Background: There has been no in vivo assay to determine mitochondrial membrane protein sorting.
Results: The Mgm1 fusion approach provides a versatile experimental tool for determining the mitochondrial sorting pathways in vivo.
Conclusion: Sorting of mitochondrial inner membrane proteins carrying moderately hydrophobic transmembrane segments are modulated, depending on the cellular environment.
Significance: Mitochondrial membrane protein sorting may be a dynamic process.
Keywords: Membrane, Membrane Proteins, Membrane Transport, Mitochondria, Yeast, Mgm1p, Conservative Sorting, Stop Transfer
Abstract
Mitochondrial inner membrane proteins that carry an N-terminal presequence are sorted by one of two pathways: stop transfer or conservative sorting. However, the sorting pathway is known for only a small number of proteins, in part due to the lack of robust experimental tools with which to study. Here we present an approach that facilitates determination of inner membrane protein sorting pathways in vivo by fusing a mitochondrial inner membrane protein to the C-terminal part of Mgm1p containing the rhomboid cleavage region. We validated the Mgm1 fusion approach using a set of proteins for which the sorting pathway is known, and determined sorting pathways of inner membrane proteins for which the sorting mode was previously uncharacterized. For Sdh4p, a multispanning membrane protein, our results suggest that both conservative sorting and stop transfer mechanisms are required for insertion. Furthermore, the sorting process of Mgm1 fusion proteins was analyzed under different growth conditions and yeast mutant strains that were defective in the import motor or the m-AAA protease function. Our results show that the sorting of mitochondrial proteins carrying moderately hydrophobic transmembrane segments is sensitive to cellular conditions, implying that mitochondrial import and membrane sorting in the physiological environment may be dynamically tuned.
Introduction
The majority of mitochondrial proteins are encoded by nuclear genes and imported into the mitochondria. These proteins usually contain mitochondrial targeting sequences (presequences) at the N terminus, which direct them to the mitochondrial matrix (1, 2). These proteins first encounter the TOM (translocase of the outer membrane) complex, followed by the TIM23 (translocase of the inner membrane) complex. Subsequently, the N-terminal presequence is cleaved by the mitochondrial processing peptidase in the matrix (2, 3).
Some mitochondrial proteins carry a hydrophobic segment in the downstream region of the presequence that is arrested at the TIM23 complex in the inner membrane and is laterally integrated into the lipid bilayer. This mechanism is referred to as the “stop transfer” pathway (4, 5). Transmembrane (TM)3 segments of some inner membrane proteins are not arrested by the TIM23 complex but are fully imported into the matrix and then integrated into the inner membrane from the matrix side. This process is called the “conservative sorting” pathway, and the Oxa1 complex, Mba1, Cox18 (6), and Bcs1 (7) have been found to mediate the insertion of membrane proteins from the matrix side. TM segments of the proteins sorted by the stop transfer mechanism tend to be more hydrophobic than those sorted by conservative sorting (8, 9).
The determination of the exact sorting pathway for each mitochondrial inner membrane protein is far from simple, in most instances involving an in vitro assay easily compromised by specific experimental conditions, which leads to conflicting results (10–14). Thus, the exact sorting preference has been determined for only a handful of inner membrane proteins, and how the majority of membrane proteins are sorted in the mitochondrial inner membrane is largely unknown.
In this report, we took advantage of the unique insertion mechanism of the yeast mitochondrial inner membrane protein, Mgm1p (15), to develop an approach that enables easy assessment of the sorting pathway for mitochondrial inner membrane proteins. Using a set of proteins for which the sorting pathways have been previously determined, we tested the feasibility of the inner membrane protein-Mgm1 fusion approach. Our results are comparable with the independently determined previous data, establishing the Mgm1 fusion approach as a robust experimental tool for determining mitochondrial inner membrane protein sorting pathways in vivo.
Further, we determined sorting of inner membrane proteins for which the sorting mode was previously unknown. For Sdh4p that carries three TM segments, our results show that the first two TM segments are sorted by the conservative sorting, whereas the last TM segment is inserted by the stop transfer mechanism. Sorting of Mgm1 fusion proteins (MFPs) was assessed under different growth conditions (e.g. differences in carbon source and temperature) and in yeast strains defective in mitochondrial import motor function or m-AAA protease activity. Our results show that the sorting of inner membrane proteins carrying moderately hydrophobic TM segments is highly dependent on growth conditions and/or the proper function of the import motor or the m-AAA function. Inner membrane proteins with moderately hydrophobic TM segments tend to be more influenced by intrinsic (import machineries) or extrinsic (growth conditions) environmental changes in the cell. These data imply that biogenesis of inner membrane proteins may be finely tuned under various cellular conditions.
EXPERIMENTAL PROCEDURES
Primer Design
Each forward primer contained 30 bases complementing the upstream region of the SmaI site in pJK110 (16) for subcloning by homologous recombination and 17–20 bases complementing the start of the gene of interest. Reverse primers contained the end of the gene of interest (or the region of truncation in the case where a truncated version of an inner membrane protein-Mgm1 fusion was prepared) and 30 bases complementing the downstream sequence of the first hydrophobic segment (HS1) in MGM1 (amino acid residues 117SSFTKDKLDR126) in pJK110.
Construction of Plasmids
Each gene was amplified by PCR using genomic yeast DNA prepared from W303-1α (MAT α, ade2, can1, his3, leu2, trp1, ura3) and synthesized primers. All plasmids were prepared by homologous recombination in yeast as described previously (16, 17).
Western Blot Analysis
Yeast transformants of W303-1α carrying each plasmid were grown overnight in 5 ml of −Leu medium at 30 °C (25 or 37 °C). Preparation of whole-cell lysates, SDS-PAGE, and Western blotting were conducted as described (16). Some whole-cell lysates were extracted by trichloroacetic acid (TCA) precipitation, where 1 A600 unit from yeast cells was harvested and resuspended with 500 μl of Milli-Q water, 75 μl of alkaline mix (for 1 ml, 641 μl of Milli-Q water, 185 μl of 10 n NaOH, 100 μl of 0.1 m PMSF, 74 μl of β-mercaptoethanol), and 575 μl of 25% TCA (w/v) and incubated for 30 min on ice. Samples were washed with 1 ml of 100% acetone (w/v), and the dried pellet was resuspended in 40 μl of SDS-PAGE sample buffer and incubated for 5 min at 95 °C before being loaded onto the SDS gels, followed by Western blotting. The relative amounts of long Mgm1 fusion protein (L-MFP) and soluble short Mgm1p (s-Mgm1p) were quantified with Image Lab version 3.0 in the Bio-Rad Chemi-doc-XRS+ system (Bio-Rad) or Multi Gauge version 3.0 in the Fuji LAS-1000 system (Fujifilm, Japan).
Isolation of Mitochondria and Proteinase K Protection Assay
Yeast transformants carrying MFP constructs were grown in 1 liter of −Leu medium containing glucose (2%, w/v) at 30 °C until reaching 1–2 A600 units/ml. Cells were harvested by centrifugation at 3,000 × g for 5 min and treated with 100 mm of Tris base, pH 11.0, and 10 mm dithiothreitol (DTT), for 20 min at 30 °C. Cells were then centrifuged at 2,000 × g for 5 min and incubated with Zymolyase-100T (5 mg/g of cells) in 1.2 m sorbitol and 20 mm potassium phosphate at 30 °C for 30 min (or up to 1 h). Cells were collected by centrifugation at 1,200 × g for 5 min at 4 °C. Afterward, the pellet was resuspended in homogenization buffer (10 mm Tris-HCl, pH 7.4, 1 mm EDTA, 0.2% BSA, 1 mm phenylmethanesulfonyl fluoride (PMSF), 0.6 m sorbitol), and cells were lysed by a glass homogenizer at 4 °C (13 strokes). The lysate was centrifuged at 1,200 × g for 5 min at 4 °C to remove unbroken cells. The mitochondrial fraction was harvested by centrifugation at 12,000 × g for 15 min at 4 °C. The pellet was resuspended in 500 μl of suspension buffer (0.6 m sorbitol, 20 mm HEPES-KOH, pH 7.4). Then 40 μg of prepared mitochondria were incubated with either 100 μl of suspension buffer (0.6 m sorbitol, 20 mm HEPES-KOH, pH 7.4) or suspension buffer with proteinase K (50 μg/ml) for 30 min on ice. To stop the proteolytic activity, 1 μl of 0.1 m PMSF was added, and the suspension was incubated for 5 min on ice. Samples were centrifuged at 20,000 × g for 10 min, and the pellets were precipitated with 12.5% (w/v) TCA as described above, followed by SDS-PAGE and Western blotting analysis. The antibodies against Tom70, Tim54, and Tim44 were graciously donated by Professor Toshiya Endo (Nagoya, Japan), and the antibodies for Tom20 and Mge1 were kindly provided by Professor Thomas Langer (Cologne, Germany).
Growth with a Non-fermentable Carbon Source
Yeast transformants of W303-1α carrying each Mgm1 fusion construct were grown at 30 °C for 2 days in 5 ml of −Leu medium containing glycerol (3%, w/v) instead of glucose as a carbon source.
