Background: Plant-specific IQD genes encode putative CaM targets of unknown functions.
Results: IQD1 interacts with KLCR1, binds to Arabidopsis CaM/CMLs, and localizes to microtubules.
Conclusion: IQD1 may act as a scaffold protein recruiting cargo to kinesin motors for directional transport along microtubules.
Significance: This work provides novel insight into IQD function and a framework to study plant kinesin regulation.
Keywords: Arabidopsis, Calcium Signaling, Calmodulin, Kinesin, Microtubules, Cell Biology, Plant Biochemistry, Plant Physiology
Abstract
Calcium (Ca2+) is a key second messenger in eukaryotes and regulates diverse cellular processes, most notably via calmodulin (CaM). In Arabidopsis thaliana, IQD1 (IQ67 domain 1) is the founding member of the IQD family of putative CaM targets. The 33 predicted IQD proteins share a conserved domain of 67 amino acids that is characterized by a unique arrangement of multiple CaM recruitment motifs, including so-called IQ motifs. Whereas IQD1 has been implicated in the regulation of defense metabolism, the biochemical functions of IQD proteins remain to be elucidated. In this study we show that IQD1 binds to multiple Arabidopsis CaM and CaM-like (CML) proteins in vitro and in yeast two-hybrid interaction assays. CaM overlay assays revealed moderate affinity of IQD1 to CaM2 (Kd ∼ 0.6 μm). Deletion mapping of IQD1 demonstrated the importance of the IQ67 domain for CaM2 binding in vitro, which is corroborated by interaction of the shortest IQD member, IQD20, with Arabidopsis CaM/CMLs in yeast. A genetic screen of a cDNA library identified Arabidopsis kinesin light chain-related protein-1 (KLCR1) as an IQD1 interactor. The subcellular localization of GFP-tagged IQD1 proteins to microtubules and the cell nucleus in transiently and stably transformed plant tissues (tobacco leaves and Arabidopsis seedlings) suggests direct interaction of IQD1 and KLCR1 in planta that is supported by GFP∼IQD1-dependent recruitment of RFP∼KLCR1 and RFP∼CaM2 to microtubules. Collectively, the prospect arises that IQD1 and related proteins provide Ca2+/CaM-regulated scaffolds for facilitating cellular transport of specific cargo along microtubular tracks via kinesin motor proteins.
Introduction
In plants, intracellular Ca2+ plays a pivotal role as a second messenger for the regulation of numerous developmental processes and responses to a myriad of abiotic cues and biotic challenges (1–3). Typically, Ca2+ signaling pathways comprise three stages. Stimulus-specific and dynamic fluctuations of cytosolic Ca2+ concentration, characterized by amplitude, frequency, and spatial distribution, are generated during the initial phase by interplay of voltage- and ligand-activated Ca2+-permeable channels (influx) and Ca2+-ATPases/antiporters (efflux) to restore resting cytosolic Ca2+ levels. The resulting Ca2+ signatures are decoded by Ca2+-sensor proteins that undergo conformational changes upon Ca2+ binding followed by dynamic interactions with target proteins to regulate downstream responses during the final stage of the transduction process (1–3).
About 250 Ca2+ sensor proteins have been identified in the reference plant Arabidopsis thaliana, which monitor Ca2+ fluctuations via their helix-loop-helix EF hand motifs and can be grouped into Ca2+ sensor responders or Ca2+ sensor relays (4, 5). The former are bifunctional proteins capable of directly transducing the Ca2+ signal via their intrinsic catalytic activity. For example, Ca2+-dependent protein kinases, a family of 34 members in Arabidopsis, couple Ca2+ binding motifs with a kinase domain to catalyze Ca2+-triggered phosphorylation of target proteins (6). In contrast, the more widespread Ca2+ sensor relays a lack catalytic activity, such as calmodulins (CaMs),7 CaM-like proteins (CMLs), and calcineurin B-like proteins. The Ca2+ sensor relays undergo conformational changes upon Ca2+ binding and transduces the signal via interaction with terminal Ca2+ responders, thereby modulating the biochemical activities of these target proteins and establishing a large number of cellular processes and subsequent plant responses (1–3). In Arabidopsis, 10 calcineurin B-like protein sensor proteins interact specifically in a Ca2+-dependent fashion with a single family of 26 SNF1-like Ser/Thr kinases, known as calcineurin B-like protein-interacting protein kinases (CIPKs). Current data indicate that calcineurin B-like protein-CIPK interaction networks provide a signaling module for integrating plant responses to an array of environmental stimuli (7, 8). On the other hand, CaMs and CMLs, encoded by 7 CaM and 50 CML genes in Arabidopsis, regulate a broad range of structurally diverse Ca2+ responders that are predicted to exceed 700 target proteins (9, 10). These include, among other functional categories, proteins implicated in generating Ca2+ signatures, enzymes in signaling and metabolic pathways, and transcriptional regulators (11, 12). The CaM-interacting domains of target proteins usually consist of a short basic amphiphilic helix (12–30 residues). Three consensus CaM recruitment motifs are currently known, although not all functionally characterized CaM binding domains contain these specific motifs: the IQ motif mediating Ca2+-independent CaM retention and two related motifs, termed 1-5-10 and 1-8-14, facilitating Ca2+-dependent CaM interaction (13–15).
We previously identified a novel class of putative CaM target proteins in A. thaliana that we called the IQD family (16, 17). The common feature of its 33 members is the presence of a plant-specific, central domain of 67 conserved amino acid residues (referred to as the IQ67 domain), which is defined by a unique and repetitive arrangement of the three consensus CaM recruitment motifs (1–3 copies each). IQD proteins are structurally diverse with respect to computed molecular mass (12–87 kDa) but are relatively uniform at the physicochemical level, sharing some properties reminiscent of RNA binding proteins, such as a basic isoelectric point (pI ∼ 10.3) and a high content of Arg/Lys (∼17%) and Ser (∼12%) residues (17). Another hallmark of the Arabidopsis IQD family is a high fraction of homeologous gene pairs (∼45% of the paralogous gene set), which is characteristic of protein families with conserved functions in regulatory multiprotein complexes (17). However, given the large size of the extant plant IQD gene families, which originated during the early evolution of land plants and their likely conserved functions (17), surprisingly little is known about the biological roles of IQD proteins.
The first described IQD gene was functionally identified in a screen for Arabidopsis mutants with altered production of glucosinolates (16), a class of secondary metabolites in crucifers whose degradation products possess profound biological activities ranging from plant defense to cancer prevention in humans (18, 19). IQD1 binds to vertebrate CaM in a Ca2+-dependent fashion, which is also true for IQD20, the smallest member of the Arabidopsis IQD family, consisting only of the IQ67 domain at its C terminus and a short N-terminal extension (16, 17). An IQD1∼GFP fusion protein localizes to the cell nucleus, and histochemical analysis of IQD1Pro::GUS lines reveals patterns of reporter gene expression consistent with tissue-specific glucosinolate biosynthesis. Overexpression of IQD1 in Arabidopsis stimulates glucosinolate accumulation and defense against herbivores. Because IQD1 expression is induced by mechanical stimuli, IQD1 was proposed to act as a nuclear factor that integrates intracellular Ca2+ signals to regulate defense metabolism in response to biotic challenge (16).
Besides IQD1, another member of the Arabidopsis family, IQD22, has been implicated as a negative regulator of the response to the plant hormone gibberellin (20), and a role for IQD12 from tomato (Solanum lycopersicum) has recently been revealed (21, 22). The sun quantitative trait locus is associated with tomato fruit shape and causes long, atypically formed fruits when compared with the wild ancestor. A genetic approach identified IQD12 to be a major gene responsible for fruit shape variation and showed that its high expression level, caused by retrotransposon-mediated IQD12 duplication, leads to extremely elongated tomato fruits via altered cell division patterns (21, 22). Whereas first biological processes regulated by IQD proteins are beginning to emerge that include aspects of plant development or defense response, their molecular functions and underlying signaling pathways remain to be elucidated.
Here, we expand our studies on Arabidopsis IQD1 and describe its direct interaction with a select set of Arabidopsis CaM/CMLs. Using a genetic yeast two-hybrid interaction screen, we identified kinesin light chain-related-1 (KLCR1) as an IQD1 binding protein in Arabidopsis, which prompted us to reinvestigate the subcellular localization of IQD1 at high resolution. Interestingly, our data demonstrate that IQD1 localizes not only to the cell nucleus but also associates with the microtubular network to which IQD1 recruits KCLR1 as well as CaM2. For the first time our work provides insight into the biochemical function of IQD1 and related proteins as putative Ca2+-CaM-regulated scaffold proteins, which may facilitate cellular transport of cargo complexes via microtubule-associated kinesin motor proteins.
EXPERIMENTAL PROCEDURES
Production of Recombinant Proteins
The bacterial vector for expression of recombinant IQD1 fused to an N-terminal T7 epitope tag was previously described (16). Similarly, full-length cDNA fragments encoding the predicted Arabidopsis IQD20 and IQD33 proteins were generated by RT-PCR using gene-specific primers and mobilized into the pET21a(+) vector (BamHI/EcoRI), which provides the N-terminal T7 tag (Novagen, Madison, WI). To allow for production of recombinant proteins with two epitope tags (N-terminal T7 tag, C-terminal His6 tag), full-length IQD1 and partial cDNAs encoding truncated IQD1 polypeptides were generated by PCR and mobilized into the pET21a(+) vector (EcoRI/HindIII). Production of recombinant Arabidopsis CaM1 (identical with CaM4), CaM2 (identical with CaM3 and CaM5), CML8, and CML9 fused to the C-terminal Strep-tag II was supported by pASK-IBA3 bacterial vectors (Genosys Biotechnologies, Inc., The Woodlands, TX), which were obtained from C. Köhler (23). The authenticity of all cloned cDNA fragments was verified by DNA sequencing. Plasmids were transformed into Escherichia coli BL21a(pLysS) DE3 cells (Novagen) for protein expression and purification using standard protocols. Expression of recombinant proteins was induced by 1 mm isopropyl β-d-thiogalactopyranoside for at least 4 h at 37 °C. Bacterial cells were lysed by sonication in buffer A (25 mm Tris-HCl, pH 7.5, 50 mm NaCl, 0.1% Triton X-100) for most experiments or in buffer B (10 mm Tris-HCl, pH 7.6, 2.5 mm MgCl2, 0.5% Triton X-100) for nucleic acid binding experiments. Both buffers contained 1 tablet of complete EDTA-free protease inhibitor mixture (Roche Applied Science) per 50 ml volume. Bacterial lysates were cleared by centrifugation at 14,000 rpm for 30 min at 4 °C. The supernatants were used for SDS-PAGE, protein purification, immunoblot analysis, and in vitro interaction assays.
