Background: d-Xylulokinase (XK), the final enzyme in the glucuronate-xylulose pathway, produces a key regulator of lipogenesis, xylulose 5-phosphate.
Results: The structure of human XK was determined, and its catalytic activity and inhibition were characterized.
Conclusion: Human XK is selective for d-xylulose and is inhibited by 5-deoxy-5-fluoro-d-xylulose.
Significance: Inhibition of XK could clarify its roles in sugar metabolism, lipogenesis, and metabolic disease.
Keywords: Carbohydrate Metabolism, Enzyme Catalysis, Enzyme Inhibitors, Lipogenesis, Protein Structure, X-ray Crystallography, Xylulose Kinase
Abstract
d-Xylulokinase (XK; EC 2.7.1.17) catalyzes the ATP-dependent phosphorylation of d-xylulose (Xu) to produce xylulose 5-phosphate (Xu5P). In mammals, XK is the last enzyme in the glucuronate-xylulose pathway, active in the liver and kidneys, and is linked through its product Xu5P to the pentose-phosphate pathway. XK may play an important role in metabolic disease, given that Xu5P is a key regulator of glucose metabolism and lipogenesis. We have expressed the product of a putative human XK gene and identified it as the authentic human d-xylulokinase (hXK). NMR studies with a variety of sugars showed that hXK acts only on d-xylulose, and a coupled photometric assay established its key kinetic parameters as Km(Xu) = 24 ± 3 μm and kcat = 35 ± 5 s−1. Crystal structures were determined for hXK, on its own and in complexes with Xu, ADP, and a fluorinated inhibitor. These reveal that hXK has a two-domain fold characteristic of the sugar kinase/hsp70/actin superfamily, with glycerol kinase as its closest relative. Xu binds to domain-I and ADP to domain-II, but in this open form of hXK they are 10 Å apart, implying that a large scale conformational change is required for catalysis. Xu binds in its linear keto-form, sandwiched between a Trp side chain and polar side chains that provide exquisite hydrogen bonding recognition. The hXK structure provides a basis for the design of specific inhibitors with which to probe its roles in sugar metabolism and metabolic disease.
Introduction
“Is life worth living? It all depends on the liver.” (William James). The rising tide of metabolic disease has focused attention on the balance between the body's requirements for glucose storage and utilization, a function carried out by the liver (1). In the post-prandial state, dietary carbohydrate in the form of glucose is converted to glycogen or further oxidized through glycolysis and converted to triglyceride fats via lipogenesis. In the fasted state, the liver supplies glucose to the body through the breakdown of glycogen stores and by gluconeogenesis. The metabolic pathways regulating glucose storage and production are hormonally regulated by insulin and glucagon, respectively (1).
In obesity, excess intake of dietary glucose leads to the accumulation of lipid in skeletal muscle (2). This is closely linked with peripheral insulin resistance and if left untreated can result in type-2 diabetes mellitus and downstream vascular complications (2). Increased liver triglyceride content, characterized by fatty liver, is also emerging as a key contributor to hepatic insulin resistance and ultimately hyperglycemia (1). Peripheral insulin resistance in skeletal muscle further increases the conversion of dietary carbohydrate into triglyceride through lipogenesis in the liver (3). In this scenario of increasing fat deposits, insulin resistance, and metabolic disease, therapies that inhibit lipogenesis are becoming increasingly attractive.
A hitherto unexplored approach is to target production of the metabolite xylulose 5-phosphate (Xu5P),4 which regulates lipogenesis and glycolysis in the liver independently of insulin (4). Xu5P acts as a signaling molecule that increases the activity of protein phosphatase 2A (PP2A) (5), leading to allosteric stimulation of the glycolytic enzyme, phosphofructokinase (4). Activation of PP2A also leads to transcriptional up-regulation of glycolytic and lipogenic pathways through carbohydrate-response element-binding protein (6). The physiological importance of carbohydrate-response element-binding protein-mediated lipogenesis is demonstrated by a reduction in glycolysis and lipogenesis in mice deficient in carbohydrate-response element-binding protein (7). Obese ob/ob mice deficient in carbohydrate-response element-binding protein expression also display reduced hepatic triglyceride content and attenuated symptoms of the metabolic syndrome (8).
Xu5P is generated as a by-product of the pentose-phosphate pathway. It is also produced as the final product of the glucuronate-xylulose (GX) pathway, which in mammals is primarily active in the kidneys and liver. This pathway converts inositol and glucose sugars to glucuronate and then to Xu5P in the following reactions: glucuronate → l-gulonate → 3-keto-gulonate → l-xylulose → xylitol → d-xylulose → Xu5P (9). Two observations suggest that the GX pathway carries significant flux and is a major source of intracellular Xu5P in vivo. First, in the benign human genetic condition of essential pentosuria, impaired conversion of l-xylulose to xylitol results in blockade of the GX pathway and increased urinary secretion of 1–2 g/day of l-xylulose (10). Second, experiments using cultured mouse-derived mhAT3F hepatocytes have shown that xylitol is a much more potent inducer of lipogenesis than is glucose (11). As xylitol is converted to Xu5P through the reactions, xylitol → d-xylulose → Xu5P within the GX pathway, these findings indicate that the pathway is constitutively active in vivo and can regulate hepatic carbohydrate and lipid metabolism through production of Xu5P.
d-Xylulokinase (XK; EC 2.7.1.17) is the final enzyme in the GX pathway (12), where it catalyzes the ATP-dependent phosphorylation of d-xylulose to produce Xu5P. It is therefore implicated as a key regulator of Xu5P levels and is of potential biomedical importance for controlling metabolic disease. XK activity is abundant in bovine liver, and the bovine enzyme has been partially characterized (13, 14). A gene encoding a putative human XK enzyme (hXK) has also been reported, with its annotation based on 22% shared amino acid identity with XK from Haemophilus influenzae (15). The functional identity of this gene has not been confirmed, however, and it remains the case that no mammalian XK has yet been fully characterized both structurally and functionally. The only known structure of an XK enzyme is that from Escherichia coli (16). In this study, we definitively establish the identity of hXK through functional characterization and x-ray crystal structures of several protein-ligand complexes. We show that hXK is highly specific for d-xylulose as its sugar substrate, and we also present kinetic and structural data on a competitive inhibitor of hXK that could be utilized to probe the role of the GX pathway in metabolic disease.
EXPERIMENTAL PROCEDURES
Protein Expression and Purification
A cDNA clone containing the putative hXK coding sequence identified by Tamari et al. (15) (clone IOH46573; NCBI accession number NM_005108.2) was obtained from Invitrogen. Full details of the expression and purification of the putative hXK gene product have been reported elsewhere (17). In brief, the coding sequence was amplified by PCR and inserted into the pProEX Htb plasmid. After verification of the correct DNA sequence, this plasmid was transformed into an E. coli BL21 (DE3) cell line supplemented with E. coli molecular chaperones. The majority of the expressed protein, which carried an N-terminal His6 affinity tag, was in a soluble form. Purification involved an initial immobilized metal affinity chromatography step, using a HisTrap FF 5-ml immobilized metal affinity chromatography column (GE Healthcare) charged with Ni2+ ions, removal of the affinity tag with recombinant tobacco etch virus protease, and a final size exclusion chromatography step on a Superdex 200 16/60 gel filtration column (GE Healthcare). The purified protein in 50 mm Tris/HCl, pH 8.0, 150 mm NaCl, 0.50 mm tris(2-carboxyethyl)phosphine, pH 8.0 (buffer A), was concentrated by ultrafiltration (2.5-ml 10-kDa MWCO Vivaspin concentrators (GE Healthcare), 1600 × g) to ∼16 mg ml−1 for crystallization or storage at −80 °C.