Mutant Strains
MFP constructs were transformed into the temperature-sensitive pam16-3 mutant or the isogenic PAM16 wild type strain (18), kindly provided by Professor Pfanner (Freiburg, Germany), and the m-AAA deletion (MATα, can1-100, ade2-1, his3-11,15, leu2-3,112, trp1-1, ura3-1, Δyta10::HIS3MX6, Δccp1::kanMX4) or the isogenic wild type strain (MATα, can1-100, ade2-1, his3-11,15, leu2-3,112, trp1-1, ura3-1,Δccp1::kanMX4) (19), kindly provided by Professor Langer (Cologne, Germany). Analysis of membrane sorting in these strains was performed as described above at 30 °C.
Analysis of Cox18(1TM) MFP and Cox18FL MFP in the pam16-3 Mutant Strain
Yeast cells carrying Cox18(1TM) MFP or Cox18FL MFP construct were grown overnight at 24 °C in medium supplemented with 2% glucose (w/v). Three A600 units of cells were harvested and resuspended in synthetic defined medium without ammonium sulfate and methionine. Cells were starved at 37 °C for 20 min, and 30 μCi (per A600 unit of cells) of [35S]Met were added to the culture for 10 min at 37 °C. Labeling was terminated by adding TCA to a final concentration of 10% (w/v). Yeast cells were disrupted with glass beads (0.5 mm), and the TCA-precipitated homogenate was spun down, resuspended in 50 μl of resuspension solution (3% SDS, 100 mm Tris base, pH 11.0, 3 mm DTT) per A600 unit of cells, and heated at 100 °C for 10 min. Insoluble debris was pelleted down, and the supernatant was transferred to 700 μl of IP mix (50 mm Tris-HCl, pH 7.5, 150 mm NaCl, 1% Triton X-100, 1 mm PMSF, 4 μl of 50× yeast protease inhibitor mixture; Roche Applied Science). A preclearing was performed by adding 10 μl of Pansorbin (Calbiochem) in IP mix and incubated with rotation for 30 min at 4 °C. After centrifugation at 20,000 × g for 1 min, the supernatant was transferred to a new tube and incubated with 2 μl of anti-HA antibody for 1 h at 4 °C. Finally, 8 μl of Pansorbin in IP mix were added and incubated with rotation at 4 °C for 2 h or longer. Samples were centrifuged at 20,000 × g for 1 min, the supernatant was discarded, and the pellet was washed three times with IP buffer (50 mm Tris-HCl, pH 7.5, 150 mm NaCl, 1% Triton X-100, 0.2% SDS) and once with 10 mm Tris-HCl, with a pH of 7.5. Moreover, 30 μl of SDS-PAGE sample buffer was added to the pellet and incubated for 5 min at 95 °C before being loaded onto the SDS gels. Protein bands were visualized in a Fuji FLA-3000 phosphor imager (Fujifilm).
Analysis of Cox5aT-HA and Cox5aFL-HA in the yta10 Deletion Strain
Labeling of yeast cells carrying Cox5aT(1–128)-HA or Cox5aFL(1–153)-HA construct with [35S]Met was performed with various chase times at 30 °C, as described above.
RESULTS
Experimental Design
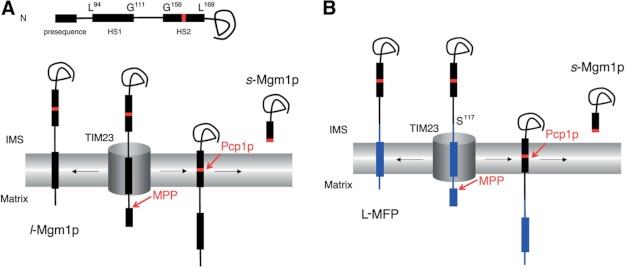
Mgm1p, a dynamin-like GTPase involved in the fusion of mitochondria in Saccharomyces cerevisiae, participates in a unique topogenic process (15, 20). After being targeted to the mitochondria by an N-terminal presequence, HS1 (amino acid residues 94–111) acts as a TM anchor for a fraction of the molecules (∼30–40%). For the remaining molecules, the HS1 slips through the TIM23 complex, and when the rhomboid cleavage region (RCR; residues between 160 and 161 or residues 162 and 163) in the second hydrophobic segment (HS2; residues 156–169) enters the inner membrane, it is cleaved by Pcp1p (a rhomboid protease), and the C-terminal end of Mgm1p is released to the IMS (Fig. 1A). Thus, two Mgm1p isoforms are generated: a membrane-anchored long isoform (l-Mgm1p) and a soluble short isoform (s-Mgm1p).
FIGURE 1.
Processing of Mgm1p and MFPs. A, alternative topogenesis of Mgm1p in yeast mitochondria (15). Residues in Mgm1p are shown for the start and the end of HS1 and HS2. RCR is shown in red. B, MFP is composed of a test protein that includes a TM domain or full-length sequence in the N terminus (blue) (Table 2) and the C terminus of Mgm1p starting from the Ser residue at position 117 (indicated) (black).
We have reasoned that the C-terminal domain of Mgm1p, which includes the RCR, could be a useful reporter for membrane insertion of the upstream TM segment of a mitochondrial inner membrane protein, as used successfully in our previous study with model TM segments (16). If the TM segment of the test membrane protein is sorted by the stop transfer mechanism, a membrane-anchored L-MFP would be generated (Fig. 1B). However, if the TM segment of the test protein is not arrested by the TIM23 complex but is instead translocated to the matrix, as in the conservative sorting pathway, the RCR of the MFP would move to the inner membrane and be cleaved by Pcp1p, and the short Mgm1p form (s-Mgm1p) would result (Fig. 1B).
To test the feasibility of this approach, we chose several mitochondrial membrane proteins with known sorting pathways (Table 1) and prepared fusion constructs containing the N-terminal part of the test protein (including the TM segment plus 10 or more downstream residues; Table 2) and the C-terminal part of the Mgm1p (residues 117–902). For proteins with more than two TM segments, the number of TM segments fused to the Mgm1p is noted in parentheses; and in case of a full-length protein, it is denoted as FL. To facilitate Western blot analysis, three copies of the hemagglutinin (HA) tag were fused to the C terminus of all MFPs.
TABLE 1.
Proteins tested in this study
| Systematic name | Gene name | Previously determined sorting pathway | Reference |
|---|---|---|---|
| YBR024w | SCO2 | Arrested | Refs. 8 and 43 |
| YBR037c | SCO1 | Arrested | Refs. 8 and 44 |
| YBR044c | TCM62 | Conservative | Refs. 8, 45. and 46 |
| YBR185c | MBA1 | Conservative | Refs. 8 and 47 |
| YDL174c | DLD1 | Arrested | Refs. 8 and 48 |
| YDR316w | OMS1 | Arrested | Refs. 8 and 49 |
| YDR393w | SHE9 | Arrested | Refs. 8 and 50 |
| YEL024w | RIP1 | Conservative | Refs. 7 and 8 |
| YER017c | YTA10 | Conservative | Refs. 8 and 14 |
| YER154w | OXA1 | Conservative | Refs. 8 and 51 |
| YGR062c | COX18 | Conservative | Refs. 8, 52, and 53 |
| YIL111w | COX5B | Arrested | Refs. 8 and 54 |
| YMR302c | YME2 | Arrested | Refs. 8 and 55 |
| YNL052w | COX5A | Arrested | Refs. 8 and 56 |
| YOR065w | CYT1a | Arrested | Refs. 1, 4, 8, 13, and 57 |
| YOR334w | MRS2 | Conservative | Refs. 8 and 14 |
| YPL063w | TIM50 | Arrested | Refs. 8 and 58 |
| YPR024w | YME1 | Arrested | Refs. 8, 59, and 60 |
a Cyt1p carries a presequence in the N terminus and a TM domain in the C terminus, forming a NIMS-Cmatrix membrane topology after cleavage of the presequence. Previously, studies have shown that the presequence (residues 1–64) of Cyt1p contains both matrix-targeting and stop transfer sorting signals; thus, the first 70-residue region of Cyt1p was fused to the C-terminal part of Mgm1p.
TABLE 2.
