Background: To characterize an unknown molecular basis of the hif-1-independent hypoxia response, we used the nematode hsp-16.1 gene as a model.
Results: HMG-1.2, together with chromatin-remodeling factors and calcium ions, is involved in the hypoxia response of hsp-16.1.
Conclusion: Chromatin modification is important for the hypoxia response of hsp-16.1 in an HIF-1-independent manner.
Significance: We report an alternative regulatory pathway for the HIF-1-independent hypoxia response.
Keywords: C. elegans, Calcium Signaling, Chromatin Remodeling, Hypoxia, Hypoxia-inducible Factor (HIF)
Abstract
Oxygen deprivation is accompanied by the coordinated expression of numerous hypoxia-responsive genes, many of which are controlled by hypoxia-inducible factor-1 (HIF-1). However, the cellular response to hypoxia is not likely to be mediated by HIF-1 alone, and little is known about HIF-1-independent hypoxia responses. To better establish the molecular mechanisms of HIF-1-independent hypoxia responses, we sought to characterize the molecular basis of the hypoxia response of the hsp-16.1 gene in the nematode Caenorhabditis elegans; this gene has been shown to be induced by hypoxia independently of hif-1. Using affinity purification followed by LC-MS/MS, we identified HMG-1.2 as a protein that binds to a specific promoter region under hypoxic conditions. By systematic prediction followed by validation of these interactions through RNAi, we identified the chromatin modifiers isw-1 and hda-1, histone H4, and NURF-1 chromatin-remodeling factors as new components of the hif-1-independent hypoxia response. These data suggest that the modulation of nucleosome positioning at the hsp-16.1 promoter may be important for the hypoxia response. In addition, we found that calcineurin acts independently of hif-1 to modulate the cellular response to hypoxia and that calcium ions are necessary for the induction of hsp-16.1 under hypoxic conditions.
Introduction
The cellular response to low oxygen levels is critical for organism survival. In normal development and in many diseased states such as cardiovascular disease and cancers involving solid tumors, cells must cope with the challenge of oxygen deprivation. Dynamic changes in the expression of specific hypoxia-responsive genes underlie the cell's metabolic adaptation to low oxygen conditions. Hypoxia-inducible factor (HIF)3 is known to be a master regulator of this response, and the roles of HIF in the cellular response to changes in the environmental oxygen concentration have been extensively studied (1–4). HIF is a heterodimeric protein composed of the HIF-1α and HIF-1β subunits. Under hypoxic conditions, HIF-1α stabilizes and subsequently binds to the promoters of target genes, thereby activating their transcription (5, 6). hif-1 is the Caenorhabditis elegans ortholog of HIF-1α, and it functions not only in the adaptive response to hypoxia but also in other biological processes, including stress responses, behavioral responses, neuronal development, and aging (7–12).
HIF-1 has been shown to be a transcription factor that is critical for the hypoxia response in animals under various conditions (13, 14); however, it has become obvious that HIF-1 is not the only regulator of this response. Furthermore, the existence of HIF-1-independent mechanisms mediating the hypoxia response has been reported. For example, c-Myc, NF-κB, and AP-1 seem to regulate hypoxia-responsive genes through an HIF-1-independent mechanism in different types of cancer (15). Nonetheless, only a few attempts have been made to elucidate the regulatory mechanism governing HIF-1-independent adaptations to low oxygen levels. In this study, using C. elegans as a model organism, we sought to identify an alternative hypoxia response pathway that is not dependent on HIF-1 function. The C. elegans homolog of the HIF-1α subunit and, importantly, most of the core signaling pathways in the hypoxia response are conserved through evolution (7, 16). In addition, feeding RNAi provides an easy way to systematically inactivate genes in C. elegans. In a study on the hypoxia-induced alteration of gene expression in C. elegans hif-1 mutants, it was reported that HIF-1-independent pathways are involved in the adaptation to hypoxia (17). Our previous study also revealed that the response of small heat shock protein hsp-16 genes (hsp-16.1 and hsp-16.2) to hypoxic conditions is HIF-1-independent and occurs via cis-acting DNA sequences (CAC(A/T)CT), hereafter referred to as “block I,” in the promoter region of these genes (18).
To establish the HIF-1-independent hypoxia response pathway, we dissected the molecular mechanism that mediates the hypoxia-inducible transcription of the C. elegans hsp-16.1 gene. We identified a potential factor that is involved in the HIF-1-independent hypoxia response of hsp-16.1 using affinity chromatography purification followed by LC-MS/MS. To further understand the mechanism, we employed an interactome network approach, which reveals direct and indirect interactions based on yeast, worm, fly, and human orthology (19, 20). Thereafter, we experimentally validated whether these genes are involved in the hypoxia response of hsp-16.1 expression. Our results indicate that chromatin-remodeling complex proteins are involved in the modulation of hsp-16.1 expression under hypoxic conditions. We also show that this hypoxia-inducible transcriptional regulation is mediated by calcium signaling. We report the existence of an alternative, HIF-1-independent mechanism by which cells adapt to hypoxic conditions.
EXPERIMENTAL PROCEDURES
Worm Strains and Culture
The standard protocol for maintaining C. elegans strains was used as described previously (21). C. elegans strains were obtained from the Caenorhabditis Genetics Center (Minneapolis, MN). The N2 strain was used as wild-type worms.
GFP Fusion Constructs and Microinjection
The full-length hsp-16.1::gfp fusion plasmid was constructed by subcloning the full-length PCR product from the genomic T27E4.8 sequence into pPD95.77 (a gift of A. Fire). To generate the hmg-1.2::gfp construct, DNA was amplified by PCR. The PCR product included the 2-kb region upstream of the F47D12.4 promoter, and the whole genomic sequence was cloned into pPD95.77. Microinjection was carried out using standard procedures. The pRF4 plasmid containing the dominant mutant rol-6 gene was used as a marker. The hsp-16.1::gfp transgene was integrated into the genome by UV irradiation using Spectrolinker XL-1000 (Spectronics, Rochester, NY).