Selenomethionine-substituted (SeMet) protein was produced in a similar manner to the native protein, except that 15 min before induction the following amino acids were added to the expression culture: 60 mg/liter l-selenomethionine, 100 mg/liter l-lysine, l-phenylalanine, and l-threonine, and 50 mg/liter l-isoleucine, l-leucine, and l-valine, following the method described by Van Duyne et al. (18). Purification was with the same protocol as for the native protein except that the tris(2-carboxyethyl)phosphine concentration was increased to 2 mm for size exclusion chromatography and storage.
Crystallization and Data Collection
Crystallization experiments were carried out at room temperature by screening protein solutions against a 480-component crystallization screen (19), using sitting drops dispensed by a Cartesian Honeybee robot (Genomic Solutions). Initial crystals were obtained from just one condition, comprising 100 nl of protein solution (16 mg ml−1 in buffer A) plus 100 nl of precipitant solution (200 mm MES-KOH, pH 6.0, 16% (w/v) polyethylene glycol 6000, 0.3 mm NaN3), as described (17). Crystals suitable for x-ray diffraction were prepared with the aid of a repetitive seeding protocol; after four rounds of seeding and optimization, crystals reached a size (∼15 × 15 × 150 μm) large enough for data collection. Crystals of SeMet-substituted hXK (Se-hXK) were obtained by cross-seeding with native crystals into solutions of Se-hXK and propagation through 5 rounds of seeding. For data collection, the crystals were treated with a cryoprotectant solution comprising 16% (w/v) polyethylene glycol 6000, 20% (v/v) ethylene glycol, 200 mm MES-KOH, pH 5.9, 25 mm Tris/HCl, pH 8.0, 75 mm NaCl. Crystals were swept through the cryoprotectant solution for 1–10 s before being flash-cooled in liquid nitrogen.
Attempts to co-crystallize hXK with various ligands produced only poorly diffracting crystals. Ligand complexes of hXK were therefore prepared by soaking native crystals in cryoprotectant solutions supplemented with the appropriate ligands. Complexes with d-xylulose (Xu) and its analog 5-deoxy-5-fluoro-d-xylulose (5FX), which are syrups at room temperature, were prepared by direct addition to the cryoprotectant to give 20% (v/v) Xu or 10% (v/v) 5FX. The Xu-ADP complex was prepared with a cryoprotectant supplement of 250 mm Xu, 10 mm ADP, 10 mm MgCl2. Soaking times were 3–30 s for the Xu and 5FX complexes and ∼5 min for the Xu-ADP complex.
All x-ray data were collected at 100 K on beamline MX2 of the Australian Synchrotron, using a Quantum 315r detector (Area Detector Systems Corp.). Data were indexed, integrated, and scaled with XDS (20). The POINTLESS package (21) was used for initial space group prediction and reindexing operations, and additional scaling and merging of the data were performed with AIMLESS (21). Averaged intensities were converted to structure factor amplitudes by TRUNCATE (22). Full details of the data collection statistics and crystal details are in Table 1.
TABLE 1.
MAD data collection and phasing statistics
| Peak | Inflection point | |
|---|---|---|
| Data collection | ||
| Space group | P32 | P32 |
| Wavelength | 0.97972 Å | 0.97962 Å |
| Cell dimensions | a = b = 102.54, c = 159.23 Å | a = b = 102.54, c = 159.23 Å |
| Resolution rangea | 19.90 to 2.50 Å (2.64 to 2.50 Å) | 19.9 to 2.50 Å (2.64 to 2.50 Å) |
| Rsym | 14.4% (99.2%) | 10.5% (57.7%) |
| Rmeas | 17.8% (99.2%) | 12.8% (70.9%) |
| Observed reflections | 369,327 (54,117) | 373,459 (54,775) |
| Unique reflections | 64,600 (9425) | 64,602 (9425) |
| Completeness | 99.8 (99.9) | 99.8 (99.9) |
| Multiplicityb | 2.8 (2.8) | 2.8 (2.8) |
| 〈I/σI〉 | 10.7 (2.3) | 12.3 (3.3) |
| Phasing statistics | ||
| No. of sites | 37 | |
| Figure of meritc | 0.381 | |
| Phasing power | ||
| Anomalous | 1.16 | 0.839 |
| Isomorphous | 0.613 | |
a Values in parentheses correspond to the highest resolution shell.
b Friedel pairs separate.
c Combined value from SHARP is shown.
Structure Determination and Refinement
The structure of hXK was determined by multiwavelength anomalous dispersion phasing using a crystal of SeMet-substituted hXK that had been soaked for 30 s in cryoprotectant supplemented with 20% (v/v) d-xylulose. Data to 2.5 Å resolution were collected at the peak and inflection wavelengths of the selenium K-edge. A total of 28 selenium sites were located with the SHELXC/D package (23), refined in autoSHARP (24), which found a further nine selenium sites, established the space group as P32 with three hXK molecules in the asymmetric unit, and gave phases to 2.5 Å resolution. After density modification with SOLOMON (25), 90% of the hXK model was built automatically with ARP/wARP (26).
Refined structures for apo-hXK and its d-xylulose complex were obtained using additional data sets to 1.68 and 1.79 Å, respectively, and collected at a high energy remote wavelength using Se-hXK crystals. In each case, the initial 2.5-Å model was positioned in the unit cell with PHASER (27), automatically rebuilt with ARP/wARP, and subjected to cycles of refinement with BUSTER-TNT (28), exploiting the 3-fold noncrystallographic symmetry with local structure similarity restraints. Manual rebuilding was with COOT (29). Potential water molecules were added automatically but were checked manually for appropriate hydrogen bond interactions and electron density before they were retained in the model. Structures for the ligand complexes were determined in the same way, positioned with PHASER, rebuilt with ARP/wARP, and refined with COOT and BUSTER-TNT. Se-hXK crystals, which tended to diffract to higher resolution, were used for the ADP and AMPPCP complexes, but native crystals were used for the 5FX complex. The inclusion of the weak data as described by Karplus and Diederichs (30) significantly improved the quality of the refined models. The free R data set used in the determination of the initial structure model was retained through refinement of all structures, with extension to higher resolution where necessary. The updated Engh-Huber geometrical restraint library (31) was used throughout the refinements, and model quality was checked with MOLPROBITY (32). Full refinement statistics are in Table 2.
TABLE 2.
Data collection and structure refinement statistics
| Apo-hXK | Xu-hXK | Xu/ADP-hXK | 5FX-hXK | |
|---|---|---|---|---|
| Data collection | ||||
| Space group | P32 | P32 | P32 | P32 |
| Wavelength | 0.8856 Å | 0.8856 Å | 0.9537 Å | 0.9537 Å |
| Cell dimensions | a = b = 99.97 Å, c = 157.54 Å | a = b = 102.80 Å, c = 159.35 Å | a = b = 102.16 Å, c = 159.39 Å | a = b = 102.10 Å, c = 159.10 Å |
| Resolution rangea | 58.3 to 1.68 Å (1.71 to 1.68) | 59.4 to 1.79 Å (1.82 to 1.79 Å) | 36.8 to 1.97 Å (2.00 to 1.97 Å) | 34.0 to 1.98 Å (2.01 to 1.98 Å) |
| Rsym | 17.1% (386.8%) | 22.4% (497.5%) | 14.4% (280.6%) | 12.7% (145.1%) |
| Rmeas (%) | 19.6% (447.1%) | 24.7% (558.8%) | 17.9% (355.5%) | 15.8% (193.8%) |
| Rp.i.m. (%) | 6.8% (156.8%) | 7.5% (174.4%) | 10.5% (216.0%) | 6.9% (113.0%) |
| CC1/2 outer shell; no. of pairsb | 0.142; n = 9926 | 0.104; n = 8492 | 0.153; n = 6389 | 0.198; n = 6079 |
| Completeness | 100 (100) | 99.8 (99.5) | 100 (99.0) | 99.9 (99.0) |
| Multiplicity | 8.5 (8.0) | 11.3 (10.1) | 5.7 (4.7) | 5.3 (3.3) |
| 〈I/σI〉 | 9.4 (0.6) | 9.7 (0.5) | 9.8 (0.5) | 6.9 (0.7) |
| Refinement | ||||
| No. of reflections | 200,085 (9928) | 177,198 (8668) | 131,532 (6488) | 129,196 (6398) |
| Rwork/Rfree | 15.9%/18.4% | 16.7%/18.7% | 17.3%/19.7% | 17.3%/19.2% |
| No. of atoms (protein/ligand/water) | 23,467/58/1187 | 23,406/90/739 | 23,622/168/732 | 23,390 /67/739 |
| r.m.s.d. bondsc | 0.010 Å | 0.010 Å | 0.010 Å | 0.010 Å |
| r.m.s.d. anglesc | 1.04° | 1.06° | 1.04° | 1.06° |
| Average B (protein/ligand/water) | 31.0 Å2 (30.4/ 36.9 /42.4) | 35.1 Å2 (35.0/30.5/41.8) | 45.8 Å2 (45.17/52.8/48.3) | 48.8 Å2 (48.7/38.3 /52.5) |
| Ramachandran plotd | ||||
| Most favored | 98.6% | 98.6% | 99.6% | 98.7% |
| Disallowed | 0.2% | 0.2% | 0.2% | 0.2% |
| PDB entry code | 4BC3 | 4BC4 | 4BC2 | 4BC5 |
Enzyme Assay Reagents
Reagents used for the assays were purchased from Sigma, except for d-ribulose and d-lyxose, which were obtained from Carbosynth. The d-xylulose syrup was described as at least 95% pure, and this was consistent with 1H NMR spectra, in which no anomalous peaks were detected. The fluorinated Xu analog, 5FX, was synthesized using a strategy based on the work of Moravcová et al. (33) and Hadwiger et al. (34).
