Abstract
It is now clear that mitochondria are an important target for oxidative stress in a broad range of pathologies including cardiovascular disease, diabetes, neurodegeneration, and cancer. Methods for assessing the impact of reactive species on isolated mitochondria are well established but constrained by the need for large amounts of material to prepare intact mitochondria for polarographic measurements. With the availability of high resolution polarography and fluorescence techniques for the measurement of oxygen concentration in solution, measurements of mitochondrial function in intact cells can be made. Recently, the development of extracellular flux methods to monitor changes in oxygen concentration and pH in cultures of adherent cells in multiple sample wells simultaneously has greatly enhanced the ability to meaure bioenergetic function in response to oxidative stress. Here we describe these methods in detail using representative cell types from the renal, cardiovascular, nervous, and tumorigenic model systems while illustrating the application of three protocols to analyze the bioenergetic response of cells to oxidative stress.
Keywords: Extracellular Flux, Oxygen Consumption Rate, Mitochondrial Profile, Reserve Capacity, Mitochondria, Oxidative Stress, Nitric Oxide, 4-Hydroxynonenal
Introduction
Assessment of mitochondrial function in cultured cells is commonly performed using either measurements of oxygen consumption of the intact isolated organelle or activity measurements of individual enzymes. While both approaches yield important information regarding mitochondrial performance, measurements of oxygen consumption in intact cells allow a physiological perspective of mitochondrial function to be achieved [1, 2]. To date, quantification of oxygen consumption in cultured cells has been largely performed using Clark electrodes with the cells suspended in stirred, buffered solution [3]. These electrodes are more commonly used for assessing mitochondrial function in isolated samples from animal studies or clinical samples. Measurement of oxygen consumption from cultured cells in a Clark-type electrode usually requires removing the cells from their growth substrate, and adding them to a stirred solution. In many cells, this detached state may result in anoikis which is associated with increased ROS and mitochondrial damage [4]. In addition, non-laminar shear, which occurs as a result of stirring in the oxygen electrode, will also result in increased oxidative stress [5, 6]. This can be partially overcome by seeding cells on micro-carrier beads to prevent anoikis, however these cells are still subject to non-laminar shear [7], and thus the effects of reactive oxygen and nitrogen species (ROS and RNS) on mitochondrial function in cells is difficult to dissect using this experimental approach. Notably, recent publications highlight the application of the Seahorse XF24 to the study of isolated mitochondria in a more high-throughput fashion [8, 9]. These publications offer a direct comparison to Clark-type electrodes in models normally measured by this method. Importantly, data derived from the Seahorse XF24 instrument compare favorably to those acquired using the Clark-type electrode. Monitoring mitochondrial oxygen consumption while the cells remain adherent is thus preferable in experiments to assess the impact of oxidative stress on cellular bioenergetics as these measurements can be related to other experimental endpoints following the intervention of interest.
This can now be achieved using recently developed fluorometric systems to detect oxygen consumption in adherent cells in culture [10–12]. These systems allow for highly sensitive and specific measurements of mitochondrial function to be made with greater throughput than possible with electrode-based systems. The most important difference between the Clark-type electrode, and these systems is in the maintenance of a cellular context within which mitochondrial dynamics are examined. In this article, several protocols describing the use of a Seahorse Bioscience XF24 Extracellular Flux Analyzer to assess the impact of oxidative stress on bioenergetics will be described.
Mitochondria and oxidative stress in health and disease
Beyond the role of mitochondria in ATP production, it is now evident that this organelle is implicated in many chronic pathologies. This is not surprising since the mitochondria are major producers of energy in the cell and deficits in energy production are readily translated to other cellular processes that require energy to function. These include ion pumping [13], which is essential for maintaining ionic homeostasis (especially critical in excitable cells such as neurons and myocytes), molecular motors such as those that enable muscular function, and the synthesis of molecules for intracellular communication. Importantly, cellular repair pathways, which are activated in response to the accumulation of proteins damaged by ROS/RNS are also energy consuming. Furthermore, many of the pathways which remove ROS/RNS use NADPH as a source of reducing equivalents, which in turn increases the metabolic demand on the cell.
At non-pathological levels mitochondria integrate signals from ROS/RNS and are therefore, uniquely poised as both regulators and integrators of intra- and inter-cellular signaling. Seminal discoveries of the necessary and permissive role of the mitochondria in diverse cellular processes have highlighted a role for mitochondria in responding to viral infection [14], growth signaling [15, 16], and death signals [17–21]. In fact, death signaling is a classic example of retrograde signaling to the nucleus [22]. This retrograde signaling is accomplished through the action of non-effector proteins. Control of the Bcl-2 family members Bax and Bak are a typical example of the mitochondrial role in apoptotic signaling [23–25], but also may include regulation of cell-cell junctions [26–29], proteosomal activation [30], kinase activation [31, 32], and oxygen sensing [33]. Thus, it is increasingly clear that mitochondrial function is not only important to the progression of a disease, are now recognized as a therapeutic target [34, 35]. This emphasizes the importance of assessing mitochondrial function in response to normal physiological and pathological stressors in a cellular setting.
The impact of oxidative stress on mitochondrial function
As mitochondria are both sources and targets of oxidants, amplification of oxidative damage can occur as the increase in ROS/RNS damage mitochondria and further exacerbate ROS production. For example, damage to mitochondria during the progression of atherosclerosis is an ROS/RNS-dependent process which causes loss of bioenergetic control and eventual vascular dysfunction [36–38]. However, an understanding of mitochondrial responses to ROS and RNS in intact cells is incomplete. Recently, our studies on the effects of ROS/RNS generators on mitochondrial function have been greatly facilitated by the Seahorse Bioscience XF24 technology (Seahorse Bioscience; N. Billerica, MA), which enables us to accurately quantify mitochondrial function in intact cells [39–42]. The non-invasive nature of this technique allows for repeated measurements of oxygen consumption and extracellular acidification rates as well as post-hoc measurements of cytosolic and mitochondrial proteins by western blot, total cellular protein levels, and other endpoints. Several models of ROS/RNS generation have now been examined in multiple cell lines. Here, examples of ROS/RNS production are used to illustrate the utility of measuring extracellular flux to monitor mitochondrial and glycolytic function.
Principles and approaches to measuring bioenergetic function by extracellular flux
The cell lines used in these studies (Table 1) have been selected to be broadly representative of the cultured cells used by investigators in the cardiovascular, cancer, neurodegeneration, and renal fields of research. In each of these cell types, an XF24 analyzer from Seahorse Bioscience which measures O2 and protons (pH) in cell culture was used to determine the effects of oxidative stress on cellular bioenergetics [11, 43]; all experiments described in this paper use plates with a 7 μl volume, termed V7 Plates. Since the assay does not involve direct manipulation of the cells apart from a transient change in oxygen tension they can be harvested at the end of the experiment for the measurement of other endpoints. The small volume and 24-well format of the XF24 allows for high throughput, real-time measurements of O2 consumption and pH change. The rate of O2 consumption (OCR) can be assigned to oxidative phosphorylation and the rate of extracellular acidification (ECAR) to glycolysis. These endpoints will be discussed in further detail below. The system is capable of measuring 20 samples at a time, and is equipped with four injection ports per well to allow for injection of a compound of interest or to add inhibitors that can aid in the elucidation of defects in individual cellular respiration pathways or enzymes [44]. A full description of the instrumentation and related methodology can be found in references [11, 12, 43].
Table 1.
Cell lines discussed in this article.
| Abbreviated Cell Name | Full Name | Species | Derivation | References |
|---|---|---|---|---|
| BAEC | Bovine aortic endothelial cells | Bos taurus | Aorta | [39] |
| NRVM | Primary Neonatal Rat Ventricular Myocytes | Rattus norvegicus | Neonatal Hearts | [40, 45, 46] |
| RASMC | Rat aortic smooth muscle cells | Rattus norvegicus | Aorta | [41,42] |
| MES13 | MES13 | Mus musculus | Mesangial Cell | |
| SH-SY5Y | SH-SY5Y | Homo sapiens | Neuroblastoma | |
| MB231 | MDA-MB231 | Homo sapiens | Breast adenocarcinoma | [47] |
| H9c2 | H9c2 Myoblast | Rattus norvegicus | Cardiac tissue |
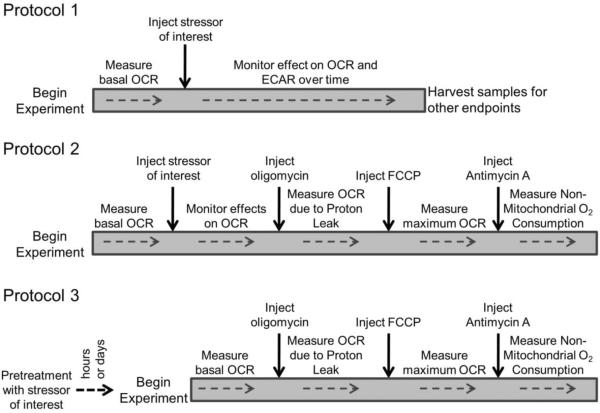
Figure 1 shows three main approaches that can be used to determine the response of cells to the effects of ROS/RNS. Protocol 1 is designed to observe the effects of oxidative stress on bioenergetic function in “real time”. It utilizes the injection ports of the XF24 to directly introduce the compound of interest into the experimental wells and follows the response in OCR and ECAR over time. Experiments of this type can be readily compared to other endpoints by harvesting the cells at the completion of the XF assay as demonstrated below. Protocol 2 extends Protocol 1 to include the measurement of a mitochondrial profile in which oxygen consumption attributed to ATP production, proton leak, reserve capacity, and non-mitochondrial sources is assigned. This procedure is performed at a defined time after injection of the stressor of interest. The third Protocol is to treat cells in culture ex machina, and assess the parameters of mitochondrial function either after the desired amount of time of pretreatment, or after removal of the compound of interest some time later. Examples utilizing each protocol to determine the response to oxidative stress are discussed in further detail below.
Figure 1. Three protocols for assessing mitochondrial function in intact cells.
Assessment of mitochondrial function is typically performed using one of three approaches. Protocol 1: After establishing baseline OCR, a compound of interest is injected. The effect on OCR and ECAR are then monitored over time. Protocol 2: After establishing baseline, the compound of interest is injected, OCR and ECAR are monitored for a predetermined amount of time, and then mitochondrial function is assessed. Protocol 3: Treatment with the compound of interest is completed outside the instrument. The cells are then washed free of the stressor, and mitochondrial function is directly assessed.
The stressors we have chosen as examples exemplify the experimental approaches discussed here. They include: A) the reactive oxygen species hydrogen peroxide generated by a redox cycling agent or added in bolus form; B) the reactive nitrogen species nitric oxide (NO) added in the form of an NO donor; and C) the reactive lipid species 4-hydroxynonenal (HNE) and 15-deoxy-Δ12,14-Prostaglandin J2 (15d-PGJ2). There is a robust literature for all of these compounds indicating their formation in vivo and their effects on mitochondrial function [48–52]. A detailed rationale for the use of each reactive species is beyond the scope of the current article and the reader is referred to the citations above for detailed discussion on the use of these oxidative stress systems.