Calmodulin Pulldown Assays
Cell extracts of bacteria expressing C-terminal Strep-tag II fusions of Arabidopsis CaM1, CaM2, CML8, or CML9 were incubated with Strep-Tactin beads according to the manufacturer's protocol (Genosys Biotechnologies) to allow for binding of recombinant CaM/CML proteins. Aliquots of Strep-Tactin beads with bound CaM/CMLs (60 μl) were washed with buffer A containing 1 mm CaCl2 or 5 mm EGTA. Bacterial lysates expressing T7-tagged IQD1 (400 μl) were added to the prewashed beads in the presence of 1 mm CaCl2 or 5 mm EGTA. The suspensions were rotated at room temperature for 30 min. After incubation, the beads were washed 4 times with 1 ml of buffer A containing 1 mm CaCl2 or 5 mm EGTA. The bound proteins were eluted by boiling the bead pellets for 2 min in 50 μl of SDS sample buffer. Proteins were resolved on 10% (w/v) SDS- polyacrylamide gels (24) and transferred to nitrocellulose membranes (Pierce). Co-sedimentation of T7-tagged IQD1 was detected by enhanced chemiluminescence using a horseradish peroxidase (HRP)-conjugated T7-Tag monoclonal antibody (Novagen) as previously described (16).
Calmodulin Overlay Assays
The coding region of Arabidopsis CaM2 was cloned into vector pENTR1A (Invitrogen) using BamHI and XhoI restriction sites. The resulting entry vector was used to mobilize the CaM2 cDNA insert by LR recombination (according to the manufacturer's protocol) into vector pEXP1-DEST (Invitrogen) to facilitate in vitro synthesis of CaM2 containing a His6-tag, the Xpress epitope tag, and the enterokinase recognition site. The cell-free protein synthesis system Expressway Cell-Free E. coli Expression System (Invitrogen) and [35S]methionine (Met) (Amersham Biosciences) were used for the production of recombinant [35S]Met-labeled CaM2 according to the manufacturer's protocol. The radiolabeled CaM2 was purified by Ni-NTA affinity chromatography (Qiagen, Valencia, CA) under native conditions and digested in situ with enterokinase EKMaxTM (Invitrogen) overnight at 37 °C. The released CaM2 was collected in the flow-through fraction. The concentration of the purified protein was determined by measuring the absorbance at 280 nm using extinction coefficient values calculated by the ExPASy ProtPARAM tool (13,980 m−1 cm−1 for the CaM2 fusion after enterokinase cleavage). Purified CaM2 was used as a probe after determination of its concentration and specific radioactivity (about 0.5 × 106 cpm/μg). Expressed T7- and His6-tagged full-length IQD proteins and truncated IQD1 polypeptides were purified by Ni-NTA affinity chromatography under native conditions (according to the manufacturer's protocol). The proteins (0.5 μg) were separated by SDS-PAGE (12% gels) and transferred to nitrocellulose membranes. The membranes were blocked overnight with 5% BSA in TBST buffer (50 mm Tris-HCl, pH 7.4, 150 mm NaCl, 0.1% Tween 20) and subsequently overlaid with 2 mm [35S]Met-labeled CaM2 in the same buffer containing 1 mm CaCl2 or 5 mm EGTA. The membranes were washed 4 times for 15 min in the respective buffers without probe. Bound CaM2 was detected by phosphorimaging analysis (GE Healthcare).
For determining saturation curves of CaM2 binding, full-length T7-IQD1-His6 recombinant protein was purified by Ni-NTA chromatography under native conditions and spotted on nitrocellulose membrane discs (100 pmol) that were sequentially incubated in blocking solution (see above) for 1 h at room temperature followed by incubation in blocking solution containing different concentrations of [35S]Met-labeled CaM2 in the presence of 1 mm CaCl2 or 5 mm EGTA (2 h at room temperature). After washing in TBST buffer without radiolabeled CaM2, the amount of bound CaM2 was measured by liquid scintillation counting. Not more than 0.1% of added [35S]Met-labeled CaM2 remained at the membrane at any condition. Specific CaM2 binding was calculated as the difference between total binding and nonspecific binding (filter discs without recombinant IQD1 protein). The Kd value was determined by Scatchard plot analysis.
Yeast Two-hybrid Assays
For yeast two-hybrid interaction assays, the coding sequences of IQD1 and IQD20 were amplified by PCR and cloned into the bait pGBT9 vector (SpeI/EcoRI) to create plasmids encoding IQD protein fusions with the Gal4 DNA binding domain (23). Plasmids encoding fusions of Arabidopsis CaMs and CMLs with the Gal4 activation domain were obtained either from C. Köhler (23) (pGAD-CaM1, pGAD-CaM2, pGAD-CML8, pGAD-CML9) or were constructed by PCR and insert mobilization into the prey pGAD424 vector for CaM6 (XhoI/EcoRI), CML5 (XhoI/EcoRI), CML11 (BamHI/XhoI), CML13 (XhoI/EcoRI), CML17 (BamHI/EcoRI), CML19 (XhoI/EcoRI), CML27 (XhoI/NotI), CML36 (XhoI/EcoRI), and CML49 (XhoI/EcoRI). Plasmid templates for Arabidopsis CaMs and CMLs cDNAs were obtained from the Arabidopsis Biological Resource Center (Columbus, OH). Full-length cDNAs of IQD1 and IQD20 were also inserted into pGAD424 (BamHI/EcoRI) to test for in vivo homodimerization. All plasmid constructs were verified by DNA sequencing. Plasmid pGAD-CNGC2 served as a positive control for CaM interaction and was provided by C. Köhler (23). The bait and prey vectors to be tested for interaction of the fusion proteins were co-transformed into yeast (Saccharomyces cerevisiae) strain HF7c using the standard lithium acetate method (25). Protein-protein interactions were scored and quantified by the ability to activate two reporter genes under control of the GAL4 promoter: HIS3 and lacZ. The HIS3 reporter activity was monitored by testing transformants on synthetic dropout (SD) medium lacking histidine (His). Yeast colonies selected for the presence of the bait and prey plasmids were grown in liquid SD medium (−Leu-Trp) to an optical density of 1 A600. Serial 10-fold dilutions (10 μl) were spotted on growth permissive (−Leu-Trp) and growth restrictive (−Leu-Trp-His) SD agar plates. LacZ reporter gene expression (β-galactosidase activity) was determined using O-nitrophenyl-β-d-galactopyranoside as a substrate (25), and the galactosidase activity was calculated as Miller units: A420 × 1000/A600 × assay time (min) × assay volume (ml).
cDNA Library Construction
An A. thaliana (ecotype Columbia) flower cDNA library was constructed using the BD-Matchmaker Library Construction and Screening Kit (Clontech, Mountain View, CA). Poly(A)+ RNA (100 ng) prepared from flower tissues was used for cDNA synthesis according to the manufacturer's instruction. First strand cDNA was amplified by long distance PCR using the BD Advantage 2 PCR kit (Clontech). The purified cDNA was then fused to the GAL4 activation domain of the bait pGADT7-Rec (Clontech) expression vector by recombination in AH109 yeast strain, which yielded more than four million original transformants.
Yeast Two-hybrid Screening
The yeast two-hybrid cDNA library screening was performed using the BD Matchmaker Two-hybrid System (BD Biosciences). Full-length cDNA of IQD1 was amplified by PCR and cloned into the GAL4 DNA binding domain of pGBKT7 (BD Biosciences), generating bait plasmid pGBKT7-IQD1, which was sequenced to ensure fidelity of the construct. The bait construct was introduced into yeast strain Y187 using the lithium acetate transformation method (BD Biosciences Yeast Protocols Handbook). Screens were carried out by mating according to the manufacturer's instructions (Clontech). Briefly, an overnight culture of strain Y187 expressing pGBKT7-IQD1 was mixed with strain AH109, harboring the flower cDNA library, for 24 h in rich medium at 30 °C in a shaker at low rotation (30–50 rpm). The mixed yeast culture (diploid cells) was then plated on restrictive SD medium lacking Trp, Leu, His, and adenine (Ade). Positive colonies appearing within 5 days of plating (presumably caused by activation of the HIS3 and ADE2 reporter genes) were subsequently streaked on restrictive SD medium and tested for lacZ expression by a liquid ONPG β-galactosidase assay (BD Biosciences Yeast Protocols Handbook) to estimate the strength of interactions. Clones generating Miller units >1.0 were regarded as candidates. After the confirmation of the growth phenotype and lacZ expression, plasmid DNA was isolated from yeast cells and transformed into E. coli strain DH5α by electroporation. Because the isolated DNA is a mixture of both plasmids (the bait and prey library clone), the transformed E. coli were plated on LB medium containing ampicillin to select for the AD-cDNA library plasmid only. The cDNA inserts were amplified using the BD Matchmaker AD LD-Insert Screening Amplimer Set (Clontech) and the BD Advantage 2 PCR kit (Clontech) to analyze the number of AD library plasmids they contain. Each positive clone generated only one PCR product (one type of AD-cDNA library plasmid). The plasmid was then sequenced, and the cDNA insert was subjected to BLAST analysis of the NCBI database. To independently confirm interaction in the yeast two-hybrid system, the yeast strains Y187 and AH109 were transformed with the respective constructs, mated, and tested for growth on restrictive (−Trp-Leu-His-Ade) SD medium in the presence of 20 μg/ml of the chromogenic β-galactosidase substrate 5-bromo-4-chloro-indolyl-β-d-galactopyranoside (X-Gal).
