Background: Calcium-dependent protein kinases (CDPKs) play essential roles in malaria parasite life cycle.
Results: Peptide P3 from the junction domain of Plasmodium falciparum CDPK1 (PfCDPK1) as well as purfalcamine inhibit PfCDPK1 activity to block microneme discharge and erythrocyte invasion.
Conclusion: PfCDPK1 regulates microneme discharge, a key process in erythrocyte invasion by malaria parasites.
Significance: PfCDPK1 is a potential target for drugs that block blood stage growth of malaria parasites.
Keywords: Calcium Signaling, Host-Pathogen Interactions, Malaria, Protein Kinases, Protein Secretion, Microneme Secretion
Abstract
Calcium-dependent protein kinases (CDPKs) play important roles in the life cycle of Plasmodium falciparum and other apicomplexan parasites. CDPKs commonly have an N-terminal kinase domain (KD) and a C-terminal calmodulin-like domain (CamLD) with calcium-binding EF hands. The KD and CamLD are separated by a junction domain (JD). Previous studies on Plasmodium and Toxoplasma CDPKs suggest a role for the JD and CamLD in the regulation of kinase activity. Here, we provide direct evidence for the binding of the CamLD with the P3 region (Leu356 to Thr370) of the JD in the presence of calcium (Ca2+). Moreover, site-directed mutagenesis of conserved hydrophobic residues in the JD (F363A/I364A, L356A, and F350A) abrogates functional activity of PfCDPK1, demonstrating the importance of these residues in PfCDPK1 function. Modeling studies suggest that these residues play a role in interaction of the CamLD with the JD. The P3 peptide, which specifically inhibits the functional activity of PfCDPK1, blocks microneme discharge and erythrocyte invasion by P. falciparum merozoites. Purfalcamine, a previously identified specific inhibitor of PfCDPK1, also inhibits microneme discharge and erythrocyte invasion, confirming a role for PfCDPK1 in this process. These studies validate PfCDPK1 as a target for drug development and demonstrate that interfering with its mechanistic regulation may provide a novel approach to design-specific PfCDPK1 inhibitors that limit blood stage parasite growth and clear malaria parasite infections.
Introduction
Malaria remains a major public health problem in the tropical world. It accounted for approximately 1.2 million deaths in 2010 (1). The clinical symptoms of malaria are associated with the blood stage of the parasite life cycle during which Plasmodium merozoites invade and multiply within host erythrocytes. Erythrocyte invasion is a complex, multistep process that is facilitated by multiple interactions between parasite ligands and host receptors (2, 3). A number of parasite proteins involved in these interactions are localized in apical organelles of Plasmodium merozoites referred to as micronemes and rhoptries (2, 3). Invasion requires the timely discharge of these proteins to the merozoite surface to enable receptor engagement.
Cytosolic calcium (Ca2+) plays an important role as a second messenger to regulate microneme secretion in Plasmodium falciparum and Toxoplasma gondii (4–7). Increase in cytosolic Ca2+ following treatment of P. falciparum merozoites and T. gondii tachyzoites with A23187 or ionomycin leads to microneme discharge (4–6). The downstream effector molecules that mediate microneme discharge in response to a rise in cytosolic Ca2+ are not completely understood. A calcium-dependent protein kinase (CDPK)4 has been implicated in this process in T. gondii (8). Pharmacological inhibitors of T. gondii CDPK1 (TgCDPK1) block microneme discharge and gliding motility, suggesting that TgCDPK1 plays a role in these processes during host cell invasion (8, 9).
CDPKs have also been shown to play important roles in the different life cycle stages of Plasmodium species. In case of Plasmodium berghei, PbCDPK3 plays a role in ookinete gliding motility and mosquito mid-gut invasion (10, 11) whereas PbCDPK4 is essential for development of sexual stages and mosquito transmission (12). P. falciparum encodes seven CDPKs. PfCDPK1 plays a role in parasite motility during merozoite egress from mature P. falciparum schizonts (13). PfCDPK1 has been shown to phosphorylate the myosin A tail domain-interacting protein and glideosome-associated protein 45 (14). These proteins are present in the motor complex at the parasite inner membrane complex and are likely to play critical roles in parasite motility. PfCDPK4 is expressed in the sexual stages of P. falciparum and may play a role in development of sexual stages similar to PbCDPK4 (15). PfCDPK5 plays a critical role in egress of P. falciparum merozoites from mature schizonts (16). These reports clearly highlight the importance of CDPKs in the biology of Plasmodium parasites at different stages.
CDPKs are a group of unique protein-Ser/Thr kinases that contain four calcium binding EF hands at the C terminus. The C-terminal domain containing EF hands is reminiscent of the calcium-binding protein calmodulin and is referred to as the calmodulin-like domain (CamLD). The N terminus of the CamLD is linked to a kinase domain (KD) by a short junction domain (JD), which plays an important role in regulation of functional activity of CDPKs in the presence of Ca2+. The N terminus of CDPKs consists of a short stretch of residues called N-terminal region (NTR) that is highly variable in terms of length and amino acid sequence. Data from crystal structures of Toxoplasma and Cryptosporidium CDPKs have shown that in the presence of Ca2+, the calcium-activated domain, which includes the JD and CamLD, undergoes a major conformational change leading to activation of kinase activity (17). Studies on CDPKs from plants as well as Plasmodium show that the JD plays a crucial role in the calcium-dependent regulation of these enzymes (18, 19). Peptides based on JD sequences inhibit PfCDPK4 activity, confirming their role in regulation of PfCDPK4 activity (15).
In this study, we have investigated the mechanistic regulation of PfCDPK1 by Ca2+ and have directly demonstrated that the CamLD interacts with the C-terminal region of the JD referred to as P3 (Leu356 to Thr370). The P3 peptide specifically blocks the functional activity of recombinant PfCDPK1 leading to inhibition of microneme discharge, a key step in erythrocyte invasion. Purfalcamine, a previously identified specific inhibitor of PfCDPK1 (13), also inhibits microneme discharge. The P3 peptide and purfalcamine also inhibit erythrocyte invasion by P. falciparum in vitro. These studies validate PfCDPK1 as a target for the development of inhibitors that block microneme release and inhibit parasite growth.
EXPERIMENTAL PROCEDURES
Culture of P. falciparum 3D7 Strain and Isolation of Viable Merozoites
Cryopreserved P. falciparum 3D7 was cultured in O+ erythrocytes at 5% hematocrit in RPMI 1640 medium (Invitrogen) supplemented with 0.5% Albumax I (Invitrogen), 25 mg liter−1 hypoxanthine (Sigma), 10 mg liter−1 gentamycin (Invitrogen), and 25 mm sodium bicarbonate (Sigma) as described previously (20). P. falciparum 3D7 cultures were tightly synchronized by two rounds of successive sorbitol treatment. Progress of synchronized schizonts was periodically monitored by light microscopy of Giemsa-stained smears. When the majority of infected erythrocytes reached the mature schizont stage with segmented merozoites, the parasite culture was resuspended in complete RPMI 1640 medium or buffer mimicking intracellular ionic conditions (IC buffer: 142 mm KCl, 5 mm NaCl, 2 mm EGTA, 1 mm MgCl2, 5.6 mm glucose, 25 mm HEPES, pH 7.2). Schizonts were allowed to rupture and release merozoites over a period of 40 min. Cultures containing unruptured schizonts and released merozoites were centrifuged at 500 × g in a Sorvall RT7 centrifuge for 5 min to separate released merozoites from unruptured schizonts and uninfected erythrocytes. Supernatant containing free merozoites was centrifuged at 3300 × g using an Eppendorf 5810R centrifuge for 5 min to collect merozoites. The merozoites were resuspended in RPMI 1640 medium (incomplete RPMI) or IC buffer for use in experiments.
Molecular Cloning of PfCDPK1 and CamLD
DNA fragments encoding the PfCDPK1 and CamLD of PfCDPK1 were amplified with the following primer sets: PfCDPK1Fwd, 5′-ATGCGCCCATGGGGTGTTCACAAAGT-3′ and PfCDPK1Rev, 5′-ATGCGCCTCGAGTGAAGATTTATTATCACAAATTTTG-3′; PfCDPK1CamLDFwd, 5′-GAACCATGGAGCTTACTGACATTTTTAAAAAGTTG-3′ and PfCDPK1CamLDRev, 5′-GCAGGTACCTGAAGATTTATTATCACAATTTTGTGC-3′, respectively, using high fidelity Phusion DNA polymerase enzyme (Finnzymes) and cDNA prepared from P. falciparum 3D7 culture as template. Amplified DNA fragments encoding PfCDPK1 and PfCDPK1-CamLD were cloned between NcoI and XhoI sites in pET-28a+ (Novagen) and NcoI and KpnI sites in pGHATA (modified pET-28a+ with a cleavable His6 tag at the C terminus). Recombinant DNA plasmids were transformed into Escherichia coli DH5α-competent cells, and positive clones (containing plasmid with insert) were confirmed by restriction digest of plasmids and sequencing of insert DNA fragments.