Information of Mgm1 fusion proteins
Residues in the predicted TM sequences by the ΔG predictor (42) are indicated. aa, amino acids.
| Systematic name | Name | Length (aa) | Predicted TM domains | Predicted TM sequence | Length of MFP (aa)a |
|---|---|---|---|---|---|
| YBR024w | Sco2 MFP | 115 | 1 | 79RWKATIALLLLSGGTYAYL97 | 944 |
| YBR037c | Sco1 MFP | 108 | 1 | 71FSTGKAIALFLAVGGALSYFF91 | 937 |
| YBR044c | Tcm62 MFP | 501 | 1 | 468FMTKVGINAVLSAVILPSEVAF489 | 1330 |
| YBR185c | Mba1 MFP | 107 | 1 | 74VFAHPLIVANALIRRLYTF92 | 936 |
| YDL174c | Dld1 MFP | 377 | 1 | 43WLKYSVIASSATLFGYLFA61 | 1206 |
| YDR178w | Sdh4 (1TM) MFP | 98 | 1 | 69WYMEKIFALSVVPLATTAMLTT90 | 927 |
| YDR178w | Sdh4 (1–2TM) MFP | 128 | 2 | 69WYMEKIFALSVVPLATTAMLTT90 | 957 |
| 93LSTAADSFFSVMLLGYCYM111 | |||||
| YDR178w | Sdh4FL MFP | 181 | 3 | 69WYMEKIFALSVVPLATTAMLTT90 | 1010 |
| 93LSTAADSFFSVMLLGYCYM111 | |||||
| 131KYAMYMLGLGSAVSLFGIYKL151 | |||||
| YDR316w | Oms1 MFP | 143 | 1 | 105MTKYMIGAYVIFLIYGLFFTKKL127 | 972 |
| YDR393w | She9 (1TM) MFP | 333 | 1 | 296TWGTFILMGMNIFLFIVLQLLL317 | 1162 |
| YDR393w | She9FL MFP | 456 | 2 | 296TWGTFILMGMNIFLFIVLQLLL317 | 1285 |
| 438FYLYSISLVSMTILVSGLI456 | |||||
| YEL024w | Rip1 MFP | 87 | 1 | 53RSYAYFMVGAMGLLSSAGA71 | 916 |
| YEL024w | Rip1FL MFP | 215 | 1 | 53RSYAYFMVGAMGLLSSAGA71 | 1044 |
| YER017c | Yta10 (1TM) MFP | 160 | 1 | 116FANTMFLTIGFTIIFTLLT134 | 989 |
| YER017c | Yta10 (1–2TM) MFP | 256 | 2 | 116FANTMFLTIGFTIIFTLLT134 | 1085 |
| 225FTFLFPFLPTIILLGGLYFITR246 | |||||
| YER154w | Oxa1 (1–4TM) MFP | 316 | 4 | 126LPWWGTIAATTILIRCLMFPLYV148 | 1145 |
| 199RWLAAPMLQIPIALGFFNALR219 | |||||
| 245PYLGLQVITAAVFISFTRL263 | |||||
| 278RLFTILPIISIPATMNLSSAVVL300 | |||||
| YGR062c | Cox18 (1TM) MFP | 88 | 1 | 51ASHIPWIVLVPLTTMTLRTLVTL73 | 917 |
| YGR062c | Cox18FL MFP | 316 | 4 | 51ASHIPWIVLVPLTTMTLRTLVTL73 | 1145 |
| 167ALLPMVQIPLWVTVSMGIRTLT188 | |||||
| 215LVAMPLLAPILVGTLAVLNVEL236 | |||||
| 273RLGCVVMLAMSSQAPFLLSLYWI295 | |||||
| YGR183c | Qcr9FL MFP | 66 | 1 | 10FFKRNAVFVGTIFAGAFVFQTVF32 | 895 |
| YHR001w-a | Qcr10FL MFP | 77 | 1 | 30LMLWGGASMLGLFVFTEGW48 | 906 |
| YIL111w | Cox5b MFP | 126 | 1 | 91FITKGVFLGLGISFGLFGLVRLL113 | 955 |
| YJL003w | Cox16FL MFP | 118 | 1 | 34FLFFGLPFCATIVLGSFWLSSFT56 | 947 |
| YJL166w | Qcr8FL MFP | 94 | 1 | 49FRRFKSQFLYVLIPAGIYWYWWK71 | 923 |
| YMR302c | Yme2 MFP | 384 | 1 | 286TRIAIPVLFALLSIFAVLVF305 | 1213 |
| YNL052w | Cox5a MFP | 128 | 1 | 94FIAKGVAAGLLFSVGLFAVVRMA116 | 957 |
| YNL052w | Cox5aFL MFP | 153 | 1 | 94FIAKGVAAGLLFSVGLFAVVRMA116 | 982 |
| YOR065w | Cyt1 MFP | 70 | 1 | 36LVTAGVAAAGITASTLLYA54 | 899 |
| YOR065w | Cyt1FL MFP | 309 | 2 | 36LVTAGVAAAGITASTLLYA54 | 1138 |
| 268RLGLKTVIILSSLYLLSIWV287 | |||||
| YOR334w | Mrs2 (1TM) MFP | 344 | 1 | 315VTIYTLGFTVASVLPAFYGMNL336 | 1173 |
| YOR334w | Mrs2 (1–2TM) MFP | 421 | 2 | 315VTIYTLGFTVASVLPAFYGMNL336 | 1250 |
| 345WGFTSVAVFSIVSALYITK363 | |||||
| YPL063w | Tim50 MFP | 151 | 1 | 112YANWFYIFSLSALTGTAIYMAR133 | 980 |
| YPR024w | Yme1 MFP | 330 | 1 | 230VSRWVKWLLVFGILTYSFS248 | 1159 |
a Length of MFP (amino acids) = length of a test protein + the C terminus of Mgm1p(117–902) + 3XHA.
Mgm1 Fusion to Inner Membrane Proteins Sorted by Stop Transfer Generated More L-MFP
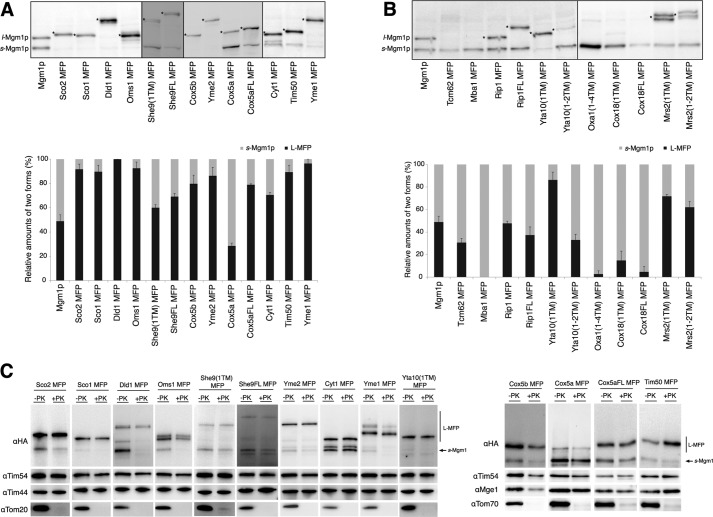
Yeast transformants carrying different MFP constructs were grown in selective medium overnight. Whole-cell lysates were subjected to SDS-PAGE and Western blotting with an anti-HA antibody. Mgm1p was used as a control, providing reference bands for l- and s-Mgm1p on Western blots. Relative amounts of L-MFP and s-Mgm1p for each sample (L-MFP plus s-Mgm1p equals 100%) were quantified (Fig. 2A).
FIGURE 2.
Membrane insertion assay with proteins sorted by the stop transfer or conservative sorting pathway. A, stop transfer proteins fused to the Mgm1p. Whole-cell lysates were prepared from W303-1α yeast strain carrying the indicated MFP constructs and analyzed by SDS-PAGE and Western blotting. Relative amounts of L-MFP and s-Mgm1p are quantified and plotted. s-Mgm1p and l-Mgm1p are shown, and the L-MFP form is indicated by asterisks. B, conservative sorting proteins fused to the Mgm1p and analyzed as in A. Experiments were carried out at least three times for each sample, and the average is shown with S.E. (error bars). C, localization of L-MFPs. Mitochondria were isolated from the indicated yeast transformants and treated with or without proteinase K (PK). Samples were analyzed by SDS-PAGE and Western blotting with the indicated antibodies, Tom70(OM), Tom20(OM), Tim54(IMS), Mge1(Matrix), Tim44(Matrix), and HA.
For Mgm1 fusions of inner membrane proteins known to be sorted by the stop transfer pathway, with the exceptions of She9(1TM) MFP and truncated Cox5a MFP, L-MFP was the major product (more than 70% of L-MFP forms) in all samples (Fig. 2A), indicating that the TM segments of these proteins were integrated into the inner membrane in a stop transfer fashion. For Cox5a and She9, both truncated and full-length MFPs were prepared and analyzed for membrane insertion. Compared with the truncated Cox5a MFP, Cox5aFL MFP resulted in an increase in the L-MFP form (30% for the truncated versus 80% for the full-length), suggesting that the downstream sequence of the TM segment is important for proper sorting of Cox5a. She9FL MFP also showed a slight increase in membrane insertion (70% of L-MFP) relative to She9(1TM) MFP (60% of L-MFP). Interestingly, membrane insertion of She9(1TM) MFP was highly sensitive to growth temperature, in that the L-MFP form was the major product (∼90%) when grown at 25 °C (Fig. 3B).