Protocol 1: Determine the response of cells to acute oxidative stress
This protocol takes advantage of the fact that the XF24 allows for the automated injection of any compound of interest into the culture well on the XF Assay plate. Since the experimental design allows for 20 individual wells to be used, up to 6 experimental groups with 3–4 replicates per group can be monitored for changes in oxygen consumption rate (OCR) and extracellular acidification rate (ECAR) over time courses as long as 6–8 h. The cell types described below have been selected to illustrate the diversity of the potential responses in different cell types to the reactive lipid species HNE.
The response of Neonatal Rat Ventricular Myocytes (NRVM) to 4-hydroxynonenal (HNE)
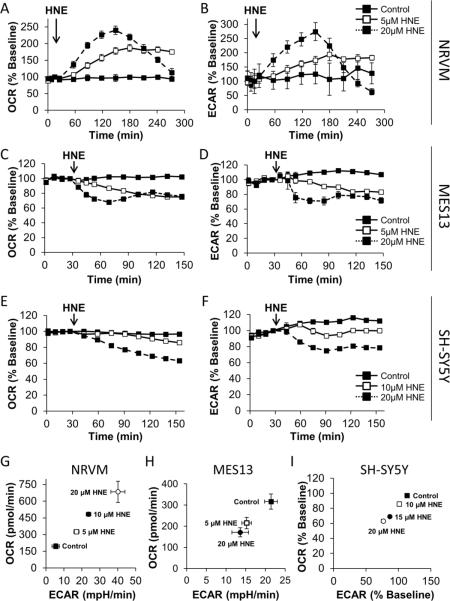
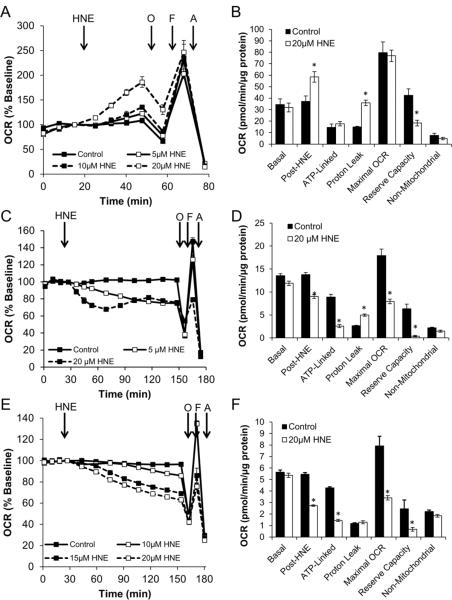
NRVM were seeded into Seahorse Bioscience XF24 microplates for 72 h at an initial density of 75,000 cells/well. The media was then changed to assay media, and the cells were loaded into the XF24. After 3 baseline measurements, 5 or 20 μM HNE was injected. The response to OCR was then monitored for 4 h. As shown in Figure 2A, these cells exhibit a dose and time dependent increase in OCR in response to HNE. Additionally, ECAR was similarly stimulated in response to HNE (Figure 2B) [40].
Figure 2. The acute effect of HNE on cultured cells.
The effect of HNE on OCR and ECAR was assessed using a Protocol 1-style strategy. Panel A: After establishing baseline OCR in NRVM seeded to 75,000 cells/well, HNE was injected to a final concentration of 5 μM (open squares, solid line) or 20 μM (closed squares, dashed line). The effect on OCR was followed for an additional 4 h. Panel B: Same as Panel A, except ECAR was measured. Panel C: MES-13 were seeded at 30,000 cells/well, baseline OCR was established then HNE was injected at 5 μM (open squares, solid line) or 20 μM (closed squares, dashed line) and OCR was followed for 2 h. Panel D: Same as Panel E, except ECAR was measured. Panel E: After establishing baseline OCR in differentiated SH-SY5Y cells, HNE was injected to a final concentration of 0, 10, 15, or 20 μM. The effect on OCR was followed for an additional 2 h. Panel F: Same as Panel E, except ECAR was measured. Panel G: NRVM were exposed to 5, 10, or 20 μM HNE for 1 h. OCR and ECAR are plotted against each other from this single time point. Panel H: MES13 cells were exposed to 5, or 20 μM HNE for 1 h. OCR and ECAR are plotted against each other from this single time point. Panel I: Differentiated SH-SY5Y cells were exposed to 10, 15, or 20 μM HNE for 1 h. OCR and ECAR are plotted against each other from this single time point. All data shown are the mean ± sem. n≥3 per treatment group. Statistical significance is omitted for clarity. Where error bars are not visible, they are smaller than the data point symbol. Data in panels A, B and G are adapted from [40].
The effect of HNE on Mesangial Cells
MES-13 cells were seeded to 30,000 cells/well in Seahorse Bioscience XF24 V7 microplates 24 h prior to assay. Four baseline measurements were recorded 1 h after changing to unbuffered media. HNE was injected to 5 and 20 μM final concentrations. Real-time OCR and ECAR were monitored for 2 h. HNE caused a decrease in MES-13 cell OCR (Figure 2C) as well as ECAR (Figure 2D) in a dose-dependent manner.
The effect of HNE on SH-SY5Y cells
In Figure 2E and F, SH-SY5Y neuroblastoma cells were seeded in Seahorse Bioscience XF24 V7 microplates to 80,000 cells/well in DMEM containing 1% FBS, and then differentiated with retinoic acid for 5 days to acquire neuron-like morphology [53]. The media was then changed to assay media specifically optimized for this cell type (unbuffered assay media as described below, supplemented with 5 mM HEPES and 1% FBS), and the cells were loaded into the XF24 Extracellular Flux Analyzer. After 4 baseline measurements, 0, 10, or 20 μM HNE was injected. The OCR was then monitored for 2 h. In contrast to NRVM and similar to the mesangial cells, SH-SY5Y cells exhibit a dose and time dependent decrease in OCR (Figure 2E), as well as a decrease in ECAR, in response to HNE (Figure 2F).
Generating a Metabolic Profile to visualize the cellular energetic response to HNE in multiple cell types
Glycolysis and oxidative phosphorylation are likely integrated during the cellular response to oxidative stress. Importantly, this integrated response will differ by cell type. This can be assessed by plotting OCR against ECAR at a single time-point after injection of the compound of interest to generate a Metabolic Profile as shown in Figure 2G, H, and I. In this series of experiments, HNE was injected as in Figure 2A in each cell type, and then OCR and ECAR data from 60–120 min post-injection were plotted against each other. As these results indicate, increasing HNE concentrations lead to stimulation of both OCR and ECAR in NRVM, but not in Mesangial cells or SH-SY5Y cells. At this point the investigator could then proceed to dissect out the mechanism leading to these overall responses including the impact on mitochondrial function as will be described in Protocol 2. Using the data obtained from ECAR and OCR the interplay between these two metabolic pathways can be examined as shown in Figure 2G–I. The changes in mitochondrial function can also be related to other biological endpoints including signaling, protein modification and cell death (see below). Taken together these data allow an integrated picture of how cellular bioenergetic function is contributes to the response of a given cell to oxidative stress. An example of how this can be achieved can be found in the following publication [40].
Relating the impact of HNE to cellular signaling in Rat Aortic Smooth Muscle Cells (RASMC)
To relate bioenergetic data to mechanistic changes in cell function, further analysis of the samples after treatment is possible (i.e. following experiments of the type shown in Figure 2). Typically there is sufficient protein to perform a western blot, assess protein concentration, or measure cell viability. It is then important to select a surrogate index to relate the mitochondrial effects to other experiments designed to probe cell function. This can be surprisingly challenging and we have found it is important to control for the following factors:
-
a)
The stoichiometric amount of a reactive lipid species such as HNE is more important than the absolute concentration of the stressor [50]. This is likely due to covalent modification of proteins by the lipid, but is characteristic of many reactions which may be studied using this technique. It is thus important to consider the ratio of media volume to total cell number when designing experiments for the XF24 which are usually different than standard tissue culture protocols with respect to the ratio of volume of media to cells.
-
b)
The culture media is changed during the XF24 analysis to media without serum, and lacking buffering capacity. This should be considered and controlled for in the interpretation if bioenergetic assays are compared to experiments in different media or standard tissue culture conditions. Further discussion of the importance of selecting proper assay media is given below in the General Considerations.
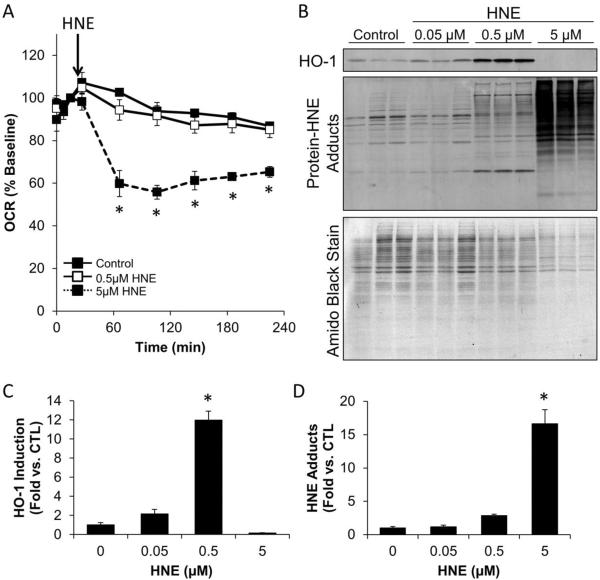
To correlate the impact of HNE on mitochondrial function with the effect on cell signaling, sample lysates were collected from individual wells of a Seahorse Bioscience V7 culture plate after the completion of mitochondrial function measurements. As shown in Figure 3A, 5 μM HNE inhibited basal OCR in Rat Aortic Smooth Muscle Cells (RASMC) to a similar extent as that seen in other cell types (Figure 2). Cell lysates were then probed for Heme-Oxygenase-1 (HO-1), an antioxidant protein known to be activated by HNE [54–56]. As shown in Figure 3B and C, HO-1 protein is increased in the presence of low levels of HNE, but not at the higher concentrations which appear to be cytotoxic. Protein-HNE adducts were also examined by western blot. These adducts increased in a concentration-dependent manner. To collect samples from Seahorse plates, the cells in each well were lysed in SDS sample buffer and loaded in their entirety onto a lane of an SDS-PAGE gel. To visualize the consistency in protein loading in these samples, the nitrocellulose membrane was stained with amido black. This stain indicates that there was less protein present in the 5 μM HNE group, consistent with HNE cytotoxicity. These Western blots were quantified, and the concentration-dependent stimulation of HO-1 is shown in Figure 3C,D. Interestingly, stimulation of HO-1 was maximal at 0.5 μM HNE, but suppressed at the higher concentration of 5 μM HNE. These data are an example of how the biphasic effects of a reactive species can be related to cell signaling, mitochondrial function and protein adduct formation. The investigator could then use this data as a basis to test the molecular mechanisms involved that link these processes together.