Expression and Purification of PfCDPK1 and CamLD
pET-28a+ and pGHATA vectors containing DNA encoding the PfCDPK1 and CamLD of PfCDPK1, respectively, were transformed into E. coli BLR (DE3) competent cells for protein expression. E. coli cells were grown to mid-log phase, and expression of recombinant PfCDPK1 and CamLD was induced with 1 mm isopropyl-β-d-1-thiogalactopyranoside at A600 nm of 0.9 for 4 h at 30 °C and for 12 h at 16 °C, respectively. E. coli cells were harvested by centrifugation and resuspended in lysis buffer (10 mm Tris, pH 8.0, 1 mm EDTA, 100 mm NaCl, 25 μg ml−1 lysozyme, 3 mm β-mercaptoethanol, and protease inhibitor mixture (Roche Applied Science)) followed by sonication for 15 min. Supernatants containing recombinant PfCDPK1 or CamLD were loaded on Ni-nitrilotriacetic acid beads (Qiagen) preequilibrated with buffer A (10 mm imidazole, 10 mm Tris, pH 8.0, 100 mm NaCl, and 3 mm β-mercaptoethanol) followed by elution in buffer A containing imidazole. Affinity-purified recombinant PfCDPK1 and CamLD were further purified by gel permeation chromatography) using a Hiload 16/60 Superdex 200 column (GE Healthcare) or Hiload 16/60 Superdex 75 column (GE Healthcare) in 50 mm Tris, pH 8.0, and 50 mm NaCl.
Site-directed Mutagenesis
Primer pairs F350AFwd (5′-TGTGGCGCTTTATCAAATATGAGGAAAGCTGAGGGCAGTCAGA-3′) and F350ARev (5′-TCTGACTGCCCTCAGCTTTCCTCATATTTGATAAAGCGCCACA3-′); L356AFwd (5′-TCAAATATGAGGAAATTTGAGGGCAGTCAGAAAGCGGCTCAAGCAGCCATATT-3′) and L356ARev (5′-AATATGGCTGCTTGAGCCGCTTTCTGACTGCCCTCAAATTTCCTCATATTTGA-3′); S353AFwd (5′-AATATGAGGAAATTTGAGGGCGCTCAGAAATTAGCTCAAGCAGC-3′)and S353ARev (5′-GCTGCTTGAGCTAATTTCTGAGCGCCCTCAAATTTCCTCATATT-3′); F363A-I364AFwd (5′-GAGGGCAGTCAGAAATTAGCTCAAGCAGCCATATTAGCTGCTGGAAGTAAATTAACAACATTAG-3′) and F363A-I364ARev (5′-CTAATGTTGTTAATTTACTTCCAGCAGCTAATATGGCTGCTTGAGCTAATTTCTGACTGCCCTC-3′) were used with pET-28a+-PfCDPK1 plasmid as template and QuikChange XL site-directed mutagenesis kit (Stratagene) to create plasmids for expression of the PfCDPK1 mutants F350A, L356A, S353A, and F363A/I364A, respectively, according to the manufacturer's instructions. Recombinant plasmids encoding the PfCDPK1 mutants were transformed into E. coli DH5α strain, and the mutations were confirmed by DNA sequencing. The plasmids were transformed into E. coli BLR(DE3) strain for expression of PfCDPK1 mutants. PfCDPK1 mutants were expressed and purified as described above for wild type PfCDPK1.
Kinase Assay
Phosphorylation experiments were performed using 40 nm recombinant PfCDPK1 or PfCDPK5 in kinase assay buffer (50 mm MgCl2, 50 mm Tris, pH 8.0, 20 mm sodium orthovanadate, 20 mm sodium fluoride, 1 mm dithiothreitol (DTT), and 2.5 mm CaCl2). 2.5 mm EGTA was added for conditions requiring absence of Ca2+ ions. Kinase assays were also performed in different concentrations of CaCl2 from 0.5 to 2 mm CaCl2. Syntide-2 and autocamtide-2 were used as substrates for PfCDPK1 and PfCDPK5, respectively. Peptides P1 (Ile325 to Thr339), P2 (Leu340 to Lys355), and P3 (Leu356 to Thr370) from the JD of PfCDPK1 were added to the reaction mixture at different concentrations and incubated on ice for 10–15 min before addition of the substrate syntide-2 or autocamtide-2. Radioactive [γ-33P]ATP was used as the source of phosphate. The reaction was allowed to take place at 30 °C for 1 h. The reactions were stopped by adding 4× sodium dodecyl sulfate (SDS) reducing dye (250 mm Tris-HCl, pH 6.8, 8% SDS, 300 mm DTT, 30% glycerol, and 0.02% bromphenol blue). Samples were separated by Tricine-polyacrylamide gel electrophoresis (PAGE), and incorporation of 33P was detected using Storage Phosphor Screen (GE Healthcare). Phosphorylation of PfCDPK1, PfCDPK5, syntide-2, and autocamtide-2 was quantified by densitometry using Typhoon scanner (GE Healthcare).
Circular Dichroism (CD)
The secondary structure of PfCDPK1 in the presence of different concentrations of calcium was investigated by CD. CD spectra were obtained on a Jasco J-710 spectropolarimeter with a constant dispersion of 1 nm. Spectra were measured with a time constant of 1 s and scan speed of 100 nm min−1. Signals were averaged five times before measurement. 50 nm protein concentration was analyzed in a cuvette of 0.1-cm path length.
Generation of Antiserum against PfCDPK1 and Western Blotting
BALB/c mice were immunized with recombinant PfCDPK1 protein formulated with complete Freund's adjuvant for the priming immunization and incomplete Freund's adjuvant for booster immunizations. The presence of PfCDPK1 in parasite lysates was detected by Western blotting. P. falciparum blood stage cultures were tightly synchronized by treatment of ring stages with 5% sorbitol in two successive cycles. Infected RBCs were treated with 0.05% saponin and washed extensively with PBS. Finally, parasites were lysed by resuspension in radioimmunoprecipitation assay buffer (50 mm Tris, pH 8.0, 150 mm NaCl, 1 mm EDTA, 1 mm EGTA, 1% Nonidet P-40, 1% sodium deoxycholate, protease inhibitor mixture (Roche Applied Science)), and incubated on ice for 1 h with intermittent shaking. Detergent-resistant and detergent-soluble fractions were separated by centrifugation at 15,000 × g for 30 min. Both fractions were separated by SDS-PAGE and transferred to nitrocellulose membrane for Western blotting. The nitrocellulose membrane was incubated with anti-PfCDPK1 serum (1:2000 dilution) followed by horseradish peroxidase (HRP)-conjugated anti-mouse IgG goat serum (Sigma) at a 1:2000 dilution for 1 h each. PfCDPK1 was detected with ECL plus Western blotting detection system kit (GE Healthcare) following the manufacturer's specifications.
Immunofluorescence Assay (IFA)
Thin smears of parasite cultures were made with different stages of synchronized P. falciparum blood stage cultures on glass slides, air-dried, and fixed in prechilled methanol for 20 min at −20 °C. The slides were blocked with 3% BSA in PBS at room temperature for 1 h, incubated with anti-PfCDPK1 mouse serum (1:1000 dilution) followed by incubation with Alexa Fluor 488-conjugated anti-mouse IgG goat antibodies (1:200 dilution) (Molecular Probes). For co-localization experiments slides with P. falciparum trophozoites were incubated with anti-PfCDPK1 rabbit serum (1:500) and anti-PfSERA5 mouse serum (1:500 dilution) followed by Alexa Fluor 488-conjugated anti-rabbit IgG goat serum (1:500) (Molecular Probes) and Alexa Fluor 594-conjugated anti-mouse IgG goat serum (1:800) (Molecular Probes). Finally, the slides were mounted with ProLong Gold antifade reagent with 4′,6-diamidino-2-phenylindole (DAPI) (Invitrogen) and analyzed using a Nikon A1 confocal microscope.
