Background: FoxM1 plays many roles in cancer development, progression, and cancer survival.
Results: FoxM1 is regulated by HSF1 and promotes cell cycle progression and cancer cell survival under heat stress conditions.
Conclusion: FoxM1 is critical for cell survival under heat stress condition.
Significance: HSF1-FoxM1 is a novel connection between heat shock proteins and stress responses and a novel pathway for cell survival under stress condition.
Keywords: Brain Tumors, Cell Cycle, Gene Regulation, Heat Shock Protein, Stress Response, Cell Survival, FoxM1, HSF1, Thermotolerance
Abstract
The forkhead box M1 (FoxM1) is a key transcription factor regulating multiple aspects of cell biology. Prior studies have shown that FoxM1 is overexpressed in a variety of human tumors, including brain tumor, and plays a critical role in cancer development and progression. In this study we found that FoxM1 was up-regulated by heat shock factor 1 (HSF1) under heat shock stress condition in multiple cell lines. Knockdown of HSF1 with HSF1 siRNA or inhibition of HSF1 with a HSF1 inhibitor abrogated heat shock-induced expression of FoxM1. Genetic deletion of HSF1 in mouse embryo fibroblast cells also abolished heat shock stress-induced FoxM1 expression. Moreover, we showed that HSF1 directly bound to FoxM1 promoter and increased FoxM1 promoter activity. Furthermore, we demonstrated that FoxM1 was required for the G2-M phase progression through regulating Cdc2, Cdc20, and Cdc25B under a mild heat shock stress but enhanced cell survival under lethal heat shock stress condition. Finally, in human glioblastoma specimens, FoxM1 overexpression correlated with elevated HSF1 expression. Our results indicate that FoxM1 is regulated by HSF1 and is critical for HSF1-mediated heat shock response. We demonstrated a novel mechanism of stress resistance controlled by HSF1 and a new HSF-FoxM1 connection that mediates cellular thermotolerance.
Introduction
Heat shock factors (HSFs)3 play critical roles in acquisition of the induced thermotolerance through inducing a set of heat shock proteins (HSPs) (1–4). In mammalians cells, upon heat shock stress HSF1 is released from Hsp90 and translocated from cytoplasm to nucleus (5, 6). HSF1 binds to heat shock elements and induces HSPs expression. HSPs induced by heat shock stress further work as the chaperones to help protein folding, transport, and degradation to prevent overaccumulation of damaged proteins and cell death (7–9). Under normal unstressed conditions, HSPs are also highly overexpressed in many types of cancers and associated with cancer development, progression, invasion, metastasis, and treatment resistance (10–12). Strategies of targeting HSPs for cancer treatment are being evaluated clinically (13). Interestingly, mice with HSF1 knock-out could not develop cancer induced by kras mutation and other carcinogens, suggesting that HSF1 plays important roles in cancer development (14, 15). However, the mechanisms underlying HSF1-mediated promotion of cancer development and progression remain to be investigated.
The forkhead box M1 (FoxM1) is a key transcription factor for cell cycle progression (16–19). Many studies including our own have demonstrated that FoxM1 is a critical molecule for tumor development and progression (20–23). Increasing evidences show that FoxM1 not only promotes cancer development but also enhances tumor invasion, angiogenesis, metastasis, and drug resistance (24–30). However, how FoxM1 is regulated in normal and malignant cell remains to be fully understood. Some studies have shown that FoxM1 can be induced by ionic radiation and oxidative stress and activated by ultraviolet exposure (31–33), suggesting that FoxM1 might be a stress response protein.
In this study we found that FoxM1 is a novel downstream target of HSF1. Induction of FoxM1 is important for cell cycle progression and protection of cells from heat-shock-induced cell death. We revealed a novel role of FoxM1 in cell survival under heat shock stress conditions.
MATERIALS AND METHODS
Cell Lines and Culture Conditions
The human glioma cell lines Hs683, HFu251, and U-87MG, human lung cancer cell lines HCC193 and H358, and immortalized monkey kidney COS-1 cell line were obtained from the American Type Culture Collection (Manassas, VA). Human papillomaviral E6/E7-transformed Hsf1−/− and Hsf+/+ MEF cells were gifts from Dr. Dr. Ivor J. Benjamin (University of Utah School of Medicine, Salt Lake City, UT) and were described previously (2). All of the cell lines were maintained in plastic flasks as adherent monolayer in DMEM supplemented with 10% fetal bovine serum, sodium pyruvate, nonessential amino acids, l-glutamine, and a vitamin solution (Flow Laboratories, Rockville, MD).
Knockdown of HSF1 and FoxM1
FoxM1 was knocked down with FoxM1-siRNA in U-87MG and Hs683 as described previously (21). For HSF1 knockdown, pre-designed siRNA was purchased from Santa Cruz Biotechnology (Santa Cruz Biotechnology, Santa Cruz, CA). U-87MG and Hs683 cells were transfected with 100 nm HSF1-siRNA and a non-targeting control siRNA using Lipofectamine 2000 (Invitrogen) for 48 h. The transfection cells were used for heat shock stress treatment. Knockdown efficiency of HSF1 and FoxM1 was determined by Western blotting.
Overexpression of FoxM1 and Constitutively Active HSF1
Hs683 cells were transfected with FoxM1 expression plasmid pcDNA3-FoxM1, CMV-hHSF1 (+) (constitutively active HSF1 plasmid), CMV-hHSF1 (−) (non-active HSF1 plasmid) (34), and control vector using Lipofectamine 2000. Expression of transfected genes was confirmed with Western blotting, and the FoxM1-transfected cells were treated with heat shock stress followed by cell cycle analysis.
Western Blotting
Whole cell lysates were extracted from the cells. Standard Western blotting was performed with polyclonal rabbit antibodies against human FoxM1 (MPP2 K-19), HSP70, p-HSF1 (S303) (Santa Cruz Biotechnology), HSF1 (Cell Signaling, Danvers, MA), Cdc2, Cdc20, and Cdc25B (BD Biosciences), and a second antibody (anti-rabbit IgG or anti-mouse IgG; GE Healthcare). The same membranes were stripped and blotted with an anti-human-β-actin antibody (Sigma) and used as loading controls. The probe proteins were detected using the Amersham Biosciences enhanced chemiluminescence system following the manufacturer's instructions.