Subcellular Localization Studies
The coding sequences of IQD1, KLCR1, and CaM2 were amplified in combination with a high fidelity DNA polymerase and subsequently inserted into pENTR/D-TOPO plasmid via directional TOPO cloning (Invitrogen) to generate IQD1-, KLCR1-, and CaM2-pENTR/D-TOPO vectors. A derivative plasmid lacking the IQD1 stop codon, IQD1*-pENTR/D-TOPO, was generated via site-directed mutagenesis (Stratagene, La Jolla, CA) according to the manufacturer's protocol. The fidelity of the inserts was verified by DNA sequencing. To study the subcellular localization of GFP- or RFP-tagged fusion proteins, IQD1-, KLCR1-, or CaM2-pENTR/D-TOPO and the IQD1*-pENTR/D-TOPO entry plasmids were recombined in LR clonase reactions with the Gateway-compatible binary destination vectors pB7WG2, pB7WGF2, or pB7FWG2 (26) to generate CaMV 35SPro::IQD1, 35SPro::GFP∼IQD1, 35SPro::GFP∼KLCR1, and 35SPro::IQD1∼GFP plasmid constructs. For generation of 35SPro::mRFP∼CaM2 and 35SPro::mRFP∼KLCR1 constructs, the pGWB455 destination vector was used (27).
Agrobacterium-mediated transient transformation of tobacco leaves (Nicotiana benthamiana) was conducted as described (28). After infiltration (1–3 days), samples were collected, and GFP fluorescence was visualized on a Zeiss LSM 710 confocal laser scanning microscope. All images were obtained using a 40× water-immersion objective. The excitation wavelength was 488 nm; emission was detected between 493 and 580 nm. For treatments, a 50 μm aqueous oryzalin solution (Sigma) was prepared from a 200 mm stock dissolved in DMSO, and tobacco leaves were infiltrated 1.5 h before microscopic analysis. For co-localization studies, tobacco leaves were co-infiltrated with the indicated plasmids, and fluorescence was visualized on a Zeiss LSM 700. All images were obtained using the sequential mode to prevent false positive signals. A 488-nm (GFP) and a 555-nm (RFP) diode laser were used for excitation; emission was detected using a short pass filter (492–555 nm) and a long pass filter (582–700 nm) for GFP and RFP, respectively. Transgenic Arabidopsis plants were generated by Agrobacterium-mediated transformation. T2 lines showing a segregation ratio of 3:1 for resistance to Basta® were selected for subsequent microscopic analysis (29). Oryzalin treatments (5 μm) on Arabidopsis plants were done for 60 min.
Whole-mount immunolocalization experiments were performed as previously described (30) using a rat anti-α-tubulin antibody (MCA78G) (AbD Serotec, Düsseldorf, Germany). After successive washings, samples were incubated with a red fluorescent rabbit anti-rat (SAB4600122) and a monoclonal green fluorescent mouse anti-GFP antibody (SAB4600051) (Sigma). Microscopy was conducted as described for co-localization studies on transiently transformed tobacco leaves.
RESULTS
IQD1 and Arabidopsis CaM/CMLs Interact in Vitro
We previously demonstrated Ca2+-dependent interaction of T7 epitope-tagged Arabidopsis IQD1 with bovine CaM (16). Here we extended these studies to include several Arabidopsis CaMs and CML proteins (Fig. 1). In Arabidopsis, 7 and 50 genes code for canonical CaMs and CML proteins, respectively. The 7 distinct CaM loci encode 4 CaM isoforms (CaM1/4, CaM2/3/5, CaM6, CaM7), which differ by 1–4 residues and share at least 89% identity to conserved vertebrate CaMs (10). We performed in vitro pulldown assays with T7-IQD1 and two Arabidopsis CaM isoforms, CaM1 and CaM2, as well as with two CML proteins, CML8 and CML9. We included CML8 and CML9 because both proteins differ considerably from conventional CaMs (<75% sequence identity). The Strep-tagged CaMs and CMLs were expressed in E. coli and purified by affinity chromatography on Strep-Tactin-Sepharose beads. After co-incubation of Strep-tagged CaMs or CMLs (bead-immobilized) and T7-epitope tagged IQD1 (bacterial extracts) in the presence of 1 mm CaCl2 or 5 mm EGTA, the Strep-Tactin-Sepharose beads were repeatedly washed. Bound proteins were eluted by boiling in SDS sample loading buffer. Proteins of all fractions were separated by SDS-PAGE, transferred to a membrane, and probed with T7-tag monoclonal antibody to detect T7-IQD1 fusion protein. As shown in Fig. 1, full-length T7-IQD1 co-sedimented with all tested CaMs and CMLs only in the presence of Ca2+, and binding was abolished when EGTA was added to the buffer. In summary, the data confirm our previous results with bovine CaM (16) and demonstrate the capacity of Arabidopsis IQD1 to interact in a Ca2+-dependent manner with various Arabidopsis CaMs and CMLs in vitro.
FIGURE 1.
Arabidopsis IQD1 interacts with Arabidopsis CaM/CMLs in vitro. Strep-Tactin beads loaded with Strep-tagged calmodulins (CaM1, CaM2) or calmodulin-like proteins (CML8, CML9) were incubated with bacterial extracts expressing full-length T7-tagged IQD1 at room temperature in the presence of 1 mm CaCl2 (+) or 5 mm EGTA (−). Proteins of the total bacterial extract (leftmost lane), of the last wash (W), and of the entire pellet (beads) fraction (P) were resolved by SDS-PAGE, transferred to a membrane, and probed with an HRP-conjugated T7-Tag monoclonal antibody, which detected the full-length T7-IQD1 protein (arrowhead) and several fragments. The rightmost three lanes demonstrate that T7-IQD1 does not bind to the Strep-Tactin matrix in the absence (No) of CaMs or CMLs.
The IQ67 Domain Mediates CaM Binding in Vitro
In a first approach to experimentally delineate the CaM binding region of IQD proteins, we took advantage of the structural diversity of the IQD family in Arabidopsis. Whereas IQD20, the smallest IQD member (103 amino acid residues), comprises the IQ67 domain at its C terminus and a short N-terminal extension of 35 amino acid residues, the IQD33 protein (442 amino acid residues) misses 51 residues of its centrally positioned IQ67 motif, which was likely caused by exon loss (17). We employed CaM overlay assays to compare the ability of IQD1, IQD20, and IQD33 to interact with CaM2 in a Ca2+-dependent manner. The T7-tagged IQD proteins were expressed in bacteria, and total protein extracts were separated by SDS-PAGE, transferred to nitrocellulose membranes, renatured, and probed with radiolabeled CaM2, which was prepared by in vitro transcription/translation in the presence of [35S]Met followed by affinity chromatography purification on Ni-NTA and proteolytic removal of the His6 epitope tag. As a positive control, we used bacterial lysates expressing Arabidopsis CNGC1 (23), which did bind to [35S]Met-labeled CaM2 in the presence of calcium (1 mm CaCl2) but not in its absence (5 mm EGTA) (supplemental Fig. S1). We also observed an interaction of the CaM2 probe with IQD1 and IQD20, but not with IQD33. In contrast to the pulldown assays (Fig. 1), CaM binding was only detected in the absence of Ca2+ (supplemental Fig. S1), which is not an uncommon observation for IQ motif-containing proteins (31, 32). In conclusion, our data point to the importance of the IQ67 domain for mediating CaM/CML interaction.
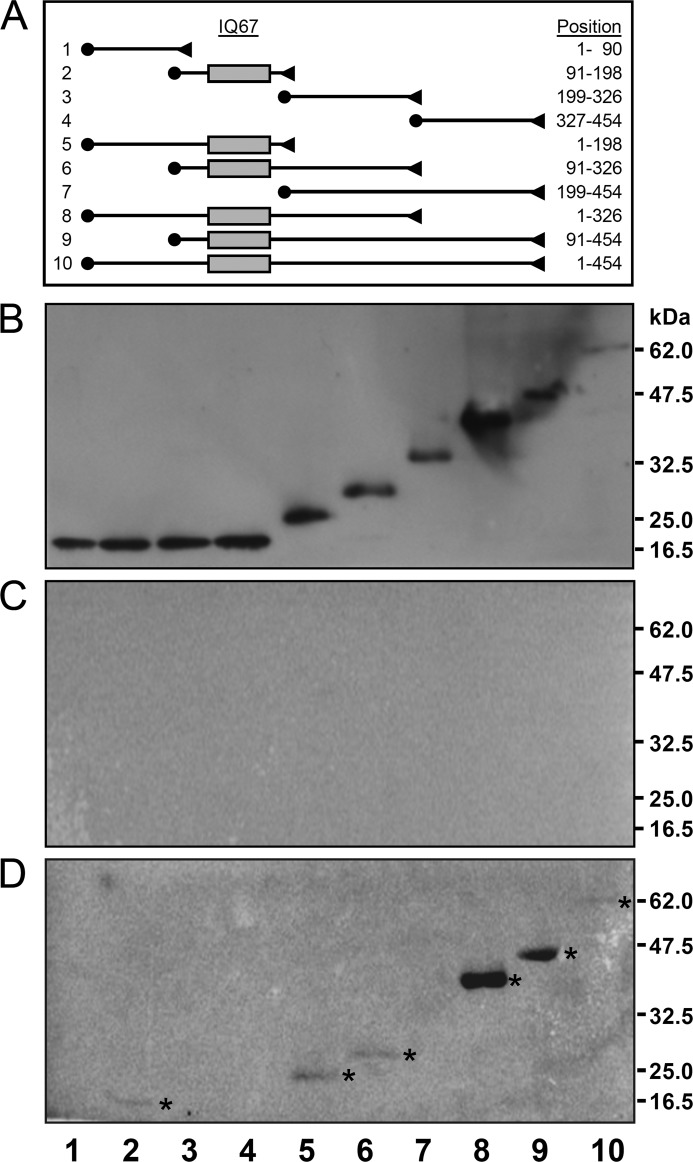
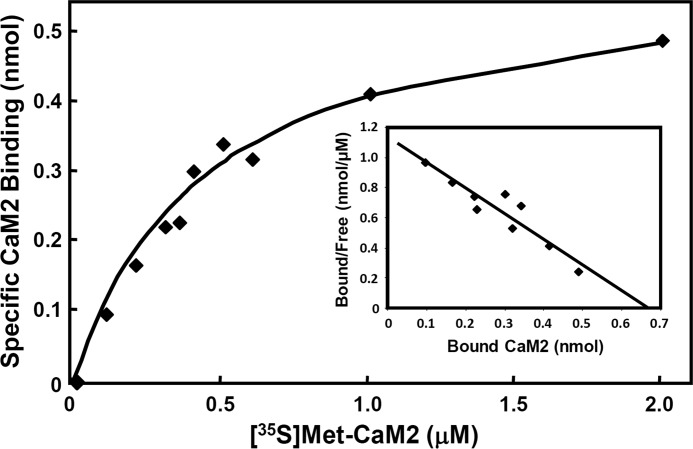
In a second approach to map the CaM interaction domain of IQD proteins, we generated a series of overlapping N- and C-terminal deletions of IQD1, each tagged with T7 and His6 epitopes (T7-IQD1-His6) that we tested for CaM binding in overlay assays (Fig. 2A). Full-length IQD1 and truncated IQD1 polypeptides were affinity-purified by Ni-NTA chromatography, separated by SDS-PAGE, transferred to nitrocellulose membranes, renatured, and probed with [35S]Met-labeled CaM2 as above. Protein loading was assessed by T7 epitope detection (Fig. 2B). As expected, CaM2 binding was only observed for IQD1 polypeptides comprising the IQ67 domain, which further supports a role of this domain for recruiting CaM (Fig. 2, C and D). Using full-length IQD1 protein, we determined by CaM overlay assays and Scatchard plot analysis the apparent dissociation constant of the interaction between IQD1 and CaM2 (Fig. 3). The calculated value (Kd ∼ 0.6 μm) indicates a relatively low affinity of IQD1 for CaM2 under the in vitro assay conditions used.
