Background: FHL2 is involved in regulation of the TGF-β signaling pathway.
Results: FHL2 increases the stability of the TGF-β pathway positive regulator Arkadia by inhibiting its ubiquitination and cooperates with Arkadia to activate TGF-β signaling.
Conclusion: FHL2 is an important regulator for Arkadia ubiquitination.
Significance: This provides novel insight into the mechanisms for the regulation of the TGF-β pathway.
Keywords: Cell Signaling, Coactivator Transcription, E3 Ubiquitin Ligase, Transforming Growth Factor β (TGFβ), Ubiquitination, Arkadia, Four and a Half LIM-only Protein 2, Really Interesting New Gene (RING) Domain E3 Ligase
Abstract
Arkadia is a RING-based ubiquitin ligase that positively regulates TGF-β signaling by targeting several pathway components for ubiquitination and degradation. However, little is known about the mechanisms controlling Arkadia activity. Here we show that the LIM-only protein FHL2 binds and synergistically cooperates with Arkadia to activate Smad3/Smad4-dependent transcription. Knockdown of FHL2 by RNA interference decreases Arkadia level and restricts the amplitude of Arkadia-induced TGF-β target gene responses. We found that Arkadia is ubiquitinated via K63- and K27-linked polyubiquitination. A single mutation at the RING domain that abolishes the E3 activity diminishes Arkadia ubiquitination, indicating that this modification partly involves autocatalytic process. Mutation of seven lysines at the C-terminal region of Arkadia severely impairs ubiquitination through the K27 but not the K63 linkage and slows down the turnover of Arkadia, suggesting that K27-linked polyubiquitination might promote proteolysis-dependent regulation of Arkadia. We show that FHL2 increases the half-life of Arkadia through inhibition of ubiquitin chain assembly on the protein, which provides a molecular basis for functional cooperation between Arkadia and FHL2 in enhancing TGF-β signaling. Our study uncovers a novel regulatory mechanism of Arkadia by ubiquitination and identifies FHL2 as important regulator of Arkadia ubiquitination and TGF-β signal transduction.
Introduction
Arkadia is a really interesting new gene (RING)5 domain E3 that plays a crucial role in the transmission of transforming growth factor β (TGF-β)/activin and bone morphogenetic protein (BMP) signaling (1). The binding of TGF-β/activin to cognate serine/threonine kinase receptors induces phosphorylation and activation of the receptor-regulated Smads (R-Smads) proteins including Smad2 and Smad3. Activated R-Smads form a heteromeric complex with their common partner Smad4, and shuttle to the nucleus where they additionally recruit transcriptional coactivators or corepressors to control the expression of target genes. The closely related transcription factors Ski and SnoN repress the function of the Smad complex in activating TGF-β target transcription. Arkadia targets Ski/SnoN for ubiquitination, but efficient degradation of ubiquinated Ski/SnoN requires the presence of activated Smad2/3 in cells. Upon TGF-β stimulation, phosphorylated Smad2/3 (P-Smad) interacts with Arkadia and Ski/SnoN, triggering the degradation of Ski/SnoN, which leads to transcriptional activation of TGF-β target genes (2, 3). Another regulatory mechanism employed by Arkadia is to target the inhibitor Smad7 for polyubiquitination and degradation (1, 4). In addition, Arkadia selectively recognizes P-Smad2/3, but not the unphosphorylated forms, leading to ubiquitination and degradation of P-Smad2/3, which switch off the TGF-β signal in the nucleus and reset the cell to the arrival of novel cues (5).
Ubiquitination is a key regulatory mechanism of cell functions. This covalent modification involves the formation of an isopeptide bond between the C-terminal glycine residue of ubiquitin and a lysine residue on the target protein. Ubiquitin is a 76 amino acid protein that contains seven lysine residues (K6, K11, K27, K29, K33, K48, and K63). Substrate-conjugated ubiquitin itself can be further ubiquitinated through one of its seven lysines to form a polyubiquitin chain (6). Ubiquitin signal supports either proteolytic or nonproteolytic functions, which are largely determined by the type of linkage through which the ubiquitin chain is attached (7). Ubiquitination reactions involve three families of enzymes, including ubiquitin-activating enzymes (E1), ubiquitin-conjugating enzymes (E2), and ubiquitin ligases (E3). Two major E3 subfamilies are identified as homologous to E6AP C terminus (HECT) domain E3 and RING domain E3. Many E3s are capable of targeting themselves for ubiquitination (8).
The four and a half LIM-only protein 2 (FHL2) contains exclusively LIM domains that serve for protein-protein interaction. This unique structure confers to this protein the ability to establish multiple interactions with cellular proteins, fostering the assembly of protein complexes with diverse functions. In the cytoplasm, FHL2 interacts with integrins and focal adhesion kinases and plays a role in transmission of extracellular matrix (ECM)/integrin receptor-mediated signals (9–11). FHL2 also mediates transduction of RANKL signaling through interaction with TRAF6 (12, 13). In the nucleus, FHL2 binds a diverse group of DNA binding factors to control a broad range of transcription programs including β-catenin/TCF, AP1, and androgen receptor (12, 14–22). Particularly, FHL2 has been shown to interact with several components of the TGF-β pathway, notably with Ski, Smad2, Smad3, and Smad4, and to increase casein kinase 1δ-mediated phosphorylation of Smad2/3 that activates TGF-β signaling (21, 23). In this study, we investigated interaction and functional cooperation between FHL2 and the RING E3 Arkadia to further shed light on the role of FHL2 in the transmission of TGF-β signaling.
EXPERIMENTAL PROCEDURES
Plasmids
Ubiquitin expression constructs were kindly supplied by Dr. Ron Kopito (24). We transferred the ubiquitin inserts into the pcDNA3 vector with N-terminal Xpress tags. Arkadia mutants C-K/R, 1–400, 400–665, 665-Cter, and 400-Cter were constructed by PCR and cloned into pCMV5. C937A and FHL2 constructs have been described previously (2, 17).
Cell Culture, Transient Transfection, and Luciferase Assay
WT and FHL2−/− mouse embryonic fibroblasts (MEFs) have been described previously (25). HeLa, HepG2, 293T, HEK293, and HaCaT cells were cultured in Dulbecco's modified Eagle's medium containing 10% fetal bovine serum, penicillin, and streptomycin. For transient transfection, cells were seeded on a 24-well plate and transfected using Lipofectamine 2000 (Invitrogen). Thirty-six hours post-transfection, cells were treated with 10 μm SB 431542 (SBI, Sigma-Aldrich) overnight and then induced by TGFβ-1 (2 ng/ml) or Activin A (20 ng/ml) (R&D Systems) for 1 or 3 h as indicated. Luciferase activity was determined using the Dual-Luciferase Reporter Assay System (Promega). The total amount DNA in each transfection was balanced by empty vector plasmid. Luciferase activities were normalized to Renilla activities under the control of thymidine kinase. All experiments were performed in duplicate and repeated at least three times.