Real-time Quantitative PCR
RNA was isolated with TRIzol (Invitrogen). After oligo(dT)-primed reverse transcription of 500 ng of total RNA was done, the resulting single stranded cDNA was amplified using TaqDNA polymerase (Promega, Madison, WI). Semiquantitative real-time PCR was performed using the LightCycler system together with the LightCycler DNA Master SYBR Green I Kit (Bio-Rad). To ensure experiment accuracy, all reactions were performed in triplicate. The primers used were 5′-tgcccagcagtctcttacct-3′ (forward primer) and 5′-ctacccaccttctggcagtc-3′ (reverse primer) for human FoxM1 and 5′-tggggaaggtgaaggtcgg-3′ (forward primer) and 5′-ctggaagatggtgatggga-3′ (reverse primer) for human glyceraldehyde-3-phosphate dehydrogenase (GAPDH). The PCR conditions consisted of an initial activation of Super-Start TaqDNA polymerase at 95 °C for 10 min followed by 40 cycles of 95 °C for 10 s, 60 °C for 1 min. The Ct (threshold cycle) value of FoxM1 amplification was normalized to that of GAPDH control. Reaction specificity was controlled by post-amplification melting curve analysis.
Promoter-Reporter Luciferase Assays
The promoter region of human FoxM1 was identified based on a previous report (35) and by analyzing the sequence of the region upstream of FoxM1 cDNA of human genomic DNA. The promoter region was amplified from human normal blood cell genomic DNA with high fidelity DNA polymerase. The promoter regions with different lengths were cut with specific restriction enzymes and cloned into the pGL3 luciferase reporter vector (Promega). The reporter assays were performed using a dual luciferase assay kit (Promega) as described (21).
Chromatin Immunoprecipitation Assays
Chromatin immunoprecipitation (ChIP) assays were performed using a kit from Upstate Cell Signaling Solutions (Charlottesville, VA). Briefly, chromatin that was cross-linked to transcription factors was immunoprecipitated using antibodies against HSF1. The immunoprecipitated chromatin was amplified with primers (forward, 5′-ccttggtcagggaatagtgtca-3′, and reverse, 5′-ggatgttgcaatactccaggca-3′) that flank the HSF1 binding sites of the FoxM1 promoter. The primers were also used for quantitative real-time PCR assay of binding of HSF1 to the FoxM1 promoter. The primers of HSP70 promoter and primers of quinone oxidoreductase (NQO1) exon 2 served as the positive control and negative control, respectively, for the quantitative real-time PCR assay. The positive primers are forward (5-cactcccccttcctctcag-3) and reverse (5-ttccttctgagccaatcac-3); negative primers are forward (5-cctgtagctgaaggtttgctgg-3, and reverse, 5-cctacctgtgatgtcctttctgg-3). One percent of total genomic DNA was used as the input DNA. The relative amount of enriched DNA was calculated by moralization with input DNA.
Analysis of Cell Cycle Distribution under Heat Shock Stress Conditions
Cells transfected with FoxM1-siRNA (100 nm) or FoxM1 expression plasmid were treated with heat shock stress at 42 °C for 6, 12, or 24 h. Cells were collected and washed with PBS once and fixed with 65% ethanol. Fixed cells were treated with RNase A and stained with propidium iodide (100 μg/ml in PBS). The stained cells were detected using flow cytometer (BD Biosciences Immunocytometry Systems). Data were analyzed with a cell cycle analysis software program (ModFit LT Version 2.0; Verity Software House) to calculate the percentage of cells at G1, S, and G2 phase.
Cell Growth and Survival Assay
U-87MG cells were transfected with FoxM1-siRNA, and 48 h after transfection cells were treated with heat shock at 42 °C for 2 h and recovered for 4 h and followed by 44 °C heat shock for 1 h. Viable Cell numbers were counted with Via-Cell counter machine at 12-, 24-, and 48-h time points. Cells were fixed with 65% ethanol, and nuclei were stained with propidium iodide for a cell viability assay under a microscope. Dead cells and apoptotic bodies were characterized by condensed or fragmented nuclei.
Immunohistochemical Staining
Brain tissue specimens were fixed by neutral-buffered formalin, embedded in paraffin, and sectioned according to standard protocols. Tissue sections were used for immunostaining with antibodies against HSF1 (Cell Signaling) and FoxM1 (Santa Cruz Biotechnology). We quantitatively scored the tissue sections according to the percentage of positive cells and staining intensity, as previously defined (30).
Statistics
Correlations between positive staining for FoxM1 and positive staining for HSF1 in the glioma specimens were assessed with Pearson's correlation test. The significance of results was determined using Student's t test (two-tailed). Statistically significant changes (p values <0.05) are indicated with asterisks.
RESULTS
Heat Shock Stress Induced FoxM1 Expression in Multiple Cell Lines
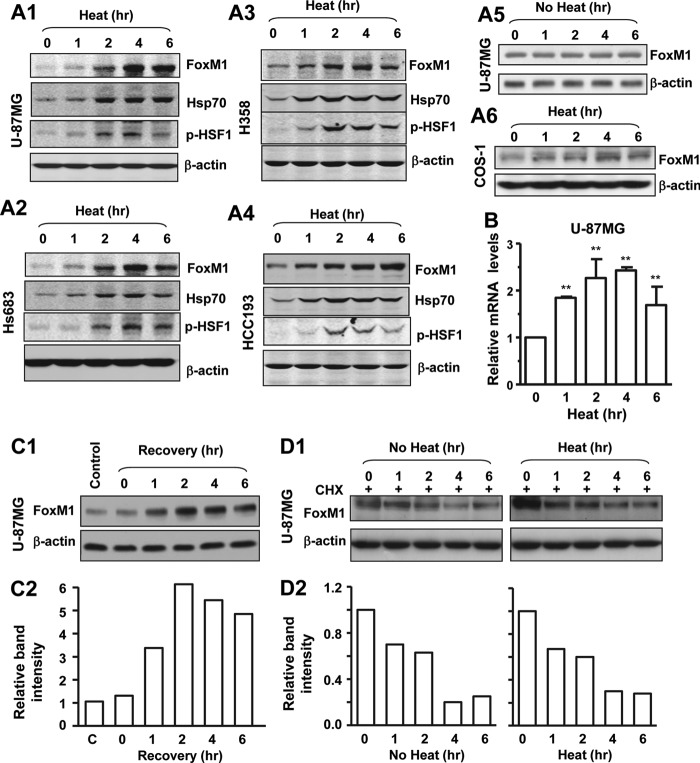
First, glioma cell lines U-87MG and Hs683, lung cancer cell lines HCC193 and H358, and COS-1 cells were treated with 42 °C for different time points and recovered for 2 h. As shown in Fig. 1A, FoxM1 was up-regulated by heat shock stress in a time-dependent manner in multiple cell lines. To determine whether induction of FoxM1 by heat shock stress is through transcriptional activation or enhanced protein stability, we performed real-time quantitative PCR to examine FoxM1 RNA levels in the cells treated with heat shock stress. The results showed that heat shock stress up-regulated FoxM1 RNA transcription in a time-dependent manner (Fig. 1B). FoxM1 was induced by heat shock after a 1-h recovery (Fig. 1C). To check the protein stability of FoxM1, U-87MG cells were treated with cycloheximide (50 μg/ml, Sigma) under heat shock stress conditions. Western blotting showed that heat shock stress did not change the protein stability (Fig. 1D). These results indicate that heat shock stress up-regulates FoxM1 expression through enhancing FoxM1 RNA transcription.