Figure 3. Integration of XF metabolic analysis with western blotting.
The impact of HNE on mitochondrial function, protein modification, and HO-1 induction was examined following OCR measurements in RASMC. Panel A: The OCR of rat aortic smooth muscle cells was measured followed by acute exposure to 0, 0.5, and 5 μM HNE. OCR was then examined for the indicated time. Panel B: At the end of the experiment, cells were lysed in detergent-containing buffer (as described in the text), and the protein was separated by SDS-PAGE, followed by western blotting for protein-HNE adducts and HO-1. The PVDF membrane was then stripped of antibodies and stained with Amido Black (0.1% Naphthol Blue-Black in 40% methanol and 10% acetic acid). The PVDF was then destained with 40% methanol and 10% acetic acid to visualize the separated proteins. Panels C and D: HO-1 expression (Panel C) and protein-HNE adducts (Panel D) were quantified and normalized to Amido Black protein stain. n = 3 per group; *, p≤0.05 vs. cells not treated with HNE.
Protocol 2: Determine the bioenergetic profile of cells exposed to acute oxidative stress
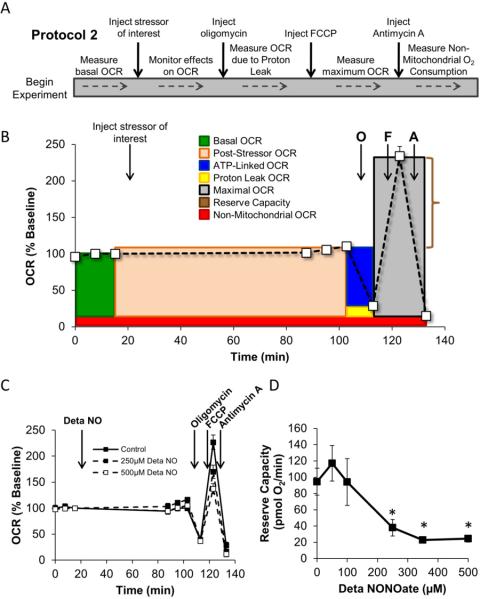
After completion of the experiment shown in Protocol 1 the investigator can decide to end the experiment at this point and measure indices of oxidative damage or cell signaling as described previously or perform a mitochondrial function test (also called a “stress test”) by following the method described here in Protocol 2. In this approach, well-characterized inhibitors of the electron transport chain (ETC) are used to dissect processes that are contributing to overall mitochondrial oxygen consumption [57]. This technique uses selected mitochondrial inhibitors which allow for determination of six main parameters that describe key aspects of mitochondrial function in a cellular context: basal OCR, ATP-linked OCR, proton leak OCR, maximal OCR, reserve capacity, and non-mitochondrial OCR. The principle of this technique is shown schematically in Supplemental Figure 1. These parameters are determined by measuring OCR after the sequential injection of oligomycin (to inhibit the ATP synthase), carbonyl cyanide p-[trifluoromethoxy]-phenyl-hydrazone (FCCP – to uncouple the mitochondrial inner membrane and allow for maximum electron flux through the ETC), and antimycin A (to inhibit complex III). The difference between the basal OCR and the oligomycin-insensitive OCR yields the amount of oxygen consumption that is ATP-linked [57]. The balance of the basal OCR is comprised of O2 consumption due to proton leak and non-mitochondrial sources.
The injection of FCCP allows for uninhibited movement of protons across the mitochondrial inner membrane, and effectively depletes the mitochondrial membrane potential (Δψm). The resulting uninhibited electron flow through the mitochondrial respiratory chain leads to an increase in oxygen consumption, and allows a determination of the maximal oxygen consumption that is possible at Cytochrome c Oxidase (Complex IV). Thus, the addition of FCCP allows for an estimate of a value for maximum OCR without dependence on ATP/ADP transport [57, 58]. The difference between the FCCP-stimulated rate and the basal OCR yields an estimate of the reserve capacity of the cells. Reserve capacity is defined as the amount of oxygen consumption that is available for cells to use in times of increased ATP demand or during other stress [39, 59, 60]. Lastly, antimycin A is injected to inhibit electron flux through Complex III. This prevents any O2 from being consumed at Complex IV, and thus yields the rate of O2 consumption from sites other than the mitochondrion [57].
The concentrations necessary to achieve the optimal result for each of these measurements must be empirically determined for each cell line as shown in Supplemental Figure 2A and B. Oligomycin injected to too low a final concentration will not fully inhibit the ATP Synthase, and inhibition of OCR will not be complete. Similarly, oligomycin at elevated concentrations will lead to maximal inhibition of OCR initially, followed by a progressive increase in OCR over time (Supplemental Figure 2A). Similarly, FCCP must be carefully titrated to achieve maximal stimulation of OCR, and we have found the concentration response to be bell-shaped (Supplemental Figure 2B).
Figure 4A demonstrates the mitochondrial function sequence schematically. The resulting changes in OCR in response to each of these metabolic modulators are also shown in Figure 4B. This graph is color-coded to demonstrate the relative contribution of each parameter listed above, and to illustrate how these calculations are made at the conclusion of the experiment. Specific examples of experiments which utilize a Protocol-2 approach are given below.
Figure 4. Principles of a Protocol-2 style analysis of mitochondrial function.
Panel A describes the approach to performing a Protocol 2-style experiment. Panel B: A typical profile of the mitochondrial function assay used to assess the acute effect of stressors on mitochondrial function. The example data is from control BAEC. After baseline OCR is established, the stressor of interest is injected and monitored for the desired amount of time. This allows the determination of the direct effect of the stressor on OCR. Then the mitochondrial function assay is performed as described. Individual parameters that can be calculated using this approach are indicated. Panel C: BAEC were seeded to 40,000 cells/well and allowed to incubate for 24 h. Basal OCR was then measured prior to injection of Deta NONOate to a final concentration of 250 or 500 μM. The cells were allowed to incubate for 1 h, then 3 additional measurements of OCR were made. This was followed by sequential injection of oligomycin (1 μg/ml), FCCP (1 μM), and antimycin A (10 μM). Panel D: From the data in Panel C, the reserve capacity of the mitochondrial can be calculated. Data shown in Panels C and D represent the mean ± sem. n≥3 per treatment group. *, p≤0.05 as compared to 0 μM Deta NONOate control. Panel C and D are adapted from Free Radical Biology and Medicine, v.48(7), Dranka BP et.al, Mitochondrial reserve capacity in endothelial cells: The impact of nitric oxide and reactive oxygen species, p. 905-14, Copyright (2010), with permission from Elsevier.
Effects of NO on the mitochondrial profile in endothelial cells
BAEC were seeded to 40,000 cells/well in Seahorse Bioscience V7 tissue culture plates. The cells were allowed to incubate for 24 h at 37°C in a humidified CO2 incubator. The culture media was then changed to assay media, and three baseline measurements of OCR were taken. The NO donor Deta NONOate was then injected to a final concentration of 250 or 500 μM. The cells were allowed to incubate for 1 h, and three additional measurements of OCR were made. As shown in Figure 4C, Deta NONOate had no effect on basal OCR following injection; however, following FCCP injection, the maximal OCR was found to be significantly decreased. This translates to a loss of reserve capacity which was dependent on Deta NONOate concentration (Figure 4D). This example demonstrates how changes in cellular mitochondrial function can be revealed by the use of the mitochondrial profile experiment. These data are adapted from a published study which also demonstrates protocols to determine the reversibility of an intervention and the effects of NO in combination with ROS [39].
The effect of HNE on NRVM mitochondrial function
Using the example of Protocol 1 shown in Figure 2, mitochondrial function was assessed after injection of HNE into cultures of NRVM. HNE was injected to a final concentration of 5, 10, or 20 μM, and OCR was assessed for 30 minutes. Time resolved data are shown in Figure 5A. For clarity, mitochondrial function parameters in control cells and those treated with 20 μM HNE are shown in Figure 5B. As this Figure shows, the post-HNE OCR was increased by 60% over baseline. This was found to be due mostly to increased proton leak, as there was little effect on ATP-linked OCR at this time point. Additionally, reserve capacity was decreased without any change in the maximal OCR stimulated by FCCP. This suggests that NRVM have a defined maximal OCR, and that the ratio of proton leak OCR to ATP-linked OCR (often referred to as the coupling efficiency [61]) may define the response of cells to oxidant stressors.
Figure 5. The effect of HNE on mitochondrial function varies according to the cell line investigated.
Employing a Protocol 2 strategy, the effect of HNE on mitochondrial function was examined in NRVM (Panels A and B), MES13 (Panels C and D), and SH-SY5Y (Panels E and F). Panel A: After establishing baseline, HNE was injected into NRVM cultures to a final concentration of 5, 10, or 20 μM. A final basal OCR measurement was taken 30 min later which was termed the post-HNE OCR. This was followed by the mitochondrial function assay as described. Panel B: Individual mitochondrial function parameters calculated from the data shown in Panel A. Panel C: Same as Panel A, except experiments were performed in SH-SY5Y cells. Panel D: Individual mitochondrial function parameters calculated from the data shown in Panel C. Panel E: Same as Panel A, except experiments were performed in MES-13 cells. Panel F: Individual mitochondrial function parameters calculated from the data shown in Panel E. All data shown are the mean ± sem. Where error bars are not visible, they are smaller than the data point symbol. n≥3 per treatment group. *, p≤0.05 as determined by two-tailed, unpaired t-test compared to the respective control measurement.
The effect of HNE on MES13 Mesangial cell mitochondrial function
Unlike the response seen in NRVM, acute HNE exposure at a final concentration of 5 and 20 μM in MES13 cells elicited a decreased OCR and maximal respiration rate (Figure 5C), opposite of the response described above for NRVM. HNE at 20 μM caused a decrease in ATP-linked OCR and maximal respiration, as well as reserve capacity (Figure 5D). Proton leak was increased with HNE exposure which could explain the decreased ATP-linked respiration, but not the change in maximal respiration which is probably a function of change in substrates from glycolysis [39].
The effect of HNE on SH-SY5Y mitochondrial function
As in NRVM and Mesangial cells, mitochondrial function was assessed after injection of HNE into cultures of differentiated SH-SY5Y cells. HNE was injected to a final concentration of 10, 15, or 20 μM, and OCR was assessed. As shown in Figure 5E, in this cell type, the post-HNE basal OCR was decreased. This was found to be due mostly to decreased ATP-linked OCR, as there was little effect on proton leak. However, in contrast to NRVM, both the maximal OCR induced by FCCP and the reserve capacity were suppressed. This suggests that differentiated SH-SY5Y cells respond to oxidative stress by reducing mitochondrial ATP production possibly by inhibition of the ATP synthase (Figure 5F), with a less substantial effect on mitochondrial electron transport. This occurs despite a decrease in glycolytic function (Figure 2F).
