Background: Calnuc is a multidomain Ca2+-binding protein with many interacting partners but whose function is still elusive.
Results: Calnuc is a serine protease with its active site catalytic triad present in the C-terminal domain.
Conclusion: The serine protease activity of calnuc is allosterically regulated by Zn2+-binding and its interaction with G protein α subunit.
Significance: Novel proteolytic function of calnuc will have vital implications in its physiological role.
Keywords: Calcium, Calcium Signaling, Protein Folding, Serine Protease, Zinc, Allosteric Regulation, G Protein
Abstract
The functions of calnuc, a novel Ca2+-binding protein with multiple structural domains and diverse interacting partners, are yet unknown. We demonstrate unknown facets of calnuc, which is a serine protease in which Ser-378 of GXSXG motif, Asp-328 of DTG motif, and His-339 form the “catalytic triad,” locating the enzyme active site in the C-terminal region. Analogous to the active site of Zn2+ carboxypeptidases, calnuc has two high affinity (Kd ∼ 20 nm), well conserved Zn2+-binding sites near its N terminus, although it is inactive as a peptidase. Zn2+ binding allosterically and negatively regulates the serine protease activity of calnuc, inhibition being caused by an “open to close” change in its conformation not seen upon Ca2+ binding. Most strikingly, interaction with G protein α subunit completely inhibits the enzymatic activity of calnuc. We thus illustrate that G proteins and Zn2+ act as two “keys” that control enzymatic activity of calnuc, arresting it in “locked” state. Calnuc, therefore, exists dynamically in two different forms, (i) as a Ca2+-binding protein in Zn2+-bound form and (ii) as a protease in Zn2+-free form, commissioning it to perform multiple functions.
Introduction
Ubiquitously expressed, multidomain Ca2+-binding proteins are vital for the physiology of cells (1, 2). Because of their complex structure, understanding their diverse functions is an arduous task. Calnuc is one such Ca2+ binding, Golgi-resident protein with multiple domains, highly conserved in species as varied as Ciona intestinalis to Homo sapiens (3–6). Earlier reports on calnuc conferred it with a DNA binding property (4), and subsequently the presence of other domains was established including EF-hand Ca2+-binding sites (3), a cyclooxygenase (COX)-binding site (7, 8), a G protein-binding site (9), and a species-specific variable C-terminal region. The presence of these domains potentiates interactions of calnuc with its cognate partners. As the list of diverse interacting partners of calnuc, such as amyloid precursor protein (APP) (10), COX-2 etc., is growing (7, 8), its involvement in Ca2+ storage in the Golgi and other pathophysiological conditions is delineated (11–16). Its central importance in the regulation of many cellular events is becoming increasingly intriguing. Co-localization of G protein and calnuc to the Golgi apparatus was followed by evidences toward co-trafficking of these two proteins and that it acts as a guanine nucleotide dissociation inhibitor (17–20). Detailed structural aspects and ion binding properties of calnuc have been recently reported (21).
The ubiquitous expression pattern of calnuc combined with its structural complexity generates a large set of varied interacting partners. We hypothesize that its complex structural design is deliberate and determinant for its probable assorted biochemical roles. In an effort to discover novel physiological functions of calnuc, we assessed the role of its predominant, non EF-hand regions. In this work we demonstrate an unknown function of calnuc, i.e. its significant inherent “serine proteolytic” activity, employing Ser-378 from the GXSXG motif, Asp-328 from the DTG motif, and His-339 in the vicinity of the DTG motif forming the active site catalytic triad. In addition, we identify calnuc as a Zn2+ sensor with high affinity Zn2+-binding (Kd = 21 nm) resulting in a conformational switch that allosterically locks its protease activity leading to a sharp inhibition. Serine protease activity of calnuc was also attenuated significantly by α subunit of G proteins, thus establishing a functional link between these two proteins. We propose that both Zn2+ and G proteins function as two different keys that lock and regulate the protease activity of calnuc. The multidomain structure of calnuc is a design to help in adopting conformational polymorphisms to be able to do multiple functions.
EXPERIMENTAL PROCEDURES
Subcloning of Calnuc Gene
The human calnuc gene lacking the first 31 amino acid (most of which code for a signal peptide) residues was subcloned from pET28a expression vector into pTYB12 vector using the restriction sites NdeI and NotI. The recombinant clone was confirmed by nucleotide sequencing after initial screening. The protein cloned into pTYB12 is expressed as a fusion to a chitin-intein tag that is self-cleavable upon incubation with dithiothreitol, releasing a “tag less” protein.
Construction of Calnuc Fragments and Mutants
The N-terminal fragment of calnuc (31 −220 amino acids) was constructed by insertion of a stop codon after amino acid number 220 in pTYB12 vector using mutagenic primers with the STOP codon. The C-terminal fragment of calnuc (residues 221–461) was generated by PCR amplification. The fragment was initially cloned into pGEMT vector with unique restriction sites (NdeI and EcoRI) and then subcloned into the expression vector pTYB12.
The point mutations of serine, aspartate, and histidine in calnuc were generated using site-directed mutagenesis. The serine double mutant (S224V and S378V) and the truncated fragment of calnuc (residues 239–461) were also generated. Leucines at positions 314 and 318 in calnuc were mutated to alanines (LL/AA mutant). Four residues in the two zinc binding sites, His-67 and His-147, and Glu-70 and Glu-150, were mutated to alanines, respectively. The primer sequences used to generate point mutation are presented in supplemental Table 1. Mutations were confirmed by sequencing, and the clones were transformed in BL21 (DE3) cells. The expression conditions and purification protocol is similar to that of wild type calnuc.
Overexpression and Purification of Calnuc, Its Mutants, and Gαi1
For overexpression in BL21(DE3) cells, calnuc and its fragments were induced with 200 μm isopropyl-thio-β-d-galactoside at A600 nm 0.6 and grown at 23 °C for 16 h. Cells expressing recombinant chitin-intein-fused calnuc, its fragments, and the mutants were lysed in buffer A (20 mm Tris, 300 mm NaCl, pH 8.0) using an ultrasonicator (Vibracell Sonics and Materials, Inc. Newtown, CT). The supernatant was loaded on to a chitin resin (New England Biolabs), equilibrated with buffer A, washed with 20 column volumes of buffer A followed by flushing with wash buffer B (20 mm Tris, 300 mm NaCl, 50 mm DTT, pH 8.0), and incubated at 4 °C for 24 h. The protein was eluted with 20 mm Tris, 50 mm NaCl, pH 8.0, and checked on SDS-PAGE (12%) gel. The concentration and yield of the protein obtained was estimated using the method of Lowry (22).
Recombinant rat Gαi1 was expressed in Escherichia coli BL21(DE3) cells that were grown at 37 °C to A600 nm of 0.5 and then induced with 100 μm isopropyl-thio-β-d-galactoside with a post induction period of 20 h at 23 °C. The purification protocol was similar to that of calnuc except for the buffers used. Lysis, equilibration, and wash buffer contains 20 mm Tris, 150 mm NaCl, 2 mm MgCl2, and 6–10 μm GDP. Protein was eluted with 20 mm Tris, 50 mm NaCl, 2 mm MgCl2, and 10 μm GDP, pH 8.0. Activity of the protein was established according to the protocol of Fahmy and Sakmar (23).
Fluorescence Spectroscopy
Protein intrinsic fluorescence measurements were carried out in fluorolog fluorimeter (HORIBA Jobin Yvon). Tryptophan emission of calnuc (0.95 μm) and its C-terminal fragment (1.5 μm) was studied in a buffer containing 20 mm Tris, 50 mm NaCl, pH 7.5, by exciting the sample at 295 nm and scanning the emission spectra from 310 to 450 nm with the emission and excitation slit widths set at 5 nm each. The effect of Zn2+ on apocalnuc and the apo form of calnuc-C fragment was monitored by titrating with ZnCl2 (up to 20 μm). All the spectra were corrected for contribution of the buffer by subtracting a “blank” spectrum of buffer alone. Tyrosine fluorescence measurements were carried out for the apo form of calnuc-N fragment by exciting the sample at 275 nm and scanning the emission spectra from 285 to 385 nm. The effect of Zn2+ on the apocalnuc-N fragment was studied by titrating with ZnCl2 (up to 20 μm). Similar experiments were also performed with calnuc containing mutations in the Zn2+-binding site.
Surface hydrophobicity changes in calnuc (apo) and the Zn2+-bound form were monitored by 8-anilino naphthalene sulfonic acid (ANS)3 fluorescence. ANS spectra were recorded by exciting 2 μm ANS (in 20 mm Tris buffer, pH 8.0, containing 50 mm NaCl) at 365 nm and recording the emission spectrum from 440 to 600 nm with the emission and excitation slits set at 5 nm each. In all of these experiments, 0.4 μm concentrations of protein and 5, 10, and 15 μm ZnCl2 were used. In the absence of the protein the fluorescence of ANS was unaffected upon the addition of the above-mentioned concentrations of ion only. The spectra were corrected for ANS fluorescence in buffer without protein. All data are representative of experiments performed several times.