Detection of CamLD Binding with Recombinant JD
Recombinant NTR and JD regions of PfCDPK1 fused with GST at their C termini (NTR-GST and JD-GST) were expressed in E. coli and purified by affinity chromatography. Binding of recombinant CamLD with recombinant NTR-GST and JD-GST was investigated as follows. Recombinant NTR-GST and JD-GST were separated by SDS-PAGE along with GST control and transferred to nitrocellulose membrane. The membrane was blocked with 5% skimmed milk in Tris buffer (10 mm Tris, pH 8.0, and 150 mm NaCl) for 1 h at 25 °C, washed with Tris buffer, and blocked again in 5% bovine serum albumin (BSA) (Sigma) in 5 mm CaCl2, 150 mm NaCl, 50 mm imidazole, pH 8.0, for 1 h at 25 °C. The membrane was washed with buffer A (5 mm CaCl2, 150 mm NaCl, 10 mm Tris, pH 8.0, 1 mm MgCl2, and 0.05% Tween 20) and incubated overnight with recombinant CamLD (1 μg ml−1) in buffer B (5 mm CaCl2, 150 mm NaCl, 10 mm Tris, pH 8.0, 1 mm MgCl2, and 1% BSA) at 25 °C. The membrane was again washed with buffer C (5 mm CaCl2, 50 mm NaCl, 10 mm Tris, pH 7.5, 1 mm MgCl2, and 0.05% Tween 20) and incubated with HRP conjugated anti-His mouse monoclonal antibody (Sigma) for 1 h at 25 °C in 5 mm CaCl2, 50 mm NaCl, 10 mm Tris, pH 7.5, 1 mm MgCl2, and 1% BSA. The blot was developed with 1 mg ml−1 3,3′-diaminobenzidine (Sigma) and 1 μl ml−1 hydrogen peroxide (H2O2) (Merck) in buffer C. To test the binding of the CamLD in the absence of Ca2+, buffers B and C were prepared without CaCl2.
Binding of Peptides from JD with CamLD
Magnetic Bioplex beads that fluoresce at different wavelengths (Bio-Rad) were conjugated with peptides P1, P2, and P3 (10 μg/50 μl of beads) using an amine coupling kit (Bio-Rad) according to the manufacturer's instructions. Different amounts of recombinant CamLD (0.02, 0.1, 0.5, and 2.5 μg) were incubated for 45 min at room temperature with beads coupled with P1, P2, and P3 with and without 1 mm CaCl2. Bound CamLD was detected with anti-His rabbit serum followed by biotinylated anti-rabbit IgG goat antibodies and streptavidin-phycoerythrin. Beads were washed three times between each step using PBS containing 0.05% Tween 20 and 1% BSA. Fluorescence signal, which indicates the amount of bound CamLD, was detected using a Bioplex 2000 (Bio-Rad) reader.
Homology Modeling
PfCDPK1 shares ∼74% similarity with Cryptosporidium parvum calcium-dependent protein kinase 3 (CpCDPK3). Active forms (Ca2+-bound forms) of wild type PfCDPK1 as well as site-directed PfCDPK1 mutants (F350A, L356A, and F363A/I364A) were modeled using the three-dimensional structure of CpCDPK3 active form (PDB ID code 3LIJ) as template. Homology modeling was done by employing Modeler9v7 program (21). All models were locally minimized for energy using Chimera software (22) and model quality was validated with PROCHECK (23). The probable intramolecular contacts of mutated residues in wild type PfCDPK1 were investigated using CONTACT program in the CCP4 package (24) with 4 Å as the cutoff distance.
Inhibition of Microneme Secretion with P3 Peptide and Purfalcamine
P. falciparum merozoites isolated in IC buffer as described above were incubated with peptide P1, P2, or P3 from the JD of PfCDPK1 or control peptide P10 (scrambled peptide P3 sequence, TALIQTKSLGIALAF) or different concentrations of purfalcamine (50–800 nm) for 15 min at 37 °C and shifted to buffer mimicking extracellular ionic conditions (EC buffer: 5 mm KCl, 142 mm NaCl, 1 mm CaCl2, 1 mm MgCl2, 5.6 mm glucose, and 25 mm HEPES, pH 7.2) containing the relevant peptide or purfalcamine. Following incubation in EC buffer for 10 min at 37 °C, merozoites and supernatants were separated by centrifugation at 3300 × g at room temperature for 5 min. The microneme protein, apical merozoite protein-1 (PfAMA1), which is proteolytically cleaved by the protease PfSub2 and released when it is translocated to the merozoite surface (25), was detected in merozoite supernatants by Western blotting using anti-PfAMA1 rabbit serum at 1:2000 dilution. Rabbit serum against the cytoplasmic P. falciparum nucleosome assembly protein-large (NAPL) was used at a 1:2000 dilution to detect NAPL in merozoite pellets and supernatants to control for number of merozoites used and merozoite lysis under different conditions, respectively. The presence of PfAMA1 on the surface of merozoites in IC buffer and upon transfer to EC buffer with and without treatment with P3 peptide (120 μm) and purfalcamine (800 nm) was detected by flow cytometry using anti-PfAMA1 rabbit serum as described previously (4).
Inhibition of Erythrocyte Invasion by P. falciparum Using P3 Peptide or Purfalcamine
P. falciparum merozoites isolated in IC buffer as described above were incubated with different concentrations of peptides P3 or control peptide P10 (15–120 μm) or peptides P1 or P2 (120 μm) or different concentrations of purfalcamine (50–800 nm). Following incubation for 15 min with peptides or purfalcamine, the merozoites were resuspended in complete RPMI 1640 medium and incubated with erythrocytes at 2% hematocrit to allow invasion. After 24–26 h of incubation at 37 °C under 5% CO2, 5% O2 and balanced with N2, newly invaded rings were scored by light microscopy of Giemsa-stained smears. Percent inhibition of invasion was calculated under each assay condition using the formula (1 − T/C) ×100, where T and C denote parasitemia in treatment and control samples, respectively.
Further cloning and purification procedures are described in the supplemental Materials and Methods.
RESULTS
PfCDPK1 Is a Bona Fide Calcium-dependent Kinase
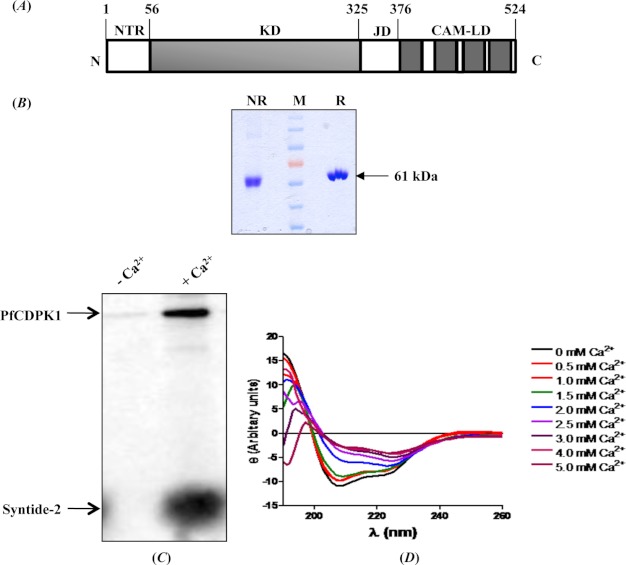
PfCDPK1 contains an N-terminal KD separated from the CamLD by a JD (Fig. 1A). Recombinant PfCDPK1 with a C-terminal His6 tag was expressed in E. coli as a soluble protein and purified to homogeneity by immobilized metal affinity chromatography and gel permeation chromatography (Fig. 1B). The calcium-dependent functional activity of recombinant PfCDPK1 was investigated by in vitro kinase assay using [γ-33P]ATP and syntide-2 as a peptide substrate. Kinase activity of recombinant PfCDPK1 increased significantly in the presence of Ca2+ both in terms of autophosphorylation of PfCDPK1 and transphosphorylation of syntide-2 (Fig. 1C), confirming that PfCDPK1 is a bona fide calcium-dependent protein kinase. A change in the secondary structure of PfCDPK1 was investigated at different concentrations of CaCl2 by CD. Overall, PfCDPK1 showed an α-helical structure (Fig. 1D). The α-helicity decreased with increasing Ca2+ concentration (Fig. 1D). In the absence of Ca2+, PfCDPK1 showed 67.4% α-helicity and 2.2% β-sheets, which changed to 44.7% α-helix and 13.4% β-sheets at 4 mm CaCl2. The kinase activity of recombinant PfCDPK1 increased with increasing Ca2+ concentration from 0.5 to 2.0 mm (supplemental Fig. 1), demonstrating that conformational change is critical for activation of kinase activity.
FIGURE 1.
PfCDPK1 is a bona fide calcium-dependent protein kinase. A, schematic diagram of PfCDPK1 domain organization. PfCDPK1 contains a stretch of residues (amino acids 1–55) at the N terminus called the NTR that is followed by a KD, a short JD, and a CamLD. B, recombinant PfCDPK1. Recombinant PfCDPK1 (∼61 kDa) shows mobility shift on SDS-PAGE under reducing (R) and nonreducing (NR) conditions. C, functional activity of recombinant PfCDPK1. Kinase activity of recombinant PfCDPK1 was tested using radioactive [γ-33P]ATP in the presence and absence of Ca2+. Incorporation of 33P in PfCDPK1 and syntide-2 was detected by Typhoon scanner. PfCDPK1 showed a significant increase in autophosphorylation and transphosphorylation of syntide-2 in the presence of Ca2+. D, secondary structure of PfCDPK1. Change in secondary structure of PfCDPK1 in the presence of different concentrations of Ca2+ was analyzed by CD. Binding of Ca2+ with PfCDPK1 induced significant change in the CD spectra, indicating changes in secondary structure.