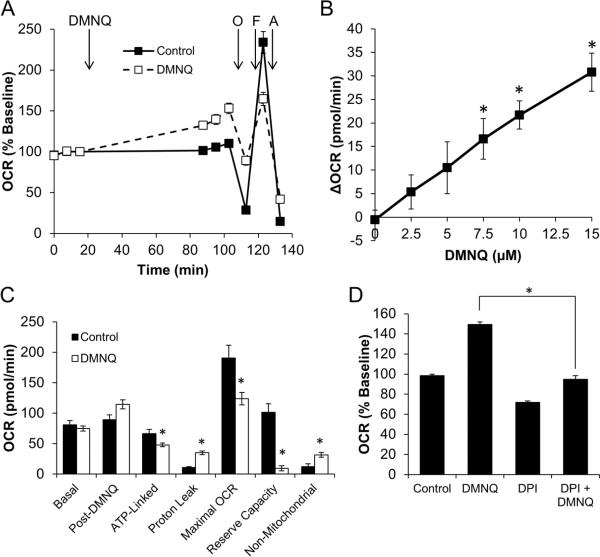
Assessing exogenous ROS production and mitochondrial function in the presence of DMNQ in BAEC
Redox cycling agents are widely used as therapeutics and as models for the production of ROS in cells. In this example, the measurement of oxygen consumption in response to the addition to the cells of the compound 2,3-dimethoxy-1,4-napthoquinone (DMNQ) is used as an example [17, 39]. As shown in Figure 6A, DMNQ stimulates basal OCR, but decreases the response to FCCP. This stimulation is concentration-dependent with respect to DMNQ (Figure 6B). Oligomycin injection allows for discrimination of ATP-linked OCR and Proton Leak OCR [39]. Using this method, it is clear that DMNQ stimulates proton leak OCR at the expense of ATP-linked OCR (Figure 6C). Additionally, the reserve capacity is decreased, and the non-mitochondrial OCR is increased. The increase in non-mitochondrial OCR suggests that a portion of DMNQ-dependent stimulation of OCR is due to redox cycling of DMNQ itself. The redox cycle of DMNQ is dependent on one of several flavoproteins. The flavoprotein inhibitor diphenyleneiodonium (DPI) was added to a final concentration of 10 μM with DMNQ (15 μM), and stimulation of OCR by DMNQ returned to the levels of untreated cells (Figure 6D). In this example the level of ROS generation of the redox cycling agent can be assessed in the same protocol used to determine the impact on bioenergetic function. These data are discussed in greater detail in reference [39].
Figure 6. The impact of DMNQ on endothelial mitochondrial function.
Using a Protocol 2 strategy, the effect of the redox-cycling compound DMNQ was examined in BAEC. After establishing baseline OCR, DMNQ was injected to a final concentration of 15 μM. Panel A depicts the effect on mitochondrial function. Basal OCR calculated from these data were then plotted as a function of DMNQ concentration (Panel B). From the data shown in Panel A, determinations of individual parameters of mitochondrial function can be made (Panel C). In some experiments, DPI (10 μM) was added at the time of DMNQ injection. DMNQ-stimulated OCR is shown in Panel D. All data shown are the mean ± sem. n=5 per treatment group. *, p≤0.05 as determined by two-tailed, unpaired t-test compared to the respective control measurement. Panels A and B are adapted from Free Radical Biology and Medicine, v.48(7), Dranka BP et.al, Mitochondrial reserve capacity in endothelial cells: The impact of nitric oxide and reactive oxygen species, p. 905-14, Copyright (2010), with permission from Elsevier.
Protocol 3: Determine the persistent effects of oxidative stress on the bioenergetic profile
The last major protocol used to assess mitochondrial function in cells exposed to oxidative stressors is to treat the cells outside the instrument for a predetermined time period, remove the stressor, and then assess mitochondrial function. This approach allows for longer treatment time periods that can be associated with additional experimental endpoints such as cell death. Importantly, the cells can be treated in their normal culture media, and incubated under conditions appropriate for the specific cell line avoiding potential artifacts due to altered cell culture conditions. After the predetermined treatment time, these cells are washed and changed to assay media for analysis of mitochondrial function. This is shown schematically in Figure 7A. A similar approach may be used for experiments where transient transfection of a gene of interest or siRNA construct is used to alter gene expression. In this case, a Protocol 3-style approach allows for the determination of stable phenotypic changes in the cell. Examples of this protocol are described below.
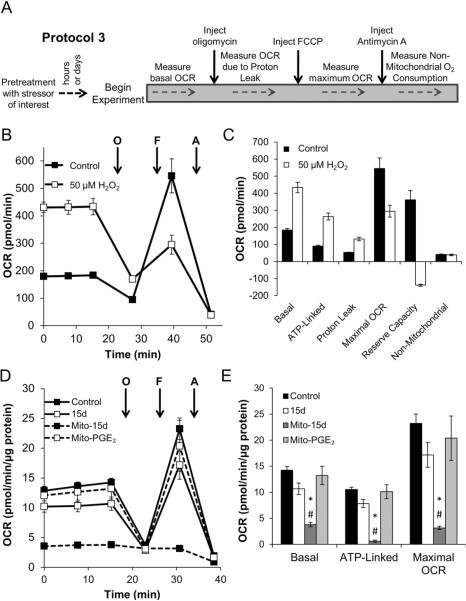
Figure 7. The impact of oxidative stressors can persist beyond their removal.
Using a Protocol 3-style approach, the effect of oxidative stressors was tested in various cell lines pretreated for a period of time. In each case, the stressor was washed out prior to assessment of mitochondrial function. Panel A describes the approach to performing a Protocol 3-style experiment. Panel B: NRVM were seeded to 75,000 cells/well for 24 h. Some cells were then treated with a bolus addition of 50 μM H2O2 for 30 min. The cells were washed, and changed to assay media for 1 h prior to assessment of mitochondrial function using 1μg/ml oligomycin, 1μM FCCP, and 10 μM antimycin A. Panel C: Individual mitochondrial function parameters calculated from data in Panel B. Data shown are the mean ± sem. n≥3 per treatment group. Panel D: MDA-MB231 cells were treated for 30 min with 10 μM 15d-PGJ2 (15d), mito-15d-PGJ2 (Mito-15d) or mito-PGE2 and mitochondrial function was assessed as described. After baseline was established, sequential injection of oligomycin (0.3 μg/ml), FCCP (1 μM) and antimycin A (10 μM) was used to determine mitochondrial function parameters. *, p≤0.05 as compared to control cells. #, p≤0.05 as compared to untargeted 15d-PGJ2 treated cells. Results represent means ± sem, n=5.
Stimulation of OCR by acute H2O2 exposure in NRVM
In this series of experiments, NRVM were seeded as described, and treated with 50 μM H2O2 for 30 min in normal culture media. The cells were then washed with XF assay media, and allowed to equilibrate at 37°C for 1 h in a non-CO2 incubator. Mitochondrial function was then assessed as described above. As shown in Figure 7B, basal OCR is increased in response to H2O2 in these cells. This occurs due to an increase in both proton leak OCR and ATP-Linked OCR (Figure 7C). Upon addition of FCCP however, the OCR is significantly inhibited, resulting in a loss of maximal OCR, and a negative apparent reserve capacity. This likely indicates that the mitochondria are damaged, and have lost their membrane potential, possibly due to opening of the mitochondrial permeability transition pore. Interestingly, these changes occur without an increase in non-mitochondrial OCR, indicating there is little additional ROS production beyond the bolus H2O2 the cells were exposed to initially.
Effect of mitochondrially-targeted electrophilic lipids on mitochondrial function in MDAMB231 cells
Beyond the direct impact of ROS/RNS on cellular bioenergetic function, the Seahorse XF technique can be used to determine effects of other reactive species. In this series of experiments, a reactive lipid species was targeted to the mitochondrion and the effects on cellular bioenergetic function were determined [47]. MDA-MB231 cells were treated for 30 min with 10 μM 15-deoxy-Δ12,14-Prostaglandin J2 (15d-PGJ2), mitochondrially-targeted-15d-PGJ2 (mito-15d-PGJ2) or mitochondrially-targeted Prostaglandin E2 (mito-PGE2) and then bioenergetic function was assessed as described above. After baseline was established, sequential injection of oligomycin (0.3 μg/ml), FCCP (1 μM) and antimycin A (10 μM) was used to determine mitochondrial function parameters. As shown in Figure 7D, 15d-PGJ2 had little effect on mitochondrial function. Interestingly, targeting the lipid to the mitochondria dramatically decreased the levels of all parameters measured. This was not due to the targeting moiety itself, as mito-PGE2 also had no effect on mitochondrial function (Figure 7E). FCCP was able to stimulate O2 consumption in the presence of 15d-PGJ2 and mito-PGE2, but not mito-15d-PGJ2 (Figure 7E). Furthermore, the loss of OCR following mito-15d-PGJ2 treatment is accompanied by a compensatory increase in ECAR (data not shown). ECAR stimulation does not occur in the presence of 15d-PGJ2 or mito-PGE2 suggesting that glycolysis is upregulated specifically in response to the inhibition of mitochondrial O2 consumption. These data show an example in which modifications to a pharmacophore can be assessed for their impact on bioenergetic function. The XF24 platform has also been used to identify specific pharmacophores which modulate the switch between mitochondrial oxidative phosphorylation and glycolysis [62].
Materials
Materials provided here are representative of what is required to perform an experiment of any of the above-described Protocols in Bovine Aortic Endothelial Cells (BAEC). The approach to using these cells is typical of what is required in most cell lines. Where empirical determination of an optimal concentration is required, we have made this notation.
Bovine Aortic Endothelial Cells, primary isolates
-
Flux Pak (Seahorse Bioscience, Product No. 100850-001)
Assay cartridge
V7 tissue culture plate
Calibration solution
Oligomycin (Sigma, Product No. O4876)
FCCP (Sigma, Product No. C2920)
Antimycin A (Sigma, Product No. A8674)
DMEM lacking sodium bicarbonate, phenol red, L-glutamine, and glucose (CellGro, Product No. 90–113)
L-Glutamine solution (Invitrogen, Product No. 25030-081)
Trypsin (Invitrogen, Product No. 25300054)
DMEM for routine cell culture containing 10% FBS, Penicillin/Streptomycin, and L-glutamine
D-Glucose (Sigma, Product No. G5400)
Sodium Pyruvate (Sigma, Product No. P5280)
0.1N NaOH, diluted from 10N (Fisher Science, Product No. SS255-1)
Tris base (Fisher Science, Product No. BP152-5)
Triton X-100 (Sigma, Product No. T-9284)
Bio-Rad Protein Assay Dye Reagent Concentrate (Bio-Rad, Product No. 500-0006)
Instruments
Seahorse Bioscience XF-24 Extracellular Flux Analyzer (Seahorse Bioscience; N. Billerica, MA)
Pipettes
Multichannel pipettes capable of 50–200 μl and 500–1000 μl volume
37°C non-CO2 incubator
pH meter
Mass balance
37°C Water bath
Centrifuge for cell culture capable of 600 × g
Hemacytometer or automated cell counter
Plate reader with filters suitable for absorbance measurement at 595 nm
Milli-Q (or equivalent) water purification system
Protocol
Seeding Bovine Aortic Endothelial Cells (BAEC) into Seahorse Bioscience XF24 Assay Plates
To ensure consistent measurements between replicate wells in an XF Assay, care must be taken to seed cells into the specialized tissue culture plates consistently and evenly. This can be achieved by careful attention to cell culture technique during seeding of the cells. An example protocol is listed below for BAEC [39].
