Background: During inflammation, the selectins engage glycosylated macromolecules expressed on blood leukocytes under fluid shear conditions.
Results: Although all three myeloid α1,3-fucosyltransferases FUT9, FUT7, and FUT4 regulate human E-selectin ligand biosynthesis, FUT7 and FUT4 are sufficient to form L/P-selectin ligands.
Conclusion: FUT9 plays a significant role during human, but not mouse, leukocyte-endothelial interactions.
Significance: This study identifies potential α(1,3)FUTs regulating inflammation in humans.
Keywords: Carbohydrate Glycoconjugate, Cell Adhesion, Cell-surface Receptor, Glycobiology, Glycoprotein, Glycosylation, Glycosyltransferases, Inflammation, Leukocyte
Abstract
Leukocyte adhesion during inflammation is initiated by the binding of sialofucosylated carbohydrates expressed on leukocytes to endothelial E/P-selectin. Although the glycosyltransferases (glycoTs) constructing selectin-ligands have largely been identified using knock-out mice, important differences may exist between humans and mice. To address this, we developed a systematic lentivirus-based shRNA delivery workflow to create human leukocytic HL-60 cell lines that lack up to three glycoTs. Using this, the contributions of all three myeloid α1,3-fucosyltransferases (FUT4, FUT7, and FUT9) to selectin-ligand biosynthesis were evaluated. The cell adhesion properties of these modified cells to L-, E-, and P-selectin under hydrodynamic shear were compared with bone marrow-derived neutrophils from Fut4−/−Fut7−/− dual knock-out mice. Results demonstrate that predominantly FUT7, and to a lesser extent FUT4, forms the selectin-ligand at the N terminus of leukocyte P-selectin glycoprotein ligand-1 (PSGL-1) in humans and mice. Here, 85% reduction in leukocyte interaction was observed in human FUT4−7− dual knockdowns on P/L-selectin substrates. Unlike Fut4−/−Fut7−/− mouse neutrophils, however, human knockdowns lacking FUT4 and FUT7 only exhibited partial reduction in rolling interaction on E-selectin. In this case, the third α1,3-fucosyltransferase FUT9 played an important role because leukocyte adhesion was reduced by 50–60% in FUT9-HL-60, 70–80% in dual knockdown FUT7−9− cells, and ∼85% in FUT4−7−9− triple knockdowns. Gene silencing results are in agreement with gain-of-function experiments where all three fucosyltransferases conferred E-selectin-mediated rolling in HEK293T cells. This study advances new tools to study human glycoT function. It suggests a species-specific role for FUT9 during the biosynthesis of human E-selectin ligands.
Introduction
Three members of the selectin family of adhesion molecules, E-(CD62E), P-(CD62P), and L-selectin (CD62L), regulate the capture of leukocytes from flowing blood to sites of inflammation under hydrodynamic/fluid shear conditions (1). Such rapid molecular bond formation at high on-rates is enabled by unique protein-carbohydrate interactions between the selectins and their sialofucosylated ligands that are expressed on the leukocytes (2). These carbohydrate ligands are post-translationally synthesized by the glycosyltransferases (glycoTs),2 a family of about 200 enzymes that constitute ∼1% of the human genome (3). Besides inflammation, selectin-mediated cell adhesion also contributes to leukocyte-platelet adhesion in circulation, hematopoietic stem cell and lymphocyte homing, and cancer metastasis (4–6).
Although much of our knowledge of glycoTs regulating selectin-ligand synthesis is derived from mice, these animals differ from humans particularly with respect to E-selectin and its ligands (1, 6). In this regard, although E-selectin binding to mouse granulocytes is completely lost upon pronase digestion, it is only partially reduced in human cells (7, 8).3 E-selectin ligand-1 (ESL-1), an important selectin-ligand for mature mouse neutrophils, is absent in humans (9). E-selectin ligands on human bone marrow CD34+ cells are distinct from mouse LSK (lin− Sca-1+ c-kit+) cells in that the CD44 variant HCELL (hematopoietic cell E- and L-selectin ligand) is the dominant ligand for the human cell type, whereas CLA (cutaneous lymphocyte antigen, epitope recognized by mAb HECA-452) plays this role in mice (10). Furthermore, unlike mice, the promoter of the human P-selectin gene (Selp) lacks binding sites for the transcription factors NF-κB and ATF-2 (11). Thus, although P-selectin expression is up-regulated by tumor necrosis factor-α (TNFα) and IL-1β in murine inflammation models, this is not the case in humans. The contributions of P/E-selectin to inflammation is thus different in murine models versus human disease (12).
Whether the selectin-ligands differ in murine versus human systems because of species-specific functions of glycoTs remains undetermined. We addressed this possibility by advancing RNA interference (RNAi) and lentiviral delivery tools to study glycoTs in human leukocytes. In particular, we evaluated the roles of all three α1,3-FUTs expressed in myeloid cells, FUT4, FUT7, and FUT9 in synthesizing the ligands for human E-, P-, and L-selectin (3). In accordance with Taniguchi et al. (3), FUT4 and FUT7 are used to designate human enzymes and Fut4 and Fut7 are used for non-human species. The strategy involved generation of a panel of stable human promyelocytic HL-60 cell lines that contain short hairpin RNA (shRNA) directed against one, two, or three different glycoTs. HL-60 cells were chosen because primary human leukocytes cannot be maintained in culture for long term assays, and because the prototypic sialofucosylated O-linked selectin-ligand located at the N terminus of human P-selectin glycoprotein ligand-1 (PSGL-1/CD162) was originally characterized using these cells (13–16). A microfluidics based cell adhesion assay under physiological flow evaluated the effect of gene silencing. Results show that, like mice, FUT7 and to a lesser extent FUT4 synthesize the selectin-ligands that mediate human leukocyte tethering and rolling on substrates bearing P- and L-selectin. Unlike mice, however, FUT9 plays a major role during the synthesis of human E-selectin ligands with FUT7 and FUT4 having smaller contributions. Overall, the study introduces RNA interference methods for studies of leukocyte glycoTs. Unlike previous studies with Fut4−/−Fut7−/− dual knock-out mice that exhibit complete loss of E-, P-, and L-selectin binding function in a variety of physiological and pathological models (17, 18), our data demonstrate a novel role for FUT9 in catalyzing human E-selectin ligand synthesis. To our best knowledge, this is the first demonstration of glycoTs in different mammalian species having distinct contributions to leukocyte rolling.
EXPERIMENTAL PROCEDURES
Cell Culture
CHO-S cells (Invitrogen), CHO-S stably expressing P-selectin (CHO-P), and HEK293T cells (ATCC, Manassas, VA) were cultured in Dulbecco's modified Eagle's medium (DMEM) containing 10% FBS. HL-60 cells were maintained in Iscove's modified Dulbecco's medium with 10% FBS. L-cells bearing E-selectin (L-E cells), kindly provided by Dr. Scott I. Simon (University of California, Davis, CA), were maintained in RPMI 1640 medium with 10% FBS, 0.25 mg/ml xanthine, sodium hypoxanthine and thymidine supplement, and 5 μg/ml mycophenolic acid. Human umbilical vein endothelial cells (HUVECs) purchased from Lonza (Walkersville, MD) were maintained in EBM2 media (Lonza). Cell adhesion studies with HUVEC were conducted within 1–3 passages.
Molecular Biology
Standard molecular biology methods were used to create lentiviral transfer plasmids that carry DNA encoding for cell-surface proteins, fusion proteins, or shRNA. Typically, cell-surface and fusion proteins were expressed using pCS-CG (plasmid 12154, Addgene, Cambridge, MA) as the transfer construct. Here, the cytomegalovirus (CMV) promoter drives gene expression. shRNAs were encoded using the pLKO.1 vector (Sigma). In this case, the human polymerase III U6 promoter drives shRNA expression, whereas a phosphoglycerokinase (PGK) promoter drives expression of either the selection marker (puromycin resistance gene) or reporter gene (DsRed or cerulean). Details are below.
Human FUT-EGFP Fusion Proteins
FUT-EGFP represents a family of fusion proteins where EGFP is fused to the C terminus of either full-length human FUT4, FUT7, or FUT9. DNA encoding for FUT-EGFP was constructed in the vector pCS-CG. To this end, first, 1.5 kb of pCS-CG (bp 2383–3882 on pCS-CG), which includes both the CMV promoter and EGFP regions, was cloned into the pUC19 cloning vector. Primers 1 and 2 (supplemental Table S1) were used to introduce an AgeI site (ACCGGT) directly prior to the EGFP in this vector, and the native start codon (ATG) of EGFP was simultaneously deleted. This pUC19 vector was digested using KpnI and AgeI to remove the 800-bp fragment encoding for the EGFP region of pCS-CG. This fragment was ligated into native pCS-CG vector using the same enzymes, to generate “pCS-CG-AgeI-EGFP.” cDNA encoding for the human fucosyltransferases FUT4, FUT7, and FUT9 were obtained in pCMVsport-6 from Open Biosystems (Huntsville, AL). Forward primers were designed to introduce either an AgeI (ACCGGT) site in the case of FUT7 and -9 or an AgeI-compatible end (BspEI, TCCGGA) in the case of FUT4, along with a Kozak sequence (CGCCACC) just prior to the start codon of the enzyme. The native stop codon was deleted, and an AgeI site was introduced using an appropriate reverse primer. Primers 3–8 (supplemental Table S1) were used for this step. PCR-amplified FUT cDNA segments thus obtained were ligated into pCS-CG-AgeI-EGFP. In summary, the above steps lead to the construction of pCS-CG that carried FUT4-EGFP, FUT7-EGFP, and FUT9-EGFP.
Human Full-length PSGL-1
Human PSGL-1 cDNA was amplified using primers 9 and 10 (supplemental Table S1) and template vector available from a previous study (19). Product digested using BamHI and KpnI was ligated into pLKO.1 TRC cloning (Addgene plasmid 10878) in place of the puromycin resistance gene. In pLKO.1, the puromycin resistance gene was located between unique BamHI and KpnI sites just downstream of the PGK promoter. shRNA was absent in the pLKO.1 TRC cloning vector.
Cloning shRNA into pLKO.1
3–5 shRNAs for each of the FUTs of interest (supplemental Table S2) were cloned into the pLKO.1 TRC cloning vector that has a puromycin selection cassette. These unique shRNA sequences were selected from the Broad Institute MISSION® shRNA library. Each shRNA sequence was introduced between unique AgeI and EcoRI sites downstream of the U6 promoter in pLKO.1.
Fluorescent Reporters in pLKO.1
In some cases, the puromycin resistance gene in pLKO.1 was replaced by either DsRed (red fluorescence protein variant) or cerulean (cyan fluorescence protein variant). Here, DsRed was isolated from mRFP-ubiquitin (Addgene plasmid 11935) using primers 11 and 12 (supplemental Table S1), and cerulean was amplified from pCerulean-VSVG (Addgene plasmid 11913) using primers 13 and 14. BamHI and KpnI present in PCR products enabled ligation into pLKO.1. The vectors resulting from this step both drive expression of shRNA for gene knockdown and express the fluorescence reporter DsRed/cerulean.
