Background: Mechanistic insight into allosteric modulation of GPCRs can facilitate antagonist design.
Results: Extracellular surface residues (ECS) of the α1B-adrenoceptor at the base of extracellular loop 3 interact with the allosteric antagonist TIA.
Conclusion: The identified ECS pharmacophore provides the first structural constraints for allosteric antagonist design at α1-adrenoceptors.
Significance: Binding to the ECS of a GPCR can allosterically inhibit agonist signaling.
Keywords: Adrenergic Receptor, Allosteric Regulation, G Protein-coupled Receptors (GPCR), Molecular Modeling, Molecular Pharmacology, Peptide Interactions, Toxins, Conotoxin
Abstract
The G protein-coupled receptor (GPCR) superfamily is an important drug target that includes over 1000 membrane receptors that functionally couple extracellular stimuli to intracellular effectors. Despite the potential of extracellular surface (ECS) residues in GPCRs to interact with subtype-specific allosteric modulators, few ECS pharmacophores for class A receptors have been identified. Using the turkey β1-adrenergic receptor crystal structure, we modeled the α1B-adrenoceptor (α1B-AR) to help identify the allosteric site for ρ-conopeptide TIA, an inverse agonist at this receptor. Combining mutational radioligand binding and inositol 1-phosphate signaling studies, together with molecular docking simulations using a refined NMR structure of ρ-TIA, we identified 14 residues on the ECS of the α1B-AR that influenced ρ-TIA binding. Double mutant cycle analysis and docking confirmed that ρ-TIA binding was dominated by a salt bridge and cation-π between Arg-4-ρ-TIA and Asp-327 and Phe-330, respectively, and a T-stacking-π interaction between Trp-3-ρ-TIA and Phe-330. Water-bridging hydrogen bonds between Asn-2-ρ-TIA and Val-197, Trp-3-ρ-TIA and Ser-318, and the positively charged N terminus and Glu-186, were also identified. These interactions reveal that peptide binding to the ECS on transmembrane helix 6 (TMH6) and TMH7 at the base of extracellular loop 3 (ECL3) is sufficient to allosterically inhibit agonist signaling at a GPCR. The ligand-accessible ECS residues identified provide the first view of an allosteric inhibitor pharmacophore for α1-adrenoceptors and mechanistic insight and a new set of structural constraints for the design of allosteric antagonists at related GPCRs.
Introduction
G protein-coupled receptors (GPCRs)2 play a major role in physiological responses to hormones and neurotransmitters, in addition to being responsible for vision, olfaction, and taste (1). The initial GPCR x-ray structure of bovine rhodopsin determined by Palczewski et al. (2) provided the first useful template for building homology models of mammalian GPCRs. However, the low level of sequence homology with other GPCR families and the uncertainty in aligning the variable loop regions have limited structure-based drug design using the bovine rhodopsin derived models (3). Recent x-ray structures of bovine opsin (4, 5), human β2-adrenergic (6), turkey β1-adrenergic (7), human A2A-adenosine (8), human dopamine D3 (9), human muscarinic M2 (10), and rat M3 (11) receptors have helped overcome many of these limitations associated with modeling class A GPCRs (3).
As the major mediators of smooth muscle contraction, the α1-adrenoceptors are important targets for the treatment of hypertension and benign prostatic hyperplasia. α1-Adrenoceptors are also present in neurons in the brain and spinal cord, as well as in the liver (12). Three α1-adrenoceptor (α1A-, α1B-, and α1D-AR) subtypes have been characterized both pharmacologically and at the gene level. In human tissue, the α1A-AR is the most abundant subtype in liver, heart, and cerebral cortex, with lower expression levels in bladder; the α1B-AR is also expressed in liver, heart, and cerebral cortex but not in bladder, although the α1D-AR is the main subtype expressed in bladder with lower expression levels in other tissues (12, 13). However, their relative roles are poorly understood given the lack of selective α1-AR antagonists that can distinguish between the different α1-subtypes. This lack of discrimination is not surprising because current small molecule inhibitors act competitively at the relatively conserved norepinephrine (NE) binding pocket and fail to take advantage of the potential offered by more structurally divergent allosteric sites. Recent NMR studies of the β2-adrenoceptor have revealed that agonists and antagonists stabilize distinct GPCR conformations, demonstrating conformational coupling between the extracellular surface (ECS) and the orthosteric binding site (14). However, GPCR structure-function studies have focused on relatively conserved 7TM residues, and the role of residues that form the ECS remains poorly defined, despite their potential to allosterically modulate GPCR signaling.
ρ-TIA, a conopeptide from the piscivorous Conus tulipa, acts as an allosteric inhibitor of the α1-AR (15, 16), with structure-activity studies identifying Arg-4 and Trp-3 as the key contributors to ρ-TIA affinity for the hamster α1B-AR (17). Given that ρ-TIA is a small and highly positively charged peptide that is unlikely to cross cell membranes, we systematically mutated residues associated with the ECS of the α1B-AR. Using a refined NMR structure of ρ-TIA and a turkey β1-AR-derived homology model of the α1B-AR to guide docking simulations and mutagenesis studies, we established the specific molecular interactions between ρ-TIA on the ECS of the α1B-AR that were confirmed by double mutant cycle experiments. These findings, which reveal that peptide binding to ECS residues on TMH6 and TMH7 at the base of ECL3 can inhibit α1-AR signaling, may prove useful in guiding the design of new classes of allosteric inhibitors.
EXPERIMENTAL PROCEDURES
Site-directed Mutagenesis
The hamster α1B-AR cDNA in the pMT2′ vector was a kind gift from Prof. Bob Graham (Victor Chang Cardiac Research Institute, Sydney, Australia). In vitro site-directed mutagenesis of the α1B-AR subunit cDNA was achieved using the QuikChangeTM mutagenesis kit (Stratagene) following the manufacturer's instructions. A series of mutation primers were designed to produce the following point mutations in the α1B-AR: V107A, L108A, G109A, Y110A, W111A, V112A, L113A, G114A, R115A, I116A, C118A, P180A, L181A, L182A, G183A, W184A, K185A, E186A, P187A, P189A, N190A, D191A, D192A, K193A, E194A, C195A, G196A, V197A, T198A, E199A, E200A, P201A, F202A, E289K, W307A, F310A, F311A, L314A, L316A, G317A, S318A, S319A, F320A, S321A, T322A, L323A, K324A, P325A, P326A, D327A, V329A, F330A, K331A, F334A, and the double mutants E186A/E199A, S318A/F330A, and D327A/F330A. Primers used to generate the mutants were from Proligo. TOP10 Escherichia coli (Invitrogen) was transformed with wild type (WT) and mutant cDNA and subsequently used for plasmid preparation using a Mini or High Speed Maxi kit (Qiagen). Purified cDNA was used to confirm all mutations by sequencing performed at the Australian Genome Research Facility.
Transient Expression of α1B-ARs and Membrane Preparation
COS-1 cells (ATCC) were cultured in Dulbecco's modified Eagle's medium (DMEM) containing glutamine and 5% fetal bovine serum. Cells were transiently transfected with purified plasmid DNA encoding WT or mutant α1B-ARs, using Lipofectamine 2000 (Invitrogen) (25 μg of DNA/145 cm2) or FuGENE HD (Roche Applied Science) (36 μg of DNA/145 cm2), following the manufacturers' protocol. Cell membranes were prepared 48 h post-transfection as follows. Cells were harvested and homogenized using a Polytron homogenizer (Brinkmann Instruments) in HEM buffer (20 mm HEPES, 1.5 mm EGTA, 12.5 mm MgCl2, pH 7.5) containing complete protease inhibitor (Roche Diagnostics). The homogenate was centrifuged at 2000 rpm for 10 min, and the resulting supernatant was centrifuged at 14,000 rpm for 30 min. The pellet was resuspended in HEM buffer containing 10% v/v glycerol and stored at −80 °C prior to use. Protein concentration was determined using the BCA protein assay kit (Pierce) following the manufacturer's protocol.
Radioligand Binding Assays
The affinity of ρ-TIA and analogs at the α1B-AR mutants were determined using the radiolabeled α1-AR antagonist [3H]prazosin (0.5 nm) or 125I-(±)β-(iodo-4-hydroxyphenyl)ethylaminomethyl-tetralone (125I-HEAT; 70 pm). Reactions containing radioligand, membranes from α1B-AR-transfected COS-1 cells (5 μg of protein), and increasing concentrations of ρ-TIA or analogs (10 pm-10 μm) in HEM buffer were established in clear round bottom 96-well plates. Saturation binding experiments were also performed to determine the Kd value for prazosin at each of the mutants. Nonspecific binding was determined in the presence of 10 μm phentolamine. Each assay was performed in triplicate in a total reaction volume of 150 μl. After incubation for 60 min at room temperature, the membranes were harvested onto Whatman GF/B filter mats (PerkinElmer Life Sciences) pretreated with 0.6% polyethyleneimine using a TomTec harvester. Beta Plate scintillant (PerkinElmer Life Sciences) was then applied, and the filter-bound radioactivity detected using a Wallac MicroBeta (PerkinElmer Life Sciences).
IP1 HTRF Assay
COS-1 cells (ATCC) were cultured and transiently transfected with WT or mutant α1B-AR DNA following the manufacturer's protocol (Lipofectamine 2000, Invitrogen or FuGENE HD, and Roche Applied Science). Assays measuring IP1 accumulation were performed 48 h post-transfection following the manufacturer's instructions (IPone HTRF assay kit, Cisbio International). In brief, increasing concentrations of NE, prazosin, or ρ-TIA were added to 30,000 transfected cells in stimulation buffer in a white 384-well plate (Optiplate, PerkinElmer Life Sciences). The plates were incubated for 1 h at 37 °C with 5% CO2. Cells were then lysed by the addition of HTRF reagents, the europium cryptate-labeled anti-IP1 antibody, and the d2-labeled IP1 analog and diluted in lysis buffer (IPone HTRF assay kit, Cisbio International), followed by incubation for 1 h at room temperature. The emission signals were measured at 590 and 665 nm after excitation at 340 nm using the Envision multilabel plate reader (PerkinElmer Life Sciences), and the signal was expressed as the HTRF ratio: F = ((fluorescence665 nm/fluorescence590 nm) × 104). The WT α1B-AR was routinely run each day to control for any batch-to-batch variation in response. In addition, antagonist-induced inhibition of the basal IP1 mediated by the constitutive active mutant E289K was also determined (three separate experiments performed in replicates of six).
Peptide Synthesis
Peptides were synthesized with standard Fmoc chemistry (18) using Fmoc-amino acid derivatives purchased from Novabiochem or Auspep P/L. The following side chain-protected amino acids were used: Cys(Trt), His(Trt), Hyp(tBu), Tyr(tBu), Lys(t-butoxycarbonyl), Trp(t-butoxycarbonyl), Arg(2,2,4,6,7-pentamethyl-dihydrobenzofuran-5-sulfonyl), Asn(Trt), Asp(OtBu), Glu(OtBu), Gln(Trt), Ser(tBu), Thr(tBu), and Tyr(tBu). All other Fmoc amino acids were unprotected. Dimethylformamide (DMF), dichloromethane, diisopropylethylamine (DIEA), and trifluoroacetic acid (TFA) were supplied by Auspep P/L (Melbourne, Australia) as peptide synthesis grade. 2-(1H-Benzotriazol-1-yl)-1,1,3,3-tetramethyluronium hexafluorophosphate, triisopropyl silane, HPLC grade acetonitrile, acetic anhydride, and methanol were supplied by Sigma. The resin used was Fmoc-Rink resin (0.65 mmol/g) supplied by Auspep P/L. Ethane dithiol was supplied by Merck.
