Background: The incorporation of CENP-A at centromeres is important for chromosome segregation during mitosis.
Results: And-1 together with HJURP regulates the assembly of new CENP-A onto centromeres.
Conclusion: And-1 facilitates the recruitment of CENP-A to centromeres.
Significance: These studies reveal a novel role of And-1 in the regulation of chromosome congression during mitosis.
Keywords: Cell Cycle, Centromeres, Chromatin, Histones, Mitosis, And-1, CENP-A, CTF4, HJURP, WDHD1
Abstract
The centromere is an epigenetically designated chromatin domain that is essential for the accurate segregation of chromosomes during mitosis. The incorporation of centromere protein A (CENP-A) into chromatin is fundamental in defining the centromeric loci. Newly synthesized CENP-A is loaded at centromeres in early G1 phase by the CENP-A-specific histone chaperone Holliday junction recognition protein (HJURP) coupled with other chromatin assembly factors. However, it is unknown whether there are additional HJURP-interacting factor(s) involving in this process. Here we identify acidic nucleoplasmic DNA-binding protein 1 (And-1) as a new factor that is required for the assembly of CENP-A nucleosomes. And-1 interacts with both CENP-A and HJURP in a prenucleosomal complex, and the association of And-1 with CENP-A is increased during the cell cycle transition from mitosis to G1 phase. And-1 down-regulation significantly compromises chromosome congression and the deposition of HJURP-CENP-A complexes at centromeres. Consistently, overexpression of And-1 enhances the assembly of CENP-A at centromeres. We conclude that And-1 is an important factor that functions together with HJURP to facilitate the cell cycle-specific recruitment of CENP-A to centromeres.
Introduction
Accurate segregation of chromosomes to each daughter cell during mitosis relies on a specialized chromatin domain called the centromere. The centromere serves as a platform for the assembly of the large multiprotein kinetochore complex that mediates chromosome attachment to spindle microtubules and regulates the spindle assembly checkpoint (1, 2). With the exception of budding yeast, where centromeres are sequence-specific, the mechanisms governing centromere specification in other eukaryotes remain elusive but appear to be epigenetic in nature (3, 4).
In all eukaryotes, a hallmark of centromeric chromatin is the incorporation of the histone H3 variant, CENP-A3 (1, 4). CENP-A-containing nucleosomes define the site of kinetochore assembly and are interspersed with canonical histone H3-containing nucleosomes (3, 5). Although constitutively present at the centromeres throughout the cell cycle, parental CENP-A is diluted after every S phase by DNA replication. Unlike canonical histones, deposition of new CENP-A onto centromeric chromatin occurs during early G1 phase (6, 7). The deposition of CENP-A at centromeres appears to be a two-part process composed of chromatin “priming” in late anaphase that prepares chromosomes to receive CENP-A followed by the recruitment of newly synthesized CENP-A to centromeres via a complex process involving multiple proteins including histone chaperones at early G1 phase (8). Chromatin priming is carried out by several proteins including the human Mis18 complex, which may prepare chromatin for CENP-A deposition by altering histone acetylation (9, 10). After centromere priming, CENP-A is loaded onto centromeres through the chaperone protein HJURP in collaboration with other chromatin assembly factors (RbAp46/48 and Npm1) (11–13). Once loaded, CENP-A is maintained at centromeres by the chromatin remodeler RSF1 and the small GTPase regulator MgcRacGAP (14, 15). Although HJURP acts as a key factor in depositing new CENP-A at centromeres, the mechanism by which HJURP is recruited to centromeric chromatin is unclear and may require interacting partners to target the HJURP-CENP-A complex.
And-1 is an acidic nucleoplasmic DNA-binding protein containing an amino-terminal WD40 domain and a carboxyl-terminal HMG motif (16). We and others have recently shown that And-1 acts as a component of the replisome to regulate DNA replication and S phase progression (16–19). Ctf4, the homolog of And-1 in Saccharomyces cerevisiae, was originally identified as a critical gene for chromosome transmission fidelity (20). Loss of Ctf4 causes a G2/M phase accumulation with elevated levels of chromosome loss and chromosome segregation defects (20, 21), suggesting a possible role of Ctf4 in centromeric chromatin. Indeed, mcl1, the homolog of And-1 in Schizosaccharomyces pombe, was shown to be required for the centromeric localization of Cnp1 (CENP-A ortholog) (22). However, how And-1/Ctf4/mcl1 regulates CENP-A/Cnp1 centromeric localization remains unknown.
Here, we report that And-1 is required for the centromere-specific deposition of new CENP-A in early G1 phase. Down-regulation of And-1 results in the accumulation of cells in early stages of mitosis with chromosome congression defects. And-1 interacts with both CENP-A and HJURP in chromatin-free extracts and is required for the centromeric localization of both CENP-A and HJURP. Consistently, overexpression of And-1 enhances the assembly of CENP-A at centromeres. Thus, And-1 is a new HJURP-CENP-A-interacting partner that is required for the assembly of new CENP-A at centromeres.
EXPERIMENTAL PROCEDURES
Immunofluorescence
Cells attached to coverslips were fixed with 4% paraformaldehyde in PBS for 10 min at room temperature, permeabilized in 0.2% Triton X-100 in PBS, and rinsed three times with PBS + 0.02% Tween 20. Alternatively, some cells were preextracted with 0.3% Triton X-100 in CSK buffer (100 mm NaCl, 300 mm sucrose, 3 mm MgCl2, 10 mm PIPES, pH 7.0). Cells were then blocked in 3% BSA in PBS and primary antibody incubations carried out in PBS + 3% BSA for 1 h at room temperature, except CENP-A primary antibody was incubated overnight at 4 °C. Afterward, the cells were washed 3 × 5 min with 0.02% Tween 20 in 1× PBS and incubated in secondary antibody (anti-rabbit Alexa Fluor 594 and anti-mouse Alexa Fluor 488, 1:1000) for 45 min. Cells were washed 3 × 5 min with 0.02% Tween 20 in 1× PBS, and the coverslips were mounted in VectaShield (Vector Laboratories) containing DAPI. Slides were imaged at room temperature using a Nikon Eclipse 80i microscope and NIS-Elements AR software. Alternatively, images were collected using a Zeiss 710 LSM confocal microscope with a 63 × 1.4 oil immersion objective and z sections acquired at 0.2-μm intervals. The intensity of CENP-A at centromeres was analyzed as described previously (14).
Immunoprecipitation
For assays involving immunoprecipitation of proteins from chromatin-free extracts, cells were harvested, washed with PBS, resuspended in 400 μl of solution A (10 mm HEPES, pH 7.9, 10 mm KCl, 1.5 mm MgCl2, 0.34 m sucrose, 10% glycerol, 1 mm DTT, 10 mm NaF, 1 mm Na2VO3, protease inhibitors, and 0.05% Nonidet P-40), and incubated on ice for 5 min. Soluble proteins were separated from nuclei by centrifugation at 1300 × g for 4 min and the resulting supernatant collected (chromatin-free extract). The pellet was washed once with solution A and the resulting supernatant collected and combined with the first collection. The samples were centrifuged at 13,000 × rpm for 10 min, and the supernatants were incubated with anti-FLAG-conjugated agarose beads for 2 h. The beads were washed three times with solution A and associated proteins were eluted with SDS loading buffer.