Viral Transduction
Following the cloning steps, lentiviral particles were generated using calcium phosphate precipitation method to co-transfect into HEK293T cells the following plasmids: 15 μg of packaging vector psPAX2, 10 μg of VSVG envelope vector pMD2.G, and 20 μg of transfer plasmid (described in the previous section). For viral preparation, 8–14 h post-transfection, 10 mm sodium butyrate was added to culture medium for 48 h. The medium with lentiviral particles was then collected, filtered through 0.45-μm syringe filters, and concentrated 100-fold by centrifugation at 50,000 × g for 2 h at 4 °C. The viral pellet was dissolved in serum-free Iscove's modified Dulbecco's medium (IMDM) and used either immediately or stored at −80 °C.
Lentivirus prepared in the above step was used to stably transduce HEK293T, CHO-S, and HL-60 cells. Substrate-adherent cells (HEK293T and CHO-S) were plated in 24-well plates overnight to achieve 70% confluence the following day. HL-60 cells were at 60,000 cells/ml at the time of transduction. In all cases, the wells had cells, 500 μl of normal cell culture media, 8 μg/ml Polybrene, and a range of viral titers during the transduction step. In the case of adherent cells, 25 μl of the 100× viral concentrate above was typically used to consistently achieve ∼100% stable transduction (multiplicity of infection ∼10). Viral titer was 2-fold greater in the case of suspension cells (HL-60). Cells were scaled up in media lacking virus and Polybrene after 24–36 h. Stable gene expression was confirmed 5–14 days post-transduction using flow cytometry.
HEK293T Cells Expressing PSGL-1 and FUT-EGFP
Stable HEK293T cell lines that express various FUTs, in the presence or absence of PSGL-1, were generated. HEK293T cells were first transduced with lentivirus encoding for human PSGL-1. Stable PSGL-1 expression in ∼100% of the cells was confirmed using a FACSCalibur flow cytometry (BD Biosciences) and phycoerythrin-conjugated anti-PSGL-1 mAb KPL-1 (BD Biosciences). Both wild-type (WT) HEK293T and HEK293T cells with PSGL-1 (“HEK-PSGL-1” or “HP” cells) were then transduced with lentivirus encoding for either FUT4-EGFP, FUT7-EGFP, or FUT9-EGFP. >95% of the transduced cells stably expressed FUT-EGFP based on EGFP fluorescence. To characterize the subcellular localization of FUT-EGFPs in these cells, immunocytochemistry was performed as described below.
Immunocytochemistry
22 × 22-mm glass coverslips were incubated overnight with 100 μg/ml poly-l-lysine to absorb the molecules. Excess poly-l-lysine was then removed, and HEK293T cells expressing one of the FUT-EGFP proteins were seeded on the substrate. Cells grown to ∼50–70% confluence overnight were washed and fixed with 1.5 ml of cold 4% paraformaldehyde for 15 min, and the membrane was permeabilized using 1 ml of 5% blocking solution (0.2% Triton X-100, 1% BSA, 5% goat serum in phosphate-buffered saline (PBS)) for 1 h. Coverslips containing cells were then incubated with 0.67 μg/ml anti-human TGN-46 antibody (Ab against trans-Golgi network protein, Sigma) for 12–16 h, washed with PBS containing 0.05% Triton X-100, and then 2 μg/ml goat-anti-rabbit Alexa 594 IgG secondary antibody (Invitrogen) was added for 1–2 h in the dark. Cells were then stained with 0.33 μg/ml 4′,6-diamidino-2-phenylindole (DAPI) for 15 min, mounted on a microscope slide, and dried overnight at 4 °C. Images were acquired using a Zeiss LSM 510 Meta NLO confocal microscope with plan-apochromat 63×/1.4 oil objective. Co-localization overlap coefficient was calculated using the ImageJ plug in JACoP, “Just Another Colocalization Plugin” (20).
FUT Enzyme Activity
Enzymology assays quantified FUT activity in HEK293T cell lysates by extending a previously published method (21). Briefly, FUT activity was quantified in a 3-μl reaction volume that contained 50 mm HEPES buffer, pH 7.5, 5 mm MnCl2, 7 mm ATP, 3 mm NaN3, 4.167 μm GDP-[14C]Fuc (0.0012 μCi), 1.25 mm synthetic acceptor, and 1 mg/ml cell lysate. Following reaction for 8 h, 5 μl of acetonitrile was added, and the sample was centrifuged at 18,000 × g for 20 min to precipitate proteins. The supernatant was collected, air-dried for 30 min to remove acetonitrile, and the remaining material resuspended in 2 μl of water. This was spotted on a reverse phase-TLC to separate radioactive product from unreacted GDP-[14C]Fuc using water containing 0.2% acetic acid as mobile phase. Reverse phase-TLC plates were scanned using phosphorimaging followed by analysis using ImageJ (National Institutes of Health). Percent of total radioactivity incorporated into various acceptors was quantified.
Quantifying glycoT shRNA Silencing Efficiency
CHO cells stably expressing one of the three FUT-EGFP fusion proteins (FUT4-EGFP, FUT7-EGFP, or FUT9-EGFP) were developed by transduction of CHO-S cells (Invitrogen) with lentivirus encoding for the fusion proteins. To determine shRNA silencing efficiency, lentivirus encoding for various FUT shRNAs (listed in supplemental Table S2) were added to these cells. Change in EGFP fluorescence was measured using flow cytometry 4–10 days after transduction. Silencing efficiency (%) = 100 × (1 −(EGFP fluorescence of FUT-EGFP cells treated with shRNA ÷ EGFP fluorescence in cells without shRNA)). Negative controls include lentivirus made using the pLKO.1 TRC cloning vector that contains a 1.9-kb stuffer in place of shRNA (Addgene plasmid 10878) and pLKO.1 TRC control vector containing 18 bp of nonhairpin sequence (Addgene plasmid 10879). Positive control was pLKO.1 EGFP shRNA vector (Addgene plasmid 12273). Although the pLKO.1 vector carrying shRNA did not have a fluorescence reporter during the initial RNAi screening studies, DsRed replaced the puromycin resistance gene in this vector in later studies that confirmed the specificity of gene silencing.
HL-60 Carrying One or More FUT shRNAs
Efficient shRNA directed against various FUTs, highlighted in bold in supplemental Table S2, were cloned into the pLKO.1 vector that had either drug selection (puromycin resistance) or reporter genes (DsRed or cerulean). In all cases, the human polymerase III U6 promoter drove shRNA expression, whereas a PGK promoter was upstream of the drug selection or reporter gene. Single knockdown HL-60 cells lacking FUT4, FUT7, or FUT9 were created by transduction of WT cells with virus encoding for corresponding shRNA, using DsRed as a reporter of stable gene expression. These cell lines are abbreviated “FUT4− HL-60,” “FUT7− HL-60,” and “FUT9− HL-60.” The pLKO.1 TRC cloning vector with DsRed cloned in place of the puromycin resistance gene was used as a negative control in the single knockdown experiments. These cells were termed “DsRed HL-60.” Dual knockdowns lacking either FUT4 and FUT7 or FUT7 and FUT9 were created by transducing FUT7− HL-60 with virus encoding for either FUT4 or FUT9 shRNA along with the puromycin resistance gene. These dual knockdown cell lines that were selected and maintained in growth media containing 0.5 μg/ml puromycin were termed FUT4−7− HL-60 and FUT7−9− HL-60. Triple knockdown HL-60s lacking all three FUTs were made by transducing FUT7−9− HL-60 cells with virus encoding for FUT4 shRNA. These cells, called FUT4−7−9− HL-60, carried an additional cerulean reporter. Virus generated using the pLKO.1 TRC cloning vector, where cerulean replaced the puromycin resistance gene, were also transduced into FUT7−9− HL-60 cells to serve as a negative control for the triple knockdown experiments. These last cells are called “FUT7−9− cerulean HL-60.”
Stable gene expression was verified using quantitative real time PCR based on a previous study (primers are listed in supplemental Table S3) (14) and flow cytometry between 4 and 20 days after viral transduction (see below). HL-60 cells were resuspended in 30 mm HEPES buffer (30 mm HEPES, 10 mm glucose, 110 mm NaCl, 10 mm KCl, 1 mm MgCl2, pH 7.2) containing 1.5 mm CaCl2 and 0.1% human serum albumin for all cytometry and cell adhesion studies described below.
Flow Cytometry
Monoclonal antibodies (mAbs) used include anti-Lewis-X (LeX)/CD15 clone HI98, anti-CLA clone HECA-452, anti-sialyl Lewis X (sLeX)/CD15s clone CSLEX-1, anti-human PSGL-1/CD162 clone KPL-1, anti-human CD65s clone VIM-2, and isotype-matched controls. All reagents were from BD Biosciences, except mAb VIM-2 was from Serotec (Oxford, UK). Among these, the binding specificity of mAb HECA-452 is similar to that of CSLEX-1 because they both recognizes sLeX and related epitopes. Fluorescent mAbs (HECA-452, HI98, and KPL-1) or isotype-matched reagents were added to cells for 15 min at room temperature (RT) prior to cytometry analysis. Binding studies with unconjugated mAb (CSLEX-1) or biotinylated Aleuria aurantia (AAL) lectin (binds α1,3/4/6 fucose, Vector Laboratories, Burlingame, CA) involved 15 min of incubation of reagents with cells before addition of 1:200 diluted secondary Ab (goat anti-mouse IgM Alexa 488 for CSLEX-1 or fluorescein-conjugated anti-biotin for AAL) for another 15 min. Cytometry analysis was then performed. Although many of the flow cytometry studies utilized a BD FACSCalibur instrument, studies with cells expressing cerulean were analyzed using a BD LSR-II cytometer with appropriate lasers.
Neutrophil Isolation from Mouse Bone Marrow
Mouse neutrophils were isolated using the protocol described elsewhere (22). Briefly, age-matched WT C57BL/6 and Fut4−/−Fut7−/− mice (The Jackson Laboratory, Bar Harbor, ME) were sacrificed by CO2 asphyxiation. Bone marrow cells collected from femur, tibia, and sternum in RPMI 1640 medium were washed and resuspended in 1 ml of PBS. These cells were layered over a Histopaque gradient consisting of a 3-ml top layer made of Histopaque 1077 (Sigma) and a 3-ml bottom layer of Histopaque 1119 in a 15-ml centrifuge tube. Following centrifugation at 700 × g for 30 min at room temperature, the white band at the junction of the two Histopaque layers was removed. Cells were washed in sterile HEPES buffer. Cytospin analysis was performed to verify neutrophil purity.
Microfluidic Cell Adhesion Assay
A custom vacuum-sealed microfluidic flow device containing a 1-cm-long flow channel with width × height dimensions of 400 × 100 μm was fabricated using standard photolithography and polydimethylsiloxane. To immobilize selectins, in some cases a 1 × 1-mm area on a 100-mm tissue culture dish was incubated with P/L/E-selectin-Fc fusion protein overnight at 4 °C. Although efforts were not undertaken to quantify the precise number of immobilized molecules, this concentration of selectin-Fc was sufficient to support robust and specific cell rolling. In other instances, CHO-P, L-E, or HUVEC cells were seeded on these same dishes and grown to confluence. HUVEC were stimulated with IL-1β for 4 h to up-regulate E-selectin. Studies with modified HEK293Ts utilized the recombinant selectin substrates due to the need to confirm binding specificity in these runs. Studies with WT and modified HL-60 used cellular substrates to mimic physiological conditions. Prior to experimentation, the microfluidic flow cell was vacuum-sealed onto either recombinant selectin fusion protein or selectin-bearing cells and mounted on the Zeiss AxioObserver microscope stage. A 10-μl pipette tip served as the inlet reservoir, and the outlet was connected to a 250-μl Hamilton syringe mounted on a Harvard Apparatus syringe pump (Pump11, Holliston, MA). HEPES buffer containing 0.5% BSA was perfused for 10 min to minimize nonspecific binding and to equilibrate the system. Various HL-60 and HEK293T cell lines at a concentration of 2 × 106 cells/ml were suspended in HEPES buffer containing 1.5 mm Ca2+. These were perfused in the flow cell at a wall shear stress of 1–2 dynes/cm2.