1H NMR Spectroscopy
1H NMR spectroscopy was used to confirm XK activity and carry out substrate screening assays. Stored hXK solution was rapidly thawed and desalted into 50 mm ammonium bicarbonate, pH 8.0, buffer using a 5-ml HiTrapTM desalting column (GE Healthcare). Sugars dissolved in water were added to substrate screening assays at a final concentration of 2 mm. Assays were carried out as 600-μl reactions and contained 3-trimethylsilylpropanoate as an internal standard. The full reaction mixture contained 50 mm NH4HCO3, pH 8.0, 10% D2O, 10 mm MgCl2, 4 mm ATP, 250 μm 3-trimethylsilylpropanoate, and 2 mm sugar. Reactions were initiated by adding 40 nmol hXK. One-dimensional 1H NMR spectra were collected from assay solutions in the absence of hXK and after a 1-h incubation with the enzyme. Spectra were collected on a 400-MHz DRX spectrometer (Bruker) at 25 °C, and the water peak was suppressed with a presaturation pulse (35). Data were acquired with XWINNMR (version 3.0, Bruker) and processed using TOPSPIN (version 1.3, Bruker).
Photometric Assay
XK activity was measured in a continuous assay in which hXK-catalyzed production of ADP was coupled to the oxidation of NADH via the action of pyruvate kinase and lactate dehydrogenase. Assay solutions consisted of 50 mm Tris/HCl, 150 mm NaCl, 250 mm NADH, 1.5 mm MgCl2, 1.8 units of pyruvate kinase (type VII from rabbit muscle, Sigma), 12 units of lactate dehydrogenase (type II from rabbit muscle, Sigma), 2 mm phosphoenolpyruvate, 600 mm ATP, 1.6 nm hXK, and 0–200 μm d-xylulose. For analysis of inhibition of hXK by 5-deoxy-5-fluoro-d-xylulose, this compound was added to the assay solution at concentrations of 1.5 to 170 μm. Reactions were initiated by the addition of d-xylulose after a 5-min preincubation of the assay solution at 25 °C. Initial rate measurements were made by following the decrease of absorbance at 340 nm (ϵNADH = 6220 m−1 cm−1) using a Cary 4000 ultraviolet-visible spectrophotometer with a temperature-controlled sample holder at 25 °C. Nonlinear fitting of the kinetic data to the Michaelis-Menten equation was carried out using GraphPad PRISM (version 5.04, Graphpad Software). The Km values for xylulose and kcat were determined from the mean values of three independent assays. The kinetic data collected in the presence of the inhibitor 5-deoxy-5-fluoro-d-xylulose were fitted to a competitive model of inhibition with respect to xylulose.
RESULTS
Structure of hXK
The hXK structure was determined by x-ray crystallography, with phasing from a multiwavelength anomalous dispersion experiment utilizing the selenium atoms of the SeMet-substituted protein (Se-hXK). Because the Se-hXK crystals diffracted better than native, the Se-hXK structure was taken as the reference structure, described here, and the Se-hXK crystals were also used for preparation of the substrate complexes. The apo-hXK structure was refined at 1.68 Å resolution, with good agreement with the x-ray data (R = 15.9%, Rfree = 18.4%) and excellent validation statistics; analysis with MOLPROBITY places it in the 100th percentile for this resolution. In each of the hXK structures reported here, the Ramachandran plot has a single outlier at Ser-246 (φ = 72°, ψ = 162°). Ser-246 is a pre-proline residue that is well defined by the electron density and whose main chain carbonyl oxygen makes a strong hydrogen bond with the indole nitrogen of Trp-299. One cis-peptide bond is also found in all structures, between Thr-383 and Pro-384.
The hXK polypeptide consists of 536 amino acid residues, with a molecular mass of 58.4 kDa. The asymmetric unit of the crystals contains three independent molecules, which show no oligomeric association. The enzyme is thus defined as monomeric, a designation supported by dynamic light scattering and size exclusion chromatography measurements (data not shown). In each chain density is absent for the chain termini, residues 1–5 and 532–536, but otherwise the polypeptides are completely modeled.
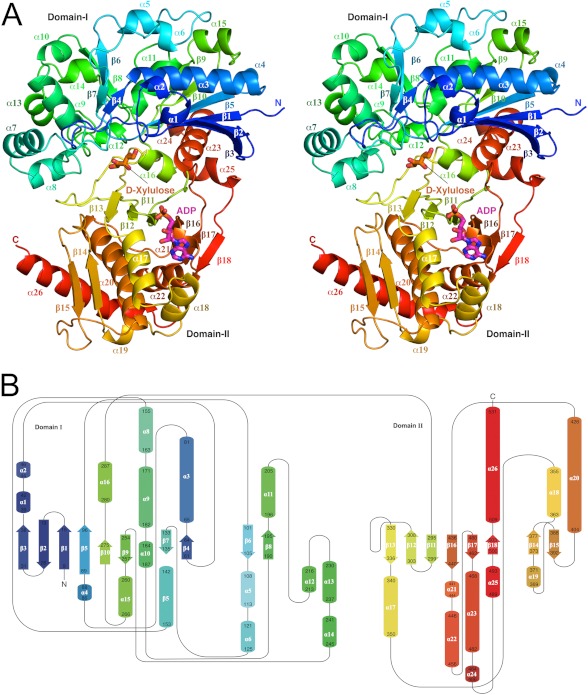
Like other members of the sugar kinase/hsp70/actin superfamily (36), hXK is folded into two domains, which here comprise residues 6–288 (domain-I) and 289–531 (domain-II) (Fig. 1). Domain-I contains one six-stranded β-sheet, two β-ribbons, and 16 α-helices, arranged in a β/α/β/α stack. The six-stranded β-sheet, sheet A, includes four major β-strands (β1, β2, β3, and β5) and two minor β-strands (β9 and β10) with 321465 topology, in which strand 2 and strand 6 are antiparallel to the rest. The two β-ribbons each have two antiparallel strands. Domain-II contains one β-sheet, and eight α-helices folded in an α/β/α stack. An extended six-stranded β-sheet (sheet B), comprising β11, β12, β13, β16, β17, and β18 runs through the middle of domain-II with 321456 topology, with an additional short β-ribbon comprising β14 and β15 arranged at the base of the α/β/α stack.