Cell Culture, Synchronization, and Transfection
HCT116, U2OS, and 293T cells were grown in DMEM supplemented with 10% FBS at 37 °C in 5% CO2 supply. HCT116 and U2OS cells expressing FLAG-And-1 or FLAG were constructed by infecting cells with retrovirus expressing FLAG or FLAG-And-1, followed by single colony selection. Cell cycle synchronization was achieved by treating cells with 100 ng/ml nocodazole for 16 h, washed three times in PBS, and then released into medium. siRNA oligonucleotides And-1-1 and And-1-2 were as described previously (16). siRNA transfections were performed with 100 nm siRNA oligonucleotide duplexes using Lipofectamine RNAiMAX (Invitrogen) according to the manufacturer's instructions.
Antibodies
Mouse anti-CENP-A and rabbit anti CENP-B were from Abcam. Rabbit anti-And-1 was described previously (23). Mouse anti-YFP/GFP was from Clontech. Rabbit anti-HJURP was raised as described (12). The secondary antibodies anti-rabbit Alexa Fluor 594 and anti-mouse Alexa Fluor 488 were from Invitrogen. Rabbit anti-CENP-A was from Cell Signaling Technology. Mouse anti-FLAG-M2 antibody and mouse anti-GAPDH were from Sigma. Rabbit anti-HJURP was from Bethyl Laboratories.
Plasmids
FLAG-And-1, FLAG-And-1 mutants (1–336), (330–1129), and (984–1129) were constructed as described previously (16). YFP-CENP-A, H3.1-YFP, H3.3-GFP, H4-GFP, and GFP-HJURP were gifts from Dr. Daniel Foltz (University of Virginia).
FACS Analysis
Flow cytometry analysis was performed as described previously (24).
Immunoblotting and Chromatin Binding Assay
To make total protein, cells were lysed in radioimmuneprecipitation assay buffer (150 mm NaCl, 1.0% IGEPAL CA-630, 0.5% sodium deoxycholate, 0.1% SDS, and 50 mm Tris, pH 8.0) followed by sonication. Chromatin binding assays were performed essentially as described previously (25).
Chromatin Immunoprecipitation Assay
A chromatin immunoprecipitation (ChIP) assay was performed using the ChIP assay kit from Millipore following the supplied protocol. Immunoprecipitations were performed using anti-And-1 antibody (16) or control IgG antibodies. DNA was purified using the QIAquick PCR purification kit. DNA samples were analyzed by quantitative real-time PCR. PCR primers for BG40.9, BG72, α-satellite (Satα), and pericentromeric (Sat2) regions were as described (26, 27).
RESULTS
And-1-depleted Cells Arrest in Mitosis with Chromosome Congression Defects
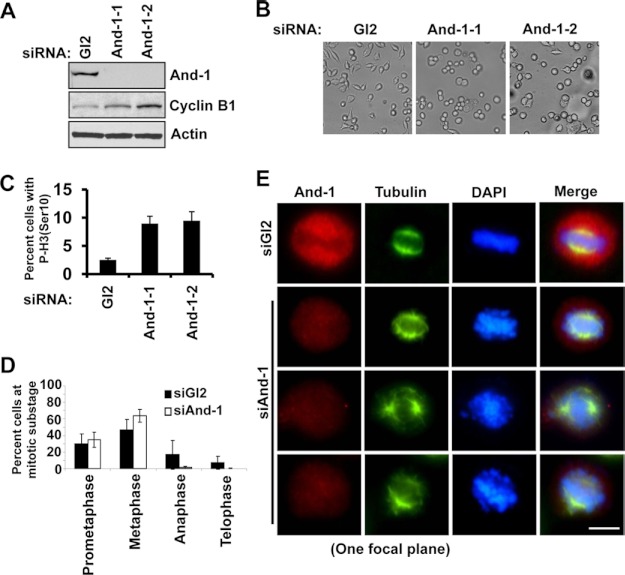
To determine the consequence of the loss of And-1 in human cells, we depleted And-1 in HCT116 cells using siRNA oligonucleotides against And-1 or firefly luciferase (Gl2 control). Transfection with two independent siRNAs significantly reduced And-1 protein levels (Fig. 1). Following And-1 depletion, the population of round mitotic cells was increased compared with control cells (Fig. 1B). To confirm and quantify mitotic arrested cells after And-1 depletion, we examined the mitotic index. The mitotic index of siAnd-1-treated cells increased ∼3-fold compared with siGl2-treated cells (Fig. 1C). We further examined the mitotic progression of And-1-depleted cells by immunofluorescence analyses. In contrast with siGl2-treated cells that distributed throughout mitosis, And-1-depleted cells accumulated in metaphase or prometaphase (Fig. 1D).
FIGURE 1.
And-1 depletion leads to mitotic arrest with chromosome congression defects. A, whole cell extracts from HCT116 cells transfected by two independent siRNAs (siAnd-1-1 and siAnd-1-2) were subjected to immunoblotting using the indicated antibodies. B, And-1-depleted cells showed two morphologically distinct cell populations: flat adherent cells and round mitotic cells. Phase-contrast imaging of HCT116 cells harvested 72 h post siRNA transfection is shown. C, mitotic index of And-1-depleted HCT116 cells was determined by FACS using phospho-histone H3 (Ser-10) antibody. The percent cells with P-H3 (Ser-10) is the ratio of the number of P-H3 W(Ser-10)-positive cells to total amount of cells. Data represent the means ± S.D. (error bars) from two independent experiments. D, distribution of mitotic stages in And-1-depleted HCT116 cells is shown. Counting was performed on cells stained for tubulin and DAPI. Two independent experiments were performed, and at least 300 cells were counted for each condition. Data represent the means ± S.D. E, chromosome congression defects appear after And-1 depletion. Two abnormal mitotic cell populations were observed: metaphase cells where most of the chromosomes aligned at the metaphase plate with few unaligned chromosomes or metaphase cells with severe chromosome alignment defects as indicated by randomly distributed chromosomes along the mitotic spindle. Cells treated as in A were stained for tubulin (green), And-1 (red), and DAPI (blue). Scale bar, 10 μm.
The arrest of cells in mitosis by And-1 depletion suggests the possibility of chromosome alignment defects. To test this possibility, we examined chromosome alignment by DAPI and α-tubulin co-staining. Strikingly, And-1-depleted cells failed to align chromosomes at the spindle equator (Fig. 1E). In addition, the metaphase spindle of And-1-depleted cells was 49% (p < 0.001) longer than that of siGl2-treated cells, indicating a deficiency of pulling forces from kinetochore fibers (Fig. 1E). Thus, we conclude that And-1 is important for proper chromosome congression.