Movies of cells interacting with selectin-bearing substrate were captured using Streampix image acquisition software (Norpix, Montreal, Canada) at 10 frames/s using a QImaging Retiga-1300 cooled CCD camera (Surrey, British Columbia, Canada). Programs utilizing ImageJ (National Institutes of Health) and the MATLAB image processing toolbox (Natick, MA) were written for data analysis. Analysis methods were similar to prior work (23, 24). Because the number of rolling cells reached steady state within 1 min after starting injection of cells, data analysis was performed in the window between 1 and 3 min. Interacting cells were defined to travel at velocities less than the theoretical free stream velocity of noninteracting 10-μm diameter spheres at the flow chamber wall (23). Among these, cells moving less than one cell diameter in a 10-s period were defined to be “adherent,” and the remaining were classified to be “rolling.” Rolling velocity was calculated by tracking individual cell trajectories.
Statistics
Error bars represent the mean ± S.E. for ≥3 independent experiments. One-way analysis of variance followed by the Tukey's post test was performed to assess statistical significance. p < 0.05 was considered significant.
RESULTS AND DISCUSSION
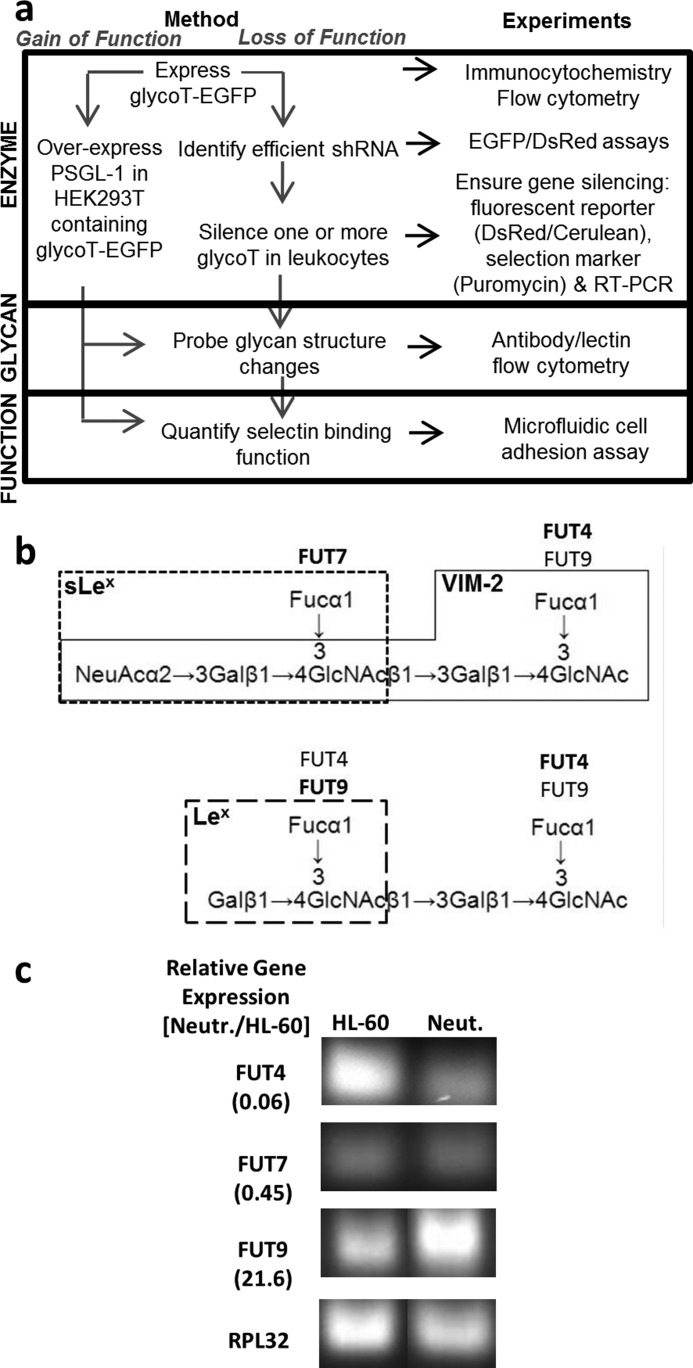
This study examines the role of human leukocyte glycoTs in regulating cell-surface glycan expression and selectin-mediated cell adhesion. It was enabled by two methodological advances as follows: (i) strategies to rapidly screen for shRNA that silence specific glycoTs, and (ii) techniques to stably express shRNA and transgenes (fluorescent reporter or drug selection marker) in hard-to-transfect human leukocytes (workflow outlined in Fig. 1a). The overall goal was to dissect the role of FUT4, FUT7, and FUT9 in regulating selectin-mediated leukocyte adhesion. Previous enzymology studies show that FUT7 exclusively fucosylates sialylated N-acetyllactosamine (sialyl-LacNAc) to form sialyl-Lewis-X (sLeX) type structures (Fig. 1b) (25). FUT9 and FUT4 fucosylate unsialylated poly-LacNAc, with FUT9 preferring the outer or nonreducing LacNAc and FUT4 acting on inner LacNAc units (26, 27). The specificity is however not absolute because either enzyme can form the VIM-2 and Lewis-X (LeX) epitopes (28, 29). All three FUTs are expressed in HL-60 cells and mature peripheral blood granulocytes (Fig. 1c) (26). FUT4 levels are high in HL-60, whereas FUT9 is expressed at high levels in mature neutrophils.
FIGURE 1.
Work flow for gain-of-function and loss-of-function studies. a, two types of stable cell systems were created using VSVG pseudotyped lentivirus to either overexpress or knock down specific genes. For the gain-of-function studies, glycoT-EGFP fusion proteins (FUTs in this case) were overexpressed in either WT HEK293T cells or the same cells bearing human PSGL-1. For the loss-of-function studies, efficient shRNA that knock down glycoT gene expression were identified using CHO-S cells that overexpress glycoT-EGFP fusion proteins. These shRNAs were then introduced into human leukocyte HL-60 cells to silence between 1 and 3 glycoTs/FUTs. In both cases, glycan structures were analyzed using flow cytometry. Cell adhesion measurements under fluid flow were performed in a microfluidic cell. b, specificity of FUT4, FUT7, and FUT9 based on previous enzymology studies. FUT7 acts on sialylated LacNAc-type structures. FUT4 and FUT9 act on nonsialylated LacNAc units with FUT9 preferring to act at the nonreducing end and FUT4 at the reducing end. Action of FUTs contribute to the synthesis of CD15/LeX, CD15s/sLeX, and CD65s/VIM-2 epitopes as indicated. c, real time PCR compares the relative mRNA levels of FUTs in human peripheral blood neutrophils versus HL-60 cells. The ratio of the two mRNA levels is presented in parentheses along the left margins. Gel images show RT-PCR products. FUT9 expression is higher in human neutrophils compared with HL-60s.
Gain-of-function Studies Show Distinct Roles for FUT4, FUT7, and FUT9 during Selectin-mediated Cell Adhesion
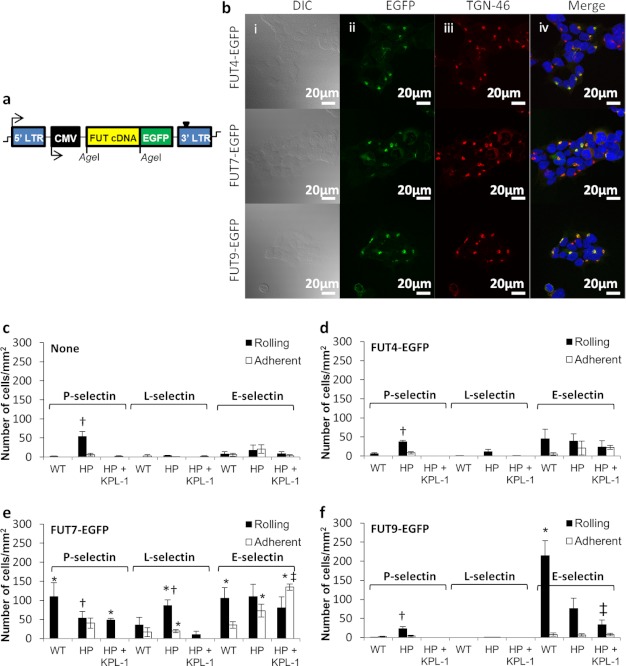
With the goal of delineating the role of the three FUTs in modifying PSGL-1-dependent and -independent cell adhesion, these enzymes were reconstituted in WT HEK293Ts and HEK293Ts bearing human PSGL-1 (HEK-PSGL-1 or HP cells) (Fig. 2). FUT4, -7, and -9 EGFPs transduced into HEK293Ts partially localized with the trans-Golgi network integral membrane glycoprotein TGN-46 (Fig. 2b). Overlap coefficient (R) between the FUT-EGFPs and TGN-46 Ab calculated using JACoP was 0.69 for FUT4, 0.76 for FUT7, and 0.67 for FUT9. High resolution confocal Z-stack for FUT7-EGFP cells is provided as supplemental Movie A.
FIGURE 2.
Gain-of-function studies using HEK cells that express individual FUTs and/or PSGL-1. a, structure of pCS-CG vector containing FUT-EGFP fusion construct driven by human cytomegalovirus (CMV) promoter. b, HEK293T FUT4-EGFP (top row), FUT7-EGFP (middle row), and FUT9-EGFP (bottom row) cells viewed using differential interference contrast (DIC) (1st column), green channel due to EGFP in intracellular compartment (2nd column), red channel due to rabbit anti-human TGN-46 mAb and Alexa 597-conjugated secondary Ab, and merged image (Merge), which also includes blue DAPI nuclear stain. c, WT HEK293T (WT) and HEK-PSGL-1 (HP) cells were perfused over substrates bearing P-selectin-Fc, L-selectin-Fc, or E-selectin-Fc fusion proteins in a microfluidic flow cell. Anti-PSGL-1 mAb KPL-1 was added in some cases to HP cells to distinguish between PSGL-1-dependent and -independent cell adhesion components. Rolling (black bar) and adherent cell number (white bar) were measured. Wall shear stress was 1 dyne/cm2 for all selectins. d–f, similar gain-of-function studies as in c were performed with HEK293T cells that overexpressed either FUT4-EGFP (d), FUT7-EGFP (e), or FUT9-EGFP (f). p < 0.05 with respect to WT alone (‡) or WT and HP+KPL-1 (†) for the same selectin interaction within the same figure panel. *, p < 0.05 with respect to the same selectin binding function measured with cells either lacking FUT (c) or having different FUTs (d–f).
Studies characterized glycan structures (Table 1), FUT enzyme activity (Table 2), and cell adhesion in a microfluidic-based cell adhesion assay (Fig. 2 and Table 3). As seen, WT HEK293Ts expressed some sLeX/CD15s and VIM-2/CD65s epitope but no native LeX/CD15 (Table 1). This is consistent with low levels of FUT enzyme activity in HEK cells (Table 2). Transfection of PSGL-1 in these cells resulted in cell rolling on substrates bearing P-selectin-Fc fusion protein (Fig. 2c). This binding was blocked by mAb against the N terminus of PSGL-1 (clone KPL-1). Thus, constitutive enzymes in HEK293Ts may contribute to the formation of sLeX-type epitopes at the N terminus of PSGL-1. Some HEK-PSGL-1 binding on E-selectin-Fc was detected, although this was not statistically significant.
