Background: MicroRNAs are differentially expressed in medulloblastoma.
Results: MicroRNA 218 expression is decreased in medulloblastoma. Re-expression of miR-218 suppresses the malignant cell phenotype in medulloblastoma cells. Unbiased HITS-CLIP analysis identified multiple oncogenic genes as miR-218 targets.
Conclusion: miR-218 inhibits medulloblastoma tumor cell phenotype by targeting multiple oncogenes.
Significance: miR-218-regulated pathways are important in medulloblastoma pathogenesis.
Keywords: Cell Invasion, Medulloblastoma, MicroRNA, Transcription Target Genes, Tumor Suppressor Gene, HITS-CLIP, miR-218
Abstract
Aberrant expression of microRNAs has been implicated in many cancers. We recently demonstrated differential expression of several microRNAs in medulloblastoma. In this study, the regulation and function of microRNA 218 (miR-218), which is significantly underexpressed in medulloblastoma, was evaluated. Re-expression of miR-218 resulted in a significant decrease in medulloblastoma cell growth, cell colony formation, cell migration, invasion, and tumor sphere size. We used C17.2 neural stem cells as a model to show that increased miR-218 expression results in increased cell differentiation and also decreased malignant transformation when transfected with the oncogene REST. These results suggest that miR-218 acts as a tumor suppressor in medulloblastoma. MicroRNAs function by down-regulating translation of target mRNAs. Targets are determined by imperfect base pairing of the microRNA to the 3′-UTR of the mRNA. To comprehensively identify actual miR-218 targets, medulloblastoma cells overexpressing miR-218 and control cells were subjected to high throughput sequencing of RNA isolated by cross-linking immunoprecipitation, a technique that identifies the mRNAs bound to the RNA-induced silencing complex component protein Argonaute 2. High throughput sequencing of mRNAs identified 618 genes as targets of miR-218 and included both previously validated targets and many targets not predicted computationally. Additional work further confirmed CDK6, RICTOR, and CTSB (cathepsin B) as targets of miR-218 and examined the functional role of one of these targets, CDK6, in medulloblastoma.
Introduction
Medulloblastoma is the most common malignant brain tumor of childhood, arising from the cerebellar granule cell precursors or neural stem cells located in the cerebellum (1, 2). Despite successful treatments, including surgery, radiation, and chemotherapy, a significant number of patients have neurocognitive and endocrinologic deficits (3). Therefore, a better treatment strategy to control cancer cell growth is needed. An emerging theme in cancer biology is that dysregulation in normal cell proliferation and differentiation contributes to malignant transformation. Novel insights into the molecular mechanism that drive this dysregulation in cell proliferation and differentiation can provide a basis for new therapeutic strategies against medulloblastoma.
MicroRNAs are known to promote or repress cell proliferation, differentiation, and apoptosis during normal cell development (4–6). MicroRNAs are small RNA molecules that bind to the target mRNAs primarily at their 3′-UTR and block translation and/or cause mRNA degradation, thereby down-regulating target gene expression (7). Therefore, alterations in microRNAs could lead to dysregulation in cell proliferation and differentiation. Differential expression of microRNAs has been observed in many cancers, suggesting that their altered expression can contribute to tumorigenesis (8, 9). We have previously shown that brain enriched microRNAs are dysregulated in medulloblastoma, including miR-1242 and miR-128 (10, 11).
The mechanisms by which microRNAs affect developmental and cancer pathways are not well understood. One reason for this is that each microRNA may target hundreds of genes. Thus, the actual targets of most microRNAs remain to be determined and necessitate additional laboratory experiments to discover the true valid microRNA targets.
In this study, we found poor expression of miR-218 consistently in many medulloblastoma patient samples and in cell lines. Forced re-expression of miR-218 decreased medulloblastoma cell growth. We therefore hypothesized that miR-218 functions as a tumor suppressor in medulloblastoma, and we investigated the biological role of miR-218. We used a high throughput method known as high throughput sequencing of RNA isolated by cross-linking immunoprecipitation (HITS-CLIP) to comprehensively identify targets of miR-218. Selected key targets were then further validated in additional experiments.