Expression of PfCDPK1 in Blood Stages
Anti-PfCDPK1 mouse serum specifically recognized PfCDPK1 from different P. falciparum blood stages by Western blotting and IFA (Fig. 2). Synchronized parasite cultures at ring, trophozoite, schizont, and merozoite stages were lysed in radioimmunoprecipitation assay buffer containing 1% Nonidet P-40 and 1% sodium deoxycholate. The presence of PfCDPK1 was examined in detergent-soluble and detergent-resistant fractions by Western blotting using anti-PfCDPK1 mouse serum. A protein of expected molecular mass of ∼61 kDa was clearly detected in detergent-resistant fractions of all blood stages (Fig. 2A), indicating that PfCDPK1 is likely associated with membranes.
FIGURE 2.
PfCDPK1 is expressed throughout erythrocytic schizogony. A, detection of PfCDPK1 in different P. falciparum blood stages by Western blotting. Different blood stages such as rings (R), trophozoites (T), schizonts (S), and merozoites (M) were lysed in radioimmunoprecipitation assay buffer containing 1% Nonidet P-40 and 1% sodium deoxycholate, fractionated into detergent-soluble and detergent-resistant fractions by centrifugation, and separated by SDS-PAGE. The presence of PfCDPK1 in detergent-soluble and detergent-resistant fractions was detected by Western blotting using anti-PfCDPK1 mouse serum. Significant amounts of PfCDPK1 were found in the detergent-resistant fractions, indicating that PfCDPK1 associates with membranes in all stages. B, localization of PfCDPK1 in different stages by IFA. PfCDPK1 was detected in blood stages of P. falciparum by IFA using anti-PfCDPK1 mouse serum and Alexa Fluor 488-conjugated anti-mouse IgG goat serum (green). Anti-SERA5 rabbit serum and Alexa Fluor 594-conjugated anti-rabbit IgG goat serum (red) were used to localize the PV protein SERA5. Nuclear DNA was counterstained with DAPI (blue). Bright field (DIC) merged with DAPI (DIC + DAPI), fluorescence images detecting PfCDPK1 (PfCDPK1), and merged fluorescence images detecting PfCDPK1 and DAPI, PfSERA5 and DAPI or PfCDPK1, PfSERA5 and DAPI (Merge) are shown. Scale bars represent 2 μm (ring, trophozoite, schizont) or 1 μm (merozoite).
P. falciparum cultures at different stages of development were smeared on glass slides and used to investigate the localization of PfCDPK1 by IFA using anti-PfCDPK1 mouse serum. PfCDPK1 localized to the rim of the infected erythrocyte and appeared to be associated with the erythrocyte membrane in rings and trophozoites (Fig. 2B). In addition, PfCDPK1 was found in the parasitophorous vacuole (PV) and co-localized with SERA5, a known PV protein (26, 27), in trophozoites (Fig. 2B). In merozoites, PfCDPK1 was found primarily at the boundary of the merozoite at the merozoite plasma membrane (Fig. 2B).
CamLD of PfCDPK1 Binds Peptide P3 from the C-terminal Region of JD
The JD plays an important role in the Ca2+-dependent activation of CDPKs (15, 18). To investigate the role of the JD in PfCDPK1 regulation, we tested the binding of recombinant JD fused to GST (JD-GST) with recombinant CamLD fused to a C-terminal His6 tag. Recombinant NTR of PfCDPK1 fused to GST (NTR-GST) was also produced and used as a control in the binding assays. Recombinant JD-GST and NTR-GST were separated by SDS-PAGE, transferred to nitrocellulose membrane, and probed for binding with recombinant CamLD in the presence and absence of Ca2+. Detection of proteins with Ponceau staining confirmed that equal amounts of JD-GST, NTR-GST, and GST were transferred to the nitrocellulose membrane used for incubation with the CamLD (data not shown). Bound CamLD was detected using an anti-His mouse monoclonal antibody. Recombinant CamLD binds specifically with JD-GST, and the binding increases in the presence of Ca2+ (supplemental Fig. 2).
The JD was dissected into three peptides, namely, P1 (Ile325 to Thr339), P2 (Leu340 to Lys355), and P3 (Leu356 to Thr370) (Fig. 3A). Peptides P1, P2, and P3 were immobilized on magnetic Bioplex beads by amine coupling and tested for binding with recombinant CamLD in the presence and absence of Ca2+. The CamLD binds specifically with P3 peptide and the binding increases in the presence of Ca2+ confirming that residues at the C terminus of the JD mediate binding to the CamLD (Fig. 3B).
FIGURE 3.
P3 peptide serves as a binding site for CamLD and inhibits kinase activity of PfCDPK1. A, schematic of PfCDPK1 and peptides from the JD. The JD of PfCDPK1 was dissected into three nonoverlapping peptides, P1, P2, and P3. B, CamLD of PfCDPK1 binding the C-terminal P3 region of the JD. Peptides P1, P2, and P3 were immobilized on Bioplex beads by amine coupling and tested for binding with different amounts of recombinant CamLD (0.02, 0.1, 0.5, and 2.5 μg) in the presence or absence of Ca2+. The mean fluorescence intensity (MFI) indicating bound CamLD was plotted against different concentrations of recombinant CamLD used in binding assay. CamLD binds specifically with P3 peptide, and the binding increases in the presence of Ca2+. C, inhibition of kinase activity of PfCDPK1 with peptides from JD. 80 μm P1, P2, and P3 peptides were tested for inhibition of kinase activity of PfCDPK1. P3 (80 μm) inhibits autophosphorylation and transphosphorylation of syntide-2 whereas peptides P1 and P2 do not. D, dose-dependent inhibition of PfCDPK1 kinase activity with P3 peptide. Different concentrations of P3 peptide were tested for inhibition of kinase activity of PfCDPK1. Percentage inhibition of autophosphorylation (left) and transphosphorylation of syntide-2 (right) by recombinant PfCDPK1 at different concentrations of P3 peptide are shown. Error bars represent S.D. from three independent experiments.
Identification of Functionally Important Residues in JD of PfCDPK1
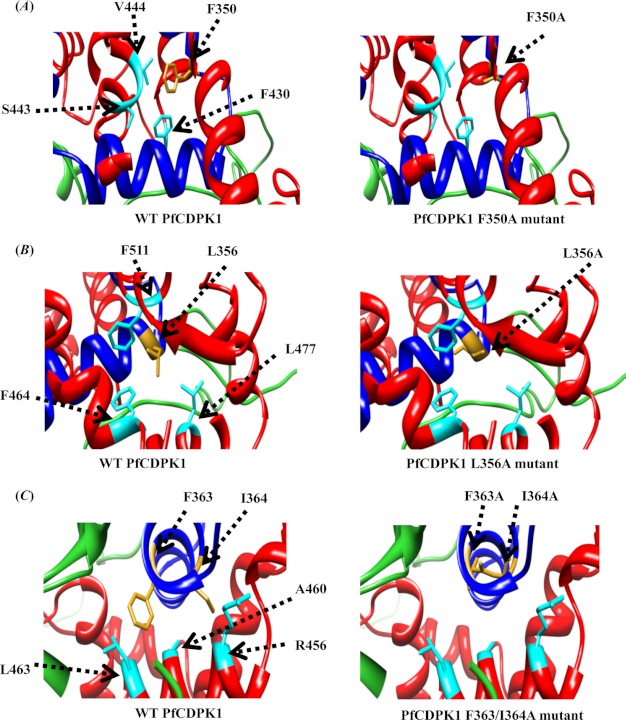
Site-directed mutagenesis was used to identify residues within the JD that may be involved in interaction with the CamLD and regulation of PfCDPK1 kinase activity. Multiple sequence alignment of the JD from PfCDPK1 with corresponding regions of PfCDPK4, TgCDPK1, TgCDPK3, and CpCDPK3 (Fig. 4A) together with previous data on key residues involved in regulation of other CDPKs identified Phe350, Leu356, and Phe363–Ile364 of the JD from PfCDPK1 as targets for site-directed mutagenesis. All PfCDPK1 alanine replacement mutants were expressed, purified to homogeneity (Fig. 4B), and tested for calcium-dependent kinase activity. The three mutants, F350A, L356A, and F363A/I364A did not have any kinase activity both in terms of autophosphorylation or transphosphorylation of syntide-2 (Fig. 4C). The CONTACT program in the CCP4 package was used to investigate the possible interactions of Phe350, Leu356, and Phe363–Ile364 with residues within the CamLD. Phe350 was found to be in close proximity (<4 Å) to Phe430, Ser443, and Val444 of the CamLD in the presence of Ca2+ (Fig. 5A), suggesting that they may engage in hydrophobic interactions. Similarly, Leu356 and Phe363–Ile364 were found to be in close proximity to Phe464, Leu477, Phe511 (Fig. 5B), and Ala460, Leu463, Arg456 (Fig. 5C), respectively. These probable interactions were absent in the mutant proteins (Fig. 5, A–C), suggesting that they may be important for activation of PfCDPK1 in the presence of Ca2+.