TABLE 1.
Glycan expression in modified HEK 293T
Mean fluorescence intensity for various cell lines was measured using a flow cytometer. Isotype control signal was in the range of 1–3 mean fluorescence intensity units.
| Cell type | PSGL-1 | LeX | sLeX | VIM-2 |
|---|---|---|---|---|
| HEK293T (WT) | 2.5 ± 1.2 | 1.1 ± 0.1 | 53.6 ± 3.7 | 23.03 ± 2.5 |
| WT-FUT4 | 2.7 ± 1.1 | 7.1 ± 0.6 | 33.0 ± 2.9 | 23.04 ± 5.4 |
| WT-FUT7 | 2.6 ± 0.9 | 5.0 ± 0.5 | 140.1 ± 13.9 | 21.70 ± 4.5 |
| WT-FUT9 | 3.6 ± 1.8 | 385.2 ± 33.2 | 126.9 ± 14.7 | 42.11 ± 5.4 |
| HEK PSGL-1 (HP) | 1440.9 ± 103.0 | 1.0 ± 0.1 | 61.8 ± 0.6 | 21.38 ± 1.6 |
| HP-FUT4 | 1445.3 ± 21.1 | 4.5 ± 3.1 | 70.2 ± 2.8 | 27.02 ± 1.7 |
| HP-FUT7 | 1123.2 ± 70.8 | 1.7 ± 0.8 | 745.1 ± 125.6 | 21.06 ± 1.3 |
| HP-FUT9 | 1071.7 ± 32.4 | 394.3 ± 23.9 | 74.7 ± 4.5 | 69.35 ± 2.6 |
TABLE 2.
FUT activity in WT and modified HEK293Ts
Percent of total GDP-Fuc radioactivity transferred to an acceptor in an 8-h reaction.
| Acceptor name | WT HEK | HEK with FUT4-GFP | HEK with FUT7-GFP | HEK with FUT9-GFP |
|---|---|---|---|---|
| di-LacNAc, Galβ1,4GlcNAcβ1,3 Galβ1,4GlcNAc-β-O-Bn | 3.2 ± 0.1 | 19.5 ± 6.9 | 5.4 ± 1.3 | 20.0 ± 2.0 |
| Core-2 tetrasaccharide, Galβ1,4 GlcNAcβ1,6(4F-Galβ1,3)GalNAc-β-O-Bn | 7.9 ± 2.2 | 18.0 ± 5.7 | 5.9 ± 2.8 | 21.7 ± 3.8 |
| Sialyl-LacNAc, Neu5Acα2,3Galβ1,4GlcNAc-biotin | 7.0 ± 5.6 | 10.8 ± 1.3 | 15.8 ± 3.3 | 7.2 ± 0.9 |
TABLE 3.
WT and modified HEK 293T rolling velocity on substrates bearing recombinant selectins (μm/s)
| P-selectin | L-selectin | E-selectin | |
|---|---|---|---|
| WT | |||
| WT-FUT4 | 9.2 ± 5.5 | ||
| WT-FUT7 | 40.32 ± 21 | 45.72 ± 21.5 | 3.81 ± 1.75 |
| WT-FUT9 | 11.6 ± 5.3 | ||
| HP | 21.9 ± 8.1 | - | |
| HP-FUT4 | 17.37 ± 7 | 9.74 ± 3.7 | |
| HP-FUT7 | 5.2 ± 4.9 | 25.62 ± 13.7 | 3.9 ± 1.4 |
| HP-FUT9 | 17 ± 11 | 14.8 ± 7 | |
| HP + KPL1 | |||
| HP-FUT4 + KPL1 | |||
| HP-FUT7 + KPL1 | 25.63 ± 21 | 3.3 ± 1.2 | |
| HP-FUT9 + KPL1 | 10.4 ± 5.7 |
Expression of FUT4 in HEK293Ts resulted in only a minor increase in LeX expression (Table 1). Consistent with this, FUT enzyme activity toward unsialylated di-LacNAc and Core-2 tetrasaccharide was elevated by 2–5-fold upon FUT4 overexpression (Table 2). Although this enzyme did not affect L- and P-selectin-dependent interactions, E-selectin binding was enhanced by FUT4 even in the absence of PSGL-1 (Fig. 2d). mAb KPL-1 did not affect this binding.
FUT7 overexpression in HEK293Ts enhanced FUT activity toward the sialyl LacNAc substrate by 2.5-fold compared with WT HEK293Ts (Table 2). This resulted in a marked up-regulation of cell-surface sLeX expression (Table 1). SLeX expression increased 2.5-fold in WT cells and 15-fold in HEK-PSGL-1 cells. FUT7 enhanced cell rolling on all selectins (Fig. 2e). In this regard, HEK cells expressing FUT7 rolled on L-selectin with the number of rolling cells being ∼2.5-fold higher for HP compared with WT HEK293Ts. This interaction was blocked by anti-PSGL-1 mAb KPL-1. HEK293T rolling on P-selectin was also robust upon co-expression of FUT7 and PSGL-1 because rolling velocity was 8-fold lower for HP compared with WT cells. Some firm adhesion on P-selectin was also detected when PSGL-1 was co-expressed with FUT7 due to the very slow rolling on the recombinant protein substrate. Blocking with anti-PSGL-1 mAb KPL-1 increased cell rolling velocity to 26 μm/s, and firm adhesion was abolished. In the case of E-selectin, slow cell rolling at 3.3–3.6 μm/s and some firm adhesion was measured in all runs performed with FUT7-EGFP cells, independent of the presence of PSGL-1 or mAb KPL-1. Overall, FUT7-expressing HEK293T cells express a major E-selectin ligand other than PSGL-1.
An important role for FUT9 in conferring E-selectin-dependent, but not L- and P-selectin-dependent, cell rolling is evident (Fig. 2f). HEK293Ts overexpressing FUT9 rolled robustly at 10–15 μm/s both in the presence and absence of PSGL-1 (Table 3). This enzyme markedly up-regulated LeX expression in HEK293Ts and it also doubled the VIM-2 epitope (Table 1). These observations are similar to that of others, who detected similar structures upon profiling glycans on the FUT9-overexpressing CHO mutant cell line LEC12 (30, 31). Enzymology results show that, to the extent tested, the fucosyltransferase acceptor specificity and activity of FUT9 overexpressing HEK293T cells were similar to that of cells overexpressing FUT4 (Table 2). The number of interacting cells in WT versus HEK-PSGL-1-overexpressing FUT9 was different (Fig. 2f), possibly due to differences in the relative expression levels of FUT9 in the two cells.
Overall, the FUT-EGFP proteins expressed in HEK293Ts were functional. These conferred a distinct pattern of selectin-mediated cell adhesion. Although FUT7 was the major enzyme necessary for P- and L-selectin-dependent binding, all three α1,3-FUTs enabled E-selectin-dependent cell adhesion. PSGL-1 was critical for P- and L-selectin but not E-selectin-mediated cell recruitment.
Determination of Efficient and Specific shRNAs That Disrupt FUT Activity
Although studies using the reconstituted HEK293Ts are informative (Fig. 2), it is more important to disrupt enzyme activity in human leukocytes because this is the relevant cell system. To this end, RNAi was applied to study leukocyte glycoT function.
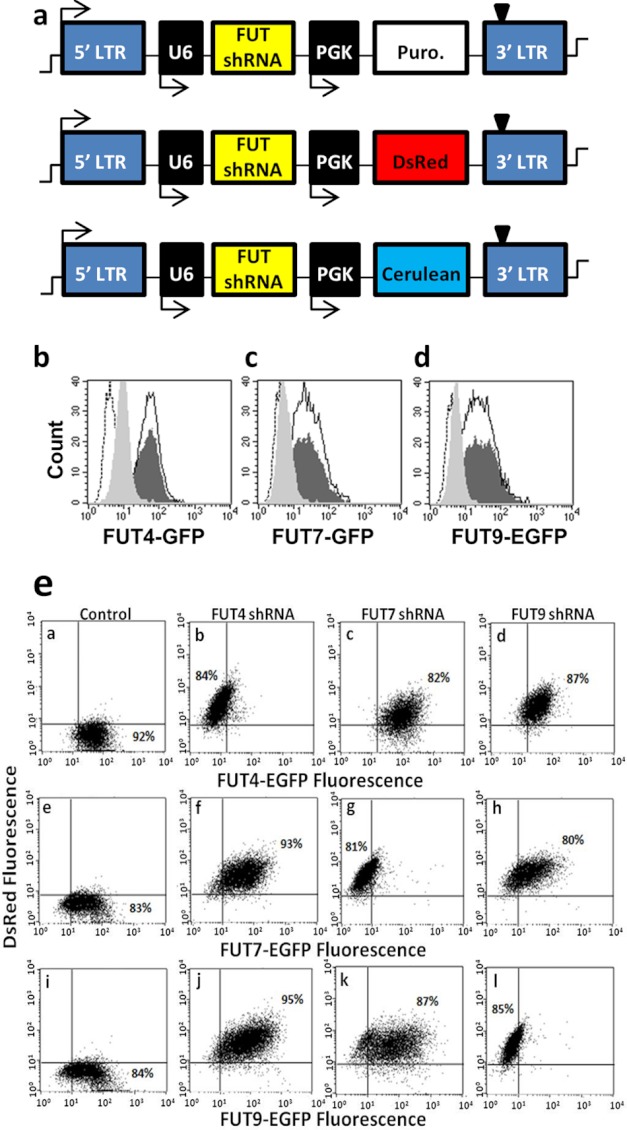
Because well characterized antibodies against individual FUTs were unavailable, the silencing efficiency of shRNA was screened using cell lines that express Golgi-localized glycoT-EGFP fusion proteins. shRNAs that reduce FUT expression were determined by screening leads identified by the RNAi Consortium (Fig. 3) (32). Virus containing 3–5 distinct shRNAs for each gene were generated (Fig. 3a). These were transduced into CHO-S cells that stably expressed either FUT4-EGFP, FUT7-EGFP, or FUT9-EGFP. shRNA silencing efficiency was defined based on the reduction in EGFP signal following transduction. The supplemental Table S2 summarizes the findings, including the results for control vectors that lack shRNA (“TRC cloning”), that carry nonhairpin DNA (“TRC control”), and that silence EGFP instead of the FUTs (“EGFP shRNA”). The most efficient shRNA identified here (highlighted in bold, supplemental Table S2) reduced target FUT-EGFP expression by ∼85–90%. Corresponding flow cytometry histograms are presented in Fig. 3, b–d. Although data are presented here using CHO-S cells, similar results were also obtained using HEK cells expressing FUT-EGFPs.
FIGURE 3.