FIGURE 1.
FoxM1 was up-regulated by heat shock stress in multiple cell lines. A, U-87MG, Hs683, Hcc193, H358, and COS1 cells were treated with heat shock stress at 42 °C for different time points as indicated. After a 2-h recovery, total cell lysates were prepared, and Western blotting was performed to examine FoxM1 and Hsp70 level. B, U-87MG cells were treated with heat shock at 42 °C with different time points as indicated, and real-time PCR was performed to examine FoxM1 mRNA level. C, U-87MG cells were treated with heat shock stress for 4 h followed by recovery for different time point as indicated, and Western blotting was performed to examine FoxM1 level. D, U-87MG cells were treated with cycloheximide (CHX) followed by heat shock stress with different time points as indicated. After 2 h of recovery, cells lysates were prepared, and Western blotting was performed. E, relative protein levels were measured using densitometry software. Error bars represent ± S.D. from experiments done in triplicate. **, p < 0.01.
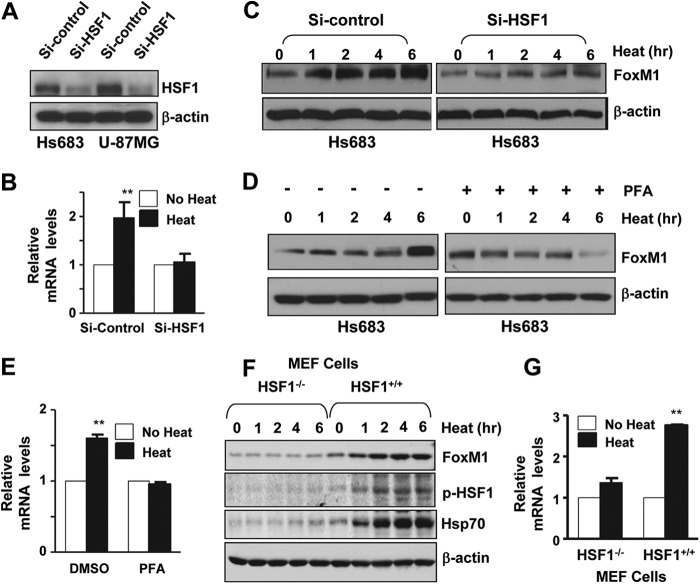
Heat Shock Factor 1 (HSF1) Is the Critical Regulator of FoxM1 Expression under Heat Shock Stress Conditions
We next analyzed FoxM1 promoter using transcription factor binding sites analysis software (Transfac)(35, 36) and revealed two putative HSF1 binding sites between −1788 and −1405 at FoxM1 promoter. Knockdown of HSF1 by siRNA in Hs683 cells (Fig. 2A) followed by heat shock for different time points blocked heat shock stress-induced FoxM1 expression (Fig. 2, B and C). Similarly, a heat shock stress inhibitor, pifithrin α (37, 38), also blocked FoxM1 induction by heat shock stress both in protein and RNA level (Fig. 2, D and E). Furthermore, in Hsf1-null mouse fibroblast cells (MEF Hsf1−/−) derived from Hsf1 knock-out mice, FoxM1 was not significantly up-regulated by heat shock stress (Fig. 2, F and G). These results indicate that FoxM1 is regulated by heat shock stress through HSF1.
FIGURE 2.
FoxM1 is regulated by HSF1 under heat shock stress. A and B, Hs683 and U-87MG were treated with HSF1 siRNA (100 nm) for 48 h, then cells were collected for Western blotting analysis. Hs683 cell were transfected with HSF1 siRNA, and 48 h after the transfection, cells were treated with heat shock stress at 42 °C with different time points. Western blotting was performed to examine the FoxM1 level. C, Hs683 cells were transfected with HSF1 siRNA, and 48 h after transfection cells were treated with heat shock stress for 2 h. Real-time PCR was performed to examine FoxM1 mRNA level. D, Hs683 cells were treated with heat shock response inhibitor, pifithrin α (PFA), for 2 h followed by heat shock stress at 42 °C with different time points as indicated. Western blotting was performed to examine FoxM1 level. E, Hs683 cells were treated with pifithrin α for 2 h followed by heat shock stress for 2 h. With 2 h recovery after the heat shock, total RNA was isolated, and real-time PCR was performed to examine FoxM1 mRNA level. F, MEF cells with Hsf1 wild type or knock-out were treated with heat shock stress at 42 °C with different time points as indicated. With a 2-h recovery after heat shock, cells were lysed with radioimmune precipitation assay buffer, and Western blotting was performed to examine FoxM1, Hsp70, and p-HSF level. G, real-time PCR was performed to check the FoxM1 mRNA expression both in Hsf1−/− and Hsf1+/+ cells. Error bars represent ± S.D. from experiments done in triplicate. **, p < 0.01.