And-1 Is Required for the Centromeric Localization of CENP-A
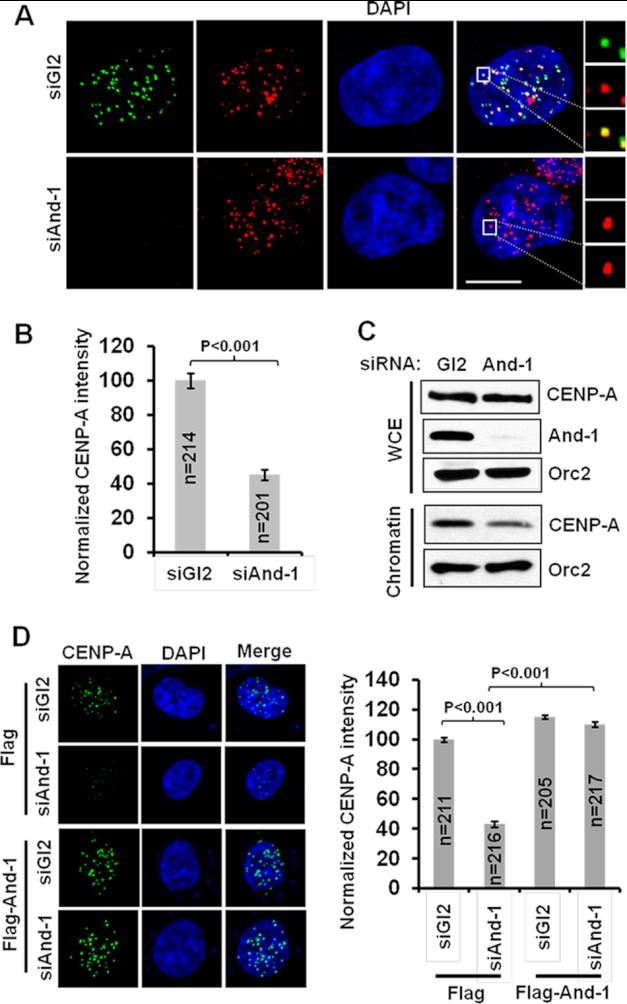
The failure to properly align chromosomes at the spindle equator in And-1-depleted cells suggests the possibility of a centromeric nucleosome defect. To test this possibility, we screened the localization of centromere proteins in And-1-depleted cells by immunostaining. Strikingly, we found that there was a dramatic reduction of endogenous CENP-A at centromeres 48 or 72 h after And-1 siRNA transfection (Fig. 2, A and B, and data not shown). Although total cellular levels of CENP-A were not affected after And-1 depletion, chromatin association of CENP-A was mildly decreased after And-1 depletion (Fig. 2C). To rule out that the reduced centromeric localization of CENP-A by And-1 depletion is due to siRNA off-target effects, we depleted And-1 using a siRNA targeting a 3′-UTR mRNA sequence in a HCT116 cell line stably expressing FLAG-And-1 from cDNA. Consistent with results shown in Fig. 2A, depletion of And-1 by this siRNA significantly reduced the localization of CENP-A at centromeres in HCT116 expressing FLAG alone (Fig. 2D). However, CENP-A centromeric assembly defects by And-1 depletion were restored in HCT116 cells expressing FLAG-And-1 (Fig. 2D). Thus, CENP-A assembly defects are specifically due to the loss of And-1. And-1 depletion also resulted in the loss of centromeric localization of CENP-A in U2OS cells (data not shown). Taken together, we conclude that And-1 is critical for CENP-A centromeric localization.
FIGURE 2.
And-1 is required for the localization of CENP-A at centromeres. A, And-1 depletion compromises the localization of CENP-A at centromeres. HCT116 cells treated as in Fig. 1A were fixed 72 h after siRNA transfection, and CENP-A was detected with α-CENP-A monoclonal antibody (green). Centromeres were identified with an anti-CENP-B (red) antibody. Scale bar, 10 μm. B, intensity of endogenous CENP-A at centromeres in siGl2- or siAnd-1-treated cells is shown. Data are means ± S.D. (error bars); n, number of cells analyzed. C, And-1 depletion mildly reduces CENP-A chromatin association. HCT116 cells treated as in Fig. 1A were harvested for chromatin binding assays or whole cell extraction (WCE) immunoblotting. D, And-1 depletion by siRNA targeting the 3′-UTR of And-1 causes a reduction of the localization of CENP-A at centromeres in HCT116 cells expressing FLAG alone but not in cells expressing FLAG-And-1 from cDNA. The quantification of CENP-A signals is in the right panel.
And-1 Forms a Complex with Prenucleosomal CENP-A
To further investigate the significance of CENP-A reduction by And-1 depletion, we examined whether And-1 and CENP-A form a complex by a co-immunoprecipitation assay using chromatin-free cell extracts from 293T cells co-expressing FLAG-And-1 and YFP or YFP-CENP-A. YFP-CENP-A but not YFP was readily detected in FLAG-And-1 immunoprecipitates (Fig. 3A), suggesting that And-1 interacts with prenucleosomal CENP-A. Having found the interaction between And-1 and CENP-A, we predicted that And-1 may associate with centromeres. To test this possibility, we performed ChIP assays. In agreement with the role of And-1 in DNA replication, we detected the accumulation of And-1 at a human β-globin replication origin BG40.9 but not a surrounding nonreplication origin BG72 (26) (Fig. 3B). Significantly, we could detect the stronger association of And-1 with α-satellite repeat region at centromere (Satα) and weaker association with pericentromeric region (Sat2).
FIGURE 3.
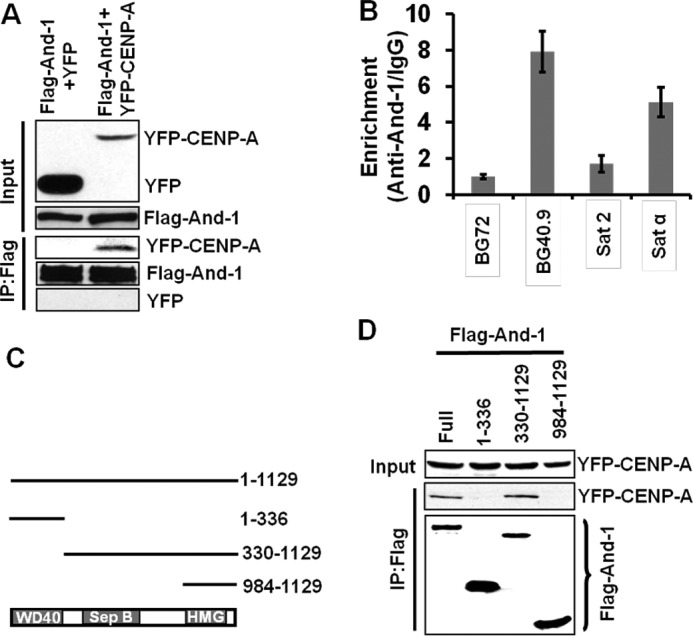
And-1 interacts with CENP-A. A, And-1 interacts with YFP-CENP-A. 293T cells co-expressing FLAG-And-1 and YFP or FLAG-And-1 and YFP-CENP-A were harvested to prepare chromatin-free fractions for co-immunoprecipitation assays. FLAG-And-1 immunoprecipitates were resolved by SDS-PAGE and immunoblotted for the indicated proteins. B, And-1 specifically associates with centromere. And-1 protein levels at replication origin BG40.9 or surrounding region BG72, α-satellite repeat (Satα) or pericentromeric region (Sat2) are shown. IgG was used as the measurement of nonspecific chromatin precipitation. Quantitative PCR was used to ascertain the And-1 levels at the indicated chromatin regions. y-axis represents the relative enrichment of the And-1 proteins compared with the IgG control (after normalization with input precipitations, data ± S.D. (error bars), n = 3). C, schematic shows And-1 protein domains and deletion mutants used for protein interactions. D, full-length And-1 and And-1(330–1129) interact with chromatin-free CENP-A. Chromatin-free extracts from 293T cells co-transfected with GFP-CENP-A and FLAG-And-1 or FLAG-And-1 mutants were immunoprecipitated with anti-FLAG and immunoblotted for the indicated antibodies.