Determination of specific and efficient shRNA that knocks down FUTs. a, pLKO.1 plasmids contain the polymerase-III U6 promoter that drives shRNA expression. The PGK promoter drives either the puromycin (Puro.) resistance gene or reporter genes (DsRed or cerulean). b–d, flow cytometry histogram showing fluorescence of CHO-S cells with either FUT4-EGFP (b), FUT7-EGFP (c), or FUT9-EGFP (d). Fluorescence decreases in each panel from dark-shaded histogram for cells without shRNA to light-shaded histogram upon gene silencing. Empty histogram with black border corresponds to TRC control virus, whereas dashed-empty histogram corresponds to WT CHO-S cells lacking FUT-EGFP. Silencing efficiency was quantified based on % reduction in EGFP fluorescence intensity upon shRNA addition in comparison with control FUT-EGFP cell fluorescence. e, two-color cross-silencing cytometry experiments were performed to confirm that each shRNA only disrupts a unique fucosyltransferase. To this end, CHO-S cells that stably express FUT4-EGFP (top row), FUT7-EGFP (middle row), and FUT9-EGFP (bottom row) were either left untransduced (1st column) or these were transduced with virus encoding for shRNA against FUT4 (2nd column), FUT7 (3rd column), or FUT9 (last column). All shRNA viruses carried the DsRed gene in addition to shRNA, and thus transduction results in an increase in DsRed signal in all cases. Silencing specificity is evident because each shRNA only reduced the EGFP fluorescence of corresponding FUT-EGFP cell lines.
To confirm the specificity of gene disruption, efficient shRNAs identified above were cloned into the pLKO.1 vector that carried the DsRed fluorescence reporter (Fig. 3a). Two-color flow cytometry dot plots performed using CHO-S cells bearing FUT-EGFPs and DsRed encoding virus confirmed the specificity of shRNA identified for each FUT (Fig. 3e). Thus, a rapid and quantitative strategy to screen for efficient and specific shRNA directed against individual glycoTs was established.
Robust Strategy for Gene Expression and Silencing in HL-60s
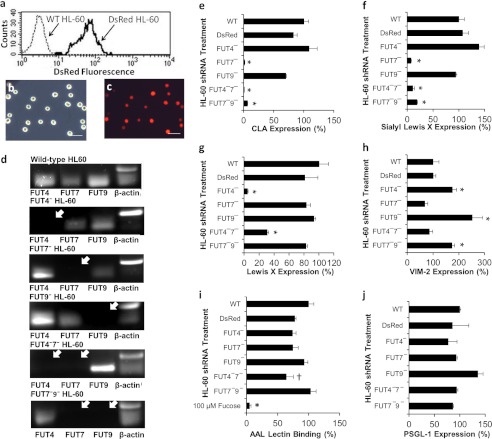
VSVG pseudotyped lentivirus carrying shRNA identified in Fig. 3 were applied to HL-60s. In contrast to previous studies that achieved only 1–4% stable transgene expression when the CMV promoter was used to drive gene expression (19), use of the PGK promoter led to stable and long term gene expression in >95% of transduced HL-60 cells (Fig. 4). Red fluorescence increased ∼40-fold in DsRed-positive cells compared with untransduced WT cells. Although data are presented 7 days after transduction, this fluorescence did not decrease appreciably through the duration of our studies (30–45 days). This key advancement eliminated the need for flow cytometry sorting or other enrichment methods when creating stable knockdown leukocytes. It also enabled the rapid construction of cells that contained more than one shRNA. Single knockdowns were created using virus that expressed the DsRed reporter. Dual knockdown FUT4−7− and FUT7−9− HL-60s were made by transducing DsRed-positive FUT7− HL-60 with a second virus encoding for either the FUT4 or FUT9 shRNA along with the puromycin resistance gene. Cells obtained from these steps were used as such in functional assays, without enrichment of any specific population based on DsRed fluorescence or cell-surface markers. FUT4−9− HL-60s were not made because the expected contribution of FUT4 to cell adhesion was small. Quantitative RT-PCR verified >95% specific gene silencing in FUT knockdown HL-60s. Gel images of PCR products from these runs are shown in Fig. 4d.
FIGURE 4.
Changes in HL-60 cell-surface glycans upon silencing FUTs. a, transduction of HL-60 cells with control DsRed lentivirus resulted in stable DsRed expression in ∼100% of the cells. WT HL-60 cells serve as negative control. b and c, microscopy images of DsRed HL-60 acquired using brightfield (b) and fluorescence (c). Scale bar, 100 μm. d, RT-PCR products showing silencing of one or more FUTs in HL-60. White arrows indicate missing transcripts. e–j, cell-surface expression of CLA/HECA-452 epitope (e), sLeX/CD15s/CSLEX-1 (f), LeX/CD15/mAb HI98 binding (g), VIM-2/CD65s epitope (h), AAL lectin binding (binds α1,3/4/6-linked fucose antigen) (i), and anti-PSGL-1 mAb KPL-1 binding (j) for WT HL-60 cells, DsRed HL-60, and cells transduced with various shRNAs. FUT4 silencing reduced LeX by 90%. FUT7 controls CLA/sLeX expression. Silencing FUT9 up-regulates the VIM-2 epitope by 2–3-fold. Overall fucosylation and PSGL-1 levels were similar in all cell types except for a modest (∼35%) reduction in AAL lectin binding to FUT4−7− dual knockdowns. 100 μm l-fucose was used to confirm the specificity of the AAL lectin. WT HL-60 expression is set to 100%. *, p < 0.05 with respect to all other treatments, except that bars marked by asterisk are not different from each other. †, p < 0.05 with respect to wild-type HL-60. Data were acquired 4–10 days post-transduction. Measured signal did not vary substantially between days.
Silencing FUTs in HL-60 resulted in a distinct change in the pattern of cell-surface glycan expression (Fig. 4, e–h). Compared with WT cells, ∼90–95% reduction in both the CLA (Fig. 4e) and sLeX epitope (Fig. 4f) was observed in single and dual knockdown cells that lacked FUT7. FUT4 silencing resulted in 90 and 75% reduction in LeX expression in single knockdown and FUT4−7− dual knockdowns, respectively (Fig. 4g). FUT9 silencing increased the VIM-2 epitope by 2–3-fold in single knockdowns and 1.8-fold in FUT7−9− HL-60 (Fig. 4h). AAL lectin binding (overall fucosylation) was unchanged except for a 35% reduction in FUT4−7− HL-60 (Fig. 4i). PSGL-1 expression was also unchanged (Fig. 4j).
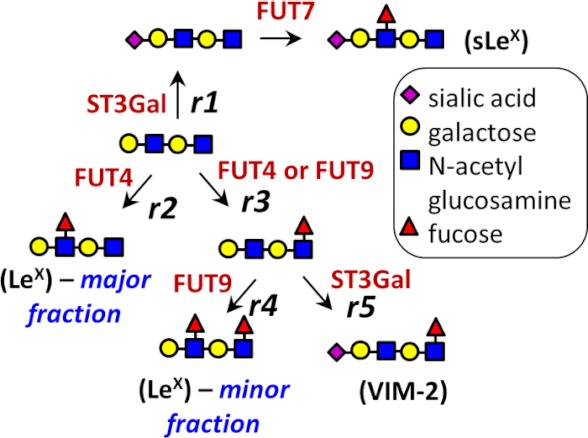
Glycan expression changes observed in FUT knockdowns can be explained based on previous literature and a model presented in Fig. 5. The down-regulation of sLeX upon silencing FUT7 is consistent with enzymatic studies (25, 33) and cell-based assays (34–36) that demonstrate a preference for FUT7 to fucosylate terminal and sialylated N-acetyllactosamine (NeuAcα2,3Galβ1,4GlcNAc) to generate sLeX-type structures (25, 33). Unlike FUT7, however, which has a distinct substrate specificity, FUT4 and FUT9 have overlapping function because they can both form LeX and VIM-2 structures (26, 27, 29, 30, 37). Both enzymes fucosylate glycolipids (29, 38), with FUT9 also acting on N-glycans (37). In this study, FUT4 generates a majority of the HL-60 LeX structure either because this enzyme is expressed at exceptionally high levels (14) or because of the presence of unique leukocyte substrates for this enzyme. Silencing FUT4 not only reduces LeX expression but it also increases VIM-2 expression possibly by diverting substrate flux from r2 to r3 (Fig. 5). Similarly, silencing FUT4 and FUT7 together diverts substrate flux from reactions r1 and r2 to r3, and this marginally increases LeX expression. Finally, FUT9 shRNA diverts formation of product in reaction r4 and increases the VIM-2 epitope due to reaction r5.
FIGURE 5.
Model for fucosylation in HL-60. Interpreting the effect of gene silencing requires consideration of the competition between reactions for a common substrate. This is illustrated here assuming the di-lactosamine substrate to be the starting material. Biochemical reactions r1–r5 are acting on this substrate. The reactions depicted in this model are based on prior literature as follows. (i) α(2,3)-Sialylation by ST3Gal followed α(1,3)-fucosylation by FUT7 results in sLeX (35) (r1). (ii) FUT4 can synthesize LeX-type antigens (28) (r2). (iii) Both FUT4 and FUT9 can fucosylate the inner GlcNAc residue (48) (r3). Sialylation of this intermediate structure results in the VIM-2 epitope (r5). Note: this is a simplified, conceptual model, and it is not inclusive of all findings reported in in vitro biochemical studies. Example, FUT7 can fucosylate the VIM-2 antigen to form sialyl-dimeric LeX structures (33).
FUT7 and FUT4 Cooperate to Form L- and P-selectin Ligands at the N Terminus of PSGL-1
Cell adhesion studies compared the binding of WT and modified HL-60 cells to WT and Fut4−/−Fut7−/− mouse neutrophils, to determine the FUTs regulating selectin binding function in both species. Such flow chamber studies evaluated the interaction of various leukocytes to P-selectin substrates bearing immobilized CHO-P cells (Fig. 6, a–c) and recombinant L-selectin-Fc (Fig. 6, d–f).
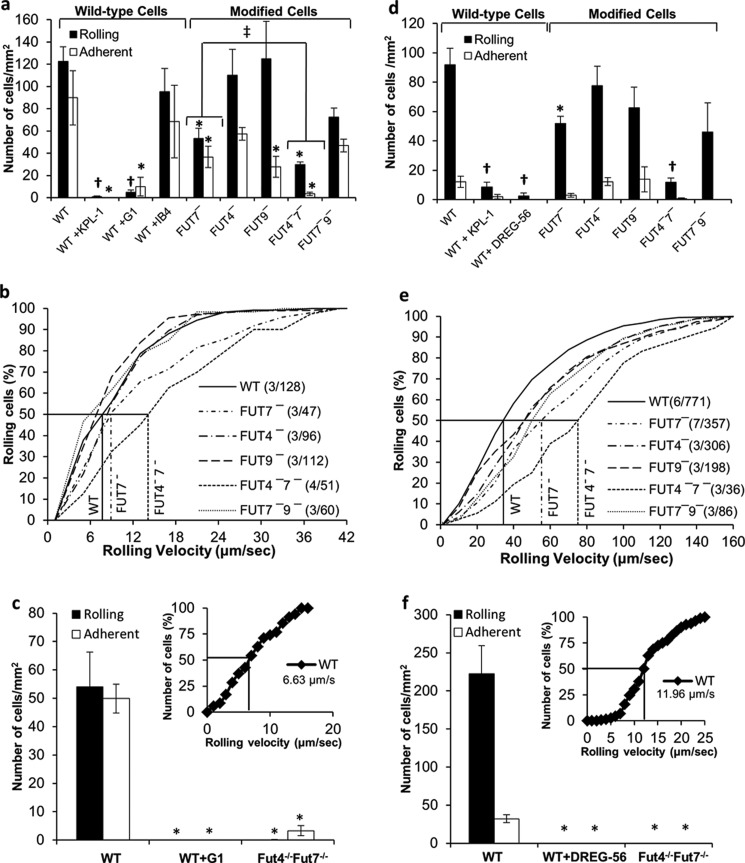
FIGURE 6.