Binding Analysis
The saturation binding of Zn2+ with calnuc and its fragments is shown by plotting the change in fluorescence emission signal along the y axis versus Zn2+ concentration on the x axis. Scatchard analysis (plot of bound/free ligand versus bound) was performed to estimate the quantitative parameters, stoichiometry (n) and affinity of interaction (24).
Zn2+ Binding by Isothermal Titration Calorimetry (ITC)
ITC measurements were carried out with a VP-ITC calorimeter (MicroCal Inc.). All the titrations were carried out at 30 °C in 20 mm Tris, pH 7.8, and 50 mm NaCl. Calnuc (18 μm) in the sample cell was titrated with 200 μm ZnCl2 solution in the syringe; each injection of 3 μl until saturation in heat changes was reached. Appropriate buffer titrations were carried out to determine the heat of dilution and subtracted from the Zn2+ binding thermograms. Data analysis was done with Microcal Origin 7.0 software.
The binding energetics of the calnuc Zn2+-binding site (His-67/147; Glu-70/150 to alanine) mutant with Zn2+ was measured at 30 °C using MicroCal ITC200. Protein sample (concentration 18 μm) was loaded in a sample cell, and 30 consecutive injections of 1 μl (duration and spacing 2 and 120 s, respectively) of 200 μm Zn2+ concentration was titrated with the help of rotator stirrer-syringe. Using same parameters, contribution of heat of dilution of buffer versus Zn2+ titration was also measured.
CD Spectroscopy
Near- and far-UV CD spectra of proteins were recorded on a Jasco-815 spectropolarimeter at room temperature in 1- and 0.1-cm path length cuvettes, respectively. All spectra were recorded in 50 mm Tris, pH 7.5, containing 50 mm NaCl in the presence or absence of Zn2+ as per the requirement of the experiment, and the appropriate buffer spectra were recorded and subtracted from the protein spectra.
Protease Assay of Calnuc Using Fluorescein Isothiocyanate (FITC)-casein
Fluorescence protease assay was carried out by incubating the fluorescent substrate (FITC-casein) with calnuc for 1 h at 37 °C in 20 mm Tris and 100 mm NaCl, pH 7.5. An increase in fluorescence intensity as a result of proteolysis of the substrate was monitored by exciting the sample at 485 nm and scanning the emission spectra from 500 to 600 nm with slits widths of 5 nm each (25). The proteolysis of FITC-casein in the presence of trypsin was used as a control for the assay.
Alternatively, the protease assay was performed by monitoring the fluorescence of the cleaved product from the FITC-casein by TCA precipitation method (25). Proteolytic activity of calnuc was also analyzed electrophoretically using Hammerstein casein as a substrate. The reaction mixture containing 20 μl of casein (2 mg/ml) and 10 μl of calnuc (2 mg/ml) or its mutants was incubated for 12 h. Samples were analyzed by 15% SDS-PAGE. Protein bands were monitored by silver staining.
Protease Activity in the Presence of Inhibitors
To elucidate the mechanism of proteolytic cleavage, we checked the activity in the presence of different inhibitors like EDTA for metalloproteases, pepstatin for aspartate protease, and PMSF and AEBSF for serine protease. Calnuc (2.7 μm) was incubated with different concentrations of inhibitors or for different time intervals and protease assay performed as mentioned above.
Methodology to Elucidate the Mode of Enzyme Action of Calnuc
N-Benzoyl-l-Arg β-naphthylamide (BANA), n-benzoyl-Phe-Val-Arg-p-nitroanilide (FVRpNA), and n-benzoyl-Val-Gly-Arg-p-nitroanilide (VGRpNA), hippuryl-l-phenylalanine (from sigma) were used to examine the mode of protease action. A fluorescence assay was performed to monitor the release of β-naphthylamine on the hydrolysis of BANA. The increase in fluorescence intensity was monitored by exciting the sample at 335 nm and the emission peak at 410 nm (26). For the substrates FVRpNA and VGRpNA, absorption spectra were recorded to monitor the release of p-nitroaniline and intensity at 405 nm used for analysis (27, 28). Calnuc and its mutants were incubated with different substrates for different time intervals to monitor the changes in their fluorescence or absorption spectra respectively. In the case of hippuryl-l-phenylalanine, the release of phenylalanine was monitored spectrophotometrically at 254 nm (29). For deducing the role of GXSXG motif of calnuc in exhibiting the esterase activity, assays were performed with phenyl acetate and p-nitrophenyl acetate to assay the release of phenol and p-nitro phenol by monitoring absorbance at 270 and 405 nm, respectively (30).
Measurement of Protease Activity of Calnuc in the Presence of Gαi
We monitored the protease activity of calnuc in the presence of G-protein α subunit (Gαi1). Equimolar concentrations of calnuc and Gαi1 (0.6 μm) were incubated for 30 min (in 20 mm Tris buffer containing 100 mm NaCl, 2 mm MgCl2, and 1 μm GDP), and this complex was further incubated with fluorescent substrate (FITC casein). The fluorescence spectra were recorded (as mentioned above for the protease assay) at different time intervals.
In Vitro Pulldown Assay
Chimera of calnuc with chitin binding domain were used to pull down purified native Gαi protein. The matrix-bound calnuc was incubated with purified Gαi (in GDP bound form) for a period of 30 min and eluted as described above. Purified calnuc without the addition of Gαi served as a negative control. The elution fractions were analyzed by SDS-PAGE for the presence of both calnuc and Gαi. The same procedure was repeated with the LL/AA double mutant of calnuc to evaluate its interaction with Gαi protein. All the steps involved in the assay were performed at 4 °C.
RESULTS
Novel Function of Calnuc as a Serine Protease Enzyme
Calnuc is a multidomain protein with several known interacting partners (Fig. 1). Our results now demonstrate a novel serine protease function of calnuc. Increase in fluorescence of a probe covalently attached to casein due to the addition of test protein is a standard assay to determine the proteolytic property of the test protein (25). Apocalnuc caused a linear increase in fluorescence of the probe FITC when incubated with FITC-casein in a time-dependent manner, thereby indicating the proteolytic activity of calnuc (Fig. 2A). Various inhibitors were tested to determine the nature and the type of protease activity exhibited by calnuc. Although EDTA (inhibitor of metalloproteases) and pepstatin (inhibitor of aspartate proteases) in the micromolar to millimolar range do not inhibit activity of calnuc, significant inhibition was observed in the presence of serine protease inhibitors, i.e. PMSF (16 μm) and AEBSF (7 μm), suggesting that calnuc possesses serine protease activity (supplemental Fig. S1).
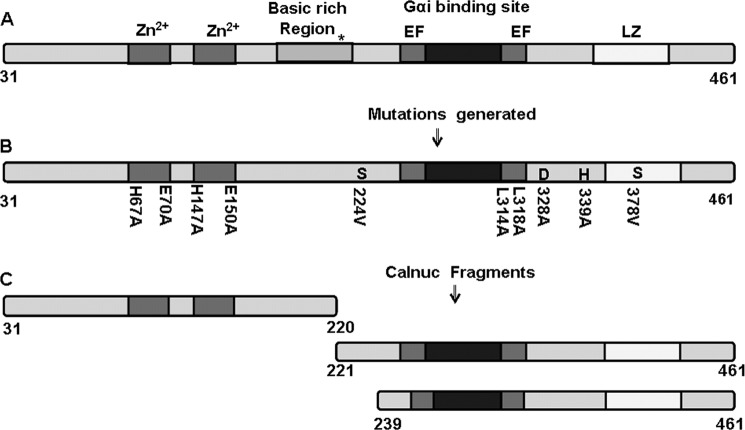
FIGURE 1.
Schematic representation of calnuc illustrating the constructed mutants and fragments. A, calnuc (31–461 amino acids), a multi-domain protein, has two Zn2+-binding sites (located near the N terminus), a basic region with a nuclear localization signal (*) and the C-terminal region consisting of two calcium binding EF-hand motifs, a Gαi binding region, and a leucine zipper region (LZ). B, the constructed mutants generated for determining the active site of protease function are illustrated. S224V, S378V, D328A, and H339A mutants were generated to evaluate their involvement in the active site for protease activity of calnuc. H67A, E70A, H147A, and E150A were generated to check the Zn2+-binding site. L314A and L318A were generated to check the binding of G protein. C, shown is the construction of N- and C-terminal fragments for examining the catalytic site of protease action in calnuc. The C-terminal (239–461) fragment was generated to investigate the involvement of the Ser-224 (conserved serine) in the active site.