And-1 contains WD40 repeats at its amino terminus, a SepB domain in the middle, and a HMG domain at its carboxyl terminus (Fig. 3C). We next tested which domains of And-1 are responsible for interacting with CENP-A by expressing CENP-A together with And-1 or its truncation mutants in 293T cells (Fig. 3D). CENP-A-And-1 interactions were detected in immunoprecipitates from full-length And-1 and the mutant And-1(330–1129), suggesting that the CENP-A binding site on And-1 covers the SepB region. No interaction was detected between CENP-A and And-1(1–336) or And-1(984–1129), suggesting that both the WD40 repeats and the HMG domain are dispensable for the interaction between And-1 and CENP-A. Taken together, our data indicate that And-1 forms a complex with prenucleosomal CENP-A.
CENP-A Centromeric Assembly Defects by And-1 Depletion Are Not Due to Defects of DNA Replication
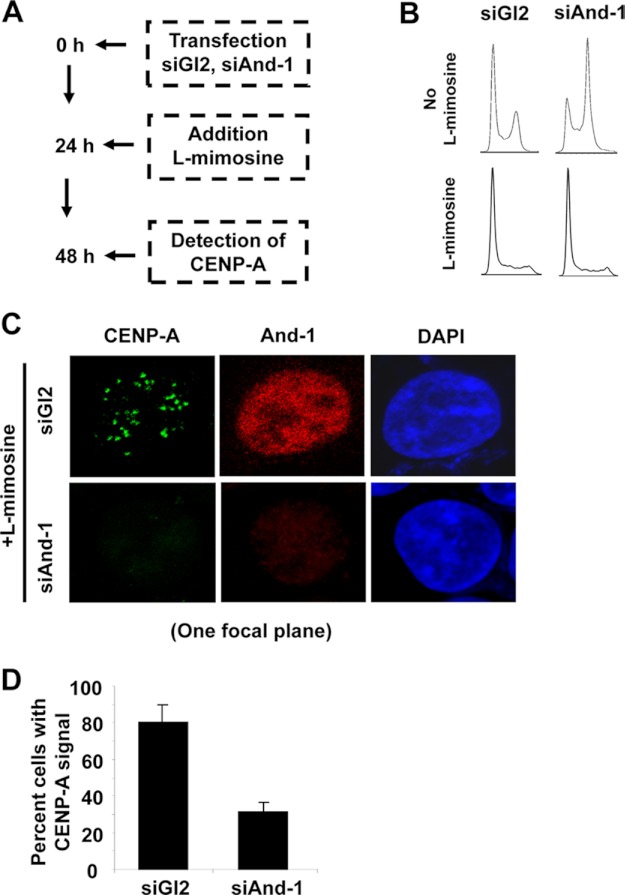
Because And-1 is involved in DNA replication (16–19), we next asked whether the CENP-A localization defects by And-1 depletion were due to DNA replication defects associated with And-1 depletion. To this end, we arrested And-1-depleted cells in G1 phase using l-mimosine, a plant amino acid that reversibly arrests cells in G1 phase (Fig. 4A) (28). Replication defects by And-1 depletion caused an S/G2/M accumulation (Fig. 4B) (16); however, addition of l-mimosine arrested siRNA-treated cells in G1 phase (Fig. 4B). Thus, we successfully obtained And-1-depleted cells which do not undergo DNA replication and therefore could not have DNA replication defects. A significant reduction of CENP-A localization at centromeres was observed in And-1-depleted G1-arrested cells (Fig. 4, C and D), suggesting that And-1 depletion but not aberrant DNA replication leads to CENP-A assembly defects. To further confirm that DNA replication impairments are not the cause of CENP-A centromere localization defects, we down-regulated two additional factors required for DNA replication, Cdc6 and Mcm7. We observed no defect in CENP-A localization when either Cdc6 or Mcm7 was down-regulated by siRNA (data not shown). Moreover, we found that DNA damage did not have effects on CENP-A localization (data not shown). These results are consistent with the observation that in contrast to the replication-dependent H3 deposition, CENP-A assembly is not a DNA replication-dependent process (29, 30). Taken together, these data suggest that And-1 plays a direct role in the CENP-A centromeric assembly.
FIGURE 4.
CENP-A localization defects are not due to defects in DNA replication. A, schematic of the experimental procedure. B, FACS analysis of cells harvested as in A. Note the addition of l-mimosine-arrested cells in G1 phase. C, CENP-A centromeric localization reduced in G1 phase cells with down-regulated And-1. CENP-A localization was detected by immunofluorescence (green) in cells harvested as in A. Scale bar, 10 μm. D, quantification of the number of positive cells for centromeric CENP-A signals for HCT116 cells treated as in A. Data represent the means ± S.D. (error bars) from two independent experiments.
And-1 Promotes the Assembly of Newly Synthesized CENP-A onto Centromeres
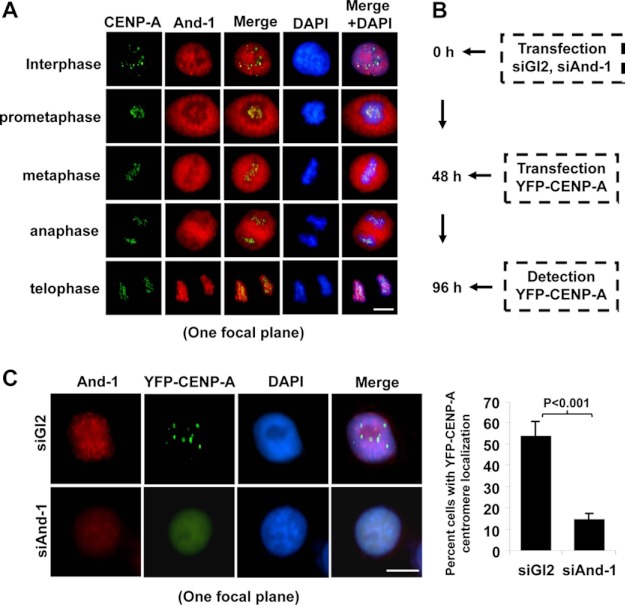
And-1 forms complexes with prenucleosomal CENP-A, suggesting that And-1 may regulate newly synthesized CENP-A assembly at centromeres. To test this possibility, we first examined the localization of And-1 and CENP-A throughout the cell cycle by co-immunostaining. And-1 was diffusely localized throughout the nucleus during interphase, dissociated from chromatin from prometaphase through anaphase, and reassociated with chromatin during telophase (Fig. 5A).
FIGURE 5.
And-1 is required for the recruitment of new CENP-A to centromeres. A, cell cycle localization of And-1 and CENP-A. HCT116 cells were fixed and stained with anti-And-1 (red) and anti-CENP-A (green) antibodies and counterstained with DAPI (blue). B, schematic of experimental procedure performed in C. C, And-1 depletion impairing the centromeric localization of new CENP-A to centromeres. Left, HCT116 cells treated as described in B, preextracted and fixed for immunostaining for And-1 proteins. Right, quantification of the percentage cells with YFP-CENP-A centromeric signals in siGl2- and siAnd-1-treated HCT116 cells. Three independent experiments were performed, and at least 300 cells were counted for each condition. Data represent the means ± S.D. (error bars). Scale bars, 10 μm.