RNA Interference and Oligonucleotides
FHL2 expression was inhibited in HepG2 cells by lentiviral transduction of FHL2 shRNA purchased from Santa Cruz Biotechnology, followed by limiting dilution onto 96-well plates and selection with puromycin. In addition, we constructed two independent FHL2 shRNA expression vectors to knock down endogenous FHL2 in HaCaT cells. Sense and antisense hairpin oligonucleotides were synthesized according to The RNAi Consortium (TRC) (Broad Institute) and cloned into pLKO.1-puro vector. shRNA lentiviral particles were produced as described previously (26). The following oligonucleotides were used for shRNA expression in pLKO.1: 5′-CCGG-CGACTGCTTTAACTGTAAGAA-CTCGAG-TTCTTACAGTTAAAGCAGTCG-TTTTT-3′ (shFHL2-5773, clone ID: TRCN0000005773); 5′-CCGG-CGAATCTCTCTTTGGCAAGAA-CTCGAG-TTCTTGCCAAAGAGAGATTCG-TTTTT-3′(shFHL2–5774, clone ID: TRCN0000005774). 5′-TCCGG-ATGAAGCAGCACGACTTCTTC-CTCGAG-GAAGAAGTCGTGCTGCTTCAT-TTTTTTG-3′ (control, clone ID: TRCN0000206279).
Ubiquitination Assay
Ubiquitination assays were carried out according to (27). Briefly, 293T cells were transfected using Lipofectamine 2000. 44 h later, cells were treated with 10 mm MG132 for 4 h and then lysed with denaturing lysis buffer (6 m guanidine-HCI/100 mm sodium phosphate buffer pH 8.0) with 5 mm imidazole. Lysates were subjected to brief sonication at the minimum setting to reduce the viscosity. Lysates were then cleared by centrifugation for 15 min at 14,000 × g at 4 °C, followed by incubation with nitrilotriacetic acid agarose beads (Ni-NTA) for 4 h at 4 °C. The beads were washed successively with denaturing lysis buffer pH 8.0, denaturing lysis buffer pH 5.8, and protein buffer (50 mm sodium phosphate buffer, pH 8.0, 100 mm KCl, 20% (v/v) glycerol, 0.2% (v/v) Nonidet P-40). Bound proteins were eluted with Laemmli buffer at 95 °C and subjected to SDS-PAGE. Proteins were transferred to the nitrocellulose membranes and analyzed by immunoblotting. For in vitro ubiquitination assay, Flag-empty vector, Flag-wt Arkadia or Flag-C937A were transfected into HEK293 cells and immunoprecipitated with anti-Flag antibody in buffer A containing 50 mm Tris (pH 8), 150 mm NaCl, 1% Nonidet P-40, 0.5% deoxycholate, 0.1% SDS, and protease inhibitors (Roche). The immunoprecipitated proteins were collected with protein A/G-Sepharose beads. The beads were washed three times with buffer A and two times with ubiquitination buffer 1× (50 mm Tris-HCl, pH 7.5, 150 mm NaCl, DTT 1 mm). Ubiquitination assay was performed with beads bound Flag-proteins in the presence of 100 ng of UbcH5b, 100 ng of UbcH5c, 5 μg of HA-Ub, 50 ng of UBE1, and 1× Energy Regenerating Solution containing ATP and MgCl2 (Boston Biochem) in ubiquitination buffer for 1h at 30 °C. Reactions were stopped by the addition of Laemmli buffer and analyzed by Western blotting.
Immunoprecipitation and Immunoblotting
For immunoprecipitation of overexpressed Arkadia and FHL2, 293T cells were transiently transfected by Lipofectamine 2000. Cells were harvested 48 h later and lysed in radio immunoprecipitation assay (RIPA) buffer (50 mm Tris, pH 7.6, 150 mm NaCl, 2 mm EDTA, 1% Nonidet P-40, complete protease inhibitor mixture, 20 μm MG132). For immunoprecipitation of endogenous proteins, HeLa cells were lysed in RIPA buffer. Lysate were pre-cleared by incubation with mouse IgG antibodies and protein A/G-Sepharose beads for 1 h. Pre-cleared lysates were immunoprecipitated with specific antibodies or control antibody. The immunoprecipitated proteins were collected with protein A/G-Sepharose beads. The beads were washed five times with RIPA buffer and bound proteins were eluted with Laemmli buffer at 95 °C and subjected to SDS-PAGE. Proteins were transferred to the nitrocellulose membranes and analyzed by immunoblotting. The following primary antibodies were used: FHL2 (MBL), Arkadia (Santa Cruz Biotechnology), Anti-Xpress (Invitrogen), hemagglutinin (HA), Flag (M2), tubulin, and β-actin (Sigma). To determine Arkadia turnover, cells were treated with 100 μg/ml cycloheximide (CHX, Sigma-Aldrich) and harvested at indicated times. Signal intensities on immunoblots were scanned and quantified by using the LI-COR/Odyssey infrared image system.
RESULTS
FHL2 Interacts and Cooperates with Arkadia in Smad3/Smad4-dependent Transcription
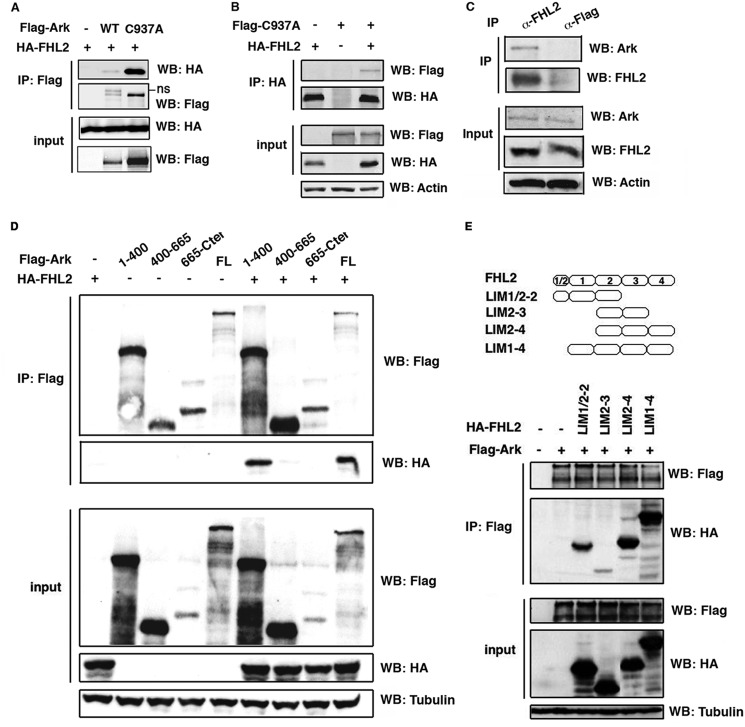
As Ski is a substrate of Arkadia and binds to FHL2 (3, 23), we searched for potential interaction between FHL2 and Arkadia. 293T cells were transfected with HA-tagged FHL2 along with wt or C937A Flag-Arkadia followed by precipitation with anti-Flag antibody. The C937A mutant results from a single amino acid substitution in the RING domain that inactivates its E3 ubiquitin ligase activity and is stably expressed (2) (Fig. 1A). Immunoblotting analysis showed that FHL2 interacted with both wt and the RING mutant (Fig. 1A). Conversely, we used 293T cells cotransfected with HA-FHL2 and the stable C937A mutant for immunoprecipitation assay with anti-HA antibody. Arkadia was detected in the immune complexes only in the presence of FHL2 (Fig. 1B). We further investigated the association of these two proteins at endogenous expression levels using HeLa cells in which both FHL2 and Arkadia are expressed at detectable levels (Fig. 1C). Cell lysates were first immunoprecipitated with specific antibody against FHL2, followed by immunoblotting with anti-Arkadia antibody. Arkadia was specifically detected in FHL2 immune complexes, but not in control anti-Flag antibody immunoprecipitated complex (Fig. 1C), showing that FHL2 can complex with Arkadia in a physiological condition.
