT, Gibson D, Painter GF, Kronenberg M, Zajonc DM. Lipid binding orientation within CD1d affects recognition of Borrelia burgorferi antigens by NKT cells. Proc Natl Acad Sci USA. 2010;107(4):1535–40. doi: 10.1073/pnas.0909479107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Yamamoto F. Molecular genetics of ABO. Vox Sang. 2000;78(Suppl 2):91–1. [PubMed] [Google Scholar]
- 47.Levery SB. Glycosphingolipid structural analysis and glycosphingolipidomics. Methods Enzymol. 2005;405:300–69. doi: 10.1016/S0076-6879(05)05012-3. [DOI] [PubMed] [Google Scholar]
- 48.Haslam SM, Julien S, Burchell JM, Monk CR, Ceroni A, Garden OA, Dell A. Characterizing the glycome of the mammalian immune system. Immunol Cell Biol. 2008;86(7):564–73. doi: 10.1038/icb.2008.54. [DOI] [PubMed] [Google Scholar]
- 49.Kornfeld R, Kornfeld S. Assembly of asparagine-linked oligosaccharides. Annu Rev Biochem. 1985;54:631–64. doi: 10.1146/annurev.bi.54.070185.003215. [DOI] [PubMed] [Google Scholar]
- 50.Zhou D. The immunological function of iGb3. Curr Protein Pept Sci. 2006;7(4):325–33. doi: 10.2174/138920306778018007. [DOI] [PubMed] [Google Scholar]
- 51.Moody DB, Briken V, Cheng TY, Roura-Mir C, Guy MR, Geho DH, Tykocinski ML, Besra GS, Porcelli SA. Lipid length controls antigen entry into endosomal and nonendosomal pathways for CD1b presentation. Nat Immunol. 2002;3(5):435–42. doi: 10.1038/ni780. [DOI] [PubMed] [Google Scholar]
- 52.Darmoise A, Maschmeyer P, Winau F. The immunological functions of saposins. Adv Immunol. 2010;105:25–62. doi: 10.1016/S0065-2776(10)05002-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kang SJ, Cresswell P. Saposins facilitate CD1d-restricted presentation of an exogenous lipid antigen to T cells. Nat Immunol. 2004;5(2):175–81. doi: 10.1038/ni1034. [DOI] [PubMed] [Google Scholar]
- 54.Li Y, Thapa P, Hawke D, Kondo Y, Furukawa K, Furukawa K, Hsu FF, Adlercreutz D, Weadge J, Palcic MM, Wang PG, Levery SB, Zhou D. Immunologic glycosphingolipidomics and NKT cell development in mouse thymus. J Proteome Res. 2009;8(6):2740–51. doi: 10.1021/pr801040h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Ashline DJ, Lapadula AJ, Liu YH, Lin M, Grace M, Pramanik B, Reinhold VN. Carbohydrate structural isomers analyzed by sequential mass spectrometry. Anal Chem. 2007;79(10):3830–42. doi: 10.1021/ac062383a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Li Y, Zhou D, Xia C, Wang PG, Levery SB. Sensitive quantitation of isoglobotriaosylceramide in the presence of isobaric components using electrospray ionization-ion trap mass spectrometry. Glycobiology. 2008;18(2):166–76. doi: 10.1093/glycob/cwm127. [DOI] [PubMed] [Google Scholar]
- 57.Li Y, Teneberg S, Thapa P, Bendelac A, Levery SB, Zhou D. Sensitive detection of isoglobo and globo series tetraglycosylceramides in human thymus by ion trap mass spectrometry. Glycobiology. 18(2):158–65. doi: 10.1093/glycob/cwm129. [DOI] [PubMed] [Google Scholar]; Glycobiology. 2008;18(8):568. Erratum in: [Google Scholar]
- 58.Zhang S, Chelius D. Characterization of protein glycosylation using chip-based infusion nanoelectrospray linear ion trap tandem mass spectrometry. J Biomol Tech. 2004;15(2):120–33. [PMC free article] [PubMed] [Google Scholar]
- 59.Sun B, Ranish JA, Utleg AG, White JT, Yan X, Lin B, Hood L. Shotgun glycopeptide capture approach coupled with mass spectrometry for comprehensive glycoproteomics. Mol Cell Proteomics. 2007;6(1):141–9. doi: 10.1074/mcp.T600046-MCP200. [DOI] [PubMed] [Google Scholar]
- 60.Deguchi K, Ito H, Baba T, Hirabayashi A, Nakagawa H, Fumoto M, Hinou H, Nishimura S. Structural analysis of O-glycopeptides employing negative- and positive-ion multi-stage mass spectra obtained by collision-induced and electron-capture dissociations in linear ion trap time-of-flight mass spectrometry. Rapid Commun Mass Spectrom. 2007;21(5):691–8. doi: 10.1002/rcm.2885. [DOI] [PubMed] [Google Scholar]