Synthetic Strategy
ρ-TIA and its analogs are peptide amides and were synthesized on a Protein Technologies Symphony automated peptide synthesizer using Rink amide resin (0.1 mmol). Assembly of the peptides was performed using 2-(1H-benzotriazol-1-yl)-1,1,3,3-tetramethyluronium hexafluorophosphate/DIEA in situ activation protocols (19) to couple the Fmoc-protected amino acid to the resin (5 eq excess, coupling time 5 min). Fmoc deprotection was performed with 30% piperidine/DMF for 1 min followed by a 2-min repeat. Washes were performed 10 times after each coupling as well as after each de-protection step. Acetylation of ρ-TIA was performed by using a 10-fold excess of acetic acid anhydride/DIEA in DMF. The success of the acetylation was monitored by the quantitative ninhydrin test (20). After chain assembly and final Fmoc deprotection, the peptide resins were washed with methanol and dichloromethane and dried in a stream of N2. The cleavage of peptide resin was performed at room temperature (RT) in TFA/H2O/triisopropyl silane/ethane dithiol (87.5:5:5:2.5) for 3 h. Cold diethyl ether (30 ml) was then added to the filtered cleavage mixture, and the peptide was precipitated. The precipitate was collected by centrifugation and subsequently washed with further cold diethyl ether to remove scavengers. The final product was dissolved in 50% aqueous acetonitrile and lyophilized to yield a white solid product. The crude reduced peptide was examined by reversed phase-HPLC for purity and the correct molecular weight confirmed by electrospray mass spectrometry (ES-MS).
Disulfide Bond Formation of Peptides 1–5
Pure reduced peptides (1 mg/ml) were oxidized by stirring at RT in 30% isopropyl alcohol, 0.1 m NH4HCO3 at pH 7.0 for 16 h. The solutions were subsequently diluted to an isopropyl alcohol concentration <5%, prior to reversed phase-HPLC purification. The obtained single main oxidized product with the cysteine connectivity (1–3, 2–4) (17) was purified to >95% purity and lyophilized.
HPLC Analysis and Purification
Analytical HPLC runs were performed using a Shimadzu HPLC system LC10A with a dual wavelength UV detector set at 214 and 254 nm. A reversed phase C-18 column (Zorbax 300-SB C-18; 4.6 × 50 mm) with a flow rate of 2 ml/min was used. Gradient elution was performed with the following buffer systems: A, 0.05% TFA in water, and B, 0.043% TFA in 90% acetonitrile in water, from 0% B to 80% B in 20 min. The crude peptides and oxidized peptides were purified by semi-preparative HPLC on a Shimadzu HPLC system LC8A associated with a reversed phase C-18 column (Vydac C18, 25 × 10 mm) at a flow rate of 5 ml/min with a 1%/min gradient of 5% B to 50% B. The purity of the final product was evaluated by analytical HPLC (Zorbax 300SB C-18: 4.6 × 100 mm) at a flow rate of 1 ml/min and a 1.67%/min gradient of B (5–45%). Purities of synthesized peptides were all >95%.
Peptide Concentration
Peptide concentrations used in in vitro screening were calculated based on peak size detected at 214 nm by HPLC. Peak size was calibrated using a peptide standard (in this case 1) with known peptide content established by amino acid analysis. Molecular extinction coefficients are calculated for the standard and the peptide of interest applying increments established by Buck et al. (21). Using Lambert Beer Law, the peptide concentration was calculated based on absorptions of standard and sample using calculated extinction coefficients. Quantitation was confirmed using averaged IC50 values for ρ-TIA displacement of [3H]prazosin determined for each batch of ρ-TIA.
ES-MS
Electrospray mass spectra were collected in line during analytical HPLC runs on an Applied Biosystems API-150 spectrometer operating in the positive ion mode with a declustering potential of 20, a focusing potential of 220, and Turbo spray of 350°. Masses between 300 and 2200 atomic mass units were detected (step 0.2 atomic mass units, dwell 0.3 ms).
NMR Analysis
Samples of ρ-TIA prepared for NMR analysis included 1 mg/ml peptide in 95% H2O, 5% D2O at pH 4.5. All data, including two-dimensional TOCSY, NOESY, and DQF-COSY, were acquired on a Bruker Avance 600 MHz spectrometer at 298 K with 4k data points in the direct dimension and 512 increments in the indirect dimension (22). The data were processed using Topspin 2.1 with the free induction decay multiplied by a 90° phase-shifted sine square window function, and zero filled to 1k data points in the indirect dimension prior to Fourier transform. The data were analyzed using the program CARA and assigned using standard two-dimensional homonuclear sequential assignment methods. Inter-proton distance restraints were derived from cross-peak intensities in a NOESY spectrum recorded with a mixing time of 250 ms. Backbone dihedral angles were derived from 3JHαHN coupling constants, with residues <5 Hz (Trp-3, Arg-4, Cys-5, Ala-10, Arg-12, Arg-13, Lys-16, Lys-17, and Cys-19) being restrained to −60 ± 30° and residues >8 Hz (Asn-2, Leu-7, Ile-8, Cys-11, Asn-14, and His-15) being restrained to −120 ± 30°. Side chain χ1 angles and stereo specific assignments of Hβ protons were derived from a combination of 3JHαHβ coupling constants and Hα-Hβ and HN-Hβ NOESY peak intensities. Residues found to be in a t2g3 conformation (Arg-4, Cys-5, Cys-6, Leu-7, His-15, and Cys-19) were restrained to −60 ± 30°, and residues in a g2t3 conformation (Cys-11 and Arg-12) were restrained to −180 ± 30°. No residues could be confirmed to be in a g2g3 conformation. The amide protons of Cys-5, Cys-6, Leu-7, and Ile-8, Cys-11, and Cys-19 have previously been reported to be slow exchanging. During preliminary structure calculations, backbone carbonyl groups acting as hydrogen bond acceptors were unambiguously identified for all these amides, and thus restraints for these hydrogen bonds could be introduced in the calculations (Cys-5 → Asn-2, Cys-6 → Trp-3, Leu-7 → Arg-4, Ile-8 → Cys-5, Cys-11 → Ile-8, and Cys-19 → His-15). Structures were calculated using restrained simulated annealing within the program CNS using the topallhdg5.3.pro/parallhdg5.3.pro force field. Initial structures were calculated using torsion angle dynamics with a high temperature phase comprising 4000 steps of 0.015 ps at 50,000 K and a cooling phase during which the temperature was lowered from 50,000 to 0 K through 4000 steps of 0.015 ps, followed by 5000 steps of Powell energy minimization. In a second step, these structures were refined through Cartesian dynamics in a water shell. This refinement involves the following: (i) a heating phase where the temperature was increased to 500 K in steps of 100 K, each comprising 50 steps of 0.005-ps dynamics; (ii) a high temperature phase comprising 2500 steps of 0.005-ps dynamics at 500 K; (iii) a cooling phase where the temperature was lowered in steps of 100 K, each comprising 2500 steps of 0.005-ps dynamics, and finally (iv) an energy minimization phase comprising 3000 steps of Powell minimization. In the final round, 50 structures were calculated, and the 20 structures with lowest overall energy were chosen to represent the solution structure of ρ-TIA. These structures were analyzed and visualized using the programs MolProbity (23) and MOLMOL (24). The coordinates of the structure and chemical shifts have been submitted to the PDB and BMRB databases and given the ID numbers 2LR9 and RCSB102733, respectively.
Modeling and Docking Simulations
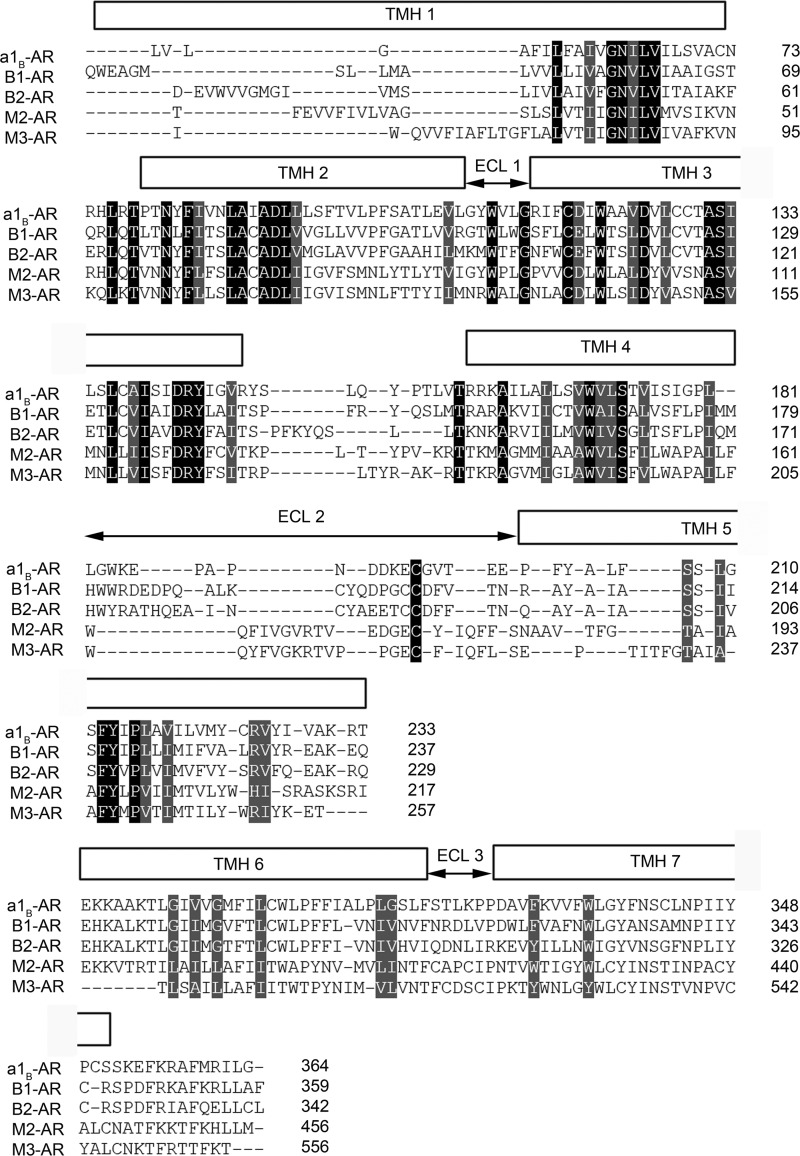
A molecular homology model of the inactive α1B-AR was built using the crystal structure of turkey β1-adrenoceptor (PDB code 2VT4 (7)) as the template in the program Modeler 9 Version 2 (25). This template was chosen because of its high degree of sequence identity to α1B-AR as determined by BLAST (26) and PHYRE (27). All sequence alignments were generated using ClustalW (28) and further corrected by hand based on the secondary structure predicted by PHYRE (27) to maximize the accuracy of sequence alignment and protein sequence identity prior to modeling (Fig. 1). The structural models were validated using the on-line server Verify3D (29) and the Ramachandran plot available from ProFunc (30) database.
FIGURE 1.