FUT4 and FUT7 mediate rolling on P- and L-selectin. 2 × 106 cells/ml of either HL-60 cells (a and b and d and e) or bone marrow-derived mouse neutrophils (c and f) were perfused over substrates bearing either CHO-P cells (wall shear stress = 1 dyne/cm2, a–c) or soluble L-selectin-Fc (2 dynes/cm2, d–f). a, c, d, and f present rolling and adherent cell density data. b, c, e, and f, cumulative rolling velocity plots for HL-60 and mouse neutrophils (inset to bottom panels). Lines at 50% rolling cell number indicate median rolling velocity. Legends include information on “number of independent experiments/number of rolling cells analyzed” in parentheses. Cell adhesion is PSGL-1-dependent and it is reduced by >85% in FUT4−7− cells. *, p < 0.05 with respect to WT cells with no blocking reagent. †, p < 0.05 with respect to WT and FUT7- HL-60. ‡, number of interacting cells (rolling plus adherent) is significantly different.
The interaction of HL-60 with CHO-P cells was specifically mediated by P-selectin binding to PSGL-1 because this could be abrogated by anti-PSGL-1 mAb KPL-1 and anti-P-selectin mAb G1 (Fig. 6, a and c). Both cell rolling and firm adhesion were observed. This is likely due to the high density of selectins on the CHO-P cell surface rather than selectin-mediated cell signaling (39). In this regard, cell adhesion was not altered upon addition of anti-CD18 blocking mAb IB4, and adherent cells could be dynamically released by perfusion with 5–10 mm EDTA at the end of the runs. Among the HL-60 single knockdowns, FUT7 shRNA reduced both the number of rolling and adherent cells by ∼65%. FUT4 and FUT9 shRNA reduced adherent cell density, although these enzymes did not affect rolling cell density. In the dual knockdowns, FUT7−9− was not different from FUT7−. FUT4 shRNA, however, acted in synergy with FUT7 to reduce the total number of interacting cells (rolling plus adherent) by >85% compared with WT cells. Similar to human cells, the robust rolling observed in WT mouse neutrophils was reduced by >95% in the Fut4−/−Fut7−/− knockouts (Fig. 6c). The rolling velocity of human HL-60 (7.7 μm/s, Fig. 6b) was similar to WT mouse neutrophils (6.6 μm/s, inset to Fig. 6c). Median rolling velocity of FUT4−7− cells (14.1 μm/s) was higher compared with WT HL-60 and FUT7− cells.
HL-60 rolling on immobilized L-selectin-Fc was specifically blocked by anti-L-selectin mAb DREG-56, and it was reduced by ∼90% by anti-PSGL-1 (Fig. 6d). Here also, FUT7 shRNA decreased rolling cell density by ∼50% (Fig. 6d). FUT4 acted in synergy with FUT7 to further reduce rolling density by 85%. FUT9 shRNA did not play a role. The median rolling velocity of FUT4−7− (75.0 μm/s) and FUT7− (55.3 μm/s) was higher than that of WT HL-60 (34.5 μm/s) (Fig. 6e). Mouse neutrophil interaction was similar to HL-60 as Fut4−/−Fut7−/− cells did not roll on L-selectin (Fig. 6f).
Overall, FUT7 is the dominant enzyme modifying the N-terminal selectin-binding epitope on human PSGL-1, with FUT4 playing a supporting role. These observations are consistent with previous co-expression studies in CHO (40), COS (41), and hematopoietic K562 cells (42) that demonstrate a role for FUT7 in fucosylating human PSGL-1 and enhancing cell adhesion. Fut7−/− mice also exhibit reduced neutrophil adhesion function in models of inflammation, and they display reduced lymphocyte homing (43). Although partial leukocyte rolling/adhesion defect is noted in Fut7−/− mice, complete function loss occurs in Fut4−/−/Fut7−/− double knockouts in noninflamed skin venules, models of microvascular inflammation, and leukocyte homing (17, 18, 44). Similarly, in our work FUT7 plays an important role during leukocyte capture, with FUT4 collaborating with FUT7 to regulate cell rolling velocity. Thus, the FUTs responsible for L/P-selectin binding in human and mouse leukocytes are similar.
FUT9 Contributes to the Formation of Human E-selectin Ligands
Studies with L-E cells quantify the direct binding of E-selectin to both PSGL-1 and non-PSGL-1 human leukocyte E-selectin ligands (Fig. 7). In this regard, because HL-60 cells do not express L-selectin on their surface, secondary leukocyte-leukocyte tethering is absent in this system (45, 46). In addition, although HL-60 binding could be completely abolished by anti-E-selectin mAb HAE-1F, anti-PSGL-1 mAb KPL-1 was only 40% effective. Firm arrest observed here is likely due to the high density of E-selectin on L-E cells because this cannot be blocked by anti-CD18 blocking mAb.
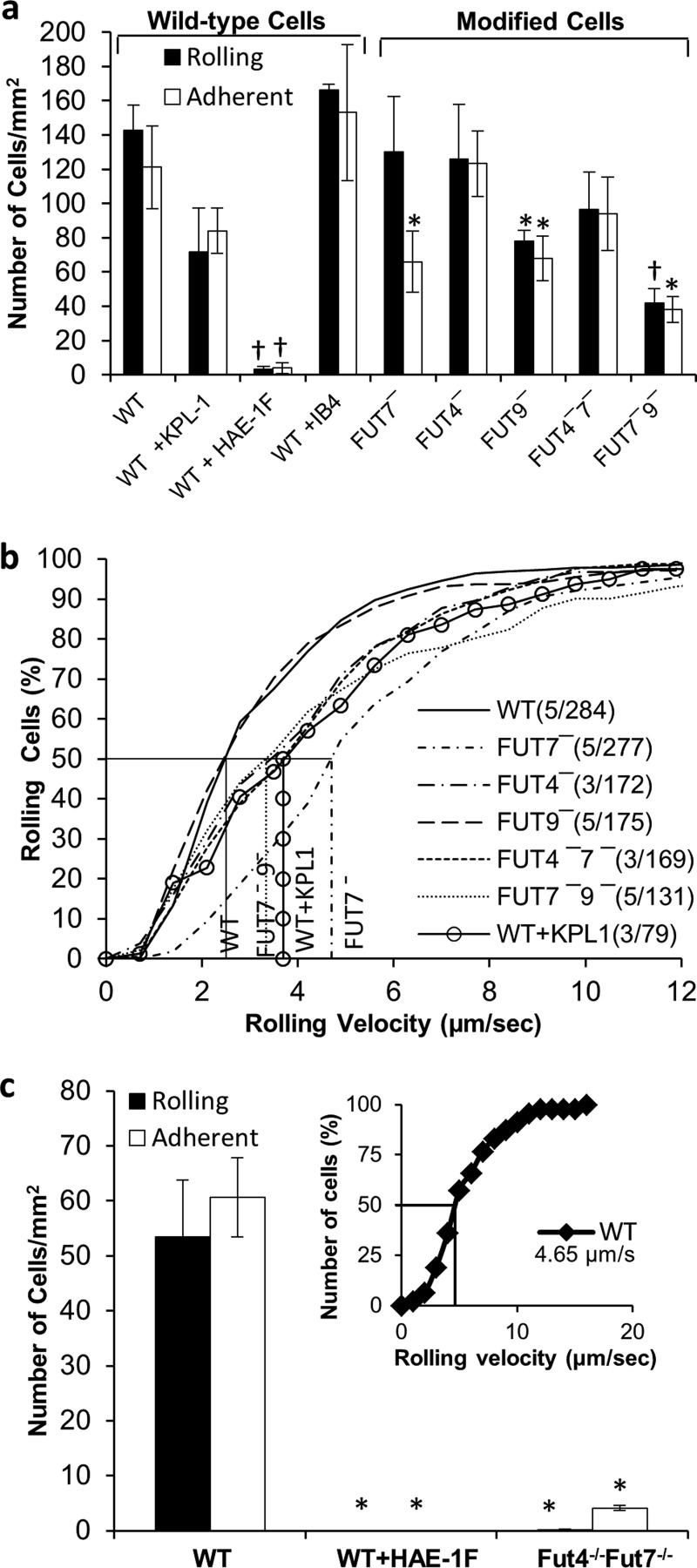
FIGURE 7.
FUT9 and FUT7 mediate E-selectin rolling. Similar to Fig. 5, 2 × 106 cells/ml were perfused over E-selectin bearing L-cells at a wall shear stress of 1 dyne/cm2. a, bar plot shows rolling and adherent cell density for each HL-60 FUT shRNA cell line. b, cumulative rolling velocity plots with information on number of cells analyzed and median rolling velocity. c, rolling, adhesion, and rolling velocity (inset) for WT and Fut4−/−Fut7−/− mouse neutrophils. Data show that FUT9 and FUT7 regulate HL-60 binding to E-selectin, whereas FUT7 and FUT4 do so in mice. Anti-PSGL-1 mAb KPL-1 only partially reduced cell rolling. *, p < 0.05 with respect to WT cells with no blocking reagent. †, p < 0.05 with respect to WT and FUT9− HL-60.
The studies with FUT knockdowns demonstrate that the FUTs regulating E-selectin binding in human leukocytes may be different from those regulating L- and P-selectin recognition. Among the single knockdowns, FUT7− reduced the number of adherent cells by 45%, whereas FUT9− displayed reduction in both rolling (∼45%) and adherent (∼45%) cell density (Fig. 7a). Unlike P- and L-selectin, FUT4−7− rolling on E-selectin substrate was only reduced by ∼30%, and this was not statistically significant. FUT7−9− cells, however, displayed ∼70% reduction in the number of rolling and adherent cells. In contrast, Fut4−/−Fut7−/− mouse neutrophil interaction with L-E cells was reduced by >95% compared with WT mouse cells (Fig. 7c). The rolling velocity of human HL-60 (Fig. 7b) was comparable with mouse neutrophils (Fig. 7c, inset) on this monolayer. FUT7− (4.7 μm/s) increased the rolling velocity by ∼2-fold compared with WT cells (2.5 μm/s). FUT4−7− had no added effect on HL-60 rolling velocity (Fig. 7b).
Together, the data suggest a species-specific difference in the function of FUTs as they synthesize physiological E-selectin ligands. In this regard, FUT9 together with FUT7 regulate the synthesis of functional human E-selectin ligands. In contrast, Fut4−/−Fut7−/− mouse neutrophils exhibit >95% reduction in E-selectin-mediated cell adhesion (17, 18). Thus, the contribution of glycoTs to E-selectin binding may be species-specific.