RNAi Assay
The bacterial feeding protocol was used in RNAi experiments as described previously (22, 23). HT115 bacteria carrying L4440, the plasmid of the empty vector pPD129.36, were used as a control. Most RNAi feeding clones were obtained from the Ahringer RNAi library (Geneservice, Cambridge, United Kingdom). Feeding vectors containing Y53F4B.3 were obtained from the Vidal RNAi feeding library (Open Biosystems, Huntsville, AL). For nurf-1, a 950-bp PCR fragment of cDNA corresponding to nucleotides 251–1201 of F26H11.2 was subcloned into pPD129.36 (a gift of A. Fire). For H20J04.2, a 900-bp sequence of the third exon was amplified by PCR and subcloned into pPD129.36. In most experiments, we placed L4 stage worms (parents or P0) on a plate seeded with a bacterial strain carrying specific RNAi plasmids. For hda-1, which is an essential gene that causes lethality when severely knocked down, we placed synchronized L1 stage worms under RNAi conditions and performed a hypoxia assay 3 days later.
Heat Shock and Hypoxia Assay
The heat shock treatment was carried out at 30 °C for 6 h. For hypoxia treatment, we soaked worms in M9 buffer without rocking as described by Hong et al. (18). The worms were allowed to recover on nematode growth medium at 20 °C before analysis.
ChIP
The ChIP experiments were adapted from Oh et al. (24) and performed using a ChIP assay kit (Upstate). Mixed-stage HMG-1.2::GFP transgenic worms were grown on NGM-lite plates and then harvested. Control worms were rocked, whereas hypoxia-treated worms were soaked in M9 medium without shaking. The worms were fixed in M9 buffer containing 2% formaldehyde at room temperature for 30 min. The reaction was quenched with 2.5 m glycine and washed three times with M9 buffer. Lysates were prepared as described above. The lysates were precleaned with salmon sperm DNA/protein G-agarose beads and incubated overnight at 4 °C with either anti-GFP antibody or IgG. The precipitates were washed, the cross-links were reversed, and the DNA was eluted. Real-time quantitative PCR was performed using Bio-Rad iQ SYBR Green Supermix in a Bio-Rad iQ5 real-time PCR machine. The following primer sets were used: sense, 5′-AGGTGCAAAGAGACGCAGAT-3′; and antisense, 5′-CTAGAACATTCGAGCTGCTT-3′.
Microscopy and Measurement of Fluorescence Intensity
All images were taken using an Axioplan 2 microscope equipped with an AxioCam HRc camera and AxioVision 4.7 software (all Zeiss). Fluorescence intensity was analyzed using ImageJ software by outlining the second intestinal cells of the worms. The density was normalized to the L4440 control. Either a one-way analysis of variance (p < 0.05) or an unpaired t test was employed to find genes that were significantly different between the control and RNAi at each time. All of the data are expressed as the mean ± S.E.
Nucleosome Preparation and Micrococcal Nuclease Assay
Mixed-stage worms were pelleted and frozen in buffer A (250 mm sucrose, 10 mm Tris-HCl (pH 8.0), 10 mm MgCl2, 1 mm EGTA, 0.2 mm PMSF, and 7 mm β-mercaptoethanol) and protease inhibitor set III (Calbiochem). The worms were ground into a fine powder in liquid nitrogen. The resultant worm dust was resuspended in buffer A. Micrococcal nuclease (MNase; Roche Applied Science), resuspended at 300 units/μl in 10 mm Tris-HCl (pH 7.4), 15 mm NaCl, 60 mm KCl, 0.15 mm spermine, and 0.5 mm spermidine, was added to the worm extract. Digestions were performed at 25 °C and stopped by the addition of worm lysis buffer (0.1 M Tris-HCl (pH 8.5), 0.1 m NaCl, 50 mm EDTA, and 1% SDS), and the sample was treated with proteinase K (20 mg/ml) for 45 min at 65 °C. Purified DNA without MNase treatment was digested with BglII (Enzynomics), and PstI, RsaI (Fermentas), or XbaI (Roche Applied Science). The same amount of DNA was loaded onto a 1.4% agarose gel, and the separated DNAs were transferred to a nylon membrane. The 200-nucleotide fragment of DNA close to the BglII site within the hsp-16.48 gene as indicated in Fig. 3 was 32P-labeled using a random prime labeling kit (Amersham Biosciences) for indirect end-label analysis.
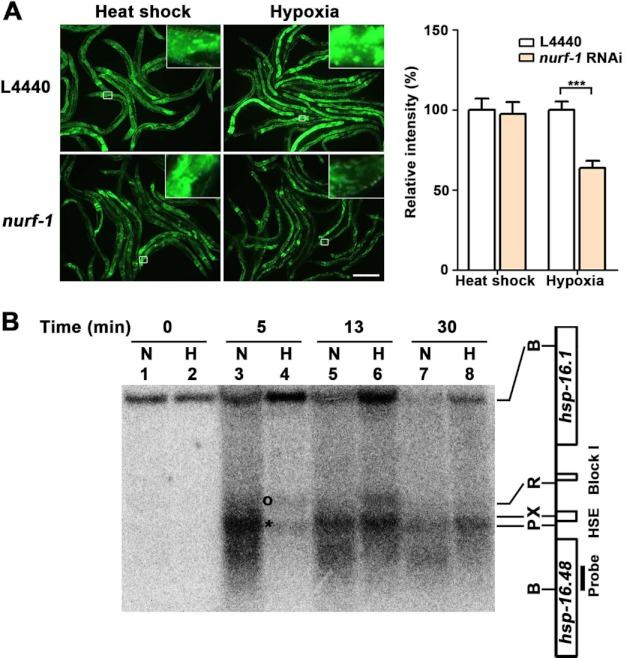
FIGURE 3.
NURF-1 chromatin-remodeling factor mediates the hypoxia response of the hsp-16.1 gene. A, decreased nurf-1 function (Drosophila NURF301) reduced the activation of hsp-16.1 by hypoxia. The results for ACF and CHRAC RNAi are presented in supplemental Fig. S7. Quantification of the relative intensity of HSP-16.1::GFP fluorescence was carried out using ImageJ software. Statistical significance was determined using an unpaired t test (***, p < 0.001). Error bars indicate S.E. Scale bar = 200 μm. B, MNase digestion patterns of the normoxic (N) and hypoxic (H) chromatins. The DNA of MNase-digested nuclei was digested by BglII and analyzed by Southern blot hybridization using a 200-nucleotide probe made from the region designated in the figure. Chromatin was incubated in the presence of MNase for 0 min (lanes 1 and 2), 5 min (lanes 3 and 4), 13 min (lanes 5 and 6), and 30 min (lanes 7 and 8). The asterisk indicates the genomic position that is more sensitive to MNase cleavage in normoxia. The circle indicates the band with increased sensitivity to MNase cleavage in hypoxia. B, BglII; P, PstI; X, XbaI; R, RsaI; HSE, heat shock response element.
