Background: Collagen prolyl 4-hydroxylases (C-P4H) are involved in the formation of extracellular matrices.
Results: The full complement of C-P4H enzymes from the human infective parasite Brugia malayi have been bioinformatically, biochemically, and functionally characterized.
Conclusion: C-P4H enzymes are essential for development in B. malayi.
Significance: Unique features of these essential enzymes may be exploited in future control mechanisms.
Keywords: C. elegans, Collagen, Extracellular matrix, Extracellular matrix proteins, Hydroxylase, Hydroxyproline, Parasitology, Post translational modification, Protein synthesis
Abstract
Collagen prolyl 4-hydroxylases (C-P4H) are required for formation of extracellular matrices in higher eukaryotes. These enzymes convert proline residues within the repeat regions of collagen polypeptides to 4-hydroxyproline, a modification essential for the stability of the final triple helix. C-P4H are most often oligomeric complexes, with enzymatic activity contributed by the α subunits, and the β subunits formed by protein disulfide isomerase (PDI). Here, we characterize this enzyme class in the important human parasitic nematode Brugia malayi. All potential C-P4H subunits were identified by detailed bioinformatic analysis of sequence databases, function was investigated both by RNAi in the parasite and heterologous expression in Caenorhabditis elegans, whereas biochemical activity and complex formation were examined via co-expression in insect cells. Simultaneous RNAi of two B. malayi C-P4H α subunit-like genes resulted in a striking, highly penetrant body morphology phenotype in parasite larvae. This was replicated by single RNAi of a B. malayi C-P4H β subunit-like PDI. Surprisingly, however, the B. malayi proteins were not capable of rescuing a C. elegans α subunit mutant, whereas the human enzymes could. In contrast, the B. malayi PDI did functionally complement the lethal phenotype of a C. elegans β subunit mutant. Comparison of recombinant and parasite derived material indicates that enzymatic activity may be dependent on a non-reducible covalent link, present only in the parasite. We therefore demonstrate that C-P4H activity is essential for development of B. malayi and uncover a novel parasite-specific feature of these collagen biosynthetic enzymes that may be exploited in future parasite control.
Introduction
The parasitic filarial nematode Brugia malayi is one of the causative agents of lymphatic filariasis, a condition affecting around 120 million people in 73 countries worldwide (1, 2). The B. malayi genome has been sequenced and is similar in size and gene number to the free-living nematode Caenorhabditis elegans (3). All nematodes display similar body plans and are encased in a collagenous exoskeleton, known as the cuticle, which covers the outermost layer of epithelial cells. We have previously demonstrated in C. elegans that collagen prolyl 4-hydroxylase (C-P4H)3 (EC 1.14.11.2) and protein disulfide isomerase (PDI) (EC 5.3.4.1) activities are essential for nematode development due to their cuticle collagen modification function (4–6). We therefore examined whether the B. malayi forms of these enzymes were similarly required for development and as such would represent effective anti-parasitic drug targets.
The nematode cuticle is a collagen-rich extracellular matrix, which is required for maintenance of body morphology and interaction with the environment (7). The collagens that constitute the cuticle are formed via a multi-step process of intra- and extracellular post-translational enzymatic modifications (reviewed in Ref. 7). An essential step in this process is catalyzed by C-P4H and occurs within the endoplasmic reticulum where secreted proteins are folded. C-P4H converts peptide bound proline residues in the X-Pro-Gly repeat region of procollagen polypeptides to 4-hydroxyproline. Mature collagen triple helices that lack this modification are thermally unstable at physiological temperatures. In most species examined, C-P4H are oligomeric complexes with the C-P4H active sites present in the α subunits and the enzyme PDI present in the β subunit (8). PDI is also found at high levels free from the C-P4H complex and functions to ensure correct disulfide bond formation and, therefore folding, of many proteins, including collagens, which transit the endoplasmic reticulum (6).
The form and function of collagen C-P4H have been reviewed in detail (8) and are summarized in supplemental Fig. S1. Humans possess three catalytically active α subunits (named P4HA1, P4HA2, and P4HA3), each of which form α2β2 complexes containing matching α subunits. The β subunit, which is common to all forms of the human complexes, is PDI (P4HB). In C. elegans, there are two α subunits (named DPY-18 and PHY-2) and a β subunit (PDI-2) that also form three C-P4H complexes. However, unlike humans, these consist of a mixed α subunit tetramer, [DPY-18][PHY-2][PDI-2]2, and two dimers, [DPY-18][PDI-2] and [PHY-2][PDI-2] (5). The diversity of complexes formed in nematodes is highlighted in Caenorhabditis briggsae, a close relative of C. elegans, where, in addition to a mixed α subunit tetramer and [DPY-18][PDI-2] dimer identical to those found in C. elegans, a [PHY-2]2[PDI-2]2 tetramer can also form (9).
Nematodes produce a new cuticle for each developmental stage, and in C. elegans, the C-P4H complex encoding genes are expressed almost exclusively in the cuticle collagen secreting cells of the hypodermis in waves of abundance that correspond to that of collagen (4). C-P4H activity is developmentally required in C. elegans; worms that have lost the α subunit DPY-18 are viable but show defects at the levels of worm body morphology, cuticle structure, and collagen localization (4). Complete combined loss of both α subunits, DPY-18 and PHY-2, or complete single loss of the β subunit, PDI-2, results in embryonic lethality (4, 6). In C. briggsae, C-P4H function is similarly essential (9). The phenotypes resulting from loss of C-P4H can be replicated using enzyme inhibitors (5).
The in vivo action of the C. elegans C-P4H β subunit PDI-2 has been examined in detail with incomplete loss of function producing sterile adults with severely abnormal body morphology, cuticle structure, and collagen localization (6). Interestingly, although it was not possible to analyze separately the C-P4H-dependent and -independent roles of PDI-2, intact PDI-2 thioredoxin active site residues are essential for correct development (6). In addition, loss of C. elegans PDI-2 could be completely rescued by human PDI (P4HB), suggesting it may be possible to heterologously express B. malayi PDIs in C. elegans to examine their function.
The likely importance of C-P4H activity during B. malayi development and cuticle collagen modification is suggested by several lines of evidence. Chemical inhibition of C-P4H produces cuticle associated defects and a reduction in 4-hydroxyproline levels in cultured B. malayi adults (10), whereas for cultured third stage larvae, the essential C-P4H co-factor ascorbate is necessary for correct molting and development (11). Furthermore, adult B. malayi cuticle collagens contain 7.9% hydroxyproline (40% of prolines are hydroxylated) (12), a level comparable with that found in C. elegans (12% hydroxyproline, 51% of prolines are hydroxylated) (13).
We have previously characterized a protein from B. malayi with homology to C-P4H α subunits, Bma-PHY-1, which showed a number of unusual properties (14). Recombinant α subunits expressed alone are in general insoluble, and therefore, complexes are usually produced by co-expression of α and β subunits in systems such as insect cells. In most cases, complexes can be formed by combining α and β subunits from different species. However, when Bma-PHY-1 was expressed in an insect cell expression system, it was soluble without a β subunit, did not form complexes with β subunits from other species, and instead formed an active homotetramer (14). Despite this in vitro activity, when Bma-PHY-1 was expressed in C. elegans, it was not capable of replacing the in vivo functions of the α subunit DPY-18 (14). We therefore speculated that either additional C-P4H α subunit-like genes were present in B. malayi, or, that only complexes containing native partner protein(s) would produce fully functional enzyme. In this study, we utilized the B. malayi genome sequence (3) to ensure identification of all potential C-P4H subunits and used a recently developed approach for RNAi in this parasite (15) to demonstrate that these genes are required for development and formation of the cuticle. In addition, we uncovered a novel parasite-specific non-reducible covalent modification required for association of this critical enzyme complex.
EXPERIMENTAL PROCEDURES
Procedures for RNA extraction, cDNA synthesis, genomic DNA isolation, plasmid construction, single worm PCR, and RT-PCR assessment of transgene expression in C. elegans are provided in the supplemental “Experimental Procedures.” All primer names, sequences, and descriptions of their uses are given in supplemental Table S1. The abbreviations Bma and Cel are used throughout before gene and protein names to indicate B. malayi and C. elegans respectively.
Nematode Strains, Culture, and Microscopy
C. elegans strains used in this study were N2 (wild type), CB364 (dpy-18(e364) III) (4), and TP69 (pdi-2(tm0689)/lon-2(e678) X) (6). The pdi-2(tm0689) allele was maintained by selecting pdi-2(tm0689) heterozygotes, which are the only phenotypically wild type progeny segregating from strain TP69. C. elegans strains were cultured as described previously (16). B. malayi nematodes were kindly provided by Rick Maizels and Yvonne Harcus (University of Edinburgh). For microscopy, live C. elegans were transferred to slides with a 2% agarose, 0.065% sodium azide pad. B. malayi were prepared for microscopy by transferring microfilaria to slides with a coverslip and heating the slides to 60 °C on a hot plate for 10 s to immobilize the larvae. All images were captured using an Axioskop 2 microscope (Zeiss) and an AxioCam MRm camera (Zeiss).