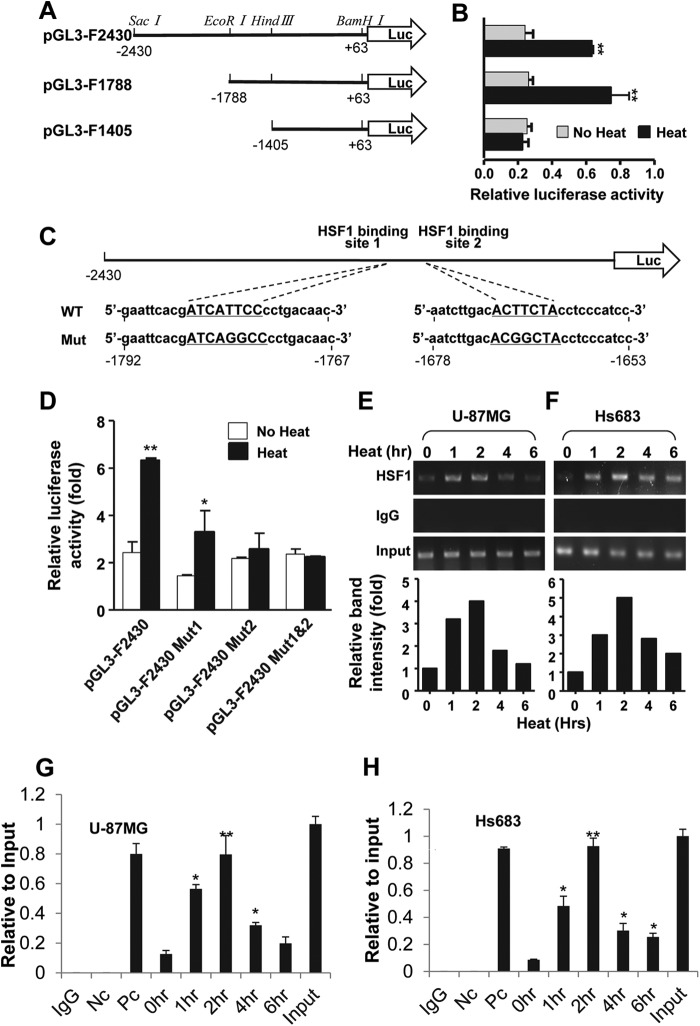
Identify HSF1 Binding Sites in FoxM1 Promoter
To define the HSF1 binding sites at FoxM1 promoter, three promoter reporters were constructed with different promoter deletion (Fig. 3A). Results of a luciferase activity assay showed that heat shock stress increased the activity of the full-length FoxM1 promoter but not the promoter with −1788 to −1405 fragment deletion, indicating that HSF1 binding sites do locate between −1788 to −1405 (Fig. 3B). To further confirm that HSF1 binding sites at FoxM1 promoter are critical for FoxM1 induction by heat shock stress, three constructs of full-length promoter were constructed with target point mutations at HSF1 putative binding sites. Mutations at the HSF1 putative binding site 1 or both HSF1 putative binding site 1 and 2 significantly blocked the heat shock-induced promoter activity (Fig. 3, C and D). However, mutation at the HSF1 putative binding site 2 alone did not significantly decrease FoxM1 promoter activity, indicating that the second putative HSF1 site might not be important for FoxM1 induction (Fig. 3, C and D). To further determine if HSF1 can directly bind to FoxM1 promoter, a ChIP assay was performed using a pair of PCR primers flanking the −1725 and −1497 sequence that containing the putative HSF1 binding sites, and the result showed that HSF1 can directly bind to FoxM1 promoter under heat shock stress conditions in both U-87MG and Hs683 cells (Fig. 3, E and F). Also, the binding of HSF1 to FoxM1 promoter was increased upon heat shock stress (Fig. 3, E and F). This observation was confirmed by the result of quantitative real-time PCR showing that binding of HSF1 to the FoxM1 promoter was significantly increased after heat shock treatment (Fig. 3, G and H).
FIGURE 3.
HSF1 activates FoxM1 promoter during heat shock stress. A, three different luciferase reporter vectors were constructed as indicated. B, luciferase reporter vectors were transfected to the U-87MG cells. Forty-eight hours after the transfection cells were treated with heat shock for 4 h followed by recovery for another 2 h. Cells were collected, and relative luciferase activity was measured. C, point mutations were generated at FoxM1 promoter reporter as show in the figure. D, promoter reporters with heat shock element mutations were transfected with Renilla luciferase control plasmid into U-87MG cells, and 48 h after the transfection cells were treated with heat shock stress for 2 h followed by 2 h recovery at 37 °C. Luciferase assay was performed with dual-luciferase assay kit. E and F, ChIP-enriched DNAs using anti-HSF1 antibody were prepared from U-87MG and Hs683 treated with heat shock at 42 °C for the indicated periods. DNA fragments of FoxM1 promoter (−1925 ∼ −1497) were amplified. G and H, relative binding activity of HSF1 to FoxM1 promoter in U-87MG and Hs683 cell with heat shock stress was analyzed by using quantitative real-time PCR with the primers flanking the HSF1 binding sites of the FoxM1 promoter (1–6 h heat shock treatments) and with negative control primers (Nc) from NQO1 (quinone oxidoreductase) exon 2 and positive control primers (Pc) from HSP70 promoter (1-h heat shock treatment). Error bars represent ± S.D. from experiments done in triplicate. *, p < 0.05; **, p < 0.01.
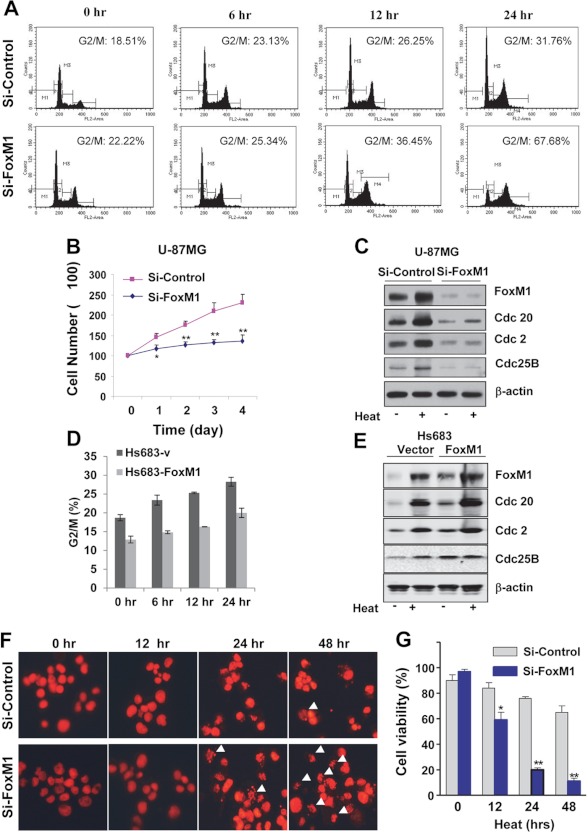
FoxM1 Is Required for G2-M Phase Progression and Protection of Cells from Lethal Heat Shock Stress
To determine the biological role of FoxM1 in heat shock stress, FoxM1 was knocked down in U-87MG cells with siRNA followed by heat shock at 42 °C for different time points. The result showed that cells with FoxM1 knockdown were arrested at G2-M phase after 24 h of heat shock stress, indicating that most cells without FoxM1 cannot progress from G2 to M phase under heat shock stress (Fig. 4A). Consistent with the cell cycle arrest, U-87MG cells transfected with FoxM1-siRNA grew much slower compared with the cell transfected with control-siRNA and treated with heat shock stress (Fig. 4B). Moreover, the expression of Cdc20, Cdc2, and Cdc25B were up-regulated under heat shock stress in control siRNA-treated cells (Fig. 4C). In contrast, knockdown of FoxM1 down-regulated the expression of Cdc20, Cdc2, and Cdc25B in normal and heat shock stress conditions (Fig. 4C). In addition, overexpression of FoxM1 in Hs683 cell dramatically induced the expression of those downstream molecules and decreased G2 arrest (Fig. 4, D and E). These results indicate that FoxM1 regulates the expression of Cdc20, Cdc2, and Cdc25B during heat shock stress and promotes G2 to M phase cell cycle progression. Furthermore, cells with FoxM1 knockdown broadly showed cell death under lethal heat shock stress, whereas more cells with siRNA control treatment survived (Fig. 4, F and G). These results indicate that induction of FoxM1 is required for cell cycle progression and important for protecting the cell from lethal heat shock stress.