EGTA and Thapsigargin Treatment
Worms were treated with EGTA under hypoxic and heat shock conditions at different concentrations (0, 100, 200, and 400 mm). Thapsigargin (Sigma) was used at a final concentration of 1.25 μm while rocking for 6 h. As a control experiment, the same amount of dimethyl sulfoxide was used in parallel.
RESULTS
Identification of HMG-1.2 as a Hypoxia-induced Promoter-binding Protein
Previously, we showed that the sequence CAC(A/T)CT (block I) is required for the HIF-1-independent hypoxia response of hsp-16 genes; we also demonstrated that regulatory proteins bind to block I-containing sequences in vitro (18). To identify the regulatory proteins that bind the hsp-16.1 promoter under hypoxic conditions, we isolated nuclear extracts from worms incubated under hypoxic conditions and applied them to streptavidin affinity columns containing biotin-labeled block I-containing sequences. As a negative control, we used a biotin-labeled probe in which the block I sequences were mutated (supplemental Fig. S1). We analyzed the isolated proteins using MALDI-TOF and quadrupole time-of-flight mass spectrometry (Fig. 1A and supplemental Fig. S2). By comparing the results from the block I-containing column with those from the block I-mutated column, we excluded most bound proteins as nonspecific binders, including HMG-1.1. This analysis showed that HMG-1.2 was the predominant protein that was bound specifically to the block I-containing sequence. We then performed ChIP to evaluate whether HMG-1.2 was bound to the block I-containing promoter in vivo and whether the binding efficiency of HMG-1.2 was affected by hypoxia. We found that HMG-1.2 binding was enriched by >20-fold under hypoxic conditions compared with normoxic conditions (Fig. 1B). Taken together, our results suggest that HMG-1.2 proteins specifically bind to the block I element of the hsp-16.1 promoter in response to hypoxic conditions.
FIGURE 1.
HMG-1.2 is a potential regulator of the hsp-16.1 hypoxia response. A, identification of block I-binding proteins. Purified proteins were separated on SDS-polyacrylamide gels and strained with Coomassie Blue. The identities of the bands were confirmed by MALDI-TOF and quadrupole time-of-flight mass spectrometry. First lane, markers (M); second lane, eluate bound to T-substituted (Tsub) sequence; third lane, eluate bound to block I. Molecular mass standards (in kilodaltons) are shown on the left. The arrowhead indicates HMG-1.2. B, ChIP with anti-GFP antibody and real-time quantitative PCR revealed that HMG-1.2 is directly associated with block I under hypoxic conditions. HSE, heat shock response element; TATA, TATA box. The arrows indicate the primers used for real-time quantitative PCR. C, a reduction in hmg-1.2 function significantly down-regulated the induction of hsp-16.1 under hypoxia, whereas hmg-1.1 RNAi did not affect the hsp-16.1 hypoxia response. Inlets are the second intestine cells that show the most consistent fluorescence intensity within worms. All of the photographs were taken under identical exposure conditions. Scale bar = 200 μm. D, the relative fluorescence intensity of stress-induced HSP-16.1::GFP was quantified using ImageJ software. Statistical significance was determined using a one-way analysis of variance and Dunnett's multiple comparison test with the hypoxia controls (***, p < 0.001). NS, not significant. Error bars indicate S.E.
Role of HMG-1.2 in the Hypoxia Response of hsp-16.1
To investigate the potential roles of HMG-1.2 in the HIF-1-independent response to hypoxia, we performed RNAi to reduce HMG-1.2 activity. L4 larvae were fed dsRNA-producing bacteria. Thereafter, we monitored the expression of hsp-16.1 fused with GFP (hsp-16.1::gfp) in F1 progeny exposed to hypoxic conditions. We also determined whether the expression of hsp-16.1 was regulated in a hypoxia-specific manner or as a general response to stress. To do so, worms were also incubated under heat shock conditions, as heat shock can induce hsp-16.1 gene expression (25, 26). Under hypoxic conditions, disruption of hmg-1.2 activity by RNAi, compared with the empty vector control, led to an ∼60% decrease in the level of HSP-16.1::GFP fusion protein. On the other hand, hmg-1.2 RNAi did not affect the induction of HSP-16.1 in response to heat shock treatment (Fig. 1, C and D). These observations suggest that induction of hsp-16.1 by hypoxia, but not by heat shock, specifically required functional HMG-1.2. As a negative control, we observed no significant change in HSP-16.1::GFP expression in response to either stress in the hmg-1.1 RNAi animals.
We also examined the effect of hmg-1.2 inactivation by RNAi on the activation of hsp-16.1 in the hif-1(ia04) background. We found that hsp-16.1 was strongly induced in hif-1(ia04) mutants and that the induction of hsp-16.1 was decreased by hmg-1.2 RNAi but not by hmg-1.1 RNAi in these mutant animals. Importantly, these observations were similar to those in wild-type animals (supplemental Fig. S3, A and B). Induction of hsp-16.1 by heat shock was not affected in hif-1(ia04) mutants. These results are consistent with the notion that the hypoxia response of hsp-16.1 is not mediated by the hif-1 pathway.