FIGURE 2.
Mapping of the CaM binding domain in IQD1. Full-length epitope-tagged T7-IQD1-His6 and nine derived truncated IQD1 polypeptides were purified by affinity chromatography on Ni-NTA, separated by SDS-PAGE, transferred to a membrane, and probed with [35S]Met-labeled CaM2 in the presence of 1 mm CaCl2 or 5 mm EGTA as described under “Experimental Procedures.” A, shown is a map of the 10 epitope-tagged IQD1constructs. The N-terminal T7 tag and the C-terminal His6 tag are indicated by solid circles and triangles, respectively. The gray box denotes the IQ67 domain, and the length of each IQD1-derived polypeptide is given on the right (position of amino acid residues). B, shown is immunodetection of SDS-PAGE, resolved Ni-NTA-affinity-purified IQD1 polypeptides with an HRP-conjugated T7-tag monoclonal antibody. C and D, overlay assays with [35S]Met-labeled CaM2 in the presence of 1 mm CaCl2 (C) or 5 mm EGTA (D) is shown. Note only IQD1-derived polypeptides containing the IQ67 domain bind to Arabidopsis CaM2 (asterisks in D).
FIGURE 3.
Binding of [35S]Met-labeled CaM2 to IQD1. [35S]Met-labeled Arabidopsis CaM2 and affinity-purified T7- IQD1-His6 protein were prepared as described under “Experimental Procedures.” The recombinant IQD1 (1 μmol) was immobilized on a nitrocellulose membrane and incubated with increasing concentrations of [35S]Met-labeled CaM2. The membranes were washed to remove unbound CaM2, and bound radioactivity was measured in a liquid scintillation counter. Each measurement was the average of three repeats. The specifically IQD1-bound [35S]Met-labeled CaM2 (y axis) at the indicated total concentrations of the [35S]Met-labeled CaM2 ligand (x axis) was graphed. The inset shows the Scatchard plots for CaM2 binding. The bound [35S]Met-labeled CaM2 was plotted against the ratio of bound to free CaM2.
IQD1and IQD20 Interact with CaM/CMLs in Yeast
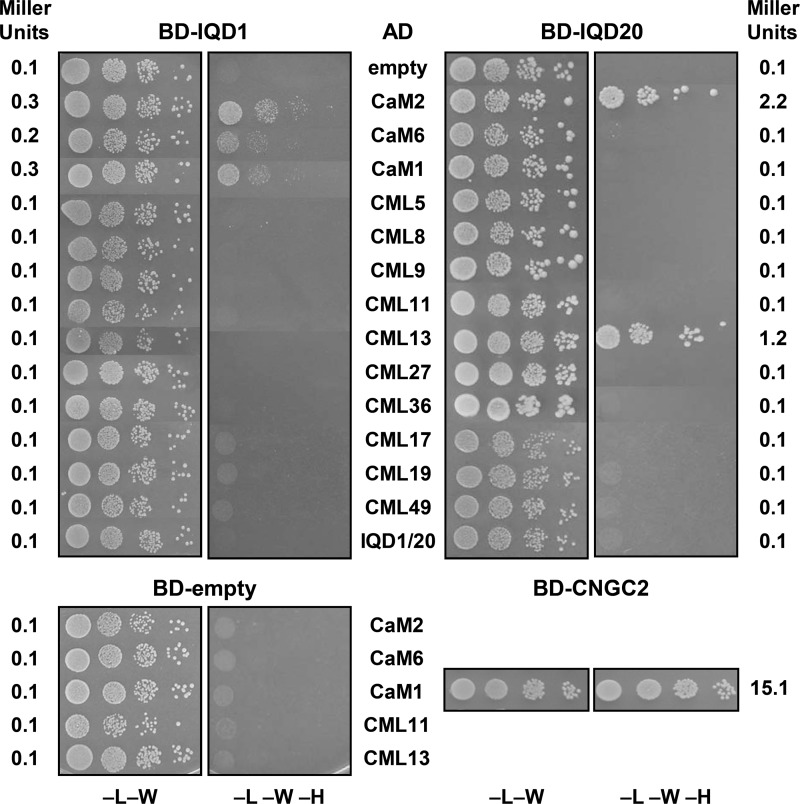
We previously showed Ca2+-dependent interaction of T7-IQD1 and T7-IQD20 with bovine CaM (16, 17). To validate in vivo the observed in vitro interactions of IQD1 and IQD20 with CaM, we performed yeast two-hybrid interaction assays. Both IQD proteins were fused to the Gal4 DNA binding domain (Gal4BD (BD)) or Gal4 activation domain (Gal4AD (AD)). We selected 13 CaM/CML proteins representing the nine groups of the Arabidopsis CaM/CML family (33) for construction of Gal4AD-CaM/CML prey vectors. A CNGC2 bait vector (Gal4DB-CNGC2) was used as a positive control for CaM interaction in yeast (23). The respective bait and prey vectors were co-transformed into the yeast reporter strain HF7c harboring two reporter genes integrated into the chromosome, His3 and LacZ. The His3 reporter activity was assayed by testing the growth of transformants on SD medium lacking His. LacZ reporter gene expression was monitored by a quantitative enzyme assay of β-galactosidase activity. As shown in Fig. 4, none of the negative controls was able to grow on restrictive (−Leu-Trp-His) SD medium (i.e. single transformations with BD-IQD1 and BD-IQD20 or AD-CaM1, AD-CaM2, AD-CaM7, AD-CML11, and AD-CML13), whereas the positive control (co-transformation with AD-CaM1/BD-CNGC2) revealed robust growth and expression of β-galactosidase activity (15.1 Miller units), which was similar to published results (23). Furthermore, there was no evidence for homotypic interaction of IQD1 or IQD20 (co-transformation with BD-IQD1/AD-IQD1 or BD-IQD20/AD-IQD20). When compared with the positive control, yeast cells transformed with plasmids encoding BD-IQD1 and AD-CaMs (CaM1, CaM2, CaM7) showed only weak growth on SD medium lacking His and low expression of β-galactosidase activity (∼0.3 Miller units). However, co-transformation with BD-IQD1 and each of the 10 different AD-CML constructs did not support yeast growth on His-deficient SC medium, and only background β-galactosidase activity was expressed (<0.1 Miller units). On the other hand, co-transformation of yeast with BD-IQD20 and AD-CaM/CMLs revealed preferential yet relatively robust interaction of IQD20 with CaM2 (2.2 Miller units) and CML13 (1.2 Miller units), whereas no interaction was observed for BD-IQD20 and the other 11 AD-CaM/CML fusion proteins (Fig. 4). In summary, although yeast two-hybrid assays may not be the ideal system to study or to identify CaM target proteins (9), our results indicate weak but differential interaction of IQD1 and IQD20 with CaM/CMLs in vivo and thus support our in vitro interaction studies.
FIGURE 4.
IQD1 and IQD20 interact with Arabidopsis CaM/CMLs in the yeast two-hybrid system. Yeast cells containing the indicated combinations of IQD1, IQD20, or CNGC2 cDNAs in the bait Gal4 DNA binding domain vector (panels BD-IQD1, BD-IQD20, BD-CNGC2, and BD-empty for control) and different CaM or CML cDNAs as well as IQD1 or IQD20 cDNAs in the prey Gal4 activation domain vector (listed in center AD column) were grown in serial 10-fold dilutions on (−L−W) SD medium to select for the presence of both vectors and on growth restrictive (−L−W−H) SD medium to test for interaction of the encoded fusion proteins (note, the upper panels are composites from different but representative agar plates). The left and right columns list the corresponding values (Miller units) of quantitative yeast two-hybrid interaction assays (β-galactosidase reporter activity). Qualitative and quantitative interaction assays were repeated 3–5 times giving similar results.