FIGURE 1.
FHL2 interacts with Arkadia. A and B, 293T cells were cotransfected with vectors encoding Flag-tagged WT or C937A mutant Arkadia and HA-tagged FHL2. Cell lysates were immunoprecipitated with anti-Flag (A) or anti-HA (B) antibodies followed by immunoblotting analysis. ns: nonspecific. C, HeLa cells were treated with MG132 for 4 h prior to immunoprecipitation with anti-FHL2 antibody and control anti-Flag antibody followed by immunoblotting with anti-Arkadia antibody. D, 293T cells were transfected with Flag-tagged fragments of Arkadia and HA-tagged FHL2. Cell lysates were immunoprecipitated with anti-Flag antibody followed by immunoblotting analysis. FL: full-length. E, FHL2 constructs are depicted on the top. 293T cells were transfected with Flag-tagged Arkadia and HA-tagged FHL2 fragments. Cell lysates were immunoprecipitated with anti-Flag antibody followed by immunoblotting analysis.
We then mapped the interaction domains of Arkadia and FHL2 using a series of expression vectors encoding different fragments of Arkadia and FHL2. After transient co-transfection of HA-FHL2 with different fragments of Flag-Arkadia in 293T cells, lysates were immunoprecipited with anti-Flag antibody, followed by immunoblotting analysis with anti-Flag and anti-HA antibodies. FHL2 was predominantly detected in the immune complexes of the Arkadia N-terminal fragment encompassing 400 amino acids (1–400), and to a minor extent with the fragment 400–665, but not the C-terminal fragment (Fig. 1D). Next, we constructed FHL2 fragments containing different LIM domains and tested their capacity to interact with Arkadia by immunoprecipitation. As shown in Fig. 1E, strong interaction was observed with LIM1/2–2, LIM2–4, and LIM1–4. Therefore, the N-terminal region of Arkadia as well as the N- and the C-terminal LIM domains of FHL2 are mainly involved in the interaction of the two proteins.
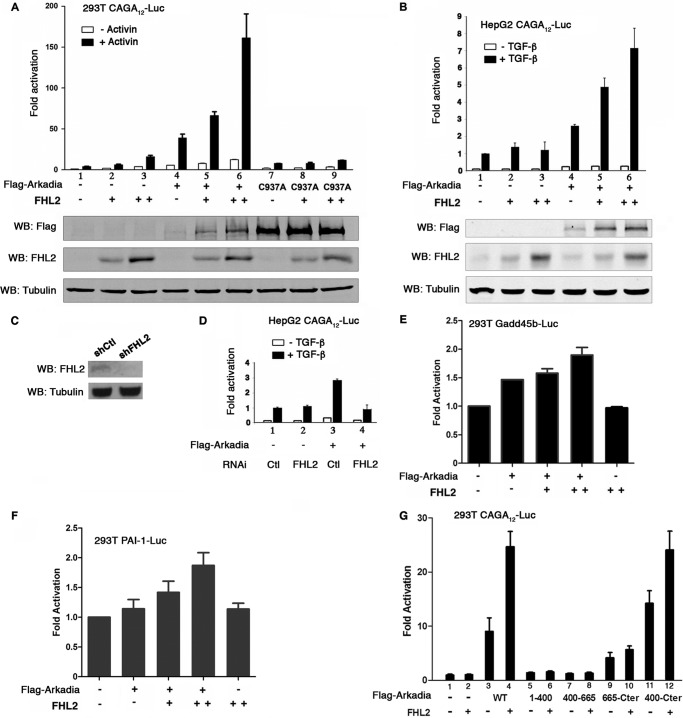
We evaluated the impact of interaction of FHL2 with Arkadia on TGF-β signal transduction in 293T and HepG2 cells treated with SB431542 (SBI) to inhibit TGF-β autocrine signaling followed by stimulation with Activin A, a homologue of TGF-β, or TGF-β for 1 h. Cells were transfected with FHL2 and the CAGA12-Luc Smad3/Smad4-dependent reporter. FHL2 moderately enhanced TGF-β-stimulated reporter activity in 293T (Fig. 2A, lanes 2–3), indicating that FHL2 can positively regulate the TGF-β signal. Arkadia is a potent activator of Smad3/Smad4-dependent transcription (Fig. 2A, lane 4 and Fig. 2B, lane 4) (2, 3). This function requires the E3 ubiquitin ligase activity, as luciferase activity was not activated by the C937A mutant (Fig. 2A, lane 7) (2, 3). Remarkably, FHL2 cooperated with Arkadia to further enhance TGF-β signaling in a dose-dependent manner in both 293T and HepG2 cells (Fig. 2A, lanes 5 and 6 and Fig. 2B, lanes 5 and 6). We knocked down FHL2 by RNA interference in HepG2 cells (Fig. 2C). Knockdown of FHL2 abolished Arkadia-mediated reporter activation (Fig. 2D, compare lane 4 with lane 3). No synergistic effect on the reporter transcription was observed when FHL2 was coexpressed with the C937A mutant (Fig. 2A, lanes 8–9), indicating that the E3 activity is required for the cooperation between FHL2 and Arkadia. We then investigated the response of natural TGF-β responsive promoters to the combined action of FHL2 and Arkadia. The growth arrest DNA damage (Gadd) 45b promoter was moderately but significantly activated by FHL2 and Arkadia (Fig. 2E). Similar results were obtained with the type-1 inhibitor of plasminogen activator (PAI-1) promoter (Fig. 2F).
FIGURE 2.