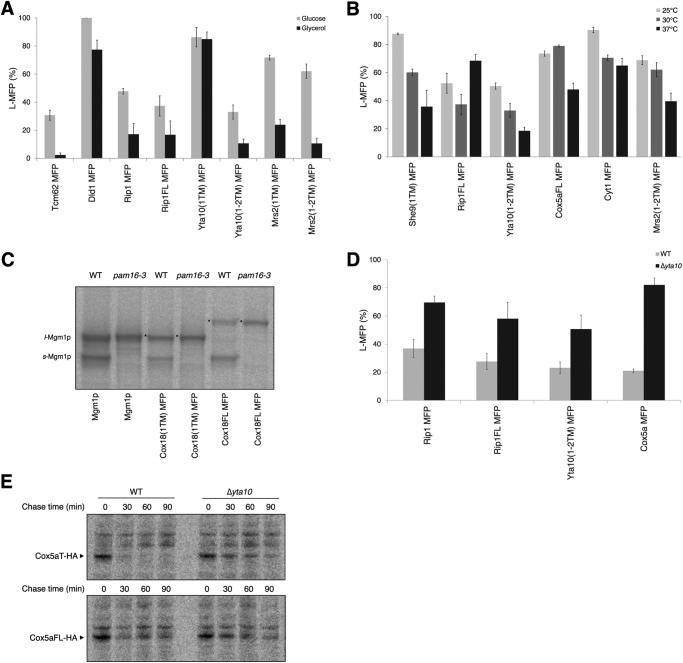
FIGURE 3.
A–D, import and membrane insertion of Mgm1 fusion proteins grown in the medium supplemented with glycerol (3%, w/v) (A), in various temperatures (B), in the yeast strain defective in Pam16 function (C), and in the yeast strain defective in m-AAA function (D). E, expression of Cox5aT-HA and Cox5aFL-HA in Δyta10 and its isogenic wild type strains. In C and E, yeast cells carrying the indicated constructs in the pam16-3 or Δyta10 and the isogenic wild type strains were radiolabeled for 10 min and chased for the indicated time (E). Proteins were immunoprecipitated and analyzed by SDS-PAGE and autoradiography. s-Mgm1p and l-Mgm1p are shown, and the L-MFP form is indicated by asterisks. Error bars, S.E.
Mgm1 Fusion to Inner Membrane Proteins Sorted by the Conservative Sorting Pathway Generated More s-Mgm1p
Mgm1 fusions of the inner membrane proteins known to be sorted by the conservative sorting route were tested. The major products of Mba1 MFP, Oxa1(1–4TM) MFP, Cox18(1TM) MFP, and Cox18FL MFP were s-Mgm1p (more than 80%), suggesting that the TM segment(s) of these proteins was translocated to the matrix, so that the downstream RCR of MFP was accessible for cleavage by Pcp1p in the inner membrane. Tcm62 MFP, Rip1 MFP, Rip1FL MFP, Yta10(1–2TM) MFP, Mrs2(1TM) MFP, and Mrs2(1–2TM) MFP showed ∼30–70% membrane insertion (Fig. 2B). However, when Tcm62 MFP, Rip1 MFP, Rip1FL MFP, Yta10(1–2TM) MFP, Mrs2(1TM) MFP, and Mrs2(1–2TM) MFP were grown in the medium supplemented with the non-fermentable carbon source, glycerol, the amounts of L-MFP were significantly decreased (<20% of total products, a 2–5-fold reduction) (Fig. 3A), indicating that the TM segment(s) of these proteins were efficiently imported into the matrix. Thus, these data suggest that, depending on growth under respiring or fermentable conditions, sorting of inner membrane proteins carrying moderately hydrophobic TM segments can significantly vary.
MFPs Are Present in Mitochondria
Pcp1p is present in the inner membrane (15); thus, the formation of s-Mgm1p is a good indication that MFPs are targeted to the mitochondria. However, to determine whether L-MFPs are present in the mitochondria, mitochondria from yeast transformants producing L-MFPs were isolated and subjected to proteinase K treatment and Western blot analysis.
Various L-MFPs were protected from proteinase K treatment, whereas mitochondrial outer membrane marker Tom20 or Tom70 was degraded, implying that L-MFPs were targeted and present in mitochondria (Fig. 2C). Compared with whole-cell lysates (Fig. 2A), data from the mitochondria preparation show somewhat increased amounts of s-Mgm1p (Fig. 2C). We suspect Pcp1 activity continued during mitochondria preparation. In the cases of Dld1 MFP and Oms1 MFP, s-Mgm1p was degraded upon proteinase K treatment, indicating that they might have leaked out from the mitochondria during mitochondria preparation.
Growth under Respiring Conditions Increases the Import of MFPs Carrying Moderately Hydrophobic TM Segments into the Matrix
When yeast cells grow in the medium supplemented with non-fermentable carbon sources, such as glycerol, cells depend on respiration by oxidative phosphorylation, and membrane potentials across the inner membrane are assumed to be stronger when compared with growth in a medium containing glucose.
To check whether a strong inner membrane potential affects the sorting of MFPs, yeast cells expressing different test proteins were grown in the medium containing glycerol (Fig. 3A). Although the MFPs sorted by the stop transfer were only slightly affected, with the exception of Dld1 MFP, the import of MFPs sorted by the conservative sorting resulted in a significant decrease in the L-MFP form (Fig. 3A and supplemental Fig. S1A). Except for Yta10(1TM) MFP, all test proteins sorted by the conservative sorting pathway produced less than 20% of the L-MFP form, indicating that the TM segment(s) of these proteins was more efficiently imported into the matrix under respiring growth conditions than under fermentable growth conditions. The hydrophobicity of the Dld1p TM segment is markedly low compared with other TM segments sorted by the stop transfer and is in the range of the hydrophobicity of TM segments sorted by conservative sorting (Table 3 and Fig. 5A). These results show that membrane insertion of moderately hydrophobic TM segments is especially susceptible to changes in growth conditions and/or the strength of the inner membrane potential. Furthermore, these data suggest that the conservative sorting process is highly sensitive to the energetic states of mitochondria in the cell.
TABLE 3.
Summary of membrane insertion of MFPs under various growth conditions, in the pam16-3 and Δyta10 strains
In comparison with samples from wild type cells grown in the glucose medium at 30 °C, increase or decrease in membrane insertion of MFPs is indicated as a positive or negative value. The table is sorted according to the predicted free energy of membrane insertion (ΔGapp) for the first TM segment.
| Systematic name | Name | Glycerol | Temperature |
pam16-3 | Δyta10 | ΔGapp | |
|---|---|---|---|---|---|---|---|
| 25 °C | 37 °C | ||||||
| YDR393w | She9 (1TM) MFP | +28 | −24 | −1.985 | |||
| YDR393w | She9FL MFP | +22 | −1.985 | ||||
| YIL111w | Cox5b MFP | −1.163 | |||||
| YMR302c | Yme2 MFP | −1.079 | |||||
| YDR316w | Oms1 MFP | −0.89 | |||||
| YER017c | Yta10 (1TM) MFP | −0.619 | |||||
| YER017c | Yta10 (1–2TM) MFP | −22 | −21 | +28 | −0.619 | ||
| YER154w | Oxa1 (1–4TM) MFP | −0.549 | |||||
| YNL052w | Cox5a MFP | +61 | −0.228 | ||||
| YNL052w | Cox5aFL MFP | −31 | −0.228 | ||||
| YBR024w | Sco2 MFP | 0.023 | |||||
| YPL063w | Tim50 MFP | 0.398 | |||||
| YBR037c | Sco1 MFP | 0.479 | |||||
| YPR024w | Yme1 MFP | 1.063 | |||||
| YOR065w | Cyt1 MFP | +20 | 1.125 | ||||
| YGR062c | Cox18 (1TM) MFP | 1.261 | |||||
| YGR062c | Cox18FL MFP | +69a | 1.261 | ||||
| YOR334w | Mrs2 (1TM) MFP | −48 | 1.334 | ||||
| YOR334w | Mrs2 (1–2TM) MFP | −51 | −22 | 1.334 | |||
| YDL174c | Dld1 MFP | −23 | 1.479 | ||||
| YEL024w | Rip1 MFP | −31 | +33 | 2.172 | |||
| YEL024w | Rip1FL MFP | −21 | +31 | −20 | +30 | 2.172 | |
| YBR185c | Mba1 MFP | 2.321 | |||||
| YBR044c | Tcm62 MFP | −28 | −21 | 2.842 | |||
a Values from radiolabeling at 37 °C (three independent experiments).
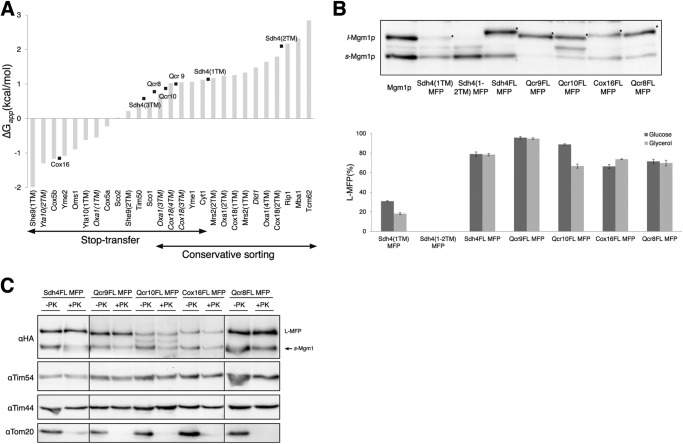
FIGURE 5.