IQD1 Interacts with KLCR1
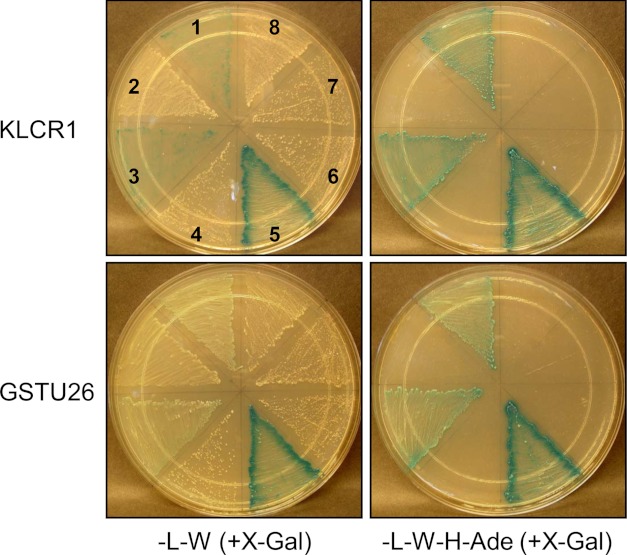
Because IQD1 is highly expressed in flowers and developing siliques of A. thaliana (16), we screened a flower cDNA library for IQD1-interacting proteins. The cDNA library was constructed by recombination with pGADT7-Rec vector in yeast strain AH109, resulting in a fusion of each cDNA with the Gal4AD. Full-length cDNA of IQD1 was cloned in-frame with the Gal4DB (pGBKT7-IQD1) bait vector and introduced into yeast strain Y187. Two-hybrid screens were carried out by mating pGBKT7-IQD1 with the pGADT7-cDNA library. The resulting diploid cells were plated on growth restrictive (−Trp, −Leu, −His, −Ade) SD medium. A total of 25 colonies were selected and tested for lacZ expression by measurement of β-galactosidase activity in liquid assays. Only 4 of the 25 yeast strains expressed robust β-galactosidase activity (>1.0 Miller units), which were used for plasmid recovery and sequencing of the prey cDNA insert. Three strains harbored cDNAs predicted to encode a tetratricopeptide repeat domain (TPR12)-containing protein (At4g10840), and the fourth library clone encoded a glutathione S-transferase (GST) belonging to the tau (U) class of GSTs (GSTU26; At1g17190). Using the predicted At4g10840 protein as a query, we identified two closely related TPR proteins in Arabidopsis (55–60% identity) with similarity to mammalian kinesin light chain (KLC) subunits (20–25% identity) of kinesin motor proteins. The three putative Arabidopsis KLCs, encoded by At4g10840, At3g27960, and At1g27500, are hereafter referred to as KLCR proteins, KLCR1-KLCR3 (supplemental Fig. S2). Superimposition of the modeled KLCR1 protein and of the experimentally solved KLC1-TPR or KLC2-TPR domains confirm presence of a structurally similar TPR12-type domain in KLCR1, which suggests functional conservation of human KLC and Arabidopsis KLCR proteins (supplemental Fig. S3).
The original diploids expressing BD-IQD1/AD-KLCR1 and BD-IQD1/AD-GSTU26 produced 13.5 ± 1.6 and 9.5 ± 1.3 Miller units of β-galactosidase activity (n = 5–6), respectively, which is indicative of a relatively robust protein-protein interaction. We confirmed these interactions by independent plasmid co-transformation of yeast with IQD1 and KLCR1 or IQD1 and GSTU26 vectors of either configuration (Gal4DB or Gal4AD) as well as with control plasmids (BD-CNGC2/AD-CAM1). After mating, the resulting diploid yeast cells expressing IQD1 and KLCR1 or IQD1 and GSTU26 showed vigorous growth and lacZ expression on restrictive (−Trp, −Leu, −His, −Ade) SD medium, which was independent of the bait or prey configuration (Fig. 5). Taken together, our results demonstrate strong and reproducible interactions between IQD1 and KLCR1 as well as between IQD1 and GSTU26, which are comparable with the interaction of the control proteins (CNGC2 and CaM1).
FIGURE 5.
IQD1 interacts with KLCR1 and GSTU26 in the yeast two-hybrid system. Confirmation of yeast two-hybrid interactions of IQD1 with KLCR1 (upper panels) and of IQD1 with GSTU26 (lower panels) is shown. Interaction assays between different test and control constructs were performed by co-transformation into yeast cells. Colony growth and lacZ expression (+X-gal) was compared on (−L−W) SD media to select for bait Gal4-BD and prey Gal4-AD vectors (left panels) and on growth restrictive (−L−W−H−Ade) SD media to test for protein interaction (right panels). Numbers (1–8, referring to all plates) indicate the following vector combinations: 1, BD-KLCR1/AD-IQD1 or BD-GSTU26/AD-IQD1; 2, BD-KLCR1/AD-empty or BD-GSTU26/AD-empty; 3, BD-IQD1/AD-KLCR1 or BD-IQD1/AD-GSTU26; 4, BD-empty/AD-KLCR1 or BD-empty/AD-GSTU26; 5, BD-CNGC2/AD-CaM1; 6, BD-empty/AD-KLCR1 or BD-empty/AD-GSTU26; 7, BD-IQD1/AD-CaM1; 8, BD-CaM1/AD-IQD1.
GFP-tagged IQD1 Proteins Localize to Microtubules and the Cell Nucleus
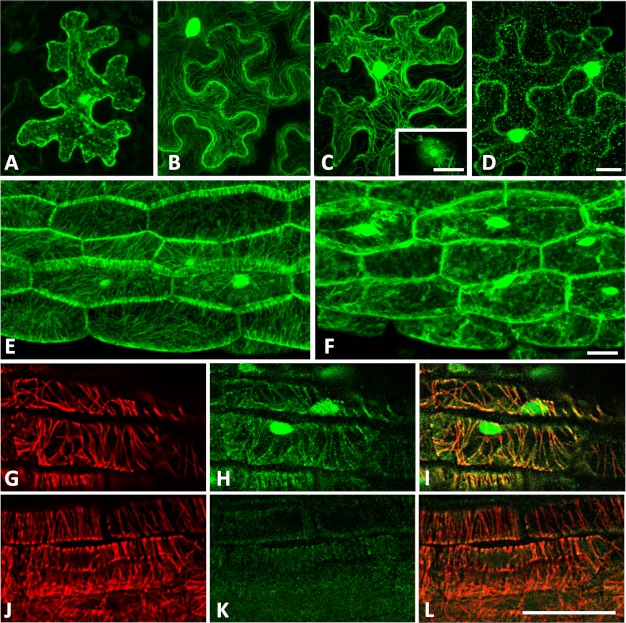
Kinesins are a family of cellular motor proteins that move along polarized microtubule tracks to transport various cargos, which are either directly attached to the KHC or to its associated KLC via the TPR12 domain (34). The putative role of KLCR proteins in microtubular transport and the verified interaction of IQD1 with KLCR1 in yeast prompted us to reinvestigate the subcellular localization of IQD1. We previously reported targeting of an IQD1∼GFP fusion protein to the cell nucleus when expressed under the control of the constitutive CaMV 35S promoter in roots of transgenic Arabidopsis plants (16). Here, we transiently expressed 35SPro::GFP, 35SPro::IQD1∼GFP or 35SPro::GFP∼IQD1 constructs in tobacco leaves (N. benthamiana) and monitored GFP fluorescence by confocal laser scanning microscopy at high resolution (Fig. 6). As expected for control transformations, the GFP protein localized to the cytosol and cell nucleus (Fig. 6A). For transient expression of C-terminal IQD1∼GFP or N-terminal GFP∼IQD1 fusion proteins, we confirmed nuclear localization of IQD1 but observed additional GFP fluorescence associated with cytoskeletal structures (Fig. 6, B and C, and supplemental Fig. S4). Because KLCR1 likely interacts with kinesin motor proteins that transport cargo along microtubule tracks, we examined the effect of oryzalin, an herbicide promoting microtubule depolymerization. As shown in Fig. 6D, oryzalin treatment caused collapse of cytoskeletal-associated GFP∼IQD1 fluorescence, indicating association of IQD1 with the microtubular network. In addition, we studied oryzalin-dependent GFP∼IQD1 localization in hypocotyl tissues of transgenic Arabidopsis seedlings (Fig. 6, E and F). The results in a stable, homologous plant transformation system are consistent with the observation in transiently transformed tobacco leaves (Fig. 6, compare panels E and F, with panels C and D). We independently analyzed the subcellular localization of GFP∼IQD1 in transgenic plants by co-immunolabeling of microtubules (anti-α-tubulin) and GFP∼IQD1 (anti-GFP), which demonstrated co-localization of both proteins to microtubules (Fig. 6, G–L).
FIGURE 6.
GFP-tagged IQD1 proteins localize to microtubules and the cell nucleus. A–D, tobacco leaves (N. benthamiana) were transiently transfected with Agrobacterium strains harboring plasmids supporting expression of CaMV 35SPro::GFP (A), CaMV 35SPro::IQD1∼GFP (B), and CaMV 35SPro::GFP∼IQD1 (C and D). Samples were collected 2 days after infiltration and visualized by confocal laser scanning microscopy after mock treatment with DMSO (A–C) or after infiltration with 50 μm oryzalin for 90 min (D). E and F, transgenic Arabidopsis seedlings (CaMV 35SPro::GFP∼IQD1) were visualized (three-dimensional projections) before (E) or after (F) treatment with 5 μm oryzalin for 60 min. G–L, GFP∼IQD1 and microtubules were co-immunolabeled in transgenic (G–I) and wild type (J–L) Arabidopsis seedlings using a combination of a rat anti-α tubulin antibody (G and J) and a green fluorescent monoclonal mouse anti-GFP antibody (H and K). Merged images are shown (I and L). Scale bars, 20 μm. Nuclear localization of GFP-tagged IQD1 was additionally demonstrated by DAPI (4′,6′ diamino-2-phenylindole·2HCl) staining (see supplemental Fig. S4).