RNAi of B. malayi by Soaking and C. elegans by Feeding
For RNAi of B. malayi, Bma-phy-1, Bma-phy-2, Bma-pdi-2, and Cel-phy-3 (negative control) were targeted following previously described methods (15). Briefly, in vitro transcription templates were produced by linearizing the Bma-phy-1, Bma-phy-2, Bma-pdi-2, and Cel-phy-3 RNAi plasmids each in two separate reactions (using the cloning restriction enzymes) and single-stranded RNAs produced using a MEGAscript kit (Ambion). Equal quantities of the complementary single stranded RNAs were then annealed and the long double-stranded RNA digested with ShortCut RNase III (New England Biolabs) to produce heterogeneous short-interfering RNAs (hsiRNAs). Freshly extracted B. malayi adult females were cultured in 24-well plates containing two females per well in 1 ml of culture medium. Medium for culture of B. malayi was RPMI 1640 (with glutamine and HEPES) (Invitrogen), 1% glucose, 100 units/ml penicillin and 100 μg/ml streptomycin (Invitrogen), and 10% heat-inactivated fetal calf serum (Invitrogen). Following an overnight culture period in medium alone, the parasites were exposed to RNAi for a total of 48 h by adding fresh medium containing hsiRNAs, at a final concentration of 0.5–2 μm, at the start of the culture and again after 24 h. For RNAi of C. elegans lines expressing Bma-pdi-2, the Bma-pdi-2 RNAi plasmid was transformed into bacterial strain HT115(DE3) (17) (HT115(DE3) available from the Caenorhabditis Genetics Center). RNAi by feeding was performed as described previously (18). As a negative control all processes were replicated using control RNAi bacteria containing the RNAi vector without an insert.
Database Searches for Additional B. malayi phy Genes
tBLASTn was performed on whole genome shotgun sequences using Bma-PHY-1 and Bma-PHY-2 (maximum values selected for “alignment” and “descriptions” options). The genomic sequences of Bma-phy-1 and -2 were then used as BLASTn queries against the same database. Those hits that showed complete nucleotide identity to each gene were considered to represent that gene (to identify any potential new Bma-phy, this analysis assumes some degree of sequence divergence). This enabled all whole genome shotgun sequences with amino acid homology to PHYs (tBLASTn results) to be assigned as being derived from either Bma-phy-1 (complete nucleotide identity to Bma-phy-1) or Bma-phy-2 (complete nucleotide identity to Bma-phy-2). The BLASTn results were then subtracted from the tBLASTn results to determine if any whole genome shotgun sequences were present which were not derived from the known genes.
Expression of B. malayi Proteins in Insect Cells
Recombinant vectors were co-transfected into Spodoptera frugiperda Sf9 cells with a modified Autographa californica nuclear polyhedrosis virus DNA (BaculoGold, BD Pharmingen) by calcium phosphate precipitation. Sf9 or High Five (Invitrogen) insect cells were cultured as monolayers in TNM-FH (modified Grace's insect cell medium, Sigma) supplemented with 10% fetal bovine serum (BioClear) or in suspension in Sf900IISFM serum-free medium (Invitrogen). The cells were seeded at a density of 5 × 106 cells/100-mm plate or 1 × 106 cells/ml and infected at a multiplicity of 5 for either single or multiple virus coding for B. malayi proteins. Cells were harvested 72 h after infection, washed with a solution of 0.15 m NaCl and 0.02 m phosphate, pH 7.4, homogenized in a 0.1 m NaCl, 0.1 m glycine, 10 μm dithiothreitol, 0.1% Triton X-100, and 0.01 m Tris buffer, pH 7.4, and centrifuged at 10,000 × g for 20 min. The pellets were further solubilized in 1% SDS. C-P4H activity was assayed by a method based on the hydroxylation-coupled decarboxylation of 2-oxo-[1-14C]glutarate, and Km and Ki values were determined as described previously (19).
Transgenic Expression in C. elegans
Transgenic C. elegans carrying extrachromosomal arrays were generated by microinjection following standard protocols (20) with at least three independent lines analyzed in all cases. Repetitive extrachromosomal arrays were produced by injecting circular plasmids. These were performed by co-injecting the test plasmid at 2–25 μg/ml, the transformation marker dpy-7prom::gfp plasmid (from Iain Johnstone, University of Glasgow) at 5 μg/ml, with pBluescript SKM (Stratagene) added to give a total plasmid concentration of 150 μg/ml. To produce complex extrachromosmal arrays (21), plasmids were linearized by restriction digest and co-injected with fragmented C. elegans genomic DNA. Genomic DNA was prepared for injection by digestion with PvuII followed by phenol/chloroform extraction and ethanol precipitation. The dpy-7prom::gfp transformation marker was digested with FspI, and plasmids for expression of B. malayi or human genes were digested with blunt cutting enzymes where possible or were subsequently treated with DNA polymerase I large (Klenow) fragment (New England Biolabs) (see supplemental Table S2). All linearized plasmids were purified using a QIAquick PCR purification kit. Injections to produce complex arrays were performed by co-injecting the linearized plasmid(s) each at 1 μg/ml, the transformation marker at 2 μg/ml, and fragmented C. elegans genomic DNA at 100 μg/ml. For unknown reasons, direct introduction of complex arrays by injection of the dpy-18 mutant strain CB364 resulted in high toxicity, even in the absence of B. malayi PHY and PDI expression constructs. Therefore, in these cases, injections were performed in the C. elegans wild type N2 strain, and the transgenic extrachromosomal array subsequently crossed into the dpy-18 mutant strain.
Nematode Extracts
B. malayi parasite extracts were made by resuspending adults in extraction buffer (0.1 m NaCl, 0.1 m glycine, 10 μm dithiothreitol, 0.1% Triton X-100, and 10 mm Tris, pH 8.0, supplemented with protease inhibitors 1 mm phenylmethylsulfonyl fluoride, 1 mm EDTA, 1 mm EGTA, 2 μm E64, and 0.1 μm pepstatin) and disrupting using a 3-ml glass hand-held homogenizer (Jencons). To examine the expression of transgenic proteins, C. elegans extracts were made by adding 50 transgenic worms to 30 μl of 1XM9 (16) with 12 μl of 4× SDS sample buffer and freezing the samples at −80 °C. For reducing gels, 5% (v/v) 2-mercaptoethanol was added, and the samples were boiled before loading.
Western Blotting
Electrophoresis and Western blotting of nematode and insect cell-derived material was carried out using Bio-Rad equipment as described by the manufacturer, with Tris-HCl gels (Bio-Rad) and Hybond-P PVDF membrane (GE Healthcare). Bma-PHY-1 was detected using an anti-peptide antibody described previously (14). An anti-peptide antibody was raised to the C-terminal sequence of Bma-PHY-2 (CLGAPEPKRHLNIRSEKARK, the underlined cysteine was added to couple the peptide to keyhole limpet hemocyanin) (Sigma). An anti-peptide antibody raised previously to Cel-PDI-2 (5) was found to cross-react with Bma-PDI-2. Secondary antibodies for PHY and PDI primaries were anti-rabbit IgG (whole molecule) peroxidase (goat) (Sigma) or anti-rabbit IgG (whole molecule) alkaline phosphatase (Sigma). Stripped blots were reprobed for β-actin using monoclonal anti-β-actin clone AC-15 (Sigma) and anti-mouse IgG HRP conjugate (Promega). Peroxidase conjugates were detected with Amersham Biosciences ECL Plus Western blotting detection system (GE Healthcare), and alkaline phosphatase conjugates with SigmaFast BCIP/NBT (Sigma).
Co-immunoprecipitation (co-IP)
The peptide used to raise the anti-Cel-PDI-2 antibody (5) was linked to a beaded agarose support, and the antibody was affinity purified using a SulfoLink immobilization kit (Thermo Scientific). Purified anti-Cel-PDI-2 antibody was dialyzed using a Slide-A-Lyzer dialysis cassette (Thermo Scientific) and the Pierce co-immunoprecipitation kit (Thermo Scientific) protocol was followed to immobilize the antibody to an agarose support and perform co-IP using adult B. malayi lysate. Lysate was generated by resuspending adult parasites in IP lysis buffer containing halt protease and phosphatase inhibitor mixture (Thermo Scientific) and disrupting using a 1-ml Radnoti Econo-Grind Micro Tissue Grinder. Control co-IP reactions were performed in parallel using isotype-matched, affinity-purified antibody reactive to C. elegans BLI-5.
RESULTS
Cloning of Bma-phy-2 and Bma-pdi-2
We had previously cloned and characterized a B. malayi protein, Bma-PHY-1, with C-P4H activity (14) (cDNA and genomic accession nos. AJ297845 and AJ421993). Prior to publication of the complete annotated B. malayi genome (3), we searched for further proteins with homology to C-P4H α subunits (known as PHY in nematodes). Sequences of an additional PHY-encoding gene, which was distinct from Bma-phy-1, were identified in the B. malayi genome preassembly using tBLASTn. The complete sequence was identified by finding overlapping sequences and identifying coding regions with homology to known PHYs. The full-length cDNA was verified by PCR amplification, cloning, and sequencing (accession no. AJ628347) with the 5′ end verified by RT-PCR utilizing the trans-spliced leader SL1 sequence (22) (data not shown).
An EST (accession no. AI784701) containing a partial sequence for a B. malayi pdi (named Bma-pdi-2 for reasons described in later sections) was identified by tBLASTn at EBI using Cel-PDI-2 as a query. The sequences identified enabled PCR screening of lysates from a BAC library (kindly provided by Mark Blaxter, University of Edinburgh) and sequencing of a positive BAC provided the full-length Bma-pdi-2 genomic sequence (accession no. AJ577085). A full-length cDNA was generated by PCR (accession no. AJ577086), and the 5′ end confirmed by SL1-RT-PCR (data not shown). In the annotated B. malayi genome (3), the genes representing Bma-pdi-2 (Bm1_39250) and Bma-phy-2 (Bm1_45460) both have inaccurately predicted splicing, and therefore, all work here refers to our experimentally confirmed sequences and the accession numbers for those that are listed above.