FHL2 cooperates with Arkadia in activation of Smad3/Smad4-dependent transcription. A, 293T cells were transfected with the TGF-β-responsive luciferase reporter (CAGA)12-Luc (0.1 μg) together with Arkadia or C937A and increasing doses of FHL2 (0.1 μg and 0.3 μg). TK-Renilla was used as internal control. Cells were treated with SBI prior to stimulation with Activin A for 1 h. The basal activity of the (CAGA)12-Luc reporter cotransfected with empty vector was arbitrarily set at 1, and data are presented as mean induction in luciferase activity ± S.D. from duplicate samples. The results shown are representative of those from more than four independent assays. Bottom: expression levels of transfected Flag-tagged Arkadia and FHL2 determined by Western blotting (WB). B, HepG2 cells were transfected with the reporter (CAGA)12-Luc (0.1 μg) together with Arkadia and increasing doses of FHL2 (0.1 μg and 0.3 μg). Cells were treated with SBI prior to stimulation with TGF-β for 1 h. Bottom: expression levels of transfected Flag-tagged Arkadia and FHL2 determined by Western blotting. C, knockdown of FHL2 in HepG2 cells. HepG2 cells were transduced with either control (shCtl) or FHL2 shRNA (shFHL2) lentiviral vectors (Santa Cruz Biotechnology). FHL2 expression was analyzed by Western blotting. D, reporter assay in HepG2 cells transduced with control shRNA (lanes 1 and 2) or FHL2 shRNA lentiviral vector (lanes 3 and 4). RNAi: RNA interference. Ctl: control. Cells were treated with SBI prior to stimulation with TGF-β for 1 h. E and F, luciferase assay in 293T cells transfected with either Gadd45b (E) or PAI-1 (F) promoter reporters with different doses of FHL2 (0.1 μg and 0.3 μg). 44 h after transfection, cells were stimulated with Activin A for 3 h. G, 293T cells were transfected with luciferase reporter (CAGA)12-Luc along with FHL2 and Arkadia subfragments as indicated. Cells were stimulated with Activin for 3 h.
Finally, we examined the requirement of the interaction between FHL2 and Arkadia in the activation of TGF-β-stimulated reporter. Different fragments of Arkadia were cotransfected with FHL2 in 293T cells followed by stimulation with Activin A for 3 h. As expected, the Arkadia N-terminal fragments (1–400 and 400–665) devoid of the RING domain failed to activate the CAGA12-Luc reporter and FHL2 had no effect on these fragments (Fig. 2G, lanes 5–8). The fragment 665-Cter, which contains the RING domain but did not bind to FHL2, activated the reporter. However, its activity could not be further augmented by FHL2 (Fig. 2G, compare lane 10 with lane 9). In stark contrast, FHL2 readily cooperated with the fragment 400-Cter to enhance the reporter activity (Fig. 2G, compare lane 12 to lane 11). These results indicate that direct interaction of FHL2 with Arkadia is important in their cooperation.
FHL2 Stabilizes Arkadia
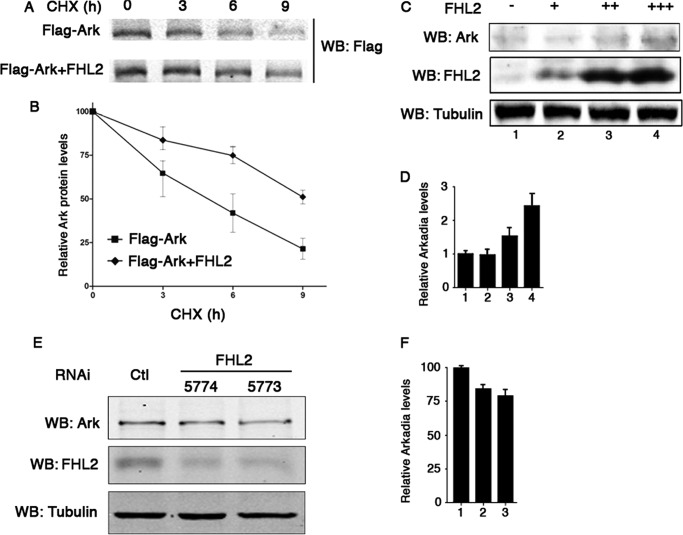
To investigate the mechanisms of cooperation between FHL2 and Arkadia, we assessed the impact of FHL2 on the stability of Arkadia by transfecting Flag-Arkadia with or without FHL2 in 293T cells, followed by blocking protein synthesis with cycloheximide. Overexpression of FHL2 resulted in a sharp increase in the stability of Arkadia (Fig. 3, A and B). This observation was further confirmed with endogenous Arkadia. As shown in Fig. 3, C and D, increasing FHL2 expression significantly augmented the levels of Arkadia in 293T cells. Conversely, knockdown of FHL2 in HaCaT cells decreased the levels of endogenous Arkadia by 15% to 20% (Fig. 3, E and F). These results suggest that FHL2 might be specifically involved in the regulation of Arkadia degradation.
FIGURE 3.
FHL2 slows down the turnover of Arkadia. A and B, 293T cells were transfected with Flag-tagged Arkadia with or without FHL2 and treated with cycloheximide (CHX) as indicated. Immunoblotting analysis was performed with anti-Flag antibody (A). Signal intensities were plotted relative to 0 h values. Data are presented as the mean values ± S.D. from three independent experiments (B). C and D, 293T cells were transfected with increasing doses of FHL2 (+: 0.5 μg, ++: 1.0 μg, +++: 1.5 μg). Endogenous Arkadia was analyzed by immunoblotting (C). Signal intensities were plotted relative to the values without transfected FHL2. Data are presented as the mean values ± standard deviation from three independent experiments (D). E and F, knockdown of FHL2 in HaCaT cell. HaCaT cells were transduced with either control (Ctl) or two independent FHL2 shRNA (shFHL2) lentiviral particles (5774 and 5773). Expression of endogenous Arkadia was analyzed by immunoblotting (E). Signal intensities were measured relative to the values transduced with control particles (F).
Arkadia Is Polyubiquitinated through K27- and K63-linked Ubiquitin
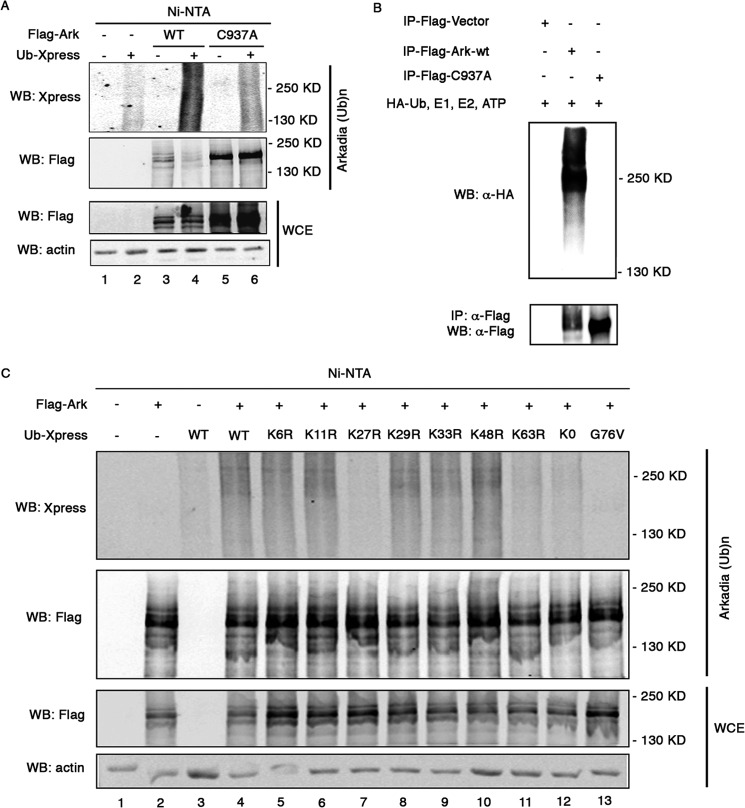
As protein degradation is closely related to ubiquitination, we tested whether Arkadia could undergo this post-translational modification. 293T cells were transfected with wt or C937A mutant Flag-Arkadia along with Xpress-tagged ubiquitin (Ub-Xpress). We took advantage of the presence of seven histidine residues (amino acid positions from 514 to 520) in Arkadia to precipitate the protein with Ni-NTA-agarose beads under denaturing condition and probed ubiquitin with anti-Xpress antibody. As shown in Fig. 4A, Arkadia was ubiquitinated, as attested by the detection of Arkadia-conjugated ubiquitin signals (lane 4). Ubiquitination of the C937A mutant was substantially reduced compared with the wt protein (Fig. 4A, compare lane 6 with lane 4), indicating that ubiquitination of Arkadia is partly catalyzed by itself. We then expressed Flag-tagged wt Arkadia and the C937A mutant in HEK293 cells, pulled down Arkadia with anti-Flag antibody and performed in vitro ubiquitination assay. As shown in Fig. 4B, ubiquitin signals were readily detected in wt Arkadia but not in C937A. This finding confirms the auto-ubiquitination ability of Arkadia and indicates that other E3 ligases are involved in C937A ubiquitination in vivo (see Fig. 4A, lane 6).