IQD1 Recruits KLCR1 and CaM2 to Microtubules
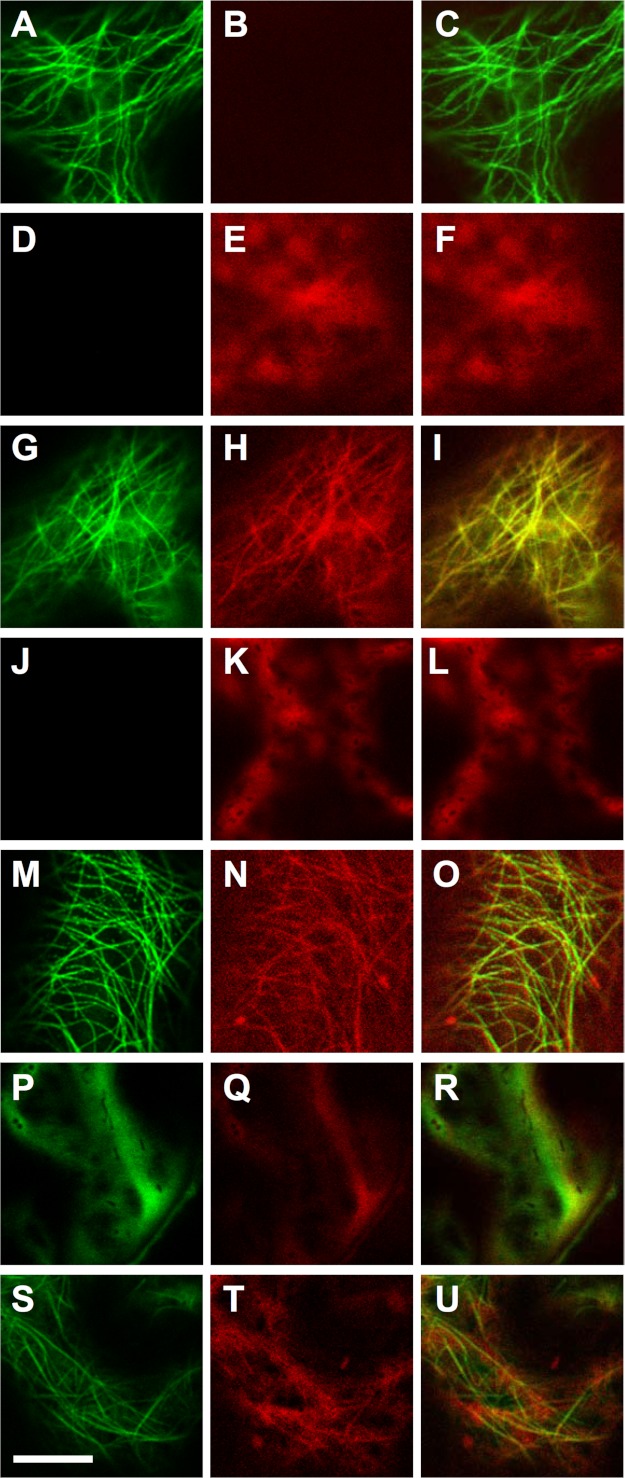
Because KLCR1 is predicted to assist kinesins in cargo transport along microtubules, we first compared the subcellular localization of GFP∼IQD1 with RFP∼KLCR1 in tobacco leaves that were transiently transfected with 35SPro::GFP∼IQD1 or 35SPro::RFP∼KLCR1 plasmids. Although GFP∼IQD1 localized to microtubules (Fig. 7, A–C), expressed RFP∼KLCR1 displayed diffuse cellular fluorescence, indicative of cytosolic localization (Fig. 7, D–F). Interestingly, when both DNA constructs were co-transfected, RFP∼KCLR1 co-localized with the microtubular pattern of GFP∼IQD1 fluorescence (Fig. 7, G–I), indicating recruitment of RFP∼KLCR1 to the microtubular network by GFP∼IQD1, most likely via direct interaction in planta. Similarly, transient expression of RFP∼CaM2 alone revealed red fluorescence consistent with cytosolic localization of tagged CaM2 (Fig. 7, J–L). Again, co-expression of RFP∼CaM2 and GFP∼IQD1 caused microtubular co-localization of both fusion proteins, suggesting IQD1-dependent recruitment of CaM2 to the microtubules (Fig. 7, M–O). Additional co-transfection experiments support a role of IQD1 for recruiting KLCR1 and CaM2 to the microtubular skeleton. Although co-expression of GFP∼KLCR1 and RFP∼CaM2 did not alter the cytosolic localization of either protein (Fig. 7, P–R), co-expression of both protein fusions together with untagged IQD1 resulted in a microtubular localization of GFP∼KLCR1 and RFP∼CaM2 (Fig. 7, S–U). These data also support the functionality of the GFP∼IQD1 protein fusion (Fig. 7, G–I, M–O, and S–U). Taken together, our results demonstrate co-localization of IQD1, KLCR1, and CaM2 to microtubules and further indicate IQD1-dependent recruitment of KLCR1 and CaM2 to the microtubular network via direct interaction.
FIGURE 7.
IQD1 recruits KLCR1 and CaM2 to microtubules. Tobacco leaves (N. benthamiana) were (co)infiltrated with Agrobacterium strains harboring plasmids supporting CaMV 35S promoter-driven expression of GFP∼IQD1 alone (A–C), mRFP∼KLCR1alone (D–F), GFP∼IQD1 and mRFP∼KLCR1 (G–I), mRFP∼CaM2 alone (J–L), GFP∼IQD1 and mRFP∼CaM2 (M–O), and GFP∼KLCR1 and mRFP∼CaM2 (P–R) as well as of untagged IQD1 together with GFP∼KLCR1 and mRFP∼CaM2 (S–U). Panels of the left column show the GFP signal, panels of the center column the RFP signal, and panels of the right column the merged GFP and RFP signals. Images were obtained on a Zeiss LSM700 confocal laser scanning microscope using sequential mode for clear separation of GFP and RFP signals. Scale bar, 10 μm. Transfection experiments were repeated at least three times with different sets of tobacco plants. Representative images of transfected leaf epidermal cells are shown.
DISCUSSION
Plant-specific IQD gene families, first annotated in Arabidopsis and rice, encode a major class of putative CaM targets, with about 30 genes expressed in either plant species (17). Although the biological roles for select IQD proteins are beginning to emerge, their mechanisms of action are elusive (16, 20–22). Here we studied the ability of IQD1 to bind to various Arabidopsis CaM/CMLs, mapped its CaM-recruitment domain, and searched for IQD1 interactors to gain first insight into its biochemical function. We used pulldown assays to probe the interaction with two canonical CaMs and two CMLs from Arabidopsis. This study revealed Ca2+-dependent recruitment of all tested CaM/CMLs by IQD1, demonstrating its potential to interact with divergent CaM/CML sensors (Fig. 1). We verified binding of IQD1 to CaM2 by gel overlay assays. In contrast to pulldown assays, we observed interaction in the absence of Ca2+ (supplemental Fig. S1, Fig. 2). Using similar overlay assays, CaM interaction in the absence but not the presence of Ca2+ was reported for OsCBT, a putative CaM binding transcription factor from rice (31), or for AtBAG6, a novel Arabidopsis CaM-binding protein (32). Deletion analysis of both proteins demonstrated a role of canonical IQ motifs for mediating their interaction with Ca2+-free CaM (31, 32). Typically, proteins containing complete canonical IQ motifs but no other CaM interaction motifs do not require Ca2+ for CaM binding, such as myosin (35), whereas proteins having incomplete IQ-like motifs tend to bind to Ca2+-CaM with higher affinity than to apoCaM (36–38). Deletion mapping of IQD1 confirmed the importance of the IQ67 domain for CaM interaction (Fig. 2), which contains one complete and one incomplete IQ consensus as well as three interspersed 1-5-10 motifs (16), suggesting complex and differential interaction of IQD1 with both apo- and Ca2+-CaM/CML sensors. It is likely that the physicochemical conditions during pulldown assays (native IQD1 in solution) and gel overlay assays (membrane-immobilized, renatured IQD1) differentially favor the accessibility of Ca2+-dependent or Ca2+-independent CaM binding motifs. Whereas high affinity binding (Kd values in the nm range) is typical for Ca2+-CaM binding proteins (39–41), the relatively low affinity of IQD1 for apoCaM2 (Kd∼ 0.6 μm) in gel overlay assays (Fig. 3) is consistent with Ca2+-independent CaM interactions reported for proteins binding to apoCaM, e.g. neuromodulin (Kd ∼ 1 μm) (13, 42). Our results of two unrelated CaM interaction assays validate the presence of functional Ca2+-independent and Ca2+-dependent CaM recruitment motifs in the IQ67 domain. Its importance for mediating CaM interaction is further supported by experiments with IQD20, which comprises only a short N-terminal extension in addition to the IQ67 domain but binds to bovine CaM (17) and Arabidopsis CaM2 (supplemental Fig. S1) in pulldown and gel overlay assays, respectively.
The capacity of IQD proteins to recruit CaM is further demonstrated by yeast two-hybrid assays (Fig. 4). These reveal weak but differential interaction of IQD1 and IQD20 with only a few of the 13 CaM/CMLs tested, suggesting regulation of IQD function by specific CaM/CML sensors. Interrogation of a recently published Arabidopsis interactome map retrieved evidence for the interaction of IQD1, IQD23, and IQD31 with CaM1 as well as of IQD31 with CML13 in yeast (43). Although the yeast system is a widely used approach to study protein interactions in vivo, the yeast system is not necessarily a suitable method for characterizing Ca2+-regulated CaM/CML-target interactions due to its difficulties to control Ca2+ concentration (9). Nonetheless, we performed yeast two-hybrid screens with IQD1 to select Arabidopsis CaM/CMLs and novel interactors. Although we did not recover any of the Ca2+ sensors, we repeatedly isolated cDNAs encoding KLCR1 and GSTU26, and we confirmed the interaction with IQD1 (Fig. 5). GSTU26 belongs to the plant-specific tau (U) class of GST enzymes of undefined metabolic functions (44–46); however, GSTUs were shown to selectively retain S-acyl-glutathione adducts, possibly to protect their unstable products upon formation in vivo (47). Interestingly, GSTU26 is closely related to GSTU20 (44). The latter was implicated in the conjugation of glutathione with activated aliphatic aldoximes to form structurally analogous S-hydroxamyl-glutathione adducts during glucosinolate biosynthesis (48). The expression of GSTU26 differentially responds to challenge with plant pathogens and treatment with defense-related hormones, jasmonic and salicylic acid (49). Thus, GSTU26 and IQD1 may both play a role for the production and regulation of glucosinolate phytoanticipins in plant defense (16).