Minor Role for FUT4 during E-selectin-mediated Leukocyte Rolling
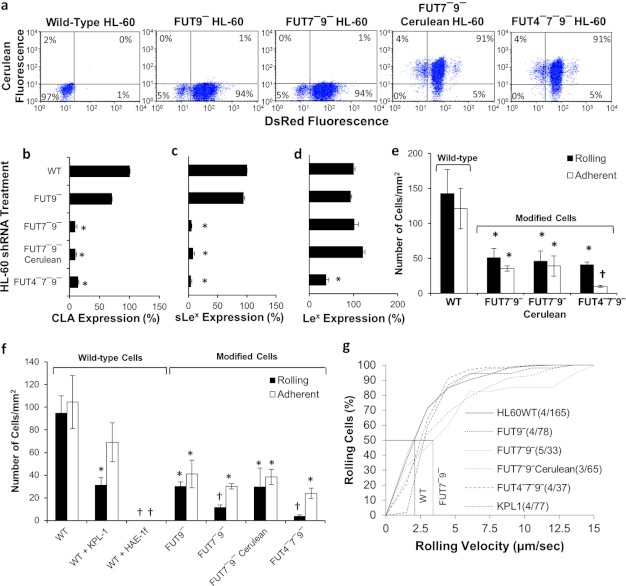
To determine whether the residual ∼30% cell adhesion observed with FUT7−9− cells during E-selectin-mediated cell adhesion is due to the role of FUT4 or if it is due to incomplete silencing by shRNA, FUT4−7−9− triple knockdown cells (Fig. 8) were created by addition of FUT4 shRNA into FUT7−9− cells using vectors carrying the cerulean reporter (Fig. 3a). TRC cloning vector (no shRNA) carrying the cerulean transgene was incorporated into FUT7−9− cells to serve as control. Knockdown was verified by RT-PCR analysis (data now shown). Cerulean incorporation into >95% of cells was verified using microscopy and also two-color flow cytometry (Fig. 8a). As expected, the triple knockdown cells had reduced expression of the CLA (Fig. 8b), sLeX (Fig. 8c), and LeX (Fig. 8d) epitopes.
FIGURE 8.
Studies with triple knockdown cells. a, flow cytometry dot plot of WT and mutant HL-60 cells lacking between one to three α(1,3)FUTs. Expression of DsRed and cerulean confirm stable gene silencing. b–d, flow cytometry readings for CLA (b), sLeX (c), and LeX (d) epitopes. e, rolling and adherent cell density data for FUT4−7−9− and FUT7−9− cerulean HL-60 cells on L-E cell substrates at 1 dyne/cm2, compared with WT and FUT7−9− dual knockdowns. f and g, cell rolling and adhesion (f), and rolling velocity data (g) for various HL-60 cell lines interacting with IL-1β-stimulated HUVEC at 1 dyne/cm2. Data indicate a minor contribution of FUT4 to E-selectin mediated leukocyte rolling. *, p < 0.05 with respect to WT HL-60. †, p < 0.05 with respect to all other treatments, except that bars with daggers are not different from each other.
In cell adhesion assays on L-E cells, the FUT7−9− cerulean cells behaved similar to the FUT7−9− dual knockdowns (Fig. 8e). FUT4−7−9− rolling density and velocity (3.7 μm/s) were similar to FUT7−9−, but adherent cell density was significantly lower.
The cell adhesion phenotype of WT and modified HL-60 rolling on IL-1β-stimulated HUVECs were similar to L-E cells (Fig. 8, f and g). This binding was partially PSGL-1-dependent. Knocking down FUT9 alone reduced cell binding by ∼60% with FUT7 acting in synergy to further reduce cell adhesion to 20% of WT levels. Consistent with this, FUT7−9− and FUT7−9− cerulean cells exhibited higher rolling velocity compared with WT cells. FUT4 silencing did not affect the nature of cell interaction. Together, the rolling experiments with L-E cells and stimulated HUVEC confirm a more significant role for FUT9 and FUT7, as opposed to FUT4, during HL-60 rolling on E-selectin.
The apparent importance of FUT9 in human leukocyte adhesion is not entirely unexpected because it is well recognized that the E-selectin ligands in human leukocytes differ from those in the mouse (1, 5). In particular, E-selectin ligands on mouse but not human neutrophils are pronase-sensitive (7, 8).3 In this regard, Nimrichter et al. (47) suggest that a relatively minor monosialylated, polyfucosylated glycolipid with poly-lactosamine units NeuAcαα2,3Galβ1,4GlcNAcβ1,3[Galβ1,4(Fucα1,3)GlcNAcβ1,3]2[Galβ1,4GlcNAcβ1,3]2Galβ1,4GlcβCer is an important functional E-selectin ligand on human granulocytes. Nishihara et al. (29) show that FUT9 can act on glycolipid acceptors to generate similar VIM-2-like fucosylated epitopes, i.e. NeuAcαα2,3Galβ1,4GlcNAcβ1,3Galβ1,4(Fucα1,3)GlcNAcβ1,3Galβ1,4GlcβCer. Thus, one possible explanation is that FUT9 contributes to sialofucosylated glycolipid structures that act as human E-selectin ligands. In addition to this, human E-selectin is promiscuous in terms of the variety of counter-receptors it binds. In addition to glycolipids, potential E-selectin ligands include sialo-fucosylated epitopes on PSGL-1, CD43, CD44, L-selectin, and CD18-integrin Mac-1 (1, 5). Whether FUT9 modifies glycans on these macromolecules remains to be determined.
Conclusion
In conclusion, this work develops gene transfer and RNA interference methods to define the role of glycoTs in human cells. It confirms that, like mice, human leukocyte/HL-60 rolling on L- and P-selectin is primarily controlled by FUT7 with a secondary role for FUT4. In contrast, all three human α(1,3)FUTs contribute to E-selectin-dependent cell binding with the relative roles varying as FUT9 > FUT7 ≫ FUT4. In support of this, overexpression of FUT9 in HEK293Ts resulted in robust E-, but not L- or P-, selectin-mediated cell rolling (Fig. 2f). This cell adhesion was PSGL-1-independent. FUT9 shRNA alone also reduced leukocyte rolling on L-cells bearing E-selectin (Fig. 7) and stimulated HUVEC (Fig. 8), with additional contributions from shRNA against FUT7 and a lesser role for FUT4 shRNA. Because primary human neutrophils express higher levels of FUT9 compared with HL-60s (Fig. 1c), a significant contribution of this enzyme to inflammation may be expected in humans.
The precise reason for the species-specific differences in E-selectin binding requires further investigation. In this regard, the protein/lipid scaffolds that carry the E-selectin ligands in mouse versus human leukocytes are different. This may be due to differences in the expression levels of these scaffolds in the two cellular backgrounds. Alternatively, the acceptor/substrate specificity of FUT9 in mouse and humans may have subtle but important differences that contribute to the species-specific effects. Finally, it is possible that other glycosyltransferases that act in coordination with FUT9 may exhibit species-specific roles that are apparent in the knockdown cells.
Acknowledgments
We thank Prof. Scott I. Simon (University California, Davis) for providing the L-E cell line, Prof. Douglas Goetz (Ohio University, Athens) for the recombinant E-selectin-Fc protein, and Chi Lo (State University of New York, Buffalo) for the pLKO.1 PSGL-1 vector.
This work was supported, in whole or in part, by National Institutes of Health Grants HL63014 (to S. N.), HL103411 (to S. N.), and HL107146 (to J. T. Y. L.).

This article contains supplemental Tables 1–3 and Movie A.
A. Buffone, Jr., N. Mondal, and S. Neelamegham, unpublished data.
- glycoT
- glycosyltransferase
- PSGL-1
- P-selectin glycoprotein ligand-1
- α(1,3)FUT
- α(1,3)-fucosyltransferase
- FUT4
- FUT7, and FUT9, these are the human α(1,3)fucosyltransferases
- Fut4 and Fut7
- these designate non-human (mouse) enzymes
- HUVEC
- human umbilical vein endothelial cell
- EGFP
- enhanced GFP
- PGK
- phosphoglycerokinase
- Ab
- antibody
- CLA
- cutaneous lymphocyte-associated antigen
- TRC
- The RNAi Consortium.
REFERENCES
- 1. Zarbock A., Ley K., McEver R. P., Hidalgo A. (2011) Leukocyte ligands for endothelial selectins. Specialized glycoconjugates that mediate rolling and signaling under flow. Blood 118, 6743–6751 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Beauharnois M. E., Lindquist K. C., Marathe D., Vanderslice P., Xia J., Matta K. L., Neelamegham S. (2005) Affinity and kinetics of sialyl Lewis-X and core-2-based oligosaccharides binding to L- and P-selectin. Biochemistry 44, 9507–9519 [DOI] [PubMed] [Google Scholar]
- 3. Taniguchi N., Honke K., Fukuda M. (eds) (2002) Handbook of Glycosyltransferases and Related Genes, Springer-Verlag, Tokyo [Google Scholar]
- 4. Konstantopoulos K., Neelamegham S., Burns A. R., Hentzen E., Kansas G. S., Snapp K. R., Berg E. L., Hellums J. D., Smith C. W., McIntire L. V., Simon S. I. (1998) Venous levels of shear support neutrophil-platelet adhesion and neutrophil aggregation in blood via P-selectin and β2-integrin. Circulation 98, 873–882 [DOI] [PubMed] [Google Scholar]
- 5. Sackstein R. (2009) Glycosyltransferase-programmed stereosubstitution (GPS) to create HCELL. Engineering a roadmap for cell migration. Immunol. Rev. 230, 51–74 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Vestweber D., Blanks J. E. (1999) Mechanisms that regulate the function of the selectins and their ligands. Physiol. Rev. 79, 181–213 [DOI] [PubMed] [Google Scholar]
- 7. Kobzdej M. M., Leppänen A., Ramachandran V., Cummings R. D., McEver R. P. (2002) Discordant expression of selectin ligands and sialyl Lewis x-related epitopes on murine myeloid cells. Blood 100, 4485–4494 [DOI] [PubMed] [Google Scholar]
- 8. Larsen G. R., Sako D., Ahern T. J., Shaffer M., Erban J., Sajer S. A., Gibson R. M., Wagner D. D., Furie B. C., Furie B. (1992) P-selectin and E-selectin. Distinct but overlapping leukocyte ligand specificities. J. Biol. Chem. 267, 11104–11110 [PubMed] [Google Scholar]
- 9. Hidalgo A., Peired A. J., Wild M. K., Vestweber D., Frenette P. S. (2007) Complete identification of E-selectin ligands on neutrophils reveals distinct functions of PSGL-1, ESL-1, and CD44. Immunity 26, 477–489 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Merzaban J. S., Burdick M. M., Gadhoum S. Z., Dagia N. M., Chu J. T., Fuhlbrigge R. C., Sackstein R. (2011) Analysis of glycoprotein E-selectin ligands on human and mouse marrow cells enriched for hematopoietic stem/progenitor cells. Blood 118, 1774–1783 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Pan J., Xia L., McEver R. P. (1998) Comparison of promoters for the murine and human P-selectin genes suggests species-specific and conserved mechanisms for transcriptional regulation in endothelial cells. J. Biol. Chem. 273, 10058–10067 [DOI] [PubMed] [Google Scholar]
- 12. Liu Z., Miner J. J., Yago T., Yao L., Lupu F., Xia L., McEver R. P. (2010) Differential regulation of human and murine P-selectin expression and function in vivo. J. Exp. Med. 207, 2975–2987 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Aeed P. A., Geng J.-G., Asa D., Raycroft L., Ma L., Elhammer A. P. (1998) Characterization of the O-linked oligosaccharide structures on P-selectin glycoprotein ligand-1 (PSGL-1). Glycoconj. J. 15, 975–985 [DOI] [PubMed] [Google Scholar]