Warm complete DMEM culture media and trypsin to 37°C in a water bath.
Lift the cells with trypsin as for other cell experiments, and resuspend in complete media equal to at least the volume of the trypsin to inactivate the enzyme.
Centrifuge the cell suspension at 600 × g for 3 min to pellet the cells.
Discard the media/trypsin supernatant and resuspend the cell pellet in 5 ml complete culture media.
Count the cells on a hemacytometer and determine the viable cell number per milliliter of culture media.
Dilute the cell suspension to the desired concentration. Typically this will be 10× the number of cells desired per well in 1 ml media. The required seeding number will vary, but in our experience should be between 10,000 and 80,000 cells per well depending on cell size and respiratory rate. Empirical optimization is required.
Add 100 μl of cell suspension to each well. Leave wells A1, B4, C3, and D6 blank as these will be used for monitoring temperature fluctuations in the plate, and for background correction. The blank wells should be filled with 100 μl media.
Incubate the cell plate at 37°C in a CO2-incubator for 4 h.
Add 150 μl complete culture media to each well, and incubate for a further 20 h.
Day prior to experiment
Seed an XF24 tissue culture plate as described above.
Prepare unbuffered assay media by mixing 8.28 g/L DMEM lacking sodium bicarbonate, 1 g/L D-Glucose, 0.11 g/L sodium pyruvate, and 4 mM L-Glutamine into Milli-Q water.
Hydrate an XF Assay Cartridge by adding 1 ml calibrant solution (included with Flux Pak) to each well of the utility plate. Place the cartridge in a 37°C non-CO2 incubator overnight.
Day of experiment
Sterile filter the media, and transfer approximately 50 ml to a clean 50 ml conical tube.
Warm the aliquot of media to 37°C in a water bath. This is now termed the “assay media.”
Adjust the pH of the media to 7.4 using 0.1 N NaOH.
Remove media from the cell plate, leaving 50 μl in the well. This can be done with careful aspiration or using a multi-channel pipette.
Rinse the cells with 1 ml of warmed assay media using a multichannel pipette.
Remove this assay media from the wells, again leaving 50 μl in the well.
Add 625 μl assay media to each well using a multichannel pipette. The final volume should be 675 μl.
Place the culture plate into a 37°C non-CO2 incubator for 1 h.
Prepare 3 ml oligomycin to a final concentration of 10 μg/ml in assay media. This concentration is 10× the final concentration desired, as this solution will be diluted 10-fold once injected into the measurement well. This concentration is suitable for BAEC. Empirical optimization is required.
Prepare 3 ml FCCP to a final concentration of 10 μM in assay media. This concentration is 10=× the final concentration desired, as this solution will be diluted 10-fold once injected into the measurement well. This concentration is suitable for BAEC. Empirical optimization is required.
Prepare 3 ml antimycin A to a final concentration of 100 μM in assay media. This concentration is 10× the final concentration desired, as this solution will be diluted 10-fold once injected into the measurement well. This concentration is suitable for BAEC. Empirical optimization is required.
Add 75 μl oligomycin solution to Port A on the XF Assay cartridge.
Add 83.3 μl FCCP solution to Port B on the XF Assay cartridge.
Add 92.6 μl antimycin A solution to Port C on the XF Assay cartridge.
Load the XF Assay cartridge into the XF24 30 min after changing the media on the cell culture plate.
After calibration has completed, exchange the utility plate for the cell culture plate and continue the measurement program. Typical mix/wait/measure cycles for BAEC are 2 min/2 min/3 min. Remove the plate and cartridge from the XF24 at the completion of the experiment.
Measurement of total protein content
After completion of the XF Assay, carefully aspirate all assay media from each well.
Gently rise the wells with 500 μl ice-cold PBS.
Carefully aspirate all PBS from each well.
Add 20 μl Lysis Buffer (10mM Tris, pH 7.4, 0.1% Triton X-100) to each well.
Allow to incubate on ice for 5 min.
Add 480 μl diluted Bradford Reagent to each well.
Prepare protein standards as for other cell experiments.
Measure absorbance at 595 nm using a plate reader.
General Considerations
Choice of media and substrates for oxidation
Knowing the appropriate substrate conditions for cell assays is fundamental to understanding the response to a given oxidant stressor. Identifying substrate preferences and interactions prior to examining the effects of oxidants on bioenergetics thus requires empirical optimization for each cell type studied. Cells may respire on multiple substrates, and this can change with duration in culture or culture conditions. The XF24 is well-suited to examine how respiratory substrates affect the response of cells to oxidative stress. The examples given below illustrate approaches to standardizing the running conditions for assessing the effects of substrate on bioenergetic responses to stress.
1. Determining optimal cell seeding conditions
Measurement of oxygen consumption and extracellular acidification in cell culture using the Seahorse XF technique requires empirical optimization of the cell seeding conditions for each cell type prior to experimental analysis. Multiple parameters must be taken into account, the most important of which are the cell seeding density, the elapsed time from seeding to analysis, the total time in culture or passage number, and the culture media conditions (discussed further below). This is especially important for primary isolated cells, as these cell types are even more sensitive to changes in their environment. Supplemental Figure 2C demonstrates the optimal cell seeding density experiment for BAEC. These cells display a linear response with regard to cell density from 20,000–50,000 cells per well both in measured OCR and ECAR. For future experiments, 40,000 cells/well was chosen as this yielded even monolayers of cells (Supplemental Figure 2D), and acceptable OCR/ECAR values.
2. Empirical determination of optimal concentrations for analysis of mitochondrial function
For each cell line used, we have found that empirical determination of the optimal concentrations for oligomycin, FCCP, and antimycin A are crucial to achieving maximal response. Supplemental Figure 2A demonstrates the response of BAEC to oligomycin. At low concentrations, oligomycin does not elicit a maximal inhibition of OCR immediately. However, at higher concentrations, the OCR trends upwards again following an initial inhibition. Thus, selection of a proper concentration is crucial for maintaining a consistent response during subsequent injections. In Supplemental Figure 2B, the stimulation of OCR in the presence of FCCP is also shown. Interestingly, FCCP displays a bell-shaped concentration response in these cells. Again, selection of the proper concentration is crucial to effectively obtain the maximal OCR in response to FCCP.
3. The effect of alternative substrates on respiration
Prior to challenging cells with an oxidant, it is important to examine how glycolytic substrates such as glucose and pyruvate affect mitochondrial respiration. The general XF assay medium contains physiological levels of glucose (5.5 mM), but supraphysiological levels of pyruvate (1 mM; blood concentration of pyruvate are generally less than 200 μM [63]). Moreover, the assay medium is generally devoid of serum (which can be a source of fatty acids), leaving the cells no choice but to use stored carbon (e.g., glycogen) or the provided extracellular substrate. Another factor to consider is whether the cells require insulin for glucose uptake (see below). Supplemental Figure 3 demonstrates why addressing these points is crucial to proper analysis of the data. Here, NRVM were provided with either no substrate, 5.5 mM glucose only, 1 mM pyruvate only, or glucose in the presence of pyruvate. Interestingly, the cardiomyocytes have a reserve bioenergetic capacity even in the absence of glucose and pyruvate, but show a surprising decrease in their energetic reserve in the presence of glucose (Supplemental Figure 3A and B). This may be due to compensatory increases in glycolysis and/or negative regulation of oxidative phosphorylation by glycolytic intermediates, e.g., the Crabtree effect [64]. Pyruvate provides a maximal response to FCCP, and this is decreased in the presence of glucose (Supplemental Figure 3A and B). Understanding substrate-specific changes such as these is therefore important for designing the appropriate experiments to examine the stressor of interest and for interpretation of results. This has been shown to be particularly important with respect to how myocytes respond to HNE [45].
In addition, there are several reports demonstrating the utility of the Seahorse XF24 to measure fatty acid oxidation [11, 65]. To do this, the fatty acids that are abundant in plasma (e.g., palmitate and oleate) are typically conjugated to bovine serum albumin (BSA) and injected via the ports of the flux pack during bioenergetic assay. The mitochondrial profile test can then be used to examine how well the mitochondria remain coupled in response to the fats and how they impact on maximal respiratory capacity. It is important to note that many oxidative and nitrosative/nitrative species can react with BSA, which could significantly affect the experiment. It is advised to use a system where oxidants are generated endogenously (e.g., DMNQ) or to use alternative approaches for examining fatty acid oxidation. These could include using inhibitors of fatty acid oxidation such as etomoxir, which inhibits carnitine palmitoyl transferase-1 [65]. For these types of assays, one can allow the cells to respire on endogenous stores of lipid (e.g., in adipocytes) or leave the serum in the medium. Etomoxir is then injected to inhibit carnitine palmitoyltransferase, and the change in OCR is monitored. The level of inhibition can be used to assess the fraction of basal oxygen consumption that was being used for fatty acid oxidation.
Glycolysis Assays
Concomitant with measurements of OCR, the XF analyzer can record ECAR and the proton production rate (PPR), both of which can be used as surrogates for lactate production formed from aerobic glycolysis [43]. Importantly, some reactive species have been shown to increase the ECAR and PPR in excitable cell types, such as primary cardiac myocytes [40, 45, 46]. As shown in Supplemental Figure 3C, exposure of NRVM to HNE results in a time-dependent increase glycolytic activity. This data is similar to that described in Figure 2B, however, the ECAR rates have been converted to PPR. While ECAR can be used to show changes in glycolytic flux, PPR is sometimes preferred because the area under the curve (AUC) can be calculated as a measure of the total increase in glycolytic activity during a given period of time. Although ECAR values may also be used for such analyses, it is difficult to represent ECAR data in AUC plots due to difficulties in understanding how a sum of mpH units relates with glycolysis; therefore, AUC values from PPR traces are preferred. For this, it is imperative that the user determine the buffer capacity of the running medium prior to the experiment using the following equation:
Some well-described cell types, such as vascular smooth muscle cells, convert >90% of their glucose to lactate [66, 67]. Such information is therefore helpful for experimental design and interpretation. In less characterized cells, the experimenter does not know a priori the metabolic fate of pyruvate, and in some cells, such as cardiomyocytes, lactate can be readily oxidized [68]; thus ECAR and PPR are not as reliable as stand-alone values for glycolytic flux. Additional experiments, measuring lactate production and/or using inhibitors of the glycolytic pathway, must then be performed to validate the XF measurements. Pharmacological inhibitors of glycolysis such as iodoacetate (IAA), koningic acid (KA), 2-deoxyglucose (2-DG), and oxamate (OA) can be used to ensure that the changes in ECAR or PPR are indeed due to glycolysis [43]. However, as with most chemical inhibitors, there are drawbacks to their use. For example, IAA inhibits glyceraldehyde-3-phosphate dehydrogenase (GAPDH) but is rather non-specific and can modify numerous other proteins that have reactive cysteine residues. Moreover, 2-DG and OA must be used in high (10–100 mM) concentrations to inhibit glycolysis and lactate dehydrogenase, respectively; this could result in experimental artifacts due to osmotic effects. A more specific inhibitor, koningic acid (KA), has been increasingly useful for validating XF glycolysis measurements. KA appears to be quite specific for GAPDH [69], inhibiting the enzyme at low μM concentrations [69–71]. Supplemental Figure 3D demonstrates that insulin stimulation of ECAR is dependent on GAPDH function. H9c2 cells were treated with 10 or 1000 nM insulin causing an increase in the measured ECAR. To demonstrate that this increase in ECAR is linked to increased glycolytic flux, KA was injected to a final concentration of 10 μg/ml. As shown in Supplemental Figure 3D, ECAR was inhibited maximally in the presence of KA. These and similar measurements are beginning to define the measurement of glycolysis in response to changes in oxidative stress in cell culture.