And-1 associates with chromatin at late mitosis, suggesting that And-1 may be involved in the assembly of newly synthesized CENP-A in early G1 phase. To test this possibility, we performed a transient transfection assay as described by two groups previously (9, 11) (Fig. 5B). Forty-eight h after YFP-CENP-A transfection, siGl2-treated cells repeatedly showed that newly synthesized YFP-CENP-A was properly targeted to centromeres (Fig. 5C). Strikingly, when And-1 was depleted the number of cells with YFP-CENP-A centromeric signals was greatly reduced, and YFP-CENP-A was detected as diffusely localized throughout the nucleus (Fig. 5C). These results indicate that And-1 is required for the recruitment of newly synthesized CENP-A to chromatin at centromeres.
And-1 Transiently Interacts with CENP-A When Cells Transit from Mitosis to G1 Phase
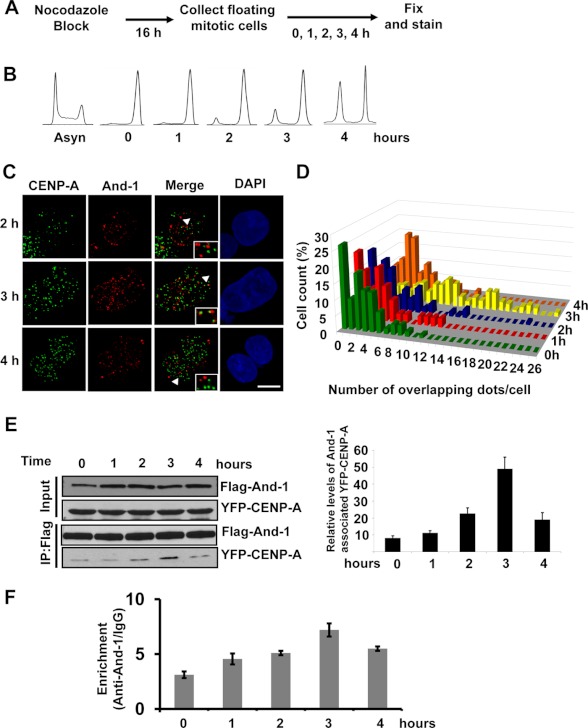
Because And-1 is required for the centromeric assembly of newly synthesized CENP-A, we hypothesized that And-1 would associate with CENP-A during late telophase or early G1 to load new CENP-A onto centromeres. To test this hypothesis, we assessed the association of And-1 with CENP-A by co-immunostaining, co-immunoprecipitation, and ChIP assays. First, we examined the co-localization of these two proteins in cells that were synchronized in mitosis with nocodazole and released back into the cell cycle followed by fixation at various time points (Fig. 6A). As shown in Fig. 6B, mitotic arrested cells reentered the cell cycle and started to enter G1 phase 2–3 h after release into nocodazole-free medium. To precisely examine the co-localization of And-1 with CENP-A, we preextracted cells with Triton X-100 to remove non-chromatin-associated And-1 (31). And-1 and CENP-A foci were observed throughout the time course. Significantly, the percentage of cells with adjacent or overlapping And-1 and CENP-A foci were increased at 3 h after nocodazole release, when cells start to transit from telophase to G1 phase (Fig. 6, C and D).
FIGURE 6.
And-1 associates with CENP-A when cells transit from mitosis to G1 phase. A, schematic showing synchronization of cells in mitosis and release into late mitosis and early G1 phases. B, FACS analysis of cells harvested as in A. C, localization of And-1 with CENP-A in late mitosis and early G1 phase. Representative images show And-1 (red) and CENP-A (green) localization in HCT116 cells during late telophase/early G1 phase. Insets are enlarged images of co-localization indicated by arrowheads. Cells were preextracted with 0.3% Triton X-100. D, quantification of cells with And-1 and CENP-A overlapping or adjacent signals in HCT116 cells for each time point evaluated after release into G1 (n = 60–150 cells for each time point). E, interaction between And-1 and CENP-A during late telophase/early G1 phase. Left, chromatin-free extracts from 293T cells co-transfected with FLAG-And-1 and YFP-CENP-A treated as in A, followed by immunoprecipitation with anti-FLAG and immunoblotting with antibodies as indicated. Right, quantification of FLAG-And-1-associated YFP-CENP-A as in left panel. The signals of FLAG-And-1-associated YFP-CENP-A and FLAG-And-1 immunoprecipitations (IP) were quantified by Scion imaging software, and the relative levels of And-1-associated YFP-CENP-A are YFP-CENP-A signal-normalized by FLAG-And-1 immunoprecipitation signals. Data represent the mean ± S.D. (error bars) from three independent experiments. F, enrichment of And-1 at α-satellite repeat (Satα) region. Cells treated as in A were harvested for ChIP assay as in Fig. 3B. y-axis represents the relative enrichment of the And-1 proteins at the α-satellite repeat region at centromere compared with the IgG control (after normalization with input precipitations, data ± S.D., n = 3).
The visualization of the overlapping of And-1 and CENP-A foci does not definitively indicate an actual association of And-1 with CENP-A. To further confirm the interaction between And-1 and CENP-A, we performed a co-immunoprecipitation assay using cells treated as above. Chromatin-free fractions were prepared from synchronized 293T cells expressing FLAG-And-1 and YFP-CENP-A at various time points following nocodazole release. The interaction between FLAG-And-1 and YFP-CENP-A could be detected in mitotic cells (0 and 1 h after nocodazole block), but the interaction increased by 5-fold at 3 h after nocodazole release and rapidly decreased by 4 h (Fig. 6E), which is consistent with the results from the co-immunostaining approach (Fig. 6, C and D).
We also performed ChIP assays to examine the association of And-1 with centromeres during late mitosis and early G1 phase. As shown in Fig. 6F, we could detect the association of And-1 with centromeric region (Satα) at metaphase cells, and this association was increased at 3 h after nocodazole release and decreased afterward. Taken altogether, our data strongly suggest that And-1 transiently interacts with CENP-A during a brief interval when cells transit from mitosis to G1 phase, a time point when CENP-A is assembled at centromeres.
And-1 Is an HJURP-interacting Partner Required for HJURP Localization at Centromeres
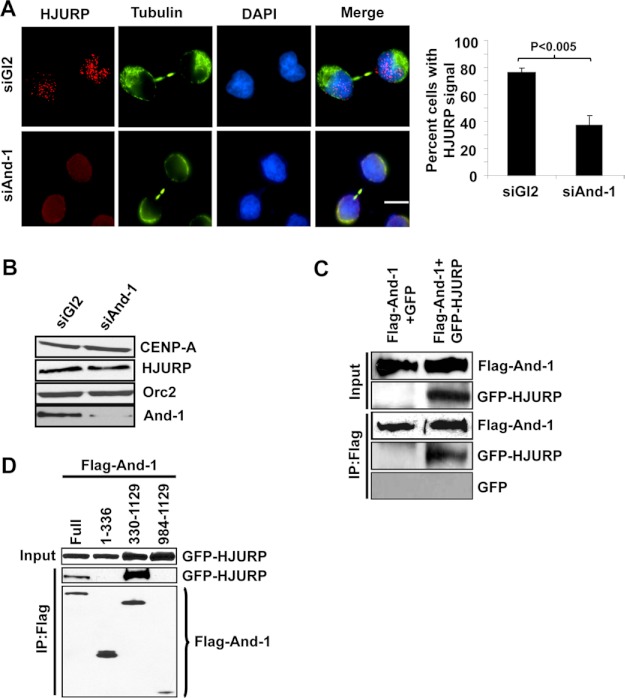
HJURP was recently identified as a CENP-A-specific chaperone required for the centromeric assembly of CENP-A in early G1 phase (11, 12). We asked whether And-1 regulates HJURP chaperone activity for the assembly of CENP-A at centromeres. As shown in Fig. 7B, And-1 depletion did not affect HJURP protein levels. We next examined the localization of HJURP at centromeres in And-1-depleted cells. Consistent with previous observations (11), HJURP centromeric localization (spotted pattern) could be detected in siGl2-treated early G1 phase cells as indicated by visible mid-bodies (Fig. 7A). However, And-1 depletion resulted in a major reduction in HJURP localization at the centromeres in early G1 phase cells.