The IQD1-KLCR1 interaction in yeast (Fig. 5) is supported by querying the Arabidopsis interactome database, which points to interactions between IQD and KLCR protein, i.e. IQD2 with At4g10840 (KLCR1) and At3g27960 (KLCR2) and IQD23 with At3g27960 (KLCR2) (43). The Arabidopsis KLCRs are similar to human KLCs (supplemental Figs. S2 and S3) and may be functionally equivalent in recruiting various cargos to KHCs. Kinesins are a class of microtubule-associated motor proteins that function in cell division and facilitate directional transport of organelles, vesicles, protein complexes, and mRNA-protein particles to specific destinations, generally toward the cell periphery (34, 50, 51). The cargo is either directly attached to the globular C-terminal tail of the KHC homodimer or indirectly via associated KLCs or via KLC binding adaptors that provide a scaffold for additional protein interactions. The KLC subunit contains an N-terminal heptad repeat region for binding to the coiled-coil stalk of the KHC dimer followed by several TPR motifs and a variable C-terminal domain that typically recognizes the C terminus of their partner scaffold protein. Mounting evidence in animals on kinesin-cargo interactions indicates that KLC-interacting scaffolds regulate cargo recruitment and kinesin activation by relieving autoinhibition. Even entire preassembled signaling modules such as mitogen-activated protein kinase cascades associated with vesicle-bound transmembrane receptors are tethered to KHC-KLC tetramers via scaffold proteins, which trigger and inform transport along microtubules to their final membrane destination. The assembly, loading, and unloading of scaffold-associated cargo complexes are tightly controlled by phosphorylation, GTPase activity, and Ca2+-signaling (50–52). Thus, scaffold proteins serve as critical nodes that integrate multiple signaling pathways to coordinate diverse cellular activities (53, 54).
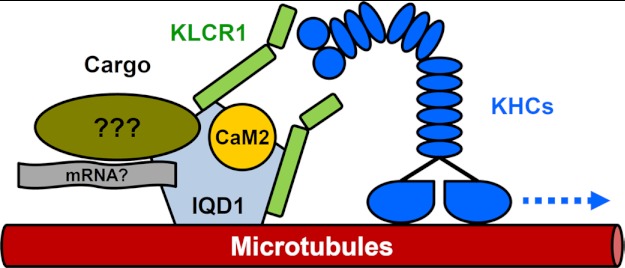
Although mammalian KHC and KLC subunits are encoded by 45 and 4 genes, respectively, the Arabidopsis kinesin family comprises 61 predicted members (34, 55, 56). The precise functions and biological roles for about two-thirds of Arabidopsis kinesins are not understood, and nothing is known about their associated light chains (55). Cargo selection, regulation of motor activity, and associated biological processes remain unexplored in plants, with the notable exception of a kinesin-like CaM-binding protein involved in the regulation of cell division and trichome morphogenesis (55, 57). Our study uncovers a novel link between Ca2+ signaling and the regulation of kinesin motor activity via direct interactions of CaM/CML, IQD, and KLRC proteins. This proposition is supported by the subcellular localization of IQD1 to microtubules (Fig. 6), a feature shared by most members of the Arabidopsis IQD family.8 Our co-transfection studies in tobacco further indicate recruitment of KLCR1 as well as CaM2 to the microtubules via IQD1 (Fig. 7). The prospect arises that IQD proteins provide an assortment of KLCR-interacting scaffolds for various kinesin-dependent transport processes (see Fig. 8). Given the affinity of IQD1 for artificial single-stranded nucleic acid substrates (supplemental Fig. S5), IQD-facilitated transport along microtubules may comprise translocation of RNA-protein complexes to distinct cellular compartments, which is an important mechanism for coupling gene expression with efficient protein sorting (58, 59).
FIGURE 8.
Working model of IQD1. The IQD1 protein localizes to the cell nucleus and microtubules and interacts with KLCR1and CaM/CML Ca2+ sensors such as CaM2. IQD1 is proposed to function as a scaffold protein that likely recruits in a Ca2+-CaM-regulated manner various cargos to kinesins (KHCs) via associated KLCR1 for unidirectional transport along microtubule tracks. Transport cargos are currently unknown and may include ribonucleoprotein complexes.
We obtained first insight into the biochemical functions of IQD1, likely having implications for other IQD proteins, but are left to speculate on the role of IQD1 in glucosinolate metabolism and plant defense. IQD1 is mainly expressed in the vascular bundles, a pattern characteristic of numerous glucosinolate-related genes, and its overexpression stimulates glucosinolate production and defense responses (16). In Arabidopsis, glucosinolates accumulate to very high concentrations (>130 mm) in the sulfur-rich S-cells, which surround the phloem zone of vascular bundles and expand to a length of >1 mm (60). Because S-cells are thought to sustain glucosinolate synthesis (60), the highly polarized cells likely necessitate active translocation of metabolons or RNA-protein complexes. Likewise, intracellular transport processes such as vesicle trafficking are crucial for the rapid deployment of defensive layers of callose at the site of pathogen contact. Interestingly, pathogen perception activates turnover of specific glucosinolates, which is necessary for triggering biosynthesis of callose and its site-specific deposition (61, 62). In conclusion, our study provides a first molecular framework for dissecting the biochemical functions of IQD1 and related proteins in A. thaliana and other plant species.
Acknowledgments
We thank Claudia Köhler (Swedish University of Agricultural Sciences, Uppsala) for CaM/CML plasmids, Petra Dietrich und Cornelia Fischer (University of Erlangen-Nürnberg) for the CaM2-pENTR/ D-TOPO vector, Kristin Eismann for technical assistance, and Claus Wasternack for critical reading of the manuscript.
This work was supported by the National Research Initiative of the United States Department of Agriculture Cooperative State Research Education and Extension Service Grant 2005-02507 (to S. A.), by a pre-doctoral fellowship from the Deutsche Forschungsgemeinschaft (GRK1026; to D. C. D.), and by core funding to the Leibniz-Institute of Plant Biochemistry from the state of Saxony-Anhalt and the Federal Republic of Germany.

This article contains supplemental Figs. S1–S5.
K. Bürstenbinder and S. Abel, unpublished work.
- CaM
- calmodulin
- CML
- CaM-like protein
- IQD1
- IQ67 domain 1
- KLCR1
- kinesin light chain-related-1
- SD
- synthetic dropout
- X-gal
- β-galactosidase substrate 5-bromo-4-chloro-indolyl-β-d-galactopyranoside
- BD
- binding domain
- AD
- activation domain
- KLC
- kinesin light chain
- KLCR
- KLC-related
- KLCR1
- KLC-related protein-1
- mRFP
- monomeric red fluorescent protein (RFP).
REFERENCES
- 1. Dodd A. N., Kudla J., Sanders D. (2010) The language of calcium signaling. Annu. Rev. Plant Biol. 61, 593–620 [DOI] [PubMed] [Google Scholar]
- 2. DeFalco T. A., Bender K. W., Snedden W. A. (2010) Breaking the code. Ca2+ sensors in plant signalling. Biochem. J. 425, 27–40 [DOI] [PubMed] [Google Scholar]
- 3. Reddy A. S., Ali G. S., Celesnik H., Day I. S. (2011) Coping with stresses. Roles of calcium- and calcium/calmodulin-regulated gene expression. Plant Cell 23, 2010–2032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Day I. S., Reddy V. S., Shad Ali G., Reddy A. S. (2002) Analysis of EF-hand-containing proteins in Arabidopsis. Genome Biol 3, RESEARCH0056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Sanders D., Pelloux J., Brownlee C., Harper J. F. (2002) Calcium at the crossroads of signaling. Plant Cell 14, S401–S417 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Hrabak E. M., Chan C. W., Gribskov M., Harper J. F., Choi J. H., Halford N., Kudla J., Luan S., Nimmo H. G., Sussman M. R., Thomas M., Walker-Simmons K., Zhu J. K., Harmon A. C. (2003) The Arabidopsis CDPK-SnRK superfamily of protein kinases. Plant Physiol 132, 666–680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Weinl S., Kudla J. (2009) The CBL-CIPK Ca2+-decoding signaling network. Function and perspectives. New Phytol. 184, 517–528 [DOI] [PubMed] [Google Scholar]
- 8. Luan S. (2009) The CBL-CIPK network in plant calcium signaling. Trends Plant Sci. 14, 37–42 [DOI] [PubMed] [Google Scholar]
- 9. Reddy A. S., Ben-Hur A., Day I. S. (2011) Experimental and computational approaches for the study of calmodulin interactions. Phytochemistry 72, 1007–1019 [DOI] [PubMed] [Google Scholar]
- 10. McCormack E., Tsai Y. C., Braam J. (2005) Handling calcium signaling. Arabidopsis CaMs and CMLs. Trends Plant Sci. 10, 383–389 [DOI] [PubMed] [Google Scholar]
- 11. Reddy V. S., Reddy A. S. (2004) Proteomics of calcium-signaling components in plants. Phytochemistry 65, 1745–1776 [DOI] [PubMed] [Google Scholar]
- 12. Bouché N., Yellin A., Snedden W. A., Fromm H. (2005) Plant-specific calmodulin-binding proteins. Annu. Rev. Plant Biol. 56, 435–466 [DOI] [PubMed] [Google Scholar]
- 13. Rhoads A. R., Friedberg F. (1997) Sequence motifs for calmodulin recognition. FASEB J. 11, 331–340 [DOI] [PubMed] [Google Scholar]
- 14. Bähler M., Rhoads A. (2002) Calmodulin signaling via the IQ motif. FEBS Lett. 513, 107–113 [DOI] [PubMed] [Google Scholar]
- 15. Hoeflich K. P., Ikura M. (2002) Calmodulin in action. Diversity in target recognition and activation mechanisms. Cell 108, 739–742 [DOI] [PubMed] [Google Scholar]
- 16. Levy M., Wang Q., Kaspi R., Parrella M. P., Abel S. (2005) Arabidopsis IQD1, a novel calmodulin-binding nuclear protein, stimulates glucosinolate accumulation and plant defense. Plant J. 43, 79–96 [DOI] [PubMed] [Google Scholar]
- 17. Abel S., Savchenko T., Levy M. (2005) Genome-wide comparative analysis of the IQD gene families in Arabidopsis thaliana and Oryza sativa. BMC Evol Biol 5, 72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Grubb C. D., Abel S. (2006) Glucosinolate metabolism and its control. Trends Plant Sci. 11, 89–100 [DOI] [PubMed] [Google Scholar]