FIGURE 4.
Arkadia is polyubiquitinated through K27- and K63-linked ubiquitin. A, 293T cells were cotransfected with vectors encoding Flag-tagged WT Arkadia or C937A mutant along with Xpress-tagged ubiqutitin. Before lysis, cells were treated with MG132 for 4 h. Whole cell extracts were subjected to precipitation with Ni-NTA-agarose beads, followed by immuoblotting with anti-Xpress and FLAG antibody. B, in vitro ubiquitination assay. Flag-tagged WT Arkadia and C937A mutant were transfected into HEK293 cells and pulled down with anti-Flag antibody. Bead-bound proteins were subjected to in vitro ubiquitination assay in the presence of HA-tagged ubiquitin, E1, E2, and ATP, followed by Western blotting analysis. C, Arkadia ubiquitination reactions with WT, K6R, K11R, K27R, K29R, K33R, K48R, K63R, K0, or G76V ubiquitin were precipitated with Ni-NTA-agarose beads and analyzed by immunoblotting. Ub: ubiquitin. WCE: whole cell extract.
We next investigated the type of linkage in Arkadia by transfecting Arkadia in 293T cells along with ubiquitin mutants containing arginine substitution at indicated position (Fig. 4C, lanes 5–13). The K0 mutant, which has all of lysines substituted by arginine, allows only monoubiquitination. The conjugation-deficient G76V mutant, which lacks the C-terminal glycine residue, was used as negative control for ubiquitination. Arkadia was firstly pulled down with Ni-NTA beads and analyzed for ubiquitination by immunoblotting with anti-Xpress antibody. Expression of the K0 mutant reduced ubiquitination of Arkadia to the background level (Fig. 4C, compare lane 12 with lane 3), indicating that polyubiquitination is mainly involved in ubiquitination of Arkadia. While the polyubiquitin linkage profiles from K6R, K11R, K29R, K33R, and K48R ubiquitin remained similar to wt ubiquitin, mutations in lysine 27 or lysine 63 of ubiquitin severely impaired Arkadia ubiquitination (Fig. 4C, lanes 7 and 11), revealing that Arkadia is predominantly modified by K27- and K63-linked ubiquitin.
K27-linked Polyubiquitination at Arkadia N-terminal Region
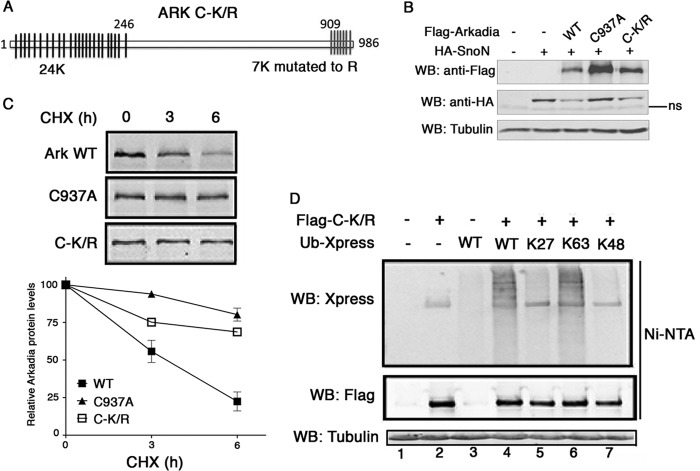
Arkadia contains two clusters of lysines including 24 lysines at the N terminus and 7 lysines at the C terminus (Fig. 5A). To determine the lysines targeted by ubiquitin, we constructed a mutant (C-K/R) in which all the seven lysines at the C terminus were changed to arginines, while keeping the N-terminal lysines. We first assessed the ability of the C-K/R mutant to degrade SnoN by transfecting HA-SnoN with wt Arkadia, C937A or C-K/R in 293T cells. After 44 h, cells were activated with Activin A for 3 h, followed by immunoblotting analysis of SnoN protein level. As shown in Fig. 5B, in contrast to the inactive mutant C937A, C-K/R had the same capacity as wt Arkadia to degrade SnoN. Thus, ubiquitination at the C terminus is not required for the E3 activity of Arkadia. Remarkably, even though the same amount of plasmids was transfected in cells, the protein level of C-K/R was much higher than that of wt (Fig. 5B), suggesting that mutations in C-K/R may enforce its stability. Indeed, blocking protein synthesis with cycloheximide showed that wt Arkadia had a much faster turnover rate than both C937A and C-K/R mutants (Fig. 5C), suggesting that self-ubiquitination of Arkadia might support proteolytic function. We then carried out ubiquitination assay using the C-K/R construct transfected with wt or ubiquitin mutants containing arginine substitutions on all lysines except the one at indicated position. Mutations in C-K/R severely abolished K27-linked modification, but had only minor effect on the K63 linkage (Fig. 5D, lanes 5 and 6), suggesting that K27-linked polyubiquitination predominantly affects the C terminus of Arkadia, while K63 polyubiquitination might be mainly at the N terminus. As control, K48-linked ubiquitination was undetectable (Fig. 5D, lane 7).
FIGURE 5.