Normalization of data
Appropriate normalization of data generated by the XF24 is paramount to proper interpretation of experimental results. There are several approaches and caveats to these data normalization, and these are discussed in detail below.
1. Temperature correction
As mentioned above, the V7 culture plates supplied with Assay Flux Paks from Seahorse Bioscience are a 24 well plate. However, in a typical experiment, 4 of these wells are used to correct for changes in temperature. This occurs because of the exquisite sensitivity of the oxygen-quenched fluorophore to temperature. The XF24 software includes functionality to assign wells as temperature controls, and automatically corrects the data to account for temperature drift.
2. Normalization to total protein content
Normalization to total protein content in the well following a XF24 style experiment is another effective way of controlling for variation in cell number. We have used this technique extensively in the data shown here and elsewhere to ensure accurate measurements of OCR with respect to known protein concentration. Importantly, caution must be used when quantifying the protein content from such a small well. At the completion of an XF Assay, the cell plate should be immediately removed, and the media carefully aspirated. For cell lines which are easily lysed, we have found it most effective to lyse the cells in 20 μl of 10 mM Tris, pH 7.4 with 0.1% Triton X-100 added. 480 μl of Bradford reagent (Bio-Rad; Hercules, CA) can then be added to the entire well, without removal of any sample. In this way, there is no loss of protein due to pipetting, and little error introduced by excess media in the well prior to lysis. Protein concentration can then be measured according to the manufacturer's instructions. An example of this is shown in Figure 2E. BAEC were lysed as described and total protein content from individual wells was determined.
3. Normalization to cell number
Another method of normalizing the data from an XF24 experiment is to count the cells following the experiment. This can be particularly useful when using Protocol 3, or when comparing cells with different rates of proliferation or treatments which can alter the proliferation rates. Furthermore, certain treatments or models can alter the cell size or mitochondrial number independent of proliferation. While normalizing to total protein is a valid means to normalize for this, it may also be of interest to look at changes in mitochondrial function or glycolytic flux per cell. In these cases, cells can be trypsinized immediately following the conclusion of an XF24 assay and counted using a hemacytometer. To specifically count viable cells, methods such as a trypan blue exclusion assay may be utilized.
Reproducibility
Seahorse Bioscience reports coefficient of variation values of 20% or less between individual wells, experiments, and experimental days and this has been our experience. Supplemental Figure 4 shows representative data from two cell lines where the average basal OCR and maximal OCR from 6 independent experiments are compared. Experiments in BAEC were performed over 8 months, and those in NRVM over 6 months. These data show that basal cellular characteristics are similar over time, and that these cells respond similarly to FCCP (used to measure maximal OCR).
Furthermore, the effect of Deta NONOate on OCR in BAEC has recently been demonstrated to be consistent when measured on a different XF24 instrument in cells from an independent isolation [72]. These observations are thus consistent with a high degree of reproducibility between experiments, and between laboratories.
Calculations and Expected Results
All data from XF Assays described here were collected using the XF Reader software from Seahorse Biosciences (North Billerica, MA). Data from these experiments were exported to Microsoft Excel 2007 or 2010 (Microsoft Corp., Redmond, WA), where they were analyzed, normalized, and graphed. Box and whisker plots in Supplemental Figure 4 were constructed using Origin 8.5. Statistical significance was determined by performing 2-tailed Student's t-tests where appropriate using Microsoft Excel 2007, or 2010, or by One-Way ANOVA using GraphPad Prism 5 (GraphPad Software, La Jolla, CA) as indicated. p≤0.05 was considered significant in all experiments.
Importantly, the serial nature of measurements using the Seahorse XF technique allows for the use of repeated measures ANOVA during some experiments. Researchers are encouraged to critically evaluate the most appropriate method for determining statistical significance when analyzing data generated using this technique.
Caveats
While the instrumentation opens new avenues for oxidative stress research, interpretation and analysis are constrained in some key areas as outlined below.
Interference of injected compounds with sensor fluorescence
The analyses described here rely on fluorescence detection for both changes in pH and O2. In some cases, compounds can interfere with the detection method due to their intrinsic ability to quench or otherwise alter the characteristics of the fluorescent probe. Controls need to be included in which compounds or stressors are added to wells without cells and if changes in fluorescence occur, then the investigator should only use Protocol 3.
Composition of assay media
There are important differences in the media used for the XF24 analysis and that generally used for growth of mammalian cells. These include a lack of serum, no bicarbonate buffering and very limited buffering capacity. We recommend that the investigator prepare their own media for these experiments if controls reveal important differences in the response of the cells due to media. It is absolutely essential that a biological endpoint is used to compare responses between the cells plated for the XF24 analysis and the standard culture media as we have described above (Figure 3). In some cases the additional information obtained from the ECAR measurement is limited and buffered media can then be prepared.
Limitations of the mitochondrial profile technique
-
1)
The FCCP dose response is biphasic probably due to the sum of the process of increased H+ permeability leading to increased OCR and the inhibitory effect on OCR due to decreased substrate availability to drive respiration.
-
2)
The FCCP concentration dependence can in some circumstances be changed by the presence of oligomycin and this should be tested in each cell type used.
-
3)
In our experience antimycin A gives a maximal inhibitory response which is not further increased by rotenone of other mitochondrial inhibitors. However, this should be assessed independently, as this result may differ between cell lines.
-
4)
The assessment of proton leak and ATP usage by the cells after the addition of oligomycin is constrained by the fact that this will increased mitochondrial membrane potential. The consequence of this effect is an underestimate of ATP-linked OCR and over-estimate of proton leak. This can be corrected for if needed by independent measurement of the mitochondrial membrane potential [58, 73].
Concluding remarks
From the experiments highlighted here, it is clear that the impact of oxidative stressors on mitochondrial function varies dependent on the cell type, stressor, and the concentration and duration of exposure of the stressor. It is then useful to approach the mitochondrial impact of a stressor of interest with a combination of the listed Protocols. To date, there are nearly 150 publications using the Seahorse XF24 analyzer to measure changes in O2 consumption and extracellular acidification. The application of the mitochondrial and glycolytic function assays described here is extending the utility of this instrument, allowing for more rigorous study of model systems where measurement of oxygen consumption was previously not possible or practical. It is the hope of the authors that the Protocols described here will aid investigators in their assessment of the impacts of oxidative stressors on metabolic function.
Supplementary Material
Highlights
The impact of oxidative stress on bioenergetics is important to assess in a cellular setting.
Protocols are reported to obtain the bioenergetic response to oxidative stress in adherent cells.
Oxidative phosphorylation and glycolysis are integrated to make a metabolic profile.
Reserve capacity is the most critical parameter in cellular bioenergetics.
Acknowledgements
This work was supported by NIH R01 Grants ES10167, AA13395, DK075867 (to VDU), HL096638 (to AL), HL101192 and HL079364 (to JCC), NS064090 (to JZ); NIH P20 Grant RR024489 (BGH); NIH Training Grant HL007918 (to BPD, ARD, and BZ); a grant from Seahorse Bioscience (to VDU); and a VA merit award to JZ.
Abbreviations
- 15d-PGJ2
15-deoxy-Δ12,14-Prostaglandin J2
- 2-DG
2-Deoxy-D-Glucose
- Anti A
Antimycin A
- AUC
Area under the curve
- Deta NO
Deta NONOate
- (Z)-1-[2-(2-Aminoethyl)-N-(2-ammonioethyl)amino]diazen-1-ium-1
2-diolate
- DMNQ
2,3-Dimethoxy-1,4-napthoquinone
- DPI
Diphenyleneiodonium
- ECAR
Extracellular Acidification Rate
- FCCP
carbonyl cyanide p-[trifluoromethoxy]-phenyl-hydrazone
- GAPDH
Glyceraldehyde-3-Phosphate Dehydrogenase
- HNE
4-hydroxy-2-nonenal
- HO-1
Heme-Oxygenase-1
- IAA
Iodoacetic Acid
- KA
Koningic Acid
- mito-15d-PGJ2
mitochondrially-targeted 15-deoxy-Δ12,14-Prostaglandin J2
- mito-PGE2
mitochondrially-targeted Prostaglandin E2
- NO
Nitric oxide
- NRVM
Neonatal Rat Ventricular Myocytes
- OCR
Oxygen Consumption Rate
- PPR
Proton Production Rate
- RASMC
Rat Aortic Smooth Muscle Cells
- RNS
Reactive Nitrogen Species
- ROS
Reactive Oxygen Species
- siRNA
short-interfering RNA
- XF
Extracellular Flux
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Conflict of Interest Statement: V. Darley-Usmar serves as a consultant for Seahorse Bioscience.