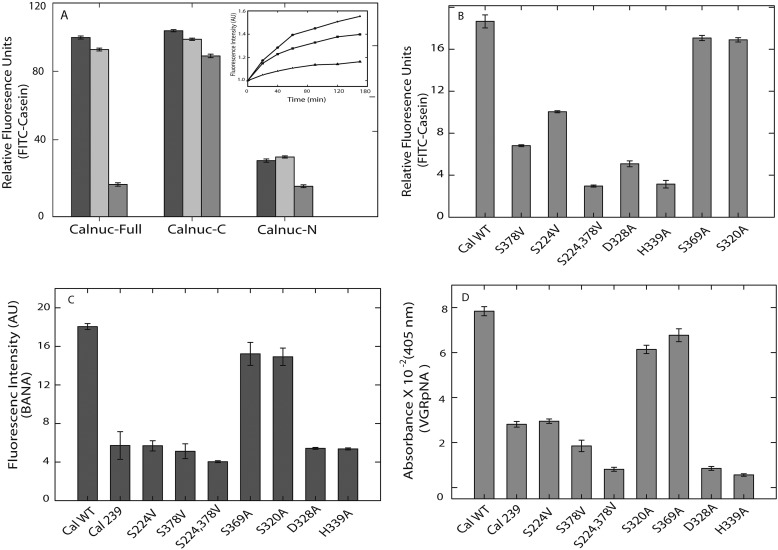
FIGURE 2.
Serine protease activity of calnuc. The protease activity of calnuc and its fragments in the presence and absence of metal ions was carried out by incubating the protein with the fluorescent substrate (FITC-casein) for 1 h at 37 °C in 20 mm Tris and 100 mm NaCl, pH 7.5. An increase in the fluorescence intensity as a result of proteolysis of the substrate was monitored by exciting the sample at 485 nm and scanning the emission spectra from 500 to 600 nm. Data represented are fluorescence intensity at 521 nm (λmax,em). Panel A, protease activity of calnuc full-length and its N and C fragments in the presence of metal ions is shown. The black bar graph (■) represents protease activity of apo form of calnuc and its fragments; the light gray bar represents ( ) activity in the presence of Ca2+ (1 mm), and the dark gray bar (■) represents activity in the presence of Zn2+ (20 μm). Inset, protease activity of calnuc full-length in the presence of metal ions is shown. The squares (■) represent protease activity of the apo form of calnuc, closed circles (●) represent protease activity upon the addition of Ca2+ (1 mm), and the triangle (▴) represents protease activity in presence of 20 μm Zn2+. Panel B, mutational analysis confirms the serine protease catalytic triad. The protease activity of calnuc mutants was assayed as mentioned above and represented in the bar diagram depicting relative fluorescence units (n = 6) using FITC-casein as the substrate. S378V and S224V mutants exhibit decrease in activity compared with that of wild type. The serine double mutant (S224V/S378V) showed further inhibition compared with that of individual serine mutants. S369A and S320A mutants did not exhibit any change in activity. D328A and H339A also demonstrate a significant inhibition compared with that of wild type and serine mutant. For the mode of protease activity of calnuc, to elucidate the mechanism of activity of calnuc, a proteolysis assay was performed with a variety of substrates. Calnuc and its mutants were incubated with respective substrates to monitor spectroscopic changes as mentioned below. Panel C, the protease activity of calnuc and its mutants using BANA (10 μm) as the substrate was monitored by an increase in fluorescence intensity (due to the release of β-napthalamide) at 410 nm. Compared with wild type, S378V and truncated calnuc (residues 239–461) fragment devoid of Ser-224 (conserved serine), the Ser-224/378V double mutant showed more inhibition (dark gray bars). AU, arbitrary units. Panel D, light gray bars (■) represent the increase in absorbance at 405 nm monitored by the release of p-nitroaniline from the substrate VGRpNA by calnuc and its mutants.
) activity in the presence of Ca2+ (1 mm), and the dark gray bar (■) represents activity in the presence of Zn2+ (20 μm). Inset, protease activity of calnuc full-length in the presence of metal ions is shown. The squares (■) represent protease activity of the apo form of calnuc, closed circles (●) represent protease activity upon the addition of Ca2+ (1 mm), and the triangle (▴) represents protease activity in presence of 20 μm Zn2+. Panel B, mutational analysis confirms the serine protease catalytic triad. The protease activity of calnuc mutants was assayed as mentioned above and represented in the bar diagram depicting relative fluorescence units (n = 6) using FITC-casein as the substrate. S378V and S224V mutants exhibit decrease in activity compared with that of wild type. The serine double mutant (S224V/S378V) showed further inhibition compared with that of individual serine mutants. S369A and S320A mutants did not exhibit any change in activity. D328A and H339A also demonstrate a significant inhibition compared with that of wild type and serine mutant. For the mode of protease activity of calnuc, to elucidate the mechanism of activity of calnuc, a proteolysis assay was performed with a variety of substrates. Calnuc and its mutants were incubated with respective substrates to monitor spectroscopic changes as mentioned below. Panel C, the protease activity of calnuc and its mutants using BANA (10 μm) as the substrate was monitored by an increase in fluorescence intensity (due to the release of β-napthalamide) at 410 nm. Compared with wild type, S378V and truncated calnuc (residues 239–461) fragment devoid of Ser-224 (conserved serine), the Ser-224/378V double mutant showed more inhibition (dark gray bars). AU, arbitrary units. Panel D, light gray bars (■) represent the increase in absorbance at 405 nm monitored by the release of p-nitroaniline from the substrate VGRpNA by calnuc and its mutants.
Mutational Analysis Reveals the Catalytic Site Residues
Comparative analysis of amino acid sequences of calnuc from various organisms established the presence of highly conserved serine, histidine, and aspartate residues (Table 1). Several mutants of serine, aspartate, and histidine were generated to determine the residues that form the catalytic site (Fig. 1).
TABLE 1.
Conserved serine in calnuc among various organisms
Conserved serine, histidine, and aspartic acid residues can be observed on the basis of the multiple sequence alignment (MSA) of calnuc from a variety of organisms. Residues forming the serine protease active site in calnuc among the different species are denoted. A, the Ser-224 residue although highly conserved among all of them except in Drosophila melanogaster may not be involved in the catalysis. B, Ser-378, Asp-238, and His-339 form the catalytic triad of the active site.

We chose serine (Ser-224) because it is highly conserved among all organisms and Ser-378 because as it is part of a well conserved GXSXG motif. Substitution of S378A or S378V exhibited a 64% decrease in the protease activity compared with that of wild type (Fig. 2B). Ser-224, the most conserved serine, when replaced either with alanine or valine exhibited a 46% decrease in enzyme activity with respective to FITC-casein.
Interestingly the S224A and S378A double mutant showed more inhibition (78–80%) compared with that of individual serine mutants. For further evaluation of the determinants of protease activity, other serine mutants, S369A and S320A, were also generated. These mutants did not exhibit significant changes compared with that of calnuc (WT) (Fig. 2, B–D). The role of several highly conserved histidines and aspartic acid residues was explored for the identification of a possible classic “serine protease catalytic triad” site in calnuc. Substitution of alanine for Asp-328 (of DTG motif) and His-339 reduced the activity by 73 and 83%, respectively (Fig. 2B). All the mutants exhibited a basal level of residual activity. From these results, we infer that Ser-378 of the GXSXG motif or Ser-224 (conserved serine), Asp-328 of DTG motif, and His-339 are involved in the formation of the catalytic triad site. Proteolytic activity was also confirmed electrophoretically wherein the extent of proteolysis of casein upon incubation with calnuc and its mutants for 16–18 h at 37 °C as observed on a SDS-PAGE matched with our fluorescence protease assay (supplemental Fig. S2).
Mechanism of Protease Activity
The mechanism of proteolytic cleavage by calnuc was assayed using a variety of substrates (Table 2) (26–29). BANA, FVRpNA, and VGRpNA were hydrolyzed significantly by calnuc monitored by the change in spectral property of the released product. An increase in fluorescence of β-naphthylamine indicating cleavage of BANA at the carboxyl side of the arginine residue suggested a trypsin-like mode of action (Fig. 2C). Further evidence for trypsin-like action by calnuc was provided by monitoring an increase in absorbance at 405 nm upon release of p-nitroaniline on the hydrolysis VGRpNA (Fig. 2D). The Ser-378, Ser-224/378 double mutant, Asp-328, and His-339 mutants showed low levels of activity with these substrates compared with that of wild type. Calnuc does not hydrolyze hippuryl-l-phenylalanine, indicating the absence of carboxypeptidase-like activity (30). The presence of GXSXG, a motif conserved in esterases, in calnuc prompted us to monitor the esterase activity using phenyl acetate and p-nitrophenyl acetate by the release of phenol and p-nitrophenol, respectively. The absence of spectral changes infers the lack of esterase activity. Our results, therefore, imply that calnuc exhibits trypsin-like protease mechanism.
TABLE 2.