FIGURE 4.
Identification of residues in the JD that mediate intramolecular interactions with CamLD and calcium-dependent activation of PfCDPK1. A, multiple sequence alignment of JDs from related CDPKs. The JD of PfCDPK1 was aligned with JDs of PfCDPK4, TgCDPK1, TgCDPK3, and CpCDPK3. Residues selected for site-directed mutagenesis, namely, Leu356 and Phe363–Ile364 (red), Phe350 (green), and Ser353 (blue) are shown. B, recombinant site-directed PfCDPK1 mutants. Phe350, Leu356, Ser353, and Phe363–Ile364 residues from the JD were mutated to Ala in PfCDPK1. Recombinant wild type PfCDPK1 and PfCDPK1 mutants were produced and used in kinase assays. C, kinase activity of wild type and mutant PfCDPK1. Kinase assay was performed with wild type and mutant PfCDPK1 using radioactive [γ-33P]ATP. Autophosphorylation and transphosphorylation of syntide-2 were quantified with a Typhoon scanner. Percentage activity of mutant PfCDPK1 was plotted with respect to activity of wild type PfCDPK1. F350A, L356A, and F363A/I364A mutations led to abrogation of kinase activity in terms of both autophosphorylation and transphosphorylation of syntide-2. Error bars represent S.D. from three independent experiments. A representative gel for autophosphorylation and transphosphorylation of syntide-2 by wild type and mutant PfCDPK1 is shown.
FIGURE 5.
Intramolecular interactions between the CamLD and JD of PfCDPK1. The CONTACT program from the CCP4 package was used to investigate the possible interactions of Phe350, Leu356, and Phe363–Ile364 with residues from CamLD in PfCDPK1 with 4 Å as cutoff. A, CamLD residues interacting with Phe350 of the JD. Phe430, Ser443, and Val444 of the CamLD region of PfCDPK1 were found to be in close proximity (<4 Å) with Phe350 of JD in wild type PfCDPK1 (left). These interactions are missing in F350A mutant (right). B, CamLD residues interacting with Leu356 of the JD. Phe464, Leu477, and Phe511 of the CamLD region of PfCDPK1 were found to be in close proximity (<4 Å) with Leu356 of the JD in wild type PfCDPK1 (left). These interactions are missing in the L356A mutant (right). C, CamLD residues interacting with F363–I364 of the JD. Ala460, Leu463, and Arg456 of CamLD were found to be in close proximity (< 4 Å) with F363–I364 of the JD in wild type PfCDPK1 (left). These interactions are missing in F363A/I364A mutant (right). Mutated residues of the JD are shown in golden yellow and interacting residues of CamLD in cyan.
P3 Peptide Specifically Inhibits the Kinase Activity of PfCDPK1 in Vitro
The kinase activity of recombinant PfCDPK1 was tested in the presence of P1, P2, and P3 peptides to investigate whether they had any inhibitory effect. Of the three peptides tested, only P3 inhibited kinase activity of PfCDPK1 (Fig. 3C). The kinase-inhibitory activity of P3 peptide showed a dose response with IC50 for autophosphorylation of 9.4 μm and IC50 for transphosphorylation of syntide-2 of 16.2 μm (Fig. 3D). Peptides P1 and P2 did not inhibit PfCDPK1 activity up to a concentration of 200 μm (data not shown). The recombinant KD of PfCDPK1 is not regulated by Ca2+ and phosphorylates syntide-2 in the absence of Ca2+ (data not shown). The kinase activity of KD was not affected by the P3 peptide up to a concentration of 80 μm (data not shown), indicating that P3 does not directly target the active site within the KD of PfCDPK1. The P3 region of PfCDPK1 was aligned with corresponding regions of PfCDPK2, 3, 4, and 5, which have similar domain architecture (supplemental Fig. 3A). The P3 region of PfCDPK1 shows maximum homology with corresponding region of PfCDPK4 and lower homology with PfCDPK2, 3, and 5. However, PfCDPK4 is not expressed in P. falciparum blood stages (15). Different concentrations of P3 peptide were thus tested for inhibition of kinase activity of the blood stage kinase PfCDPK5 using autocamtide-2 as peptide substrate. Functional activity of PfCDPK5, as determined by estimating transphosphorylation of autocamtide-2, did not show any decrease in the presence of P3 (supplemental Fig. 3B). Thus, the P3 peptide specifically inhibits kinase activity of PfCDPK1 and does not inhibit the closely related PfCDPK5.
P3 Inhibits Microneme Secretion and Invasion of Erythrocytes by P. falciparum Merozoites
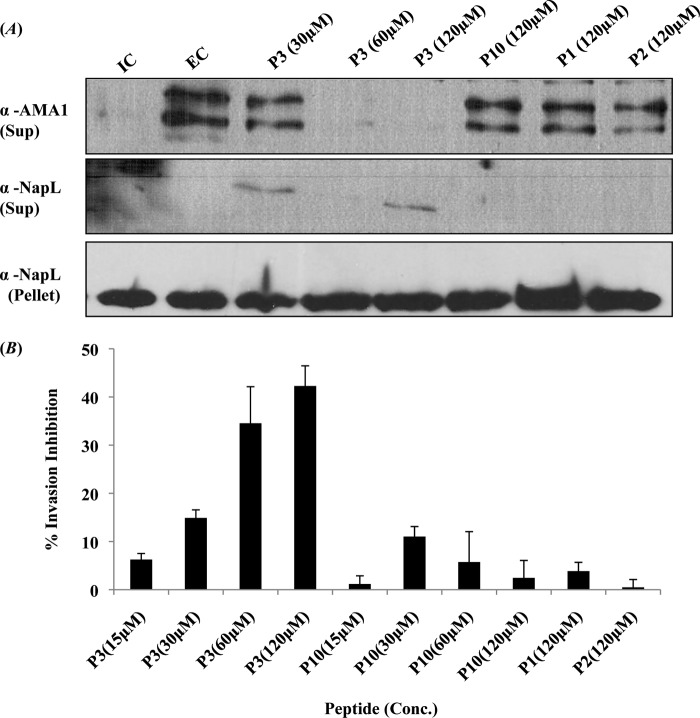
Discharge of micronemes is an essential step in the invasion of erythrocytes by P. falciparum merozoites. Singh et al. have shown that exposure of P. falciparum merozoites to an EC domain that mimics the ionic environment that merozoites are exposed to in blood plasma leads to a rise in cytosolic Ca2+ levels and discharge of microneme proteins such as EBA175 and PfAMA1 to the merozoite surface (4). PfAMA1 is proteolytically cleaved by the protease PfSub2 and released into the supernatant following translocation to the merozoite surface (25). Secretion of PfAMA1 following transfer of merozoites from an IC to EC buffer was thus tested by detecting the presence of PfAMA1 in the supernatant of merozoites by Western blotting using anti-PfAMA1 rabbit serum (Fig. 6A). Pretreatment of merozoites with P3 peptide prior to transfer from IC buffer to EC buffer blocks the secretion of PfAMA1 (Fig. 6A). The control peptide, P10 (scrambled P3 sequence), as well as peptides P1 and P2 do not block secretion of PfAMA1 (Fig. 6A). The presence of PfAMA1 on the surface of merozoites following transfer to EC buffer with or without pretreatment with peptide P3 was also detected by flow cytometry using anti-PfAMA1 rabbit serum (Fig. 7). Transfer to EC buffer resulted in an increase in PfAMA1 detected on the surface of merozoites (Fig. 7). Pretreatment with peptide P3 inhibited the increase in PfAMA1 on the merozoite surface following transfer to EC buffer (Fig. 7).
FIGURE 6.