FIGURE 1.
Three-dimensional structure and folding of hXK. A, molecule is shown as a ribbon diagram, in stereo, colored in rainbow style from the N terminus (dark blue) to the C terminus (red). The binding sites for the two substrates are indicated, with d-xylulose shown in orange and the nucleotide (modeled as AMP) shown in magenta, both in stick mode. B, topology diagram, with secondary structural elements colored as in A and labeled.
The two domains are oriented roughly perpendicular to each other with respect to their α/β stacking and are linked by a loop consisting of 11 residues (amino acids 289–299). A prominent cleft is formed at the center of the molecule between the domains. The domains have folds characteristic of the N- and C-terminal domains of the FGGY sugar kinase fold, as defined in the structural characterization databases (37), in which they are both considered to have a ribonuclease H-like structural motif, an α/β/α stack with a mixed 5-stranded β-sheet with 32145 topology.
Structural Comparisons
Searches for structural homologs are complicated by variations in domain orientations in proteins belonging to this superfamily. Nevertheless, the top hits in searches with SSM (38) are all authenticated or putative XKs. The closest structural homolog of hXK in this analysis was the apo-form of E. coli d-xylulokinase (ecXK) (PDB code 2nlx (16)), which shares 19% sequence identity and for which 433 Cα atoms can be superimposed with a root-mean-square (r.m.s.) difference of 2.17 Å. Other close relationships were found for the putative bacterial XKs from Bifidobacterium adolescintis (PDB code 3i8b),5 Rhodospirillum rubrum (PDB code 3ifr),5 and Chromobacterium violaceum (PDB code 3hz6).5 For these latter proteins r.m.s. differences are 2.34 Å (429 Cα), 2.42 Å (446 Cα), and 2.64 Å (430 Cα), respectively, and sequence identities are ∼20%. There is no clear discrimination from other members of the sugar kinase family, however, with glycerol kinases from several species having r.m.s. differences of 2.7–3.0 Å for 430–440 Cα atoms when compared with hXK.
A notable feature is that when single domain comparisons are carried out, the N-terminal domains superimpose better than the C-terminal domains. For hXK and ecXK, the N-terminal domains superimpose with an r.m.s. difference of 1.53 Å (236 Cα), whereas the C-terminal domains have an r.m.s. difference of 2.91 Å (230 Cα). In the latter, the β-sheets and several helices match well, but there are many insertions, deletions, and altered loops around the domain periphery; the sequences are also less conserved (17% identity compared with 23% identity). These differences may be linked to the fact that the E. coli XK forms dimers through its C-terminal domain, whereas hXK is monomeric. A similar pattern is seen for the other (putative) bacterial XKs, which associate into similar dimers in the crystal.
Demonstration of Authentic XK Activity
The annotation of this enzyme as a human d-xylulokinase was tested through two functional assays. One-dimensional 1H NMR spectroscopy was carried out to verify its ability to convert d-xylulose (Xu) to the expected product, Xu5P, and to test its activity on other potential sugar substrates. The d-xylulokinase activity was confirmed by observing the loss of 1H NMR peaks corresponding to Xu in a reaction mixture and the evolution of peaks that match those in a spectrum of a commercial preparation of Xu5P, as shown in Fig. 2. Assignment of the 1H NMR peaks for the predominant β-furanose form of xylulose, and those of xylulose 5-phosphate, was based on published NMR characterization of these compounds (39, 40). Because of overlap in other regions of the spectrum, the appearance of a peak with a chemical shift of 3.77 ppm in the presence of hXK, corresponding to the C5 protons of Xu5P, is particularly distinctive. Similar NMR experiments with related sugars indicate that hXK is specific for Xu. Specifically, the sugars tested were d-ribulose (C3 epimer of Xu,), xylitol (five-carbon polyol), and d-arabinose, d-lyxose, and d-xylose (aldopentoses). Assays using native and SeMet-substituted hXK showed that both forms are equally active as xylulokinases.
FIGURE 2.
Analysis of the d-xylulose 5-kinase reaction catalyzed by hXK, using 1H NMR spectroscopy. A,1H NMR spectrum of an assay solution containing ATP and d-xylulose in the absence of hXK; B, after incubation with hXK. C, 1H NMR spectrum of xylulose 5-phosphate.
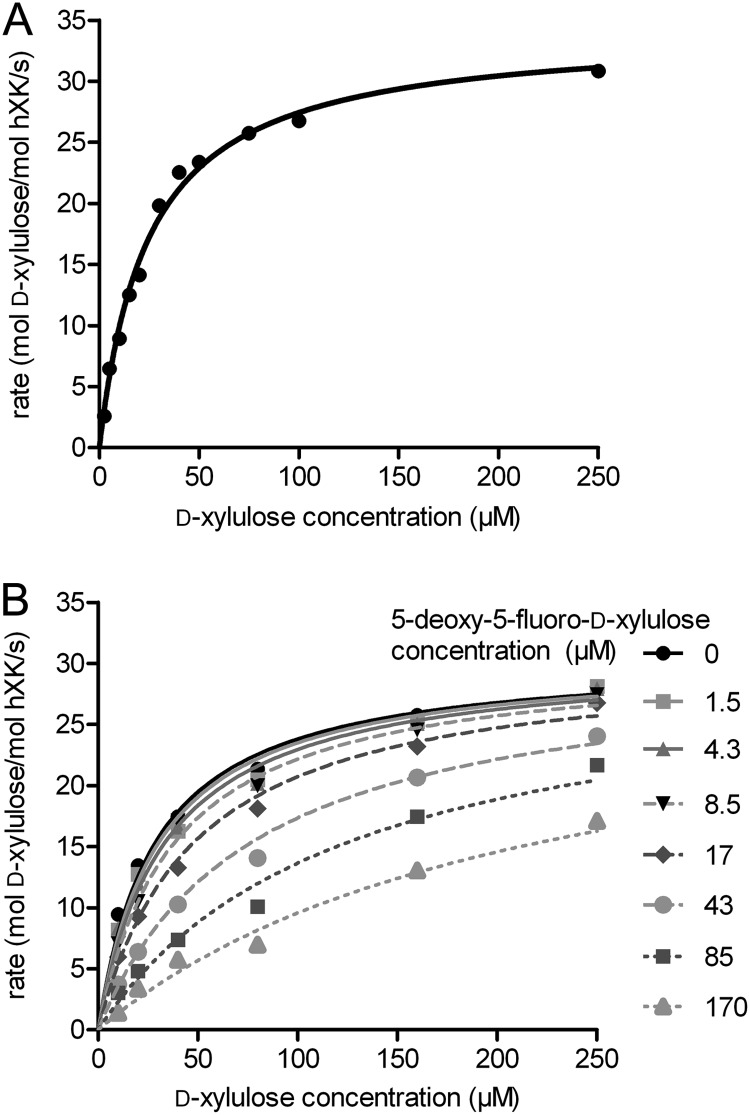
The more sensitive coupled photometric assay, in which hXK-catalyzed production of ADP was coupled to the oxidation of NADH via the action of pyruvate kinase and lactate dehydrogenase, was used to determine the apparent kinetic parameters for the XK reaction (Fig. 3A). The experimental kinetic constants for the turnover of Xu reaction were as follows: Km(Xu) = 24 ± 3 μm, kcat = 35 ± 5 s−1, and kcat/Km = 1500 ± 400 s−1 mm−1 (Table 3). The Xu C3-epimer, d-ribulose, was tested at a series of concentrations up to 20 mm with varied XK concentrations, but it was again found not to be a substrate, confirming the results of the 1H NMR assay. Experiments also showed that XK activity is substrate-inhibited by Xu concentrations greater than 200 μm, as judged by a lag in the initial rate of the reaction (data not shown), but is unaffected by the presence of Xu5P, the reaction product, up to a concentration of 100 μm. ATP hydrolysis was not detected in the absence of Xu, indicating that uncoupled ATPase activity was absent for XK.