Bma-phy-2 encodes a 551-amino acid protein with a predicted signal peptide (23) occurring between residues 17 and 18 (VSA-DS) and a single predicted N-glycosylated asparagine. The mature protein shows 51.9% identity and 69.1% similarity with Bma-PHY-1 (Table 1), and is most similar to Onchocerca volvulus PHY-1 (10), with which it shows 69.3% identity and 77.0% similarity. As described for Bma-PHY-1 (14), all residues identified as being required for protein structure, complex formation, and enzyme activity (24–26) are conserved in Bma-PHY-2 (supplemental Fig. S2). Bma-pdi-2 encodes a 503-amino acid protein with a predicted signal peptide occurring between residues 22 and 23 (AHD-AS) and an endoplasmic reticulum retention signal at the C terminus. Bma-PDI-2 shows higher homology to C-P4H β subunit PDIs (Table 2) than to other PDIs. The degree of conservation of residues defined previously as the active sites (27), active site role charged residues (28), hydrophobic ligand binding site (29), and residues involved in C-P4H complex formation (30), is shown in supplemental Fig. S3.
TABLE 1.
Amino acid similarity and identity of C-P4H α subunits
| Cel-PHY-2 | Bma-PHY-1 | Bma-PHY-2 | Hsa-P4HA1 | Hsa-P4HA2 | |
|---|---|---|---|---|---|
| Cel-DPY-18 | 53.7/65.6a | 57.1/70.9 | 53.1/66.3 | 40.7/56.1 | 39.4/54.7 |
| Cel-PHY-2 | 55.1/67.5 | 47.6/63.3 | 44.8/58.8 | 43.9/58.9 | |
| Bma-PHY-1 | 51.9/69.1 | 43.0/55.5 | 42.3/56.7 | ||
| Bma-PHY-2 | 39.1/55.1 | 39.1/54.7 | |||
| Hsa-C-P4HA1 | 63.4/77.0 |
a Identity/similarity.
TABLE 2.
Amino acid similarity and identity of C-P4H β subunits
| Bma-PDI-2 | Hsa-PDI (P4HB) | |
|---|---|---|
| Cel-PDI-2 | 71.7/82.7a | 54.3/68.9 |
| Bma-PDI-2 | 56.7/67.7 |
a Identity/similarity.
Database Searches Indicate Bma-phy-1 and Bma-phy-2 Are the Only B. malayi phy Genes
Analyzing the available completed genome sequences for other parasitic nematodes we noted that although the Trichinella spiralis genome (31) is likely to encode only two PHYs (results not shown), an expanded family of five PHY-encoding genes exists in the compact genome of the plant parasite Meloidogyne hapla (32),4 and Ascaris suum, which, similar to B. malayi, is an animal parasite in nematode clade III, has three PHY-encoding genes (accession nos. ADY44523, ADY43128, and ADY44875) (33, 34). A careful analysis of the B. malayi genome was therefore undertaken to determine whether any additional PHY-encoding genes, other than Bma-phy-1 and -2, were present in this organism. tBLASTn analysis of the assembled B. malayi scaffolds using PHY protein sequences did not reveal any additional Bma-phy genes, other than Bma-phy-1 and -2 and a Bma-phy gene fragment. Bma-phy-1 and -2 are arranged in tandem in the genome with the Bma-phy gene fragment situated between them (supplemental Fig. S4 and supplemental “Experimental Procedures”). tBLASTn and BLASTn was then carried out using the whole genome shotgun database so that sequences that may not have assembled into scaffolds could also be examined. The BLASTn results were used to identify sequences that showed complete nucleotide identity with the known genes (and were therefore assumed to represent these genes) and were subtracted from the tBLASTn results. This analysis did not reveal any sequences with homology to PHY proteins that were not completely identical to the known Bma-phy genes at the nucleotide level. Similar analysis of the B. malayi BAC end sequences held at TIGR and the Sanger Institute, EST sequences at EBI, and NematodeNet, and analysis of genomic scaffolds from a resequenced B. malayi genome (kindly provided prepublication by Elodie Ghedin, University of Pittsburgh) also failed to uncover any sequences considered to represent new genes. In addition, sequencing of an ∼2-kb genome assembly gap, situated between the Bma-phy-1 and -2 genes and downstream of the Bma-phy gene fragment region, revealed no further PHY-encoding genes (see supplemental “Experimental Procedures”). Therefore, as exhaustive searches of multiple databases revealed no additional genes encoding proteins with homology to C-P4H α subunits, Bma-phy-1 and Bma-phy-2 represent the only identifiable genes of this type in all databases and are very likely to encode the only PHYs in this organism.
Expression Profiling
The expression of Bma-phy and Bma-pdi-2 genes was examined in a range of developmental stages by RT-PCR. In each case, the test gene was amplified simultaneously along with the constitutively expressed tubulin gene Bma-tub-1. Expression in the first larval stage (termed microfilaria) of Bma-phy-2, Bma-pdi-2, and Bma-phy-1 is shown in Fig. 1A, with Bma-phy-2 found at a lower level than Bma-pdi-2 and Bma-phy-1. A panel of cDNAs used previously to investigate the expression of Bma-phy-1 (14), representing stages from post-infective third stage larvae (L3) to juvenile adult, was then employed for Bma-phy-2 and Bma-pdi-2. The Bma-phy-2 message was predominantly found in the early and mid L3 stages and was barely detectable in the late fourth stage larvae (L4) and adult stages (Fig. 1B). In contrast, the Bma-pdi-2 message was abundant in all stages examined, displaying a very similar profile to that shown previously for Bma-phy-1 (14).
FIGURE 1.
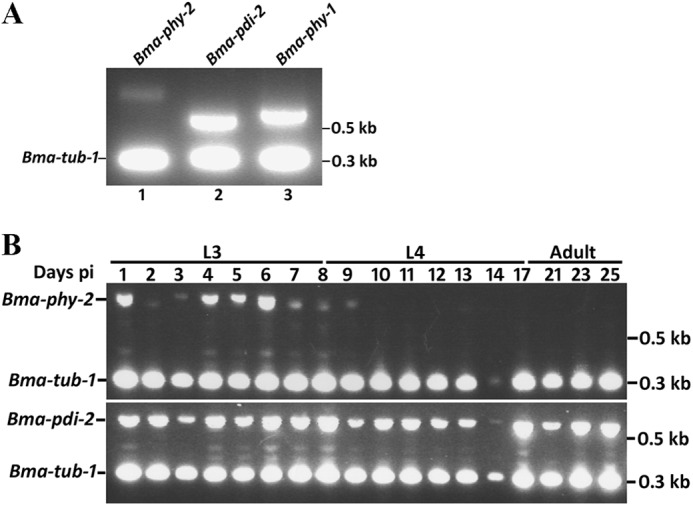
Developmental expression profiles of B. malayi C-P4H genes assessed by RT-PCR. For each gene, simultaneous amplifications were performed using gene-specific primers and primers to B. malayi tubulin (Bma-tub-1) (accession no. AJ551180) as an internal control. A, expression of Bma-phy-2, Bma-pdi-2, and Bma-phy-1 in microfilaria (first stage larvae). B, expression of Bma-phy-2 and Bma-pdi-2 in post-infective stages. Parasite material was taken from the time points indicated post-infection (days pi) from infected jirds. L3 and L4 denote larval stages 3 and 4, and Adult indicates juvenile adults. Sizes of cDNA amplicons were as follows: Bma-phy-2, 808 bp; Bma-pdi-2, 597 bp; Bma-phy-1, 655 bp; Bma-tub-1, 308 bp. Sizes of genomic amplicons, Bma-phy-2, 3529 bp; Bma-pdi-2, 1301 bp; Bma-phy-1, 1909 bp; Bma-tub-1, 390 bp.
RNAi of Cultured B. malayi
To investigate the functional roles of our identified genes during B. malayi development, we employed a recently described RNAi approach (15). In contrast to previously described RNAi studies in animal parasitic nematodes (reviewed in Ref. 35), this approach uses a pool of hsiRNAs generated enzymatically from a long double-stranded RNA template, to give robust, reproducible RNAi. The effect of targeting Bma-phy-1, Bma-phy-2, and Bma-pdi-2 by RNAi was determined by culturing adult females in the presence of hsiRNAs corresponding to each gene singly, as well as Bma-phy-1 and Bma-phy-2 in combination. For each RNAi, multiple pairs of adult females (at least four groups of two females over two experiments) were treated, and the viability and morphology of the first stage larvae (microfilaria) produced was analyzed. Applying either Bma-phy-1 or Bma-phy-2 RNAi singly at 1 μm produced no visible effects on either the females or the microfilaria produced (Fig. 2, A and B). Bma-pdi-2 RNAi at 1 μm (data not shown) and especially 2 μm produced mutant microfilaria that were immotile and presented with a coiled lumpy appearance (Fig. 2C). The first larval cuticle surrounding the microfilaria is highly irregular, whereas the extracuticular microfilarial sheath, a remnant of the eggshell, remained intact. This phenotype was more penetrant after the second day of culture. A practically identical highly penetrant microfilarial mutant phenotype was found by simultaneous RNAi of Bma-phy-1/Bma-phy-2 at a combined concentration of 1 μm (each gene at 0.5 μm) (Fig. 2, D and E) and was also observed at a combined concentration of 0.5 μm and at 2 μm (data not shown). Untreated controls with no hsiRNA added and controls treated with 2 μm hsiRNA from Cel-phy-3 (Fig. 2F) (36), for which no homologous sequences were identifiable in the B. malayi genome, were included in all cases.