FIGURE 7.
And-1 is a HJURP-interacting partner required for HJURP localization at centromeres. A, And-1 depletion leads to the major reduction of HJURP centromeric localization. Left, immunostaining of cells for HJURP and α-tubulin marks the mid-body that appears at late telophase/early G1. Left, cells transfected with siRNA (siGl2 or siAnd-1) were harvested 72 h after transfection and immunostained with α-HJURP (red) and α-tubulin (green) antibodies. Right, quantification of the percentage cells with HJURP centromeric signals in early G1 phase cells is indicated by a visible mid-body. Three independent experiments were performed. At least 100 siGl2- and 40 siAnd-1-transfected cells with visible mid-body were analyzed in each experiment. Data represent the means ± S.D. (error bars). B, HCT116 cells treated as in Fig. 1A were harvested, and the whole cell extracts were immunoblotted with the indicated antibodies. C, And-1 interacts with HJURP. Chromatin-free extracts from 293T cells co-expressing FLAG-And-1 and GFP-HJURP were immunoprecipitated (IP) with anti-FLAG and immunoblotted with the indicated antibodies. D, And-1 (Full) and fragment (330–1129) interact with chromatin-free HJURP. Chromatin-free extracts from 293T cells co-transfected with GFP-HJURP and FLAG-And-1 or FLAG-And-1 mutants were immunoprecipitated with anti-FLAG and immunoblotted for the indicated antibodies.
The fact that And-1 is required for the centromeric localization of HJURP at centromeres prompted us to examine the interaction between And-1 and HJURP. To this end, we performed a co-immunoprecipitation assay using the chromatin-free extracts from 293T cells co-expressing FLAG-And-1 and GFP or GFP-HJURP. GFP-HJURP but not GFP was readily detected in FLAG-And-1 immunoprecipitates (Fig. 7C), indicating that And-1 forms a complex with prenucleosomal HJURP. Next, we examined which domains of And-1 interact with prenucleosomal HJURP by co-expressing GFP-HJURP with full-length And-1 and its truncated mutants (Fig. 3B). Similar to the interactions between And-1 and CENP-A (Fig. 3B), And-1-HJURP interactions were only detected in immunoprecipitates from full-length And-1 and the And-1(330–1129) truncation mutant (Fig. 7D). Taken together, we conclude that And-1 interacts with HJURP and is required for the centromeric association of HJURP in early G1 phase.
Overexpression of And-1 Enhances the Assembly of CENP-A at Centromeres
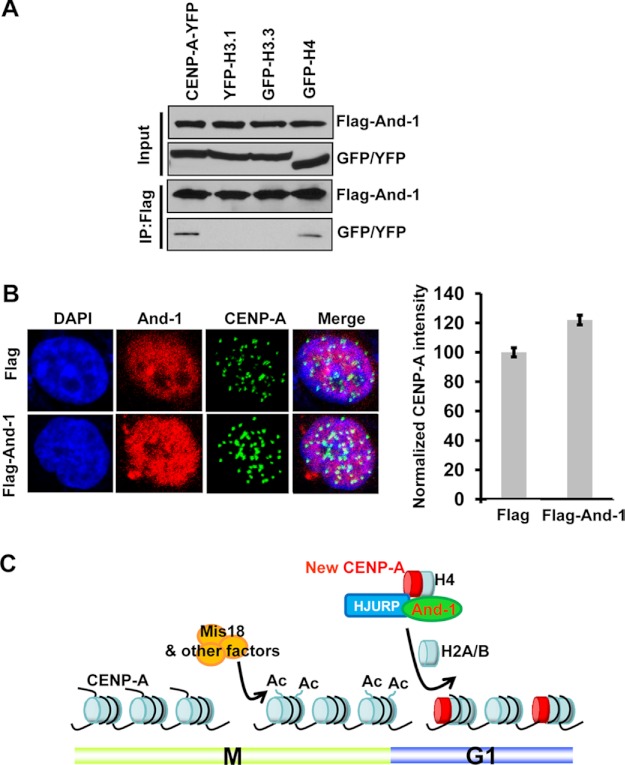
If And-1 specifically promotes the assembly of CENP-A at centromeres, we expect that And-1 interacts with histone H4 but not H3 in chromatin-free extractions. To test this possibility, we examined the interactions between And-1 and H3 or H4 using chromatin-free extracts from 293T cells co-expressing FLAG-And-1 and YFP-CENP-A, H3.1-YFP, H3.3-GFP, or H4-GFP. Although we have reported that And-1 interacts with H3 in whole cell extracts or chromatin extracts (23), we failed to detect the interaction between And-1 and H3.1-YFP or H3.3-GFP in chromatin-free extractions (Fig. 8A). Significantly, And-1 was found to interact with prenucleosomal YFP-CENP-A and H4-GFP, suggesting an important role of And-1 in the regulation of the CENP-A-H4 complex.
FIGURE 8.
And-1 overexpression enhances the intensity of CENP-A at centromeres. A, And-1 interacts with CENP-A and H4 but not H3.1 or H3.3. 293T cells co-expressing FLAG-And-1 and either YFP-CENP-A, H3.1-YFP, H3.3-GFP, or H4-GFP were harvested and chromatin-free fractions prepared for co-immunoprecipitation (IP) assays. FLAG-And-1 immunoprecipitates were resolved by SDS-PAGE and immunoblotted for the indicated proteins. B, CENP-A intensity is increased in U2OS (FLAG-And-1) cells compared with that in U2OS (FLAG) cells. Asynchronous cells were harvested and immunostained for And-1 and CENP-A proteins. The intensity of CENP-A at centromeres is shown in the right panel. C, model shows the role of And-1 in the HJURP-mediated deposition of CENP-A at centromeres. Replication of sister chromatids during S phase dilutes the amount of CENP-A at the centromeres, which is interspersed with H3-containing nucleosomes. During late anaphase/telophase centromeric chromatin is primed for the deposition of new CENP-A by the Mis18 complex and other factors possibly through changing the acetylation status of the chromatin. At late mitosis or early G1 phase, And-1 interacts with HJURP-CENP-A-H4 complexes and facilitates their assembly at centromeres in a process involving multiple proteins such as RbAp48/46 and Npm1 (data not shown).