EXPERIMENTAL PROCEDURES
Biological Samples
Medulloblastoma Cell Lines
The Daoy, D341, and D283 cell lines were purchased from the ATCC (Manassas, VA). The ONS76 cell line was kindly provided by Dr. James T. Rutka (University of Toronto, Canada). The UW228 cell line was kindly provided by Dr. John Silber (University of Washington, Seattle, WA). D425 and D458 cell lines were kindly provided by Dr. Darell D. Bigner (Duke University Medical Center, Durham, NC). HEK293T cells were obtained from David Ginsburg (University of Michigan, Ann Arbor, MI). All cell lines except for D341 were cultured in DMEM (Invitrogen) supplemented with 10% fetal bovine serum (Atlanta Biologicals, Lawrenceville, GA). D341 was cultured in DMEM supplemented with 20% fetal bovine serum. D425, D458, D283, and D341 cells were grown in suspension in their respective culture medium.
Patient Samples
The first cohort of eight primary patient samples was obtained from Children's Hospital Colorado and conducted in accordance with local and federal human research protection guidelines and Institutional Review Board regulations. Normal brain tissue was collected from autopsy and purchased from Stratagene (Santa Clara, CA) and Clontech (Mountain View, CA). Two normal cerebellar samples were obtained from non-malignant brain autopsies at the Children's Hospital Colorado under Institutional Review Board guidelines. These two samples (labeled UPN 514 and UPN 605 in Fig. 5B) are from 4-year-old and 5-year-old patients, respectively. The second large cohort of 90 tumor specimens was obtained in accordance with the Research Ethics Board at the Hospital for Sick Children (Toronto, Canada) and the N. N. Burdenko Neurosurgical Institute (Moscow, Russia) as described (12).
FIGURE 5.
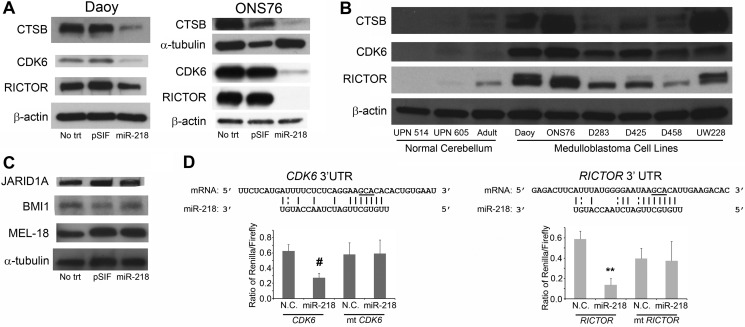
Validation of targets. A, targets identified by HITS-CLIP. miR-218 down-regulates the expression of CTSB, CDK6, and RICTOR protein levels in both the Daoy and ONS76 cell lines. B, verification of HITS-CLIP-identified target protein levels. Shown is Western blot analysis of CTSB, CDK6, and RICTOR protein levels in different well characterized medulloblastoma cell lines compared with pediatric and adult normal cerebellum. UPN 514 and UPN 605 are normal pediatric cerebellum samples. CDK6 and RICTOR are consistently up-regulated in all low miR-218-expressing cell lines, and CTSB is differently up-regulated in different medulloblastoma cell lines. C, validation of targets predicted by TargetScan alone by Western blot. miR-218 overexpression in Daoy cells did not down-regulate the protein expression of JARID1A, BMI1, or MEL-18. D, validation by luciferase assay. Top, base pairing between miR-218 and the mRNA seed sequence of CDK6 and RICTOR. The seed sequences of the CDK6 and RICTOR shown are the ones that were pulled down with AGO2 using HITS-CLIP. The mutated nucleotides are underlined. Bottom, siCHECK plasmid containing the plasmid sequences of either CDK6 or RICTOR (as indicated) downstream of Renilla luciferase was co-transfected with control miR (N.C.) or miR-218 oligonucleotide in HEK293T cells. There was a significant decrease in the ratio of Renilla/firefly luciferase activity in cells transfected with either CDK6 or RICTOR 3′-UTR compared with N.C. #, p < 0.05; **, p < 0.01. Mutated sequence of both CDK6 and RICTOR showed no regulation. Error bars, S.E.