FIGURE 3.
Kinetic analysis of the hXK reaction. A, relationship between initial reaction velocity and d-xylulose concentration in a single hXK activity assay. B, effect of the competitive inhibitor 5FX on the initial reaction velocity as a function of d-xylulose concentration. The assays were carried out with 600 mm ATP and 1.6 nm hXK and with varied d-xylulose and 5FX concentrations.
TABLE 3.
Apparent kinetic parameters determined for hXK
| Km for d-xylulosea | kcata | kcat/Km | Ki for 5-deoxy-5-fluoro-d-xylulose |
|---|---|---|---|
| μm | s−1 | s−1mm−1 | μm |
| 24 ± 3 | 35 ± 5 | 1500 ± 400 | 25 ± 2 |
a Mean value was from three independent assays. Errors are the means ± S.D.
The photometric assay was also used to validate 5FX as an inhibitor of hXK (Fig. 3B). This compound has been shown to be a dead-end competitive inhibitor of ecXK and was used by Di Luccio et al. (16) to deduce the preferred substrate binding order for this enzyme. For hXK, the value of Ki for 5FX was 25 ± 2 μm, comparable with the Km for Xu.
Structural Basis of Substrate Binding
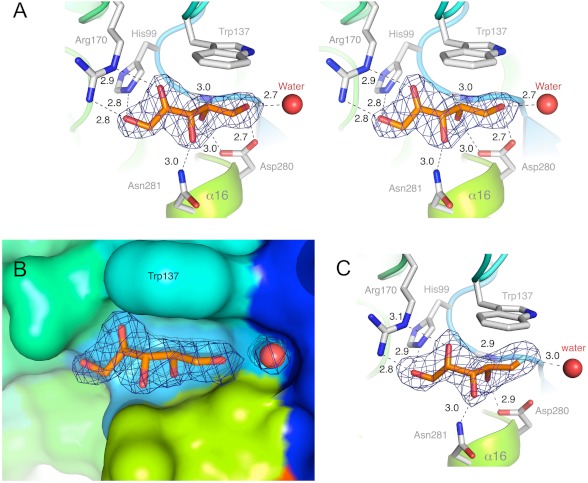
As the biochemical assays demonstrated a high specificity for Xu as the sugar substrate, we examined its binding mode through the crystal structure of its complex with hXK. Xu binds in a small cavity on domain-I, directly opposite the domain-II β-sheet, which is about 7 Å away in this open form structure. The site is formed entirely by residues on domain-I, bounded by the β5-β6 loop, which contributes Gln-98 and His-99, the β7-α7 loop contributing Trp-137, the α8-α9 loop contributing Arg-170, and the N terminus of helix α16, to which Asp-280 and Asn-281 belong. The same mode of Xu binding is seen for all three molecules in the asymmetric unit, and in both the Xu and Xu-ADP complexes, the electron density is excellent for the whole Xu molecule (Fig. 4), and the 10 atoms have B-factors in the range 30–37 Å2 (average 33.5 Å2), which compare well with the B-factors of the surrounding structure, 28–40 Å2. The site appears to be pre-formed for Xu binding, as the residues that directly engage the Xu molecule are identically arranged in the apo-hXK structure; taking molecule C as the reference, the atoms of the Xu-binding residues superimpose with an r.m.s. difference of 0.47 Å (61 atoms), compared with 0.45 Å for the whole domain.
FIGURE 4.
Binding of the substrate d-xylulose to hXK. A, stereo view of d-xylulose (Xu) modeled into its electron density. The electron density is from the initial 2mFo − DFc electron density map (blue mesh), calculated before Xu was included in the model, and is contoured at 1.0 σ. Hydrogen bonds are shown as broken lines, with distances in Å. The water molecule that is a consistent feature of the binding site is shown as a red sphere. B, fit of Xu into its binding site on domain-I, shown as a molecular surface and colored by chain succession as in Fig. 1. C, competitive inhibitor 5FX modeled into the initial 2mFo − DFc electron density map (blue mesh), calculated before it was included in the model, and contoured at 1.0 σ. The binding mode is virtually identical to that of Xu.
The d-xylulose substrate binds in its linear keto-form. The ketone oxygen O2 (readily identified by the planar geometry at C2) and the O1 hydroxyl are hydrogen bonded to Arg-170 NE and NH2, respectively (2.9 and 2.7 Å), with O1 additionally hydrogen bonded to His-99 NE2 (2.8 Å). The other Xu oxygens also form highly favorable interactions, in which O3 is hydrogen bonded to Asn-281 ND2 (2.9 Å), O4 to Asp-280 OD1 (2.7 Å), Gln-98 NH (3.1 Å), and O5 to Asp-280 OD2 (2.8 Å) and a water molecule (2.6 Å) (Fig. 4A). The indole ring of Trp-137 stacks (3.8 Å away) against the C3, C4, and C5 atoms of Xu, thus covering the non-hydrogen bonding side of the substrate (Fig. 4, A and B). This completes a highly specific set of bonding interactions. By analogy with the mechanisms advanced for ecXK (16) and glycerol kinase (41), Asp-280 should be the general base that extracts a proton from the O5 hydroxyl and activates it for nucleophilic attack on the incoming ATP phosphate.
The nucleotide-binding site, however, provides far fewer interactions. In the Xu-ADP complex, a nucleotide is present in each of the three molecules but can only be incompletely modeled (Fig. 5). The binding site, a cleft between helices α17, α18, and α21 of domain-II, provides stable binding only for the adenine ring, which fits into a slot, sandwiched between the indole ring of Trp-355, 3.6 Å away on one side, and the Gly-441–Ala-442 peptide, ∼4.0 Å away on the other side. There are few hydrogen bonds; N1 of the adenine ring is hydrogen bonded to Asn-455 ND2 and N3 to a water molecule that bridges to the ribose O2′ hydroxyl. The most complete nucleotide is on molecule B and is modeled as AMP, whereas on molecules A and C only an adenosine can be modeled. The ribose and phosphates project out into solution, and their greater flexibility is reflected in the mean B-factors, 56 Å2 for the adenine moiety, 70 Å2 for the ribose, and 88 Å2 for the α-phosphate for the molecule B nucleotide. When a complete ATP molecule is modeled on to the bound nucleotide, the γ-phosphate is positioned at least 10 Å from the reactive O5 of the Xu substrate across the interdomain cleft, implying that considerable movement is required to bring the two substrates together.
FIGURE 5.
Nucleotide binding to hXK. Stereo view showing ADP (modeled as AMP) fitted to the 2mFo − DFc electron density map (blue mesh) calculated by BUSTER-TNT prior to inclusion of the ligand in the model and contoured at 1.0 σ. Relevant side chains (gray) and α-helices (yellow and orange) are labeled, and hydrogen bonds are drawn as dashed lines and distances shown in Å. A bridging water molecule is shown with a red sphere.
Conformational Change
Enzymes of the sugar kinase/hsp70/actin superfamily are known to undergo large scale domain movements that enable catalysis to occur; these movements have been well described, for example, for hexokinase (42) and glycerol kinase (43). The hXK structures determined here are all in an “open” form, in which the two domains are widely separated and the two substrates at least 10 Å apart. There is evidence, however, of some domain movement when the three molecules of the asymmetric unit are compared across the various structures. All have slightly different degrees of opening. Between the most open (apo-hXK molecule C) and the most closed (hXK/5FX molecule A), there is a rigid body rotation of 7° of domain-I relative to domain-II, about a hinge lying across the interdomain cleft. The range of conformers is smoothly varied through this 7° range, with no preferred subgrouping.