Structural alignments of hamster α1B-AR, human β1-AR (PDB code 2VT4), human β2-AR (PDB code 2RH1), human M2-AR (PDB code 3UON), and rat M3-AR (PDB code 4DAJ). The three-dimensional structural alignments were performed using program TOPOFIT (55) with a joint distance cutoff of 3 Å between the Cα atoms of the residues in the α1B-AR model and each crystal structure. The alignment represents the degree of conservation of residues spatially. The poorly aligned regions indicate residues are not topologically close, although the sequence may be conserved.
Blind (global) docking was applied initially to search for possible binding sites and used to assess the quality of our homology model by comparing the literature with our experimental data. In blind docking simulation, all side chains on ρ-TIA (PDB code 2LR9), prazosin (DrugBank), and the entire ECS on α1B-AR were set as active residues using Haddock (31). The passive residues, which are solvent-exposed and surround the active residues, were defined by Haddock. No inter- or intramolecular restraints were defined. Semi-flexible and fully flexible regions were automatically detected by Haddock, and sampling parameters were set as default. The blind docking solutions for ρ-TIA and prazosin were examined manually in PyMOL to define the predicted ligand-binding sites. The predicted binding sites for ρ-TIA and prazosin from blind docking were in agreement with our experimental data, suggesting that the α1B-AR homology model was predictive.
Flexible solvated docking simulations were performed using Haddock (31), with experimentally determined restraints used to refine the binding mode. Specifically, Trp-3 and Arg-4 on ρ-TIA (17) and Ser-318, Asp-327, and Phe-330 on α1B-AR were set as active residues, and the neighboring solvent-exposed residues were set as passive restraints and semi- and fully flexible fragments were automatically detected by Haddock. Sampling and cluster parameters were default values, except that 2000 structures were calculated for rigid body docking, with the best 400 solutions then subjected to semi-flexible simulated annealing and explicit solvent refinement. The Haddock score, minimal energy, existing experimental data, and the multiplicity of the same docking orientation (r.m.s.d. <7.5 Å) were used as selection criteria for choosing possible binding modes. The final 200 best solutions generated by Haddock were confirmed by visual inspection using PyMOL. Comparing the three approached solutions derived from Haddock provided the best fit for the available ρ-TIA-α1B-AR structure-activity data. Slight outward conformational movements at TMH6 and TMH7 and ECL3 (1.5–2.2 Å) were observed in the model following ρ-TIA docking, although the structure of ρ-TIA was little altered upon docking (Fig. 2). Additional α1B-AR mutants (D327A/F330A and S318A/F330A) were modeled following residue replacement and docked against ρ-TIA and its W3A and R4A analogs, as described above, to provide additional support for the predicted intermolecular interactions.
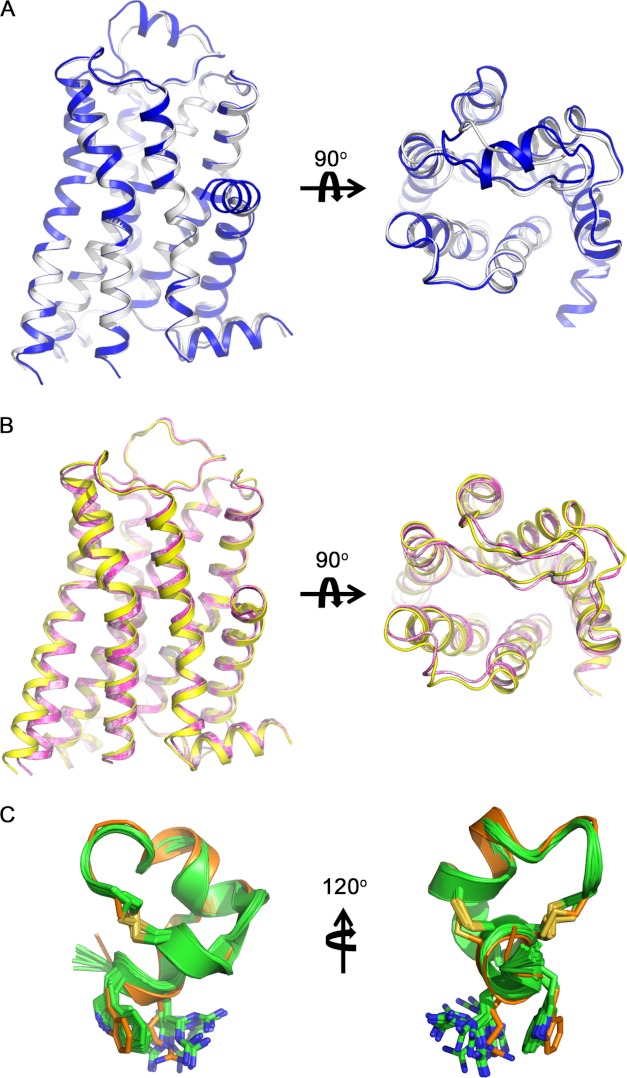
FIGURE 2.
Structural comparison of α1B-AR molecular models and template turkey β1-AR (PDB code 2VT4). A, superimposition of α1B-AR model (white) to the template turkey β1-AR crystal structure (blue) (r.m.s.d. = 0.334 Å), showing that ECL2 is the only region that significantly differs from the template. B, comparison of ρ-TIA-bound (yellow) and prazosin-bound (magenta) conformations to the initial α1B-AR model (white), showing that the orthosteric ligand binding does not perturb the overall structure, which is in agreement with the literature. Allosteric peptide binding introduces slightly conformational movements (<1Å between Cα atoms) in TMH6, TMH7, ECL2, and ECL3 extracellularly and in TMH5 intracellularly. C, superimposition of the docked ρ-TIA (orange) to its NMR structures (green) shows that the conformation of ρ-TIA was little altered during docking.
Statistics and Data Analysis
Data are presented as means ± S.E. of results obtained from 2 to 52 separate experiments, as indicated. Sigmoidal curves for the calculation of the half-maximal excitatory concentration (EC50) and half-maximal inhibitory concentration (IC50) values were fitted to individual data points by nonlinear regression using the software package Prism (GraphPad Software). For multiple comparisons, one-way analysis of variance was used with post hoc t tests performed by Dunnett's method. Values of p < 0.05 were considered significant.
RESULTS
Characterization of ρ-TIA and Prazosin Affinity at ECS Mutants of the α1B-AR
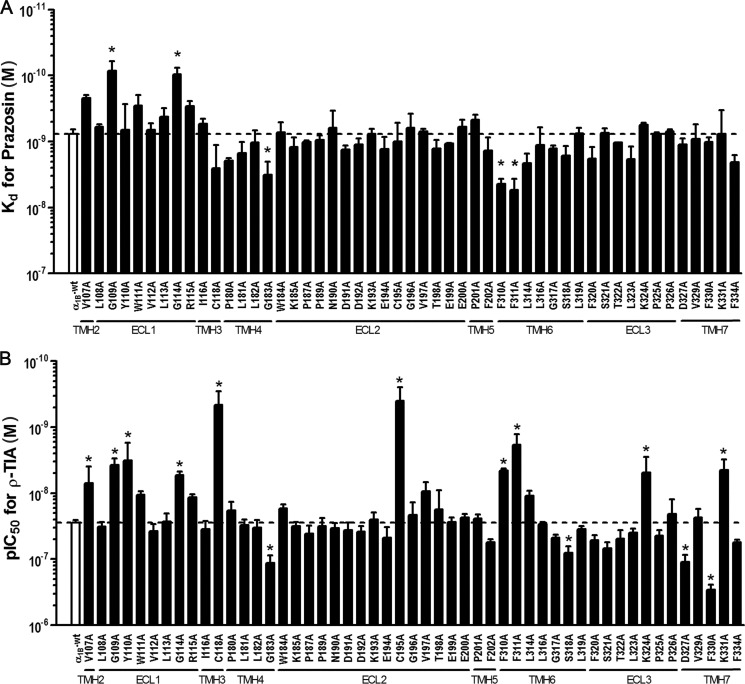
Given ρ-TIA is a relatively hydrophilic noncompetitive inhibitor, we hypothesized that this peptide does not partition into or across the cell membrane and likely binds to ECS residues above and separate from the orthosteric site that is deeper in the transmembrane helical bundle of α1-ARs. To determine the contribution of individual ECS residues to ρ-TIA binding, we constructed 53 alanine-substituted point mutations of the α1B-AR across ECL1–3 and the extracellular portions of associated transmembrane helices (Fig. 3 and Table 1). Wild type and mutant α1B-ARs were expressed in COS-1 cells and first tested for their ability to bind the inverse agonist prazosin. Saturation binding experiments generated prazosin Kd values for all mutants except E186A and W307A (Fig. 3A). The E186A mutant showed readily detectable 125I-HEAT binding allowing any potency shift for ρ-TIA to be determined, whereas the W307A mutant failed to bind detectable levels of either 125I-HEAT or [3H]prazosin. Saturation binding experiments indicated that the G109A and G114A mutants increased the affinity of prazosin 9- and 8-fold, respectively, compared with that of WT (Kd = 0.76 ± 0.05 nm; n = 29), whereas the remaining mutations either did not significantly affect or slightly decreased (4-, 5-, and 7-fold changes for mutants G183A, F310A, and F311A, respectively) the Kd value for prazosin (Fig. 3A). In contrast, ρ-TIA affinity was significantly affected by 14 receptor mutants (Fig. 3B). Ten mutants V107A (4-fold), G109A (7-fold), Y110A (9-fold), G114A (5-fold), C118A (60-fold), C195A (70-fold), F310A (6-fold), F311A (15-fold), K324A (6-fold), and K331A (6-fold) enhanced affinity, whereas four mutants G183A (4-fold), S318A (3-fold), D327A (4-fold), and F330A (10-fold) reduced affinity compared with the WT receptor (IC50 = 27.8 ± 1.1 nm; n = 52) (Fig. 3B).
FIGURE 3.
Effect of α1B-AR mutants on prazosin Kd and the IC50 values of ρ-TIA. A, comparison of WT and α1B-AR mutants Kd for prazosin determined from saturation binding curves, where nonspecific binding was determined in the presence of 10 nm phentolamine. Values are means ± S.E. of 2–4 separate experiments for mutants and n = 29 for WT, each performed in triplicate. B, comparison of WT and α1B-AR mutants IC50 for ρ-TIA determined using the radiolabeled α1-AR antagonist [3H]prazosin (0.5 nm) or 125I-HEAT (70 pm) and increasing concentrations of ρ-TIA (10 pm to 10 nm). Values are means ± S.E. of 52 separate experiments for WT and 2–10 separate experiments for the mutants, each performed in triplicate. All mutants produced for this study are shown, except for mutant E186A and W307A, where prazosin binding was not detectable. *, values of p < 0.05 were considered significant.
TABLE 1.