K27-linked polyubiquitination at the N-terminal region of Arkadia. A, schematic representation of lysine positions in Arkadia and location of lysine to arginine mutations in C-K/R mutant. B, C-K/R mutant conserves the E3 ligase activity. 293T cells were co-transfected with HA-SnoN and Flag-Arkadia (WT, C937A, and C-K/R). SnoN expression was determined by Western blotting. ns: nonspecific. C, turnover of Arkadia is dependent on the E3 activity and C-terminal ubiqutitination. 293T cells were transfected with Flag-tagged WT Arkadia, C937A, or C-K/R and treated with cycloheximide as indicated. Immunoblotting analysis was performed with anti-Flag antibody. Signal intensities were plotted relative to 0 h values. Data are presented as the mean values ± standard deviation from three independent experiments. CHX: cycloheximide. D, 293T cells were cotransfected with vectors encoding Flag-tagged C-K/R along with Xpress-tagged WT, K27, K63, or K48 ubiqutitin. Ubiquitination of C-K/R was analyzed by immunoblotting with anti-Xpress and FLAG antibody. Ub: ubiquitin. WCE: whole cell extract.
Taken together, these findings demonstrate that specific linkages involve distinct regions of Arkadia. Ubiquitination through K27 of the ubiquitin chain, which is not required for Arkadia-mediated SnoN degradation, might function as a degradation signal in vivo.
FHL2 Inhibits Arkadia Ubiquitination
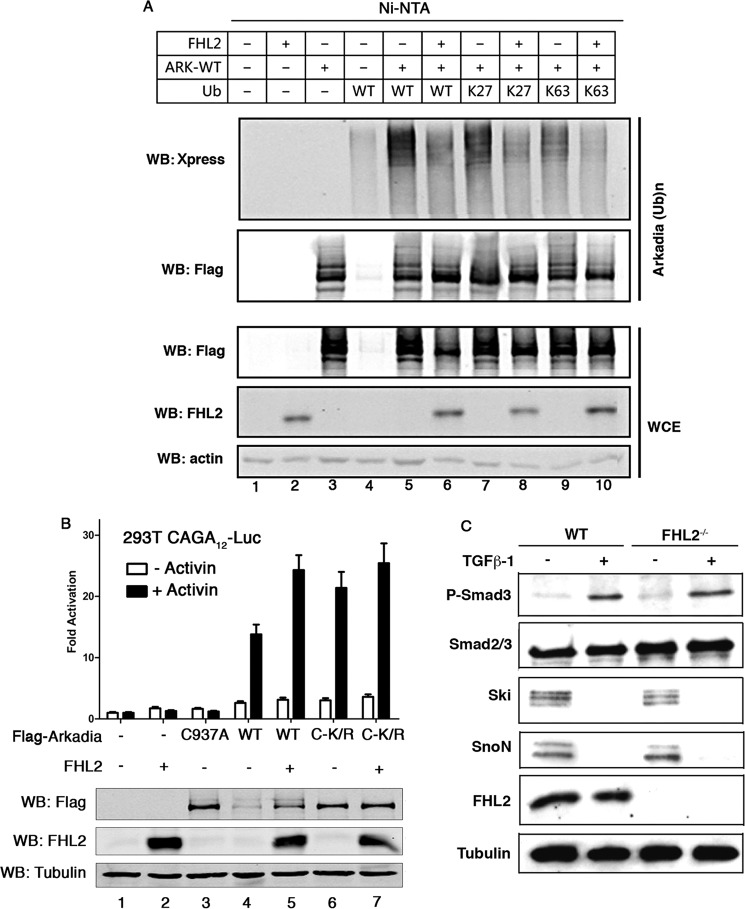
To assess whether FHL2 could play a role in ubiquitination of Arkadia, we cotransfected Arkadia with FHL2 as well as wt ubiquitin or K27 and K63 mutants in 293T cells followed by ubiquitination assay. Immunoblot analysis of lysates after Ni-NTA pull-down revealed that FHL2 caused a striking decrease in Arkadia ubiquitination, affecting the abundance of both K27- and K63-linked chains (Fig. 6A, compare lane 6 with lane 5, lane 8 with lane 7 and lane 10 with lane 9). These findings correlate with the observations that FHL2 stabilizes Arkadia.
FIGURE 6.
FHL2 decreases Arkadia ubiquitination. A, 293T cells were transfected with different combination of Flag-tagged Arkadia, FHL2 and Xpress-tagged WT, K27, and K63 ubiqutitin as indicated. Before lysis, cells were treated with MG132 for 4 h. Whole cell extracts were subjected to precipitation with Ni-NTA-agarose beads, followed by immuoblotting with anti-Xpress and FLAG antibody. Ub: ubiquitin. WCE: whole cell extract. B, FHL2 had moderate effects on the stabilized C-K/R in activating luciferase activity. 293T cells were transfected with the luciferase reporter (CAGA)12-Luc together with Arkadia, C937A, C-K/R, and FHL2 as indicated. Cells were treated with SBI prior to stimulation with Activin A for 3 h. Bottom: expression levels of transfected Flag-tagged Arkadia and FHL2 determined by Western blotting. C, WT and FHL2−/− MEFs were treated with SBI prior to stimulation with TGF-β for 1 h. Expression levels of phospho-Smad3, SnoN, and Ski were analyzed by immunoblotting.
The ability of FHL2 to stabilize Arkadia might account for the cooperation between FHL2 and Arkadia in activation of TGF-β targets. This hypothesis was investigated in a reporter assay comparing wt and C-K/R Arkadia in combination with FHL2. As shown in Fig. 6B, after treatment with Activin for 3 h, C-K/R showed higher activity than wt in activating the reporter (compare lane 6 with lane 4), correlating with the observation that C-K/R was more stable than wt Arkadia. Interestingly, whereas FHL2 readily enhanced wt Arkadia activating activity, it only had moderate effects on the C-K/R mutant (Fig. 6B, compare lane 7 on lane 6 with lane 5 on lane 4). Finally, we examined if FHL2 could have impact on the stability of Arkadia targets. WT and FHL2−/− mouse embryonic fibroblasts (MEFs) (25) were analyzed for expression of P-Smad3, Smad7, SnoN, and Ski after treatment with TGF-β. As shown in Fig. 6C, except for Smad7 for which expression was not detectable in these cells (data not shown), similar expression levels were observed for P-Smad3, SnoN, and Ski between wt and FHL2−/− MEFs, showing that deficiency of FHL2 had no effect on the stability of these factors of the TGF-β pathway. Taken together, these data suggest that FHL2 can interfere in Arkadia ubiquitination, resulting in stabilization of Arkadia and activation of TGF-β signaling.
DISCUSSION
Although several substrates of Arkadia have been identified in the recent years, little is known about the composition, regulation and function of this E3 ligase. We show here that FHL2 is a binding partner of Arkadia and stabilizes the protein by regulating its ubiquitination, leading to enhanced transduction of TGF-β signals. FHL2 is composed of four and a half LIM domains, which share primary sequence pattern of cysteine and histidine with the RING domain, but are devoid of E3 ubiquitin ligase activity (28). Instead, the LIM domains confer to FHL2 the ability to engage in protein-protein interactions with a large variety of proteins. Recently, binding proteins are emerging as important players in tight control over the dynamic ubiquitin ligase complexes; e.g. the cullin-binding protein CAND1 sequesters cullins in an inactive state, thus blocking the activity of the E3 complexes (29, 30). Similar mechanism could be employed by FHL2 to inhibit ubiquitination of Arkadia. Alternatively, FHL2 could compete with a specific E2 in the binding with Arkadia, thus depriving the RING E3 ligase of the access to conjugated ubiquitin. FHL2 could also bring deubiquitinases or inhibitors of ubiquitination to the Arkadia complex. Further studies are needed to elucidate the mechanisms underlying Arkadia ubiquitination and its regulation by FHL2. Interestingly, FHL2 is known to interact with other RING domain E3s or their interacting proteins such as BRCA1, TRAF6 and DNA damage-binding protein 1 (DDB1) (12, 31),6 suggesting that FHL2 might be part of distinct RING ubiquitin ligases complexes and have diverse functions in ubiquitin signaling.