- 61.Stalnaker SH, Hashmi S, Lim JM, Aoki K, Porterfield M, Gutierrez-Sanchez G, Wheeler J, Ervasti JM, Bergmann C, Tiemeyer M, Wells L. Site mapping and characterization of O-glycan structures on alpha-dystroglycan isolated from rabbit skeletal muscle. J Biol Chem. 2010;285(32):24882–91. doi: 10.1074/jbc.M110.126474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Nilsson J, Nilsson J, Larson G, Grahn A. Characterization of site-specific O-glycan structures within the mucin-like domain of alpha-dystroglycan from human skeletal muscle. Glycobiology. 2010;20(9):1160–9. doi: 10.1093/glycob/cwq082. [DOI] [PubMed] [Google Scholar]
- 63.Wang Y, Wu SL, Hancock WS. Approaches to the study of N-linked glycoproteins in human plasma using lectin affinity chromatography and nano-HPLC coupled to electrospray linear ion trap--Fourier transform mass spectrometry. Glycobiology. 2006;16(6):514–23. doi: 10.1093/glycob/cwj091. [DOI] [PubMed] [Google Scholar]
- 64.Wada Y, Tajiri M, Yoshida S. Hydrophilic affinity isolation and MALDI multiple-stage tandem mass spectrometry of glycopeptides for glycoproteomics. Anal Chem. 2004;76(22):6560–5. doi: 10.1021/ac049062o. [DOI] [PubMed] [Google Scholar]
- 65.Taguchi R, Houjou T, Nakanishi H, Yamazaki T, Ishida M, Imagawa M, Shimizu T. Focused lipidomics by tandem mass spectrometry. J Chromatogr B Analyt Technol Biomed Life Sci. 2005;823(1):26–36. doi: 10.1016/j.jchromb.2005.06.005. [DOI] [PubMed] [Google Scholar]
- 66.Crul M, Mathôt RA, Giaccone G, Punt CA, Rosing H, Hillebrand MX, Ando Y, Nishi N, Tanaka H, Schellens JM, Beijnen JH. Population pharmacokinetics of the novel anticancer agent KRN7000. Cancer Chemother Pharmacol. 2002;49(4):287–93. doi: 10.1007/s00280-001-0413-3. [DOI] [PubMed] [Google Scholar]
- 67.Holmberg LA, Sandmaier BM. Vaccination with Theratope (STn-KLH) as treatment for breast cancer. Expert Rev Vaccines. 2004;3(6):655–63. doi: 10.1586/14760584.3.6.655. [DOI] [PubMed] [Google Scholar]
- 68.Ragupathi G. Carbohydrate antigens as targets for active specific immunotherapy. Cancer Immunol Immunother. 1996;43(3):152–7. doi: 10.1007/s002620050316. [DOI] [PubMed] [Google Scholar]
- 69.Livingston P. Ganglioside vaccines with emphasis on GM2. Semin Oncol. 1998;25(6):636–45. [PubMed] [Google Scholar]
- 70.Zhu J, Wan Q, Lee D, Yang G, Spassova MK, Ouerfelli O, Ragupathi G, Damani P, Livingston PO, Danishefsky SJ. From synthesis to biologics: preclinical data on a chemistry derived anticancer vaccine. J Am Chem Soc. 2009;131(26):9298–303. doi: 10.1021/ja901415s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Vasan M, Rauvolfova J, Wolfert MA, Leoff C, Kannenberg EL, Quinn CP, Carlson RW, Boons GJ. Chemical synthesis and immunological properties of oligosaccharides derived from the vegetative cell wall of Bacillus anthracis. Chembiochem. 2008;9(11):1716–20. doi: 10.1002/cbic.200800210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Werz DB, Seeberger PH. Total synthesis of antigen bacillus anthracis tetrasaccharide--creation of an anthrax vaccine candidate. Angew Chem Int Ed Engl. 2005;44(39):6315–8. doi: 10.1002/anie.200502615. [DOI] [PubMed] [Google Scholar]
- 73.Almeida IC, Covas DT, Soussumi LM, Travassos LR. A highly sensitive and specific chemiluminescent enzyme-linked immunosorbent assay for diagnosis of active Trypanosoma cruzi infection. Transfusion. 1997;37(8):850–7. doi: 10.1046/j.1537-2995.1997.37897424410.x. [DOI] [PubMed] [Google Scholar]
- 74.Nelson BC, Roddy T, Araghi S, Wilkens D, Thomas JJ, Zhang K, Sung CC, Richards SM. Globotriaosylceramide isoform profiles in human plasma by liquid chromatography-tandem mass spectrometry. J Chromatogr B Analyt Technol Biomed Life Sci. 2004;805(1):127–34. doi: 10.1016/j.jchromb.2004.02.032. [DOI] [PubMed] [Google Scholar]