Prazosin Kd, ρ-TIA IC50, and NE EC50 values for α1B-AR mutants
Values are the mean ± S.E. for (n) separate experiments; ND means not detected, and – means not determined.
| Mutant | Prazosin Kd | ρ-TIA IC50 | NE EC50 |
|---|---|---|---|
| nm | nm | nm | |
| α1B WT | 0.76 ± 0.48 (29) | 27.81 ± 1.05 (52) | 11.67 ± 0.61 (35) |
| V107A | 0.22 ± 0.0088 (2) | 6.99 ± 1.76 (3) | 11.64 ± 1.45 (3) |
| L108A | 0.60 ± 0.024 (2) | 32.11 ± 2.31 (3) | – |
| G109A | 0.085 ± 0.012 (2) | 3.71 ± 0.36 (3) | 13.72 ± 1.65 (3) |
| Y110A | 0.67 ± 0.26 (3) | 3.18 ± 0.86 (3) | 3840.02 ± 995.14 (3) |
| W111A | 0.29 ± 0.047 (2) | 10.54 ± 0.54 (3) | – |
| V112A | 0.66 ± 0.066 (2) | 37.55 ± 4.09 (3) | – |
| L113A | 0.42 ± 0.054 (2) | 26.87 ± 3.31 (3) | – |
| G114A | 0.096 ± 0.0097 (2) | 5.33 ± 0.31 (3) | 188.22 ± 9.51 (3) |
| R115A | 0.29 ± 0.022 (2) | 11.52 ± 0.54 (3) | – |
| I116A | 0.54 ± 0.043 (2) | 35.10 ± 4.40 (3) | – |
| C118A | 2.53 ± 0.89 (3) | 45.59 ± 0.092 (2) | 3336.83 ± 843.23 (3) |
| P180A | 1.94 ± 0.077 (3) | 18.10 ± 2.25 (4) | – |
| L181A | 1.48 ± 0.24 (2) | 30.20 ± 2.48 (3) | – |
| L182A | 1.04 ± 0.19 (2) | 33.37 ± 3.81 (3) | – |
| G183A | 3.17 ± 0.61 (3) | 114.72 ± 13.60 (10) | 3280.64 ± 312.35 (4) |
| W184A | 0.72 ± 0.11 (2) | 16.88 ± 0.99 (4) | – |
| K185A | 1.21 ± 0.18 (2) | 31.45 ± 1.97 (3) | – |
| E186A | ND | 20.56 ± 2.05 (4) | 3065.49 ± 1120.50 (4) |
| P187A | 1.02 ± 0.026 (3) | 40.83 ± 4.79 (3) | – |
| P189A | 0.94 ± 0.064 (3) | 31.78 ± 4.12 (4) | – |
| N190A | 0.62 ± 0.16 (2) | 34.07 ± 2.77 (3) | – |
| D191A | 1.31 ± 0.080 (2) | 36.39 ± 4.20 (3) | – |
| D192A | 1.10 ± 0.098 (3) | 38.06 ± 3.08 (4) | – |
| K193A | 0.77 ± 0.063 (2) | 25.18 ± 2.70 (3) | – |
| E194A | 1.29 ± 0.24 (4) | 47.59 ± 7.60 (4) | – |
| C195A | 1.00 ± 0.29 (2) | 0.40 ± 0.080 (5) | 19,195.40 ± 846.22 (3) |
| G196A | 0.62 ± 0.13 (2) | 21.46 ± 4.19 (5) | – |
| V197A | 0.70 ± 0.026 (2) | 9.40 ± 1.29 (3) | – |
| T198A | 1.27 ± 0.16 (4) | 17.71 ± 5.25 (4) | – |
| E199A | 1.07 ± 0.0028 (2) | 27.48 ± 1.90 (3) | – |
| E200A | 0.60 ± 0.062 (2) | 23.51 ± 1.32 (3) | – |
| P201A | 0.47 ± 0.037 (2) | 24.21 ± 1.55 (3) | – |
| F202A | 1.38 ± 0.28 (4) | 55.76 ± 2.68 (3) | – |
| W307A | ND | ND | 1394.13 ± 622.07 (2) |
| F310A | 4.36 ± 0.31 (3) | 4.54 ± 0.15 (3) | 24,951.90 ± 5055.00 (4) |
| F311A | 5.45 ± 0.92 (3) | 1.83 ± 0.29 (5) | 1337.03 ± 182.22 (5) |
| L314A | 2.13 ± 0.30 (4) | 10.99 ± 0.82 (3) | – |
| L316A | 1.13 ± 0.30 (2) | 29.31 ± 0.95 (3) | – |
| G317A | 1.27 ± 0.059 (2) | 46.88 ± 1.98 (3) | – |
| S318A | 1.63 ± 0.23 (2) | 80.61 ± 7.94 (7) | 19.03 ± 3.17 (3) |
| L319A | 0.75 ± 0.063 (2) | 34.93 ± 1.62 (4) | – |
| F320A | 1.83 ± 0.33 (4) | 51.67 ± 3.90 (4) | – |
| S321A | 0.74 ± 0.054 (2) | 68.35 ± 6.23 (4) | – |
| T322A | 1.04 ± 0.0043 (2) | 48.81 ± 6.31 (4) | – |
| L323A | 1.85 ± 0.35 (4) | 39.63 ± 2.53 (4) | – |
| K324A | 0.56 ± 0.017 (2) | 4.85 ± 1.14 (3) | 34.52 ± 3.97 (3) |
| P325A | 0.77 ± 0.022 (2) | 44.18 ± 3.86 (4) | – |
| P326A | 0.70 ± 0.024 (2) | 20.64 ± 4.53 (3) | – |
| D327A | 1.11 ± 0.11 (2) | 109.53 ± 11.43 (9) | 6.85 ± 1.97 (3) |
| V329A | 0.92 ± 0.21 (4) | 23.57 ± 3.07 (3) | – |
| F330A | 1.02 ± 0.071 (2) | 289.72 ± 22.07 (6) | 38.54 ± 5.49 (3) |
| K331A | 0.76 ± 0.27 (3) | 4.49 ± 0.74 (3) | 9.85 ± 1.19 (3) |
| F334A | 2.07 ± 0.22 (3) | 55.72 ± 2.27 (3) | – |
Signaling of α1B-AR Mutants in Response to NE
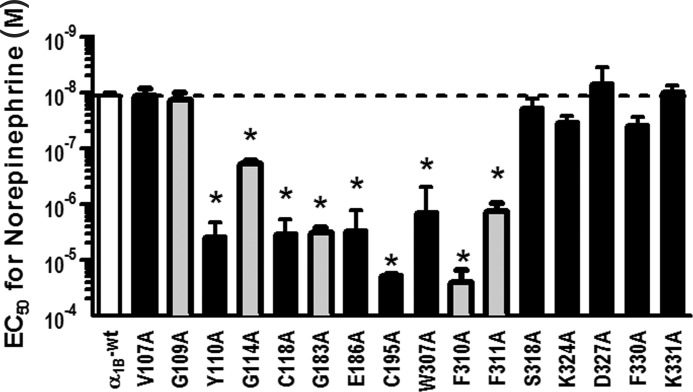
The α1B-AR signals via Gq, which induces phospholipase C activation that triggers the inositol phosphate (IP) cascade. IP1, a downstream metabolite of inositol 1,4,5-trisphosphate, accumulates in cells following Gq receptor activation, making it a useful marker of receptor activation. To confirm that each mutant affecting ρ-TIA and/or prazosin binding (V107A, G109A, Y110A, G114A, C118A, E186A, G183A, C195A, F310A, F311A, S318A, K324A, W307A, D327A, F330A, and K331A) remained functional, we determined their effect on NE signaling to IP1. NE had significantly decreased potency at the Y110A and G114A mutants in ECL1 (330- and 16-fold, respectively), C118A in TMH3 (285-fold), the G183A mutant in THM4 (280-fold), the C195A mutant in ECL2 (1645-fold), and the W307, F310A, and F311A mutants in TMH6 (120-, 2140-, and 115-fold, respectively) compared with WT (EC50 = 11.7 ± 0.61 nm; n = 35; Fig. 4). The other mutants affecting ρ-TIA binding (V107A, G109A, S318A, K324A, D327A, F330A, and K331A) and prazosin binding (G109A) had no significant effect on NE signaling (Fig. 4).
FIGURE 4.
Comparison of NE EC50 values for α1B-AR mutants measuring IP1 accumulation in response to increasing concentrations of NE. Mutants affecting both prazosin and ρ-TIA binding are cross-hatched, and mutants affecting only ρ-TIA affinity are black bars. Values are means ± S.E. of 35 separate experiments for WT and 3–5 separate experiments for the mutants, each performed in triplicate. Results for mutant W307A, which had no detectable prazosin binding, are also shown (n = 2). *, values of p < 0.05 were considered significant.
Inverse Agonist Activity of ρ-TIA
To further characterize the pharmacological properties of ρ-TIA, we determined whether it acted as an inverse agonist or a neutral antagonist using the constitutively active E289K mutant of the α1B-AR (32). Using the IP1 assay, we confirmed the E289K mutant was constitutively active (220% over basal) and inhibited both by prazosin (EC50 = 11.6 ± 7.5 nm; n = 3), a known inverse agonist (33), and ρ-TIA (EC50 = 19.0 ± 5.8 nm; n = 3). In contrast, basal signaling was unaffected by prazosin or ρ-TIA in WT α1B-AR.
Molecular Model of the α1B-Adrenoceptor
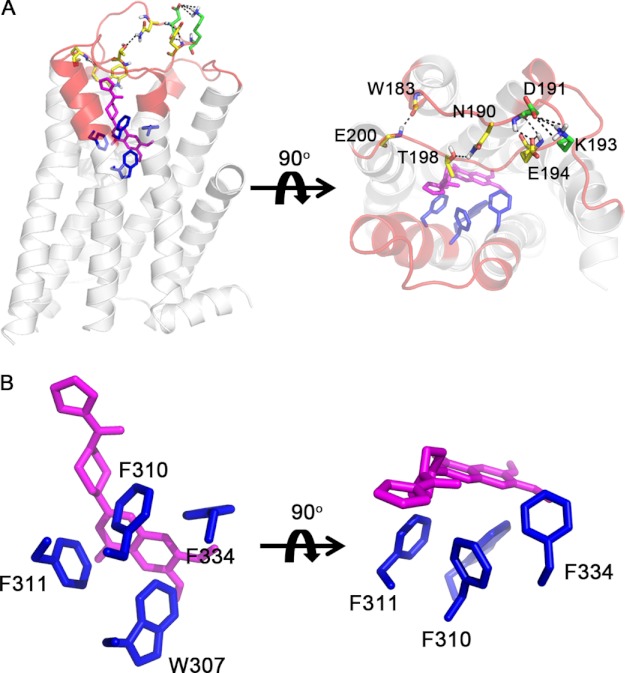
To better understand the mechanism of allosteric inhibition by ρ-TIA, we constructed a homology model for the α1B-adrenoceptor using the crystal structure of the turkey β1-adrenoceptor (7) as a template using the alignment shown in Fig. 1. An overlay of the α1B-AR model with turkey β1-AR is shown in Fig. 2. The model includes seven transmembrane helices, forming a barrel-like structure with a large ordered hairpin ECL2, blocking half of the substrate accessible area. Several intramolecular interactions, including four hydrogen bonds (Asn-190 side chain to Thr-198 side chain; Asp-191 main chain to Glu-194 main chain; Asp-191 side chain to Glu-194 main chain; and Trp-183 main chain to Glu-200 main chain) and a salt bridge (Asp-191 to Lys-193), were identified within ECL2, which is likely further stabilized by the conserved disulfide bond between Cys-195 and Cys-118, anchoring ECL2 to TMH3 (Fig. 6A). Remarkably, the alanine substitutions C118A or C195A improved ρ-TIA affinity 60- and 70-fold, respectively, but produced a dramatic 286- and 1645-fold reduction in the potency of NE to signal to IP1, while having no significant effect on prazosin binding (Figs. 3 and 4).
FIGURE 6.