The output of FHL2 interaction with Arkadia is the activation of TGF-β signaling. FHL2 also interacts with the inactive mutant C937A, but this interaction lacks functional cooperation. Thus, ubiquitin signaling might be a key factor in the mechanisms of cooperation between Arkadia and FHL2. Curiously, deficiency of FHL2 did not modify the degradation of P-Smad3, SnoN, and Ski by Arkadia, raising the possibility that stabilization of Arkadia by FHL2 may affect yet unidentified substrates. Furthermore, Arkadia has been found in the Smad complex associated with DNA (3). It is possible that stabilized Arkadia might act directly on the target promoters as transcriptional coactivator. In sum, FHL2 can contribute to many segments of the intracellular TGF-β signaling cascade through implication in either Smad phosphorylation (21) or Arkadia ubiquitination. Furthermore, FHL2 is itself a target of TGF-β (32), indicating that FHL2 can monitor the amplitude of TGF-β response through positive feedback links that would be determinant for outcome of the signal.
Despite the critical role of Arkadia in TGF-β signaling, many aspects of this RING-based ligase remain poorly understood. The current study demonstrates ubiquitination of Arkadia as a crucial regulatory mechanism. Two types of linkage are identified in the Arkadia ubiquitin conjugates, including K27- and K63-linked ubiquitin chains. Polyubiquitin chains linked through K27 have been identified in Parkin and voltage-dependent anion channel, which target the proteins for mitophagy (33), whereas K27-linked ubiquitination in c-Jun leads to lysosomal localization of c-Jun (34). In Arkadia, mutations that blunt ubiquitination through K27 linkage stabilize the protein, suggesting that K27-linked chains provide signals to drive Arkadia for degradation. Like many E3s, Arkadia can be self-ubiquitinated. Inactivating mutation in the RING domain delays Arkadia turnover in vivo, indicating that autocatalytic ubiquitination might be involved in K27-linked chain synthesis. Although direct evidence of how K63-linked chains regulate Arkadia is lacking, K63-mediated ubiquitin conjugation has been shown to play key roles in protein-protein interaction and protein kinase activation (35). Elucidating these functions in further studies is crucial for understanding the biochemical mechanisms operating in TGF-β signal transmission.
Acknowledgments
We thank Dr. Vincent Deubel for support of the project. We thank Dr. Ron R. Kopito for providing the ubiquitin plasmids. We are grateful to Drs. Yves Jacob and Marie-Louise Michel for insightful discussion.
This work was supported in part by the French Ligue contre le Cancer, Comité d'Ile-de-France, the Association pour la Recherche sur le Cancer, Total-IPS scientific partnership program, and National Science and Technology Major Projects of China (2008ZX10002006, 2012ZX10002007).
Y. Wei, unpublished data.
- RING
- really interesting new gene
- BMP
- bone morphogenetic protein
- FHL2
- four and a half LIM-only protein 2
- ECM
- extracellular matrix
- MEF
- mouse embryonic fibroblasts.
REFERENCES
- 1. Koinuma D., Shinozaki M., Komuro A., Goto K., Saitoh M., Hanyu A., Ebina M., Nukiwa T., Miyazawa K., Imamura T., Miyazono K. (2003) Arkadia amplifies TGF-β superfamily signaling through degradation of Smad7. EMBO J. 22, 6458–6470 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Levy L., Howell M., Das D., Harkin S., Episkopou V., Hill C. S. (2007) Arkadia activates Smad3/Smad4-dependent transcription by triggering signal-induced SnoN degradation. Mol. Cell. Biol. 27, 6068–6083 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Nagano Y., Mavrakis K. J., Lee K. L., Fujii T., Koinuma D., Sase H., Yuki K., Isogaya K., Saitoh M., Imamura T., Episkopou V., Miyazono K., Miyazawa K. (2007) Arkadia induces degradation of SnoN and c-Ski to enhance transforming growth factor-β signaling. J. Biol. Chem. 282, 20492–20501 [DOI] [PubMed] [Google Scholar]
- 4. Liu W., Rui H., Wang J., Lin S., He Y., Chen M., Li Q., Ye Z., Zhang S., Chan S. C., Chen Y. G., Han J., Lin S. C. (2006) Axin is a scaffold protein in TGF-β signaling that promotes degradation of Smad7 by Arkadia. EMBO J. 25, 1646–1658 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Mavrakis K. J., Andrew R. L., Lee K. L., Petropoulou C., Dixon J. E., Navaratnam N., Norris D. P., Episkopou V. (2007) Arkadia enhances Nodal/TGF-β signaling by coupling phospho-Smad2/3 activity and turnover. PLoS Biol. 5, e67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Peng J., Schwartz D., Elias J. E., Thoreen C. C., Cheng D., Marsischky G., Roelofs J., Finley D., Gygi S. P. (2003) A proteomics approach to understanding protein ubiquitination. Nat. Biotechnol. 21, 921–926 [DOI] [PubMed] [Google Scholar]
- 7. Ikeda F., Dikic I. (2008) Atypical ubiquitin chains: new molecular signals. 'Protein Modifications: Beyond the Usual Suspects' review series. EMBO Rep. 9, 536–542 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Weissman A. M., Shabek N., Ciechanover A. (2011) The predator becomes the prey: regulating the ubiquitin system by ubiquitylation and degradation. Nat. Rev. Mol. Cell Biol. 12, 605–620 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Park J., Will C., Martin B., Gullotti L., Friedrichs N., Buettner R., Schneider H., Ludwig S., Wixler V. (2008) Deficiency in the LIM-only protein FHL2 impairs assembly of extracellular matrix proteins. Faseb J. 22, 2508–2520 [DOI] [PubMed] [Google Scholar]
- 10. Samson T., Smyth N., Janetzky S., Wendler O., Müller J. M., Schüle R., von der Mark H., von der Mark K., Wixler V. (2004) The LIM-only proteins FHL2 and FHL3 interact with α- and β-subunits of the muscle α7β1 integrin receptor. J. Biol. Chem. 279, 28641–28652 [DOI] [PubMed] [Google Scholar]
- 11. Wixler V., Geerts D., Laplantine E., Westhoff D., Smyth N., Aumailley M., Sonnenberg A., Paulsson M. (2000) The LIM-only protein DRAL/FHL2 binds to the cytoplasmic domain of several α and β integrin chains and is recruited to adhesion complexes. J. Biol. Chem. 275, 33669–33678 [DOI] [PubMed] [Google Scholar]