FIGURE 4.
FoxM1 is required for cell cycle progression and survival in glioma cells with heat shock stress. A, U-87MG cells were transfected with FoxM1-specific siRNA, and 48 h after the transfection cells were treated with heat shock stress at 42 °C for the indicated times. Cell cycle analysis was performed with FACS. B, U-87MG cells were transfected with FoxM1-specific siRNA, and 48 h after the transfection cells were grows for the 4 days, and cell numbers were counted at each time point to generate cell growth curve. C, U-87MG cells were transfected with FoxM1-specific siRNA and 48 h after the transfection cells were treated with heat shock at 42 °C for 2 h followed by recovery for 2 h; total cell lysates were used for Western blotting. D, Hs683 cell were transfected with FoxM1 expression vector and control plasmid. 24 h after the transfection, cells were treated with heat shock stress at 42 °C for the indicated times, and cell cycle analysis was performed. E, Hs683 cells were transfected with FoxM1 expression plasmid and control vector, and 48 h after the transfection the cells were treated with heat shock at 42 °C for 2 h and recovered for 2 h. Western blotting was performed to examine the expression of FoxM1 and downstream molecules. F, U-87MG cells were transfected with FoxM1-specific siRNA, and 48 h after the transfection cells were treated with lethal heat shock stress (44 °C) for the indicated time points. Cells were stained with propidium iodide and examined under fluorescence microscope. Dead cells and apoptotic bodies were characterized by condensed or fragmented nuclei. G, U-87MG cells were transfected with FoxM1-specific siRNA. 48 h after the transfection cells were treated with lethal heat shock stress at 44 °C for different time points, viable cell numbers were counted. *, p < 0.05; **, p < 0.01.
HSF1 Expression Correlates with FoxM1 Overexpression in Human Primary Glioma Specimens
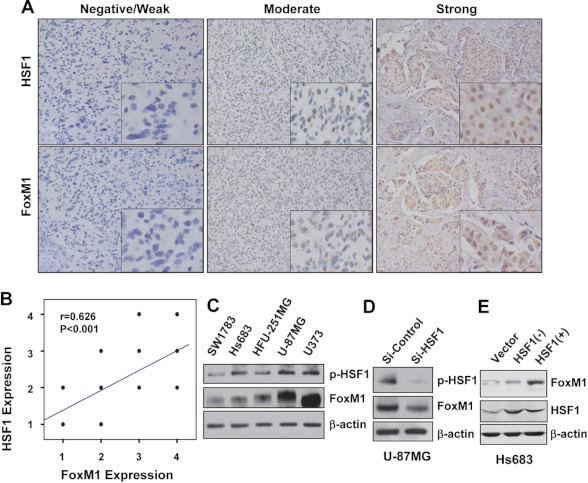
Because heat shock factors play many roles in cancer development, progression, and treatment resistance, we further examined if HSF1 regulates FoxM1 expression in glioma cell without heat shock stress. We first determined the HSF1 expression patterns in serial sections of 34 human glioblastoma (grade 4) specimens by immunohistochemical analyses. Fig. 5A shows tissue sections displaying strong staining (+++), moderate staining (++), and negative staining (0/+) FoxM1 or HSF1. Immunohistochemical staining of HSF1 and FoxM1 in the glioma specimens were quantified on the 1–4 scale. We analyzed the scores and found a significant correlation between the FoxM1 and HSF1 expression levels (Fig. 5B, r = 0.626, p < 0.001). Western blotting showed that glioma cells with high basal level of p-HSF1 also have high FoxM1 expression and that HSF1 knockdown down-regulated FoxM1 expression in glioma cells (Fig. 5C and 5D). In addition, overexpression of a constitutively active HSF1 up-regulated FoxM1 expression in Hs683 cell (Fig. 5E). These results suggest that HSF1 may regulate FoxM1 expression in glioma cell lines under normal condition.
FIGURE 5.
Levels of FoxM1 expression correlate with HSF1 expression in human glioblastoma specimens. A, immunohistochemical staining with specific anti-FoxM1 and anti-HSF1 antibodies was done on 34 glioblastoma tissues. B, the percentage and intensity scores were combined to obtain a total score (range, 0–4). FoxM1 expression levels correlated positively with HSF expression levels in glioblastoma samples (Spearman's correlation test r = 0.626; p < 0.001. Note that some of the dots on the graphs represented more than one specimen (some scores overlapped). C, levels of p-HSF correlate with FoxM1 expression in brain tumor cell lines. Western blotting was performed to check the expression of p-HSF1, HSF1, and FoxM1 in five brain tumor cell lines, and the level of β-actin was used as the loading control. D, knockdown of HSF1 reduced FoxM1 expression in U87 cell lines. E, overexpression of constitutively active HSF1 up-regulated FoxM1 expression in Hs683 cells.
DISCUSSION
Previous studies showed that FoxM1 can be activated during radiation treatment or UV exposure and regulate DNA damage repair genes, suggesting that FoxM1 plays an important role in protecting cell from stress induced damage (33). Recent studies also reported that FoxM1 can be induced by reactive oxygen species and is a key regulator of oxidative stress during tumorigenesis (31, 32). In this study we showed that FoxM1 was up-regulated with a time point-dependent manner during heat shock stress condition in glioma, lung cancer, and COS-1 cells. Using real-time PCR and promoter reporter assays we found that induction of FoxM1 by heat shock stress was through transcriptional regulation. Furthermore, promoter activity assay suggest that the promoter sequence between the −1788 and −1405 is critical for promoter activation induced by heat shock. Through targeted point mutation we further defined that the HSF1 binding site located at −1670 to −1662 was important for FoxM1 promoter activation induced by heat shock. Our results indicate that FoxM1 is a heat shock response gene.