FIGURE 2.
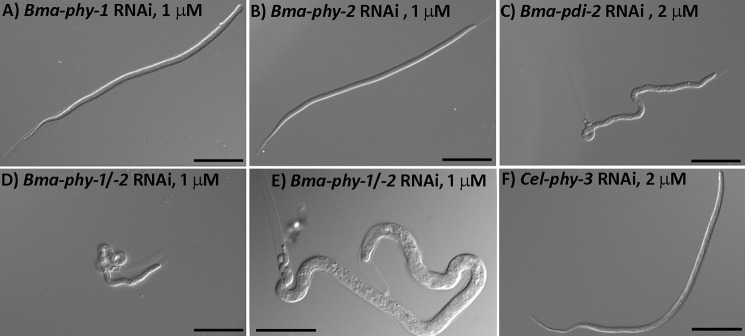
RNAi effects in B. malayi microfilariae (first stage larvae). Cultured adult female B. malayi were treated with hsiRNA derived from Bma-phy-1 (A), Bma-phy-2 (B), and Bma-pdi-2 (C), and the effects were determined in the microfilariae produced. Single targeting of Bma-pdi-2 shows severe morphological defects. D and E depict the severe morphological defects resulting from the simultaneous targeting of Bma-phy-1 and -2. F, Cel-phy-3 (negative control). All images are of microfilariae produced from females treated with hsiRNA at a concentration of 1–2 μm over a 48-h period. Scale bars represent 50 μm in A–D and F and 25 μm in E.
Expression of B. malayi Proteins in Insect Cells
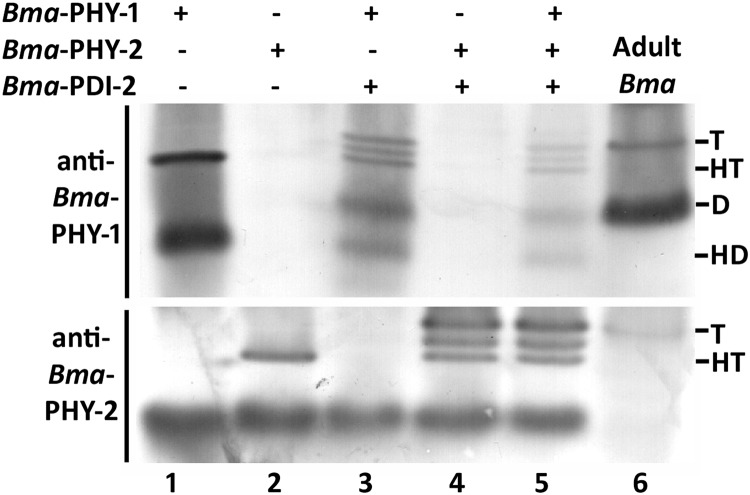
Recombinant baculoviruses were produced containing Bma-phy-2 and Bma-pdi-2 singly. These were used, along with a recombinant virus for Bma-phy-1 generated previously (14), to investigate the enzymatic activity and protein-protein interactions of these presumed C-P4H subunits. Insect cells were then infected either with a single recombinant virus or with combinations of viruses. Cells were harvested 72 h after infection and extracts analyzed by Western blotting of native PAGE gels with peptide-specific antibodies. C-P4H activity of extracted material was assessed using a method based on the hydroxylation-coupled decarboxylation of radiolabeled co-substrate and a defined artificial substrate (19). These methods have been used to determine recombinant C-P4H activities from a range of species (8). The insect cell extracts were compared with parasite-derived extracts (Fig. 3, lane 6) with the designation of the type of complex being based on comparisons with previously characterized forms of the enzyme (5, 14). Sole expression of Bma-PHY-1 produced a soluble homotetramer and homodimer as described previously (Fig. 3, lane 1) (14), whereas single expression of Bma-PHY-2 resulted in a soluble protein that formed a higher order structure, considered to be a homotetramer (Fig. 3, lane 2). The homotetramer was found to be enzymatically inactive (data not shown). To investigate whether Bma-PHY-1 and -2 required a PDI from B. malayi to produce a fully active C-P4H enzyme complex, each subunit was co-expressed with Bma-PDI-2. For Bma-PHY-1, co-expression with Bma-PDI-2 resulted in an additional dimer and a band running above the homotetramer (Fig. 3, lane 3), both of which show the same mobility as the two bands from the parasite extract (Fig. 3, upper panel, lane 6). For Bma-PHY-2, co-expression with Bma-PDI-2 produced a band running above the homotetramer that was equivalent to the band in parasite extract (Fig. 3, lower panel, lane 6). Co-expression of Bma-PDI-2 along with either Bma-PHY-1 or -2 had no effect on enzymatic activity (data not shown). Finally, to determine whether, similar to C. elegans, C-P4H tetramer formation required the presence of all three proteins, Bma-PHY-1, -2 and Bma-PDI-2 were all co-expressed. Surprisingly, the availability of all identified potential C-P4H subunits had no additional effect on complex formation (Fig. 3, lane 5) or enzymatic activity (data not shown). A low molecular weight nonspecific band was noted in Western blots using the anti-Bma-PHY-2 antibody in insect cell derived material but not in the parasite extracts (Fig. 3).
FIGURE 3.
Western blot (native PAGE) of Triton X-100 extracts of B. malayi proteins expressed in insect cells. Bma-PHY-1 and Bma-PHY-2 were expressed singly (lanes 1 and 2), co-expressed with Bma-PDI-2 (lanes 3 and 4), and triple-expressed (lane 5). The complexes produced are compared with B. malayi adult parasite material (lane 6, Adult Bma). The size of homotetramer (HT), homodimer (HD), heterodimer (D), and higher molecular weight heterotetramer (T) bands are indicated.
Bma-pdi-2 Can Replace the Essential Function of C. elegans pdi-2
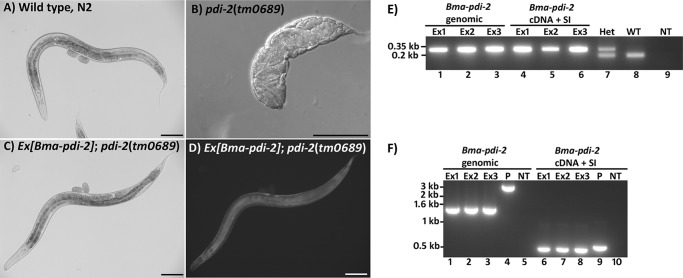
In vertebrates and C. elegans, a single PDI is the C-P4H β subunit. We have previously shown that the human C-P4H β subunit PDI (P4HB) could functionally substitute for the C. elegans C-P4H β subunit PDI (Cel-PDI-2) (6). Although examination of the B. malayi genome suggested that Bma-PDI-2 was the only likely orthologue of Cel-PDI-2 (data not shown), we looked to confirm this in vivo. A C. elegans pdi-2 mutant strain described previously (6) has severe body morphology defects and results in sterile adults. The ability of Bma-pdi-2 to functionally substitute for loss of Cel-pdi-2 was tested by expressing Bma-pdi-2 in this mutant background. Two Bma-pdi-2 expression plasmids, one genomic and one cDNA (with synthetic intron, SI), were made using vector pAW2 (Cel-pdi-2 promoter and Cel-dpy-18 3′ UTR (6)). Striking rescue of the severe phenotype of the Cel-pdi-2 mutant was achieved using complex extrachromosomal arrays carrying either of the Bma-pdi-2 expression plasmids (Fig. 4, A–D). In C. elegans, extrachromosmal arrays are lost naturally in a percentage of the progeny, and viability of the Bma-pdi-2 rescued transgenic lines was completely dependent on the presence of the extrachromosmal array, with animals that lost the array all dying during embryogenesis. SW-PCR was performed to ensure the Bma-pdi-2 expression plasmids were present (data not shown), and the transgenic lines were genotyped to confirm they were Cel-pdi-2(tm0689) mutant homozygotes (Fig. 4E). Genotyping also ensured that no wild type Cel-pdi-2 sequences had been introduced with the co-injected fragmented C. elegans genomic DNA used to generate the complex arrays. RT-PCR was performed to verify Bma-pdi-2 expression (Fig. 4F). Transgenic lines carrying only the transformation marker and fragmented genomic DNA failed to rescue. Finally, the rescued transgenic lines were RNAi-treated with a construct targeting Bma-pdi-2. As shown in Table 3, viability was completely dependent on transgenic Bma-pdi-2, confirming that rescue was due entirely to the introduced B. malayi gene.
FIGURE 4.