Peptide P3 blocks discharge of micronemes and inhibits invasion of erythrocytes by P. falciparum merozoites. A, inhibition of microneme discharge by peptide P3. Merozoites isolated in IC buffer (mimicking intracellular ionic environment) were transferred to EC buffer (mimicking ionic environment in blood plasma) either with or without pretreatment with peptides P1, P2, P3, and control P10 peptide (scrambled P3 sequence). Discharge of microneme protein PfAMA1 was detected in merozoite supernatants by Western blotting. Merozoite supernatants and pellets were also probed with antiserum raised against cytoplasmic protein PfNAPL to detect merozoite lysis and confirm that similar numbers of merozoites were used for different conditions respectively. Transfer of merozoites from IC to EC buffer leads to discharge of PfAMA1. Pretreatment of merozoites with P3 peptide blocked microneme discharge in a dose-dependent manner. Peptides P1, P2, and control P10 peptide (scrambled P3 sequence) did not inhibit secretion of PfAMA1. B, inhibition of erythrocyte invasion by P. falciparum merozoites by peptide P3. P. falciparum merozoites were pretreated with different concentrations (15–120 μm) of peptides P3 or P10, or P1 and P2 at the highest concentration (120 μm) in IC buffer and then incubated with erythrocytes in RPMI 1640 medium to allow invasion. Newly invaded rings were scored by Giemsa staining 24–26 h after invasion. Percentage inhibition of invasion with peptides P1, P2, P3, and control peptide P10 is shown. Error bars represent S.D. from three independent experiments. P3 peptide inhibits erythrocyte invasion by P. falciparum merozoites in a dose-dependent manner. Peptides P1, P2, and control peptide P10 (scrambled P3 sequence) did not inhibit erythrocyte invasion.
FIGURE 7.
Peptide P3 and purfalcamine block microneme discharge by P. falciparum merozoites. P. falciparum merozoites isolated in IC buffer (mimicking intracellular ionic environment) were transferred to EC buffer (mimicking ionic environment in blood plasma) either with or without pre-treatment with peptide P3 (120 μm) or purfalcamine (800 nm). The presence of microneme protein PfAMA1 on the surface of P. falciparum merozoites in IC (red) and EC (blue) buffer was detected by flow cytometry using anti-PfAMA1 serum. Preimmune serum was used as control (black). Transfer of merozoites from IC to EC buffer leads to an increase in PfAMA1 detected on the merozoite surface. Pretreatment of merozoites with peptide P3 (dark green) or purfalcamine (light green) prior to transfer to EC buffer blocks increase in PfAMA1 on the merozoite surface.
To test whether P3 peptide can block erythrocyte invasion by P. falciparum, merozoites were pretreated with P3 peptide in IC buffer prior to transfer to complete RPMI 1640 medium containing erythrocytes. Invasion rates were determined by scoring for newly formed rings following invasion by Giemsa staining. P3 peptide showed dose-dependent invasion inhibitory activity (Fig. 6B). The control P10 peptide as well as P1 and P2 peptides had no effect on invasion (Fig. 6B).
PfCDPK1 Inhibitor Purfalcamine Blocks Microneme Discharge and Erythrocyte Invasion by P. falciparum
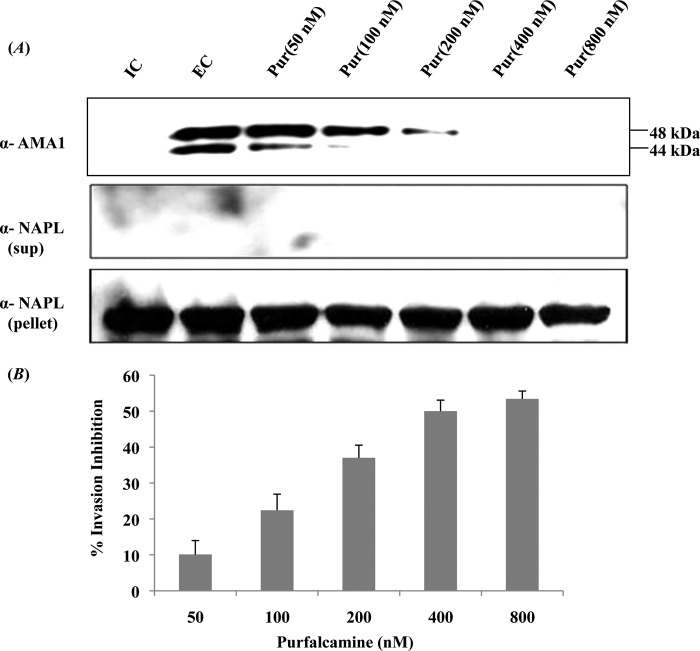
Purfalcamine, a previously identified specific inhibitor of PfCDPK1, was also tested for inhibition of microneme secretion and erythrocyte invasion by P. falciparum merozoites. Pretreatment of P. falciparum merozoites with purfalcamine before transfer from IC to EC buffer inhibited discharge of PfAMA1 (Figs. 7 and 8A). Moreover, pretreatment of P. falciparum merozoites with purfalcamine prior to addition to erythrocytes inhibited erythrocyte invasion in a dose-dependent manner with an IC50 of 585 nm (Fig. 8B).
FIGURE 8.
PfCDPK1 inhibitor purfalcamine blocks microneme discharge and erythrocyte invasion by P. falciparum merozoites. A, inhibition of microneme discharge by purfalcamine. Merozoites isolated in IC buffer (mimicking intracellular ionic environment) were transferred to EC buffer (mimicking ionic environment in blood plasma) either with or without pretreatment with different concentrations of purfalcamine (50-800 nm). Discharge of microneme protein PfAMA1 was detected in merozoite supernatants by Western blotting. Merozoite supernatants and pellets were also probed with antiserum raised against cytoplasmic protein PfNAPL to detect merozoite lysis and confirm that similar numbers of merozoites were used for different conditions respectively. Transfer of merozoites from IC to EC buffer leads to discharge of PfAMA1. Pretreatment of merozoites with purfalcamine blocked microneme discharge in a dose-dependent manner. B, inhibition of erythrocyte invasion by P. falciparum merozoites by purfalcamine. P. falciparum merozoites were pretreated with different concentrations of purfalcamine (50-800 nm) in IC buffer and were transferred to erythrocytes resuspended in RPMI 1640 medium to allow invasion. Newly invaded rings were scored by Giemsa staining 24–26 h after invasion. Percentage inhibition of invasion with different concentrations of purfalcamine is shown. Error bars represent S.D. from three independent experiments. Purfalcamine inhibits erythrocyte invasion by P. falciparum merozoites in a dose-dependent manner.
DISCUSSION
CDPKs have been reported to play essential roles in controlling various aspects of the Plasmodium life cycle. Here, we have investigated the mechanism of regulation of kinase activity of PfCDPK1 and have for the first time demonstrated that it plays a role in discharge of microneme proteins during erythrocyte invasion by P. falciparum merozoites. PfCDPK1 is expressed in all blood stages including rings, trophozoites, schizonts, and merozoites. PfCDPK1 primarily partitions in detergent-resistant fractions of parasite lysates made with different blood stages (Fig. 2A), suggesting that it may be associated with membranes. Localization by IFA suggests that PfCDPK1 may be associated with the erythrocyte membrane in rings and trophozoites and with the plasma membrane in merozoites (Fig. 2B). PfCDPK1 also co-localizes with SERA5, which is localized in the PV in trophozoites (Fig. 2B). These observations are consistent with previous studies that have demonstrated that PfCDPK1 contains distinct signals at the N terminus for myristoylation at a glycine residue (Gly2) and palmitoylation at a cysteine residue (Cys3) that contribute toward its attachment to membranes (28, 29). These signals have also been shown to be responsible for the translocation of PfCDPK1 into the PV and tubulovesicular network (28), enabling its translocation to the erythrocyte membrane by a nonclassical pathway (Refs. 29, 30 and Fig. 2B). The precise mechanism of translocation of PfCDPK1 to the PV and erythrocyte membrane by such a pathway is not understood.