Mgm1 fusion assay is used to determine sorting of inner membrane proteins for which the sorting pathway is unknown. A, hydrophobicity of TM segments of the proteins inserted by stop transfer or conservative sorting mechanism are plotted according to ΔGapp (42). Hydrophobicity of TM segments of proteins of unknown sorting pathway is indicated in black squares. The TM segments of proteins that fall out of the expected range are labeled in italic type. B, whole-cell lysates were prepared from yeast cells carrying the indicated MFP construct grown in the medium supplemented with glucose (2%) or glycerol (3%), analyzed by SDS-PAGE and Western blotting. Relative amounts of L-MFP compared with s-Mgm1p are quantified and plotted. s-Mgm1p and l-Mgm1p are shown, and the L-MFP form is indicated by asterisks. C, localization of L-MFPs. Samples were analyzed as in Fig. 2C. Error bars, S.E.
Growth at Different Temperatures Shows Varying Membrane Insertion Efficiency for MFPs
An earlier study by Gaume et al. (21) has shown that the import rate of a precursor protein into the mitochondria could increase at higher temperatures due to the unfolding of protein domains. Recently, Harner et al. (22) have also shown that the N terminus of GFP-tagged Tim23p is localized differently, depending on growth temperatures. These studies demonstrate that mitochondrial protein import efficiency can vary depending on growth temperatures. However, it is unknown if membrane insertion efficiency also varies depending on growth temperatures.
To test the effects of temperature on translocation and membrane insertion of MFPs, cells carrying various MFP constructs were grown at 25, 30, or 37 °C. Whole-cell lysates were prepared and then analyzed by SDS-PAGE and Western blotting (Fig. 3B and supplemental Fig. S1B). She9(1TM) MFP, Yta10(1–2TM) MFP, Cox5aFL MFP, Cyt1 MFP, and Mrs2(1–2TM) MFP showed varying degrees of decrease in the generation of L-MFP at 37 °C, compared with 25 °C (Fig. 3B), indicating the facilitation of import at 37 °C. In contrast, Rip1FL MFP showed an increase in the generation of L-MFP at 37 °C when compared with 30 °C, implying that membrane insertion was increased whereas translocation to the matrix was reduced at 37 °C. This observation agrees with the recent report by Wagener et al. (7) that Rip1 import increased at lower temperatures. Interestingly, membrane insertion of Rip1FL MFP, but not Rip1 MFP, increased at 37 °C, suggesting that the C-terminal portion of Rip1 plays a role in temperature-dependent import. Recently, Mzm1 has been found to mediate the C-terminal folding of Rip1 in the matrix (23). Deletion of Mzm1 markedly reduced protein levels of Rip1 at 37 °C, which is indicative of a labile nature for the Rip1 C-terminal domain at 37 °C. It is speculated that the C-terminal portion of Rip1 may form folding intermediates or aggregates in the cytosol or IMS at higher temperatures, which would prevent further import into the matrix. In sum, these data show that import and insertion of some membrane proteins in mitochondria are sensitive to growth temperatures, suggesting that membrane protein sorting can be modulated according to the cellular environment.
Defective Import Motor Function Increases Membrane Insertion of Some MFPs
It has been previously shown that the formation of Mgm1p isoforms is especially sensitive to the activity of the mitochondrial import motor. When it was defective, l-Mgm1p was produced more than s-Mgm1p (15, 16), implying that the formation of Mgm1p isoforms depends on the balance between membrane insertion of the moderately hydrophobic HS1 into the inner membrane by the stop transfer mechanism and translocation into the matrix mediated by the import motor. In a similar way, we reasoned that moderately hydrophobic TM segment(s) in proteins sorted by the conservative sorting mechanism might be sensitive to impaired function of the import motor.
To elucidate the possible effects of the import motor on the sorting of MFPs, MFP constructs were transformed with the yeast mutant strain defective in the function of Pam16 (18), which is one of the subunits of the import motor (PAM complex). Although proteins sorted by stop transfer were not affected, Mba1 MFP, Cox18(1TM) MFP, Cox18FL MFP, and Mrs2(1–2TM) MFP, sorted by the conservative sorting pathway, showed a 17–69% increase in production of the L-MFP form in the Pam16 mutant strain (Table 3 and supplemental Fig. S1C), suggesting that TM segment(s) might be arrested in the inner membrane en route to the matrix when the import motor is impaired. This effect was especially noticeable for Cox18FL MFP because it displayed a significant increase in generation of L-MFP in the Pam16 mutant strain (Fig. 3C), indicating that the PAM complex is required to properly sort Cox18p. It has been shown previously that mtHsp70, which is the central component of the PAM complex, is required for import of Cox18 into the matrix (24). Yta10(1–2TM) MFP and Rip1FL MFP produced slightly less of the L-MFP form in the Pam16 mutant strain grown at a non-permissive temperature (S1C). Although the import of Cox18 is severely impaired in the pam16-3 mutant strain, other conservative sorting proteins showed only modest defects (Table 3).
Pam16 is one of the subunits of the PAM complex. Recent studies by Schilke et al. (25) analyzed interactions between the PAM and the TIM23 complexes and reported that Pam16/Tim44 mediate interaction with the TIM23 complex in the matrix and that Pam18 mediates interaction in the IMS. Disruption of either interaction has only modest effects on protein import. Furthermore, the authors showed that when the interaction between Pam16 and Tim44 was defective, higher amounts of Pam17, a newly found component, were associated with the TIM23 complex. Therefore, it may be that either the defect in Pam16 is somewhat mitigated by other PAM subunits or that Pam16 selectively assists the import of a subset of proteins, such as Cox18.
Defective m-AAA Function Increases Membrane Insertion of Some Test Proteins
The m-AAA protease is a hetero-oligomeric complex in the mitochondrial inner membrane shown to be involved in the mitochondrial protein quality control system by degrading misfolded proteins (26, 27). However, recent reports have revealed that the m-AAA protease is also involved in biogenesis of Ccp1p, a heme-binding protein (19), and MrpL32, a ribosomal subunit (28) in S. cerevisiae. When MrpL32 maturation is defective in the m-AAA deletion strain, mitochondrial translation is impaired, leading to deletions in the mitochondrial DNA (29). Consequently, it is assumed that these mitochondria have weakened inner membrane potential, compared with the yeast cells containing mitochondrial DNA.
To test whether the m-AAA protease affects biogenesis of MFPs, as observed in Ccp1p (19), all MFPs were expressed in a yeast strain deleted with one of the m-AAA subunits, YTA10. This strain also lacks the CCP1 gene, so the isogenic wild type strain is Δccp1. Whole-cell lysates were prepared from these strains and analyzed by SDS-PAGE and Western blotting. Membrane insertions of Rip1 MFP, Rip1FL MFP, Yta10(1–2TM) MFP, and Cox5a MFP were more than 2-fold increased in m-AAA mutant strain, compared with the isogenic wild type strain (Fig. 3D and supplemental Fig. S1D). Membrane insertion of Rip1 MFP, Rip1FL MFP, and Yta10(1–2TM) MFP was dependent on growth conditions (glucose versus glycerol in the medium) (Fig. 3A). Although it is uncertain, increased membrane insertion of these fusion proteins may be due to protein import defects caused by reduced membrane potential in the inner membrane of the m-AAA-defective strain.
Membrane insertion of Cox5a MFP, but not the Cox5aFL MFP, was 4-fold increased for the m-AAA defective strain (Fig. 3D and supplemental Fig. S1D). Previous studies have shown that the m-AAA protease is involved in the degradation of misfolded proteins in the mitochondrial inner membrane for quality control (26, 27). Thus, we suspected that the Cox5a C-terminal residues might mark this protein for recognition and degradation by the m-AAA protease. In this situation, Cox5a MFP would be pulled into the matrix by the m-AAA protease, and consequently more s-Mgm1p would be formed. To test this possibility, HA-tagged Cox5a truncated (Cox5aT-HA) and full-length (Cox5aFL-HA) proteins were prepared, and the stability of these proteins in the yta10 deletion strain was determined by the pulse-chase experiment (Fig. 3E). After 30 min of chase, the band corresponding to the Cox5aT-HA was greatly decreased in the wild type strain, whereas it was stable in the yta10 deletion strain. In comparison, Cox5aFL-HA expression was stable over 30 min for both the yta10 deletion and wild type strains. Therefore, these results suggest a role of the m-AAA protease in sorting or in quality control of Cox5a in mitochondria.
