Background: Auxin is essential for plant growth, but its biosynthesis in plants has not been biochemically defined.
Results: Key features of the catalytic mechanism for the YUCCA flavoprotein, the rate-limiting enzyme of auxin biosynthesis, are determined.
Conclusion: YUCs generate an observable though relatively short lived C4a-(hydro)peroxyflavin intermediate for catalysis in auxin biosynthesis.
Significance: This work establishes the previously unknown biochemical mechanism of auxin biosynthesis.
Keywords: Arabidopsis, Auxin, Biosynthesis, Enzyme Catalysis, Enzyme Mechanisms, Flavin, Plant Biochemistry, Plant Hormones
Abstract
Auxin regulates every aspect of plant growth and development. Previous genetic studies demonstrated that YUCCA (YUC) flavin-containing monooxygenases (FMOs) catalyze a rate-limiting step in auxin biosynthesis and that YUCs are essential for many developmental processes. We proposed that YUCs convert indole-3-pyruvate (IPA) to indole-3-acetate (IAA). However, the exact biochemical mechanism of YUCs has remained elusive. Here we present the biochemical characterization of recombinant Arabidopsis YUC6. Expressed in and purified from Escherichia coli, YUC6 contains FAD as a cofactor, which has peaks at 448 nm and 376 nm in the UV-visible spectrum. We show that YUC6 uses NADPH and oxygen to convert IPA to IAA. The first step of the YUC6-catalyzed reaction is the reduction of the FAD cofactor to FADH− by NADPH. Subsequently, FADH− reacts with oxygen to form a flavin-C4a-(hydro)peroxy intermediate, which we show has a maximum absorbance at 381 nm in its UV-visible spectrum. The final chemical step is the reaction of the C4a-intermediate with IPA to produce IAA. Although the sequences of the YUC enzymes are related to those of the mammalian FMOs, which oxygenate nucleophilic substrates, YUC6 oxygenates an electrophilic substrate (IPA). Nevertheless, both classes of enzymes form quasi-stable C4a-(hydro)peroxyl FAD intermediates. The YUC6 intermediate has a half-life of ∼20 s whereas that of some FMOs is >30 min. This work reveals the catalytic mechanism of the first known plant flavin monooxygenase and provides a foundation for further investigating how YUC activities are regulated in plants.
Introduction
Auxin is an essential hormone for many aspects of plant growth and development (1). Indole-3-acetic acid (IAA),3 the main auxin in plants, is synthesized using tryptophan (Trp) as a precursor. Recently, a two-step pathway that converts Trp to IAA has been proposed as the main auxin biosynthesis pathway in plants (Fig. 1) (2–5). Trp is first converted to indole-3-pyruvate (IPA) by the TAA1 family of aminotransferases (6). Subsequently, the YUC family flavin-containing monooxygenases (FMOs) catalyze the production of IAA using IPA as a substrate (Fig. 1) (4, 7, 8). The YUC-catalyzed reaction is suggested as the rate-limiting step in auxin biosynthesis (9).
FIGURE 1.
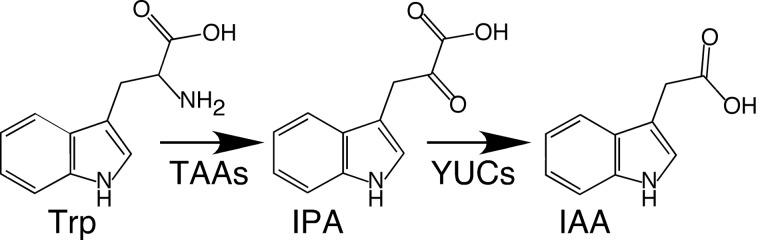
The main auxin biosynthesis pathway in plants. Tryptophan is first converted by the TAA transaminase to IPA, which undergoes oxidative decarboxylation catalyzed by the YUC flavin monooxygenase.
YUC genes were initially linked to auxin biosynthesis because their overexpression in Arabidopsis caused auxin overproduction phenotypes (9). YUC genes have been identified in all of the plant genomes that have been sequenced, suggesting that YUC-mediated auxin biosynthesis is widely used by plants. It has been shown that overexpression of YUC genes in petunia (10), tobacco (9), and rice (11) also leads to auxin overproduction. In Arabidopsis, there are 11 YUC genes. Inactivation of a single YUC gene in Arabidopsis does not cause obvious developmental defects because of the overlapping functions among YUC genes. However, simultaneous inactivation of YUC1 and YUC4 causes dramatic defects in vascular and floral development (7, 8). Detailed analysis of various yuc mutant combinations has demonstrated that localized auxin biosynthesis is important for all of the major developmental processes in plants, including embryogenesis, seedling growth, root elongation, gravitropism, vascular pattern formation, and flower development (7, 8). Furthermore, some loss-of-function yuc mutants are rescued by producing IAA in situ, which was achieved by expressing the bacterial auxin biosynthesis gene iaaM under the control of a YUC promoter (7, 8). Mutations in YUC genes in rice (12), corn (13), and petunia (10) also lead to severe developmental defects, indicating that YUC genes are essential for plant development in both monocots and dicots.
Although genetic studies have unambiguously demonstrated that YUCs are key auxin biosynthesis genes, the exact biochemical roles of YUCs in auxin biosynthesis have still been a subject of debate. It was initially suggested that YUCs catalyze the hydroxylation of tryptamine (9, 14–16). Recent studies have indicated that YUCs use IPA as a substrate in auxin biosynthesis (Fig. 1) (5). It was shown that yuc mutants and taa mutants can phenocopy each other (4). Mutations in TAA genes caused IPA deficiency whereas yuc mutants accumulate IPA (2, 4). Furthermore, auxin overproduction phenotypes in YUC overexpression lines are dependent on TAA functions (4). In vitro biochemical assays have demonstrated that YUCs have the ability to catalyze the oxidative decarboxylation of IPA to produce IAA directly (2). Therefore, the TAA/YUC-mediated two-step auxin pathway shown in Fig. 1 is consistent with the available genetic and biochemical evidence.
Although the reaction catalyzed by YUCs in Arabidopsis now appears clear, their biochemical properties or mechanisms have not been explored until the present work. Prior to this study, for example, it was not even clear what cofactors and co-substrates are needed for YUC-catalyzed reactions, although the YUC sequence homology to the mammalian liver FMOs suggests that YUCs are likely to be FMOs. Biochemical characterization of the YUCs, as with some other subclasses of FMOs, has been limited by a series of difficulties in handling either native or recombinantly expressed forms of the proteins (17, 18). In this paper, we show the successful expression in Escherichia coli and the purification to near homogeneity of YUC6, 1 of the 11 YUCs in Arabidopsis. YUC6 was chosen because it was the first YUC protein for which we were able to generate sufficient quantities of stable purified enzyme for detailed analysis. We demonstrate that YUC6 contains oxidized FAD when expressed in and purified from E. coli without adding FAD to the purification buffers. The purified YUC6 was readily reduced by NADPH and showed significant NADPH oxidase activity. Reduced YUC6 reacted with molecular oxygen to form an observable but short lived C4a-(hydro)peroxyflavin intermediate,4 which reacted with IPA to generate IAA. We were able to spectroscopically characterize the oxidized form, the reduced form, and the C4a-(hydro)peroxyflavin intermediate of YUC6. Concurrent analysis of several thousand FMO sequences identified relationships between YUC proteins and the greater FMO superfamily. This work establishes key features of the catalytic mechanism through which YUC6 converts IPA to IAA, providing the final proof for the TAA/YUC-mediated two-step auxin biosynthesis pathway in Arabidopsis. A scheme for the chemical mechanism is also proposed.