Prazosin docking to the α1B-AR. The residues studied in this paper are highlighted in red. A, side and top view of prazosin (magenta) binding to the α1B-AR. B shows a detailed view of intermolecular interactions between prazosin and binding site residues (blue) on α1B-AR. Residues that form stabilizing intramolecular interactions (black dashed lines) in ECL2 are colored yellow (hydrogen bonds) or green (salt bridge).
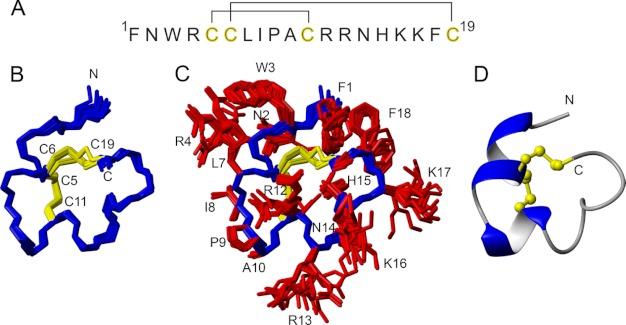
Refinement of the Solution NMR Structure of ρ-TIA
Although an initial solution NMR structure of ρ-TIA has been reported (15), detailed analysis revealed inconsistencies between this structure and the NMR data that might compromise docking studies. Thus, we further refined the NMR structure of ρ-TIA using an explicit solvent force field and additional restraints, including χ1 dihedral angles. New NMR data were recorded at 600 MHz, and complete resonance assignments were achieved using standard homonuclear sequential assignment methods. In all cases, the resonance assignments were consistent with those previously reported (15). Structural restraints for ρ-TIA included inter-proton distances, backbone and side chain dihedral angles, and hydrogen bonds, which were based on NOESY cross-peak intensities, coupling constants derived from one-dimensional 1H and DQF-COSY spectra, and amide exchange data. These restraints were used as input for the generation of a refined ρ-TIA NMR structure through simulated annealing and energy minimization in a water shell. The derived structural family shown in Fig. 5 (see Table 2 for structural statistics) is of high quality in terms of resolution of both backbone and side chains, and it is in excellent agreement with both experimental data and ideal covalent geometry, as evident from minimal violations and overall MolProbity scores of >90. The overall fold, which is dominated by two helical regions, is consistent with the previously reported structure (15), but some significant differences are seen in the N-terminal region critical for high affinity. In particular, in the new structure the Trp-3 and Arg-4 side chains stack against each other, which is consistent with the observed upfield shifts of the Arg-4 Hβ and Hγ resonances. Finally, the structure shows an improved correlation between the hydrogen bonding pattern, which includes a well defined 310 helical region between residues 3 and 7, and the amide exchange data. Based on these differences, we have used the new structure of ρ-TIA (PDB code 2LR9) to establish its molecular complementarity with surface residues on the α1B-AR.
FIGURE 5.
Sequence and structure of ρ-TIA. A, primary sequence and disulfide connectivities. B and C show a superposition of the final 20 structures representing the three-dimensional NMR structure of ρ-TIA (PDB code 2LR9) without and with side chains, respectively. Residues are labeled with single letter amino acid codes and residue numbers. D shows the lowest energy structure in ribbon style, illustrating the elements of secondary structure, and the disulfide bonds in ball-and-stick representation.
TABLE 2.
Energies and structural statistics for the family of 20 lowest energy structures with highest overall MolProbity score
| Energies (kcal/mol) | |
| Overall | −602.9 ± 12.56 |
| Bonds | 2.83 ± 0.243 |
| Angles | 13.1 ± 1.23 |
| Improper | 3.45 ± 0.553 |
| van der Waals | −78.1 ± 4.52 |
| NOE | 8.05 ± 0.781 |
| cDih | 0.351 ± 0.233 |
| Dihedral | 103.6 ± 2.959 |
| Electrostatic | −656.2 ± 15.43 |
| MolProbity statistics | |
| Clashes (>0.4 Å/1000 atoms) | 6.76 ± 3.76 |
| Poor rotamers | 0.278 ± 1.24 |
| Ramachandran outliers | 0.00 ± 0.00% |
| Ramachandran favored | 100.00 ± 0.00% |
| MolProbity score | 1.32 ± 0.266 |
| MolProbity score percentilea | 97.2 ± 2.43 |
| Residues with bad bonds | 0.00 ± 0.00 |
| Residues with bad angles | 0.00 ± 0.00 |
| Atomic r.m.s.d. | |
| Mean global backbone | 0.34 ± 0.14 Å |
| Mean global heavy | 1.54 ± 0.28 Å |
| Distance restraints | |
| Intraresidue (i − j = 0) | 0 |
| Sequential (|i − j| = 1) | 72 |
| Medium range (|i − j| <5) | 36 |
| Long range (|i − j| >) | 11 |
| Hydrogen bonds | 12 (for 6 H-bonds) |
| Total | 131 |
| Dihedral angle restraints | |
| φ | 16 |
| χ1 | 8 |
| Total | 24 |
| Violations from experimental restraints | |
| Total NOE violations exceeding 0.2 Å | 0 |
| Total dihedral violations exceeding 3.0° | 3 (highest 3.33) |
a 100th percentile is the best among structures of comparable resolution; 0th percentile is the worst.
Blind Docking of Prazosin
To validate our model, we performed a blind docking simulation of prazosin binding to our α1B-AR model. Prazosin was predicted to form extensive π-π interactions with Trp-307, Phe-310, and Phe-311 in TMH6 and a strong hydrophobic interaction between the methyl tail with Phe-334 in TMH7 of the α1B-AR (Fig. 6, A and B). The final docking pose chosen was consistent with these observations, the F310A and F311A mutants increased prazosin Kd 6- and 7-fold (4.4 ± 0.31, n = 3, and 5.4 ± 0.92 nm, n = 3, respectively) compared with WT (0.76 ± 0.05 nm, n = 29) (Fig. 3), although the F334A mutant also slightly increased prazosin Kd (2.1 ± 0.22, n = 3), although this change did not reach significance (Fig. 3A). Unfortunately, the W307A caused a dramatic drop in receptor expression levels (∼1% of WT) that precluded determination of prazosin affinity.
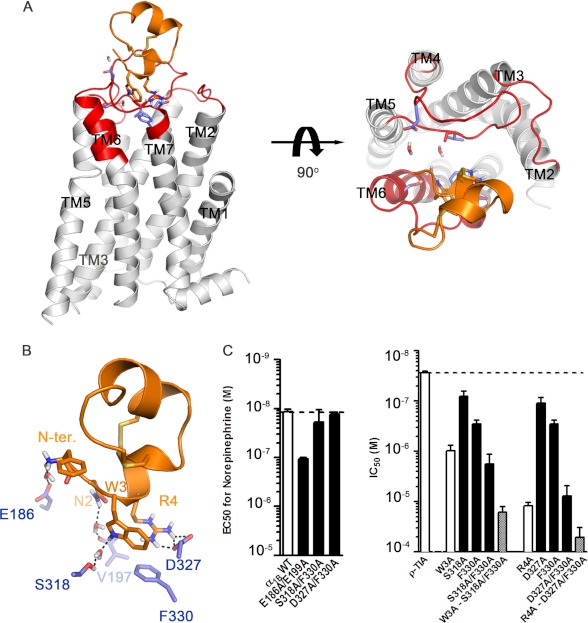
Experimentally Guided Docking of ρ-TIA
Interactions identified from our mutational studies were used to guide the docking of ρ-TIA with the program Haddock. This docking revealed that the binding site for ρ-TIA was located on top of ECL3 and TMH7 (Fig. 7, A and B). The final docking pose for ρ-TIA binding to the α1B-AR reveals it binds high on the α1B-AR, with the N terminus facing down toward ECL3 of the receptor (Fig. 7A). ρ-TIA binding is predicted to be dominated by a salt bridge between Arg-4-ρ-TIA and Asp-327, a cation-π interaction between Arg-4-ρ-TIA and Phe-330, and a predicted T-stacking-π interaction between Trp-3-ρ-TIA and Phe-330, consistent with our mutational and previous SAR studies (17). Three hydrogen bonds bridged by water molecules were also observed between N-terminal positive charge of ρ-TIA and Glu-186, Asn-2-ρ-TIA and the backbone Val-197, and Trp-3-ρ-TIA and Ser-318. In this conformation, Asn-2-ρ-TIA forms a hydrogen bond with the backbone of Val-197, which could not be validated via mutational approaches. Surprisingly, ECL2 contributed little to ρ-TIA binding, despite its high electronegativity that was expected to complement the positive charges on ρ-TIA. Docking simulation with or without solvation yielded similar solutions (r.m.s.d. < 3 Å), except that the N-terminal positive charge of ρ-TIA formed a direct ionic interaction with Glu-186 under nonsolvated conditions, whereas a weaker hydrogen bond through water to Glu-186 was predicted under solvated conditions. To better understand the role of water in allosteric binding, the N-terminal acetylated ρ-TIA analog and the E186A mutant were constructed and tested. As Glu-199 is in close proximity to Glu-186 and may compensate a lost interaction in the E186A mutant, the E199A and E186A/E199A mutants were also constructed and tested. A significant 7-fold enhancement in affinity was obtained for acetylated ρ-TIA at the α1B-AR (IC50 = 3.6 ± 0.49 nm; n = 3), whereas a significant 5-fold decrease in ρ-TIA affinity was obtained at the E186A/E199A mutant (IC50 = 139 ± 22 nm; n = 4). In contrast, acetylated ρ-TIA at the E186A/E199A mutant had intermediate potency (IC50 = 9.4 ± 2.2 nm; n = 3), and ρ-TIA affinity at mutants E186A (IC50 = 20.6 ± 2.1 nm; n = 4) and E199A (IC50 = 27.5 ± 1.9 nm; n = 3) was unchanged from WT receptor (IC50 = 27.8 ± 1.1 nm; n = 52). Together, these results indicate only a modest interaction between the N-terminal positive charge and Glu-186, supporting the solvated docking model that we used to generate the final docking pose for ρ-TIA.
FIGURE 7.
ρ-TIA (orange) docking to the α1B-AR. A, side and top view of ρ-TIA binding to the α1B-AR with the residues studied in this paper colored in red and predicted allosteric binding site residues shown in slate. B shows a detailed view of ρ-TIA interactions with α1B-AR indicated by dashed lines. C, left panel shows the WT and α1B-AR double mutant EC50 values for NE. Values are means ± S.E. of 35 separate experiments each performed in triplicate for WT and 3–5 separate experiments each performed in triplicate for mutants. The right panel shows the IC50 values of ρ-TIA and analogs at the α1B-AR and mutants. Values are means ± S.E. of 52 separate experiments each performed in triplicate for WT and 3–9 separate experiments each performed in triplicate for analogs and mutants.