References
- [1].Will Y, Hynes J, Ogurtsov VI, Papkovsky DB. Analysis of mitochondrial function using phosphorescent oxygen-sensitive probes. Nat Protoc. 2006;1:2563–2572. doi: 10.1038/nprot.2006.351. [DOI] [PubMed] [Google Scholar]
- [2].Brand MD, Nicholls DG. Assessing mitochondrial dysfunction in cells. Biochem J. 2011;435:297–312. doi: 10.1042/BJ20110162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Clark LC, Jr., Wolf R, Granger D, Taylor Z. Continuous recording of blood oxygen tensions by polarography. J Appl Physiol. 1953;6:189–193. doi: 10.1152/jappl.1953.6.3.189. [DOI] [PubMed] [Google Scholar]
- [4].Li AE, Ito H, Rovira II, Kim K-S, Takeda K, Yu Z-Y, Ferrans VJ, Finkel T. A Role for Reactive Oxygen Species in Endothelial Cell Anoikis. Circ Res. 1999;85:304–310. doi: 10.1161/01.res.85.4.304. [DOI] [PubMed] [Google Scholar]
- [5].De Keulenaer GW, Chappell DC, Ishizaka N, Nerem RM, Alexander RW, Griendling KK. Oscillatory and steady laminar shear stress differentially affect human endothelial redox state: role of a superoxide-producing NADH oxidase. Circ Res. 1998;82:1094–1101. doi: 10.1161/01.res.82.10.1094. [DOI] [PubMed] [Google Scholar]
- [6].Hwang J, Saha A, Boo YC, Sorescu GP, McNally JS, Holland SM, Dikalov S, Giddens DP, Griendling KK, Harrison DG, Jo H. Oscillatory shear stress stimulates endothelial production of O2- from p47phox-dependent NAD(P)H oxidases, leading to monocyte adhesion. J Biol Chem. 2003;278:47291–47298. doi: 10.1074/jbc.M305150200. [DOI] [PubMed] [Google Scholar]
- [7].Clementi E, Brown GC, Foxwell N, Moncada S. On the mechanism by which vascular endothelial cells regulate their oxygen consumption. PNAS. 1999;96:1559–1562. doi: 10.1073/pnas.96.4.1559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Sauerbeck A, Pandya J, Singh I, Bittman K, Readnower R, Bing G, Sullivan P. Analysis of regional brain mitochondrial bioenergetics and susceptibility to mitochondrial inhibition utilizing a microplate based system. J Neurosci Methods. 2011;198:36–43. doi: 10.1016/j.jneumeth.2011.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Rogers GW, Brand MD, Petrosyan S, Ashok D, Elorza AA, Ferrick DA, Murphy AN. High throughput microplate respiratory measurements using minimal quantities of isolated mitochondria. PLoS One. 2011;6:e21746. doi: 10.1371/journal.pone.0021746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Wu Y, Song P, Xu J, Zhang M, Zou MH. Activation of protein phosphatase 2A by palmitate inhibits AMP-activated protein kinase. J Biol Chem. 2007;282:9777–9788. doi: 10.1074/jbc.M608310200. [DOI] [PubMed] [Google Scholar]
- [11].Ferrick DA, Neilson A, Beeson C. Advances in measuring cellular bioenergetics using extracellular flux. Drug Discov Today. 2008;13:268–274. doi: 10.1016/j.drudis.2007.12.008. [DOI] [PubMed] [Google Scholar]
- [12].Gerencser AA, Neilson A, Choi SW, Edman U, Yadava N, Oh RJ, Ferrick DA, Nicholls DG, Brand MD. Quantitative Microplate-Based Respirometry with Correction for Oxygen Diffusion. Anal Chem. 2009 doi: 10.1021/ac900881z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Scott ID, Nicholls DG. Energy transduction in intact synaptosomes. Influence of plasma-membrane depolarization on the respiration and membrane potential of internal mitochondria determined in situ. Biochem J. 1980;186:21–33. doi: 10.1042/bj1860021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Seth RB, Sun L, Ea CK, Chen ZJ. Identification and characterization of MAVS, a mitochondrial antiviral signaling protein that activates NF-kappaB and IRF 3. Cell. 2005;122:669–682. doi: 10.1016/j.cell.2005.08.012. [DOI] [PubMed] [Google Scholar]
- [15].Gutierrez J, Ballinger SW, Darley-Usmar VM, Landar A. Free Radicals, Mitochondria, and Oxidized Lipids: The Emerging Role in Signal Transduction in Vascular Cells. Circ Res. 2006;99:924–932. doi: 10.1161/01.RES.0000248212.86638.e9. [DOI] [PubMed] [Google Scholar]
- [16].Quintero M, Colombo SL, Godfrey A, Moncada S. Mitochondria as signaling organelles in the vascular endothelium. PNAS. 2006;103:5379–5384. doi: 10.1073/pnas.0601026103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Ramachandran A, Moellering D, Go YM, Shiva S, Levonen AL, Jo H, Patel RP, Parthasarathy S, Darley-Usmar VM. Activation of c-Jun N-Terminal Kinase and Apoptosis in Endothelial Cells Mediated by Endogenous Generation of Hydrogen Peroxide. Biological Chemistry. 2002;383:693. doi: 10.1515/BC.2002.071. [DOI] [PubMed] [Google Scholar]
- [18].Ramachandran A, Moellering DR, Ceaser E, Shiva S, Xu J, Darley-Usmar V. Inhibition of mitochondrial protein synthesis results in increased endothelial cell susceptibility to nitric oxide-induced apoptosis. PNAS. 2002;99:6643–6648. doi: 10.1073/pnas.102019899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Borutaite V, Brown GC. Nitric oxide induces apoptosis via hydrogen peroxide, but necrosis via energy and thiol depletion. Free Radic Biol Med. 2003;35:1457–1468. doi: 10.1016/j.freeradbiomed.2003.08.003. [DOI] [PubMed] [Google Scholar]
- [20].Landar A, Shiva S, Levonen AL, Oh JY, Zaragoza C, Johnson MS, Darley-Usmar VM. Induction of the permeability transition and cytochrome c release by 15-deoxy-Delta12,14-prostaglandin J2 in mitochondria. Biochem J. 2006;394:185–195. doi: 10.1042/BJ20051259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Ow YP, Green DR, Hao Z, Mak TW. Cytochrome c: functions beyond respiration. Nat Rev Mol Cell Biol. 2008;9:532–542. doi: 10.1038/nrm2434. [DOI] [PubMed] [Google Scholar]
- [22].Liu Z, Butow RA. Mitochondrial retrograde signaling. Annu Rev Genet. 2006;40:159–185. doi: 10.1146/annurev.genet.40.110405.090613. [DOI] [PubMed] [Google Scholar]
- [23].Scorrano L, Korsmeyer SJ. Mechanisms of cytochrome c release by proapoptotic BCL-2 family members. Biochem Biophys Res Commun. 2003;304:437–444. doi: 10.1016/s0006-291x(03)00615-6. [DOI] [PubMed] [Google Scholar]
- [24].Tsujimoto Y. Cell death regulation by the Bcl-2 protein family in the mitochondria. J Cell Physiol. 2003;195:158–167. doi: 10.1002/jcp.10254. [DOI] [PubMed] [Google Scholar]
- [25].Karbowski M, Norris KL, Cleland MM, Jeong SY, Youle RJ. Role of Bax and Bak in mitochondrial morphogenesis. Nature. 2006;443:658–662. doi: 10.1038/nature05111. [DOI] [PubMed] [Google Scholar]
- [26].Naitoh K, Ichikawa Y, Miura T, Nakamura Y, Miki T, Ikeda Y, Kobayashi H, Nishihara M, Ohori K, Shimamoto K. MitoKATP channel activation suppresses gap junction permeability in the ischemic myocardium by an ERK-dependent mechanism. Cardiovasc Res. 2006;70:374–383. doi: 10.1016/j.cardiores.2006.01.023. [DOI] [PubMed] [Google Scholar]
- [27].Rodriguez-Sinovas A, Boengler K, Cabestrero A, Gres P, Morente M, Ruiz-Meana M, Konietzka I, Miro E, Totzeck A, Heusch G, Schulz R, Garcia-Dorado D. Translocation of connexin 43 to the inner mitochondrial membrane of cardiomyocytes through the heat shock protein 90-dependent TOM pathway and its importance for cardioprotection. Circ Res. 2006;99:93–101. doi: 10.1161/01.RES.0000230315.56904.de. [DOI] [PubMed] [Google Scholar]
- [28].Rodriguez-Sinovas A, Garcia-Dorado D, Ruiz-Meana M, Soler-Soler J. Protective effect of gap junction uncouplers given during hypoxia against reoxygenation injury in isolated rat hearts. Am J Physiol Heart Circ Physiol. 2006;290:H648–656. doi: 10.1152/ajpheart.00439.2005. [DOI] [PubMed] [Google Scholar]
- [29].Boengler K, Konietzka I, Buechert A, Heinen Y, Garcia-Dorado D, Heusch G, Schulz R. Loss of ischemic preconditioning's cardioprotection in aged mouse hearts is associated with reduced gap junctional and mitochondrial levels of connexin 43. Am J Physiol Heart Circ Physiol. 2007;292:H1764–1769. doi: 10.1152/ajpheart.01071.2006. [DOI] [PubMed] [Google Scholar]
- [30].Cohen MM, Leboucher GP, Livnat-Levanon N, Glickman MH, Weissman AM. Ubiquitin-proteasome-dependent degradation of a mitofusin, a critical regulator of mitochondrial fusion. Mol Biol Cell. 2008;19:2457–2464. doi: 10.1091/mbc.E08-02-0227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Miyashita K, Itoh H, Tsujimoto H, Tamura N, Fukunaga Y, Sone M, Yamahara K, Taura D, Inuzuka M, Sonoyama T, Nakao K. Natriuretic Peptides/cGMP/cGMP-dependent Protein Kinase Cascades Promote Muscle Mitochondrial Biogenesis and Prevent Obesity. Diabetes. 2009 doi: 10.2337/db09-0393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Southworth R. Hexokinase-mitochondrial interaction in cardiac tissue: implications for cardiac glucose uptake, the 18FDG lumped constant and cardiac protection. J Bioenerg Biomembr. 2009;41:187–193. doi: 10.1007/s10863-009-9207-9. [DOI] [PubMed] [Google Scholar]
- [33].Olson KR. Hydrogen Sulfide and Oxygen Sensing in the Cardiovascular System. Antioxid Redox Signal. 2009 doi: 10.1089/ars.2009.2921. [DOI] [PubMed] [Google Scholar]
- [34].Thomas B, Beal MF. Mitochondrial therapies for Parkinson's disease. Movement Disorders. 2010;25:S155–S160. doi: 10.1002/mds.22781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Wallace DC, Fan W, Procaccio V. Mitochondrial energetics and therapeutics. Annu Rev Pathol. 2010;5:297–348. doi: 10.1146/annurev.pathol.4.110807.092314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Ramachandran A, Levonen AL, Brookes PS, Ceaser E, Shiva S, Barone MC, Darley-Usmar V. Mitochondria, nitric oxide, and cardiovascular dysfunction. Free Radic Biol Med. 2002;33:1465–1474. doi: 10.1016/s0891-5849(02)01142-5. [DOI] [PubMed] [Google Scholar]
- [37].Ballinger SW. Mitochondrial dysfunction in cardiovascular disease. Free Radic Biol Med. 2005;38:1278–1295. doi: 10.1016/j.freeradbiomed.2005.02.014. [DOI] [PubMed] [Google Scholar]
- [38].Madamanchi NR, Runge MS. Mitochondrial dysfunction in atherosclerosis. Circ Res. 2007;100:460–473. doi: 10.1161/01.RES.0000258450.44413.96. [DOI] [PubMed] [Google Scholar]