Various substrates used in to elucidate the mode of cleavage by calnuc
Activities were monitored as mentioned in the parentheses.
| Serial no. | Substrate (product monitored) | Mode of enzyme action | Activity |
|---|---|---|---|
| 1 | FITC-casein (increase in fluorescence emission at 520 nm) | Trypsin | Yes |
| 2 | N-Benzoyl-l-Arg-β-napthalamide (increase in fluorescence emission at 410 nm) | Trypsin cathepsin | Yes |
| 3 | N-Benzoyl-Phe-Val-Arg-p-nitroaniline (release of p-nitroaniline at 405 nm) | Trypsin | Yes |
| 4 | N-Benzoyl-Val-Gly-Arg-p-nitroaniline (release of p-nitroaniline at 405 nm) | Trypsin | Yes |
| 5 | Hippuryl-l-phenylalanine (release of phenylalanine at 254 nm) | Carboxypeptidase | No |
| 6 | Phenyl acetate (release of phenol at 270 nm) | Esterase | No |
| 7 | p-Nitrophenyl acetate (release of p-nitro phenol at 405 nm) | Esterase | No |
Prediction of Zn2+-binding Sites in Calnuc
Comparative analysis of calnuc sequences revealed the presence of two putative Zn2+-binding sites. Among the Zn2+ binding motifs known to exist in proteins, we noticed two putative carboxypeptidase-like (HFREXnH and HFTEXnH (n = 108 to 135 residues)) Zn2+ binding motifs in the sequence of human calnuc located near the N terminus of the protein. The distribution of these sites among different species follows a distinct pattern. The second Zn2+ binding motif is largely conserved in lower organisms (only exception being Caenorhabditis elegans). In higher organisms both the Zn2+ binding motifs are conserved in isoform-1, whereas only the second Zn2+ binding motif was present in isoform-2 (Fig. 3). To pinpoint both Zn2+-binding site and the active site of the protease function, separate N- and C-terminal fragments of calnuc were generated. The N-terminal fragment (first 220 amino acids) comprised the putative DNA binding region. On the other hand, the C-terminal fragment contained the two EF-hand motifs, G protein-binding site, and the leucine zipper domain (Fig. 1).
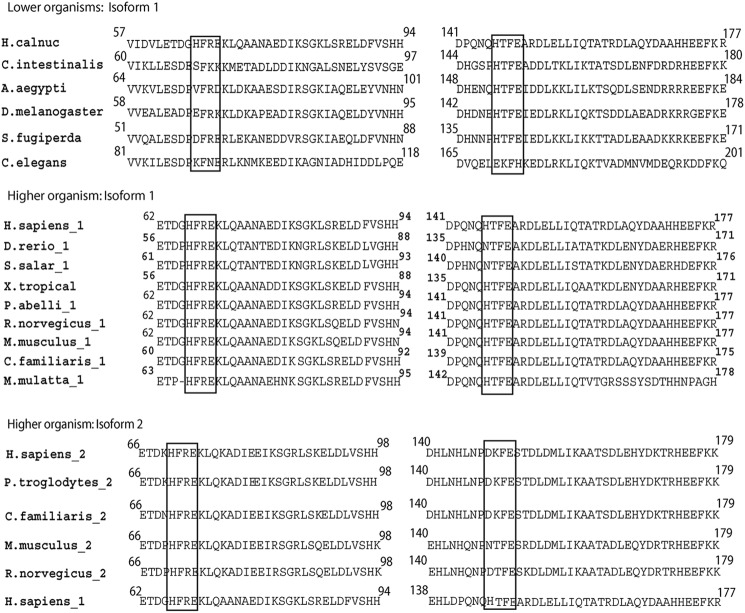
FIGURE 3.
Zn2+-binding sites (HXXEXnH) in calnuc. On the basis of the multiple sequence alignment of calnuc, organisms can be conveniently grouped as lower organisms and higher organisms. Among higher organisms, two different isoforms of calnuc are found. The lower organisms include Ciona, Caenorhabditis, Drosophila, Aedes, and Spodoptera, compared here with human calnuc (H. calnuc). In higher organisms, isoform 1 includes calnuc from Macaca, Danio, pongo, salmon, and Xenopus in addition to Rattus, Mus, Canis, and Homo. Isoform 2 includes calnuc from Rattus, Mus, Pan, Canis, and Homo. The two Zn2+-binding sites are compared among these species with respect to human calnuc isoform-1. Regions enclosed in blocks demonstrate the conserved residues in the upper and middle panel, whereas the right side of lower panel highlights lack of the conserved histidine residue (see Table 3). In lower organisms the second Zn2+-binding site is conserved in all species except in C. elegans. In the case of isoform-1 from higher organisms, both zinc binding motifs are conserved. In isoform-2 only the first zinc binding motif is conserved.
High Affinity Zn2+ Binding to Calnuc
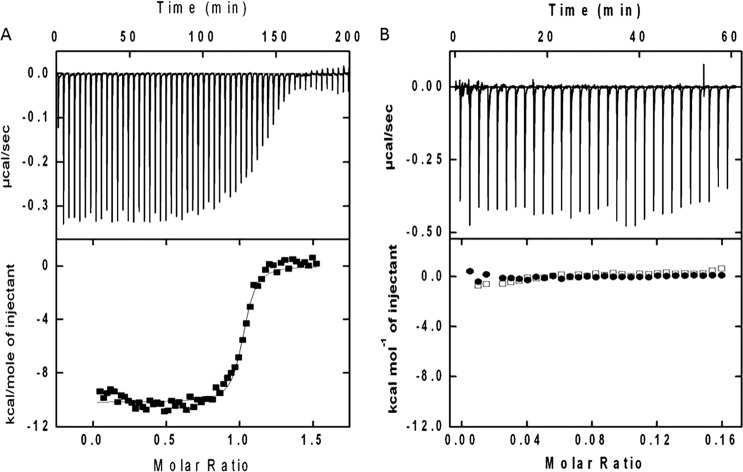
The data of heat change upon Zn2+ titration to calnuc, monitored by ITC (Fig. 4), were best fitted to the “one set of sites” model (supplemental Table S2). Zn2+ binding to calnuc is an exothermic reaction with favorable enthalpy (ΔH = −1.05 × 104 kcal/mol) and entropy (ΔS = 0.73 cal·mol−1). The dissociation constant (Kd = 1/Ka) of Zn2+ binding was 32 nm with one set of binding sites (n = 2.13 ± 0.007), suggesting that both the binding sites are almost identical (macroscopic binding constants for both Zn2+ binding motifs are similar). Mutations in the Zn2+-binding site (of residues that coordinate with the ion, H67A/147A; E70A/150A) did not elucidate any heat changes, signifying the total loss of binding. Thus, calnuc can be classified as a Zn2+-binding protein. Similarly we checked the binding affinity of the calnuc-N fragment by Scatchard plot analysis from fluorescence data. The calculated binding affinity (Kd) of Zn2+ to the N fragment is 20 nm, identical to calnuc (supplemental Fig. S3).
FIGURE 4.
Calnuc is a high affinity Zn2+ binding protein. Panel A, ITC data demonstrate heat changes upon titration of 18 μm apo calnuc with 3-μl aliquots of 200 μm ZnCl2 solution at 30 °C. All solutions were prepared in 20 mm Tris, pH 7.5, buffer containing 50 mm NaCl. A plot of heat change (kcal/mol) per injection of ZnCl2 as a function of molar ratio (metal/protein) is shown in the lower trace (heat differences obtained per injections). The best least-squares fit of the data (■) to a one set of sites model is given by the solid line. Results obtained are represented in Table 2. Panel B, mutations pinpoint Zn2+-binding site. The upper panel of the thermogram demonstrates integrated heat change is due to heat of dilution during each injection. In the lower panel, ITC profiles (□) correspond to the Zn2+-binding site mutant calnuc (H67A, E70A, H147A, and E150A) with Zn2+, whereas (●) represents Zn2+ titration in buffer alone at 30 °C. Concentrations of protein, Zn2+, and buffer conditions are same as in panel A.
Zn2+-mediated Conformational Changes in Calnuc; Evidence for Being a Zn2+ Sensor
Our results reveal Zn2+-dependent changes in conformation of calnuc. Incremental addition of Zn2+ exhibited a dose-dependent (5–20 μm Zn2+ to 1.6 μm calnuc) increase in tryptophan fluorescence emission spectra with a blue shift of 5 nm in the λmax,em (from 351 to 346 nm) as a result of conformational changes in the protein (Fig. 5A). Incidentally, Ca2+ does not influence the Zn2+-dependent changes in fluorescence.
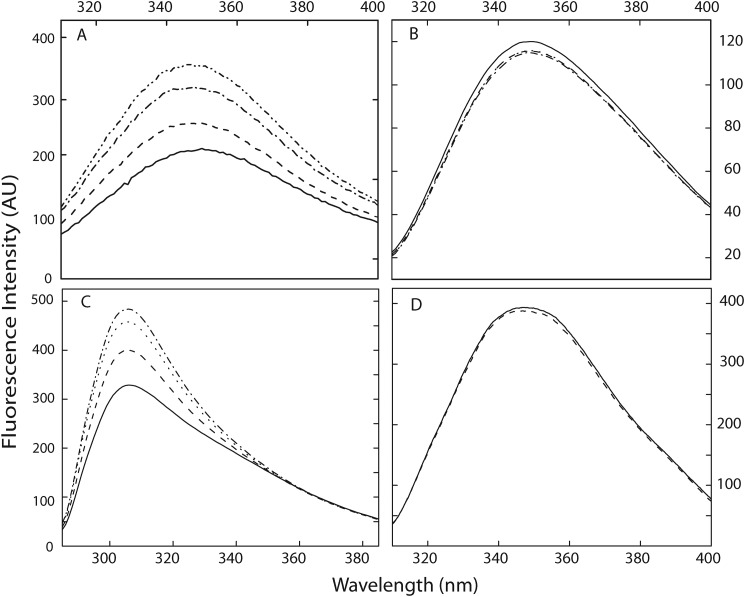
FIGURE 5.