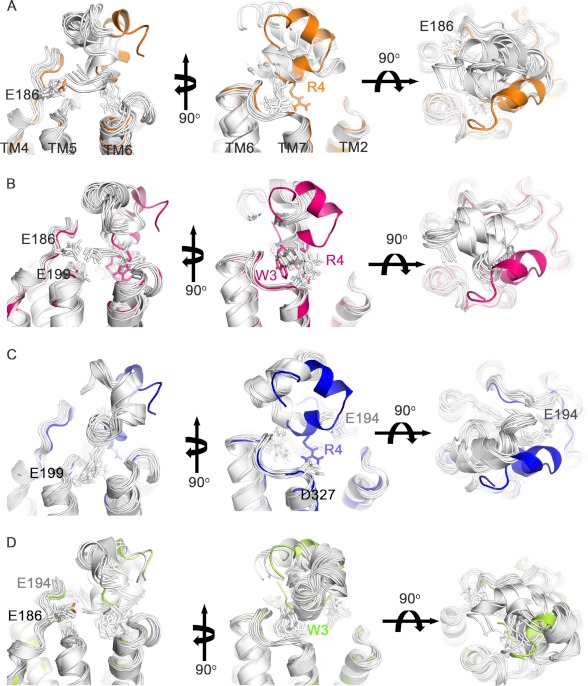
The predicted hydrogen bond and T-stacking π interaction between Trp-3-ρ-TIA and Ser-318 and Trp-3-ρ-TIA and Phe-330, respectively, were supported by the reductions in affinity associated with the single mutations S318A (IC50 = 80.6 ± 7.9 nm, n = 7), F330A (290 ± 22 nm, n = 6) that approached the drop in affinity observed for W3A-ρ-TIA (IC50 = 984 ± 106 nm, n = 8) and the double mutant S318A/F330A (IC50 = 1794 ± 351 nm, n = 8) (Fig. 7C). Testing W3A-ρ-TIA against the double mutant receptor S318A/F330A resulted in a further weakening of the interaction (IC50 = 16349 ± 1909 nm, n = 6) (Fig. 7C). To better understand the roles of the allosteric binding site residues and to identify any compensatory interactions that might arise in these mutants, solvated docking simulations of W3A-ρ-TIA to α1B-AR, ρ-TIA to S318A/F330A, and W3A-ρ-TIA to S318A/F330A were performed. These docking simulations for W3A-ρ-TIA to α1B-AR and ρ-TIA to S318A/F330A revealed the potential for compensatory shifts in binding associated with additional interactions with highly negatively charged ECL2 (Glu-186, Asp-191, Glu-194, and Glu-199) that arise from a small rotation along the y and/or z axis of ∼25° (Fig. 8), which could explain the less than expected affinity reductions observed.
FIGURE 8.
Docking of ρ-TIA analogs and ρ-TIA to the α1B-AR and mutants. The 10 lowest energy docking complexes for each simulation are shown. All panels are prepared in the same orientation for ease of comparison, with water molecules not shown for clarity. A, docking of W3A-ρ-TIA to α1B-AR showed ∼25° rotation along the y axis (left and right) and/or z axis (middle), compared with that of ρ-TIA in α1B-AR. An additional salt bridge is observed between the positively charged N termini of W3A-ρ-TIA and Glu-186 of α1B-AR. B, docking complexes of ρ-TIA to α1B-AR-S318A/F330A showed a similar orientation to that of W3A-ρ-TIA to α1B-AR. A potential salt bridge is predicted between the positively charged N termini of W3A-ρ-TIA and Glu-186 or Glu-199 of α1B-AR. C, docking complexes of W3A-ρ-TIA and α1B-AR-S318A/F330A showed a different binding mode (∼180° rotation along the y axis) to that of ρ-TIA in α1B-AR, with Arg-4 of ρ-TIA-W3A forming a salt bridge with either Glu-199 or Glu-194, instead of Asp-327. D, R4A-ρ-TIA to α1B-AR showed rotation angles of ∼20° and ∼50° along the y and/or z axis, respectively, with the N terminus forming a salt bridge with either Glu-186 or Glu-194 on α1B-AR.
The predicted salt bridge between Arg-4-ρ-TIA and Asp-327 and cation-π interaction between Arg-4-ρ-TIA and Phe-330 were assessed by examining ρ-TIA binding to receptor mutants D327A (IC50 = 110 ± 11 nm, n = 9) and F330A (IC50 = 290 ± 22 nm, n = 6) (Figs. 3B and 7C). Single mutations at these positions produced a less pronounced reduction in ρ-TIA affinity compared with the reduction in affinity for R4A-ρ-TIA at the α1B-AR (IC50 = 12178 ± 813 nm, n = 4). Interestingly, ρ-TIA binding to the double mutant D327A/F330A exhibited a comparable reduction (IC50 = 7732 ± 1539 nm, n = 7) in ρ-TIA affinity to that of R4A-ρ-TIA to α1B-AR, whereas R4A-ρ-TIA against the double mutant D327A/F330A resulted in a further reduction in affinity (51,286 ± 9906 nm, n = 4) (Fig. 7C). Solvated docking simulations of R4A-ρ-TIA to α1B-AR and ρ-TIA to D327A/F330A were again performed to better understand these differences. R4A-ρ-TIA docking to α1B-AR showed a compensatory rotation along the x and z axis to a maximum angle of ∼20° and ∼50°, respectively (Fig. 8), reminiscent of our observations for W3A-ρ-TIA docking to α1B-AR and ρ-TIA to S318A/F330A. However, attempts to dock ρ-TIA to D327A/F330A were unsuccessful, presumably due to the removal of the key binding residues Asp-327 and Phe-330, but were consistent with the large reduction in affinity observed for R4A-ρ-TIA binding to the D327A/F330A mutant (1800-fold). To confirm their structural integrity, we determined the prazosin Kd and NE EC50 values for the double mutants E186A/E199A, S318A/F330A, and D327A/F330A. No significant changes in prazosin affinity were observed for either E186A/E199A (0.4 ± 0.01 nm, n = 3), S318A/F330A (0.8 ± 0.23 nm, n = 3), or D327A/F330A (2.4 ± 0.17 nm, n = 3), whereas the E186A/E199A mutant produced a 9-fold decrease in NE potency compared with WT (EC50 = 11.7 ± 0.61 nm, n = 35) (Fig. 7C).
DISCUSSION
Allosteric ligands have the potential to be more selective modulators of class A GPCRs due to the structural divergence typically observed beyond the structurally and functionally conserved transmembrane region (3, 34, 35). In this study, we performed mutational and docking studies to determine the sites on the α1B-AR, where the orthosteric antagonist prazosin and the allosteric antagonist ρ-TIA bind. Our studies establish that ρ-TIA binds to the ECS at the base of ECL3 to inhibit GPCR signaling at a site that is distinct from the prazosin-binding site. Despite the differences in their binding sites, both ρ-TIA and prazosin acted as inverse agonists at the α1B-AR.
Docking studies indicated that prazosin binds to the α1B-AR through extensive hydrophobic interactions with Trp-307, Phe-310, and Phe-311 in TMH6, and Phe-334 in TMH7. With the exception of Phe-334, which produced only a modest reduction in prazosin affinity, direct involvement of these residues in prazosin binding was supported by mutational studies. The significant reduction in the affinity of prazosin for the F310A-α1B-AR and F311A-α1B-AR mutants was observed previously in the corresponding alanine mutants of closely related subtypes (36–38). The W307A mutant, which forms part of the conserved CWXP motif and is proposed to function as an activation switch (39), reduced NE potency to signal to IP1 but almost abolished specific prazosin binding, indicating this residue also contributes to prazosin binding. Because the hydrophobic residues identified as interacting with prazosin are highly conserved across all class A GPCRs, it appears that prazosin selectivity for α1-ARs is determined by either the specific shape of this binding pocket or by adjacent residues in ECL2 and ECL3 that define the shape of the entrance to the orthosteric binding pocket and are highly divergent across class A GPCRs (3).
Based on its size, polarity, and noncompetitive pharmacology, ρ-TIA was predicted to bind to the ECS at a site that was topologically distinct from the prazosin-binding site. Blind docking of ρ-TIA to α1B-AR indicated a major docking cluster dominated by interactions with Asp-327. A refined docking simulation incorporating the structural restraints obtained from mutational studies and the SAR of ρ-TIA (17) using Haddock revealed that ρ-TIA formed a salt bridge with Asp-327 in TMH7, T-stacking-π and cation-π interactions with Phe-330 in TMH7, and a hydrogen bond with Ser-318 in TMH6. These interactions reveal that the allosteric binding site for ρ-TIA is located on the ECS of the α1-AR at the base of ECL3. These interactions were supported by mutational studies that confirmed ρ-TIA and prazosin-binding sites were separate and distinct. Phe-330 is conserved across the α1-ARs, whereas position 327 is either a negatively charged Asp in α1B-AR or Glu in α1A- and α1D-AR, explaining the shallow selectivity profile for ρ-TIA at α1-ARs (17). Interestingly, Phe-330 on the α1-AR is at an equivalent position to the Trp in muscarinic receptors that contribute to allosteric small molecule inhibitor (40–42) and three-fingered snake toxin MT7 (41) binding (Fig. 9), suggesting that this position may play a central role in the allosteric inhibition of GPCRs.
FIGURE 9.
Sequence frequency plot of the D(E)XXF(WY) motif in six different class A human GPCRs. Shown are the corresponding sequences α1-ARs (α1A–D), α2-ARs (α2A–C), β-ARs (β1–3), dopamine receptors (D1A, D1B, and D2–5), histamine receptors (H1–4), and muscarinic receptors (M1–5). The negative, positive, and aromatic residues are colored red, blue, and orange, respectively, with letter size proportional to frequency of each position. Residue numbering refers to the first subtype in each group. The D(E)XXF motif present in the hamster α1B-AR is Asp-327, Ala-328, Val-329, and Phe-330. The figure was prepared using WebLogo3 (56).
The identification of the intramolecular cation-π interaction between Trp-3 and Arg-4 within ρ-TIA, and the combination of intermolecular interactions between Trp-3 and Arg-4 on ρ-TIA with Phe-330 and/or Asp-327 on α1B-AR complicate the interpretation of mutational data investigating the influence of these residues on ρ-TIA affinity. For the double mutant S318A/F330A, the interactions between α1B-AR and Trp-3-ρ-TIA were removed, resulting in a significant 65-fold drop in affinity. A comparable reduction in affinity (35-fold) was also observed for W3A-ρ-TIA at the WT α1B-AR, as observed previously (17). However, interactions between S318A/F330A and W3A-ρ-TIA decreased nearly 10-fold more than expected (590-fold) for a strictly complementary interaction. The adjacent Arg-4 in ρ-TIA was also predicted to interact strongly with the α1B-AR through Asp-327 and Phe-330. Here, the single mutants D327A and F330A decreased ρ-TIA affinity only 4- and 10-fold, respectively, although the double mutant D327A/F330A decreased the affinity ∼280-fold, comparable with the ∼440-fold loss in affinity for R4A-TIA at the WT α1B-AR. Again, R4A-ρ-TIA binding to the double mutant D327A/F330A caused a larger than expected drop in affinity (∼1845-fold). To address these discrepancies, docking studies of these mutant cycle experiments were performed that revealed ∼20° shifts in the binding orientation of ρ-TIA (Fig. 8). These larger than expected drops in ρ-TIA affinity do not appear to arise from broader structural effects, because the double mutants E186A/E199A, S318A/F330A, and D327A/F330A that affected ρ-TIA affinity had no effect on the affinity of prazosin, with only the E186A/E199A mutant decreasing NE potency. Thus, we conclude that compensatory shifts in binding for the single mutant interactions resulted in an underestimate of the strength of individual binding interactions.
N-terminal truncated ρ-TIA significantly (∼5-fold) reduced ρ-TIA affinity for the α1B-AR (17), suggesting that the N-terminal positive charge might also contribute to ρ-TIA binding. However, our solvated docking studies identified only a hydrogen bond bridged by water between Glu-186 and the N-terminal positive charge. This relatively weak hydrogen-bonding interaction was supported by studies on acetylated ρ-TIA, which maintained WT or better affinity at E186A-α1B-AR, the adjacent E199A-α1B-AR mutant, and E186A/E199A-α1B-AR double mutants.