Yta10(1–2TM) MFP Is Inserted Into the Inner Membrane from the IMS
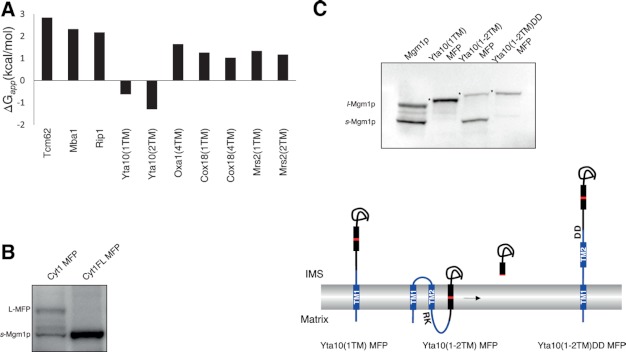
Previously, Baumann et al. (14) have reported that Yta10p is integrated into the mitochondrial inner membrane via the conservative sorting pathway. Yta10p has two TM segments with both N and C termini facing the matrix. Comparing the hydrophobicity of TM segments sorted by conservative sorting, we noticed that the TM segments of Yta10p are markedly more hydrophobic than others (Fig. 4A and Table 3).
FIGURE 4.
Yta10(1–2TM) MFP is inserted into the inner membrane from the IMS. A, ΔGapp (42) of TM segments of proteins sorted by the conservative sorting pathway. B, membrane insertion of Cyt1 MFP and Cyt1FL MFP was assessed by SDS-PAGE and Western blotting. C, whole-cell lysates were prepared from yeast transformants carrying Yta10(1TM) MFP, Yta10(1–2TM) MFP, or Yta10(1–2TM)DD MFP and analyzed by SDS-PAGE and Western blotting. s-Mgm1p and l-Mgm1p are shown, and L-MFP form is indicated by asterisks. Schematics of MFP sorting mechanisms are illustrated. Yta10p is shown in blue, and Mgm1p is shown in black, with the RCR in red. TM domains are shown in rectangles.
For Yta10(1TM) MFP, the major product was L-MFP (Figs. 2B and 4C), indicating that the TM1 was anchored in the inner membrane. Not only under the normal growth conditions but under all growth conditions and in the Pam16 and m-AAA mutant strains, Yta10(1TM) MFP was present as the L-MFP form (Figs. 2B and 3A and supplemental Fig. S1), suggesting that the TM1 of Yta10p is firmly anchored in the inner membrane. Such results were observed only with the stop transfer TM segments, suggesting that the TM1 of Yta10p is sorted by the stop transfer mechanism.
For Yta10(1–2TM) MFP, s-Mgm1p was ∼70% of the product (Figs. 2B and 4C). This observation raised two possibilities: 1) the presence of the downstream TM2 interferes with the membrane insertion of the upstream TM1, or 2) the TM1, inserted by the stop transfer mechanism, and the TM2, inserted as positively charged residues downstream of the TM2 are pulled into the matrix, as observed for Cyt1p (13). First, to check whether the Mgm1 fusion approach is suitable for the assessment of the latter mode of membrane insertion, Cyt1 MFP and Cyt1FL MFP were prepared and analyzed for their import and membrane insertion. Whereas Cyt1 MFP produced the L-MFP form, Cyt1FL MFP generated mainly s-Mgm1p (Fig. 4B), indicating that the C terminus of Cyt1FL MFP might be pulled into the matrix as a loop between the C-terminal TM segment of Cyt1p and the HS2 of Mgm1p. Subsequently, the RCR of Mgm1p was positioned in the inner membrane for Pcp1p cleavage.
We reasoned that if the TM2 of Yta10p is inserted in the inner membrane by pulling the positively charged residues downstream of the TM2 into the matrix, mutations in those residues would prevent membrane insertion of the TM2. Positively charged residues (246RK247) in the downstream region of the TM2 were mutated to negatively charged Asp residues (DD), and relative amounts of L-MFP and s-Mgm1p were determined. Yta10(1–2TM) MFP carrying a DD mutation resulted in formation of the L-MFP form (Fig. 4C). These results strongly support the mode of Yta10p insertion from the IMS as proposed previously (13).
Mgm1 Fusion Assay Is Used to Determine Sorting Pathway for Inner Membrane Proteins with Unknown Sorting Mechanisms
Next, we selected a number of inner membrane proteins that carry an N-terminal targeting sequence and whose sorting pathways are unknown. Qcr8p (30), Qcr9p, and Qcr10p (31–33) are subunits of cytochrome bc1 complex, Sdh4p (34, 35) is a subunit of the succinate dehydrogenase complex, and Cox16p (36) is a cytochrome oxidase assembly factor. Except for Sdh4p, which contains three TM segments, all are single-spanning membrane proteins with the C terminus oriented to the IMS. The hydrophobicity of the TM segments of these proteins is shown in Fig. 5A. Membrane insertion of MFPs was analyzed by SDS-PAGE and Western blotting. Sdh4FL MFP, Qcr9FL MFP, and Qcr10FL MFP gave rise to a majority of the L-MFP forms (>80%), and Cox16FL MFP and Qcr8FL MFP resulted in 65–70% L-MFP forms (Fig. 5B). Membrane insertion of Qcr10FL MFP resulted in a 22% reduction when grown in glycerol medium compared with grown in glucose medium, but membrane insertion of the other MFPs did not show much difference between fermentable and respiring growth conditions (Fig. 5B). Localization of MFPs in the mitochondria was confirmed by proteinase K protection of isolated mitochondria (Fig. 5C).
In Fig. 5A, the TM segments of the known pathway proteins are sorted according to ΔG. In the range of ΔG where both pathways overlap, the presence or absence of the proline residue(s) in the TM domain seems to be an important determinant as suggested previously (8, 9); Oxa1(3TM), Cox18(4TM), and Cox18(3TM) contain Pro residues, whereas Yme1 and Cyt1 do not. The hydrophobicity of Qcr8, Qcr9, and Qcr10 TM segments falls within the overlapping range for both pathways. Qcr9 and Qcr10 do not have Pro residues, but Qcr8 contains one Pro residue in the TM domain. Examining the sequences near the TM domain of Qcr8, positively charged residues flanking the TM domain were found. Our previous study (16) has shown that positively charged flanking residues significantly enhance membrane insertion efficiency. Thus, if the hydrophobicity of the TM domain of inner membrane proteins falls within the overlapping range of ΔG for both pathways, the presence of Pro and/or flanking charged residues influences whether the TM segment is laterally integrated by the stop transfer mechanism or imported into the matrix. The hydrophobicity of Cox16 and Shd4(3TM) TM segments falls within the range of ΔG for the stop transfer pathway. We conclude that the TM segments of Qcr10p, Qcr9p, Cox16p, Qcr8p, and TM3 of Sdh4p are probably sorted by the stop transfer mechanism.
The hydrophobicity of Sdh4(1TM) and Sdh4(2TM) TM segments falls within the range of ΔG for the conservative sorting pathway (Fig. 5A). Sdh4(1TM) MFP and Sdh4(1–2TM) MFP resulted predominantly in s-Mgm1p (Fig. 5B), suggesting that the TM1 and TM2 of Sdh4p were imported into the matrix. These results suggest that whereas the TM1 and TM2 of Sdh4p are sorted by conservative sorting, the TM3 is inserted into the inner membrane by stop transfer. Cooperation of stop transfer and conservative sorting mechanisms has been previously observed for Mdl1p, a multispanning ABC transporter (37). We propose that Sdh4p is also integrated into the inner membrane by coordination of both conservative and stop transfer sorting mechanisms.
DISCUSSION
Compared with the endoplasmic reticulum, where the Sec61 complex mediates protein translocation and membrane insertion, mitochondria are more complex, with various sorting pathways and different machineries for import and membrane sorting. Nucleus-encoded mitochondrial inner membrane proteins that carry an N-terminal presequence either are integrated into the inner membrane by the stop transfer mechanism or are fully imported into the matrix and then integrated into the inner membrane from the matrix by the conservative sorting pathway. The major point of divergence of these two sorting pathways is at the TIM23 complex in the inner membrane.
The Mgm1 fusion approach enabled the assessment of the sorting of mitochondrial inner membrane proteins in various growth conditions and in the mutant yeast strain defective in its import motor function or m-AAA activity (Table 3). Not only conservative sorting or stop transfer mechanisms but also the mode of membrane insertion for proteins such as Cyt1p and Yta10p were capable of being validated by the Mgm1 fusion method. Testing in different growth conditions and with different yeast mutant strains shows that the sorting of mitochondrial inner membrane proteins that contain moderately hydrophobic TM segments and follow the conservative sorting pathway is especially sensitive to the cellular environment.
Most proteins that follow the conservative sorting pathway are directly or indirectly involved in the assembly of respiratory chain complexes in the inner membrane. When cells ferment under fermentable growth conditions, the electron transfer chain may not function fully; thus, these protein subunits may be less susceptible to damage. In turn, they may undergo slower protein turnover, and mitochondria may not need to import them actively. Recently, Schmidt et al. (38) have reported that Tom70, a receptor subunit of the TOM complex in the outer membrane, becomes phosphorylated under non-respiring conditions, inhibiting the import of carrier proteins. Carrier proteins include major subunits of the import machineries, such as Tim23, Tim17, and Tim22. Both that study (38) and this study thus show that the import of some mitochondrial proteins is reduced or inhibited under non-respiring conditions. However, it is unclear whether the import of only a subset of proteins, such as those involved in mitochondrial respiration, is reduced or if mitochondria reduce the overall import of proteins gradually (i.e. only after the number of inner membrane import machineries is reduced are the matrix and inner membrane proteins consequently reduced as well).