A–D, Bma-pdi-2 introduced into a C. elegans pdi-2 null mutant results in complete repair of the mutant phenotypes. A, wild type C. elegans N2 strain; B, C. elegans pdi-2 homozygote mutant (from a heterozygous mother); C and D, C. elegans pdi-2 homozygote mutant carrying Bma-pdi-2 on an extrachromosomal array with D showing GFP expression from the transformation marker dpy-7prom::gfp carried on the same extrachromosomal array. Scale bars represent 100 μm. E, genotyping by single-worm PCR. A set of three primers designed around the Cel-pdi-2(tm0689) deletion were use to genotype all transgenic lines to ensure mutant homozygosity. The upper 320-bp band identifies the deleted region, and the 185-bp band indicates the wild type. The three transgenic lines (Ex1–Ex3) generated for each construct, Bma-pdi-2 genomic and Bma-pdi-2 cDNA + synthetic intron (SI), all produce only the deletion sized amplicon. Lane 7, the larger deletion-derived amplicon and smaller wild type-derived amplicon from a pdi-2(tm0689) heterozygote (Het). Lane 8 is wild type (WT), and lane 9 represents no template control (NT). F, RT-PCR showing expression of Bma-pdi-2 in transgenic C. elegans. Expression is shown for three transgenic lines (Ex1–Ex3) for each Bma-pdi-2 construct and Bma-pdi-2 genomic and Bma-pdi-2 cDNA + synthetic intron (SI). For the Bma-pdi-2 genomic construct, primers amplifying the full-length Bma-pdi-2 sequence were used. Lanes 1–3 show expression of the 1512-bp spliced Bma-pdi-2 cDNA in lines carrying the genomic construct. Lane 4, the 3579-bp amplicon generated using the Bma-pdi-2 genomic plasmid (P) as template. Lane 5, no template (NT) control. For the Bma-pdi-2 cDNA + synthetic intron (SI) construct, primers corresponding to Bma-pdi-2 sequences situated either side of the 51-bp synthetic intron were used. Lanes 6–8 show the 486-bp spliced amplicon derived from three transgenic C. elegans lines compared with the 537-bp plasmid-derived product (P, lane 9) and no template (NT) (lane 10). Bma-pdi-2 from genomic construct; spliced, 1512 bp; unspliced, 3579 bp. Bma-pdi-2 from construct with cDNA and a synthetic intron; spliced, 486 bp; unspliced, 537 bp.
TABLE 3.
RNAi of C. elegans pdi-2 mutants carrying Bma-pdi-2 transgenes
| Construct/line |
Bma-pdi-2 RNAi |
Control |
||
|---|---|---|---|---|
| Hatched | Unhatcheda | Hatchedb | Unhatchedc | |
| Bma-pdi-2 genomic | ||||
| Ex1 | 0 | 222 | 57 | 181 |
| Ex2 | 0 | 215 | 78 | 96 |
| Ex3 | 0 | 80 | 63 | 39 |
| Bma-pdi-2 cDNA + SI | ||||
| Ex1 | 0 | 184 | 150 | 80 |
| Ex2 | 0 | 181 | 62 | 119 |
| Ex3 | 0 | 23 | 1 | 15 |
| Total | 0 (0%) | 905 (100%) | 411 (43.7%) | 530 (56.3%) |
a Unhatched consisted of GFP-positive and -negative embryos for all lines.
b All hatched larvae were GFP-positive for all lines.
c Some GFP-positive dead embryos were present, possibly due to expression levels being either borderline too high or too low.
Rescue of a C. elegans C-P4H α Subunit Mutant with Human but Not B. malayi Proteins
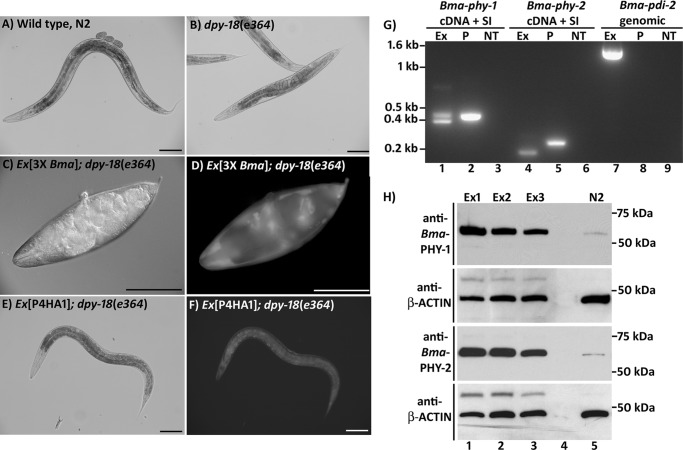
We again used C. elegans as an expression system to examine the possibility that formation of active B. malayi C-P4H complexes might require a factor, such as a specific post-translational modification or auxillary protein(s) required for folding, which was present in C. elegans but not in insect cells. A previously characterized homozygous viable C. elegans C-P4H α subunit mutant strain, Cel-dpy-18(e364), shows a marked reduction in C-P4H activity and has a malformed cuticle resulting in a readily identifiable body morphology defect (4, 5). We therefore expressed heterologous protein(s) in this mutant and observed whether they were able to repair the mutant body morphology, thereby providing a simple assay for enzyme activity and the ability of these proteins to functionally substitute for C. elegans dpy-18. Previously, this approach was successful when using C. briggsae sequences (9) but not with Bma-phy-1 introduced singly (14). Bma-phy-2 was expressed from a vector, pAW1 (14), which contains the promoter and 3′ UTR from Cel-dpy-18. To express multiple genes simultaneously, a similar construct for Bma-phy-1 (generated previously (14)) and a Bma-pdi-2 expression construct (described above) were also used. Bma-phy-2 was introduced singly and in combination with Bma-phy-1 and Bma-pdi-2 into the C. elegans dpy-18 mutant strain on repetitive extrachromosomal arrays. Neither Bma-phy-2 alone, in multiple pairings of the three genes, nor all three genes in combination was able to rescue the body morphology defect of Cel-dpy-18(e364) (results not shown). However, RT-PCR of transgenic lines carrying the three B. malayi genes highlighted the difficulty of achieving robust expression of multiple transgenes from repetitive arrays (results not shown). We therefore next attempted rescue of the Cel-dpy-18 mutant strain using complex extrachromosomal arrays. As explained under “Experimental Procedures,” complex arrays to be tested in the Cel-dpy-18 strain were first generated in the C. elegans wild type N2 strain and then crossed into the mutant. Three constructs for expression of B. malayi genes in C. elegans were used (Bma-phy-1 cDNA + SI, Bma-phy-2 cDNA + SI, and Bma-pdi-2 genomic). The rescue results above for Bma-pdi-2 demonstrate the production of functional protein in transgenic C. elegans using this construct. Surprisingly, introduction of an array carrying the three B. malayi genes into the Cel-dpy-18 mutant strain resulted in an exacerbation of the existing body morphology defect (Fig. 5, A–D) and could not be maintained in the homozygote mutant genetic background. Control experiments where complex arrays bearing only the transformation marker and fragmented genomic DNA were crossed into Cel-dpy-18 strain did not result in an enhanced mutant phenotype. The presence of the three B. malayi constructs on the array was verified by SW-PCR (data not shown), RT-PCR indicated that all three transgenes were expressed (Fig. 5G), and production of Bma-PHY-1 and Bma-PHY-2 protein was demonstrated by Western blotting (Fig. 5H). Due to the toxicity of these arrays in the Cel-dpy-18 mutant, RT-PCR and Western blotting had to be performed with the array in the wild type genetic background.
FIGURE 5.
A–F, introduction of transgenes into a C. elegans C-P4H mutant strain, dpy-18. A, wild type C. elegans N2 strain; B, C. elegans dpy-18(e364) homozygote mutant; C and D, introduction of an extrachromosomal array carrying three B. malayi genes (Bma-phy-1, Bma-phy-2, and Bma-pdi-2) results in an exacerbation of the body morphology defects of the dpy-18 mutant strain, with D showing GFP expression from the transformation marker dpy-7prom::gfp carried on the same extrachromosomal array. E and F, introduction of an extrachromosomal array carrying the human C-P4H α subunit gene P4HA1 partially repairs the body morphology defect of the dpy-18 strain. Scale bars represent 100 μm. G, RT-PCR demonstrating expression of all transgenes from the extrachromosomal array carrying Bma-phy-1, Bma-phy-2 and Bma-pdi-2. The results from one transgenic line are shown with other lines showing comparable expression levels. For the cDNA + synthetic intron (SI) construct, primers corresponding to the B. malayi gene sequences situated on either side of the 51-bp synthetic intron were used, whereas expression from the Bma-pdi-2 genomic construct was detected using primers amplifying the full-length Bma-pdi-2 sequence. Lanes 2 and 5 show the unspliced amplicon size derived using the plasmid (P) carrying the cDNA + synthetic intron, in lane 7 the 3.5-kb amplicon from the Bma-pdi-2 genomic plasmid (P) is not amplified under the conditions used. Lanes 3, 6, and 9 are the no template (NT) controls for each primer set. Bma-phy-1 from construct with cDNA and a synthetic intron; spliced, 369 bp; unspliced, 420 bp. Bma-phy-2 from construct with cDNA and a synthetic intron; spliced, 178 bp; unspliced, 229 bp. Bma-pdi-2 from genomic construct; spliced, 1512 bp; unspliced, 3579 bp. H, Western blots (reducing SDS-PAGE) of transgenic C. elegans with complex extrachromosomal arrays carrying Bma-phy-1, Bma-phy-2, and Bma-pdi-2. Expression of Bma-PHY-1 and Bma-PHY-2 protein was demonstrated in three transgenic lines (Ex1–Ex3). Blots were stripped and reprobed for β-actin as a loading control. Lane 4 is blank, and lane 5 is wild type N2; the faint band produced with both Bma-PHY-1 and -2 antibodies is likely to be a cross-reaction with the C. elegans proteins. The sizes of the mature (minus signal peptide) proteins are as follows: Bma-PHY-1, 60 kDa; Bma-PHY-2, 62 kDa.