In addition to identifying three closely positioned residues that contribute directly to ρ-TIA binding, 11 more widely distributed residues were found that either enhanced the affinity of ρ-TIA (V107A, G109A, Y110A, G114A, C118A, C195A, F310A, F311A, K324A, and K331A) or reduced ρ-TIA affinity (Gly-183). These residues either had no effect, reduced (Gly-183, F310A, and F311A), or enhanced (Gly-109 and Gly-114) prazosin affinity and were predicted to play a structural role as they were topologically distant from the predicted allosteric binding site. To further characterize the effect of these mutations on the α1B-AR conformation, we determined their effect on NE signaling. V107A, Gly-109, K324A, and K331A had no effect, whereas Y110A, Gly-114, C118A, Gly-183, C195A, F310A, and F311A reduced NE potency to signal to IP1. Phe-310 has previously been reported to bind NE, and the F311A mutation has been suggested to cause global changes in receptor conformation, including folding and expression (36), consistent with the reduced expression observed for the Phe-311 mutant and reduced NE potency for the Phe-310 mutant. From our model, Lys-331 in TMH7 is predicted to form a salt bridge with Glu-106 in TMH2 and thus contribute to the α1B-AR structure by stabilizing TMH2 and TMH7. It appears that disrupting this salt bridge in α1B-AR favors conformations with enhanced ρ-TIA affinity. However, the involvement of Lys-324 (ECL3) in ρ-TIA binding remains unclear, and the increased ρ-TIA affinity and decreased NE potency at the Y110A (ECL1) mutant cannot be easily explained from our model. Two glycine residues in ECL1 (Gly-109 and Gly-114) increased both ρ-TIA and prazosin affinity when mutated to alanine. Interestingly, the G114A mutant reduced NE potency, whereas the G109A mutant had no effect. In contrast, the glycine mutant G183A in TMH4 showed decreased affinity for ρ-TIA, prazosin, and NE, despite its distant position from the binding cavity. Because these five mutations are at positions distant from the binding pocket for ρ-TIA and orthosteric ligands, it is likely that mutating these positions influences the structure of the α1B-AR to affect ligand binding at both the allosteric and orthosteric binding sites.
To better understand the role of extracellular surface residues in ligand binding and signaling, we compared the secondary structure of our model to the GPCR crystal structures determined (27). Although ECL2 varies widely across class A GPCR families, it invariably presents as a highly ordered hairpin-like conformation stabilized by a network of intramolecular interactions (4–11, 43, 44) despite the lacking of an extensive well defined secondary structure. Consistent with this view obtained directly from x-ray crystallography, our homology model also showed a highly ordered hairpin-like conformation stabilized by a network of intramolecular interactions. The stability and integrity of ECL2 have been shown to affect ligand binding and receptor activation in many GPCRs by altering the molecular shape and size of the entrance of ligand-binding site (14, 45–54). Interestingly, disrupting the disulfide bond between Cys-118 and Cys-195 significantly enhanced ρ-TIA affinity ∼60-fold, had no effect on prazosin affinity, and reduced NE potency ∼300-fold. Surprisingly, [3H]prazosin binding revealed that the C118A mutant had only ∼1% of the expression level of the C195A mutant, indicating that these positions are not equivalent. A decrease in agonist potency of up to 4400-fold has been observed previously in other class A GPCRs when this disulfide bond was disrupted by mutations or exposure to reducing agents (45, 47–54).
In conclusion, this study provides the first analysis of an allosteric binding site on α1-ARs. Through extensive pharmacological characterization and computational simulations, we determined that ρ-TIA binds to an allosteric modulatory site on TMH6 and TMH7 at the base of ECL3. ρ-TIA binding was dominated by a salt bridge and cation-π interaction between Arg-4-ρ-TIA and Asp-327 and Phe-330 in TMH7, respectively, and a hydrogen bond and T-stacking-π interaction between Trp-3-ρ-TIA and Phe-330 in TMH7. The ECS pharmacophore identified in this study provides the first set of structural constraints for the design of novel selective allosteric antagonists acting at the α1-ARs. A number of ECS mutations also affected NE potency, confirming the important role played by the ECS in regulating agonist signaling. The D(E)XXF diad in the α1B-AR varies across related GPCRs (Fig. 9), being mostly charged at the first position (DEKRH) and typically an aromatic residue at the second position (FWY), revealing opportunities to exploit this allosteric site for the development of new classes of subtype-selective allosteric inhibitors of GPCRs.
Acknowledgment
We thank Prof W. Thomas for helpful comments on a draft of the manuscript.
This work was supported by National Health and Medical Research Council Project Grants 011246 and 351446 and a National Health and Medical Research Council Principal Research Fellowship (to R. J. L.).
This article was selected as a Paper of the Week.
The atomic coordinates and structure factors (code 2LR9) have been deposited in the Protein Data Bank (http://wwpdb.org/).
Submitted to the Biological Magnetic Resonance Bank Database under code RCSB102733.
- GPCR
- G protein-coupled receptor
- ECS
- extracellular surface
- α1B-AR
- α1B-adrenoceptor
- TMH
- transmembrane helix
- ECL
- extracellular loop
- NE
- norepinephrine
- r.m.s.d.
- root mean square deviation
- PDB
- Protein Data Bank
- HEAT
- (±)β-(iodo-4-hydroxyphenyl)ethylaminomethyl-tetralone
- Fmoc
- N-(9-fluorenyl)methoxycarbonyl
- DIEA
- diisopropylethylamine
- DMF
- dimethylformamide
- IP1
- inositol 1-phosphate
- HTRF
- homogeneous time-resolved fluorescence
- tBu
- t-butyl
- Trt
- trityl.
REFERENCES
- 1. Rosenbaum D. M., Rasmussen S. G., Kobilka B. K. (2009) The structure and function of G-protein-coupled receptors. Nature 459, 356–363 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Palczewski K., Kumasaka T., Hori T., Behnke C. A., Motoshima H., Fox B. A., Le Trong I., Teller D. C., Okada T., Stenkamp R. E., Yamamoto M., Miyano M. (2000) Crystal structure of rhodopsin. A G protein-coupled receptor. Science 289, 739–745 [DOI] [PubMed] [Google Scholar]
- 3. Wang C. I., Lewis R. J. (2012) Emerging opportunities for allosteric modulation of G-protein coupled receptors. Biochem. Pharmacol., In Press [DOI] [PubMed] [Google Scholar]
- 4. Park J. H., Scheerer P., Hofmann K. P., Choe H. W., Ernst O. P. (2008) Crystal structure of the ligand-free G-protein-coupled receptor opsin. Nature 454, 183–187 [DOI] [PubMed] [Google Scholar]
- 5. Scheerer P., Park J. H., Hildebrand P. W., Kim Y. J., Krauss N., Choe H. W., Hofmann K. P., Ernst O. P. (2008) Crystal structure of opsin in its G-protein-interacting conformation. Nature 455, 497–502 [DOI] [PubMed] [Google Scholar]
- 6. Cherezov V., Rosenbaum D. M., Hanson M. A., Rasmussen S. G., Thian F. S., Kobilka T. S., Choi H. J., Kuhn P., Weis W. I., Kobilka B. K., Stevens R. C. (2007) High-resolution crystal structure of an engineered human β2-adrenergic G protein-coupled receptor. Science 318, 1258–1265 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Warne T., Serrano-Vega M. J., Baker J. G., Moukhametzianov R., Edwards P. C., Henderson R., Leslie A. G., Tate C. G., Schertler G. F. (2008) Structure of a β1-adrenergic G-protein-coupled receptor. Nature 454, 486–491 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Jaakola V. P., Griffith M. T., Hanson M. A., Cherezov V., Chien E. Y., Lane J. R., Ijzerman A. P., Stevens R. C. (2008) The 2.6 angstrom crystal structure of a human A2A adenosine receptor bound to an antagonist. Science 322, 1211–1217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Chien E. Y., Liu W., Zhao Q., Katritch V., Han G. W., Hanson M. A., Shi L., Newman A. H., Javitch J. A., Cherezov V., Stevens R. C. (2010) Structure of the human dopamine D3 receptor in complex with a D2/D3 selective antagonist. Science 330, 1091–1095 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Haga K., Kruse A. C., Asada H., Yurugi-Kobayashi T., Shiroishi M., Zhang C., Weis W. I., Okada T., Kobilka B. K., Haga T., Kobayashi T. (2012) Structure of the human M2 muscarinic acetylcholine receptor bound to an antagonist. Nature 482, 547–551 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Kruse A. C., Hu J., Pan A. C., Arlow D. H., Rosenbaum D. M., Rosemond E., Green H. F., Liu T., Chae P. S., Dror R. O., Shaw D. E., Weis W. I., Wess J., Kobilka B. K. (2012) Structure and dynamics of the M3 muscarinic acetylcholine receptor. Nature 482, 552–556 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Michelotti G. A., Price D. T., Schwinn D. A. (2000) α1-Adrenergic receptor regulation. Basic science and clinical implications. Pharmacol. Ther. 88, 281–309 [DOI] [PubMed] [Google Scholar]
- 13. Zhong H., Minneman K. P. (1999) α1-Adrenoceptor subtypes. Eur. J. Pharmacol. 375, 261–276 [DOI] [PubMed] [Google Scholar]
- 14. Bokoch M. P., Zou Y., Rasmussen S. G., Liu C. W., Nygaard R., Rosenbaum D. M., Fung J. J., Choi H. J., Thian F. S., Kobilka T. S., Puglisi J. D., Weis W. I., Pardo L., Prosser R. S., Mueller L., Kobilka B. K. (2010) Ligand-specific regulation of the extracellular surface of a G-protein-coupled receptor. Nature 463, 108–112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Sharpe I. A., Gehrmann J., Loughnan M. L., Thomas L., Adams D. A., Atkins A., Palant E., Craik D. J., Adams D. J., Alewood P. F., Lewis R. J. (2001) Two new classes of conopeptides inhibit the α1-adrenoceptor and noradrenaline transporter. Nat. Neurosci. 4, 902–907 [DOI] [PubMed] [Google Scholar]
- 16. Chen Z., Rogge G., Hague C., Alewood D., Colless B., Lewis R. J., Minneman K. P. (2004) Subtype-selective noncompetitive or competitive inhibition of human α1-adrenergic receptors by ρ-TIA. J. Biol. Chem. 279, 35326–35333 [DOI] [PubMed] [Google Scholar]
- 17. Sharpe I. A., Thomas L., Loughnan M., Motin L., Palant E., Croker D. E., Alewood D., Chen S., Graham R. M., Alewood P. F., Adams D. J., Lewis R. J. (2003) Allosteric α1-adrenoceptor antagonism by the conopeptide ρ-TIA. J. Biol. Chem. 278, 34451–34457 [DOI] [PubMed] [Google Scholar]