- 14. Marathe D. D., Chandrasekaran E. V., Lau J. T., Matta K. L., Neelamegham S. (2008) Systems-level studies of glycosyltransferase gene expression and enzyme activity that are associated with the selectin binding function of human leukocytes. FASEB J. 22, 4154–4167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Sako D., Chang X. J., Barone K. M., Vachino G., White H. M., Shaw G., Veldman G. M., Bean K. M., Ahern T. J., Furie B. (1993) Expression cloning of a functional glycoprotein ligand for P-selectin. Cell 75, 1179–1186 [DOI] [PubMed] [Google Scholar]
- 16. Wilkins P. P., McEver R. P., Cummings R. D. (1996) Structures of the O-glycans on P-selectin glycoprotein ligand-1 from HL-60 cells. J. Biol. Chem. 271, 18732–18742 [DOI] [PubMed] [Google Scholar]
- 17. Weninger W., Ulfman L. H., Cheng G., Souchkova N., Quackenbush E. J., Lowe J. B., von Andrian U. H. (2000) Specialized contributions by α(1,3)-fucosyltransferase-IV and FucT-VII during leukocyte rolling in dermal microvessels. Immunity 12, 665–676 [DOI] [PubMed] [Google Scholar]
- 18. Homeister J. W., Thall A. D., Petryniak B., Malý P., Rogers C. E., Smith P. L., Kelly R. J., Gersten K. M., Askari S. W., Cheng G., Smithson G., Marks R. M., Misra A. K., Hindsgaul O., von Andrian U. H., Lowe J. B. (2001) The α(1,3)-fucosyltransferases FucT-IV and FucT-VII exert collaborative control over selectin-dependent leukocyte recruitment and lymphocyte homing. Immunity 15, 115–126 [DOI] [PubMed] [Google Scholar]
- 19. Jayakumar D., Marathe D. D., Neelamegham S. (2009) Detection of site-specific glycosylation in proteins using flow cytometry. Cytometry 75, 866–873 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Bolte S., Cordelières F. P. (2006) A guided tour into subcellular colocalization analysis in light microscopy. J. Microsc. 224, 213–232 [DOI] [PubMed] [Google Scholar]
- 21. Patil S. A., Chandrasekaran E. V., Matta K. L., Parikh A., Tzanakakis E. S., Neelamegham S. (2012) Scaling down the size and increasing the throughput of glycosyltransferase assays. Activity changes on stem cell differentiation. Anal. Biochem. 425, 135–144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Luo Y., Dorf M. E. (2001) Isolation of mouse neutrophils. Curr. Protoc. Immunol. Chapter 3, Unit 3.20 [DOI] [PubMed] [Google Scholar]
- 23. Zhang Y., Neelamegham S. (2002) Estimating the efficiency of cell capture and arrest in flow chambers. Study of neutrophil binding via E-selectin and ICAM-1. Biophys. J. 83, 1934–1952 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Marathe D. D., Buffone A., Jr., Chandrasekaran E. V., Xue J., Locke R. D., Nasirikenari M., Lau J. T., Matta K. L., Neelamegham S. (2010) Fluorinated per-acetylated GalNAc metabolically alters glycan structures on leukocyte PSGL-1 and reduces cell binding to selectins. Blood 115, 1303–1312 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Niemelä R., Natunen J., Majuri M.-L., Maaheimo H., Helin J., Lowe J. B., Renkonen O., Renkonen R. (1998) Complementary acceptor and site specificities of Fuc-TIV and Fuc-TVII allow effective biosynthesis of sialyl-TriLex and related polylactosamines present on glycoprotein counterreceptors of selectins. J. Biol. Chem. 273, 4021–4026 [DOI] [PubMed] [Google Scholar]
- 26. Nakayama F., Nishihara S., Iwasaki H., Kudo T., Okubo R., Kaneko M., Nakamura M., Karube M., Sasaki K., Narimatsu H. (2001) CD15 expression in mature granulocytes is determined by α1,3-fucosyltransferase IX, but in promyelocytes and monocytes by α1,3-fucosyltransferase IV. J. Biol. Chem. 276, 16100–16106 [DOI] [PubMed] [Google Scholar]
- 27. Toivonen S., Nishihara S., Narimatsu H., Renkonen O., Renkonen R. (2002) Fuc-TIX. A versatile α1,3-fucosyltransferase with a distinct acceptor- and site-specificity profile. Glycobiology 12, 361–368 [DOI] [PubMed] [Google Scholar]
- 28. Lowe J. B., Kukowska-Latallo J. F., Nair R. P., Larsen R. D., Marks R. M., Macher B. A., Kelly R. J., Ernst L. K. (1991) Molecular cloning of a human fucosyltransferase gene that determines expression of the Lewis x and VIM-2 epitopes but not ELAM-1-dependent cell adhesion. J. Biol. Chem. 266, 17467–17477 [PubMed] [Google Scholar]
- 29. Nishihara S., Iwasaki H., Nakajima K., Togayachi A., Ikehara Y., Kudo T., Kushi Y., Furuya A., Shitara K., Narimatsu H. (2003) α1,3-Fucosyltransferase IX (Fut9) determines Lewis X expression in brain. Glycobiology 13, 445–455 [DOI] [PubMed] [Google Scholar]
- 30. Patnaik S. K., Potvin B., Stanley P. (2004) LEC12 and LEC29 gain-of-function Chinese hamster ovary mutants reveal mechanisms for regulating VIM-2 antigen synthesis and E-selectin binding. J. Biol. Chem. 279, 49716–49726 [DOI] [PubMed] [Google Scholar]
- 31. North S. J., Huang H. H., Sundaram S., Jang-Lee J., Etienne A. T., Trollope A., Chalabi S., Dell A., Stanley P., Haslam S. M. (2010) Glycomics profiling of Chinese hamster ovary cell glycosylation mutants reveals N-glycans of a novel size and complexity. J. Biol. Chem. 285, 5759–5775 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Root D. E., Hacohen N., Hahn W. C., Lander E. S., Sabatini D. M. (2006) Genome-scale loss-of-function screening with a lentiviral RNAi library. Nat Methods 3, 715–719 [DOI] [PubMed] [Google Scholar]
- 33. Britten C. J., van den Eijnden D. H., McDowell W., Kelly V. A., Witham S. J., Edbrooke M. R., Bird M. I., de Vries T., Smithers N. (1998) Acceptor specificity of the human leukocyte α3-fucosyltransferase. Role of FucT-VII in the generation of selectin ligands. Glycobiology 8, 321–327 [DOI] [PubMed] [Google Scholar]
- 34. Sasaki K., Kurata K., Funayama K., Nagata M., Watanabe E., Ohta S., Hanai N., Nishi T. (1994) Expression cloning of a novel α1,3-fucosyltransferase that is involved in biosynthesis of the sialyl Lewis x carbohydrate determinants in leukocytes. J. Biol. Chem. 269, 14730–14737 [PubMed] [Google Scholar]
- 35. Natsuka S., Gersten K. M., Zenita K., Kannagi R., Lowe J. B. (1994) Molecular cloning of a cDNA encoding a novel human leukocyte α1,3-fucosyltransferase capable of synthesizing the sialyl Lewis x determinant. J. Biol. Chem. 269, 16789–16794 [PubMed] [Google Scholar]
- 36. Wagers A. J., Stoolman L. M., Kannagi R., Craig R., Kansas G. S. (1997) Expression of leukocyte fucosyltransferases regulates binding to E-selectin: relationship to previously implicated carbohydrate epitopes. J. Immunol. 159, 1917–1929 [PubMed] [Google Scholar]
- 37. Brito C., Kandzia S., Graça T., Conradt H. S., Costa J. (2008) Human fucosyltransferase IX. Specificity toward N-linked glycoproteins and relevance of the cytoplasmic domain in intra-Golgi localization. Biochimie 90, 1279–1290 [DOI] [PubMed] [Google Scholar]
- 38. Huang M. C., Laskowska A., Vestweber D., Wild M. K. (2002) The α(1,3)-fucosyltransferase Fuc-TIV, but not Fuc-TVII, generates sialyl Lewis X-like epitopes preferentially on glycolipids. J. Biol. Chem. 277, 47786–47795 [DOI] [PubMed] [Google Scholar]
- 39. Schaff U. Y., Dixit N., Procyk E., Yamayoshi I., Tse T., Simon S. I. (2010) Orai1 regulates intracellular calcium, arrest, and shape polarization during neutrophil recruitment in shear flow. Blood 115, 657–666 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Martinez M., Joffraud M., Giraud S., Baïsse B., Bernimoulin M. P., Schapira M., Spertini O. (2005) Regulation of PSGL-1 interactions with L-selectin, P-selectin, and E-selectin. J. Biol. Chem. 280, 5378–5390 [DOI] [PubMed] [Google Scholar]
- 41. Pouyani T., Seed B. (1995) PSGL-1 recognition of P-selectin is controlled by a tyrosine sulfation consensus at the PSGL-1 amino terminus. Cell 83, 333–343 [DOI] [PubMed] [Google Scholar]
- 42. Snapp K. R., Wagers A. J., Craig R., Stoolman L. M., Kansas G. S. (1997) P-selectin glycoprotein ligand-1 is essential for adhesion to P-selectin but not E-selectin in stably transfected hematopoietic cell lines. Blood 89, 896–901 [PubMed] [Google Scholar]
- 43. Malý P., Thall A., Petryniak B., Rogers C. E., Smith P. L., Marks R. M., Kelly R. J., Gersten K. M., Cheng G., Saunders T. L., Camper S. A., Camphausen R. T., Sullivan F. X., Isogai Y., Hindsgaul O., von Andrian U. H., Lowe J. B. (1996) The α(1,3)-fucosyltransferase Fuc-TVII controls leukocyte trafficking through an essential role in L-, E-, and P-selectin ligand biosynthesis. Cell 86, 643–653 [DOI] [PubMed] [Google Scholar]
- 44. M'Rini C., Cheng G., Schweitzer C., Cavanagh L. L., Palframan R. T., Mempel T. R., Warnock R. A., Lowe J. B., Quackenbush E. J., von Andrian U. H. (2003) A novel endothelial L-selectin ligand activity in lymph node medulla that is regulated by α(1,3)-fucosyltransferase-IV. J. Exp. Med. 198, 1301–1312 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Taylor A. D., Neelamegham S., Hellums J. D., Smith C. W., Simon S. I. (1996) Molecular dynamics of the transition from L-selectin- to β2-integrin-dependent neutrophil adhesion under defined hydrodynamic shear. Biophys. J. 71, 3488–3500 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Walcheck B., Moore K. L., McEver R. P., Kishimoto T. K. (1996) Neutrophil-neutrophil interactions under hydrodynamic shear stress involve L-selectin and PSGL-1. A mechanism that amplifies initial leukocyte accumulation of P-selectin in vitro. J. Clin. Invest. 98, 1081–1087 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Nimrichter L., Burdick M. M., Aoki K., Laroy W., Fierro M. A., Hudson S. A., Von Seggern C. E., Cotter R. J., Bochner B. S., Tiemeyer M., Konstantopoulos K., Schnaar R. L. (2008) E-selectin receptors on human leukocytes. Blood 112, 3744–3752 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Nishihara S., Iwasaki H., Kaneko M., Tawada A., Ito M., Narimatsu H. (1999) α1,3-Fucosyltransferase 9 (FUT9; Fuc-TIX) preferentially fucosylates the distal GlcNAc residue of polylactosamine chain whereasthe other four α1,3-FUT members preferentially fucosylate the inner GlcNAc residue. FEBS Lett. 462, 289–294 5 5 [DOI] [PubMed] [Google Scholar]