Hypoxia-inducible Expression of hsp-16.1 through Chromatin-remodeling Factors and Histone 4
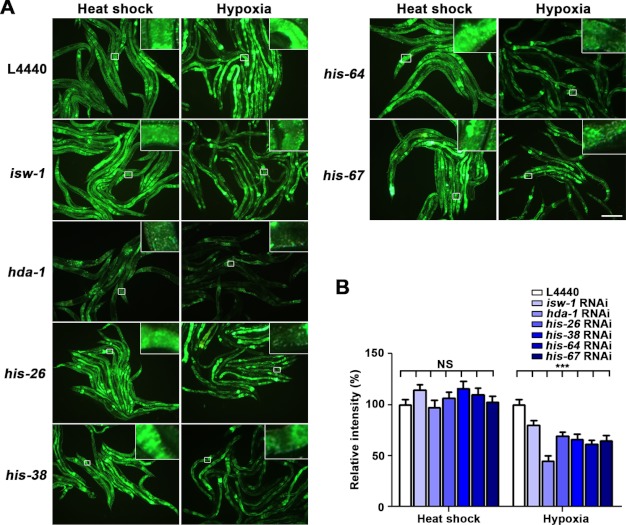
It has been suggested that if two genes are connected in any way, they are likely to participate in a common biological process (27). Therefore, we used functional gene networks to further characterize the mechanism by which hmg-1.2 mediates the HIF-1-independent hypoxia response. We used tools provided by the WormNet v.2 (19, 20) that can predict the genetic or physical interaction of a gene with a gene of interest, in our case, hmg-1.2. WormNet provides functional gene networks by combining different types of data such as co-cited gene associations, mRNA coexpression, protein-protein interaction, physical interaction, and genetic interaction among C. elegans, yeast, fly, and human proteins (19, 20, 28). Probability of interactions is scored with a Bayesian network (28). Genes predicted to interact with hmg-1.2 included a transcription factor (mab-5); a nuclear transport factor (28); a protein kinase (hpk-1); and several chromatin-associated genes such as a predicted subunit of FACT (facilitates chromatin transcription; hmg-4), components of the chromatin-remodeling complex (psa-4 and isw-1), histone H4-coding genes, an H2A.Z histone variant (htz-1), and histone deacetylase (hda-1) (supplemental Table S1). We then inactivated the candidate genes using RNAi to test whether they were involved in the hypoxia response of hsp-16.1. Although we found no effect for some genes, including mab-5, ima-3, hpk-1, hmg-4, psa-4, and htz-1, we did find that the inhibition of his-26, his-38, his-64, his-67, isw-1, and hda-1 by RNAi decreased the expression of the hsp-16.1 gene under hypoxic conditions (Fig. 2, A and B). Given the sequence similarity between his-26, his-38, his-64, and his-67 and their similarity to other histone H4 genes (supplemental Fig. S4), it is conceivable that the inactivation of the four histone H4 genes by RNAi may also have resulted in the silencing of other histone H4 genes. ISW-1 is an ortholog of Drosophila ISWI and acts as an ATPase in chromatin-remodeling complexes. HDA-1 is a histone deacetylase involved in chromatin modification. Our results thus suggest that chromatin modifiers are involved in the hypoxia response of hsp-16.1. Furthermore, the inactivation of isw-1 and histone H4-coding genes by RNAi in hif-1(ia04) deletion mutants showed results that were consistent with the HIF-1 independence of hsp-16.1 in wild-type worms (supplemental Fig. S5, A and B).
FIGURE 2.
Chromatin-remodeling components histone deacetylase and histone H4 are involved in the hsp-16.1 hypoxia response. A, a reduction in isw-1, hda-1, his-26, his-38, his-64, and his-67 RNAi suppressed the hypoxia up-regulation of hsp-16.1. Scale bar = 200 μm. B, quantification of the relative intensity of HSP-16.1::GFP fluorescence was carried out using ImageJ software. ***, values that differ from the L4440 hypoxia controls at the p < 0.001 significance level (Dunnett's test). NS, not significant. Error bars indicate S.E.
The NURF Chromatin-remodeling Complex May Mediate the hif-1-independent Hypoxia Response of hsp-16.1
There are three chromatin-remodeling complexes that contain the ATPase subunit ISWI: ACF (ATP-dependent chromatin assembly and remodeling factor) (29), CHRAC (chromatin accessibility complex) (30), and NURF (nucleosome-remodeling factor) (31). Genes encoding these components of chromatin-remodeling complexes are well conserved in C. elegans: ACF-1 (flt-1 and athp-2), CHRAC-14 (T27A5.8), and CHRAC-16 (Y53F4B.3) (32). In the Drosophila NURF complex, ISWI is composed of NURF301, NURF38, and NURF55 (33, 34); these subunits are homologous to nurf-1, pyp-1, and rba-1 in C. elegans, respectively. We determined whether the C. elegans orthologs of the ACF, CHRAC, and NURF proteins were required for the hypoxia response of hsp-16.1, similar to isw-1. Inactivation of flt-1, athp-2, T7A5.8, and Y53F4B.3 did not affect the induction of hsp-16.1 in response to hypoxia (supplemental Fig. S6). Only RNAi inhibition of the gene encoding the NURF301 ortholog failed to fully induce hsp-16.1 by hypoxia, but not by heat shock (Fig. 3A). Furthermore, we found that NURF-1 regulation of hsp-16.1 was independent of HIF-1 (supplemental Fig. S7, A and B). Our results indicate that ISW-1 likely functions as a component of the NURF complex to regulate the expression of hsp-16.1 in response to hypoxia and that this activity is independent of HIF-1.
Hypoxia Response Occurs through Modification of the Chromatin Environment
Next, we investigated the roles of ISW-1 and NURF-1 during hypoxia. Because NURF has been shown to have nucleosome disruption activity in vitro (35, 36) and because the chromatin structure of hsp-16 genes has been shown to markedly differ upon heat shock treatment (37), we decided to examine whether hypoxia affected the chromatin structure of the hsp-16.1 promoter in vivo. We purified nuclei from control and hypoxia-treated worms and subsequently performed MNase digestion coupled with Southern blot analysis to map nucleosome positioning. Purified nuclei were free from endonucleases, as no degraded bands were observed (Fig. 3B, lanes 1 and 2). The differences in nucleosome arrays were obvious when nuclei were partially digested by MNase (lanes 3–6). To map the relative positions, we digested naked DNA with restricted enzymes (data not shown). An MNase-hypersensitive site (which marks the heat shock response element) was detected under normoxic conditions near the PstI and XbaI sites (asterisk, lanes 3, 5, and 7), and this was attenuated in hypoxia (lanes 4 and 8). In contrast, an MNase-sensitive site (which corresponds to the block I region) appeared in hypoxia-stressed nuclei (circle, lanes 4 and 6). These data suggest that nucleosome remodeling has occurred along the block I region in response to hypoxia and that the block I region is exposed outside of nucleosomes, which in turn results in the transcriptional activation of hsp-16.1.