Zn2+ induced conformational changes in calnuc and its fragments. Conformational changes induced by Zn2+ in calnuc and its fragments were monitored by the intrinsic fluorescence emission spectra of the protein. All spectra were recorded in 20 mm Tris buffer containing 50 mm NaCl, pH 7.5. Spectra are representative of independent experiments repeated several times at 25 °C. Panel A, shown are tryptophan fluorescence spectra of calnuc WT, recorded by exciting the protein at 295 nm. The solid line represents apocalnuc (0.04 μm). Dashed, dotted, and dash-dot-dash lines represent the changes upon the addition of 5, 10, and 20 μm Zn2+, respectively. AU, arbitrary units. Panel B, mutations in the Zn2+-binding site in calnuc led to a lack of spectral changes upon titration with Zn2+. The solid line represents the apo form of the Zn2+-binding site mutant (0.04 μm). Dashed, dotted, and dash-dot-dash lines represent the presence of 5, 10, and 20 μm Zn2+, respectively. Panel C, tyrosine fluorescence spectra of the calnuc N fragment were recorded by exciting sample at 275 nm. The solid line represents apocalnuc N fragment (0.04 μm). Dashed, dotted, and dash-dot-dash lines represent the changes upon the addition of 5, 10, and 20 μm Zn2+, respectively. Panel D, tryptophan fluorescence spectra of calnuc C-terminal fragment were recorded by exciting the protein at 295 nm. The solid line represents apocalnuc C fragment (0.04 μm). Dashed, dotted, and dash-dot-dash lines represent the changes upon the addition of 5, 10, and 20 μm Zn2+, respectively.
An increase in tyrosine emission fluorescence intensity (∼65%) of the N-terminal fragment was observed upon Zn2+ binding, similar to the full-length protein, whereas the tryptophan fluorescence signature of the C-terminal fragment remained unaffected by Zn2+ (Fig. 5, C and D). Further confirmation of the binding site was obtained by alanine-scanning mutants of the histidine and glutamate residues in the HXXE motif, which did not exhibited any change in fluorescence spectra (Fig. 5B).
Zn2+ binding (5 - 20 μm) to calnuc (1.6 μm) (Fig. 6A) and its N-terminal fragment (Fig. 6C) also led to increased ellipticity (in far-UV CD spectra), confirming marked structural changes in the protein. The calnuc Zn2+ motif mutant and C-terminal fragment (that lacks the zinc binding motif) did not exhibit any spectral changes upon titration with in presence of Zn2+ (Fig. 6, B and C).
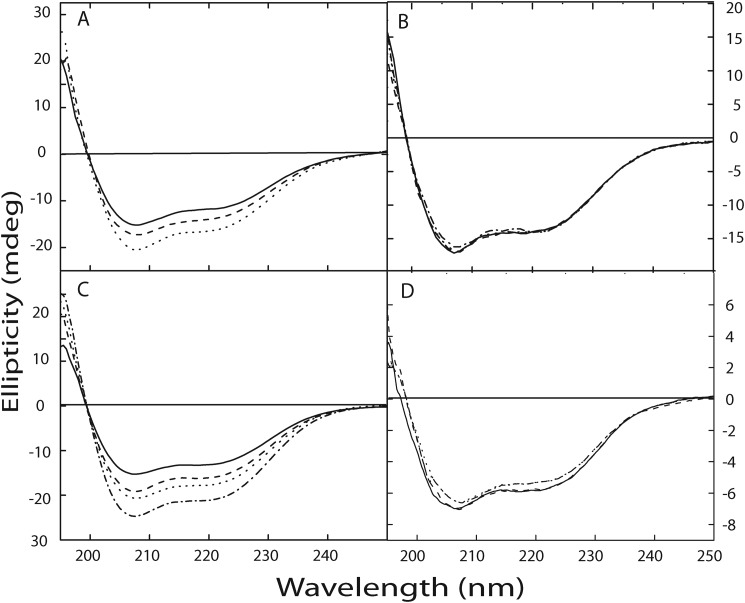
FIGURE 6.
Zn2+ induced changes in secondary structure of calnuc. Changes in the secondary structure of the protein upon interaction with Zn2+ were determined using far-UV CD spectra. The far-UV CD spectrum of calnuc and its mutants, illustrating a largely α-helical structure, were recorded in 20 mm Tris buffer containing 50 mm NaCl, pH 7.5. Spectra are representative of independent experiments repeated several times at 25 °C. Panel A, changes in the secondary structure of apocalnuc full-length upon Zn2+ binding are shown. The solid line denotes the spectra corresponding to apocalnuc only (0.4 μm), and the dashed and dotted lines represent the changes upon the addition of 10 and 20 μm Zn2+, respectively. Panel B illustrates that mutation at the Zn2+-binding site does not exhibit any changes in the secondary structure upon Zn2+ addition. The solid line denotes the spectra corresponding to the apo form of the Zn2+-binding site mutant (0.4 μm), and dashed and dotted lines represent the presence of 10 μm and 20 μm Zn2+, respectively. Panel C, changes in the secondary structure of the apocalnuc N fragment upon Zn2+ binding. The solid line denotes the spectra corresponding to apocalnuc N fragment (0.8 μm), and the dashed, dotted, and dash-dot-dashed lines represent the changes upon the addition of 10, 15, and 20 μm Zn2+, respectively. Panel D shows changes in the secondary structure of apocalnuc C fragment upon titration with Zn2+. The solid line denotes the spectra corresponding to apocalnuc C fragment (0.6 μm), and the dashed, dotted, and dash-dot-dashed lines represent the changes upon the addition of 10, 15, and 20 μm Zn2+, respectively.
Zn2+-induced Changes Leads to Exposure of the Hydrophobic Surface in Calnuc
Dramatic changes were observed in the surface hydrophobicity in calnuc upon Zn2+ binding, measured using the fluorescence signature of ANS, namely, enhancement and blue shift in spectra (λmax,em) of the probe (0.5 μm) bound to calnuc (1.6 μm) or its N-terminal fragment upon titration with Zn2+ (5 μm to 20 μm) (supplemental Fig. S4, A and B). Spectral properties of the probe bound to C fragment were not affected upon the addition of Zn2+. These spectral changes imply the presence of a Zn2+-operated conformational switch located at the N-terminal region and that calnuc could be defined as a Zn2+ sensor.
Protease Activity Is Dependent on Zn2+ but Not on Ca2+
The fact that the Zn2+ binding motif in calnuc is identical to the active site of family of carboxypeptidases initially led us to explore for the protease activity in the protein. Protease assay was performed with both the N and the C fragments. The N-terminal fragment, with the two Zn2+ binding motifs (lacking Ca2+-binding motif), did not exhibit protease activity. In contrast, proteolytic activity demonstrated by the C-terminal fragment is comparable with the activity shown by the full-length protein and is unaffected by Ca2+ binding (Fig. 2A). Studies to check the effect of metal ions on the protease activity established that Zn2+ exhibits significant inhibition. The two Zn2+ binding motifs (HXXE-AXXA) mutants did not show inhibition of enzyme activity upon the addition of increasing concentrations of Zn2+ (Fig. 7A). From these observations we hypothesize that Zn2+-induced structural changes in the N terminus of calnuc affect the formation of the catalytic triad and hence its proteolytic activity.