EXPERIMENTAL PROCEDURES
Plasmid Construction
The open reading frame of the YUC6 cDNA from Arabidopsis was amplified using the primers (5′-CACCATGGATTTCTGTTGGAAGAG-3′) and (5′-TCAGATTTTTTTTACTTGCTCG-3′). The PCR product was cloned into pENTR/d-TOPO vector (Invitrogen). After verifying the sequences, the YUC6 cDNA was transferred into pET-53-DEST destination vector (Novagen) using LR Clonase II enzyme mix (Invitrogen) to generate pET-53-DEST-YUC6.
Expression and Purification of YUC6
E. coli BL21 Star (DE3) from Invitrogen was transformed with the plasmid pET-53-DEST-YUC6. Cultures were grown in Terrific Broth supplemented with 50 μg/ml ampicillin at 18 °C until the A600 reached 1.0. Isopropyl 1-thio-β-d-galactopyranoside was then added to a final concentration of 1 mm to induce YUC6 expression. The cells were harvested 48 h after induction by centrifugation (7000 × g, 8 min, 4 °C) and cell pellets were stored at −80 °C.
The cell pellet was resuspended in buffer I (50 mm sodium phosphate, pH 8.0, 500 mm NaCl, 30 mm imidazole, 1% Tween 20, and 30% glycerol) using 4 ml of buffer I/g of pellet. Cells were sonicated for 5 min using a Branson ultrasonifier. The cell lysate from a 4-liter culture was centrifuged at 16,000 × g, 30 min, 4 °C. The supernatant was applied to a 5 ml of Ni2+-agarose resin column preequilibrated with buffer II (50 mm sodium phosphate, pH 8.0, 500 mm NaCl, 30 mm imidazole, and 30% glycerol). The column was washed with 12 column volumes of buffer II. YUC6 was eluted with 12 ml of buffer III (50 mm sodium phosphate, pH 8.0, 500 mm NaCl, 300 mm imidazole, and 30% glycerol). The purified YUC6 was quickly frozen in liquid N2 and stored at −80 °C for further use. Protein concentrations were calculated using the molar extinction coefficient of 57,410 at 280 nm (19). FAD concentration was determined using the extinction coefficient of 11,410 m−1 cm−1 at 450 nm. A typical purification of YUC6 from 4-liter culture usually yielded about 10 mg of protein.
Enzymatic Assays
Frozen YUC6 from −80 °C was thawed on ice and changed to a working buffer (50 mm sodium phosphate, pH 8.0, 500 mm NaCl, and 30% glycerol) using a disposable PD-10 desalting column from GE Healthcare. The buffer exchange was conducted according to the procedure provided by GE. For a typical assay, 1 μm YUC6 (based on FAD concentration) was used to initiate the reaction in 350 μl. NADPH oxidation was monitored continuously using an Eppendorf Kinetic BioSpectrometer. Product analyses of IAA and PAA were conducted as described previously (2).
Hydrogen peroxide resulting from the uncoupled YUC6 reactions was determined using the Amplex® Red Hydrogen Peroxide/Peroxidase Assay Kit from Invitrogen. Coupling ratios were measured in the presence of limiting amounts of NADPH while the phenyl pyruvate (PPA) substrate was in excess.
Transient Kinetics of Reductive and Oxidative Reactions
Transient reactions of YUC6 with NADPH and O2 were monitored using a TgK HiTech stopped-flow spectrophotometer with diode array detection at 25 °C. For reactions with NADPH, YUC6 was first made anaerobic in a custom-designed glass tonometer by repeated cycles of vacuum and purging with O2-scrubbed argon gas. NADPH in the same buffer (with and without substrates added) was made anaerobic in a second tonometer by vacuum/argon purging cycles. YUC6 was mixed with NADPH in the stopped-flow instrument, and the reaction was monitored over time.
To study the reaction of reduced YUC6 with oxygen, YUC6 was first made anaerobic and then titrated with anaerobic NADPH to full reduction. Reduction of the flavin was monitored spectroscopically using a cuvette affixed to a side arm of the tonometer. Using the stopped-flow instrument, the reduced YUC6 was mixed with buffer saturated with 100% O2 or saturated with air, and the reactions were subsequently monitored over time.
Informatics Analysis of YUCs and Other FMOs
A representative YUC (NP_171955), a liver microsomal flavin monooxygenase (CAA87633.1), and a siderophore-associated N-hydroxylating monooxygenase (NP_251076.1) were used as seed sequences for the NCBI Blast program to select 11283 homologous protein sequences. Sequences with E-values less than 1e−5 were selected and were filtered via different pairwise sequence identities using CD-Hit. Networks were generated using previously described methodologies (20), and the relationships among these sequences were visualized using the Cytoscape network program. Nodes in the network represent sequences and edges represent Blast E-values. An edge is drawn between two sequences only if the statistical significance of the similarity score between them is less than (i.e. better than) a defined E-value cutoff. The “organic layout” method, which treats two sequences as two masses attached by a spring and E-values as force constants, was used to generate the final groupings.
RESULTS
YUC6 Is an FAD-containing Enzyme
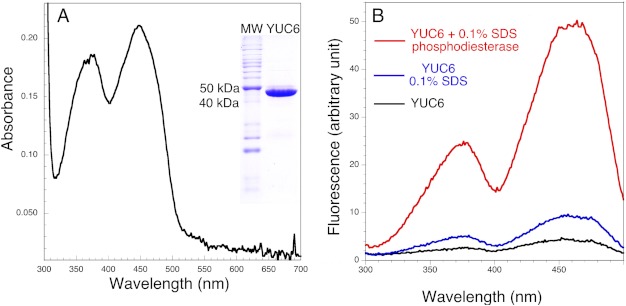
YUC proteins are predicted to be FAD-containing enzymes on the basis of their sequence homology to class B FMOs (21), including mammalian liver microsome FMOs, bacterial and fungal Baeyer-Villiger monooxygenases (BVMOs), and siderophore-associated monooxygenases. However, the proposed YUC-catalyzed reactions (Fig. 1) are very similar to that of lactate 2-monooxygenase, which catalyzes the oxidative decarboxylation of lactate using FMN as a cofactor (22). It was therefore important to establish the cofactor for YUCs. We overexpressed YUC6 with an N-terminal His tag and purified the recombinant YUC6 to near homogeneity (Fig. 2A). The purified YUC was bright yellow with UV-visible absorbance maxima at 448 nm and 376 nm (Fig. 2A). This spectrum resembles those of oxidized FMN and FAD (23), indicating that YUC6 contained oxidized flavin as a cofactor.
FIGURE 2.