Calcium Signaling May Mediate the Hypoxia Response in an HIF-1-independent Manner
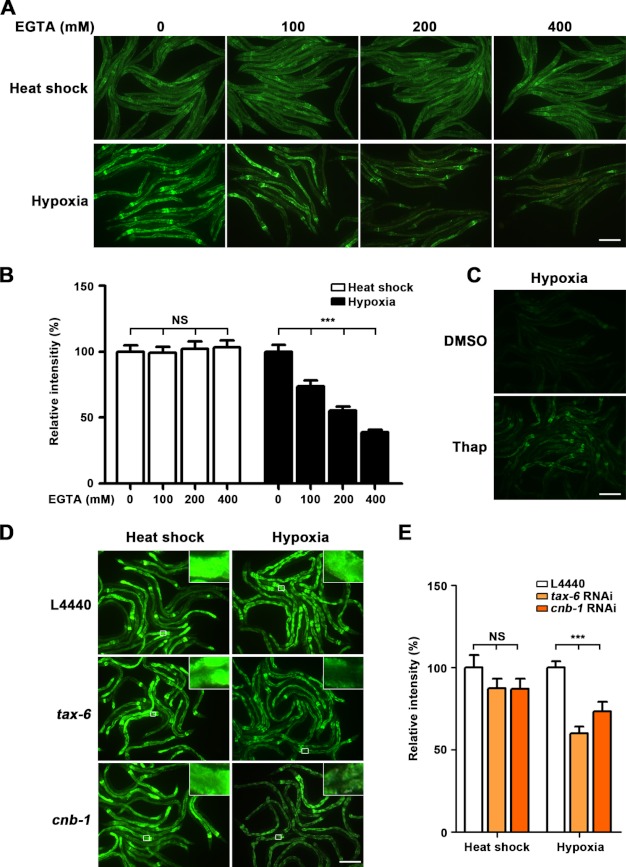
Cells are known to increase intracellular calcium levels as a primary response to hypoxia (38). Furthermore, the binding of calcium to the C-terminal domain of HMGB1, another mammalian homolog of HMG-1.2, is known to modulate the DNA-binding properties HMGB1 (39, 40). Therefore, we investigated whether the disruption of the cellular calcium balance affected the hypoxia response of hsp-16.1. We used chemical reagents to manipulate the endoplasmic reticulum release of calcium: thapsigargin, which induces the release of endoplasmic reticulum calcium, and EGTA, which specifically chelates calcium (41). We found that treatment with EGTA decreased the induction of hsp-16.1 in response to hypoxia in a concentration-dependent manner, whereas heat shock-responsive activation remained unaffected (Fig. 4, A and B). In contrast, thapsigargin treatment was sufficient to activate hsp-16.1 at normal oxygen concentrations (Fig. 4C). Interestingly, tax-6, which encodes the catalytic subunit of calcineurin, was up-regulated in the hif-1(ia04) mutant animals under hypoxic conditions (17). Calcineurin contains a catalytic subunit, calcineurin A, and a regulatory subunit, calcineurin B, which are encoded by the C. elegans genes tax-6 and cnb-1, respectively (41, 51). We found that inactivating tax-6 or cnb-1 resulted in failure to induce hsp-16.1 expression under hypoxia (Fig. 4, D and E). This phenotype is similar to that of hif-1(ia04) mutants (supplemental Fig. S7, A and B). Taken together, our results indicate that changes in cellular calcium levels generate a primary signal that induces the expression of hsp-16.1 in response to hypoxia.
FIGURE 4.
Manipulation of calcium balance affects hypoxia-induced hsp-16.1 expression. A, treatment with the calcium chelator EGTA at different concentrations (0, 100, 200, and 400 mm). EGTA-driven depletion of calcium decreased the induction of hsp-16.1 under hypoxia but not with heat shock treatment. However, EGTA did not affect the heat shock-inducible expression of hsp-16.1. B, quantification of the relative intensity of HSP-16.1::GFP. ***, p < 0.001 (Dunnett's test). NS, not significant. C, thapsigargin (Thap)-induced Ca2+ release mimicked the effect of hypoxia to activate the expression of hsp-16.1. DMSO, dimethyl sulfoxide. D, inactivation of calcineurin decreased the activation of hsp-16.1 under hypoxia. E, quantification of the relative intensity of HSP-16.1::GFP fluorescence was carried out using ImageJ software. ***, values that differ from the L4440 hypoxia controls at the p < 0.001 significance level (Dunnett's test). Error bars indicate S.E. Scale bars = 200 μm.
DISCUSSION
In this study, we sought to better establish the molecular mechanisms of HIF-1-independent hypoxia responses in C. elegans. We have shown that chromatin-remodeling factors, including HMG-1.2, are involved in the hif-1-independent hypoxia response of hsp-16.1 expression. In addition, we have shown that calcium is important for induction of hsp-16.1 transcription in response to hypoxia. Taking all of our data and previous studies together, we propose a mechanism for the HIF-1-independent hypoxia response (Fig. 5). As this regulatory mechanism may not act specifically on hsp-16.1, future study will be needed to investigate the extent to which this mechanism is involved in hypoxia-responsive regulation. Because chromatin-remodeling factors are well conserved in evolution, it is conceivable that a similar mechanism of hif-1-independent hypoxia response may occur in mammals.
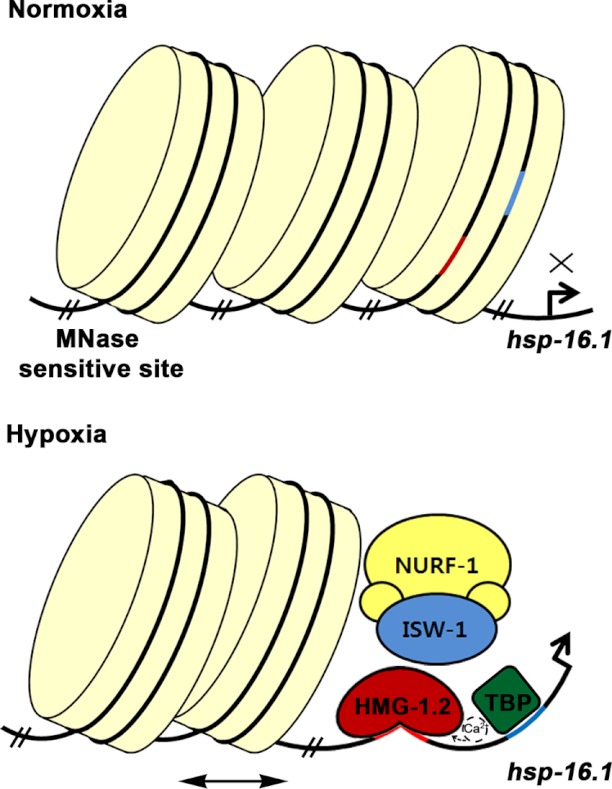
FIGURE 5.