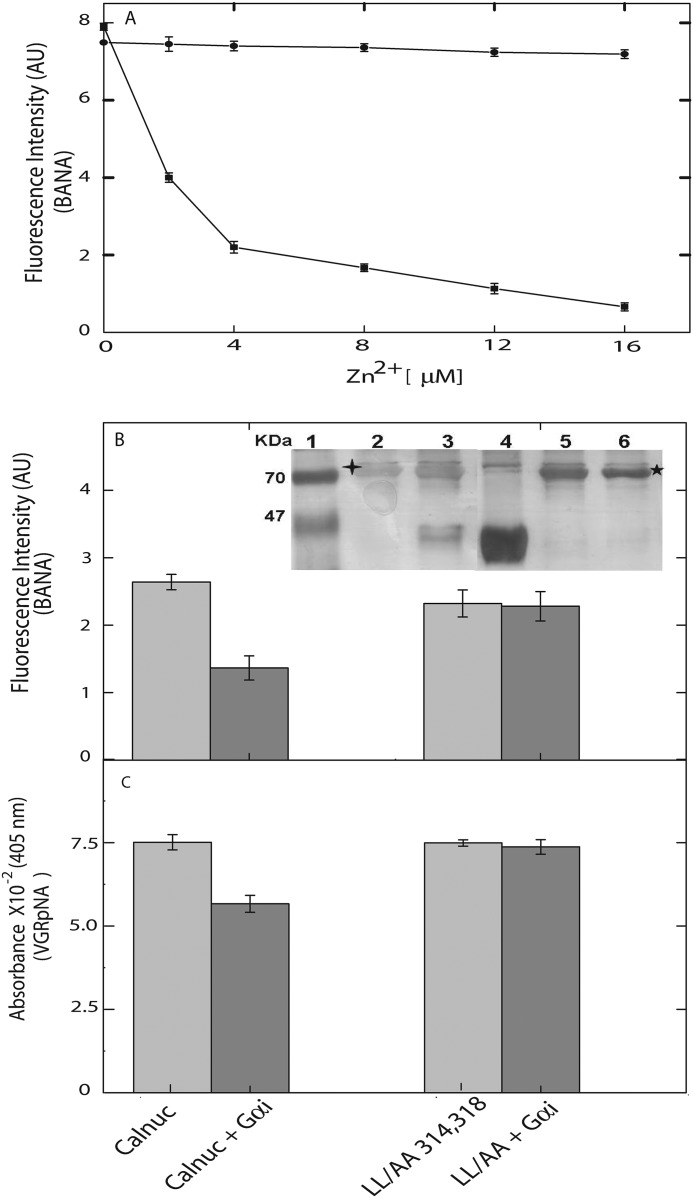
FIGURE 7.
G protein α subunit and Zn2+ modulate the protease activity calnuc. In a two-way locking system, G protein α subunit and Zn2+ allosterically inhibit the protease activity of calnuc. A, dose-dependent inhibition of enzyme activity of calnuc by Zn2+ was observed in wild type (closed squares) and not exhibited by the calnuc in which the Zn2+-binding site has been mutated (closed circles). AU, arbitrary units. B, calnuc (0.6 μm) was incubated with Gαi1 (0.6 μm). The sample was further incubated with fluorescent substrate (BANA) and then monitored by the increase in fluorescence intensity. The protease activity of calnuc (light gray bar) was reduced upon its interaction with Gαi (GDP)-bound form (gray bar). C, VGRpNA was used as the substrate, and activity was monitored by checking the absorbance at 405 nm. All other conditions were same as in panel B. In the case of the LL/AA calnuc mutant (light gray bar) the protease activity is not affected in the presence of the Gαi (GDP)-bound form (gray bar). An in vitro pulldown assay demonstrated calnuc interaction with G-protein. The LL/AA calnuc mutant did not interact with the G protein (inset). Lane 1, marker; lane 2, calnuc (WT); lane 3, calnuc (WT) + Gαi (GDP); lane 4, Gαi alone; lane 5, LL/AA double mutant alone; lane 6, LL/AA double mutant + Gαi (GDP).
G Protein Modulates the Protease Activity of Calnuc
To further understand the possible role of calnuc in signal transduction, mainly implicating its interactions with G proteins, we probed the involvement of these interactions on its protease activity. Protein-protein interactions with G protein α subunit (Gαi1) led to a sharp inhibition of protease activity of calnuc. Significant inhibition in the protease activity was elicited by G protein α subunit (Fig. 7, B and C). To ensure the effect of the Gαi protein on the protease activity of calnuc, L314A/L318A (LL to AA) double mutant, which has been demonstrated to lose its ability to bind to G protein (20), was generated. Protease activity of this double mutant is not attenuated by Gαi protein. Furthermore, pulldown assay results reveal the interaction of calnuc with Gαi, whereas the LL/AA mutant did not show any interaction (Fig. 7B, insert).
DISCUSSION
Calnuc is a structurally intricate Ca2+-binding protein implicated to be vital for many cellular processes that are attributed to its highly modular structure designed strategically to generate multiple interacting partners (for a recent review, see Ref. 31). Despite the variety of interactions with important molecules, physiological functions of calnuc are elusive. The multidomain structure of calnuc led us to search for the role of its non EF-hand regions. We report here several hitherto unknown properties of calnuc, i.e. its inherent serine protease activity and ability to bind Zn2+. Our data project a regulatory role for both Zn2+ and G proteins, that of holding the protease activity of calnuc in a locked state, and hence attributing a possible physiological function resulting from these interactions.
Calnuc Has Inactive Carboxypeptidase-type Zn2+-binding Sites
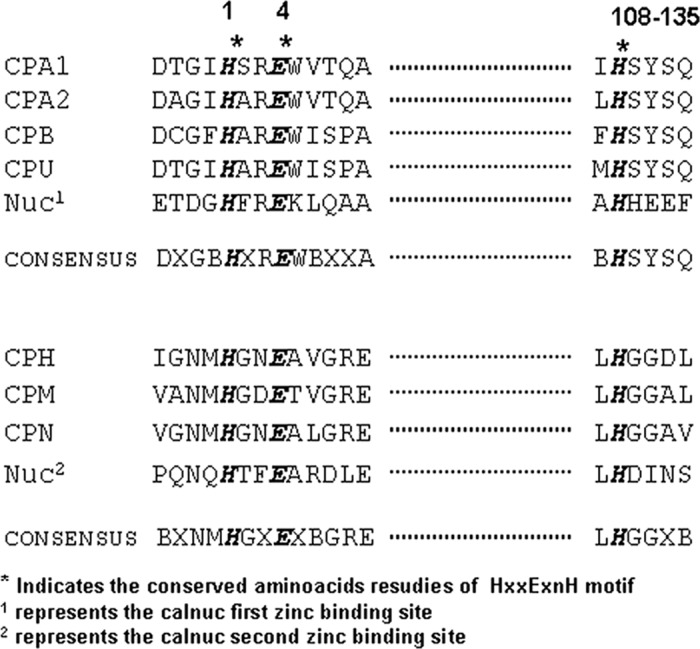
Calnuc (NUCB1 or isoform-1) has two putative Zn2+ binding motifs, HFREXnH and HFTEXnH (n = 108 to 135 residues), that are well conserved among various organisms (Table 3). Both the Zn2+-binding sites in calnuc, located in the vicinity of N terminus (Fig. 1), are similar to the active site of carboxypeptidase A family of proteases (24). Examination of the HXXEXnH (the two histidine and the glutamate residues act as ligands for Zn2+) type of active sites among carboxypeptidases revealed the requirement of additional conservation of amino acid residues around the histidine that acts as the third ligand for coordinating Zn2+ (either LHSYSQ or LHGGD) at a distance of 108–135 amino acids from the HFREX site. This sequence signature seems to be an absolute requirement to perform peptidase activity (32) (Table 3). Calnuc lacks the above-mentioned residues around the third coordinating histidine and, therefore, is unable to cleave substrate proteins by the Zn2+-bound HFREXnH and HFTEXnH sites. This renders the site to be proteolytically inactive even as two histidine and glutamate residues coordinate and bind Zn2+. We have compared and contrasted the Zn2+-binding sites in the isoform-2 of calnuc (NUCB2) among different species in addition to its comparison with NUCB1 (calnuc). Sequence analysis demonstrates that NUCB2, in most of the organisms, possesses only one Zn2+-binding motif (HXXEXnH) (Fig. 2B) (see below for more discussion).
TABLE 3.
Comparison of the calnuc HXXE zinc motif with consensus sequences of carboxypeptidase family of enzymes
Comparison of zinc binding motif (HXXEXnH) present in calnuc with the consensus sequences of carboxypeptidases is shown. The table shows the zinc binding motif, marked in bold, is conserved, whereas the sequence involved in the protease activity of carboxypeptidases is not conserved. Calnuc therefore binds Zn2+ but does not exhibit protease activity.
Zn2+ Sensing and Protease Active Site Are Exclusive of Each Other
Fragment-based approach also revealed that the N-terminal fragment, containing the Zn2+-binding sites, did not exhibit protease activity (Fig. 2A). On the other hand, the C-terminal fragment with two Ca2+-binding sites exhibited inherent protease activity. The extent of activity from the C-terminal fragment was similar to that of the full-length calnuc. Full-length protein and the N-terminal fragment exhibited specific Zn2+-dependent (Kd = 32 nm) structural changes (Fig. 5, A and C). These results establish that the Zn2+-binding site, located in the N-terminal region, is distant to the protease active site in the C-terminal region.