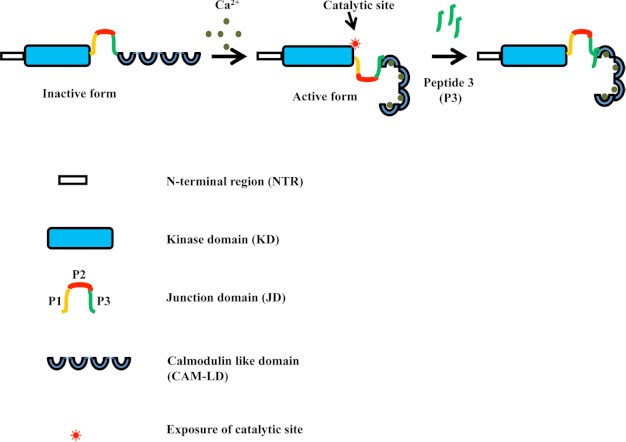
PfCDPK1 shows a significant increase in kinase activity in the presence of Ca2+ thus classifying as a genuine calcium-dependent protein kinase (Fig. 1C and supplemental Fig. 1). α-Helicity of PfCDPK1 was found to decrease in the presence of Ca2+, suggesting significant conformational change upon binding to Ca2+ (Fig. 1D). Recent data from crystal structure of active and inactive forms of TgCDPK1 show that the two α-helices, C1 and C2, which run antiparallel to each other in the inactive form, are disassembled in the presence of Ca2+, leading to change in the overall secondary structure of TgCDPK1 (17). PfCDPK1 has typical domain architecture with an N-terminal KD separated from the CamLD, having calcium-binding EF hands by a JD. JDs of various CDPKs have been shown to play essential roles in regulating the Ca2+-dependent activity of the enzyme (15, 18). In case of PfCDPK4, different truncation mutants were used to map the CamLD binding site to a C-terminal region of the JD (15). Synthetic peptides corresponding to different sections of the JD of PfCDPK4 were used in kinase assays with full-length and truncated PfCDPK4. The C-terminal PIII peptide from the JD of PfCDPK4 was shown to block kinase activity of PfCDPK4 (15). In this study we have provided direct evidence for the interaction of the CamLD with peptide P3 from the C-terminal region of the JD of PfCDPK1 in the presence of Ca2+. The P3 peptide specifically inhibited kinase activity of full-length PfCDPK1. Data from crystal structures of active and inactive forms of Toxoplasma and Cryptosporidium CDPKs as well as our structural model of PfCDPK1 suggest that binding of Ca2+ with EF hands of the CamLD induces substantial conformational changes. Based on the present study, we propose that in the presence of Ca2+ ions, PfCDPK1 undergoes a conformational change leading to interaction of the CamLD with the P3 region of the JD and activation of PfCDPK1 kinase activity (Fig. 9). The P3 peptide competes with the intramolecular P3 region of the JD for binding with the CamLD and inhibits the activation of kinase activity of PfCDPK1 in the presence of Ca2+ (Fig. 9).
FIGURE 9.
A model for PfCDPK1 activation and inhibition of its activity by P3 peptide. In presence of Ca2+, PfCDPK1 undergoes a conformational change so that the CamLD interacts with the P3 region of the JD and exposes the substrate binding catalytic site in the KD, leading to activation of kinase activity. The P3 peptide competes with the intramolecular P3 region in the JD that binds with the CamLD, preventing exposure of the PfCDPK1 catalytic site and activation of kinase activity.
Two mutations, L356A and F363A/I364A in the P3 region of PfCDPK1 were designed based on multiple sequence alignment with CDPKs from other apicomplexan parasites and reports on analysis of PfCDPK4 and Arabidopsis thaliana CDPK1(AtCDPK1) (15, 19). A conserved residue, Phe350, from the P2 region was also included in the analysis. The effect of these mutations on kinase activity of PfCDPK1 was investigated. The mutants L356A, F363A/364A, and F350A lost PfCDPK1 kinase activity both in terms of autophosphorylation of PfCDPK1 and transphosphorylation of syntide-2. L360A and F436A mutations in PfCDPK4 and AtCDPK1, respectively, which correspond to Leu356 in PfCDPK1, also led to abrogation of kinase activity (15, 19). Similarly, Phe363–Ile364 in PfCDPK1, which corresponds to the V444A/I445A mutation in AtCDPK1, led to abrogation of kinase activity highlighting the importance of these residues in CDPK activation.
Using the CONTACT program of the CCP4 package (24) with 4 Å cutoff, the probable residues interacting with Phe350, Leu356, and Phe363–Ile364 in the JD of PfCDPK1 were identified. Phe430, Ser443, and Val444 in the CamLD region of PfCDPK1 were found to be in close proximity (<4 Å) to Phe350 (Fig. 5A). Similarly, Leu356 was found to be in close proximity to Phe464, Leu477, and Phe511 of the CamLD (Fig. 5B). Likewise, Phe363–Ile364 were found to be in close proximity to Ala460, Leu463, and Arg456 of the CamLD (Fig. 5C). Interestingly, the guanidino group of Arg456 and hydroxyl group of Ser443 were found to be directed away from the hydrophobic core formed by other interacting residues (Fig. 5, A and C). Based on this model we predict that alanine replacement mutations at positions Phe350, Leu356, and Phe363–Ile364 in the JD abolish these interactions with the CamLD, leading to abrogation of PfCDPK1 activity.
The P3 region of PfCDPK1 was aligned with corresponding regions of other PfCDPKs such as PfCDPK2, PfCDPK3, PfCDPK4, and PfCDPK5 that have similar domain architecture. The P3 region of PfCDPK1 was most identical (∼80%) to PfCDPK4, but PfCDPK4 is reported to be present only in sexual stages of the parasite (15). The P3 region of PfCDPK1 is only 20–26% identical to the corresponding region in PfCDPK2, PfCDPK3, and PfCDPK5 (supplemental Fig. 3A). P3 peptide did not inhibit the kinase activity of PfCDPK5, which is expressed in blood stages (supplemental Fig. 3B). These results show that P3 can be used to specifically inhibit PfCDPK1 in merozoites. Singh et al. have shown that free cytosolic Ca2+ levels increase in merozoites when they are shifted from buffer mimicking IC to buffer mimicking EC (4). This rise in cytosolic Ca2+ levels in merozoites leads to discharge of micronemal proteins such as EBA-175 and PfAMA1 to the merozoite surface (4). PfAMA1 is proteolytically cleaved by the subtilisin-like protease PfSub2 when it is translocated to the merozoite surface (25). The discharge of PfAMA1 from micronemes can thus be studied by detecting PfAMA1 in the supernatant of P. falciparum merozoites by Western blotting. In addition, the presence of PfAMA1 on the merozoite surface can be detected by flow cytometry using anti-PfAMA1 serum. We have used both methods to follow the secretion of PfAMA1 from micronemes. The effector molecules that are involved in triggering microneme discharge in response to increase in cytosolic Ca2+ have not yet been identified. Here, we have demonstrated that prior treatment of merozoites in IC buffer with P3 peptide, which specifically inhibits PfCDPK1, blocks the secretion of microneme protein PfAMA1 when merozoites are shifted to EC buffer (Figs. 6A and 7). Also, P3 inhibits erythrocyte invasion by P. falciparum merozoites (Fig. 6B), suggesting that PfCDPK1 activity is required for microneme secretion and successful invasion of erythrocytes. Given that we cannot completely rule out the possibility that P3 might inhibit kinases other than PfCDPK1, we tested the ability of purfalcamine, a previously identified specific small molecular inhibitor of PfCDPK1 (13), to inhibit microneme discharge. Pretreatment of P. falciparum merozoites in IC buffer with purfalcamine inhibited the secretion of PfAMA1 from micronemes upon transfer of merozoites from IC to EC buffer (Figs. 7 and 8A). Purfalcamine also inhibited erythrocyte invasion by P. falciparum merozoites (Fig. 8B). These observations implicate PfCDPK1 as a Ca2+-dependent effector that plays a role in microneme secretion. They also validate PfCDPK1 as a potential target for development of specific inhibitors that inhibit erythrocyte invasion to block parasite growth and clear malaria parasite infections. Moreover, this study has explored the mechanism for the regulation of the kinase activity of PfCDPK1 and has demonstrated that one could design inhibitors that interfere with the regulatory mechanism that activates the calcium-dependent kinase activity of PfCDPK1 to develop novel specific inhibitors of this key kinase to block the growth of blood stage parasites.
Acknowledgments
We thank Dr. Case McNamara, Genomics Institute of the Novartis Research Foundation, San Diego, and Dr. Elizabeth Winzeler, University of San Diego, for the purfalcamine for these studies.
This work was supported by European Commission Framework Program 7 (FP7) projects titled “Signaling in Malaria” (MALSIG, HEALTH-F3-2009-223044) and “Network of Excellence: European Virtual Institute for Malaria Research” (EviMalaR, Health-2009-2.3.2-1-242095) and a Program Support grant from Department of Biotechnology, Government of India.

This article contains supplemental Materials and Methods and Figs. 1–3.
- CDPK
- calcium-dependent protein kinase
- AMA1
- apical membrane antigen 1
- AtCDPK
- A thaliana CDPK
- CamLD
- calmodulin-like domain
- CpCDPK3
- C. parvum CDPK
- EC
- extracellular buffer
- IC
- intracellular buffer
- IFA
- immunofluorescence assay
- JD
- junction domain
- KD
- kinase domain
- NAPL
- nucleosome assembly protein-large
- NTR
- N-terminal domain
- PbCDPK
- P. berghei CDPK
- PfCDPK
- P. falciparum CDPK
- PV
- parasitophorous vacuole
- TgCDPK
- T. gondii CDPK
- Tricine
- N-[2-hydroxy-1,1-bis(hydroxymethyl)ethyl]glycine.