Model of HIF-1-independent hypoxia regulation of hsp-16.1. Under normal oxygen conditions, the block I region (red line), surrounded by nucleosomes, may not activate the transcription of hsp-16.1. Hypoxia induces the release of calcium from the endoplasmic reticulum and influences HMG-1.2 in a way that HMG-1.2 has a higher affinity for DNA. In concert with HMG-1.2, ISW-1 and the NURF-1 remodeling complex are involved in remodeling the positioning of the nucleosomes on block I. The horizontal bidirectional arrow indicates a possible nucleosome sliding. Once HMG-1.2 binds to DNA, it can bend the DNA to allow efficient binding by several elements of the transcription machinery such as TATA-binding protein (TBP). The TATA box is indicated by the blue line.
HMG-1.2 is a homolog of the human HMGB2 protein and exhibits ∼55% identity within the HMG boxes (42). HMGB proteins are non-histone chromatin-binding proteins that are generally thought to have limited sequence-specific DNA recognition ability, preferring to bind to bent DNA and four-way junction DNA (43). Nevertheless, evidence was found in C. elegans that HMG-1.2 might also have site-specific sequence recognition abilities, e.g. in Wnt signaling in specific developmental processes (42). The sequence specificity of HMG-1.2 may allow it to regulate transcription of the hsp-16.1 gene under oxygen-deprived conditions independently of HIF-1. It would be of a great interest to determine whether genes other than hsp-16.1 and hsp-16.2 that contain the block I sequences at their promoter regions also require HMG-1.2 for their response to hypoxia.
A functional gene network enabled us to predict interplays between HMG-1.2 and chromatin-remodeling factors. It is known that HMGB proteins bind to nucleosomes, thereby loosening the wrapped DNA and enhancing its accessibility to chromatin-remodeling complexes (43, 44). HMGB proteins have also been suggested to accelerate the sliding activities of chromatin remodeling and to enhance the binding of chromatin-remodeling factors to nucleosomal DNA (44). In addition, recent genetic evidence has shown that HMG-1.2 is up-regulated by hypoxia and that knockdown of hmg-1.2, isw-1, and hda-1 significantly increases sensitivity to hypoxia (45). These studies support a relationship between HMG-1.2 and chromatin modifiers under hypoxic conditions. Moreover, the N terminus of histone H4 is essential for stimulating ISWI ATPase activity and inducing nucleosome sliding (46, 47). This suggests that histone H4 may act to modulate the activity of chromatin remodelers in the hypoxia response. It would be interesting to investigate whether the deacetylation at the N-terminal region of histone H4 proteins by HDA-1 is involved in this modulation.
Our data suggest that ISW-1 may act with NURF chromatin-remodeling factors to modulate the hypoxia response of hsp-16.1 independently of HIF-1 and that NURF-mediated unwrapping of the block I region by nucleosome movement may contribute to the activation of hsp-16 under hypoxic conditions. Additionally, nurf-1 is up-regulated by hypoxia in hif-1-deficient animals (17). This implies that NURF-1 is regulated under hypoxia independently of HIF-1. Unfortunately, because the null mutants of hmg-1.2, isw-1, and nurf-1 showed severe defects in both development and fertility, we could not determine whether the nucleosome remodeling was abolished in these null mutants in vivo. However, ISWI and NURF are known to promote nucleosome sliding at promoters, which leads to the disruption of regularly ordered arrays and thereby activates gene transcription (36, 48). Given this knowledge, we propose that the chromatin-remodeling complex of ISW-1 and NURF-1 catalyzes the nucleosome sliding at the hsp-16.1 promoter to facilitate transcriptional activation and that transcriptional regulation via nucleosome and chromatin modifications is important for the regulation of the hypoxia response. As knockdown of pyp-1 and rba-1 resulted in embryonic lethality, we could not test whether these genes were involved in the hypoxia response.
Our finding that the hypoxia response of hsp-16.1 is calcium-dependent is consistent with the idea that calcium release is a landmark of early hypoxia response. There are several possible explanations for how hsp-16.1 expression is regulated by calcium signaling under hypoxic conditions. First, an increase in the intracellular calcium levels increases the affinity of HMG-1.2 for DNA, which is necessary to induce the hypoxia response of hsp-16.1. Another possibility is that calcineurin activated by a high cytosolic calcium level may dephosphorylate unidentified substrates, which in turn positively modulate the HIF-1-independent hypoxia response of hsp-16.1. It would be of interest to pursue the issue of the calcium action mechanism in hypoxia response.
HIF-1α is overexpressed in common human cancers, in cells in the center of a solid tumor, which experience hypoxia (49). Although HIF-1α has been considered to be a major target for tumor therapy, HIF-1 is not the only regulator of the hypoxia response in cancer cells (50). Our findings propose a novel alternative mechanism that regulates gene induction in hypoxia in an HIF-1-independent manner. Because C. elegans cellular adaptations to hypoxia are homologous to those found in mammals, HMG-1.2 and the ISW-1 chromatin factor may be therapeutic targets, in addition to HIF-1, and could block hypoxia-induced responses in a combinatorial manner in cancer cells.
Acknowledgment
We thank the Caenorhabditis Genetics Center for the nematode strains.
This work was supported in part by the World Class University Program and the Research Center for Functional Cellulomics.

This article contains supplemental “Experimental Procedures,” Figs. S1–S7, Table S1, and additional references.
- HIF
- hypoxia-inducible factor
- MNase
- micrococcal nuclease.