The protease function of calnuc is inhibited by PMSF and AEBSF, confirming it to be a serine protease (supplemental Fig. S1). Examination of the amino acid sequences of calnuc from various organisms revealed a highly conserved serine residue (Ser-224) (Table 1). Mutation of the calnuc Ser-224 either to alanine or valine caused significant inhibition but retention of 40–50% of the activity compared with wild type, whereas a Ser-378 mutation exhibited up to a 65% drop in activity. The involvement of Ser-224 in catalysis is indicated by the protease assay of the calnuc 239–461 fragment, which exhibited 60–65% inhibition. The S224A/S378A double mutant showed a 80–85% decrease in activity, implicating that both serines have the capacity to be a part of the enzymatic active site. This observation is supported by our preliminary structural analysis of the protein by homology modeling (result not shown). We observe that calnuc adopts a long coiled-coil-like structure. This probably leads to the two serines being close to each other, and therefore, either of them is able to partially fit in to the active site. The probable structure emanating from the modeling is also supported by the tryptophan fluorescence signature of the protein, with the λmax,em 351 nm due to the tryptophans experiencing a polar environment. This would happen only if the protein is not globular and adopts a coiled-coil extended structure. The partial activity exhibited by the mutants is also similar to many other serine proteases known to exhibit the residual enzyme activity even when the residues forming the catalytic triad are mutated (33). Distinctly sharp differences between the wild type and mutants are observed when well defined substrates are used (Fig. 2, C and D). Based on a detailed analysis, we inferred that Ser-378 of GXSXG motif and/or conserved Ser-224, Asp-328 of the DTG motif, and the His-339 construct the catalytic triad site. These residues are conserved among the spectrum of calnuc isofom-1 sequences from diverse species, suggesting that irrespective of the species calnuc, (NUCB1) should be classified as an EF-hand containing Ca2+ binding serine protease. Furthermore, we have confirmed the site at which calnuc cleaves other proteins. Using a variety of substrates, we demonstrate that the cleavage site is C terminus to an arginine residue, similar to the mechanism of trypsin.
It is interesting to note that, among the amino acid residues required for protease active site, NUCB2 possesses only the conserved Ser-224 and lacks Ser-378 (GXSXG motif), Asp-328 (DTG), and His-339 in those positions. NUCB2, however, possesses other conserved aspartate and histidine residues that might be involved in catalysis. As the structure of the two isoforms is still unknown, it is difficult to predict the role of Nucb2 as a protease based on the sequence alignment (experiments in this direction are under way in our laboratory).
Allosteric Regulation of the Protease Activity
In general, Zn2+ bound to proteins plays either a structural role or a catalytic role (34). Metal ion binding studies and their effect on the protease activity of calnuc have yielded extremely interesting results. Although Ca2+ binding does not affect the protease activity, Zn2+ causes a dramatic inhibition of the activity of calnuc. Our results, based on ITC and spectroscopy, demonstrate that calnuc is a high affinity Zn2+-binding protein in addition to being a Ca2+-binding protein. Zn2+ and Ca2+ binding processes are independent of each other. Because Zn2+ binding changes the protein conformation from open-to-close state, this might be a mode by which Zn2+ modulates the proteolytic activity of calnuc. A conformational switch caused by Zn2+ arrests the protease activity in a locked state; on the other hand, Ca2+ binding, known to cause minimal changes in protein conformation (21) thus does not affect its enzyme activity. Therefore, we propose a plausible model that calnuc has the ability to “sense” Zn2+ that mediates its enzymatic activity. Zn2+ elucidates its inhibitory action by switching the conformation of the protein, probably distorting the geometry of the serine protease active site. Additional structural analysis of Zn2+ binding to the fragments also validates that the serine protease activity of calnuc (located in its C-terminal region) is allosterically regulated by a conformational switch due to Zn2+ binding at the N-terminal HFREXnH and HFTEXnH sites.
Distribution of Zn2+-binding sites in calnuc among diverse organisms revealed a discrete pattern of conservation displaying both isoform and organism dependence. It would be interesting to analyze the comparative activity of calnuc with variability in its Zn2+ binding sequences. It is tempting to propose that although calnuc from all the organisms would exhibit serine protease activity, there would be significant differences in the activity based on the number and pattern of distribution of the Zn2+-binding sites.
Interaction of calnuc with G proteins, both organellar (on the Golgi) and cytoplasmic, and the ensuing biochemical effects are its best understood properties (17–21). In this study we demonstrate an important feature of this interaction. G protein α subunit attenuates the protease activity of calnuc. Interestingly, the G protein-binding site is located in the vicinity of the active site serine; thus, this interaction holds the enzymatic activity in a locked state similar to the Zn2+-bound form. Both calnuc (having many interacting partners) and G proteins are very important molecules for the physiological functioning of a cell. Interactions between calnuc and G proteins assume vital importance due to the already known fact that they are involved in regulating each other's trafficking behavior in cells (18). The fact that these two proteins are targeted to the Golgi and the cytoplasm and that calnuc is also secreted to the outside suggests that the interaction might play a role in keeping the enzymatic activity in the off state until calnuc is sequestered from the G protein for its enzymatic function.
The functions of the “Zn2+ key” in calnuc, which locks the enzymatic activity, is similar to that observed in several other proteases that are regulated by Zn2+-mediated conformational switches (35–40). The intracellular concentration of Zn2+ varies from the picomolar (in the cytosol) to 100 mm in various organelles (e.g. Zn2+ storage vacuoles), whereas the extracellular concentrations are in the micromolar range (34). The low cellular levels of Zn2+ results in acquiring the high affinities of proteins toward Zn2+ varying from picomolar (classic zinc fingers) to nanomolar (Zn2+-binding proteins like metallothionein) (34). Calnuc possesses a high affinity for Zn2+ in the nanomolar range that suggests its importance in cellular physiology. From our studies we hypothesize that Zn2+ probably plays a structural role in calnuc, and the varying concentrations of the metal ion in different compartments of the cell effect the function of the protein differently.
It will be interesting to understand the exact physiological and functional role of interaction between calnuc and the Zn2+ ions. Interestingly, Zn2+ has a vital link in the accumulation of amyloid-β peptide (Aβ) and formation of plaques in Alzheimer disease wherein calnuc involvement has been demonstrated (10). Zn2+ plays a protective role, causing a conformational change in the Aβ peptide and preventing the association of other metals and consequent oxidative damage associated with the metal-Aβ complex (41). Oxidative and nitrosative stress led to a release of Zn2+ from the vesicular zinc pool, and the excess Zn2+ promotes aggregation of Aβ (42, 43). It will be interesting to look into the role of the tripartite association of Aβ:calnuc:Zn2+ in the pathophysiology of the disease.
We hypothesize that in a tissue- and isoform expression-dependent manner, G proteins and Zn2+ fine-tune the enzyme activity of calnuc either to adjust to the condition of the cell or as a physiological response to a molecular signal. Involvement of G proteins in regulating activity also generates the possibility of selectivity based on expression pattern of the G proteins and extracellular signals. The complex and multidomain structure has evolved in to gain conformational plasticity to be able to perform multiple functions.
Acknowledgments
We acknowledge Dr. Yogendra Sharma (Centre for Cellular and Molecular Biology) for his critique and support. We also thank A. Sai Krishna for help in initial cloning experiments and Rama Ramesh for help in ITC experiments.
This work was supported by Indian Institute of Technology Madras (IITM), Council of Scientific and Industrial Research (India), Department of Science and Technology (DST) and Department of Biotechnology (DBT).

This article contains supplemental Tables 1 and 2 and Figs. 1–4.
- ANS
- 8-anilino-1-naphthalenesulfonic acid
- AEBSF
- 4-(2-aminoethyl) benzene sulfonyl fluoride hydrochloride
- BANA
- N-benzoyl-l-Arg β-naphthylamide
- FVRpNA
- n-benzoyl-Phe-Val-Arg-p-nitroanilide
- VGRpNA
- n-benzoyl-Val-Gly-Arg-p-nitroanilide
- ITC
- isothermal titration calorimetry
- Aβ
- amyloid-β peptide.