The possibility that failure to rescue the C. elegans C-P4H α subunit mutant using B. malayi proteins might be a result of evolutionary distance was next investigated by similarly analyzing the human C-P4H α subunits P4HA1 and P4HA2. Introduction of either of these genes into the Cel-dpy-18 mutant using complex arrays resulted in partial rescue with a clear return to wild type body shape being evident (shown for P4HA1 in Fig. 5, E and F). The human proteins are therefore able to compensate to a large degree for loss of Cel-DPY-18.
Comparison of Parasite-derived and Insect Cell-expressed Protein Identified a Non-reducible Covalent Complex Association
Due to the difficulties encountered producing active B. malayi C-P4H both in insect cells and C. elegans systems, we next re-examined the insect cell expressed B. malayi proteins and compared these with parasite-derived material by reducing SDS-PAGE followed by Western blotting (Fig. 6). In addition to the monomer-sized band, a band running at the size predicted for a dimer was found in the parasite material, whereas B. malayi proteins produced in insect cell displayed only the monomer size for all three proteins. The same sized band was observed using three different antibodies reactive to the three B. malayi proteins (Fig. 6) and was replicated in three independent extracts of parasite material (results not shown). We also note that this band was absent from extracts of C. elegans expressing the B. malayi proteins (result not shown). These findings suggested that a non-reducible post-translational modification was present in the parasite material. We suspected that such a linkage would be most likely to occur between Bma-PDI-2 and each Bma-PHY to produce linked dimers. This was further investigated by co-IP using affinity-purified anti-Cel-PDI-2 antibody to isolate proteins that interact with Bma-PDI-2 from adult parasite extract. Testing the parasite extract via sensitive Western blotting methods prior to co-IP confirmed the presence of dimer-sized bands and shows they are present at high abundance (Fig. 7A). Co-IP was carried out on these extracts and analyzed by reducing SDS-PAGE and sensitive Western blotting using antibodies against Bma-PHY-1 and -2. This showed both Bma-PHYs were co-immunoprecipiated using the anti-PDI-2 antibody, whereas control co-IPs using isotype-matched antibody, reactive to C. elegans BLI-5, did not result in co-IP of either Bma-PHY (Fig. 7B). As well as monomer forms, Bma-PHY-1 and Bma-PHY-2 are both detected as dimer-sized bands after reducing SDS-PAGE, confirming that Bma-PHY-1 and -2 are each individually linked to Bma-PDI-2 via a non-reducible covalent linkage.
FIGURE 6.
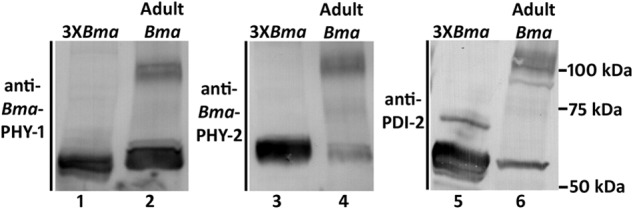
Western blots (reducing SDS-PAGE) of insect cell extracts compared with B. malayi adult worms. Lanes 1, 3, and 5 are extracts from insect cells co-expressing Bma-PHY-1, Bma-PHY-2, and Bma-PDI-2 (3XBma). Lanes 2, 4, and 6 are B. malayi extracts (Adult Bma). The antibodies used are indicated at the side of each blot. All three antibodies detect a band of the same size, running at approximately the size expected of a dimer (migrating at approximately 100 kDa), in the B. malayi worm extracts only. The sizes of the mature (minus signal peptide) proteins are as follows: Bma-PHY-1, 60 kDa; Bma-PHY-2, 62 kDa; Bma-PDI-2, 54 kDa.
FIGURE 7.
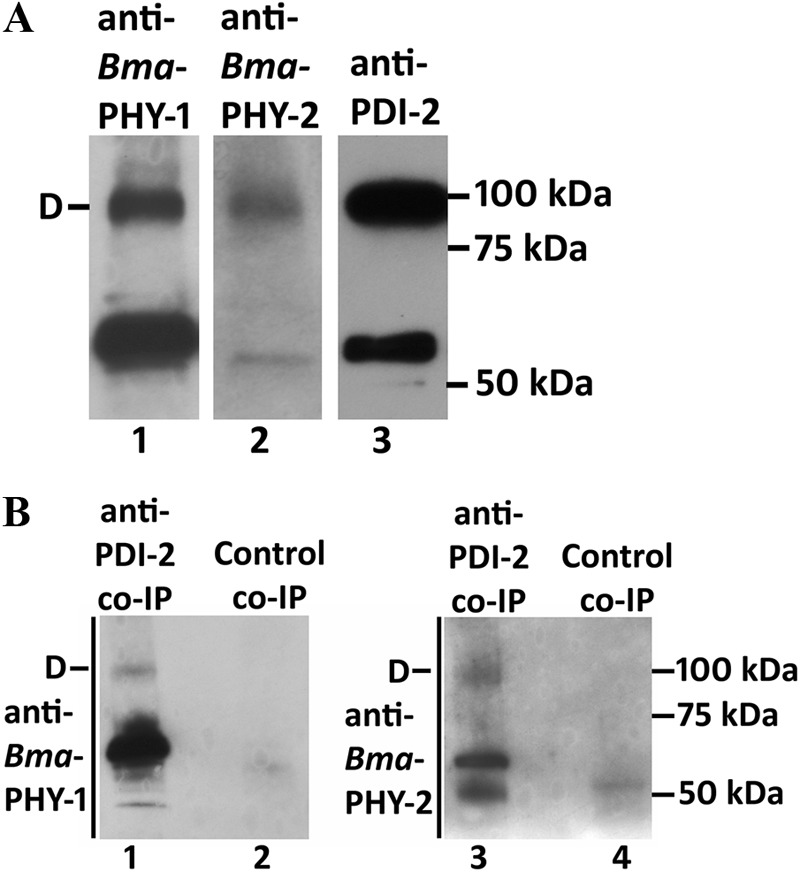
A, Western blots (reducing SDS-PAGE) of B. malayi adult material extracted in cell lysis buffer for co-IP, detected using the sensitive ECL plus system. The position of the dimer (D) is indicated. B, co-IP using an anti-PDI-2 antibody on native B. malayi extracts. Eluted complexes were separated by reducing SDS-PAGE and detected by the sensitive ECL Plus Western blotting system with anti-Bma-PHY-1 antibody (lanes 1 and 2), anti-Bma-PHY-2 antibody (lanes 3 and 4). The position of the dimer (D) in lanes 1 and 3 is indicated. The results of a control co-IP performed using anti-Cel-BLI-5 antibody are shown in lanes 2 and 4.
DISCUSSION
The C-P4H enzymes play critical roles in matrix formation and development in metazoans (8). Our previous studies in C. elegans had shown that C-P4H function was vital for development and collagen biogenesis in this species (4–6), whereas an analogous function remained to be established in any parasitic nematode species. We therefore investigated the role of C-P4H in B. malayi and in this study demonstrate that these enzymes are developmentally essential in this important human parasite.
The size of the phy gene family in B. malayi was examined in detail revealing that Bma-PHY-1 (14) and Bma-PHY-2 are the only proteins with homology to C-P4H α subunits in this organism. The temporal expression profile of Bma-phy-2 is more restricted than that of either Bma-pdi-2 or the previously described Bma-phy-1 (14). Bma-phy-1 and Bma-pdi-2 are both abundantly expressed in all stages examined, whereas Bma-phy-2 is expressed only in the first larval stage (microfilaria) and the early and mid-L3 stages. The lower abundance of Bma-phy-2 in microfilaria may reflect a more restricted cell/tissue expression pattern.
C-P4H Is Essential for Larval Development in B. malayi
RNAi approaches to examine gene function in parasitic nematodes of animals have, to date, met with very limited success (35). However, in this study, we employed a recently adapted RNAi approach (15) to examine the B. malayi phy and pdi genes in cultured parasites. Pronounced mutant body morphology phenotypes of stunted, lumpy immotile larvae, were found in first stage larvae when Bma-phy-1 and Bma-phy-2 were disrupted simultaneously, but not singly, or when Bma-pdi-2 was targeted singly. The severity of the phenotype from simultaneous RNAi of Bma-phy-1 and -2 also supports the contention that other Bma-PHYs are unlikely to be present in this organism. These results echo those of C. elegans where single RNAi of either Cel-dpy-18 or Cel-phy-2 are viable but combined RNAi results in mutant larvae with body morphology defects and malformed cuticles (4). Interestingly, in C. elegans, single disruption of Cel-pdi-2 results in phenotypes equivalent to the combined disruption of Cel-dpy-18 with Cel-phy-2 (4, 6). Likewise, in B. malayi, Bma-pdi-2 RNAi produces a similar phenotype to that of combined Bma-phy-1/Bma-phy-2 RNAi. In C. elegans, complete removal of both Cel-dpy-18 and Cel-phy-2 by genetic mutation is completely embryonic lethal (4, 6), suggesting that this could also be the case in B. malayi.
That Bma-pdi-2 RNAi resulted in a phenotype suggests that the Bma-PHY-1 complex we described previously (14) is either not sufficiently active in vivo for normal development or, as it does not possess a recognizable endoplasmic reticulum retention signal, becomes mislocalized. Our RNAi results in B. malayi represent one of a limited number of successful RNAi experiments reported in animal parasitic nematode species. In addition, the striking, highly penetrant morphological mutant phenotypes found have, to our knowledge, not been described previously in this species.