REFERENCES
- 1. Weidemann A., Johnson R. S. (2008) Biology of HIF-1α. Cell Death Differ. 15, 621–627 [DOI] [PubMed] [Google Scholar]
- 2. Semenza G. L. (1998) Hypoxia-inducible factor 1: master regulator of O2 homeostasis. Curr. Opin. Genet. Dev. 8, 588–594 [DOI] [PubMed] [Google Scholar]
- 3. Semenza G. L. (2001) Hypoxia-inducible factor 1: oxygen homeostasis and disease pathophysiology. Trends Mol. Med. 7, 345–350 [DOI] [PubMed] [Google Scholar]
- 4. Wenger R. H. (2002) Cellular adaptation to hypoxia: O2-sensing protein hydroxylases, hypoxia-inducible transcription factors, and O2-regulated gene expression. FASEB J. 16, 1151–1162 [DOI] [PubMed] [Google Scholar]
- 5. Kim W., Kaelin W. G., Jr. (2003) The von Hippel-Lindau tumor suppressor protein: new insights into oxygen sensing and cancer. Curr. Opin. Genet. Dev. 13, 55–60 [DOI] [PubMed] [Google Scholar]
- 6. Semenza G. L. (2003) Targeting HIF-1 for cancer therapy. Nat. Rev. Cancer 3, 721–732 [DOI] [PubMed] [Google Scholar]
- 7. Jiang H., Guo R., Powell-Coffman J. A. (2001) The Caenorhabditis elegans hif-1 gene encodes a bHLH-PAS protein that is required for adaptation to hypoxia. Proc. Natl. Acad. Sci. U.S.A. 98, 7916–7921 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Treinin M., Shliar J., Jiang H., Powell-Coffman J. A., Bromberg Z., Horowitz M. (2003) HIF-1 is required for heat acclimation in the nematode Caenorhabditis elegans. Physiol. Genomics 14, 17–24 [DOI] [PubMed] [Google Scholar]
- 9. Bretscher A. J., Busch K. E., de Bono M. (2008) A carbon dioxide avoidance behavior is integrated with responses to ambient oxygen and food in Caenorhabditis elegans. Proc. Natl. Acad. Sci. U.S.A. 105, 8044–8049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Chang A. J., Bargmann C. I. (2008) Hypoxia and the HIF-1 transcriptional pathway reorganize a neuronal circuit for oxygen-dependent behavior in Caenorhabditis elegans. Proc. Natl. Acad. Sci. U.S.A. 105, 7321–7326 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Pocock R., Hobert O. (2008) Oxygen levels affect axon guidance and neuronal migration in Caenorhabditis elegans. Nat. Neurosci. 11, 894–900 [DOI] [PubMed] [Google Scholar]
- 12. Chen D., Thomas E. L., Kapahi P. (2009) HIF-1 modulates dietary restriction-mediated lifespan extension via IRE-1 in Caenorhabditis elegans. PLoS Genet. 5, e1000486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Iyer N. V., Kotch L. E., Agani F., Leung S. W., Laughner E., Wenger R. H., Gassmann M., Gearhart J. D., Lawler A. M., Yu A. Y., Semenza G. L. (1998) Cellular and developmental control of O2 homeostasis by hypoxia-inducible factor 1α. Genes Dev. 12, 149–162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Wang G. L., Semenza G. L. (1995) Purification and characterization of hypoxia-inducible factor 1. J. Biol. Chem. 270, 1230–1237 [DOI] [PubMed] [Google Scholar]
- 15. Mizukami Y., Kohgo Y., Chung D. C. (2007) Hypoxia inducible factor-1 independent pathways in tumor angiogenesis. Clin. Cancer Res. 13, 5670–5674 [DOI] [PubMed] [Google Scholar]
- 16. Epstein A. C., Gleadle J. M., McNeill L. A., Hewitson K. S., O'Rourke J., Mole D. R., Mukherji M., Metzen E., Wilson M. I., Dhanda A., Tian Y. M., Masson N., Hamilton D. L., Jaakkola P., Barstead R., Hodgkin J., Maxwell P. H., Pugh C. W., Schofield C. J., Ratcliffe P. J. (2001) C. elegans EGL-9 and mammalian homologs define a family of dioxygenases that regulate HIF by prolyl hydroxylation. Cell 107, 43–54 [DOI] [PubMed] [Google Scholar]
- 17. Shen C., Nettleton D., Jiang M., Kim S. K., Powell-Coffman J. A. (2005) Roles of the HIF-1 hypoxia-inducible factor during hypoxia response in Caenorhabditis elegans. J. Biol. Chem. 280, 20580–20588 [DOI] [PubMed] [Google Scholar]
- 18. Hong M., Kwon J. Y., Shim J., Lee J. (2004) Differential hypoxia response of hsp-16 genes in the nematode. J. Mol. Biol. 344, 369–381 [DOI] [PubMed] [Google Scholar]
- 19. Lee I., Lehner B., Crombie C., Wong W., Fraser A. G., Marcotte E. M. (2008) A single gene network accurately predicts phenotypic effects of gene perturbation in Caenorhabditis elegans. Nat. Genet. 40, 181–188 [DOI] [PubMed] [Google Scholar]
- 20. Lee I., Lehner B., Vavouri T., Shin J., Fraser A. G., Marcotte E. M. (2010) Predicting genetic modifier loci using functional gene networks. Genome Res. 20, 1143–1153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Brenner S. (1974) The genetics of Caenorhabditis elegans. Genetics 77, 71–94 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Timmons L., Court D. L., Fire A. (2001) Ingestion of bacterially expressed dsRNAs can produce specific and potent genetic interference in Caenorhabditis elegans. Gene 263, 103–112 [DOI] [PubMed] [Google Scholar]
- 23. Kamath R. S., Fraser A. G., Dong Y., Poulin G., Durbin R., Gotta M., Kanapin A., Le Bot N., Moreno S., Sohrmann M., Welchman D. P., Zipperlen P., Ahringer J. (2003) Systematic functional analysis of the Caenorhabditis elegans genome using RNAi. Nature 421, 231–237 [DOI] [PubMed] [Google Scholar]
- 24. Oh S. W., Mukhopadhyay A., Dixit B. L., Raha T., Green M. R., Tissenbaum H. A. (2006) Identification of direct DAF-16 targets controlling longevity, metabolism and diapause by chromatin immunoprecipitation. Nat. Genet. 38, 251–257 [DOI] [PubMed] [Google Scholar]
- 25. Jones D., Dixon D. K., Graham R. W., Candido E. P. (1989) Differential regulation of closely related members of the hsp-16 gene family in Caenorhabditis elegans. DNA 8, 481–490 [DOI] [PubMed] [Google Scholar]