In Vitro Experiments to Determine the Functional Role of miR-218 in Medulloblastoma Cell Lines
Cell Transfections
Medulloblastoma cell lines were transfected with plasmid DNA using Lipofectamine 2000 (Invitrogen) according to the manufacturer's instructions. Briefly 1 × 105 cells were seeded onto a 6-well tissue culture plate 24 h before transfection. Cells were transfected with 1 μg of plasmid DNA. Cells were harvested, and assays were performed 48 h after transfection.
MicroRNA Extraction, Quantitative PCR Analysis, and MicroRNA Microarrays
Total RNA, including microRNAs, were extracted using the Qiagen miReasy isolation kit (Valencia, CA). MicroRNA expression was measured by quantitative PCR using an ABI-StepOnePlus real-time PCR machine. MicroRNA TaqMan primers and probes were purchased from Applied Biosystems (Foster City, CA). The small RNA U6b was used as an endogenous control, and relative microRNA quantity was calculated by the ΔΔCt method. The microRNA expression profiling in the large cohort of 90 tumor specimens was performed using a version 3.0 microRNA microarray (13).
Growth Analysis by xCELLigence
Cell growth was measured in real time using the xCELLigence system and an E 96 plate (Roche Applied Science). The xCELLigence system continuously records cellular response by measuring the cell impedance and provides quantitative information about cell status. Cells were trypsinized after 48 h of transfection, and 2100 cells were seeded onto the 96-well E plate, and the rate of cell growth was measured over a period of time (14).
Colony Focus Assay
Non-transfected or pSIF- or miR-218-transfected cells were seeded into 6-well tissue plates in triplicate at a density of 500 cells/well in 3 ml of medium containing 10% FBS. Cells were grown for 7–10 days in a 37 °C humidified atmosphere containing 95% air and 5% CO2. The cell colonies were stained for 15 min with a solution containing 0.5% crystal violet and 25% methanol. The colony numbers (>50 cells/colony) were counted using a dissecting microscope. For validation experiments, Daoy cells were co-transfected with 1 μg each of miR-218 and CDK6 vector that lacks 3′-UTR using Lipofectamine 2000.
Tumor Sphere Assay
Daoy cells (104 cells/well) after 48 h of plasmid transfection were seeded onto ultra-low attachment 24-well plates (Costar, Corning, Inc.) in triplicate using neurobasal medium (Invitrogen) supplemented with fibroblast growth factor (20 ng/ml) (Sigma-Aldrich), B-27 (Invitrogen), epidermal growth factor (20 ng/ml) (Sigma-Aldrich), and l-glutamine (Invitrogen). After 7 days, the spheres were visualized and counted using an inverted microscope (Olympus CKX41). Images of the spheres were captured using a high resolution camera fitted to the microscope. The sphere diameters were measured using the Qcapture version 6.0 software with ×4 magnification. The data calculated for the size of the tumor spheres were the average of three experiments. For validation experiments, Daoy cells were co-transfected with 1 μg each of plasmids, miR-218, and CDK6 vector that lacks 3′-UTR using Lipofectamine 2000.
RNA Isolation and Quantitative PCR Analysis of Genes
RNA from the tumor spheres was isolated using the Qiagen RNA Easy kit (Valencia, CA). Gene expression assays were performed in triplicate on an ABI StepOnePlus real-time PCR system using Taqman probes. The ABI assay probe IDs for NANOG, MAP2, and GAPDH are Hs002387400_g1, Hs01103239_ml, and Hs99999905_m1, respectively. GAPDH was used as the endogenous control, and the gene expression was calculated using the ΔΔCt method.