REFERENCES
- 1. Murray C. J., Rosenfeld L. C., Lim S. S., Andrews K. G., Foreman K. J., Haring D., Fullman N., Naghavi M., Lozano R., Lopez A. D. (2012) Global malaria mortality between 1980 and 2010: a systematic analysis. Lancet 379, 413–431 [DOI] [PubMed] [Google Scholar]
- 2. Cowman A. F., Crabb B. S. (2006) Invasion of red blood cells by malaria parasites. Cell 124, 755–766 [DOI] [PubMed] [Google Scholar]
- 3. Gaur D., Chitnis C. E. (2011) Molecular interactions and signaling mechanisms during erythrocyte invasion by malaria parasites. Curr. Opin. Microbiol. 14, 422–428 [DOI] [PubMed] [Google Scholar]
- 4. Singh S., Alam M. M., Pal-Bhowmick I., Brzostowski J. A., Chitnis C. E. (2010). Distinct external signals trigger sequential release of apical organelles during erythrocyte invasion by malaria parasites. PLoS Pathog. 6, e1000746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Carruthers V. B., Sibley L. D. (1999) Mobilization of intracellular calcium stimulates microneme discharge in Toxoplasma gondii. Mol. Microbiol. 31, 421–428 [DOI] [PubMed] [Google Scholar]
- 6. Lovett J. L., Marchesini N., Moreno S. N., Sibley L. D. (2002) Toxoplasma gondii microneme secretion involves intracellular Ca2+ release from inositol 1,4,5-trisphosphate (IP3)/ryanodine-sensitive stores. J. Biol. Chem. 277, 25870–25876 [DOI] [PubMed] [Google Scholar]
- 7. Carruthers V. B., Giddings O. K., Sibley L. D. (1999) Secretion of micronemal proteins is associated with Toxoplasma invasion of host cells. Cell Microbiol. 1, 225–235 [DOI] [PubMed] [Google Scholar]
- 8. Lourido S., Shuman J., Zhang C., Shokat K. M., Hui R., Sibley L. D. (2010) Calcium-dependent protein kinase 1 is an essential regulator of exocytosis in Toxoplasma. Nature 465, 359–362 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Sugi T., Kato K., Kobayashi K., Watanabe S., Kurokawa H., Gong H., Pandey K., Takemae H., Akashi H. (2010) Use of the kinase inhibitor analog 1NM-PP1 reveals a role for Toxoplasma gondii CDPK1 in the invasion step. Eukaryot. Cell 9, 667–670 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Ishino T., Orito Y., Chinzei Y., Yuda M. (2006) A calcium-dependent protein kinase regulates Plasmodium ookinete access to the midgut epithelial cell. Mol. Microbiol. 59, 1175–1184 [DOI] [PubMed] [Google Scholar]
- 11. Siden-Kiamos I., Ecker A., Nybäck S., Louis C., Sinden R. E., Billker O. (2006) Plasmodium berghei calcium-dependent protein kinase 3 is required for ookinete gliding motility and mosquito midgut invasion. Mol. Microbiol. 60, 1355–1363 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Billker O., Dechamps S., Tewari R., Wenig G., Franke-Fayard B., Brinkmann V. (2004) Calcium and a calcium-dependent protein kinase regulate gamete formation and mosquito transmission in a malaria parasite. Cell 117, 503–514 [DOI] [PubMed] [Google Scholar]
- 13. Kato N., Sakata T., Breton G., Le Roch K. G., Nagle A., Andersen C., Bursulaya B., Henson K., Johnson J., Kumar K. A., Marr F., Mason D., McNamara C., Plouffe D., Ramachandran V., Spooner M., Tuntland T., Zhou Y., Peters E. C., Chatterjee A., Schultz P. G., Ward G. E., Gray N., Harper J., Winzeler E. A. (2008) Gene expression signatures and small-molecule compounds link a protein kinase to Plasmodium falciparum motility. Nat. Chem. Biol. 4, 347–356 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Green J. L., Rees-Channer R. R., Howell S. A., Martin S. R., Knuepfer E., Taylor H. M., Grainger M., Holder A. A. (2008) The motor complex of Plasmodium falciparum: phosphorylation by a calcium-dependent protein kinase. J. Biol. Chem. 283, 30980–30989 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Ranjan R., Ahmed A., Gourinath S., Sharma P. (2009) Dissection of mechanisms involved in the regulation of Plasmodium falciparum calcium-dependent protein kinase 4. J. Biol. Chem. 284, 15267–15276 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Dvorin J. D., Martyn D. C., Patel S. D., Grimley J. S., Collins C. R., Hopp C. S., Bright A. T., Westenberger S., Winzeler E., Blackman M. J., Baker D. A., Wandless T. J., Duraisingh M. T. (2010) A plant-like kinase in Plasmodium falciparum regulates parasite egress from erythrocytes. Science 328, 910–912 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Wernimont A. K., Artz J. D., Finerty P., Jr., Lin Y. H., Amani M., Allali-Hassani A., Senisterra G., Vedadi M., Tempel W., Mackenzie F., Chau I., Lourido S., Sibley L. D., Hui R. (2010) Structures of apicomplexan calcium-dependent protein kinases reveal mechanism of activation by calcium. Nat. Struct. Mol. Biol. 17, 596–601 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Huang J. F., Teyton L., Harper J. F. (1996) Activation of a Ca2+-dependent protein kinase involves intramolecular binding of a calmodulin-like regulatory domain. Biochemistry 35, 13222–13230 [DOI] [PubMed] [Google Scholar]
- 19. Vitart V., Christodoulou J., Huang J. F., Chazin W. J., Harper J. F. (2000) Intramolecular activation of a Ca2+-dependent protein kinase is disrupted by insertions in the tether that connects the calmodulin-like domain to the kinase. Biochemistry 39, 4004–4011 [DOI] [PubMed] [Google Scholar]
- 20. Trager W., Jensen J. B. (1976) Human malaria parasites in continuous culture. Science 193, 673–675 [DOI] [PubMed] [Google Scholar]
- 21. Eswar N., Eramian D., Webb B., Shen M. Y., Sali A. (2008) Protein structure modeling with MODELLER. Methods Mol. Biol. 426, 145–159 [DOI] [PubMed] [Google Scholar]
- 22. Pettersen E. F., Goddard T. D., Huang C. C., Couch G. S., Greenblatt D. M., Meng E. C., Ferrin T. E. (2004) UCSF Chimera: a visualization system for exploratory research and analysis. J. Comput. Chem. 25, 1605–1612 [DOI] [PubMed] [Google Scholar]
- 23. Laskowski R. A., Rullmannn J. A., MacArthur M. W., Kaptein R., Thornton J. M. (1996) AQUA and PROCHECK-NMR: programs for checking the quality of protein structures solved by NMR. J. Biomol. NMR 8, 477–486 [DOI] [PubMed] [Google Scholar]
- 24. Collaborative Computational Project, Number 4 (1994) The CCP4 suite: programs for protein crystallography. Acta Crystallogr. D Biol. Crystallogr. 50, 760–763 [DOI] [PubMed] [Google Scholar]
- 25. Harris P. K., Yeoh S., Dluzewski A. R., O'Donnell R. A., Withers-Martinez C., Hackett F., Bannister L. H., Mitchell G. H., Blackman M. J. (2005) Molecular identification of a malaria merozoite surface sheddase. PLoS Pathogens 1, 241–251 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Delplace P., Bhatia A., Cagnard M., Camus D., Colombet G., Debrabant A., Dubremetz J. F., Dubreuil N., Prensier G., Fortier B. (1988) Protein p126: a parasitophorous vacuole antigen associated with the release of Plasmodium falciparum merozoites. Biol. Cell 64, 215–221 [DOI] [PubMed] [Google Scholar]
- 27. Blackman M. J. (2008) Malarial proteases and host cell egress: an “emerging” cascade. Cell Microbiol. 10, 1925–1934 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Möskes C., Burghaus P. A., Wernli B., Sauder U., Dürrenberger M., Kappes B. (2004) Export of Plasmodium falciparum calcium-dependent protein kinase 1 to the parasitophorous vacuole is dependent on three N-terminal membrane anchor motifs. Mol. Microbiol. 54, 676–691 [DOI] [PubMed] [Google Scholar]
- 29. Jones M. L., Collins M. O., Goulding D., Choudhary J. S., Rayner J. C. (2012) Analysis of protein palmitoylation reveals a pervasive role in Plasmodium development and pathogenesis. Cell Host Microbe 12, 246–258 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Zhao Y., Franklin R. M., Kappes B. (1994) Plasmodium falciparum calcium-dependent protein kinase phosphorylates proteins of the host erythrocytic membrane. Mol. Biochem. Parasitol. 66, 329–343 [DOI] [PubMed] [Google Scholar]