Mitochondria constantly undergo fusion and fission to mix their contents, to rescue and restore damaged mitochondria, or to sort out irreversibly damaged mitochondria (39). Varying degrees of import and membrane insertion of MFPs in vivo may thus reflect the energetic state of the individual mitochondrion. High sensitivity to the cellular environment in the sorting of MFPs suggests two possibilities for dynamic sorting processes in mitochondria: 1) the inner membrane protein sorting may be modulated and finely tuned, depending on environmental changes (e.g. carbon source and temperature), or 2) healthy mitochondria may be fully capable of, whereas damaged mitochondria have limited capacity for, proper sorting (e.g. changes in the inner membrane potential and/or impaired function of the import motor or m-AAA function). The latter possibility could also be one way to mark the mitochondrion for repair or degradation. Recently, an example of missorting in defective mitochondria has been shown for PINK1, a serine/threonine kinase involved in Parkinson disease (40, 41).
Acknowledgments
We thank laboratory members for critical reading of the manuscript and Professors Langer (Cologne, Germany), Pfanner (Freiburg, Germany), and Endo (Nagoya, Japan) for the yeast strains and antibodies.
This work was supported by Basic Research Grant 2012-0001935 and Global Research Network Grant C00048 from the National Research Foundation of Korea (to H. K.).

This article contains supplemental Fig. S1.
- TM
- transmembrane
- MFP
- Mgm1 fusion protein
- L-MFP
- long MFP
- s-Mgm1p
- soluble short Mgm1p
- l-Mgm1p
- long Mgm1p
- IP
- immunoprecipitation
- RCR
- rhomboid cleavage region
- HS
- hydrophobic segment
- IMS
- intermembrane space.
REFERENCES
- 1. van Loon A. P., Brändli A. W., Schatz G. (1986) The presequences of two imported mitochondrial proteins contain information for intracellular and intramitochondrial sorting. Cell 44, 801–812 [DOI] [PubMed] [Google Scholar]
- 2. Neupert W., Herrmann J. M. (2007) Translocation of proteins into mitochondria. Annu. Rev. Biochem. 76, 723–749 [DOI] [PubMed] [Google Scholar]
- 3. Bolender N., Sickmann A., Wagner R., Meisinger C., Pfanner N. (2008) Multiple pathways for sorting mitochondrial precursor proteins. EMBO Rep. 9, 42–49 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Glick B. S., Brandt A., Cunningham K., Müller S., Hallberg R. L., Schatz G. (1992) Cytochromes c1 and b2 are sorted to the intermembrane space of yeast mitochondria by a stop-transfer mechanism. Cell 69, 809–822 [DOI] [PubMed] [Google Scholar]
- 5. van der Laan M., Hutu D. P., Rehling P. (2010) On the mechanism of preprotein import by the mitochondrial presequence translocase. Biochim. Biophys. Acta 1803, 732–739 [DOI] [PubMed] [Google Scholar]
- 6. Herrmann J. M., Funes S. (2005) Biogenesis of cytochrome oxidase-sophisticated assembly lines in the mitochondrial inner membrane. Gene 354, 43–52 [DOI] [PubMed] [Google Scholar]
- 7. Wagener N., Ackermann M., Funes S., Neupert W. (2011) A pathway of protein translocation in mitochondria mediated by the AAA-ATPase Bcs1. Mol. Cell 44, 191–202 [DOI] [PubMed] [Google Scholar]
- 8. Meier S., Neupert W., Herrmann J. M. (2005) Proline residues of transmembrane domains determine the sorting of inner membrane proteins in mitochondria. J. Cell Biol. 170, 881–888 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Herrmann J. M., Neupert W. (2003) Protein insertion into the inner membrane of mitochondria. IUBMB Life 55, 219–225 [DOI] [PubMed] [Google Scholar]
- 10. Beasley E. M., Müller S., Schatz G. (1993) The signal that sorts yeast cytochrome b2 to the mitochondrial intermembrane space contains three distinct functional regions. EMBO J. 12, 2303–2311 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Ono H., Gruhler A., Stuart R. A., Guiard B., Schwarz E., Neupert W. (1995) Sorting of cytochrome b2 to the intermembrane space of mitochondria. Kinetic analysis of intermediates demonstrates passage through the matrix. J. Biol. Chem. 270, 16932–16938 [DOI] [PubMed] [Google Scholar]
- 12. Schwarz E., Seytter T., Guiard B., Neupert W. (1993) Targeting of cytochrome b2 into the mitochondrial intermembrane space. Specific recognition of the sorting signal. EMBO J. 12, 2295–2302 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Arnold I., Fölsch H., Neupert W., Stuart R. A. (1998) Two distinct and independent mitochondrial targeting signals function in the sorting of an inner membrane protein, cytochrome c1. J. Biol. Chem. 273, 1469–1476 [DOI] [PubMed] [Google Scholar]
- 14. Baumann F., Neupert W., Herrmann J. M. (2002) Insertion of bitopic membrane proteins into the inner membrane of mitochondria involves an export step from the matrix. J. Biol. Chem. 277, 21405–21413 [DOI] [PubMed] [Google Scholar]
- 15. Herlan M., Bornhövd C., Hell K., Neupert W., Reichert A. S. (2004) Alternative topogenesis of Mgm1 and mitochondrial morphology depend on ATP and a functional import motor. J. Cell Biol. 165, 167–173 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Botelho S. C., Osterberg M., Reichert A. S., Yamano K., Björkholm P., Endo T., von Heijne G., Kim H. (2011) TIM23-mediated insertion of transmembrane α-helices into the mitochondrial inner membrane. EMBO J. 30, 1003–1011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Oldenburg K. R., Vo K. T., Michaelis S., Paddon C. (1997) Recombination-mediated PCR-directed plasmid construction in vivo in yeast. Nucleic Acids Res. 25, 451–452 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Frazier A. E., Dudek J., Guiard B., Voos W., Li Y., Lind M., Meisinger C., Geissler A., Sickmann A., Meyer H. E., Bilanchone V., Cumsky M. G., Truscott K. N., Pfanner N., Rehling P. (2004) Pam16 has an essential role in the mitochondrial protein import motor. Nat. Struct. Mol. Biol. 11, 226–233 [DOI] [PubMed] [Google Scholar]
- 19. Tatsuta T., Augustin S., Nolden M., Friedrichs B., Langer T. (2007) m-AAA protease-driven membrane dislocation allows intramembrane cleavage by rhomboid in mitochondria. EMBO J. 26, 325–335 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Herlan M., Vogel F., Bornhovd C., Neupert W., Reichert A. S. (2003) Processing of Mgm1 by the rhomboid-type protease Pcp1 is required for maintenance of mitochondrial morphology and of mitochondrial DNA. J. Biol. Chem. 278, 27781–27788 [DOI] [PubMed] [Google Scholar]
- 21. Gaume B., Klaus C., Ungermann C., Guiard B., Neupert W., Brunner M. (1998) Unfolding of preproteins upon import into mitochondria. EMBO J. 17, 6497–6507 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Harner M., Neupert W., Deponte M. (2011) Lateral release of proteins from the TOM complex into the outer membrane of mitochondria. EMBO J. 30, 3232–3241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Cui T. Z., Smith P. M., Fox J. L., Khalimonchuk O., Winge D. R. (2012) Late stage maturation of the Rieske Fe/S protein. Mzm1 stabilizes Rip1 but does not facilitate its translocation by the AAA ATPase Bcs1. Mol. Cell Biol. 32, 4400–4409 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Frazier A. E., Chacinska A., Truscott K. N., Guiard B., Pfanner N., Rehling P. (2003) Mitochondria use different mechanisms for transport of multispanning membrane proteins through the intermembrane space. Mol. Cell Biol. 23, 7818–7828 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Schilke B. A., Hayashi M., Craig E. A. (2012) Genetic analysis of complex interactions among components of the mitochondrial import motor and translocon in Saccharomyces cerevisiae. Genetics 190, 1341–1353 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Korbel D., Wurth S., Käser M., Langer T. (2004) Membrane protein turnover by the m-AAA protease in mitochondria depends on the transmembrane domains of its subunits. EMBO Rep. 5, 698–703 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Leonhard K., Guiard B., Pellecchia G., Tzagoloff A., Neupert W., Langer T. (2000) Membrane protein degradation by AAA proteases in mitochondria. Extraction of substrates from either membrane surface. Mol. Cell 5, 629–638 [DOI] [PubMed] [Google Scholar]