To model the rotation required to bring the two substrates together, we used a closed form of the homologous E. coli glycerol kinase (ecGK), as seen in an ecGK mutant with a nonhydrolysable ATP analog, β,γ-difluoromethyleneadenosine 5′-triphosphate (PDB entry 1glj) (44). The XK monomer with the most open conformation (molecule C of the hXK/5FX complex) was modeled onto ecGK using a molecular dynamics approximation calculated with CHIMERA (45). This showed that domain closure of hXK involves a 47° rotation of domain-II relative to domain-I (Fig. 6) about an axis through strands β11, β12, and β13 and two hinge loops that precede helix α23 (hinge loop-1) and strand β17 (hinge loop-2). This predicted movement is greater than in ecGK (22.3°) (43) and in Plasmodium falciparum glycerol kinase (27°) (46); superpositions show that the domains are wider apart in the open form of hXK. The modeled closure of hXK buries the Xu substrate and brings the γ-phosphate of ATP in contact with the O5 hydroxyl of Xu.
FIGURE 6.
Modeling the closed form of hXK. The experimentally determined open form of hXK is shown in blue, superimposed on a modeled closed form of hXK in green, generated with Chimera (44), and based on a closed E. coli glycerol kinase structure (PDB code 1glj, see Ref. 43). The hinge axis, indicated by the purple circle, is normal to the page.
Inhibition of hXK
The inhibition assay described earlier established 5FX as a competitive inhibitor of Xu turnover, with an inhibition constant Ki of 25 ± 2 μm. We also examined the binding of 5FX through the crystal structure of its complex with hXK. Electron density maps following the first cycle of refinement unequivocally showed one 5FX molecule bound to each XK monomer (Fig. 4C). Xu and 5FX bind in the same mode, in which 5FX is in its linear keto-form. A comparison of Xu-bound complexes of hXK reveals them to be essentially indistinguishable from the 5FX complex at the resolution of the analysis. Unlike Xu, however, 5FX cannot hydrogen bond to a deprotonated Asp-280 side chain, leaving six hydrogen bonds involved in the protein-inhibitor interaction. The closest protein hydrogen bond donor to the fluorine atom, with which it could potentially form a hydrogen bond, is Thr-17 OG1, situated 3.7 Å away. A water molecule is, however, 2.9–3.0 Å from the fluorine, bridging to Ser-16 OG.
DISCUSSION
Much of the previous interest in the enzyme XK has been driven by its potential use for enhancing the production of ethanol as a renewable fuel from plant material. Xylan is the major component of plant hemicellulose and, after cellulose, is the second most abundant polysaccharide in plants, consisting of ∼30% of the biomass. XK is regarded as the rate-limiting enzyme in the catabolism of d-xylose in bacteria and fungi (47), prompting efforts to engineer enhanced activity (48).
In contrast, the role of XK in mammals has been under-explored, although it is potentially of great importance because of the strategic position this enzyme occupies at the intersection of two pathways with key roles in sugar metabolism. As the final enzyme in the GX pathway, XK produces the important signaling molecule Xu5P, ultimately from the metabolism of glucose and inositol sugars. Xu5P in turn links to the pentose-phosphate pathway, and hence to an important role in energy metabolism. Xu5P is further proposed to control both acute and long term regulation of glucose metabolism and fat synthesis (5). Excessive intake of sugars in human diet is thus linked through XK to lipogenesis, fatty liver disease, insulin resistance, and type 2 diabetes.
Despite its metabolic importance, XK has only been characterized functionally from one mammalian source, bovine liver. No structural information is available for any mammalian XK, and no human XK has been definitively identified. In this work, we have characterized the product of a human gene that had been tentatively identified as XK from its 22% sequence identity with XK from Salmonella typhimurium (15). Similar levels of sequence identity are seen across the wider sugar kinase family, however, between enzymes with quite distinct substrate specificities, making the annotation as hXK far from certain.
We have shown here that the gene identified by Tamari et al. (15) does indeed encode human XK. Moreover, the enzyme is highly specific for Xu, which appears to be its only sugar substrate. It is inactive against the C3 epimer of Xu, d-ribulose, as well as the 5-carbon polyol xylitol, and the aldopentoses d-xylose, d-arabinose, and d-lyxose. In this respect, it differs from the E. coli XK (ecXK), which appears to have a weaker affinity for Xu (Km of 290 μm compared with 24 μm for hXK) and a more relaxed specificity; ecXK is active against d-ribulose, albeit with 50-fold lower kcat/Km, and also has measurable activity against xylitol and d-arabitol. It should be noted, however, that the metabolic context of XK in bacteria is very different from that in humans, and the bacterial XKs differ in other respects as well; all appear to be dimers, whereas hXK is clearly monomeric.
The strict substrate specificity of hXK is clearly explained by its crystal structure. As anticipated, hXK shares the 2-domain fold characteristic of the sugar kinase/hsp70/actin superfamily. The Xu substrate is bound in its linear keto-form, in a cleft in domain-I, tightly sandwiched between the indole ring of Trp-137 on one side and specific hydrogen bonding side chains on the other side. All the polar groups on Xu make short, geometrically favorable hydrogen bonds; the electron density is excellent, and the B-factors are low, indicative of stable bonding. One end of the binding cleft is closed by Arg-170, which hydrogen bonds to O1 and O2 (Fig. 4A), whereas at the other end the C5 hydroxyl sits between Asp-280 and Trp-137, in perfect position for phosphorylation by ATP. The exclusion of other sugars as substrates may be largely determined by an inability of hXK to bind sugars that strongly favor the cyclic furanose configuration. Thus, whereas Xu is present at ∼20% in its linear form in solution, aldopentoses such as d-xylose, d-arabinose, and d-lyxose are almost exclusively present in the cyclic configuration (49). In addition, the tight fit of the Xu substrate determines the allowable configurations at C2 and C3. Thus, the inability of hXK to act on d-ribulose is explained by the fact that the C3 hydroxyl of d-ribulose would approach to less than ∼2.5 Å from the indole ring of Trp-137. The latter cannot adjust to avoid the clash because it is packed against Met-138 and hydrogen bonded through its indole NH to the side chain hydroxyl of Thr-17 (2.7 Å). In contrast, in the Xu complex with ecXK (16), the binding appears much looser; the electron density is poor, and half of the substrate atoms have B-factors greater than 90 Å2. Whether the substrate is correctly modeled or not, it is apparent that the binding cleft is somewhat wider in ecXK and in particular has no equivalent to Arg-170 at the inner end; the equivalent residue in ecXK is Gly-126. This is consistent with its apparent weaker affinity for Xu and more relaxed substrate specificity.
The strict specificity and catalytic efficiency of human XK is consistent with its role in a high flux biochemical pathway. Based on our enzyme kinetic data, the Xu5P route of Xu metabolism is heavily favored over a potentially harmful alternative reaction in which fructokinase converts Xu to d-xylulose 1-phosphate; the Km value of hXK for Xu is 250-fold lower than that of fructokinase for Xu (50). This alternative pathway (51) was discovered as a result of catastrophic outcomes that followed the clinical use of xylitol infusions (52). Xylitol is converted to Xu in the GX pathway, and saturation of this pathway led to the formation of d-xylulose 1-phosphate and then to an intracellular build up of oxalate and in severe cases multiple organ failure and death (52). Our observation that hXK shows evidence of substrate inhibition at Xu concentrations greater than 200 μm may explain the fact that, despite the high flux nature of the GX pathway, it can still be saturated by excessive sugar flux.
The strategic position of XK as the final enzyme in the XK pathway of sugar metabolism (9), and the recognition that its enzymatic product Xu5P is a major regulator of lipogenesis (5, 53), suggests that there could be value in developing specific inhibitors of XK to better understand its physiological roles. The inhibitor tested in our study, 5FX, differs from the natural Xu substrate only in substitution of the 5-hydroxy group by fluorine. This prevents phosphorylation. The affinity of hXK for 5FX as an inhibitor closely matches its affinity for Xu as substrate (both 25 μm), probably because although 5FX makes one fewer hydrogen bond, it is fully linear, whereas the linear form of Xu is a relatively minor (20%) conformer (49). The crystal structure of the 5FX complex suggests, however, that more potent substrate mimics could be developed by replacing the fluorine by chemical groups that are able to displace two adjacent, well defined water molecules that are present in all the hXK structures, hydrogen bonded to Asp-14, Ser-16, Thr-17, and Asp-280. This approach may enhance inhibitor potency and provide a useful tool for probing the physiological roles of XK and the GX pathway.