REFERENCES
- 1. Kawasaki H., Nakayama S., Kretsinger R. H. (1998) Classification and evolution of EF-hand proteins. Biometals 11, 277–295 [DOI] [PubMed] [Google Scholar]
- 2. Schaub M. C., Heizmann C. W. (2008) Calcium, troponin, calmodulin, S100 proteins. From myocardial basics to new therapeutic strategies. Biochem. Biophys. Res. Commun. 369, 247–264 [DOI] [PubMed] [Google Scholar]
- 3. Miura K., Kurosawa Y., Kanai Y. (1994) Calcium binding activity of nucleobindin mediated by an EF hand moiety. Biochem. Biophys. Res. Commun. 199, 1388–1393 [DOI] [PubMed] [Google Scholar]
- 4. Miura K., Titani K., Kurosawa Y., Kanai Y. (1992) Molecular cloning of nucleobindin, a novel DNA-binding protein that contains both a signal peptide and a leucine zipper structure. Biochem. Biophys. Res. Commun. 187, 375–380 [DOI] [PubMed] [Google Scholar]
- 5. Karabinos A., Bhattacharya D., Morys-Wortmann C., Kroll K., Hirschfeld G., Kratzin H. D., Barnikol-Watanabe S., Hilschmann N. (1996) The divergent domains of the NEFA and nucleobindin proteins are derived from an EF-hand ancestor. Mol. Biol. Evol. 13, 990–998 [DOI] [PubMed] [Google Scholar]
- 6. Karabinos A., Bhattacharya D., Kratzin H. D., Hilschmann N. (1998) Origin of the NEFA and Nuc signal sequences. J. Mol. Evol. 46, 327–333 [DOI] [PubMed] [Google Scholar]
- 7. Ballif B. A., Mincek N. V., Barratt J. T., Wilson M. L., Simmons D. L. (1996) Interaction of cyclooxygenases with an apoptosis- and autoimmunity-associated protein. Proc. Natl. Acad. Sci. USA 93, 5544–5549 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Leclerc P., Biarc J., St-Onge M., Gilbert C., Dussault A. A., Laflamme C., Pouliot M. (2008) Nucleobindin co-localizes and associates with cyclooxygenase, COX-2, in human neutrophils. PLoS One. 3, e2229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Mochizuki N., Hibi M., Kanai Y., Insel P. A. (1995) Interaction of the protein nucleobindin with G alpha i2, as revealed by the yeast two-hybrid system. FEBS Lett. 373, 155–158 [DOI] [PubMed] [Google Scholar]
- 10. Lin P., Li F., Zhang Y. W., Huang H., Tong G., Farquhar M. G., Xu H. (2007) Calnuc binds to Alzheimer's β-amyloid precursor protein and affects its biogenesis. J. Neurochem. 100, 1505–1514 [DOI] [PubMed] [Google Scholar]
- 11. Lin P., Yao Y., Hofmeister R., Tsien R. Y., Farquhar M. G. (1999) Overexpression of calnuc (Nucleobindin) increases agonist and thapsigargin releaseable calcium storage in the Golgi. J. Cell Biol. 145, 279–289 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Kanai Y., Tanuma S. (1992) Purification of a novel B cell growth and differentiation factor associated with lupus syndrome. Immunol. Lett. 32, 43–48 [DOI] [PubMed] [Google Scholar]
- 13. Kanai Y., Miura K., Uehara T., Amagai M., Takeda O., Tanuma S., Kurosawa Y. (1993) Natural occurrence of Nuc in the sera of autoimmune-prone MRL/lpr mice. Biochem. Biophys. Res. Commun. 196, 729–736 [DOI] [PubMed] [Google Scholar]
- 14. Tsukumo Y., Tomida A., Kitahara O., Nakamura Y., Asada S., Mori K., Tsuruo T. (2007) Nucleobindin 1 controls the unfolded protein response by inhibiting ATF6 activation. J. Biol. Chem. 282, 29264–29272 [DOI] [PubMed] [Google Scholar]
- 15. Valencia C. A., Cotten S. W., Duan J., Liu R. (2008) Modulation of nucleobindin-1 and nucleobindin-2 by caspases. FEBS Lett. 582, 286–290 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Chen Y., Lin P., Qiu S., Peng X. X., Looi K., Farquhar M. G., Zhang J. Y. (2007) Autoantibodies to Ca2+ binding protein Calnuc is a potential marker in colon cancer detection. Int. J. Oncol. 30, 1137–1144 [PubMed] [Google Scholar]
- 17. Weiss T. S., Chamberlain C. E., Takeda T., Lin P., Hahn K. M., Farquhar M. G. (2001) Gαi3 binding to calnuc on Golgi membranes in living cells monitored by fluorescence resonance energy transfer of green fluorescent protein fusion proteins. Proc. Natl. Acad. Sci. U. S. A. 98, 14961–14966 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Lin P., Fischer T., Lavoie C., Huang H., Farquhar M. G. (2009) Calnuc plays a role in dynamic distribution of Gαi but not Gβ subunits and modulates ACTH secretion in AtT-20 neuroendocrine secretory cells. Mol. Neurodegener. 4, 15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Kapoor N., Gupta R., Menon S. T., Folta-Stogniew E., Raleigh D. P., Sakmar T. P. (2010) Nucleobindin 1 is a calcium-regulated guanine nucleotide dissociation inhibitor of Gαi1. J. Biol. Chem. 285, 31647–31660 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Garcia-Marcos M., Kietrsunthorn P. S., Wang H., Ghosh P., Farquhar M. G. (2011) G protein binding sites on Calnuc (nucleobindin 1) and NUCB2 (nucleobindin 2) define a new class of Gαi-regulatory motifs. J. Biol. Chem. 286, 28138–28149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Kanuru M., Samuel J. J., Balivada L. M., Aradhyam G. K. (2009) Ion binding properties of Calnuc, Ca2+ versus Mg2+-calnuc adopts additional and unusual Ca2+-binding sites upon interaction with G-protein. FEBS J. 276, 2529–2546 [DOI] [PubMed] [Google Scholar]
- 22. Lowry O. H., Rosenbrough N. J., Farr A. L., Randall R. J. (1951) Protein measurement with the Folin phenol reagent. J. Biol. Chem. 193, 265–275 [PubMed] [Google Scholar]
- 23. Fahmy K., Sakmar T. P. (1993) Regulation of the rhodopsin-transducin interaction by a highly conserved carboxylic acid group. Biochemistry 32, 7229–7236 [DOI] [PubMed] [Google Scholar]
- 24. Scatchard G. (1949) The attractions of proteins for small molecules and ions. Ann. N.Y. Acad. Sci. 51, 660–672 [Google Scholar]
- 25. Twining S. S. (1984) Fluorescein isothiocyanate-labeled casein assay for proteolytic enzymes. Anal. Biochem. 143, 30–34 [DOI] [PubMed] [Google Scholar]
- 26. McDonald J. K., Ellis S., Reilly T. J. (1966) Properties of dipeptidyl arylamidase I of the pituitary. J. Biol. Chem. 241, 1494–1501 [PubMed] [Google Scholar]
- 27. Sarath G., Dela Motte R. S., Wagner W. (1989) Protease Assay: Methods in Proteolytic Enzymes. A Practical Approach, pp. 22–55, Oxford IRI Press, Oxford, UK [Google Scholar]
- 28. Tunlid A., Rosén S., Ek B., Rask L. (1994) Purification and characterization of an extracellular serine protease from the nematode-trapping fungus. Arthrobotrys oligospora. Microbiology 140, 1687–1695 [DOI] [PubMed] [Google Scholar]
- 29. Bergmeyer H. U., Gawehn K., Grassl M. (1974) Methods of Enzymatic Analysis, pp. 436–437, Academic Press Inc., New York [Google Scholar]
- 30. Taylor E. L., Meriwether B. P., Park J. H. (1963) The hydrolysis of p-nitrophenyl acetate catalyzed by 3-phosphoglyceraldehyde dehydrogenase crystallized from yeast. J. Biol. Chem. 238, 734–740 [PubMed] [Google Scholar]
- 31. Aradhyam G. K., Balivada L. M., Kanuru M., Vadivel P., Vidhya B. S. (2010) Calnuc. Emerging roles in calcium signaling and human diseases. IUBMB Life 62, 436–446 [DOI] [PubMed] [Google Scholar]
- 32. Hooper N. M. (1994) Families of zinc metalloproteases. FEBS Lett. 354, 1–6 [DOI] [PubMed] [Google Scholar]
- 33. Carter P., Wells J. A. (1988) Dissecting the catalytic triad of a serine protease. Nature 332, 564–568 [DOI] [PubMed] [Google Scholar]
- 34. Maret W., Li Y. (2009) Coordination dynamics of zinc in proteins. Chem. Rev. 109, 4682–4707 [DOI] [PubMed] [Google Scholar]
- 35. Geiger S. R., Böttcher T., Sieber S. A., Cramer P. (2011) A conformational switch underlies ClpP protease function. Angew. Chem. Int. Ed. Engl. 50, 5749–5752 [DOI] [PubMed] [Google Scholar]
- 36. Ostermeier M. (2009) Designing switchable enzymes. Curr. Opin. Struct. Biol. 19, 442–448 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Guntas G., Mansell T. J., Kim J. R., Ostermeier M. (2005) Directed evolution of protein switches and their application to the creation of ligand-binding proteins. Proc. Natl. Acad. Sci. U. S. A. 102, 11224–11229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Abian O., Neira J. L., Velazquez-Campoy A. (2009) Thermodynamics of zinc binding to hepatitis C virus NS3 protease. A folding by binding event. Proteins 77, 624–636 [DOI] [PubMed] [Google Scholar]
- 39. Liang J., Kim J. R., Boock J. T., Mansell T. J., Ostermeier M. (2007) Ligand binding and allostery can emerge simultaneously. Protein Sci. 16, 929–937 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Bauer M. C., Nilsson H., Thulin E., Frohm B., Malm J., Linse S. (2008) Zn2+ binding to human calbindin D28k and the role of histidine residues. Protein Sci. 17, 760–767 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Cuajungco M. P., Fagét K. Y., Huang X., Tanzi R. E., Bush A. I. (2000) Metal chelation as a potential therapy for Alzheimer's disease. Ann. N.Y. Acad. Sci. 920, 292–304 [DOI] [PubMed] [Google Scholar]
- 42. Cuajungco M. P., Lees G. J. (1997) Zinc metabolism in the brain. Relevance to human neurodegenerative disorders. Neurobiol. Dis. 4, 137–169 [DOI] [PubMed] [Google Scholar]
- 43. Bleackley M. R., Macgillivray R. T. (2011) Transition metal homeostasis. From yeast to human disease. Biometals 24, 785–809 [DOI] [PubMed] [Google Scholar]