- 12. Bai S., Kitaura H., Zhao H., Chen J., Müller J. M., Schüle R., Darnay B., Novack D. V., Ross F. P., Teitelbaum S. L. (2005) FHL2 inhibits the activated osteoclast in a TRAF6-dependent manner. J. Clin. Invest. 115, 2742–2751 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Bai S., Zha J., Zhao H., Ross F. P., Teitelbaum S. L. (2008) Tumor necrosis factor receptor-associated factor 6 is an intranuclear transcriptional coactivator in osteoclasts. J. Biol. Chem. 283, 30861–30867 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Müller J. M., Isele U., Metzger E., Rempel A., Moser M., Pscherer A., Breyer T., Holubarsch C., Buettner R., Schüle R. (2000) FHL2, a novel tissue-specific coactivator of the androgen receptor. EMBO J. 19, 359–369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Fimia G. M., De Cesare D., Sassone-Corsi P. (2000) A family of LIM-only transcriptional coactivators: tissue-specific expression and selective activation of CREB and CREM. Mol. Cell. Biol. 20, 8613–8622 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Martin B., Schneider R., Janetzky S., Waibler Z., Pandur P., Kühl M., Behrens J., von der Mark K., Starzinski-Powitz A., Wixler V. (2002) The LIM-only protein FHL2 interacts with β-catenin and promotes differentiation of mouse myoblasts. J. Cell Biol. 159, 113–122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Wei Y., Renard C. A., Labalette C., Wu Y., Lévy L., Neuveut C., Prieur X., Flajolet M., Prigent S., Buendia M. A. (2003) Identification of the LIM protein FHL2 as a coactivator of β-catenin. J. Biol. Chem. 278, 5188–5194 [DOI] [PubMed] [Google Scholar]
- 18. Morlon A., Sassone-Corsi P. (2003) The LIM-only protein FHL2 is a serum-inducible transcriptional coactivator of AP-1. Proc. Natl. Acad. Sci. U.S.A. 100, 3977–3982 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Purcell N. H., Darwis D., Bueno O. F., Müller J. M., Schüle R., Molkentin J. D. (2004) Extracellular signal-regulated kinase 2 interacts with and is negatively regulated by the LIM-only protein FHL2 in cardiomyocytes. Mol. Cell. Biol. 24, 1081–1095 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Paul C., Lacroix M., Iankova I., Julien E., Schäfer B. W., Labalette C., Wei Y., Cam A. L., Le Cam A., Sardet C. (2006) The LIM-only protein FHL2 is a negative regulator of E4F1. Oncogene 25, 5475–5484 [DOI] [PubMed] [Google Scholar]
- 21. Ding L., Wang Z., Yan J., Yang X., Liu A., Qiu W., Zhu J., Han J., Zhang H., Lin J., Cheng L., Qin X., Niu C., Yuan B., Wang X., Zhu C., Zhou Y., Li J., Song H., Huang C., Ye Q. (2009) Human four-and-a-half LIM family members suppress tumor cell growth through a TGF-β-like signaling pathway. J. Clin. Invest. 119, 349–361 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Maetzel D., Denzel S., Mack B., Canis M., Went P., Benk M., Kieu C., Papior P., Baeuerle P. A., Munz M., Gires O. (2009) Nuclear signalling by tumour-associated antigen EpCAM. Nat. Cell Biol. 11, 162–171 [DOI] [PubMed] [Google Scholar]
- 23. Chen D., Xu W., Bales E., Colmenares C., Conacci-Sorrell M., Ishii S., Stavnezer E., Campisi J., Fisher D. E., Ben-Ze'ev A., Medrano E. E. (2003) SKI activates Wnt/β-catenin signaling in human melanoma. Cancer Res. 63, 6626–6634 [PubMed] [Google Scholar]
- 24. Ward C. L., Omura S., Kopito R. R. (1995) Degradation of CFTR by the ubiquitin-proteasome pathway. Cell 83, 121–127 [DOI] [PubMed] [Google Scholar]
- 25. Labalette C., Nouët Y., Sobczak-Thepot J., Armengol C., Levillayer F., Gendron M. C., Renard C. A., Regnault B., Chen J., Buendia M. A., Wei Y. (2008) The LIM-only protein FHL2 regulates cyclin D1 expression and cell proliferation. J. Biol. Chem. 283, 15201–15208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Root D. E., Hacohen N., Hahn W. C., Lander E. S., Sabatini D. M. (2006) Genome-scale loss-of-function screening with a lentiviral RNAi library. Nat. Methods 3, 715–719 [DOI] [PubMed] [Google Scholar]
- 27. Laney J. D., Hochstrasser M. (2011) Analysis of protein ubiquitination. Curr. Protoc Protein Sci. Chapter 14, Unit14 15 [DOI] [PubMed] [Google Scholar]
- 28. Deshaies R. J., Joazeiro C. A. (2009) RING domain E3 ubiquitin ligases. Annu. Rev. Biochem. 78, 399–434 [DOI] [PubMed] [Google Scholar]
- 29. Liu J., Furukawa M., Matsumoto T., Xiong Y. (2002) NEDD8 modification of CUL1 dissociates p120(CAND1), an inhibitor of CUL1-SKP1 binding and SCF ligases. Mol. Cell 10, 1511–1518 [DOI] [PubMed] [Google Scholar]
- 30. Zheng J., Yang X., Harrell J. M., Ryzhikov S., Shim E. H., Lykke-Andersen K., Wei N., Sun H., Kobayashi R., Zhang H. (2002) CAND1 binds to unneddylated CUL1 and regulates the formation of SCF ubiquitin E3 ligase complex. Mol. Cell 10, 1519–1526 [DOI] [PubMed] [Google Scholar]
- 31. Yan J., Zhu J., Zhong H., Lu Q., Huang C., Ye Q. (2003) BRCA1 interacts with FHL2 and enhances FHL2 transactivation function. FEBS Lett. 553, 183–189 [DOI] [PubMed] [Google Scholar]
- 32. Levy L., Hill C. S. (2005) Smad4 dependency defines two classes of transforming growth factor {β} (TGF-{β}) target genes and distinguishes TGF-{β}-induced epithelial-mesenchymal transition from its antiproliferative and migratory responses. Mol. Cell. Biol. 25, 8108–8125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Geisler S., Holmström K. M., Skujat D., Fiesel F. C., Rothfuss O. C., Kahle P. J., Springer W. (2010) PINK1/Parkin-mediated mitophagy is dependent on VDAC1 and p62/SQSTM1. Nat. Cell Biol. 12, 119–131 [DOI] [PubMed] [Google Scholar]
- 34. Ikeda H., Kerppola T. K. (2008) Lysosomal localization of ubiquitinated Jun requires multiple determinants in a lysine-27-linked polyubiquitin conjugate. Mol. Biol. Cell 19, 4588–4601 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Chen Z. J., Sun L. J. (2009) Nonproteolytic functions of ubiquitin in cell signaling. Mol. Cell 33, 275–286 [DOI] [PubMed] [Google Scholar]