It has been shown that during heat shock stress conditions, HSF1 can be activated and translocated to the nucleus to further activate heat shock response proteins (HSPs) such as Hsp70 and Hsp90 (8, 9). HSPs prevent the disruption of normal cellular mitosis, meiosis, or differentiation by environmental stressors. In a mild heat shock stress condition, such as 42 °C, cells can overcome heat shock stress through HSF1 by regulating downstream genes expression, whereas targeted deletion of heat shock factor1 can induce cell cycle arrest (4, 39). Because FoxM1 can also be induced during heat shock stress by HSF1 and FoxM1 has been well known as a cell cycle regulator for G1-S and G2-M cell cycle progression, we proposed that FoxM1 may play a role in cell cycle progression under heat shock stress conditions. Our results showed that cells with FoxM1 knockdown were arrested at G2-M phase after heat shock treatment at 42 °C for 24 h. U-87MG cells treated with control siRNA had less G2-M cell arrest, indicating that without FoxM1 most cells cannot progress from G2 to M phase under heat shock stress. U-87MG cells with FoxM1 knockdown grew much slower compared with the cells treated with siRNA control. These results were consistent with a previous report that Hsf1 knock-out cells were accumulated in G2-M phase during heat shock stress, whereas Hsf1 wild type cells could overcome the cell cycle arrest induced by heat shock stress (39).
A previous study showed that Cdc20, Cdc2, and Cdc25B are downstream targets of FoxM1 in normal condition (40, 41). Because FoxM1 is induced by heat shock stress, we want to know if FoxM1 also regulates these molecules under heat shock stress on transcriptional level and thus promotes G2-M progression. Our results showed that the expressions of Cdc20, Cdc2, and Cdc25B were increased with heat shock stress, whereas knockdown of FoxM1 down-regulated expression of those molecules both in normal and heat shock stress conditions. These results indicate that FoxM1 regulates Cdc20 and Cdc2 expression during heat shock condition, which is important for G2 to M phase cell cycle progression. Interestingly, Polo like kinase 1 (Plk1) can phosphorylate both HSF1 and FoxM1 promoting G2-M cell cycle progression (42, 43). Our results provide a link between the functions of HSF1 and FoxM1 in cell cycle progression under stress condition. Moreover, under lethal heat shock stress conditions, cells with FoxM1 knockdown broadly showed cell death compared with the cells transfected with control siRNA. Our results indicate that FoxM1 is required for cell cycle progression and important for protecting the cell from lethal heat shock stress. A previous study showed that Hsf1-knock-out mouse fibroblast cells (Hsf1−/− MEF) are sensitive to lethal heat shock challenge, whereas Hsf1 wild type MEF can acquire thermotolerant capability during heat shock stress (2, 4, 39). Our result provided evidence that FoxM1, a cell cycle regulator and oncogenic protein, is one of the components for HSF1-mediated thermotolerance.
Most previous studies concentrated on the oncogenic functions of FoxM1 during cancer development and its regulation on the protein level (44–46). Here we show a new mechanism of FoxM1 regulation on the transcriptional level under the stress condition. We showed that FoxM1 is a HSF1 target and plays a critical role for cell survival under the heat stress condition. We found that FoxM1 promotes cell cycle progression through regulating Cdc2, Cdc20, and Cdc25B under heat shock stress, which might be one of the mechanisms of how FoxM1 protects cell from lethal heat shock stress; however, we could not exclude that FoxM1 may enhance the cell survival ability through regulating other molecules. Hsp90 and Hsp70 are also the downstream targets of HSF1 and have the function of protecting cells from lethal heat shock stress. Whether FoxM1 also regulates Hsp90 and Hsp70 during heat shock stress or normal condition deserves further study.
More and more studies have shown that heat shock factors and HSPs play important roles in cancer initiation and progression and treatment resistance (47, 48). Targeting HSF1- HSPs has shown potential theoretical benefit for cancer patients (14, 15). Results in this study showed that the levels of HSF1 correlated with FoxM1 expression in glioma tissue samples, whereas knockdown of HSF1 down-regulated FoxM1 expression in glioma cells, suggesting that HSF1 regulates the FoxM1 expression in both heat shock stress and normal conditions. Further studies to determine whether FoxM1 can cooperate with other HSPs to promote cancer progression will be interesting, and targeting these mechanisms might provide potential cancer treatment strategies.
In summary, in this study for the first time we found that FoxM1 is regulated by HSF1 under heat shock stress conditions and the induction of FoxM1 by HSF1 is required for cell cycle progression through regulating the expression of downstream Cdc20, Cdc2, and Cdc25B. Our results demonstrate that FoxM1 plays an important role in enhancing cell survival ability under heat shock stress conditions. This study also provided a novel HSF1-FoxM1 connection, which plays an important role in protecting the cell from the stresses and possibly in cancer development.
Acknowledgments
We thank Dr. Ivor J. Benjamin from the University of Utah School of Medicine for the MEF-hsf1+/+ and MEF-hsf−/− cells, Dr. Richard Voellmy from HSF Pharmaceuticals SA, Switzerland, and Dr. Neil L. Harrison from Weill Cornell Medical College for CMV-HSF1 (+) and CMV-HSF1 (−) plasmids. We thank Don Norwood for editorial comments.
This work was supported, in whole or in part, by National Institutes of Health Grants R01CA157933, R21CA152623, P50CA127001, and CA-16672 (NCI). This work was also supported by research grants from the Center for Targeted Therapy and the Multidisciplinary Research Program at M.D. Anderson Cancer Center.
- HSF
- heat shock factor
- HSP
- heat shock protein
- FoxM1
- forkhead box M1.