- 18. Brust A., Palant E., Croker D. E., Colless B., Drinkwater R., Patterson B., Schroeder C. I., Wilson D., Nielsen C. K., Smith M. T., Alewood D., Alewood P. F., Lewis R. J. (2009) χ-Conopeptide pharmacophore development. Toward a novel class of norepinephrine transporter inhibitor (Xen2174) for pain. J. Med. Chem. 52, 6991–7002 [DOI] [PubMed] [Google Scholar]
- 19. Schnölzer M., Alewood P., Jones A., Alewood D., Kent S. B. (1992) In situ neutralization in Boc-chemistry solid phase peptide synthesis. Rapid, high yield assembly of difficult sequences. Int. J. Pept. Protein Res. 40, 180–193 [DOI] [PubMed] [Google Scholar]
- 20. Sarin V. K., Kent S. B., Tam J. P., Merrifield R. B. (1981) Quantitative monitoring of solid-phase peptide synthesis by the ninhydrin reaction. Anal. Biochem. 117, 147–157 [DOI] [PubMed] [Google Scholar]
- 21. Buck M. A., Olah T. A., Weitzmann C. J., Cooperman B. S. (1989) Protein estimation by the product of integrated peak area and flow rate. Anal. Biochem. 182, 295–299 [DOI] [PubMed] [Google Scholar]
- 22. Andersson H. S., Figueredo S. M., Haugaard-Kedstrom L. M., Bengtsson E., Daly N. L., Qu X., Craik D. J., Ouellette A. J., Rosengren K. J. (2012) The α-defensin salt-bridge induces backbone stability to facilitate folding and confer proteolytic resistance. Amino Acids, 43, 1471–1483 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Davis I. W., Leaver-Fay A., Chen V. B., Block J. N., Kapral G. J., Wang X., Murray L. W., Arendall W. B., 3rd, Snoeyink J., Richardson J. S., Richardson D. C. (2007) MolProbity. All-atom contacts and structure validation for proteins and nucleic acids. Nucleic Acids Res. 35, W375–383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Koradi R., Billeter M., Wüthrich K. (1996) MOLMOL. A program for display and analysis of macromolecular structures. J. Mol. Graph. 14, 51–55 [DOI] [PubMed] [Google Scholar]
- 25. Fiser A., Sali A. (2003) Modeller. Generation and refinement of homology-based protein structure models. Methods Enzymol. 374, 461–491 [DOI] [PubMed] [Google Scholar]
- 26. Altschul S. F., Gish W., Miller W., Myers E. W., Lipman D. J. (1990) Basic local alignment search tool. J. Mol. Biol. 215, 403–410 [DOI] [PubMed] [Google Scholar]
- 27. Kelley L. A., Sternberg M. J. (2009) Protein structure prediction on the Web. A case study using the Phyre server. Nat. Protoc. 4, 363–371 [DOI] [PubMed] [Google Scholar]
- 28. Larkin M. A., Blackshields G., Brown N. P., Chenna R., McGettigan P. A., McWilliam H., Valentin F., Wallace I. M., Wilm A., Lopez R., Thompson J. D., Gibson T. J., Higgins D. G. (2007) Clustal W and Clustal X version 2.0. Bioinformatics 23, 2947–2948 [DOI] [PubMed] [Google Scholar]
- 29. Bowie J. U., Lüthy R., Eisenberg D. (1991) A method to identify protein sequences that fold into a known three-dimensional structure. Science 253, 164–170 [DOI] [PubMed] [Google Scholar]
- 30. Laskowski R. A., Watson J. D., Thornton J. M. (2005) ProFunc. A server for predicting protein function from 3D structure. Nucleic Acids Res. 33, W89–W93 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. de Vries S. J., van Dijk M., Bonvin A. M. (2010) The HADDOCK web server for data-driven biomolecular docking. Nat. Protoc. 5, 883–897 [DOI] [PubMed] [Google Scholar]
- 32. Greasley P. J., Fanelli F., Rossier O., Abuin L., Cotecchia S. (2002) Mutagenesis and modelling of the α(1b)-adrenergic receptor highlight the role of the helix 3/helix 6 interface in receptor activation. Mol. Pharmacol. 61, 1025–1032 [DOI] [PubMed] [Google Scholar]
- 33. McCune D. F., Edelmann S. E., Olges J. R., Post G. R., Waldrop B. A., Waugh D. J., Perez D. M., Piascik M. T. (2000) Regulation of the cellular localization and signaling properties of the α(1B)- and α(1D)-adrenoceptors by agonists and inverse agonists. Mol. Pharmacol. 57, 659–666 [DOI] [PubMed] [Google Scholar]
- 34. Kenakin T., Miller L. J. (2010) Seven transmembrane receptors as shape-shifting proteins: the impact of allosteric modulation and functional selectivity on new drug discovery. Pharmacol. Rev. 62, 265–304 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Keov P., Sexton P. M., Christopoulos A. (2011) Allosteric modulation of G protein-coupled receptors. A pharmacological perspective. Neuropharmacology 60, 24–35 [DOI] [PubMed] [Google Scholar]
- 36. Chen S., Xu M., Lin F., Lee D., Riek P., Graham R. M. (1999) Phe310 in transmembrane VI of the α1B-adrenergic receptor is a key switch residue involved in activation and catecholamine ring aromatic bonding. J. Biol. Chem. 274, 16320–16330 [DOI] [PubMed] [Google Scholar]
- 37. Asher W. B., Hoskins S. N., Slasor L. A., Morris D. H., Cook E. M., Bautista D. L. (2007) Two model system of the α1A-adrenoceptor docked with selected ligands. J. Chem. Inf. Model. 47, 1906–1912 [DOI] [PubMed] [Google Scholar]
- 38. Waugh D. J., Gaivin R. J., Zuscik M. J., Gonzalez-Cabrera P., Ross S. A., Yun J., Perez D. M. (2001) Phe-308 and Phe-312 in transmembrane domain 7 are major sites of α1-adrenergic receptor antagonist binding. Imidazoline agonists bind like antagonists. J. Biol. Chem. 276, 25366–25371 [DOI] [PubMed] [Google Scholar]
- 39. Nygaard R., Frimurer T. M., Holst B., Rosenkilde M. M., Schwartz T. W. (2009) Ligand binding and micro-switches in 7TM receptor structures. Trends Pharmacol. Sci. 30, 249–259 [DOI] [PubMed] [Google Scholar]
- 40. Huang X. P., Prilla S., Mohr K., Ellis J. (2005) Critical amino acid residues of the common allosteric site on the M2 muscarinic acetylcholine receptor. More similarities than differences between the structurally divergent agents gallamine and bis(ammonio)alkane-type hexamethylene-bis-[dimethyl-(3-phthalimidopropyl)ammonium]dibromide. Mol. Pharmacol. 68, 769–778 [DOI] [PubMed] [Google Scholar]
- 41. Marquer C., Fruchart-Gaillard C., Letellier G., Marcon E., Mourier G., Zinn-Justin S., Ménez A., Servent D., Gilquin B. (2011) Structural model of ligand-G protein-coupled receptor (GPCR) complex based on experimental double mutant cycle data. MT7 snake toxin bound to dimeric hM1 muscarinic receptor. J. Biol. Chem. 286, 31661–31675 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Voigtländer U., Jöhren K., Mohr M., Raasch A., Tränkle C., Buller S., Ellis J., Höltje H. D., Mohr K. (2003) Allosteric site on muscarinic acetylcholine receptors. Identification of two amino acids in the muscarinic M2 receptor that account entirely for the M2/M5 subtype selectivities of some structurally diverse allosteric ligands in N-methylscopolamine-occupied receptors. Mol. Pharmacol. 64, 21–31 [DOI] [PubMed] [Google Scholar]
- 43. Hanson M. A., Roth C. B., Jo E., Griffith M. T., Scott F. L., Reinhart G., Desale H., Clemons B., Cahalan S. M., Schuerer S. C., Sanna M. G., Han G. W., Kuhn P., Rosen H., Stevens R. C. (2012) Crystal structure of a lipid G protein-coupled receptor. Science 335, 851–855 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Wu B., Chien E. Y., Mol C. D., Fenalti G., Liu W., Katritch V., Abagyan R., Brooun A., Wells P., Bi F. C., Hamel D. J., Kuhn P., Handel T. M., Cherezov V., Stevens R. C. (2010) Structures of the CXCR4 chemokine GPCR with small-molecule and cyclic peptide antagonists. Science 330, 1066–1071 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Cook J. V., Eidne K. A. (1997) An intramolecular disulfide bond between conserved extracellular cysteines in the gonadotropin-releasing hormone receptor is essential for binding and activation. Endocrinology 138, 2800–2806 [DOI] [PubMed] [Google Scholar]
- 46. Correa S. A., Pignatari G. C., Ferro E. S., Pacheco N. A., Costa-Neto C. M., Pesquero J. B., Oliveira L., Paiva A. C., Shimuta S. I. (2006) Role of the Cys18–Cys274 disulfide bond and of the third extracellular loop in the constitutive activation and internalization of angiotensin II type 1 receptor. Regul. Pept. 134, 132–140 [DOI] [PubMed] [Google Scholar]
- 47. Dohlman H. G., Caron M. G., DeBlasi A., Frielle T., Lefkowitz R. J. (1990) Role of extracellular disulfide-bonded cysteines in the ligand binding function of the β2-adrenergic receptor. Biochemistry 29, 2335–2342 [DOI] [PubMed] [Google Scholar]
- 48. Fraser C. M. (1989) Site-directed mutagenesis of β-adrenergic receptors. Identification of conserved cysteine residues that independently affect ligand binding and receptor activation. J. Biol. Chem. 264, 9266–9270 [PubMed] [Google Scholar]
- 49. Karnik S. S., Khorana H. G. (1990) Assembly of functional rhodopsin requires a disulfide bond between cysteine residues 110 and 187. J. Biol. Chem. 265, 17520–17524 [PubMed] [Google Scholar]
- 50. Karnik S. S., Sakmar T. P., Chen H. B., Khorana H. G. (1988) Cysteine residues 110 and 187 are essential for the formation of correct structure in bovine rhodopsin. Proc. Natl. Acad. Sci. U.S.A. 85, 8459–8463 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Kurtenbach E., Curtis C. A., Pedder E. K., Aitken A., Harris A. C., Hulme E. C. (1990) Muscarinic acetylcholine receptors. Peptide sequencing identifies residues involved in antagonist binding and disulfide bond formation. J. Biol. Chem. 265, 13702–13708 [PubMed] [Google Scholar]
- 52. Lin S. W., Sakmar T. P. (1996) Specific tryptophan UV-absorbance changes are probes of the transition of rhodopsin to its active state. Biochemistry 35, 11149–11159 [DOI] [PubMed] [Google Scholar]
- 53. Perlman J. H., Wang W., Nussenzveig D. R., Gershengorn M. C. (1995) A disulfide bond between conserved extracellular cysteines in the thyrotropin-releasing hormone receptor is critical for binding. J. Biol. Chem. 270, 24682–24685 [DOI] [PubMed] [Google Scholar]
- 54. Zhou H., Tai H. H. (2000) Expression and functional characterization of mutant human CXCR4 in insect cells. Role of cysteinyl and negatively charged residues in ligand binding. Arch. Biochem. Biophys. 373, 211–217 [DOI] [PubMed] [Google Scholar]
- 55. Ilyin V. A., Abyzov A., Leslin C. M. (2004) Structural alignment of proteins by a novel TOPOFIT method, as a superimposition of common volumes at a topomax point. Protein Sci. 13, 1865–1874 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Crooks G. E., Hon G., Chandonia J. M., Brenner S. E. (2004) WebLogo. A sequence logo generator. Genome Res. 14, 1188–1190 [DOI] [PMC free article] [PubMed] [Google Scholar]