YUC6 contains FAD as a cofactor. A, UV-visible spectrum of YUC6, which has absorbance peaks at 448 nm and 376 nm. The inset is a SDS-polyacrylamide image indicating that YUC6 was purified to near homogeneity. B, fluorescence excitation spectra of YUC6 before and after various treatments. Emission was recorded at 525 nm. The black fluorescence spectrum is of the as-isolated His-tagged YUC6. Treatment with 0.1% SDS causes a slight increase in fluorescence (blue spectrum). Phosphodiesterase treatment of denatured (SDS-treated) YUC6 yields the red spectrum, which exhibits a large increase of fluorescence, indicating that the native flavin cofactor is FAD.
A classic method was used to determine whether the flavin cofactor in YUC6 is FAD or FMN (24). The fluorescence of FAD usually is only approximately 10% of that of an equal molar amount of FMN because the ADP moiety in FAD quenches the fluorescence from the flavin ring (24). YUC6 as isolated showed very little fluorescence (Fig. 2B), suggesting that the cofactor is buried in a pocket and quenched by nearby residues. Incubation of YUC6 with 0.1% SDS led to the release of the cofactor from YUC6 with a concomitant more than doubling of fluorescence (Fig. 2B). Addition of a phosphodiesterase, which hydrolyzes FAD to FMN, led to a dramatic increase of fluorescence (Fig. 2B), demonstrating that the cofactor for YUC6 is FAD, not FMN.
HPLC analysis was also used to confirm the identity of the cofactor in YUC6. YUC6 was denatured by adding SDS to 0.1% final concentration. As shown in supplemental Fig. 1, the cofactor had the same retention time as the standard FAD, further demonstrating that YUC6 uses FAD as a bound cofactor. We determined that the extinction coefficient of FAD in YUC6 was 11,410 m−1 cm−1, which is slightly higher than that of free FAD (11,300 m−1 cm−1) (25).
YUC6 Readily Reacts with NADPH
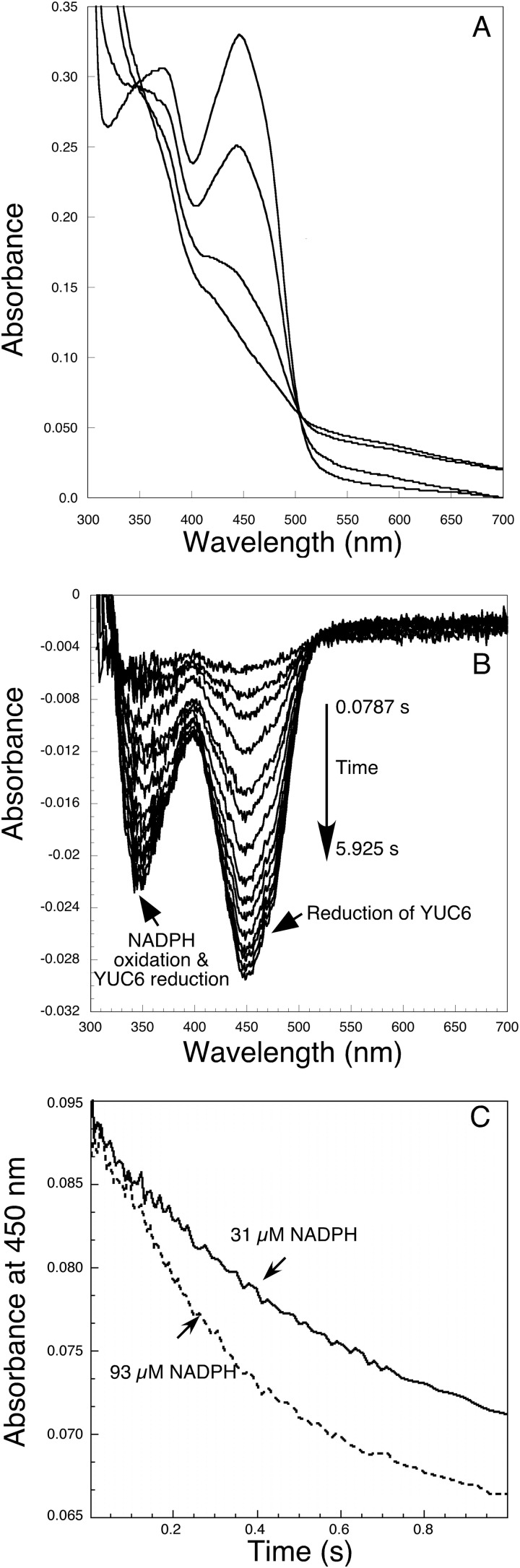
The enzyme-bound FAD cofactor served as an effective tool for analysis of YUC6 activity. When NADPH was titrated into YUC6 anaerobically, YUC6 became reduced (Fig. 3A) as evidenced by the loss of absorbance maxima at 448 nm and 376 nm (Fig. 3A). This was accompanied by a small but significant increase in absorbance at wavelengths longer than 500 nm (Fig. 3A), which could be the charge transfer band caused by bound NADP+. Such charge-transfer complexes are typical for the class B flavin-dependent monooxygenases (21). However, turbidity is also known to cause absorbance increases in the wavelength range. Although the enzyme appeared to be clear, we could not rule out the possibility of turbidity. It is likely that a combination of charge transfer and turbidity contributed to the observed absorbance increase. The reaction of YUC6 with NADPH is essentially irreversible, and complete oxidation of NADPH was observed when [NADPH] was <[YUC6]. The reduced and oxidized YUC6 shared two isosbestic points at 350 nm and 505 nm, consistent with direct interconversion of the oxidized and reduced species (Fig. 3A). NADPH was able to reduce YUC6 regardless of the presence of IPA or oxygen or both (data not shown).
FIGURE 3.
Reduction of FAD in YUC6 by NADPH. A, titration of YUC6 (28 μm) with NADPH under anaerobic conditions. Reduction of YUC6 leads to a decrease of absorbance at 450 nm. The NADPH concentrations for the four spectra are 0, 7, 20, and 28.5 μm. B, reaction after mixing in the stopped-flow instrument equal volumes of NADPH (62 μm) with YUC6 (13 μm), as shown by difference spectra generated by subtracting the first spectrum at 0.00474 s from each successive spectrum. C, kinetics of YUC6 reduction monitored at 450 nm. The final concentrations of NADPH were 31 μm (solid line) and 93 μm (dotted line). YUC6 was 6.5 μm.
We measured the rate of the reduction of YUC6 by NADPH after mixing them anaerobically in the stopped-flow instrument. The reduction of YUC6 was clearly observed as the absorbance at 450 nm decreased over time (Fig. 3B). The decrease in absorbance at 340 nm is due to both flavin reduction and NADPH oxidation (Fig. 3B). The reduction of YUC6 appeared to be biphasic, with the major phase being the faster of the two. A sum of two exponentials fit well to the traces. The faster phase probably represents the reduction of active YUC6, and the second phase may represent the reaction of a small population of less active YUC6. With YUC6 at 6.5 μm, when the final NADPH concentration was 93 μm, the average rate constants from three experiments were: k1 = 2.67 ± 0.04 s−1 and k2 = 0.14 ± 0.02 s−1. When the NADPH concentration was 31 μm, average rate constants were: k1 = 1.50 ± 0.21 s−1 and k2 = 0.12 ± 0.03 s−1. The presence of the substrate PPA (see below) did not alter the overall kinetics of the reduction of YUC6. When YUC6 (13 μm) was mixed with 186 μm NADPH and 500 μm PPA, the rate constants for YUC6 reduction were: k1 = 2.7 s−1 and k2 = 0.03 s−1.