REFERENCES
- 1. Lindquist S. (1986) The heat-shock response. Annu. Rev. Biochem. 55, 1151–1191 [DOI] [PubMed] [Google Scholar]
- 2. McMillan D. R., Xiao X., Shao L., Graves K., Benjamin I. J. (1998) Targeted disruption of heat shock transcription factor 1 abolishes thermotolerance and protection against heat-inducible apoptosis. J. Biol. Chem. 273, 7523–7528 [DOI] [PubMed] [Google Scholar]
- 3. Pirkkala L., Nykänen P., Sistonen L. (2001) Roles of the heat shock transcription factors in regulation of the heat shock response and beyond. FASEB J. 15, 1118–1131 [DOI] [PubMed] [Google Scholar]
- 4. Zhang Y., Huang L., Zhang J., Moskophidis D., Mivechi N. F. (2002) Targeted disruption of hsf1 leads to lack of thermotolerance and defines tissue-specific regulation for stress-inducible Hsp molecular chaperones. J. Cell. Biochem. 86, 376–393 [DOI] [PubMed] [Google Scholar]
- 5. Baler R., Dahl G., Voellmy R. (1993) Activation of human heat shock genes is accompanied by oligomerization, modification, and rapid translocation of heat shock transcription factor HSF1. Mol. Cell. Biol. 13, 2486–2496 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Knauf U., Newton E. M., Kyriakis J., Kingston R. E. (1996) Repression of human heat shock factor 1 activity at control temperature by phosphorylation. Genes Dev. 10, 2782–2793 [DOI] [PubMed] [Google Scholar]
- 7. Ahn S. G., Thiele D. J. (2003) Redox regulation of mammalian heat shock factor 1 is essential for Hsp gene activation and protection from stress. Genes Dev. 17, 516–528 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Liberek K., Lewandowska A., Zietkiewicz S. (2008) Chaperones in control of protein disaggregation. EMBO J. 27, 328–335 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Nollen E. A., Brunsting J. F., Roelofsen H., Weber L. A., Kampinga H. H. (1999) In vivo chaperone activity of heat shock protein 70 and thermotolerance. Mol. Cell. Biol. 19, 2069–2079 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Calderwood S. K., Khaleque M. A., Sawyer D. B., Ciocca D. R. (2006) Heat shock proteins in cancer. Chaperones of tumorigenesis. Trends Biochem. Sci. 31, 164–172 [DOI] [PubMed] [Google Scholar]
- 11. Ciocca D. R., Calderwood S. K. (2005) Heat shock proteins in cancer. Diagnostic, prognostic, predictive, and treatment implications. Cell Stress Chaperones 10, 86–103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Neckers L., Neckers K. (2002) Heat-shock protein 90 inhibitors as novel cancer chemotherapeutic agents. Expert Opin. Emerg. Drugs 7, 277–288 [DOI] [PubMed] [Google Scholar]
- 13. Karapanagiotou E. M., Syrigos K., Saif M. W. (2009) Heat shock protein inhibitors and vaccines as new agents in cancer treatment. Expert Opin. Investig. Drugs 18, 161–174 [DOI] [PubMed] [Google Scholar]
- 14. Dai C., Whitesell L., Rogers A. B., Lindquist S. (2007) Heat shock factor 1 is a powerful multifaceted modifier of carcinogenesis. Cell 130, 1005–1018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Whitesell L., Lindquist S. (2009) Inhibiting the transcription factor HSF1 as an anticancer strategy. Expert Opin. Ther. Targets 13, 469–478 [DOI] [PubMed] [Google Scholar]
- 16. Laoukili J., Kooistra M. R., Brás A., Kauw J., Kerkhoven R. M., Morrison A., Clevers H., Medema R. H. (2005) FoxM1 is required for execution of the mitotic programme and chromosome stability. Nat. Cell Biol. 7, 126–136 [DOI] [PubMed] [Google Scholar]
- 17. Wang I. C., Chen Y. J., Hughes D., Petrovic V., Major M. L., Park H. J., Tan Y., Ackerson T., Costa R. H. (2005) Forkhead box M1 regulates the transcriptional network of genes essential for mitotic progression and genes encoding the SCF (Skp2-Cks1) ubiquitin ligase. Mol. Cell. Biol. 25, 10875–10894 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Westendorf J. M., Rao P. N., Gerace L. (1994) Cloning of cDNAs for M-phase phosphoproteins recognized by the MPM2 monoclonal antibody and determination of the phosphorylated epitope. Proc. Natl. Acad. Sci. U.S.A. 91, 714–718 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Ye H., Holterman A. X., Yoo K. W., Franks R. R., Costa R. H. (1999) Premature expression of the winged helix transcription factor HFH-11B in regenerating mouse liver accelerates hepatocyte entry into S phase. Mol. Cell. Biol. 19, 8570–8580 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Kalinichenko V. V., Major M. L., Wang X., Petrovic V., Kuechle J., Yoder H. M., Dennewitz M. B., Shin B., Datta A., Raychaudhuri P., Costa R. H. (2004) Foxm1b transcription factor is essential for development of hepatocellular carcinomas and is negatively regulated by the p19ARF tumor suppressor. Genes Dev. 18, 830–850 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Liu M., Dai B., Kang S. H., Ban K., Huang F. J., Lang F. F., Aldape K. D., Xie T. X., Pelloski C. E., Xie K., Sawaya R., Huang S. (2006) FoxM1B is overexpressed in human glioblastomas and critically regulates the tumorigenicity of glioma cells. Cancer Res. 66, 3593–3602 [DOI] [PubMed] [Google Scholar]
- 22. Wang I. C., Chen Y. J., Hughes D. E., Ackerson T., Major M. L., Kalinichenko V. V., Costa R. H., Raychaudhuri P., Tyner A. L., Lau L. F. (2008) FoxM1 regulates transcription of JNK1 to promote the G1/S transition and tumor cell invasiveness. J. Biol. Chem. 283, 20770–20778 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Wang I. C., Meliton L., Tretiakova M., Costa R. H., Kalinichenko V. V., Kalin T. V. (2008) Transgenic expression of the forkhead box M1 transcription factor induces formation of lung tumors. Oncogene 27, 4137–4149 [DOI] [PubMed] [Google Scholar]
- 24. Behren A., Mühlen S., Acuna Sanhueza G. A., Schwager C., Plinkert P. K., Huber P. E., Abdollahi A., Simon C. (2010) Phenotype-assisted transcriptome analysis identifies FOXM1 downstream from Ras-MKK3-p38 to regulate in vitro cellular invasion. Oncogene 29, 1519–1530 [DOI] [PubMed] [Google Scholar]
- 25. Dai B., Kang S. H., Gong W., Liu M., Aldape K. D., Sawaya R., Huang S. (2007) Aberrant FoxM1B expression increases matrix metalloproteinase-2 transcription and enhances the invasion of glioma cells. Oncogene 26, 6212–6219 [DOI] [PubMed] [Google Scholar]