We next asked whether overexpression of And-1 could increase CENP-A assembly at centromeres. To this goal, we carefully measured the intensity of CENP-A foci signal at centromeres in U2OS cell lines which express FLAG control or FLAG-And-1. The FLAG-And-1 protein levels are an approximately 2-fold increase in U2OS (FLAG-And-1) cells compared with endogenous And-1 in U2OS (FLAG) cells (data not shown). Significantly, we could detect an ∼18% increase of CENP-A foci intensity in U2OS (FLAG-And-1) cells (Fig. 8B). These data strongly suggest a positive role of And-1 in the assembly of CENP-A at centromeres.
DISCUSSION
And-1 Plays a Direct Role in the Centromere-specific Assembly of CENP-A
Centromere-specific assembly of CENP-A nucleosomes is the most important event that ensures the epigenetic inheritance of centromeres. The histone chaperone HJURP appears to play a key role in the assembly of CENP-A at centromeres (11–13, 32). However, whether other HJURP- or CENP-A-interacting factors are involved in CENP-A assembly remains largely unknown. Here, we provide evidence to demonstrate that And-1 is essential for the HJURP-mediated assembly of newly synthesized CENP-A at centromeres. And-1 interacts with both CENP-A and HJURP in chromatin-free extracts, and the loss of And-1 significantly reduces the association of HJURP and CENP-A at centromeres in early G1 phase. Moreover, overexpression of And-1 can specifically enhance the assembly of CENP-A at centromeres.
And-1 contains both WD40 and HMG domains. The HMG domain is a DNA binding domain that usually facilitates the formation of nucleoprotein complexes on the chromatin via its DNA binding capability (33, 34). HMG domain is found in histone chaperones such as SSRP1 (one subunit of the FACT complex) which is responsible for the assembly/disassembly of histone H2A-H2B dimers (35). WD40 domain has also been found in many chromatin assembly factors such as CAF-1 p60, RbAp46, RbAp48, and HIRA. Unlike many replisome components, which are loaded onto chromatin at the onset of S phase for DNA replication, human And-1 is loaded onto chromatin at late telophase (Fig. 5A), suggesting that And-1 may play other roles in the regulation of chromatin function during late telophase/early G1 phase. Indeed, our findings reveal a novel role of And-1 in the assembly of CENP-A at centromeres during late mitosis and early G1 phase. We have ruled out the possibility that CENPA-centromeric assembly defects are due to the failure of DNA replication associated with loss of And-1 (Fig. 4). We recently reported that And-1 is required for Gcn5 protein stability and thereby histone H3 acetylation (23). Thus, it is possible that And-1 may regulate CENP-A assembly by impacting histone H3 acetylation. However, this is not the case because we could still detect the significant reduction of CENP-A foci at centromeres in siAnd-1-transfected cells after treatment with the HDAC inhibitor tricostatin A that could restore histone acetylation (data not shown). Consistently, depletion of Gcn5 has no effect on CENP-A localization at centromeres (data not shown). Several other lines of evidence from our study strongly suggest that And-1 plays a direct role in the assembly of CENP-A at centromeres. First, And-1 interacts with both CENP-A and HJURP in chromatin-free fractions (Figs. 3 and 7). Second, the interaction between And-1 and prenucleosomal CENP-A is increased when cells transit from mitosis to G1 phase, which is the time point when newly synthesized CENP-A is loaded onto centromeres (Fig. 6). Third, And-1 specifically interacts with CENP-A, HJURP, and H4 but not H3 in chromatin-free fractions and is required for the localization of CENP-A-HJURP at centromeres (Figs. 2, 7A, and 8A). Finally, overexpression of And-1 specifically enhances the CENP-A intensity at centromeres (Fig. 8B). Taken together, we propose that And-1 is a new factor required for the assembly of CENP-A at centromeres.
And-1 Is an Important Factor Required for New CENP-A Centromeric Assembly
The assembly of newly synthesized CENP-A at centromeres occurs during a brief interval in early G1 phase via the events involving multiple proteins such as HJURP, RbAp48/46, Npm1, and the Mis18 complex (6, 9, 11, 12, 36, 37). HJURP interacts with prenucleosomal CENP-A and is required for the centromeric assembly of new CENP-A in a process after centromere priming. Whether or not other HJURP-interacting partners are required for the loading of CENP-A-HJURP complexes onto centromeres is unknown. Our data demonstrate that And-1 plays such a role. Like HJURP, And-1 interacts with prenucleosomal CENP-A and exhibits a centromeric association pattern similar to that of HJURP in the cell cycle. However, unlike HJURP, which is required for both CENP-A protein stability and deposition at centromeres (11, 12), the role of And-1 is restricted to the delivery and deposition of HJURP-CENP-A complexes onto centromeres. And-1 is not required for the stability of CENP-A or HJURP but is critical for the centromeric localization of these two proteins in early G1 phase. We therefore propose that And-1 acts as a key factor involving the assembly of HJURP-CENP-A-H4 complexes at centromeres (Fig. 8C). At this moment, we could not rule out that And-1 may also regulate other factors involving CENP-A centromere assembly, such as Mis18 complex. In the future, it will be intriguing to determine the relationship of And-1 with these CENP-A-specific loading factors.
Acknowledgments
We thank Dr. Daniel Foltz for CENP-A, HJURP, H3.1, and H4 plasmids. Images collected with the Zeiss 710 microscope were supported by National Center for Research Resources Grant S10RR025565-01.
This work was supported, in whole or in part, by National Institutes of Health Grant CA136555 (to W. Z).

This article contains supplemental Figs. S1–S4 and an additional reference.
- CENP-A
- centromere protein A
- And-1
- acidic nucleoplasmic DNA-binding protein 1
- HMG
- high mobility group
- HJURP
- Holliday junction recognition protein.