YUC6 Forms a C4a-(hydro)peroxyflavin Intermediate
It is known that some flavin-dependent monooxygenases form fairly stable C4a-(hydro)peroxyflavin intermediates, which are the hydroxylating species for these enzymes. For some of the mammalian liver microsomal FMOs, these C4a-intermediates can persist for hours (17). This property minimizes the uncoupled oxidation of NADPH and the production of H2O2 by these enzymes, which is referred to as oxidase activity. Other flavin-dependent monooxygenases form unstable C4a-intermediates, which can release H2O2. In those hydroxylases for which the C4a-intermediate is only observed to exist for a few milliseconds, rampant NADPH oxidase activity is often minimized by not permitting rapid reduction of the flavin unless substrate is bound. Therefore, the stability of the C4a-(hydro)peroxyflavin reveals mechanistic insights and has implications for how enzyme activities are regulated. Because reduction of YUC proteins by NADPH takes place regardless of the presence of substrates, it was anticipated that like FMOs, YUC enzymes form a detectable (and somewhat stable) C4a-(hydro)peroxyflavin as a key catalytic intermediate.
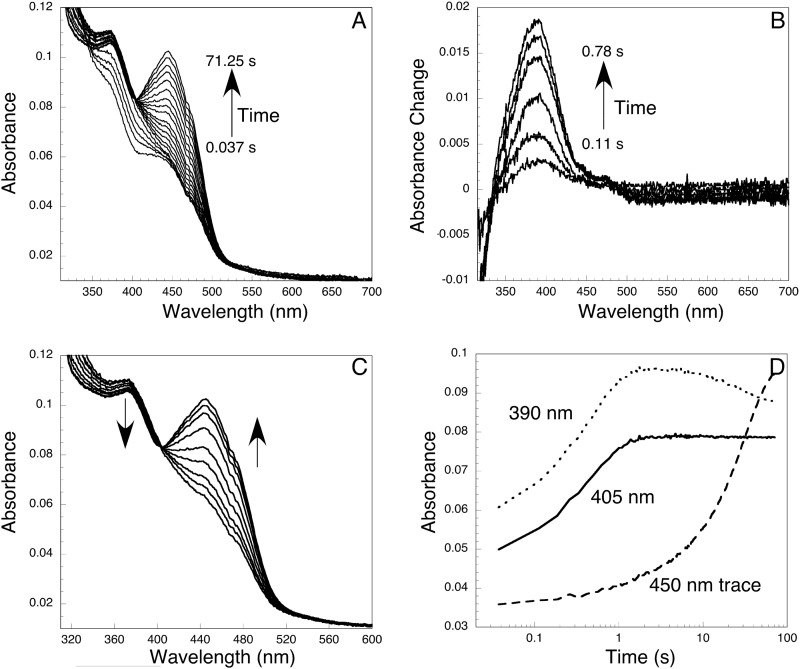
To test this notion, YUC6, stoichiometrically reduced with NADPH, was mixed with oxygen-saturated buffer in the stopped-flow instrument. As shown in Fig. 4A, within the first second after mixing, an intermediate appeared with a peak at ∼381 nm. This intermediate is assigned as the C4a-(hydro)peroxyflavin on the basis of its spectroscopic resemblance to known C4a-(hydro)peroxyl intermediates, which normally have absorbance peaks between 370 and 390 nm (26). The intermediate converted to the FAD (oxidized) form (λmax = 448 nm) over the following ∼70 s. The characteristics of the spectrum of the C4a-intermediate are more clearly shown in the difference spectra generated by subtracting the first spectrum, obtained at 0.037 s (this spectrum corresponds closely to that of fully reduced YUC6), from each subsequent spectrum over the first 0.78 s of the reaction (Fig. 4B). Note also that the absorbance at wavelengths longer than 480 nm decreased slightly during the formation of the intermediate as the charge-transfer interaction between FADH− and NADP was lost (Fig. 4B). An isosbestic point is observed at ∼480 nm (Fig. 4B).
FIGURE 4.
Formation and decomposition of the C4a-(hydro)peroxyflavin intermediate. Equal volumes of reduced YUC6 (18 μm) and oxygen-saturated buffer were mixed in the stopped-flow instrument. A, spectra recorded after mixing reduced YUC6 with oxygen. The first spectrum was taken at 0.037 s, and the last one was taken at 71.25 s. B, formation of the C4a-(hydro)peroxyflavin as shown by an increase of absorbance at 381 nm. These difference spectra, which clarify the appearance of the C4a-(hydro)peroxyflavin, were calculated by subtracting the initial spectrum of reduced YUC6 from each successive spectrum out to 0.78 s. Within the first second, decomposition of the intermediate was minimal. No absorbance change at 450 nm was observed within this period. C, decomposition of C4a-(hydro)peroxyflavin to FAD. Approximately 1 s after mixing, the intermediate started to decompose. Spectra shown were recorded in the time window of 2–4 s. D, kinetics of the formation and decomposition of the C4a-(hydro)peroxyflavin. The formation of the intermediate was evident in the traces recorded at 390 nm and 405 nm. The decomposition was followed at 450 nm. The isosbestic point between C4a-intermediate and oxidized FAD was at 405 nm.
The C4a-intermediate was moderately stable and started to decay within 1 s after mixing the reduced YUC6 with oxygen (Fig. 4C). The conversion of the C4a-intermediate to oxidized FAD exhibited a clear isosbestic point at 405 nm and was completed within 2 min (Fig. 4C). The kinetics of formation and decay of the C4a-(hydro)peroxyflavin can be seen more clearly in the single wavelength traces in Fig. 4D. The absorbance at 390 nm, which increased within the first second and decreased afterward, clearly marks the formation of the C4a-intermediate. The 405 nm trace also shows the formation of the intermediate. After the first second, this trace stayed flat because 405 nm is an isosbestic point for the C4a-intermediate and oxidized FAD (Fig. 4C). The 450 nm trace shows the formation of the oxidized FAD, which did not take place until after the first second (Fig. 4D).
A sum of two exponentials fit the 390 nm trace well. The first phase represents the formation of the C4a-intermediate,and the second phase shows its conversion to oxidized FAD. When reduced YUC6 was rapidly mixed with O2-saturated buffer that contained ∼1 mm dissolved oxygen, the rate constant for the formation of C4a-intermediate was 2.22 s−1 whereas mixing with air-saturated buffer (240 μm O2) gave a rate constant of 0.75 s−1. Using these rate constants, we estimated the second-order rate constant for the formation of C4a-intermediate to be ∼4.5 × 103 m−1 s−1. This value is approximately 1 order of magnitude slower than that for pig liver FMO (27, 28) or for ornithine monooxygenase (25).
The kinetics of C4a-intermediate decay as followed at 450 nm (Fig. 4D) required two exponentials to fit the 450-m trace well. However, The first phase with a rate constant of 2.17 s−1 only accounted for 5% of the absorbance change and could be due to a small fraction of less active enzyme. The second phase with a rate constant of 0.035 s−1 represented 95% of the absorbance change and corresponds to a half-life for the C4a-intermediate of ∼20 s (Fig. 4D).