- 26. Dai B., Pieper R. O., Li D., Wei P., Liu M., Woo S. Y., Aldape K. D., Sawaya R., Xie K., Huang S. (2010) FoxM1B regulates NEDD4–1 expression, leading to cellular transformation and full malignant phenotype in immortalized human astrocytes. Cancer Res. 70, 2951–2961 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Li Q., Zhang N., Jia Z., Le X., Dai B., Wei D., Huang S., Tan D., Xie K. (2009) Critical role and regulation of transcription factor FoxM1 in human gastric cancer angiogenesis and progression. Cancer Res. 69, 3501–3509 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Millour J., Constantinidou D., Stavropoulou A. V., Wilson M. S., Myatt S. S., Kwok J. M., Sivanandan K., Coombes R. C., Medema R. H., Hartman J., Lykkesfeldt A. E., Lam E. W. (2010) FOXM1 is a transcriptional target of ERα and has a critical role in breast cancer endocrine sensitivity and resistance. Oncogene 29, 2983–2995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Wang Z., Banerjee S., Kong D., Li Y., Sarkar F. H. (2007) Down-regulation of Forkhead Box M1 transcription factor leads to the inhibition of invasion and angiogenesis of pancreatic cancer cells. Cancer Res 67, 8293–8300 [DOI] [PubMed] [Google Scholar]
- 30. Zhang Y., Zhang N., Dai B., Liu M., Sawaya R., Xie K., Huang S. (2008) FoxM1B transcriptionally regulates vascular endothelial growth factor expression and promotes the angiogenesis and growth of glioma cells. Cancer Res 68, 8733–8742 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Li S. K., Smith D. K., Leung W. Y., Cheung A. M., Lam E. W., Dimri G. P., Yao K. M. (2008) FoxM1c counteracts oxidative stress-induced senescence and stimulates Bmi-1 expression. J. Biol. Chem. 283, 16545–16553 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Park H. J., Carr J. R., Wang Z., Nogueira V., Hay N., Tyner A. L., Lau L. F., Costa R. H., Raychaudhuri P. (2009) FoxM1, a critical regulator of oxidative stress during oncogenesis. EMBO J. 28, 2908–2918 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Tan Y., Raychaudhuri P., Costa R. H. (2007) Chk2 mediates stabilization of the FoxM1 transcription factor to stimulate expression of DNA repair genes. Mol. Cell. Biol. 27, 1007–1016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Jones T. J., Li D., Wolf I. M., Wadekar S. A., Periyasamy S., Sánchez E. R. (2004) Enhancement of glucocorticoid receptor-mediated gene expression by constitutively active heat shock factor 1. Mol. Endocrinol. 18, 509–520 [DOI] [PubMed] [Google Scholar]
- 35. Korver W., Roose J., Heinen K., Weghuis D. O., de Bruijn D., van Kessel A. G., Clevers H. (1997) The human TRIDENT/HFH-11/FKHL16 gene. Structure, localization, and promoter characterization. Genomics 46, 435–442 [DOI] [PubMed] [Google Scholar]
- 36. Wingender E., Chen X., Hehl R., Karas H., Liebich I., Matys V., Meinhardt T., Prüss M., Reuter I., Schacherer F. (2000) TRANSFAC. An integrated system for gene expression regulation. Nucleic Acids Res. 28, 316–319 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Komarova E. A., Neznanov N., Komarov P. G., Chernov M. V., Wang K., Gudkov A. V. (2003) p53 inhibitor pifithrin α can suppress heat shock and glucocorticoid signaling pathways. J. Biol. Chem. 278, 15465–15468 [DOI] [PubMed] [Google Scholar]
- 38. Wang J., He H., Yu L., Xia H. H., Lin M. C., Gu Q., Li M., Zou B., An X., Jiang B., Kung H. F., Wong B. C. (2006) HSF1 down-regulates XAF1 through transcriptional regulation. J. Biol. Chem. 281, 2451–2459 [DOI] [PubMed] [Google Scholar]
- 39. Luft J. C., Benjamin I. J., Mestril R., Dix D. J. (2001) Heat shock factor 1-mediated thermotolerance prevents cell death and results in G2/M cell cycle arrest. Cell Stress Chaperones 6, 326–336 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Leung T. W., Lin S. S., Tsang A. C., Tong C. S., Ching J. C., Leung W. Y., Gimlich R., Wong G. G., Yao K. M. (2001) Over-expression of FoxM1 stimulates cyclin B1 expression. FEBS Lett. 507, 59–66 [DOI] [PubMed] [Google Scholar]
- 41. Zhao Y. Y., Gao X. P., Zhao Y. D., Mirza M. K., Frey R. S., Kalinichenko V. V., Wang I. C., Costa R. H., Malik A. B. (2006) Endothelial cell-restricted disruption of FoxM1 impairs endothelial repair following LPS-induced vascular injury. J. Clin. Invest. 116, 2333–2343 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Fu Z., Malureanu L., Huang J., Wang W., Li H., van Deursen J. M., Tindall D. J., Chen J. (2008) Plk1-dependent phosphorylation of FoxM1 regulates a transcriptional programme required for mitotic progression. Nat. Cell Biol. 10, 1076–1082 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Lee Y. J., Kim E. H., Lee J. S., Jeoung D., Bae S., Kwon S. H., Lee Y. S. (2008) HSF1 as a mitotic regulator: phosphorylation of HSF1 by Plk1 is essential for mitotic progression. Cancer Res. 68, 7550–7560 [DOI] [PubMed] [Google Scholar]
- 44. Laoukili J., Alvarez M., Meijer L. A., Stahl M., Mohammed S., Kleij L., Heck A. J., Medema R. H. (2008) Activation of FoxM1 during G2 requires cyclin A/Cdk-dependent relief of autorepression by the FoxM1 N-terminal domain. Mol. Cell. Biol. 28, 3076–3087 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Laoukili J., Alvarez-Fernandez M., Stahl M., Medema R. H. (2008) FoxM1 is degraded at mitotic exit in a Cdh1-dependent manner. Cell Cycle 7, 2720–2726 [DOI] [PubMed] [Google Scholar]
- 46. Park H. J., Costa R. H., Lau L. F., Tyner A. L., Raychaudhuri P. (2008) Anaphase-promoting complex/cyclosome-CDH1-mediated proteolysis of the forkhead box M1 transcription factor is critical for regulated entry into S phase. Mol. Cell. Biol. 28, 5162–5171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Powers M. V., Workman P. (2007) Inhibitors of the heat shock response. Biology and pharmacology. FEBS Lett. 581, 3758–3769 [DOI] [PubMed] [Google Scholar]
- 48. Sherman M., Multhoff G. (2007) Heat shock proteins in cancer. Ann. N. Y. Acad. Sci. 1113, 192–201 [DOI] [PubMed] [Google Scholar]