- 19. Halkier B. A., Gershenzon J. (2006) Biology and biochemistry of glucosinolates. Annu. Rev. Plant Biol. 57, 303–333 [DOI] [PubMed] [Google Scholar]
- 20. Zentella R., Zhang Z. L., Park M., Thomas S. G., Endo A., Murase K., Fleet C. M., Jikumaru Y., Nambara E., Kamiya Y., Sun T. P. (2007) Global analysis of della direct targets in early gibberellin signaling in Arabidopsis. Plant Cell 19, 3037–3057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Xiao H., Jiang N., Schaffner E., Stockinger E. J., van der Knaap E. (2008) A retrotransposon-mediated gene duplication underlies morphological variation of tomato fruit. Science 319, 1527–1530 [DOI] [PubMed] [Google Scholar]
- 22. Wu S., Xiao H., Cabrera A., Meulia T., van der Knaap E. (2011) SUN regulates vegetative and reproductive organ shape by changing cell division patterns. Plant Physiol. 157, 1175–1186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Köhler C., Neuhaus G. (2000) Characterisation of calmodulin binding to cyclic nucleotide-gated ion channels from Arabidopsis thaliana. FEBS Lett. 471, 133–136 [DOI] [PubMed] [Google Scholar]
- 24. Laemmli U. K. (1970) Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227, 680–685 [DOI] [PubMed] [Google Scholar]
- 25. Kim J., Harter K., Theologis A. (1997) Protein-protein interactions among the Aux/IAA proteins. Proc. Natl. Acad. Sci. U.S.A. 94, 11786–11791 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Karimi M., Inzé D., Depicker A. (2002) GATEWAY vectors for Agrobacterium-mediated plant transformation. Trends Plant Sci. 7, 193–195 [DOI] [PubMed] [Google Scholar]
- 27. Nakagawa T., Suzuki T., Murata S., Nakamura S., Hino T., Maeo K., Tabata R., Kawai T., Tanaka K., Niwa Y., Watanabe Y., Nakamura K., Kimura T., Ishiguro S. (2007) Improved Gateway binary vectors. High performance vectors for creation of fusion constructs in transgenic analysis of plants. Biosci. Biotechnol. Biochem. 71, 2095–2100 [DOI] [PubMed] [Google Scholar]
- 28. Voinnet O., Rivas S., Mestre P., Baulcombe D. (2003) An enhanced transient expression system in plants based on suppression of gene silencing by the p19 protein of tomato bushy stunt virus. Plant J 33, 949–956 [DOI] [PubMed] [Google Scholar]
- 29. Clough S. J. (2005) Floral dip, Agrobacterium-mediated germ line transformation. Methods Mol. Biol. 286, 91–102 [DOI] [PubMed] [Google Scholar]
- 30. Müller J., Beck M., Mettbach U., Komis G., Hause G., Menzel D., Samaj J. (2010) Arabidopsis MPK6 is involved in cell division plane control during early root development and localizes to the pre-prophase band, phragmoplast, trans-Golgi network, and plasma membrane. Plant J 61, 234–248 [DOI] [PubMed] [Google Scholar]
- 31. Choi M. S., Kim M. C., Yoo J. H., Moon B. C., Koo S. C., Park B. O., Lee J. H., Koo Y. D., Han H. J., Lee S. Y., Chung W. S., Lim C. O., Cho M. J. (2005) Isolation of a calmodulin-binding transcription factor from rice (Oryza sativa L.). J. Biol. Chem. 280, 40820–40831 [DOI] [PubMed] [Google Scholar]
- 32. Kang C. H., Jung W. Y., Kang Y. H., Kim J. Y., Kim D. G., Jeong J. C., Baek D. W., Jin J. B., Lee J. Y., Kim M. O., Chung W. S., Mengiste T., Koiwa H., Kwak S. S., Bahk J. D., Lee S. Y., Nam J. S., Yun D. J., Cho M. J. (2006) AtBAG6, a novel calmodulin-binding protein, induces programmed cell death in yeast and plants. Cell Death Differ. 13, 84–95 [DOI] [PubMed] [Google Scholar]
- 33. McCormack E., Braam J. (2003) Calmodulin and related potential calcium sensors of Arabidopsis. New Phytol. 159, 585–598 [DOI] [PubMed] [Google Scholar]
- 34. Verhey K. J., Kaul N., Soppina V. (2011) Kinesin assembly and movement in cells. Annu. Rev. Biophys. 40, 267–288 [DOI] [PubMed] [Google Scholar]
- 35. Houdusse A., Gaucher J. F., Krementsova E., Mui S., Trybus K. M., Cohen C. (2006) Crystal structure of apocalmodulin bound to the first two IQ motifs of myosin V reveals essential recognition features. Proc. Natl. Acad. Sci. U.S.A. 103, 19326–19331 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Houdusse A., Cohen C. (1995) Target sequence recognition by the calmodulin superfamily. Implications from light chain binding to the regulatory domain of scallop myosin. Proc. Natl. Acad. Sci. U.S.A. 92, 10644–10647 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Munshi H. G., Burks D. J., Joyal J. L., White M. F., Sacks D. B. (1996) Ca2+ regulates calmodulin binding to IQ motifs in IRS-1. Biochemistry 35, 15883–15889 [DOI] [PubMed] [Google Scholar]
- 38. DeFalco T. A., Chiasson D., Munro K., Kaiser B. N., Snedden W. A. (2010) Characterization of GmCaMK1, a member of a soybean calmodulin-binding receptor-like kinase family. FEBS Lett. 584, 4717–4724 [DOI] [PubMed] [Google Scholar]
- 39. Bouché N., Scharlat A., Snedden W., Bouchez D., Fromm H. (2002) A novel family of calmodulin binding transcription activators in multicellular organisms. J. Biol. Chem. 277, 21851–21861 [DOI] [PubMed] [Google Scholar]
- 40. Yang T., Poovaiah B. W. (2000) An early ethylene up-regulated gene encoding a calmodulin-binding protein involved in plant senescence and death. J. Biol. Chem. 275, 38467–38473 [DOI] [PubMed] [Google Scholar]
- 41. Reddy V. S., Safadi F., Zielinski R. E., Reddy A. S. (1999) Interaction of a kinesin-like protein with calmodulin isoforms from Arabidopsis. J. Biol. Chem. 274, 31727–31733 [DOI] [PubMed] [Google Scholar]
- 42. Black D. J., LaMartina D., Persechini A. (2009) The IQ domains in neuromodulin and PEP19 represent two major functional classes. Biochemistry 48, 11766–11772 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Arabidopsis Interactome Mapping Consortium (2011) Evidence for network evolution in an Arabidopsis interactome map. Science 333, 601–607 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Dixon D. P., Lapthorn A., Edwards R. (2002) Plant glutathione transferases. Genome Biol. 3, REVIEWS3004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Wagner U., Edwards R., Dixon D. P., Mauch F. (2002) Probing the diversity of the Arabidopsis glutathione S-transferase gene family. Plant Mol. Biol. 49, 515–532 [DOI] [PubMed] [Google Scholar]
- 46. Dixon D. P., Skipsey M., Edwards R. (2010) Roles for glutathione transferases in plant secondary metabolism. Phytochemistry 71, 338–350 [DOI] [PubMed] [Google Scholar]
- 47. Dixon D. P., Edwards R. (2009) Selective binding of glutathione conjugates of fatty acid derivatives by plant glutathione transferases. J. Biol. Chem. 284, 21249–21256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Sønderby I. E., Geu-Flores F., Halkier B. A. (2010) Biosynthesis of glucosinolates. Gene discovery and beyond. Trends Plant Sci. 15, 283–290 [DOI] [PubMed] [Google Scholar]
- 49. Nutricati E., Miceli A., Blando F., De Bellis L. (2006) Characterization of two Arabidopsis thaliana glutathione S-transferases. Plant Cell Rep. 25, 997–1005 [DOI] [PubMed] [Google Scholar]
- 50. Akhmanova A., Hammer J. A., 3rd. (2010) Linking molecular motors to membrane cargo. Curr. Opin. Cell Biol. 22, 479–487 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Hirokawa N., Noda Y., Tanaka Y., Niwa S. (2009) Kinesin superfamily motor proteins and intracellular transport. Nat. Rev. Mol. Cell Biol. 10, 682–696 [DOI] [PubMed] [Google Scholar]
- 52. Schnapp B. J. (2003) Trafficking of signaling modules by kinesin motors. J. Cell Sci. 116, 2125–2135 [DOI] [PubMed] [Google Scholar]
- 53. Brown M. D., Sacks D. B. (2009) Protein scaffolds in MAP kinase signalling. Cell Signal 21, 462–469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. White C. D., Brown M. D., Sacks D. B. (2009) IQGAPs in cancer. A family of scaffold proteins underlying tumorigenesis. FEBS Lett. 583, 1817–1824 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Zhu C., Dixit R. (2012) Functions of the Arabidopsis kinesin superfamily of microtubule-based motor proteins. Protoplasma 249, 887–899 [DOI] [PubMed] [Google Scholar]
- 56. Reddy A. S., Day I. S. (2001) Kinesins in the Arabidopsis genome. A comparative analysis among eukaryotes. BMC Genomics 2, 2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Reddy V. S., Day I. S., Thomas T., Reddy A. S. (2004) KIC, a novel Ca2+ binding protein with one EF-hand motif, interacts with a microtubule motor protein and regulates trichome morphogenesis. Plant Cell 16, 185–200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Kanai Y., Dohmae N., Hirokawa N. (2004) Kinesin transports RNA. Isolation and characterization of an RNA-transporting granule. Neuron 43, 513–525 [DOI] [PubMed] [Google Scholar]
- 59. Kindler S., Wang H., Richter D., Tiedge H. (2005) RNA transport and local control of translation. Annu. Rev. Cell Dev. Biol. 21, 223–245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Koroleva O. A., Gibson T. M., Cramer R., Stain C. (2010) Glucosinolate-accumulating S-cells in Arabidopsis leaves and flower stalks undergo programmed cell death at early stages of differentiation. Plant J 64, 456–469 [DOI] [PubMed] [Google Scholar]
- 61. Clay N. K., Adio A. M., Denoux C., Jander G., Ausubel F. M. (2009) Glucosinolate metabolites required for an Arabidopsis innate immune response. Science 323, 95–101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Bednarek P., Pislewska-Bednarek M., Svatos A., Schneider B., Doubsky J., Mansurova M., Humphry M., Consonni C., Panstruga R., Sanchez-Vallet A., Molina A., Schulze-Lefert P. (2009) A glucosinolate metabolism pathway in living plant cells mediates broad-spectrum antifungal defense. Science 323, 101–106 [DOI] [PubMed] [Google Scholar]