Reaction of C4a-(hydro)peroxyflavin with IPA
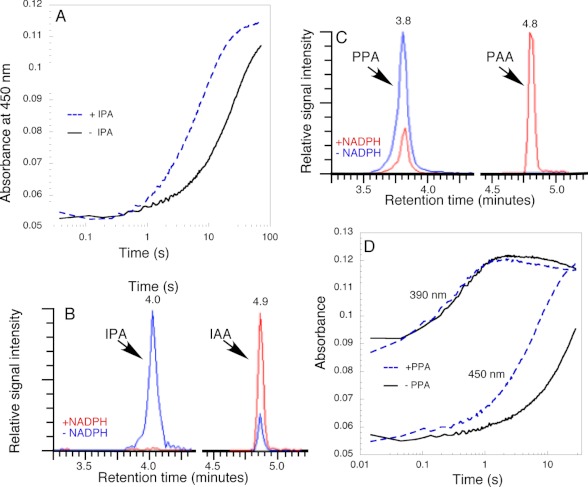
In the absence of the substrate IPA, the C4a-(hydro)peroxyflavin decayed into oxidized FAD and hydrogen peroxide. In the presence of IPA (250 μm) the rate of conversion of the C4a-intermediate to FAD was stimulated (Fig. 5A) with a rate constant for return to oxidized FAD of ∼ 0.14 s−1, approximately 4-fold faster than in the absence of substrate (see above). Product analysis using HPLC and LC-MS confirmed the conversion of IPA to IAA (Fig. 5B). In the absence of NADPH and in the presence of YUC6, a small amount IPA was converted into IAA nonenzymatically. IPA is known for its instability and spontaneous conversion to IAA (29). When NADPH was added to the reaction, a substantially larger fraction of the IPA was converted into IAA (Fig. 5B).
FIGURE 5.
Reaction of C4a-(hydro)peroxyflavin with IPA and PPA. A, comparison of the conversion of C4a-intermediate to oxidized FAD with and without 250 μm IPA. Other conditions are the same as in Fig. 4A. B, HPLC analysis of the products generated by the reaction of YUC6 with IPA. IPA is converted to IAA in the presence of NADPH. Note that there was a small amount nonenzymatic conversion of IPA to IAA in the absence of NADPH. C, YUC6 catalyzing the conversion of PPA to PAA. Note that the reaction is cleaner than that with IPA, and no nonenzymatic conversion of PPA to PAA is observed. D, comparison of the formation and decomposition of the C4a-intermediate with and without the substrate PPA (250 μm). Other conditions are the same as in Fig. 4A. The formation of the C4a-intermediate was followed at the first part of the 390-nm traces. Note that the substrate PPA did not affect the formation of the C4a-intermediate. The decomposition of the C4a-intermediate was followed at 450 nm. Note that PPA substantially accelerated the rate of conversion of the C4a-intermediate to FAD.
YUC6 Can Also Catalyze the Conversion of PPA to Phenyl Acetic Acid (PAA)
YUC6 was examined for its ability to use other α-keto acids as a substrate. As shown in Fig. 5C, YUC6 catalyzed the NADPH-dependent conversion of PPA to PAA. In the presence of PPA, the C4a-(hydro)peroxyflavin intermediate was also observed (supplemental Fig. 2). However, PPA did not affect the rates of the formation of the C4a-intermediate, as shown by the 390-nm traces (Fig. 5D), but it did accelerate the oxidation of C4a-(hydro)peroxyflavin to FAD (Fig. 5D). In the presence of 250 μm PPA the conversion of the C4a-(hydro)peroxyflavin to FAD was ∼0.18 s−1, or approximately 5-fold faster than in the absence of substrate. In the absence of PPA, there was no decay of the C4a-intermediate within 0.78 s as indicated by the lack of absorbance change at 450 nm (Fig. 4B). However, a clear shoulder at 450 nm was observed in the difference spectrum at 0.76 s when PPA was present because PPA stimulates the decomposition of the C4a-intermediate (supplemental Fig. 2).
Steady-state Kinetics of YUC6 using PPA as a Substrate
Because PPA is much more stable in aqueous solution than IPA, PPA is a more reliable substrate for in vitro assays of YUC activity. Moreover, IPA has significant absorbance at 340 nm with an extinction coefficient of 4770 m−1cm−1 and thus can interfere with measurements of NADPH, whereas PPA has no significant absorbance above 300 nm (30). In the absence of PPA and at saturating concentrations of NADPH, YUC6 displayed basal NADPH oxidase activity with a turnover number of 0.04 s−1. When PPA was used as a substrate, the kcat for oxidation of NADPH was 0.31 s−1, which is approximately 8-fold more active than the basal NADPH oxidase activity. In the presence of PPA, the Km for NADPH was measured at 26 μm, and Km for PPA at 43 μm. In the presence of PPA, there was a small amount of uncoupled reaction that led to the production of hydrogen peroxide. The uncoupling ratio was measured at 3.8% ± 0.2% at room temperature.
Global Sequence Analysis Shows That YUC Proteins Are Most Closely Related to Liver FMOs
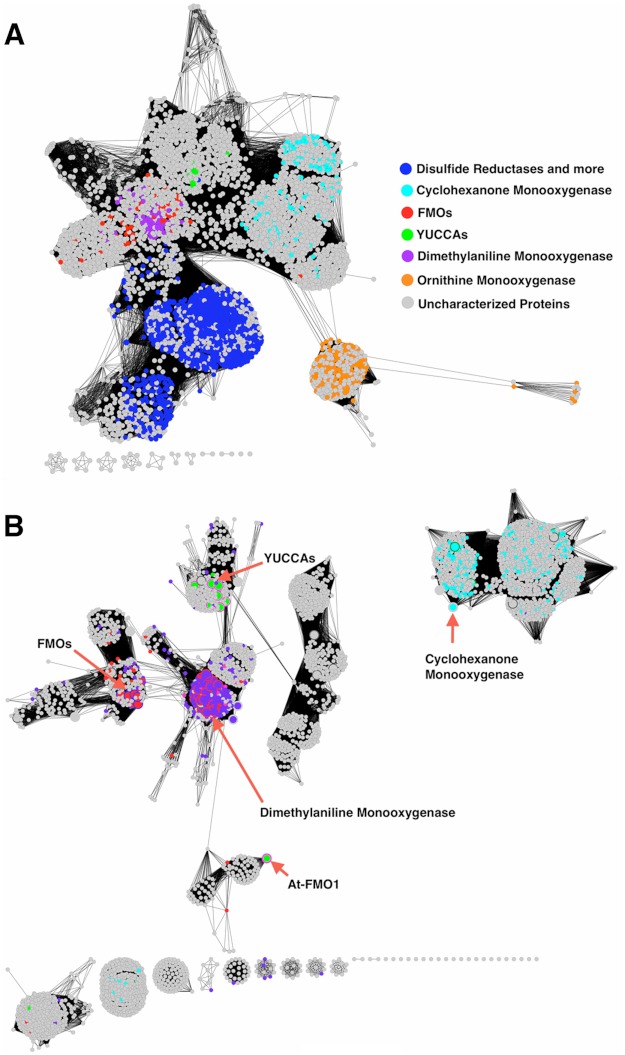
Pairwise alignments have indicated homology between YUC proteins and each of the various groups of class B FMOs named above. To gain a more sophisticated view of the relationships between YUC and its sequence relatives, the 11 YUC sequences from Arabidopsis were subjected to a sequence similarity network informatics analysis using the Cytoscape program. The approach uses sequence similarity (e.g. NCBI Blast E-values) among multiple sequences as a matrix to group them. The output is analogous to a dendrogram (a tree without distances); however, unlike a tree diagram, which is limited in complexity, the sequence similarity network allows thousands of sequences to be considered at a time and connects nearest neighbor relatives. Different types of relationships among the input sequences emerge depending on the stringency of the similarity threshold used. Because new enzymatic functions evolve through gene duplication followed by mutations, functions that are more similar to the parental function require fewer mutations and consequently retain a higher degree of mutual sequence similarity. Groups generated at very stringent confidence cutoffs where only highly similar sequences are grouped together therefore generally represent one function.