Functional Analysis of B. malayi Proteins in C. elegans
In parasitic nematodes such as B. malayi, experimental options using the parasite itself are limited, with no genetic approaches available and transgenesis only possible to a very limited extent (37). The function of the B. malayi proteins was therefore further investigated utilizing C. elegans mutants and transgenically introducing proteins to interrogate their functional roles. Surprisingly, despite confirmation of expression of multiple transgenic B. malayi genes in C. elegans, we were unable to rescue the phenotype associated with the C. elegans C-P4H α subunit mutant strain, dpy-18(e364). This result is all the more intriguing as we were able to rescue the same mutant using the human enzymes. However, at the amino acid level, Cel-PHY-1 and -2 are more similar to Bma-PHY-1 and -2 than they are to the human C-P4H α subunits P4HA1 and P4HA2 (Table 1). It therefore seemed unlikely that evolutionary distance would explain the inability of the B. malayi proteins to rescue the C. elegans mutant. In addition, we found that the cuticle collagen gene families of B. malayi were very similar to those of C. elegans (see supplemental “Experimental Procedures”), making it unlikely that differences in the substrate might account for our results. In contrast, the natural collagen substrates of human P4HA1 and P4HA2 differ from C. elegans cuticle collagens in a number of respects; yet, as reasonable repair was found by expressing the human enzymes in C. elegans, they must be capable of correctly modifying worm collagen substrates. Importantly, we also find that the failure to rescue the C. elegans dpy-18 mutant strain is not a reflection of the general lack of complementation using B. malayi genes because we conclusively demonstrated that B. malayi pdi-2 can rescue the more extreme phenotypes associated with loss of C. elegans pdi-2. The highlighted differences in C-P4H complex formation between the host and parasite may represent novel drug targets in otherwise highly conserved enzymes.
Bma-PDI-2 Is a C-P4H β Subunit
Oligomeric C-P4H in every species examined contain only one type of PDI that is present in all forms of the complex. That we had correctly identified the C-P4H forming PDI in B. malayi and that this represented the only PDI with this function was suggested by sequence analysis, which showed that only Bma-PDI-2 had high homology to C-P4H forming PDIs (data not shown). That Bma-PDI-2 was the sole protein involved in C-P4H complex formation was also shown by the effect of single Bma-pdi-2 RNAi being equivalent to combined loss of Bma-phy-1 and -2, suggesting the PDI was present in complexes with both Bma-PHYs. This hypothesis is also supported by the fact that the PDI-2 antibody immunoprecitated complexes containing Bma-PHY-1 and Bma-PHY-2. In addition, transgenic rescue of a C. elegans pdi-2 mutant with Bma-PDI-2 demonstrated this was the orthologue of the C-P4H forming PDI in C. elegans. All these lines of evidence indicate that Bma-PDI-2 is the C-P4H β subunit in B. malayi.
Dominant Negative Phenotype from Expression of B. malayi Proteins in dpy-18 Mutant
Interestingly, expression of the three B. malayi genes in the C. elegans dpy-18 mutant strain caused a pronounced enhancement of the aberrant body morphology phenotype. However, in the wild type genetic background, the same transgenic arrays did not disrupt worm body morphology. In the wild type background, all C-P4H complexes are able to form. Where the three B. malayi genes were expressed in the wild type background, assuming Cel-PDI-2 is not preferentially incorporated into complexes, some of these will contain Bma-PDI-2 and others Cel-PDI-2. In the wild type background, where all forms of the C-P4H complexes can be produced, if any reduction to C-P4H activity resulted from Bma-PDI-2-containing complexes, it must not have been enough to affect worm morphology. That Bma-PDI-2-containing C-P4H complexes generate sufficient activity is supported by expression of Bma-PDI-2 in the Cel-PDI-2 mutant; here, full rescue was found when all C-P4H complexes would have contained Bma-PDI-2. However, in the dpy-18 mutant strain, the only C-P4H complex present is the (Cel-PHY-2)(Cel-PDI-2) dimer (5). This dimer does not form naturally and is found only after loss of DPY-18, where its low C-P4H activity must be enough to permit survival (5). In the dpy-18 mutant strain expressing the three B. malayi genes, Bma-PDI-2 most likely competes with Cel-PDI-2 for association with Cel-PHY-2 but forms a complex that is less active or inactive. As Bma-PHY-1 and Bma-PHY-2 containing complexes provide no or minimal C-P4H activity, due to lack of the required linkage, the net result is a reduction in the already low level of C-P4H activity, causing it to drop below the threshold required for viability.
Subunit Linkage via a Non-reducible Covalent Bond
The subunit associations and enzymatic activity of C-P4H complexes are usually assessed in an insect cell co-expression system, an approach that has been invaluable in defining these enzymes in organisms as diverse as Drosophila melanogaster, C. elegans, mouse, and human (8). The ability of the PHY and PDI proteins from B. malayi to form active C-P4H complexes was therefore also examined using this recombinant system. Bma-PHY-2 was soluble when expressed alone in the absence of a PDI and formed what is likely to be a homotetramer, a similar finding to that described previously for Bma-PHY-1 (14). However, unlike Bma-PHY-1, the Bma-PHY-2 homotetramer was not enzymatically active. In addition, Bma-PHY-2, again similar to Bma-PHY-1, failed to form an active complex with C-P4H β subunit PDIs from other species.4 Surprisingly, however, co-expression of all combinations of Bma-PHY-1, Bma-PHY-2, and Bma-PDI-2, including all three together, also resulted in no enzyme activity, despite the formation of complexes analogous to those found in parasite-derived material. Potential explanations for our findings in insect cells, such as additional Bma-PHYs, with Bma-PDI-2 not being the correct and only C-P4H β subunit, or B. malayi enzymes evolving to process unusual collagen substrates, were ruled out by our systematic examination described above.
Previously described nematode C-P4H subunits combine in unique fashions (7), and our results in B. malayi indicate a further level of complexity in this species. Our co-IP analysis showed that in vivo Bma-PHY-1 and -2 are each found in two types of complex with Bma-PDI-2. In the first type, each Bma-PHY protein is linked to Bma-PDI-2 via a non-reducible covalent linkage, and in the second, each Bma-PHY is associated with Bma-PDI-2 but not linked by such a bond (see supplemental Fig. S6). We suspect that Bma-PDI-2 epitope may be partially masked in the covalently linked complex, resulting in this binding less efficiently during co-IP than the non-linked form. We therefore predict based on our combined results that in B. malayi C-P4H activity is generated by the covalent linked form of the complex. This novel modification has not been found in C-P4H identified from any other species to date. The enzyme generating this modification is currently unknown but could potentially be identified by RNAi screening of candidate enzymes to identify a similar phenotype to that produced by knockdown of B. malayi PHY-1/-2 and PDI-2.
Acknowledgments
Some nematode strains used in this work were provided by the Caenorhabditis Genetics Center, which is funded by the National Institutes of Health National Center for Research Resources. We thank the following people for reagents and materials: Rick Maizels and Yvonne Harcus (University of Edinburgh) for B. malayi nematodes and staged cDNA, Steven Williams (NIAID/National Institutes of Health FR3, Smith College) for EST clones, Mark Blaxter (University of Edinburgh) for BAC library lysates and clones, and Elodie Ghedin (University of Pittsburgh) for access to the resequenced B. malayi genome data.
This work was supported by grants from the Medical Research Council and the Biotechnology and Biological Sciences Research Council of Great Britain.

This article contains supplemental “Experimental Procedures,” Tables S1 and S2, Figs. S1–S6, and additional references.
A. D. Winter and A. P. Page, unpublished results.
- C-P4H
- collagen prolyl 4-hydroxylase(s)
- PDI
- protein disulfide isomerase
- hsiRNA
- heterogeneous short-interfering RNA
- co-IP
- co-immunoprecipitation.