Acknowledgments
We thank Andrew Wadsworth and Margaret Brimble (Chemistry Department, University of Auckland) for their synthesis of the 5-deoxy-5-fluoro-d-xylulose. Access to the Australian Synchrotron was supported by the New Zealand Synchrotron Group, Ltd.
This work was supported by the Tertiary Education Commission of New Zealand through funding of the Maurice Wilkins Centre for Molecular Biodiscovery.
The atomic coordinates and structure factors (codes 4BC2, 4BC3, 4BC4, and 4BC5) have been deposited in the Protein Data Bank (http://wwpdb.org/).
Z. Zhang, S. K. Burley, and S. Swaminathan, unpublished data.
- Xu5P
- d-xylulose 5-phosphate
- XK
- d-xylulokinase
- hXK
- human XK
- ecHK
- E. coli XK
- Xu
- d-xylulose
- 5FX
- 5-deoxy-5-fluoro-d-xylulose
- GX pathway
- glucuronate-xylulose pathway
- SeMet
- selenomethionine
- Se-hXK
- selenomethionine-substituted hXK
- PDB
- Protein Data Bank
- r.m.s.
- root-mean-square
- AMPPCP
- adenosine 5′-(β,γ-methylenetriphosphate).
REFERENCES
- 1. Home P. D., Pacini G. (2008) Hepatic dysfunction and insulin insensitivity in type 2 diabetes mellitus. A critical target for insulin-sensitizing agents. Diabetes Obes. Metab. 10, 699–718 [DOI] [PubMed] [Google Scholar]
- 2. McGarry J. D. (2002) Banting lecture 2001. Dysregulation of fatty acid metabolism in the etiology of type 2 diabetes. Diabetes 51, 7–18 [DOI] [PubMed] [Google Scholar]
- 3. Petersen K. F., Dufour S., Savage D. B., Bilz S., Solomon G., Yonemitsu S., Cline G. W., Befroy D., Zemany L., Kahn B. B., Papademetris X., Rothman D. L., Shulman G. I. (2007) The role of skeletal muscle insulin resistance in the pathogenesis of the metabolic syndrome. Proc. Natl. Acad. Sci. U.S.A. 104, 12587–12594 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Uyeda K., Repa J. (2006) Carbohydrate-response element-binding protein, ChREBP, a transcription factor coupling hepatic glucose utilization and lipid synthesis. Cell Metab. 4, 107–110 [DOI] [PubMed] [Google Scholar]
- 5. Kabashima T., Kawaguchi T., Wadzinski B. E., Uyeda K. (2003) Xylulose 5-phosphate mediates glucose-induced lipogenesis by xylulose 5-phosphate-activated protein phosphatase in rat liver. Proc. Natl. Acad. Sci. U.S.A. 100, 5107–5112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Kawaguchi T., Takenoshita M., Kabashima T., Uyeda K. (2001) Glucose and cAMP regulate L-type pyruvate kinase gene by phosphorylation/dephosphorylation of the carbohydrate-response element-binding protein. Proc. Natl. Acad. Sci. U.S.A. 98, 13710–13715 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Iizuka K., Bruick R. K., Liang G., Horton J. D., Uyeda K. (2004) Deficiency of carbohydrate-response element-binding protein (ChREBP) reduces lipogenesis as well as glycolysis. Proc. Natl. Acad. Sci. U.S.A. 101, 7281–7286 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Iizuka K., Miller B., Uyeda K. (2006) Deficiency of carbohydrate-activated transcription factor ChREBP prevents obesity and improves plasma glucose control in leptin-deficient (ob/ob) mice. Am. J. Physiol. Endocrinol. Metab. 291, E358–E364 [DOI] [PubMed] [Google Scholar]
- 9. Touster O. (1960) Essential pentosuria and the glucuronate-xylulose pathway. Fed. Proc. 19, 977–983 [PubMed] [Google Scholar]
- 10. Bozian R. C., Touster O. (1959) Essential pentosuria. Renal or enzymic disorder? Nature 184, 463–464 [DOI] [PubMed] [Google Scholar]
- 11. Doiron B., Cuif M. H., Chen R., Kahn A. (1996) Transcriptional glucose signaling through the glucose response element is mediated by the pentose-phosphate pathway. J. Biol. Chem. 271, 5321–5324 [DOI] [PubMed] [Google Scholar]
- 12. Touster O. (1959) Pentose metabolism and pentosuria. Am. J. Med. 26, 724–739 [DOI] [PubMed] [Google Scholar]
- 13. Dills W. L., Jr., Parsons P. D., Westgate C. L., Komplin N. J. A. (1994) Assay, purification, and properties of bovine liver d-xylulokinase. Protein Exp. Purif. 5, 259–265 [DOI] [PubMed] [Google Scholar]
- 14. Hickman J., Ashwell G. (1958) Purification and properties of d-xylulokinase in liver. J. Biol. Chem. 232, 737–748 [PubMed] [Google Scholar]
- 15. Tamari M., Daigo Y., Ishikawa S., Nakamura Y. (1998) Genomic structure of a novel human gene (XYLB) on chromosome 3p22→p21.3 encoding a xylulokinase-like protein. Cytogenet. Cell Genet. 82, 101–104 [DOI] [PubMed] [Google Scholar]
- 16. Di Luccio E., Petschacher B., Voegtli J., Chou H. T., Stahlberg H., Nidetzky B., Wilson D. K. (2007) Structural and kinetic studies of induced fit in xylulose kinase from Escherichia coli. J. Mol. Biol. 365, 783–798 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Bunker R. D., Dickson J. M., Caradoc-Davies T. T., Loomes K. M., Baker E. N. (2012) Use of a repetitive seeding protocol to obtain diffraction-quality crystals of a putative human d-xylulokinase. Acta Crystallogr. Sect. F Struct. Biol. Cryst. Commun. 68, 1259–1262 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Van Duyne G. D., Standaert R. F., Karplus P. A., Schreiber S. L., Clardy J. (1993) Atomic structures of the human immunophilin FKBP-12 complexes with FK506 and rapamycin. J. Mol. Biol. 229, 105–124 [DOI] [PubMed] [Google Scholar]
- 19. Moreland N., Ashton R., Baker H. M., Ivanovic I., Patterson S., Arcus V. L., Baker E. N., Lott J. S. (2005) A flexible and economical medium-throughput strategy for protein production and crystallization. Acta Crystallogr. D Biol. Crystallogr. 61, 1378–1385 [DOI] [PubMed] [Google Scholar]
- 20. Kabsch W. (2010) XDS. Acta Crystallogr. D Biol. Crystallogr. 66, 125–132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Evans P. (2006) Scaling and assessment of data quality. Acta Crystallogr. D Biol. Crystallogr. 62, 72–82 [DOI] [PubMed] [Google Scholar]
- 22. French S., Wilson K. (1978) On the treatment of negative intensity observations. Acta Crystallogr. A 34, 517–525 [Google Scholar]
- 23. Sheldrick G. M. (2010) Experimental phasing with SHELXC/D/E. Combining chain tracing with density modification. Acta Crystallogr. D Biol. Crystallogr. 66, 479–485 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Vonrhein C., Blanc E., Roversi P., Bricogne G. (2007) Automated structure solution with autoSHARP. Methods Mol. Biol. 364, 215–230 [DOI] [PubMed] [Google Scholar]
- 25. Abrahams J. P., Leslie A. G. (1996) Methods used in the structure determination of bovine mitochondrial F1 ATPase. Acta Crystallogr. D Biol. Crystallogr. 52, 30–42 [DOI] [PubMed] [Google Scholar]