- [39].Dranka BP, Hill BG, Darley-Usmar VM. Mitochondrial reserve capacity in endothelial cells: The impact of nitric oxide and reactive oxygen species. Free Radic Biol Med. 2010;48:905–914. doi: 10.1016/j.freeradbiomed.2010.01.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Hill BG, Dranka BP, Zou L, Chatham JC, Darley-Usmar VM. Importance of the bioenergetic reserve capacity in response to cardiomyocyte stress induced by 4-hydroxynonenal. Biochem J. 2009;424:99–107. doi: 10.1042/BJ20090934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Hill BG, Higdon AN, Dranka BP, Darley-Usmar VM. Regulation of vascular smooth muscle cell bioenergetic function by protein glutathiolation. Biochim Biophys Acta. 2010;1797:285–295. doi: 10.1016/j.bbabio.2009.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Perez J, Hill BG, Benavides GA, Dranka BP, Darley-Usmar VM. Role of cellular bioenergetics in smooth muscle cell proliferation induced by platelet-derived growth factor. Biochem J. 2010;428:255–267. doi: 10.1042/BJ20100090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Wu M, Neilson A, Swift AL, Moran R, Tamagnine J, Parslow D, Armistead S, Lemire K, Orrell J, Teich J, Chomicz S, Ferrick DA. Multiparameter metabolic analysis reveals a close link between attenuated mitochondrial bioenergetic function and enhanced glycolysis dependency in human tumor cells. Am J Physiol Cell Physiol. 2007;292:C125–136. doi: 10.1152/ajpcell.00247.2006. [DOI] [PubMed] [Google Scholar]
- [44].Sridharan V, Guichard J, Li CY, Muise-Helmericks R, Beeson CC, Wright GL. O(2)-sensing signal cascade: clamping of O(2) respiration, reduced ATP utilization, and inducible fumarate respiration. Am J Physiol Cell Physiol. 2008;295:C29–37. doi: 10.1152/ajpcell.00466.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Sansbury BE, Jones SP, Riggs DW, Darley-Usmar VM, Hill BG. Bioenergetic function in cardiovascular cells: the importance of the reserve capacity and its biological regulation. Chem Biol Interact. 2011;191:288–295. doi: 10.1016/j.cbi.2010.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Sansbury BE, Riggs DW, Brainard RE, Salabei JK, Jones SP, Hill BG. Responses of hypertrophied myocytes to reactive species: implications for glycolysis and electrophile metabolism. Biochem J. 2011;435:519–528. doi: 10.1042/BJ20101390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Diers AR, Higdon AN, Ricart KC, Johnson MS, Agarwal A, Kalyanaraman B, Landar A, Darley-Usmar VM. Mitochondrial targeting of the electrophilic lipid 15-deoxy-Delta12,14-prostaglandin J2 increases apoptotic efficacy via redox cell signalling mechanisms. Biochem J. 2010;426:31–41. doi: 10.1042/BJ20091293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Cleeter MW, Cooper JM, Darley-Usmar VM, Moncada S, Schapira AH. Reversible inhibition of cytochrome c oxidase, the terminal enzyme of the mitochondrial respiratory chain, by nitric oxide. Implications for neurodegenerative diseases. FEBS Lett. 1994;345:50–54. doi: 10.1016/0014-5793(94)00424-2. [DOI] [PubMed] [Google Scholar]
- [49].Esterbauer H, Schaur RJ, Zollner H. Chemistry and biochemistry of 4-hydroxynonenal, malonaldehyde and related aldehydes. Free Radic Biol Med. 1991;11:81–128. doi: 10.1016/0891-5849(91)90192-6. [DOI] [PubMed] [Google Scholar]
- [50].Oh JY, Giles N, Landar A, Darley-Usmar V. Accumulation of 15-deoxy-delta(12,14)-prostaglandin J2 adduct formation with Keap1 over time: effects on potency for intracellular antioxidant defence induction. Biochem J. 2008;411:297–306. doi: 10.1042/bj20071189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [51].Hill BG, Dranka BP, Bailey SM, Lancaster JR, Jr., Darley-Usmar VM. What part of NO don't you understand? Some answers to the cardinal questions in nitric oxide biology. J Biol Chem. 2010;285:19699–19704. doi: 10.1074/jbc.R110.101618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [52].Kansanen E, Kivela AM, Levonen AL. Regulation of Nrf2-dependent gene expression by 15-deoxy-Delta12,14-prostaglandin J2. Free Radic Biol Med. 2009;47:1310–1317. doi: 10.1016/j.freeradbiomed.2009.06.030. [DOI] [PubMed] [Google Scholar]
- [53].Xie HR, Hu LS, Li GY. SH-SY5Y human neuroblastoma cell line: in vitro cell model of dopaminergic neurons in Parkinson's disease. Chin Med J (Engl) 2010;123:1086–1092. [PubMed] [Google Scholar]
- [54].Landar A, Zmijewski JW, Dickinson DA, Le Goffe C, Johnson MS, Milne GL, Zanoni G, Vidari G, Morrow JD, Darley-Usmar VM. Interaction of electrophilic lipid oxidation products with mitochondria in endothelial cells and formation of reactive oxygen species. Am J Physiol Heart Circ Physiol. 2006;290:H1777–1787. doi: 10.1152/ajpheart.01087.2005. [DOI] [PubMed] [Google Scholar]
- [55].Ishii T, Itoh K, Ruiz E, Leake DS, Unoki H, Yamamoto M, Mann GE. Role of Nrf2 in the regulation of CD36 and stress protein expression in murine macrophages: activation by oxidatively modified LDL and 4-hydroxynonenal. Circ Res. 2004;94:609–616. doi: 10.1161/01.RES.0000119171.44657.45. [DOI] [PubMed] [Google Scholar]
- [56].Iles KE, Dickinson DA, Wigley AF, Welty NE, Blank V, Forman HJ. HNE increases HO-1 through activation of the ERK pathway in pulmonary epithelial cells. Free Radic Biol Med. 2005;39:355–364. doi: 10.1016/j.freeradbiomed.2005.03.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [57].Jekabsons MB, Nicholls DG. In situ respiration and bioenergetic status of mitochondria in primary cerebellar granule neuronal cultures exposed continuously to glutamate. J Biol Chem. 2004;279:32989–33000. doi: 10.1074/jbc.M401540200. [DOI] [PubMed] [Google Scholar]
- [58].Nicholls DG, Ferguson SJ. Bioenergetics 3. Academic Press; New York: 2002. [Google Scholar]
- [59].Choi SW, Gerencser AA, Nicholls DG. Bioenergetic analysis of isolated cerebrocortical nerve terminals on a microgram scale: spare respiratory capacity and stochastic mitochondrial failure. J Neurochem. 2009;109:1179–1191. doi: 10.1111/j.1471-4159.2009.06055.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [60].Fern R. Variations in spare electron transport chain capacity: The answer to an old riddle? J Neurosci Res. 2003;71:759–762. doi: 10.1002/jnr.10553. [DOI] [PubMed] [Google Scholar]
- [61].Brand MD, Evans SM, Mendes-Mourao J, Chappell JB. Fluorocitrate inhibition of aconitate hydratase and the tricarboxylate carrier of rat liver mitochondria. Biochem J. 1973;134:217–224. doi: 10.1042/bj1340217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [62].Gohil VM, Sheth SA, Nilsson R, Wojtovich AP, Lee JH, Perocchi F, Chen W, Clish CB, Ayata C, Brookes PS, Mootha VK. Nutrient-sensitized screening for drugs that shift energy metabolism from mitochondrial respiration to glycolysis. Nat Biotechnol. 2010;28:249–255. doi: 10.1038/nbt.1606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [63].Chuang CK, Wang TJ, Yeung CY, Hsieh WS, Lin DS, Ho SC, Lin SP. Interference and blood sample preparation for a pyruvate enzymatic assay. Clin Biochem. 2006;39:74–77. doi: 10.1016/j.clinbiochem.2005.10.007. [DOI] [PubMed] [Google Scholar]
- [64].Diaz-Ruiz R, Averet N, Araiza D, Pinson B, Uribe-Carvajal S, Devin A, Rigoulet M. Mitochondrial oxidative phosphorylation is regulated by fructose 1,6-bisphosphate. A possible role in Crabtree effect induction? J Biol Chem. 2008;283:26948–26955. doi: 10.1074/jbc.M800408200. [DOI] [PubMed] [Google Scholar]
- [65].Pike LS, Smift AL, Croteau NJ, Ferrick DA, Wu M. Inhibition of fatty acid oxidation by etomoxir impairs NADPH production and increases reactive oxygen species resulting in ATP depletion and cell death in human glioblastoma cells. Biochim Biophys Acta. 2011;1807:726–734. doi: 10.1016/j.bbabio.2010.10.022. [DOI] [PubMed] [Google Scholar]
- [66].Lynch RM, Paul RJ. Compartmentation of glycolytic and glycogenolytic metabolism in vascular smooth muscle. Science. 1983;222:1344–1346. doi: 10.1126/science.6658455. [DOI] [PubMed] [Google Scholar]
- [67].Michelakis ED, Weir EK. The metabolic basis of vascular oxygen sensing: diversity, compartmentalization, and lessons from cancer. Am J Physiol Heart Circ Physiol. 2008;295:H928–H930. doi: 10.1152/ajpheart.00697.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [68].Wen Y, Li W, Poteet EC, Xie L, Tan C, Yan LJ, Ju X, Liu R, Qian H, Marvin MA, Goldberg MS, She H, Mao Z, Simpkins JW, Yang SH. Alternative mitochondrial electron transfer as a novel strategy for neuroprotection. J Biol Chem. 2011 doi: 10.1074/jbc.M110.208447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [69].Sakai K, Hasumi K, Endo A. Identification of koningic acid (heptelidic acid)-modified site in rabbit muscle glyceraldehyde-3-phosphate dehydrogenase. Biochim Biophys Acta. 1991;1077:192–196. doi: 10.1016/0167-4838(91)90058-8. [DOI] [PubMed] [Google Scholar]
- [70].Endo A, Hasumi K, Sakai K, Kanbe T. Specific inhibition of glyceraldehyde-3-phosphate dehydrogenase by koningic acid (heptelidic acid) J Antibiot (Tokyo) 1985;38:920–925. doi: 10.7164/antibiotics.38.920. [DOI] [PubMed] [Google Scholar]
- [71].Sakai K, Hasumi K, Endo A. Inactivation of rabbit muscle glyceraldehyde-3-phosphate dehydrogenase by koningic acid. Biochim Biophys Acta. 1988;952:297–303. doi: 10.1016/0167-4838(88)90130-6. [DOI] [PubMed] [Google Scholar]
- [72].Diers AR, Broniowska KA, Darley-Usmar VM, Hogg N. Differential regulation of metabolism by nitric oxide and S-nitrosothiols in endothelial cells. Am J Physiol Heart Circ Physiol. 2011 doi: 10.1152/ajpheart.00210.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [73].Nicholls DG, Ward MW. Mitochondrial membrane potential and neuronal glutamate excitotoxicity: mortality and millivolts. Trends Neurosci. 2000;23:166–174. doi: 10.1016/s0166-2236(99)01534-9. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.