Fig. 6A shows the sequence similarity network generated at an E-value of 1e−10. At this significance level, YUC sequences cluster together with several other groups of flavoenzymes, including liver FMOs, BVMOs, siderophore-associated NMOs, and even bacterial flavin-dependent reductases. Biochemically characterized FMOs are labeled on Fig. 6 and are used to indicate the likely locations of the different sequence subgroups. As the stringency is raised, the group of siderophore-associated NMOs separates from the YUCs even before the reductases (data not shown), suggesting that some aspect of the YUC function is more similar to reductases, even though the siderophore-associated NMOs have functionally more in common with the YUCs. Fig. 6B shows the sequence similarity network of the group that includes representative FMOs, YUCs, and BVMOs generated at an E-value of 1e−45. The network at this level of stringency shows that the YUC proteins are much more closely related to liver FMOs and less closely related to BVMOs despite having a predicted mechanism more closely allied to the latter (see below). The YUC paralog At-FMO1 is particularly distant from the YUC group. This protein is expected to be involved in host-pathogen interaction and not in auxin biosynthesis.
FIGURE 6.
Sequence similarity networks showing relationships of YUCs to their homologs. The names of protein classes are as shown in the legend and colored as annotated in the Uniprot database. Nodes in gray color are proteins with unknown or un-annotated functions. The 11 YUCCA proteins from Arabidopsis form a cluster at the center of the network. Cyclohexanone monooxygenases are representative BVMOs. Dimethylaniline monooxygenase is encoded by the mammalian liver fmo3 gene. Ornithine monooxygenase is a representative siderophore-associated N-monooxygenase. The islands at the bottom of each panel represent small, isolated clusters of distantly related sequences. A, the network was generated with a low stringency E-value cutoff at 1e−10. At this level of stringency, YUCs are connected to several flavoprotein families including reductases. A total of 4306 sequences with <70% pairwise sequence identity are shown. B, the network generated at a significantly more stringent E-value cutoff of 1e−45 (4188 sequences) includes the sequences shown in the red brackets from A. As the stringency was incrementally raised from 1e−10, the ornithine monooxygenase cluster separated first from the network (less closely related to the YUCs), followed by the reductase cluster (more closely related). The lack of connections between the YUCs and cyclohexanone monooxygenases indicates that they are more distantly related than the YUCs and liver FMOs. The large nodes with red borders in this figure are experimentally characterized proteins.
DISCUSSION
Monooxygenases catalyze the insertion of an oxygen atom into an organic compound as part of the biosynthesis of many physiologically important molecules, the degradation of a large variety of aromatic and heteroatom-containing compounds and the detoxification of xenobiotics (31). The family of flavin-dependent monooxygenases is one of the largest groups of monooxygenase that has been identified. Plants appear to have a greatly expanded family of FMOs (32). For example, the Arabidopsis genome has 29 genes encoding proteins with significant homology to the well characterized mammalian liver microsome FMOs. In contrast, humans only have five well defined FMOs. Although flavin monooxygenases are identified in all sequenced plant genomes, until now no plant FMO has been biochemically characterized. Unlike their mammalian counterparts that mainly function in detoxification, at least some of the plant flavin monooxygenases are involved in the biosynthesis of signaling molecules. Eleven of the 29 putative flavin-dependent monooxygenases in Arabidopsis belong to the YUC family and have been shown genetically to participate in the biosynthesis of the essential hormone auxin (7, 8). In this paper, an initial kinetic and spectroscopic characterization is presented for YUC6, which synthesizes auxin in pollen and other tissues (7). The oxidized, the reduced, and the catalytic intermediate forms of YUC6 are identified, providing a foundation for biochemically analyzing other YUC family members and flavin monooxygenases from a variety of plant species.
A major obstacle in characterizing plant flavin monooxygenases to date has been obtaining sufficient quantities of active enzyme. In earlier studies, YUC proteins expressed in and purified from E. coli lacked color, indicating that the recombinant YUC protein did not contain the FAD cofactor and that the YUC protein was probably not folded correctly. Early biochemical assays of YUCs required the addition of free FAD to the reaction mixture, probably causing off-enzyme reactions. Recently, we managed to purify a small amount of GST-tagged YUC2 to demonstrate that YUC2 had the capacity to convert IPA to IAA in vitro (2). However, the amount of GST-YUC2 was insufficient for detailed biochemical studies. The His-tagged YUC6 has now been successfully expressed in E. coli. Growing E. coli at low temperature and including a high concentration of glycerol (30%) in purification buffers has enabled us to obtain useful quantities of stable recombinant YUC6. Purified YUC6 contains good quantities of its FAD cofactor (Fig. 2) and displays both NADPH oxidase activity and oxidative decarboxylase activity against IPA and PPA α-keto acids in vitro (Fig. 5). The bound FAD cofactor in YUC6 allowed us to use transient kinetic methods to track crucial catalytic intermediates in the YUC6 reactions.
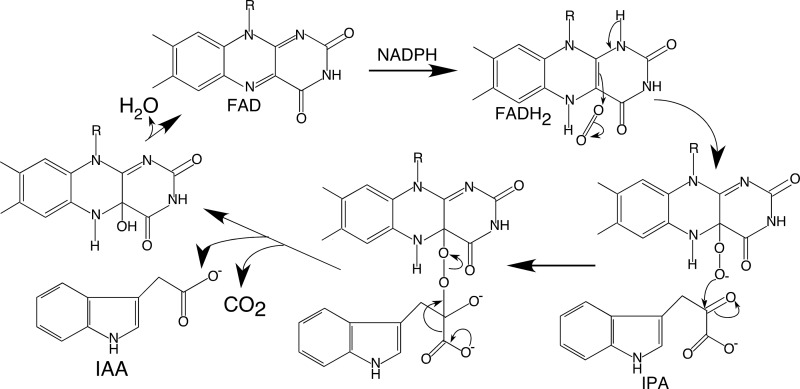
Based on sequence homology, YUCs clearly belong to the group B flavin monooxygenase family (21), which includes the mammalian liver FMOs, the siderophore-associated FMOs, and the flavin-containing BVMOs (Fig. 6). The mammalian liver FMOs and the siderophore-associated FMOs utilize the hydroperoxy intermediate to catalyze N-hydroxylations, whereas the BVMOs utilize the peroxyl intermediate to catalyze the transfer of an oxygen atom to an electrophilic carbon center (33). The YUC-catalyzed reaction (Fig. 7) appears to be more similar to the BVMO-catalyzed oxidation of a ketone into an ester because in both cases the substrate is an electrophile (34). Thus, the oxygenating intermediate is likely to be the nucleophilic C4a-peroxyflavin (35). In the case of the BVMO enzymes, attack of the C4a-peroxyflavin intermediate on the ketone generates a Criegee complex, which undergoes rearrangements to yield the final ester product. In the YUC-catalyzed reaction, the postulated first step in the conversion of IPA to IAA is also a nucleophilic attack at the ketone group of IPA (Fig. 7). In contrast, the N-hydroxylating flavin monooxygenases use the electrophilic C4a-hydroperoxyflavin as an agent. The comprehensive, nearest neighbor sequence similarity analysis (see above) showed the strongest connection between YUCs and the liver FMOs (Fig. 6). This is despite the fact that the physiological function of YUC is to catalyze oxidative decarboxylation of α-keto acids rather than N-hydroxylation of a substrate. It is still not clear what characteristics of the proteins lead to the mechanistic differences between YUCs and N-hydroxylating monooxygenases.