- 26. Langer G., Cohen S. X., Lamzin V. S., Perrakis A. (2008) Automated macromolecular model building for x-ray crystallography using ARP/wARP version 7. Nat. Protoc. 3, 1171–1179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. McCoy A. J., Grosse-Kunstleve R. W., Adams P. D., Winn M. D., Storoni L. C., Read R. J. (2007) Phaser crystallographic software. J. Appl. Crystallogr. 40, 658–674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Bricogne G., Blanc E., Brandl M., Flensburg C., Keller P., Paciorek C., Roversi P., Sharff A., Smart O. S., Vonrhein C., Womack T. O. (2011) BUSTER, Version 2.11.1, Global Phasing Ltd., Cambridge, UK [Google Scholar]
- 29. Emsley P., Cowtan K. (2004) Coot. Model-building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr. 60, 2126–2132 [DOI] [PubMed] [Google Scholar]
- 30. Karplus P. A., Diederichs K. (2012) Linking crystallographic model and data quality. Science 336, 1030–1033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Engh R., Huber R. (2006) in International Tables for X-ray Crystallography (Rossmann M. G., Arnold E., eds) Vol. F, pp. 382–392, Kluwer Academic Publishers, Dordrecht [Google Scholar]
- 32. Chen V. B., Arendall W. B., 3rd, Headd J. J., Keedy D. A., Immormino R. M., Kapral G. J., Murray L. W., Richardson J. S., Richardson D. C. (2010) Molprobity. All-atom structure validation for macromolecular crystallography. Acta Crystallogr. D Biol. Crystallogr. 66, 12–21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Moravcova J., Capkova J., Stanek J. (1994) One-pot synthesis of 1,2-O-isopropylidene-α-d-xylofuranose. Carbohydr. Res. 263, 61–66 [Google Scholar]
- 34. Hadwiger P., Mayr P., Nidetzky B., Stutz A. E., Tauss A. (2000) Synthesis of 5,6-dimodified open-chain d-fructose derivatives and their properties as substrates of bacterial polyol dehydrogenase. Tetrahedron 11, 607–620 [Google Scholar]
- 35. Stutz A. E. (1976) Solvent peak saturation with single phase and quadrature Fourier transformation. J. Magn. Reson. 21, 337–347 [Google Scholar]
- 36. Hurley J. H. (1996) The sugar kinase/heat shock protein 70/actin superfamily. Implications of conserved structure for mechanism. Annu. Rev. Biophys. Biomol. Struct. 25, 137–162 [DOI] [PubMed] [Google Scholar]
- 37. Apweiler R., Attwood T. K., Bairoch A., Bateman A., Birney E., Biswas M., Bucher P., Cerutti L., Corpet F., Croning M. D., Durbin R., Falquet L., Fleischmann W., Gouzy J., Hermjakob H., Hulo N., Jonassen I., Kahn D., Kanapin A., Karavidopoulou Y., Lopez R., Marx B., Mulder N. J., Oinn T. M., Pagni M., Servant F., Sigrist C. J., Zdobnov E. M. (2001) The InterPro database, an integrated documentation resource for protein families, domains, and functional sites. Nucleic Acids Res. 29, 37–40 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Krissinel E., Henrick K. (2004) Secondary-structure matching (SSM), a new tool for fast protein structure alignment in three dimensions. Acta Crystallogr. D Biol. Crystallogr. 60, 2256–2268 [DOI] [PubMed] [Google Scholar]
- 39. Vuorinen T., Serianni A. S. (1991) Synthesis of d-erythro-2-pentulose and d-threo-2-pentulose and analysis of the 13C and 1H n.m.r. spectra of the 1-13C- and 2-13C-substituted sugars. Carbohydr. Res. 209, 13–31 [DOI] [PubMed] [Google Scholar]
- 40. Yew W. S., Wise E. L., Rayment I., Gerlt J. A. (2004) Evolution of enzymatic activities in the orotidine 5′-monophosphate decarboxylase superfamily: mechanistic evidence for a proton relay system in the active site of 3-keto-l-gulonate-6-phosphate decarboxylase. Biochemistry 43, 6427–6437 [DOI] [PubMed] [Google Scholar]
- 41. Mao C., Ozer Z., Zhou M., Uckun F. M. (1999) X-ray structure of glycerol kinase complexed with an ATP analog implies a novel mechanism for the ATP-dependent glycerol phosphorylation by glycerol kinase. Biochem. Biophys. Res. Commun. 259, 640–644 [DOI] [PubMed] [Google Scholar]
- 42. Steitz T. A., Shoham M., Bennett W. S. (1981) Structural dynamics of yeast hexokinase during catalysis. Philos. Trans. R. Soc. Lond. B Biol. Sci. 293, 43–52 [DOI] [PubMed] [Google Scholar]
- 43. Feese M. D., Faber H. R., Bystrom C. E., Pettigrew D. W., Remington S. J. (1998) Glycerol kinase from Escherichia coli and an Ala-65 → Thr mutant: the crystal structures reveal conformational changes with implications for allosteric regulation. Structure 6, 1407–1418 [DOI] [PubMed] [Google Scholar]
- 44. Bystrom C. E., Pettigrew D. W., Branchaud B. P., O'Brien P., Remington S. J. (1999) Crystal structures of Escherichia coli glycerol kinase variant S58→W in complex with nonhydrolyzable ATP analogues reveal a putative active conformation of the enzyme as a result of domain motion. Biochemistry 38, 3508–3518 [DOI] [PubMed] [Google Scholar]
- 45. Pettersen E. F., Goddard T. D., Huang C. C., Couch G. S., Greenblatt D. M., Meng E. C., Ferrin T. E. (2004) UCSF Chimera–a visualization system for exploratory research and analysis. J. Comput. Chem. 25, 1605–1612 [DOI] [PubMed] [Google Scholar]
- 46. Schnick C., Polley S. D., Fivelman Q. L., Ranford-Cartwright L. C., Wilkinson S. R., Brannigan J. A., Wilkinson A. J., Baker D. A. (2009) Structure and nonessential function of glycerol kinase in Plasmodium falciparum blood stages. Mol. Microbiol. 71, 533–545 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Richard P., Toivari M. H., Penttilä M. (2000) The role of xylulokinase in Saccharomyces cerevisiae xylulose catabolism. FEMS Microbiol. Lett. 190, 39–43 [DOI] [PubMed] [Google Scholar]
- 48. Hahn-Hägerdal B., Wahlbom C. F., Gárdonyi M., van Zyl W. H., Cordero Otero R. R., Jönsson L. J. (2001) Metabolic engineering of Saccharomyces cerevisiae for xylose utilization. Adv. Biochem. Eng. Biotechnol. 73, 53–84 [DOI] [PubMed] [Google Scholar]
- 49. Wu J., Serianni A. S., Vuorinen T. (1990) Furanose ring anomerization: kinetic and thermodynamic studies of the d-2-pentuloses by 13C n.m.r. spectroscopy. Carbohydr. Res. 206, 1–12 [DOI] [PubMed] [Google Scholar]
- 50. Bais R., James H. M., Rofe A. M., Conyers R. A. (1985) The purification and properties of human liver ketohexokinase, a role for ketohexokinase and fructose bisphosphatase aldolase in the metabolic production of oxalate from xylitol. Biochem. J. 230, 53–60 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Holmes R. P., Assimos D. G. (1998) Glyoxylate synthesis and its modulation and influence on oxalate synthesis. J. Urol. 160, 1617–1624 [PubMed] [Google Scholar]
- 52. Conyers R. A., Bais R., Rofe A. M. (1990) The relation of clinical catastrophes, endogenous oxalate production, and urolithiasis. Clin. Chem. 36, 1717–1730 [PubMed] [Google Scholar]
- 53. Veech R. L. (2003) A humble hexose monophosphate pathway metabolite regulates short- and long-term control of lipogenesis. Proc. Natl. Acad. Sci. U.S.A. 100, 5578–5580 [DOI] [PMC free article] [PubMed] [Google Scholar]