REFERENCES
- 1. Ottesen E. A., Duke B. O., Karam M., Behbehani K. (1997) Strategies and tools for the control/elimination of lymphatic filariasis. Bull. W.H.O 75, 491–503 [PMC free article] [PubMed] [Google Scholar]
- 2. Hotez P. J., Brindley P. J., Bethony J. M., King C. H., Pearce E. J., Jacobson J. (2008) Helminth infections: the great neglected tropical diseases. J. Clin. Investig. 118, 1311–1321 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Ghedin E., Wang S., Spiro D., Caler E., Zhao Q., Crabtree J., Allen J. E., Delcher A. L., Guiliano D. B., Miranda-Saavedra D., Angiuoli SV, Creasy T., Amedeo P., Haas B., El-Sayed N. M., Wortman J. R., Feldblyum T., Tallon L., Schatz M., Shumway M., Koo H., Salzberg S. L., Schobel S., Pertea M., Pop M., White O., Barton G. J., Carlow C. K., Crawford M. J., Daub J., Dimmic M. W., Estes C. F., Foster J. M., Ganatra M., Gregory W. F., Johnson N. M., Jin J., Komuniecki R., Korf I., Kumar S., Laney S., Li B. W., Li W., Lindblom T. H., Lustigman S., Ma D., Maina C. V., Martin D. M., McCarter J. P., McReynolds L., Mitreva M. (2007) Draft genome of the filarial nematode parasite Brugia malayi. Science 317, 1756–1760 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Winter A. D., Page A. P. (2000) Prolyl 4-hydroxylase is an essential procollagen-modifying enzyme required for exoskeleton formation and the maintenance of body shape in the nematode Caenorhabditis elegans. Mol. Cell Biol. 20, 4084–4093 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Myllyharju J., Kukkola L., Winter A. D., Page A. P. (2002) The exoskeleton collagens in Caenorhabditis elegans are modified by prolyl 4-hydroxylase with unique combinations of subunits. J. Biol. Chem. 277, 29187–29196 [DOI] [PubMed] [Google Scholar]
- 6. Winter A. D., McCormack G., Page A. P. (2007) Protein disulfide isomerase activity is essential for viability and extracellular matrix formation in the nematode Caenorhabditis elegans. Dev. Biol. 308, 449–461 [DOI] [PubMed] [Google Scholar]
- 7. Page A. P., Winter A. D. (2003) Enzymes involved in the biogenesis of the nematode cuticle. Adv. Parasitol. 53, 85–148 [DOI] [PubMed] [Google Scholar]
- 8. Myllyharju J. (2008) Prolyl 4-hydroxylases, key enzymes in the synthesis of collagens and regulation of the response to hypoxia, and their roles as treatment targets. Ann. Med. 40, 402–417 [DOI] [PubMed] [Google Scholar]
- 9. Winter A. D., Keskiaho K., Kukkola L., McCormack G., Felix M. A., Myllyharju J., Page A. P. (2007) Differences in collagen prolyl 4-hydroxylase assembly between two Caenorhabditis nematode species despite high amino acid sequence identity of the enzyme subunits. Matrix Biol. 26, 382–395 [DOI] [PubMed] [Google Scholar]
- 10. Merriweather A., Guenzler V., Brenner M., Unnasch T. R. (2001) Characterization and expression of enzymatically active recombinant filarial prolyl 4-hydroxylase. Mol. Biochem. Parasitol. 116, 185–197 [DOI] [PubMed] [Google Scholar]
- 11. Rajan T. V., Paciorkowski N., Kalajzic I., McGuiness C. (2003) Ascorbic acid is a requirement for the morphogenesis of the human filarial parasite Brugia malayi. J. Parasitol. 89, 868–870 [DOI] [PubMed] [Google Scholar]
- 12. Petralanda I., Piessens W. F. (1991) Onchocerca volvulus, O. gutturosa, Brugia malayi, and Dirofilaria immitis: A comparative study of the immunochemical properties of cuticular proteins from filarial parasites. Exp. Parasitol. 72, 164–173 [DOI] [PubMed] [Google Scholar]
- 13. Cox G. N., Kusch M., Edgar R. S. (1981) Cuticle of Caenorhabditis elegans: its isolation and partial characterization. J. Cell Biol. 90, 7–17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Winter A. D., Myllyharju J., Page A. P. (2003) A hypodermally expressed prolyl 4-hydroxylase from the filarial nematode Brugia malayi is soluble and active in the absence of protein disulphide isomerase. J. Biol. Chem. 278, 2554–2562 [DOI] [PubMed] [Google Scholar]
- 15. Landmann F., Foster J. M., Slatko B. E., Sullivan W. (2012) Efficient in vitro RNA interference and immunofluorescence-based phenotype analysis in a human parasitic nematode, Brugia malayi. Parasit. Vectors 5, 16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Stiernagle T. (2006) Maintenance of C. elegans. WormBook, doi/ 10.1895/wormbook.1.101.1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Timmons L., Court D. L., Fire A. (2001) Ingestion of bacterially expressed dRNAs can produce specific and potent genetic interference in Caenorhabditis elegans. Gene 263, 103–112 [DOI] [PubMed] [Google Scholar]
- 18. Kamath R. S., Martinez-Campos M., Zipperlen P., Fraser A. G., Ahringer J. (2001) Effectiveness of specific RNA-mediated interference through ingested double-stranded RNA in Caenorhabditis elegans. Genome Biol. 2, RESEARCH0002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Kivirikko K. I., Myllylä R. (1982) Posttranslational enzymes in the biosynthesis of collagen: intracellular enzymes. Methods Enzymol. 82, 245–304 [DOI] [PubMed] [Google Scholar]
- 20. Evans T. C. (2006) Transformation and microinjection. WormBook, doi/ 10.1895/wormbook.1.108.1 [DOI] [Google Scholar]
- 21. Kelly W. G., Xu S., Montgomery M. K., Fire A. (1997) Distinct requirements for somatic and germline expression of a generally expressed Caernorhabditis elegans gene. Genetics 146, 227–238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Takacs A. M., Denker J. A., Perrine K. G., Maroney P. A., Nilsen T. W. (1988) A 22-nucleotide spliced leader sequence in the human parasitic nematode Brugia malayi is identical to the trans-spliced leader exon in Caenorhabditis elegans. Proc. Natl. Acad. Sci. U.S.A. 85, 7932–7936 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Bendtsen J. D., Nielsen H., von Heijne G., Brunak S. (2004) Improved prediction of signal peptides: SignalP 3.0. J. Mol. Biol. 340, 783–795 [DOI] [PubMed] [Google Scholar]
- 24. John D. C., Bulleid N. J. (1994) Prolyl 4-hydroxylase: defective assembly of a-subunit mutants indicates that assembled a-subunits are intramolecularly disulfide bonded. Biochemistry 33, 14018–14025 [DOI] [PubMed] [Google Scholar]
- 25. Lamberg A., Pihlajaniemi T., Kivirikko K. I. (1995) Site-directed mutagenesis of the a-subunit of human prolyl 4-hydroxylase. Identification of three histidine residues critical for catalytic activity. J. Biol. Chem. 270, 9926–9931 [DOI] [PubMed] [Google Scholar]
- 26. Myllyharju J., Kivirikko K. I. (1997) Characterization of the iron- and 2-oxoglutarate-binding sites of human prolyl 4-hydroxylase. EMBO J. 16, 1173–1180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Lappi A. K., Lensink M. F., Alanen H. I., Salo K. E., Lobell M., Juffer A. H., Ruddock L. W. (2004) A conserved arginine plays a role in the catalytic cycle of the protein disulphide isomerases. J. Mol. Biol. 335, 283–295 [DOI] [PubMed] [Google Scholar]
- 28. Ellgaard L., Ruddock L. W. (2005) The human protein disulphide isomerase family: substrate interactions and functional properties. EMBO Rep. 6, 28–32 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Pirneskoski A., Klappa P., Lobell M., Williamson R. A., Byrne L., Alanen H. I., Salo K. E., Kivirikko K. I., Freedman R. B., Ruddock L. W. (2004) Molecular characterization of the principal substrate binding site of the ubiquitous folding catalyst protein disulfide isomerase. J. Biol. Chem. 279, 10374–10381 [DOI] [PubMed] [Google Scholar]
- 30. Koivunen P., Pirneskoski A., Karvonen P., Ljung J., Helaakoski T., Notbohm H., Kivirikko K. I. (1999) The acidic C-terminal domain of protein disulfide isomerase is not critical for the enzyme subunit function or for the chaperone or disulfide isomerase activities of the polypeptide. EMBO J. 18, 65–74 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Mitreva M., Jasmer D. P., Zarlenga D. S., Wang Z., Abubucker S., Martin J., Taylor C. M., Yin Y., Fulton L., Minx P., Yang S. P., Warren W. C., Fulton R. S., Bhonagiri V., Zhang X., Hallsworth-Pepin K., Clifton S. W., McCarter J. P., Appleton J., Mardis E. R., Wilson R. K. (2011) The draft genome of the parasitic nematode Trichinella spiralis. Nat. Genet. 43, 228–235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Opperman C. H., Bird D. M., Williamson V. M., Rokhsar D. S., Burke M., Cohn J., Cromer J., Diener S., Gajan J., Graham S., Houfek T. D., Liu Q., Mitros T., Schaff J., Schaffer R., Scholl E., Sosinski B. R., Thomas V. P., Windham E. (2008) Sequence and genetic map of Meloidogyne hapla: A compact nematode genome for plant parasitism. Proc. Natl. Acad. Sci. U.S.A. 105, 14802–14807 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Wang J., Czech B., Crunk A., Wallace A., Mitreva M., Hannon G. J., Davis R. E. (2011) Deep small RNA sequencing from the nematode Ascaris reveals conservation, functional diversification, and novel developmental profiles. Genome Res. 21, 1462–1477 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Jex A. R., Liu S., Li B., Young N. D., Hall R. S., Li Y., Yang L., Zeng N., Xu X., Xiong Z., Chen F., Wu X., Zhang G., Fang X., Kang Y., Anderson G. A., Harris T. W., Campbell B. E., Vlaminck J., Wang T., Cantacessi C., Schwarz E. M., Ranganathan S., Geldhof P., Nejsum P., Sternberg P. W., Yang H., Wang J., Wang J., Gasser R. B. (2011) Ascaris suum draft genome. Nature 479, 529–533 [DOI] [PubMed] [Google Scholar]
- 35. Maule A. G., McVeigh P., Dalzell J. J., Atkinson L., Mousley A., Marks N. J. (2011) An eye on RNAi in nematode parasites. Trends Parasitol. 27, 505–513 [DOI] [PubMed] [Google Scholar]
- 36. Riihimaa P., Nissi R., Page A. P., Winter A. D., Keskiaho K., Kivirikko K. I., Myllyharju J. (2002) Egg shell collagen formation in Caenorhabditis elegans involves a novel prolyl 4-hydroxylase expressed in spermatheca and embryos and possessing many unique properties. J. Biol. Chem. 277, 18238–18243 [DOI] [PubMed] [Google Scholar]
- 37. Lok J. B. (2012) Nucleic acid transfection and transgenesis in parasitic nematodes. Parasitology 139, 574–588 [DOI] [PMC free article] [PubMed] [Google Scholar]