FIGURE 7.
Catalytic mechanism of YUC flavin monooxygenases. The YUC catalyzed reaction can be divided into a reductive half-reaction and an oxidative half-reaction. The FAD cofactor in YUC6 is first reduced by NADPH, and then it reacts with oxygen to form the C4a-(hydro)peroxy FAD. The C4a-intermediate reacts with IPA to produce IAA.
We also noticed that, although spectroscopically observable, the C4a-(hydro)peroxyflavin in YUC6 was much less stable than the counterpart intermediate in N-hydroxylating monooxygenases in the absence of substrates. The half-life (t½) of the C4a-intermediate of YUC6 was about 20 s in the absence of IPA whereas the C4a-(hydro)peroxyflavin of N-hydroxylating monooxygenases usually have t½ values of >30 min (25). However, some of the measured half-lives for the C4a-intermediates of BVMOs are similar to that of YUC6. For example, in the absence of the substrate, the C4a-peroxy form of phenylacetone monooxygenase from Thermobifida fusca decays at 0.01 s−1 (t½ = 69.3 s) to form the oxidized enzyme and hydrogen peroxide (36). This behavior is consistent with our hypothesis that YUC6 uses a mechanism similar to that of BVMO.
Mammalian FMOs have broad substrate tolerance whereas the siderophore-associated N-hydroxylating monooxygenases are very specific (17, 25). YUC6 can use either PPA or IPA as in vitro substrates, suggesting that YUCs do not have strict substrate specificity. However, YUC6 did not catalyze the decarboxylation reaction of benzoylformic acid, which does not have the methylene group in the side chain common to IPA and PPA (data not shown). It will be interesting to test whether other physiological α-keto acids such as pyruvate and α-ketoglutarate can serve as YUC substrates in vitro. It is not clear whether the conversion of PPA to PAA by YUC6 has any physiological significance. However, we recognize that PAA has auxin activities when applied to plants.
In summary, we clearly show that YUC6 catalyzes the oxidative decarboxylation of α-keto acids including IPA and PPA. Furthermore, we present initial spectroscopic and kinetic analyses of YUC6, establishing key details of the catalytic mechanism for the YUC family flavin-dependent monooxygenases (Fig. 7). This first biochemical characterization of a flavin-dependent monooxygenase involved in auxin biosynthesis in plants provides a framework for understanding other flavin monooxygenases in plants. YUC6 displays kinetic properties that are distinct from N-hydroxylating flavin monooxygenases, despite the strong sequence similarities between them. Rather, YUCs are more similar to BVMOs in terms of catalytic mechanisms. Therefore, studies on the catalytic mechanisms of YUCs not only help us to understand auxin biosynthesis, but also to gain insights into how the various activities of flavin-dependent monooxygenases evolved and are regulated. Future work will examine how these differences in catalytic behavior map onto the differences in their sequences and structures and how YUC activities may be regulated in plants.
Acknowledgments
We thank Drs. Zuyu Zheng, Barrie Entsch, and Yongxia Guo and the members of the Zhao laboratory for comments on this manuscript.
This work was supported, in whole or in part, by National Institutes of Health Grant R01GM68631 (to Y. Z.). This work was also supported by Japan Society for the Promotion of Science KAKENHI Grant 24370027 (to H. K.).

This article contains supplemental Figs. 1 and 2.
Mechanistically, flavin-dependent monooxygenases use either a C4a-hydroperoxyflavin or a C4a-peroxyflavin as the species that carries out the oxygenation reaction. It is generally difficult to distinguish these two forms spectroscopically, so we have used C4a-(hydro)peroxyflavin to indicate our uncertainty about the actual form of the intermediate in YUC6.
- IAA
- indole-3-acetic acid
- BVMO
- Baeyer-Villiger monooxygenase
- FMO
- flavin-containing monooxygenase
- IPA
- indole-3-pyruvate
- PAA
- phenyl acetic acid
- PPA
- phenyl pyruvate
- YUC
- YUCCA.
REFERENCES
- 1. Zhao Y. (2010) Auxin biosynthesis and its role in plant development. Annu. Rev. Plant Biol. 61, 49–64 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Mashiguchi K., Tanaka K., Sakai T., Sugawara S., Kawaide H., Natsume M., Hanada A., Yaeno T., Shirasu K., Yao H., McSteen P., Zhao Y., Hayashi K., Kamiya Y., Kasahara H. (2011) The main auxin biosynthesis pathway in Arabidopsis. Proc. Natl. Acad. Sci. U.S.A. 108, 18512–18517 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Stepanova A. N., Yun J., Robles L. M., Novak O., He W., Guo H., Ljung K., Alonso J. M. (2011) The Arabidopsis YUCCA1 flavin monooxygenase functions in the indole-3-pyruvic acid branch of auxin biosynthesis. Plant Cell 23, 3961–3973 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Won C., Shen X., Mashiguchi K., Zheng Z., Dai X., Cheng Y., Kasahara H., Kamiya Y., Chory J., Zhao Y. (2011) Conversion of tryptophan to indole-3-acetic acid by tryptophan aminotransferases of Arabidopsis and YUCCAs in Arabidopsis. Proc. Natl. Acad. Sci. U.S.A. 108, 18518–18523 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Zhao Y. (2012) Auxin biosynthesis: a simple two-step pathway converts tryptophan to indole-3-acetic acid in plants. Mol. Plant 5, 334–338 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Tao Y., Ferrer J. L., Ljung K., Pojer F., Hong F., Long J. A., Li L., Moreno J. E., Bowman M. E., Ivans L. J., Cheng Y., Lim J., Zhao Y., Ballaré C. L., Sandberg G., Noel J. P., Chory J. (2008) Rapid synthesis of auxin via a new tryptophan-dependent pathway is required for shade avoidance in plants. Cell 133, 164–176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Cheng Y., Dai X., Zhao Y. (2006) Auxin biosynthesis by the YUCCA flavin monooxygenases controls the formation of floral organs and vascular tissues in Arabidopsis. Genes Dev. 20, 1790–1799 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Cheng Y., Dai X., Zhao Y. (2007) Auxin synthesized by the YUCCA flavin monooxygenases is essential for embryogenesis and leaf formation in Arabidopsis. Plant Cell 19, 2430–2439 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Zhao Y., Christensen S. K., Fankhauser C., Cashman J. R., Cohen J. D., Weigel D., Chory J. (2001) A role for flavin monooxygenase-like enzymes in auxin biosynthesis. Science 291, 306–309 [DOI] [PubMed] [Google Scholar]