- 28. Bonn F., Tatsuta T., Petrungaro C., Riemer J., Langer T. (2011) Presequence-dependent folding ensures MrpL32 processing by the m-AAA protease in mitochondria. EMBO J. 30, 2545–2556 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Myers A. M., Pape L. K., Tzagoloff A. (1985) Mitochondrial protein synthesis is required for maintenance of intact mitochondrial genomes in Saccharomyces cerevisiae. EMBO J. 4, 2087–2092 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Maarse A. C., Grivell L. A. (1987) Nucleotide sequence of the gene encoding the 11-kDa subunit of the ubiquinol-cytochrome c oxidoreductase in Saccharomyces cerevisiae. Eur. J. Biochem. 165, 419–425 [DOI] [PubMed] [Google Scholar]
- 31. Brandt U., Uribe S., Schägger H., Trumpower B. L. (1994) Isolation and characterization of QCR10, the nuclear gene encoding the 8.5-kDa subunit 10 of the Saccharomyces cerevisiae cytochrome bc1 complex. J. Biol. Chem. 269, 12947–12953 [PubMed] [Google Scholar]
- 32. Lange C., Hunte C. (2002) Crystal structure of the yeast cytochrome bc1 complex with its bound substrate cytochrome c. Proc. Natl. Acad. Sci. U.S.A. 99, 2800–2805 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Smith P. M., Fox J. L., Winge D. R. (2012) Biogenesis of the cytochrome bc1 complex and role of assembly factors. Biochim. Biophys. Acta 1817, 276–286 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Bullis B. L., Lemire B. D. (1994) Isolation and characterization of the Saccharomyces cerevisiae SDH4 gene encoding a membrane anchor subunit of succinate dehydrogenase. J. Biol. Chem. 269, 6543–6549 [PubMed] [Google Scholar]
- 35. Oyedotun K. S., Lemire B. D. (1997) The carboxyl terminus of the Saccharomyces cerevisiae succinate dehydrogenase membrane subunit, SDH4p, is necessary for ubiquinone reduction and enzyme stability. J. Biol. Chem. 272, 31382–31388 [DOI] [PubMed] [Google Scholar]
- 36. Carlson C. G., Barrientos A., Tzagoloff A., Glerum D. M. (2003) COX16 encodes a novel protein required for the assembly of cytochrome oxidase in Saccharomyces cerevisiae. J. Biol. Chem. 278, 3770–3775 [DOI] [PubMed] [Google Scholar]
- 37. Bohnert M., Rehling P., Guiard B., Herrmann J. M., Pfanner N., van der Laan M. (2010) Cooperation of stop-transfer and conservative sorting mechanisms in mitochondrial protein transport. Curr. Biol. 20, 1227–1232 [DOI] [PubMed] [Google Scholar]
- 38. Schmidt O., Harbauer A. B., Rao S., Eyrich B., Zahedi R. P., Stojanovski D., Schönfisch B., Guiard B., Sickmann A., Pfanner N., Meisinger C. (2011) Regulation of mitochondrial protein import by cytosolic kinases. Cell 144, 227–239 [DOI] [PubMed] [Google Scholar]
- 39. Westermann B. (2010) Mitochondrial fusion and fission in cell life and death. Nat. Rev. Mol. Cell Biol. 11, 872–884 [DOI] [PubMed] [Google Scholar]
- 40. Jin S. M., Lazarou M., Wang C., Kane L. A., Narendra D. P., Youle R. J. (2010) Mitochondrial membrane potential regulates PINK1 import and proteolytic destabilization by PARL. J. Cell Biol. 191, 933–942 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Meissner C., Lorenz H., Weihofen A., Selkoe D. J., Lemberg M. K. (2011) The mitochondrial intramembrane protease PARL cleaves human Pink1 to regulate Pink1 trafficking. J. Neurochem. 117, 856–867 [DOI] [PubMed] [Google Scholar]
- 42. Hessa T., Meindl-Beinker N. M., Bernsel A., Kim H., Sato Y., Lerch-Bader M., Nilsson I., White S. H., von Heijne G. (2007) Molecular code for transmembrane-helix recognition by the Sec61 translocon. Nature 450, 1026–1030 [DOI] [PubMed] [Google Scholar]
- 43. Lode A., Paret C., Rödel G. (2002) Molecular characterization of Saccharomyces cerevisiae Sco2p reveals a high degree of redundancy with Sco1p. Yeast 19, 909–922 [DOI] [PubMed] [Google Scholar]
- 44. Buchwald P., Krummeck G., Rödel G. (1991) Immunological identification of yeast SCO1 protein as a component of the inner mitochondrial membrane. Mol. Gen. Genet. 229, 413–420 [DOI] [PubMed] [Google Scholar]
- 45. Dibrov E., Fu S., Lemire B. D. (1998) The Saccharomyces cerevisiae TCM62 gene encodes a chaperone necessary for the assembly of the mitochondrial succinate dehydrogenase (complex II). J. Biol. Chem. 273, 32042–32048 [DOI] [PubMed] [Google Scholar]
- 46. Klanner C., Neupert W., Langer T. (2000) The chaperonin-related protein Tcm62p ensures mitochondrial gene expression under heat stress. FEBS Lett. 470, 365–369 [DOI] [PubMed] [Google Scholar]
- 47. Preuss M., Leonhard K., Hell K., Stuart R. A., Neupert W., Herrmann J. M. (2001) Mba1, a novel component of the mitochondrial protein export machinery of the yeast Saccharomyces cerevisiae. J. Cell Biol. 153, 1085–1096 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Rojo E. E., Guiard B., Neupert W., Stuart R. A. (1998) Sorting of d-lactate dehydrogenase to the inner membrane of mitochondria. Analysis of topogenic signal and energetic requirements. J. Biol. Chem. 273, 8040–8047 [DOI] [PubMed] [Google Scholar]
- 49. Lemaire C., Guibet-Grandmougin F., Angles D., Dujardin G., Bonnefoy N. (2004) A yeast mitochondrial membrane methyltransferase-like protein can compensate for oxa1 mutations. J. Biol. Chem. 279, 47464–47472 [DOI] [PubMed] [Google Scholar]
- 50. Messerschmitt M., Jakobs S., Vogel F., Fritz S., Dimmer K. S., Neupert W., Westermann B. (2003) The inner membrane protein Mdm33 controls mitochondrial morphology in yeast. J. Cell Biol. 160, 553–564 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Herrmann J. M., Neupert W., Stuart R. A. (1997) Insertion into the mitochondrial inner membrane of a polytopic protein, the nuclear-encoded Oxa1p. EMBO J. 16, 2217–2226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Saracco S. A., Fox T. D. (2002) Cox18p is required for export of the mitochondrially encoded Saccharomyces cerevisiae Cox2p C-tail and interacts with Pnt1p and Mss2p in the inner membrane. Mol. Biol. Cell 13, 1122–1131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Funes S., Gerdes L., Inaba M., Soll J., Herrmann J. M. (2004) The Arabidopsis thaliana chloroplast inner envelope protein ARTEMIS is a functional member of the Alb3/Oxa1/YidC family of proteins. FEBS Lett. 569, 89–93 [DOI] [PubMed] [Google Scholar]
- 54. Cumsky M. G., Trueblood C. E., Ko C., Poyton R. O. (1987) Structural analysis of two genes encoding divergent forms of yeast cytochrome c oxidase subunit V. Mol. Cell Biol. 7, 3511–3519 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Hanekamp T., Thorsness P. E. (1996) Inactivation of YME2/RNA12, which encodes an integral inner mitochondrial membrane protein, causes increased escape of DNA from mitochondria to the nucleus in Saccharomyces cerevisiae. Mol. Cell Biol. 16, 2764–2771 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Miller B. R., Cumsky M. G. (1993) Intramitochondrial sorting of the precursor to yeast cytochrome c oxidase subunit Va. J. Cell Biol. 121, 1021–1029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. van Loon A. P., Brändli A. W., Pesold-Hurt B., Blank D., Schatz G. (1987) Transport of proteins to the mitochondrial intermembrane space. The “matrix-targeting” and the “sorting” domains in the cytochrome c1 presequence. EMBO J. 6, 2433–2439 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Mokranjac D., Sichting M., Popov-Celeketić D., Mapa K., Gevorkyan-Airapetov L., Zohary K., Hell K., Azem A., Neupert W. (2009) Role of Tim50 in the transfer of precursor proteins from the outer to the inner membrane of mitochondria. Mol. Biol. Cell 20, 1400–1407 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Leonhard K., Herrmann J. M., Stuart R. A., Mannhaupt G., Neupert W., Langer T. (1996) AAA proteases with catalytic sites on opposite membrane surfaces comprise a proteolytic system for the ATP-dependent degradation of inner membrane proteins in mitochondria. EMBO J. 15, 4218–4229 [PMC free article] [PubMed] [Google Scholar]
- 60. Graef M., Seewald G., Langer T. (2007) Substrate recognition by AAA+ ATPases. Distinct substrate binding modes in ATP-dependent protease Yme1 of the mitochondrial intermembrane space. Mol. Cell Biol. 27, 2476–2485 [DOI] [PMC free article] [PubMed] [Google Scholar]