REFERENCES
- 1. Cleveland D. W., Mao Y., Sullivan K. F. (2003) Centromeres and kinetochores: from epigenetics to mitotic checkpoint signaling. Cell 112, 407–421 [DOI] [PubMed] [Google Scholar]
- 2. Musacchio A., Salmon E. D. (2007) The spindle-assembly checkpoint in space and time. Nat. Rev. Mol. Cell Biol. 8, 379–393 [DOI] [PubMed] [Google Scholar]
- 3. Black B. E., Bassett E. A. (2008) The histone variant CENP-A and centromere specification. Curr. Opin. Cell Biol. 20, 91–100 [DOI] [PubMed] [Google Scholar]
- 4. Sullivan K. F. (2001) A solid foundation: functional specialization of centromeric chromatin. Curr. Opin. Genet. Dev. 11, 182–188 [DOI] [PubMed] [Google Scholar]
- 5. Blower M. D., Sullivan B. A., Karpen G. H. (2002) Conserved organization of centromeric chromatin in flies and humans. Dev. Cell 2, 319–330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Jansen L. E., Black B. E., Foltz D. R., Cleveland D. W. (2007) Propagation of centromeric chromatin requires exit from mitosis. J. Cell Biol. 176, 795–805 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Schuh M., Lehner C. F., Heidmann S. (2007) Incorporation of Drosophila CID/CENP-A and CENP-C into centromeres during early embryonic anaphase. Curr. Biol. 17, 237–243 [DOI] [PubMed] [Google Scholar]
- 8. Black B. E., Jansen L. E., Foltz D. R., Cleveland D. W. (2010) Centromere identity, function, and epigenetic propagation across cell divisions. Cold Spring Harb. Symp. Quant. Biol. 75, 403–418 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Fujita Y., Hayashi T., Kiyomitsu T., Toyoda Y., Kokubu A., Obuse C., Yanagida M. (2007) Priming of centromere for CENP-A recruitment by human hMis18α, hMis18β, and M18BP1. Dev. Cell 12, 17–30 [DOI] [PubMed] [Google Scholar]
- 10. Hayashi T., Fujita Y., Iwasaki O., Adachi Y., Takahashi K., Yanagida M. (2004) Mis16 and Mis18 are required for CENP-A loading and histone deacetylation at centromeres. Cell 118, 715–729 [DOI] [PubMed] [Google Scholar]
- 11. Dunleavy E. M., Roche D., Tagami H., Lacoste N., Ray-Gallet D., Nakamura Y., Daigo Y., Nakatani Y., Almouzni-Pettinotti G. (2009) HJURP is a cell-cycle-dependent maintenance and deposition factor of CENP-A at centromeres. Cell 137, 485–497 [DOI] [PubMed] [Google Scholar]
- 12. Foltz D. R., Jansen L. E., Bailey A. O., Yates J. R., 3rd, Bassett E. A., Wood S., Black B. E., Cleveland D. W. (2009) Centromere-specific assembly of CENP-a nucleosomes is mediated by HJURP. Cell 137, 472–484 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Shuaib M., Ouararhni K., Dimitrov S., Hamiche A. (2010) HJURP binds CENP-A via a highly conserved N-terminal domain and mediates its deposition at centromeres. Proc. Natl. Acad. Sci. U.S.A. 107, 1349–1354 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Lagana A., Dorn J. F., De Rop V., Ladouceur A. M., Maddox A. S., Maddox P. S. (2010) A small GTPase molecular switch regulates epigenetic centromere maintenance by stabilizing newly incorporated CENP-A. Nat. Cell Biol. 12, 1186–1193 [DOI] [PubMed] [Google Scholar]
- 15. Perpelescu M., Nozaki N., Obuse C., Yang H., Yoda K. (2009) Active establishment of centromeric CENP-A chromatin by RSF complex. J. Cell Biol. 185, 397–407 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Zhu W., Ukomadu C., Jha S., Senga T., Dhar S. K., Wohlschlegel J. A., Nutt L. K., Kornbluth S., Dutta A. (2007) Mcm10 and And-1/CTF4 recruit DNA polymerase α to chromatin for initiation of DNA replication. Genes Dev. 21, 2288–2299 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Bermudez V. P., Farina A., Tappin I., Hurwitz J. (2010) Influence of the human cohesion establishment factor Ctf4/AND-1 on DNA replication. J. Biol. Chem. 285, 9493–9505 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Gambus A., van Deursen F., Polychronopoulos D., Foltman M., Jones R. C., Edmondson R. D., Calzada A., Labib K. (2009) A key role for Ctf4 in coupling the MCM2–7 helicase to DNA polymerase α within the eukaryotic replisome. EMBO J. 28, 2992–3004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Im J. S., Ki S. H., Farina A., Jung D. S., Hurwitz J., Lee J. K. (2009) Assembly of the Cdc45-Mcm2–7-GINS complex in human cells requires the Ctf4/And-1, RecQL4, and Mcm10 proteins. Proc. Natl. Acad. Sci. U.S.A. 106, 15628–15632 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Miles J., Formosa T. (1992) Evidence that POB1, a Saccharomyces cerevisiae protein that binds to DNA polymerase α, acts in DNA metabolism in vivo. Mol. Cell Biol. 12, 5724–5735 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Kouprina N., Kroll E., Bannikov V., Bliskovsky V., Gizatullin R., Kirillov A., Shestopalov B., Zakharyev V., Hieter P., Spencer F. (1992) CTF4 (CHL15) mutants exhibit defective DNA metabolism in the yeast Saccharomyces cerevisiae. Mol. Cell Biol. 12, 5736–5747 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Mamnun Y. M., Katayama S., Toda T. (2006) Fission yeast Mcl1 interacts with SCF(Pof3) and is required for centromere formation. Biochem. Biophys. Res. Commun. 350, 125–130 [DOI] [PubMed] [Google Scholar]
- 23. Li Y., Jaramillo-Lambert A. N., Yang Y., Williams R., Lee N. H., Zhu W. (2012) And-1 is required for the stability of histone acetyltransferase Gcn5. Oncogene 31, 643–652 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Zhu W., Chen Y., Dutta A. (2004) Rereplication by depletion of geminin is seen regardless of p53 status and activates a G2/M checkpoint. Mol. Cell. Biol. 24, 7140–7150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Zou L., Cortez D., Elledge S. J. (2002) Regulation of ATR substrate selection by Rad17-dependent loading of Rad9 complexes onto chromatin. Genes Dev. 16, 198–208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Sibani S., Price G. B., Zannis-Hadjopoulos M. (2005) Ku80 binds to human replication origins prior to the assembly of the ORC complex. Biochemistry 44, 7885–7896 [DOI] [PubMed] [Google Scholar]
- 27. Alexiadis V., Ballestas M. E., Sanchez C., Winokur S., Vedanarayanan V., Warren M., Ehrlich M. (2007) RNAPol-ChIP analysis of transcription from FSHD-linked tandem repeats and satellite DNA. Biochim. Biophys. Acta 1769, 29–40 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Lalande M. (1990) A reversible arrest point in the late G1 phase of the mammalian cell cycle. Exp. Cell Res. 186, 332–339 [DOI] [PubMed] [Google Scholar]
- 29. Shelby R. D., Monier K., Sullivan K. F. (2000) Chromatin assembly at kinetochores is uncoupled from DNA replication. J. Cell Biol. 151, 1113–1118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Ahmad K., Henikoff S. (2001) Centromeres are specialized replication domains in heterochromatin. J. Cell Biol. 153, 101–110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Martini E., Roche D. M., Marheineke K., Verreault A., Almouzni G. (1998) Recruitment of phosphorylated chromatin assembly factor 1 to chromatin after UV irradiation of human cells. J. Cell Biol. 143, 563–575 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Bernad R., Sánchez P., Rivera T., Rodríguez-Corsino M., Boyarchuk E., Vassias I., Ray-Gallet D., Arnaoutov A., Dasso M., Almouzni G., Losada A. (2011) Xenopus HJURP and condensin II are required for CENP-A assembly. J. Cell Biol. 192, 569–582 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Bustin M. (1999) Regulation of DNA-dependent activities by the functional motifs of the high-mobility-group chromosomal proteins. Mol. Cell. Biol. 19, 5237–5246 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Grosschedl R., Giese K., Pagel J. (1994) HMG domain proteins: architectural elements in the assembly of nucleoprotein structures. Trends Genet. 10, 94–100 [DOI] [PubMed] [Google Scholar]
- 35. Ransom M., Dennehey B. K., Tyler J. K. (2010) Chaperoning histones during DNA replication and repair. Cell 140, 183–195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Hemmerich P., Weidtkamp-Peters S., Hoischen C., Schmiedeberg L., Erliandri I., Diekmann S. (2008) Dynamics of inner kinetochore assembly and maintenance in living cells. J. Cell Biol. 180, 1101–1114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Maddox P. S., Hyndman F., Monen J., Oegema K., Desai A. (2007) Functional genomics identifies a Myb domain-containing protein family required for assembly of CENP-A chromatin. J. Cell Biol. 176, 757–763 [DOI] [PMC free article] [PubMed] [Google Scholar]