- 10. Tobeña-Santamaria R., Bliek M., Ljung K., Sandberg G., Mol J. N., Souer E., Koes R. (2002) FLOOZY of petunia is a flavin monooxygenase-like protein required for the specification of leaf and flower architecture. Genes Dev. 16, 753–763 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Yamamoto Y., Kamiya N., Morinaka Y., Matsuoka M., Sazuka T. (2007) Auxin biosynthesis by the YUCCA genes in rice. Plant Physiol. 143, 1362–1371 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Woo Y. M., Park H. J., Su'udi M., Yang J. I., Park J. J., Back K., Park Y. M., An G. (2007) Constitutively wilted 1, a member of the rice YUCCA gene family, is required for maintaining water homeostasis and an appropriate root to shoot ratio. Plant Mol. Biol. 65, 125–136 [DOI] [PubMed] [Google Scholar]
- 13. Gallavotti A., Barazesh S., Malcomber S., Hall D., Jackson D., Schmidt R. J., McSteen P. (2008) sparse inflorescence1 encodes a monocot-specific YUCCA-like gene required for vegetative and reproductive development in maize. Proc. Natl. Acad. Sci. U.S.A. 105, 15196–15201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Exposito-Rodrıguez M., Borges A. A., Borges-Perez A. B., Hernandez M., Perez J. A. (2007) Cloning and biochemical characterization of ToFZY, a tomato gene encoding a flavin monooxygenase involved in a Tryptophan-dependent auxin biosynthesis pathway. J. Plant Growth Regul. 26, 329–340 [Google Scholar]
- 15. Kim J. I., Sharkhuu A., Jin J. B., Li P., Jeong J. C., Baek D., Lee S. Y., Blakeslee J. J., Murphy A. S., Bohnert H. J., Hasegawa P. M., Yun D. J., Bressan R. A. (2007) yucca6, a dominant mutation in Arabidopsis, affects auxin accumulation and auxin-related phenotypes. Plant Physiol. 145, 722–735 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. LeClere S., Schmelz E. A., Chourey P. S. (2010) Sugar levels regulate tryptophan-dependent auxin biosynthesis in developing maize kernels. Plant Physiol. 153, 306–318 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Ziegler D. M. (1990) Flavin-containing monooxygenases: enzymes adapted for multisubstrate specificity. Trends Pharmacol. Sci. 11, 321–324 [DOI] [PubMed] [Google Scholar]
- 18. Ziegler D. M. (2002) An overview of the mechanism, substrate specificities, and structure of FMOs. Drug Metab. Rev. 34, 503–511 [DOI] [PubMed] [Google Scholar]
- 19. Gasteiger E., Gattiker A., Hoogland C., Ivanyi I., Appel R. D., Bairoch A. (2003) ExPASy: the proteomics server for in-depth protein knowledge and analysis. Nucleic Acids Res. 31, 3784–3788 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Atkinson H. J., Morris J. H., Ferrin T. E., Babbitt P. C. (2009) Using sequence similarity networks for visualization of relationships across diverse protein superfamilies. PLoS One 4, e4345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. van Berkel W. J., Kamerbeek N. M., Fraaije M. W. (2006) Flavoprotein monooxygenases, a diverse class of oxidative biocatalysts. J. Biotechnol. 124, 670–689 [DOI] [PubMed] [Google Scholar]
- 22. Müh U., Williams C. H., Jr., Massey V. (1994) Lactate monooxygenase. II. Site-directed mutagenesis of the postulated active site base histidine 290. J. Biol. Chem. 269, 7989–7993 [PubMed] [Google Scholar]
- 23. Ooi T., Ogata D., Matsumoto K. Ä., Yu J., Yao M., Kitamura M., Taguchi S. (2012) Flavin-binding of azoreductase: Direct evidences for dual-binding property of apo-azoreductase with FMN and FAD. J. Mol. Catalysis B Enzymatic 74, 204–208 [Google Scholar]
- 24. Bessey O. A., Lowry O. H., Love R. H. (1949) The fluorometric measurement of the nucleotides of riboflavin and their concentration in tissues. J. Biol. Chem. 180, 755–769 [PubMed] [Google Scholar]
- 25. Mayfield J. A., Frederick R. E., Streit B. R., Wencewicz T. A., Ballou D. P., DuBois J. L. (2010) Comprehensive spectroscopic, steady state, and transient kinetic studies of a representative siderophore-associated flavin monooxygenase. J. Biol. Chem. 285, 30375–30388 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Ballou D. P., Entsch B., Cole L. J. (2005) Dynamics involved in catalysis by single-component and two-component flavin-dependent aromatic hydroxylases. Biochem. Biophys. Res. Commun. 338, 590–598 [DOI] [PubMed] [Google Scholar]
- 27. Beaty N. B., Ballou D. P. (1981) The oxidative half-reaction of liver microsomal FAD-containing monooxygenase. J. Biol. Chem. 256, 4619–4625 [PubMed] [Google Scholar]
- 28. Beaty N. B., Ballou D. P. (1981) The reductive half-reaction of liver microsomal FAD-containing monooxygenase. J. Biol. Chem. 256, 4611–4618 [PubMed] [Google Scholar]
- 29. Bentley J. A., Farrar K. R., Housley S., Smith G. F., Taylor W. C. (1956) Some chemical and physiological properties of 3-indolylpyruvic acid. Biochem. J. 64, 44–49 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Jean M., DeMoss R. D. (1968) Indolelactate dehydrogenase from Clostridium sporogenes. Can J. Microbiol. 14, 429–435 [DOI] [PubMed] [Google Scholar]
- 31. Cashman J. R., Zhang J. (2006) Human flavin-containing monooxygenases. Annu. Rev. Pharmacol. Toxicol. 46, 65–100 [DOI] [PubMed] [Google Scholar]
- 32. Schlaich N. L. (2007) Flavin-containing monooxygenases in plants: looking beyond detox. Trends Plant Sci. 12, 412–418 [DOI] [PubMed] [Google Scholar]
- 33. Palfey B. A., McDonald C. A. (2010) Control of catalysis in flavin-dependent monooxygenases. Arch. Biochem. Biophys. 493, 26–36 [DOI] [PubMed] [Google Scholar]
- 34. Walsh C. T., Chen Y. C. (1988) Enzymic Baeyer-Villiger oxidations by Flavin-dependent monooxygenases. Angew. Chem. Int. Ed. Engl. 27, 333–343 [Google Scholar]
- 35. Sheng D., Ballou D. P., Massey V. (2001) Mechanistic studies of cyclohexanone monooxygenase: chemical properties of intermediates involved in catalysis. Biochemistry 40, 11156–11167 [DOI] [PubMed] [Google Scholar]
- 36. Torres Pazmiño D. E., Baas B. J., Janssen D. B., Fraaije M. W. (2008) Kinetic mechanism of phenylacetone monooxygenase from Thermobifida fusca. Biochemistry 47, 4082–4093 [DOI] [PubMed] [Google Scholar]