- 26. Stringham E. G., Dixon D. K., Jones D., Candido E. P. (1992) Temporal and spatial expression patterns of the small heat shock (hsp-16) genes in transgenic Caenorhabditis elegans. Mol. Biol. Cell 3, 221–233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Von Stetina S. E., Mango S. E. (2008) Wormnet: a crystal ball for Caenorhabditis elegans. Genome Biol. 9, 226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Lehner B., Lee I. (2008) Network-guided genetic screening: building, testing and using gene networks to predict gene function. Brief. Funct. Genomic Proteomic 7, 217–227 [DOI] [PubMed] [Google Scholar]
- 29. Ito T., Bulger M., Pazin M. J., Kobayashi R., Kadonaga J. T. (1997) ACF, an ISWI-containing and ATP-utilizing chromatin assembly and remodeling factor. Cell 90, 145–155 [DOI] [PubMed] [Google Scholar]
- 30. Varga-Weisz P. D., Wilm M., Bonte E., Dumas K., Mann M., Becker P. B. (1997) Chromatin-remodelling factor CHRAC contains the ATPases ISWI and topoisomerase II. Nature 388, 598–602 [DOI] [PubMed] [Google Scholar]
- 31. Tsukiyama T., Wu C. (1995) Purification and properties of an ATP-dependent nucleosome remodeling factor. Cell 83, 1011–1020 [DOI] [PubMed] [Google Scholar]
- 32. Andersen E. C., Lu X., Horvitz H. R. (2006) C. elegans ISWI and NURF301 antagonize an Rb-like pathway in the determination of multiple cell fates. Development 133, 2695–2704 [DOI] [PubMed] [Google Scholar]
- 33. Martínez-Balbás M. A., Tsukiyama T., Gdula D., Wu C. (1998) Drosophila NURF-55, a WD repeat protein involved in histone metabolism. Proc. Natl. Acad. Sci. U.S.A. 95, 132–137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Gdula D. A., Sandaltzopoulos R., Tsukiyama T., Ossipow V., Wu C. (1998) Inorganic pyrophosphatase is a component of the Drosophila nucleosome remodeling factor complex. Genes Dev. 12, 3206–3216 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Xiao H., Sandaltzopoulos R., Wang H. M., Hamiche A., Ranallo R., Lee K. M., Fu D., Wu C. (2001) Dual functions of largest NURF subunit NURF301 in nucleosome sliding and transcription factor interactions. Mol. Cell 8, 531–543 [DOI] [PubMed] [Google Scholar]
- 36. Mizuguchi G., Tsukiyama T., Wisniewski J., Wu C. (1997) Role of nucleosome remodeling factor NURF in transcriptional activation of chromatin. Mol. Cell 1, 141–150 [DOI] [PubMed] [Google Scholar]
- 37. Dixon D. K., Jones D., Candido E. P. (1990) The differentially expressed 16-kD heat shock genes of Caenorhabditis elegans exhibit differential changes in chromatin structure during heat shock. DNA Cell Biol. 9, 177–191 [DOI] [PubMed] [Google Scholar]
- 38. Buckler K. J., Vaughan-Jones R. D. (1994) Effects of hypoxia on membrane potential and intracellular calcium in rat neonatal carotid body type I cells. J. Physiol. 476, 423–428 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Stros M., Reich J., Kolíbalová A. (1994) Calcium binding to HMG1 protein induces DNA looping by the HMG-box domains. FEBS Lett. 344, 201–206 [DOI] [PubMed] [Google Scholar]
- 40. Stros M., Bernués J., Querol E. (1990) Calcium modulates the binding of high-mobility-group protein 1 to DNA. Biochem. Int. 21, 891–899 [PubMed] [Google Scholar]
- 41. Bandyopadhyay J., Lee J., Lee J., Lee J. I., Yu J.-R., Jee C., Cho J.-H., Jung S., Lee M. H., Zannoni S., Singson A., Kim D. H., Koo H.-S., Ahnn J. (2002) Calcineurin, a calcium/calmodulin-dependent protein phosphatase, is involved in movement, fertility, egg laying, and growth in Caenorhabditis elegans. Mol. Biol. Cell 13, 3281–3293 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Jiang L. I., Sternberg P. W. (1999) An HMG1-like protein facilitates Wnt signaling in Caenorhabditis elegans. Genes Dev. 13, 877–889 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Bustin M., Reeves R. (1996) High-mobility-group chromosomal proteins: architectural components that facilitate chromatin function. Prog. Nucleic Acid Res. Mol. Biol. 54, 35–100 [DOI] [PubMed] [Google Scholar]
- 44. Bonaldi T., Längst G., Strohner R., Becker P. B., Bianchi M. E. (2002) The DNA chaperone HMGB1 facilitates ACF/CHRAC-dependent nucleosome sliding. EMBO J. 21, 6865–6873 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Mabon M. E., Mao X., Jiao Y., Scott B. A., Crowder C. M. (2009) Systematic identification of gene activities promoting hypoxic death. Genetics 181, 483–496 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Clapier C. R., Längst G., Corona D. F., Becker P. B., Nightingale K. P. (2001) Critical role for the histone H4 N terminus in nucleosome remodeling by ISWI. Mol. Cell Biol. 21, 875–883 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Georgel P. T., Tsukiyama T., Wu C. (1997) Role of histone tails in nucleosome remodeling by Drosophila NURF. EMBO J. 16, 4717–4726 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Hamiche A., Sandaltzopoulos R., Gdula D. A., Wu C. (1999) ATP-dependent histone octamer sliding mediated by the chromatin remodeling complex NURF. Cell 97, 833–842 [DOI] [PubMed] [Google Scholar]
- 49. Zhong H., De Marzo A. M., Laughner E., Lim M., Hilton D. A., Zagzag D., Buechler P., Isaacs W. B., Semenza G. L., Simons J. W. (1999) Overexpression of hypoxia-inducible factor 1α in common human cancers and their metastases. Cancer Res. 59, 5830–5835 [PubMed] [Google Scholar]
- 50. Mizukami Y., Fujiki K., Duerr E. M., Gala M., Jo W. S., Zhang X., Chung D. C. (2006) Hypoxic regulation of vascular endothelial growth factor through the induction of phosphatidylinositol 3-kinase/Rho/ROCK and c-Myc. J. Biol. Chem. 281, 13957–13963 [DOI] [PubMed] [Google Scholar]
- 51. Kuhara A., Inada H., Katsura I., Mori I. (2002) Negative regulation and gain control of sensory neurons by the C. elegans calcineurin TAX-6. Neuron 33, 751–763 [DOI] [PubMed] [Google Scholar]