Cell Migration and Invasion Assays
Real-time impedance-based cell migration and cell invasion assays were carried out using the xCELLigence system and E 16 plates (Roche Applied Science). The impedance measurement gives quantitative information about cell migration and invasion ability. Cells transfected with pSIF or miR-218 for 44 h and non-transfected cells were all serum-starved for 4 h. The cells were then trypsinized, washed with PBS, and resuspended in serum-free medium. For the cell migration assay ∼40,000 cells were seeded onto the top chamber of the E plate. The bottom chamber wells contained medium with 10% FBS, which is used as a chemoattractant, and the wells with no serum were used as the negative controls. Cells were allowed to migrate, and cell migration was measured over a period of time. For the cell invasion assay, the top chamber wells of the E 16 plate were first coated with Matrigel (1:20) and incubated for 4 h. Cells (∼40,000) were transfected and serum-starved as described for the cell migration assay and then seeded onto Matrigel (BD Biosciences)-coated wells, and cell invasion was measured. These experiments were repeated at least three times.
Experiments Using Neural Stem Cells to Study the Functional Effect of miR-218
Cells
C17.2 murine neural stem cells were kindly provided by Dr. Snyder (Burnham Institute, La Jolla, CA), and cells were established in DMEM supplemented with 10% FBS, 5% equine horse serum, l-glutamine, and penicillin/streptomycin (15).
Colony Focus Assay
C17.2 cells were transfected with miR-218 plasmid to overexpress miR-218 and with the corresponding empty vector pSIF. To knock down miR-218, C17.2 cells were transfected with anti-miR-218 plasmid or with the corresponding scrambled control plasmid, miRZip (Systems Bioscience Inc). After 48 h of transfection, 1000 cells/well were plated in a 6-well plate. Cells were allowed to form colonies for 7 days. The cell colonies were stained and counted as described earlier.
Immunofluorescence
C17.2, murine neural stem cells were transfected with either control vectors or miR-218 or anti-miR-218 plasmid. After 48 h of transfection, cells were trypsinized and seeded (3000 cells/well) on poly-d-lysine-coated chamber slides. Transfected C17.2 cells were grown in differentiation medium (neurobasal medium consisting of B27, 2 mm l-glutamine, penicillin/streptomycin, and 200 ng/ml nerve growth factor (NGF). After 5–7 days in differentiation medium, cells were processed for immunofluorescence. Briefly, cells were washed with PBS, fixed with 4% paraformaldehyde, and permeabilized with 0.05% Triton X-100. Cells were then blocked at room temperature with blocking buffer (0.05% Triton X-100 containing 5% milk) for 30 min. Slides were incubated overnight at 4 °C with either anti-NESTIN (Abcam) to label undifferentiated cells or anti-β-tubulin, Tuj1 (Cell Signaling Technologies) to label immature neurons. Antibodies were diluted 1:200 with 0.05% Triton X-100. Slides were washed with PBS and incubated with Alexafluor 488-conjugated goat anti-mouse (1:500) (Molecular Probes) antibody. Cells not incubated with primary antibodies were processed the same way and treated as controls for false positives. Slides were washed three times with PBS and counterstained with anti-fading reagent containing DAPI (Invitrogen) to visualize cell nuclei. Coverslips were mounted, and the cells were visualized using confocal microscopy with a ×40 oil objective. The confocal images were taken using the 3I Marianas inverted spinning disk imaging system built on a Zeiss Axio Observer Z1 microscope equipped with Yokogawa CSU-X1 and Photometrics Evolve 16-bit EM-CCD camera. Image analysis was performed using 3I Slidebook version 5.0 software.
Soft Agar Assay
The effect of miR-218 on the oncogenic transformation of C17.2 cells by the oncogene REST (repressor element 1-silencing transcription factor) was studied by performing a soft agar assay. C17.2 cells were co-transfected with REST and control vector or REST and miR-218 plasmids and plated on soft agar. The plates were incubated for 3 weeks, and colonies were stained overnight with nitro blue tetrazolium. Images of the wells containing colonies were taken using Chemdoc (Bio-Rad), and the colonies were counted using QuantityoneTM software.
Identification of miR-218 Targets in Medulloblastoma Cells
HITS-CLIP
We performed HITS-CLIP to identify direct targets of miR-218 by following the detailed protocol reported earlier with minimum modifications (16). HITS-CLIP is a genome-wide means of mapping protein-RNA binding sites in vivo based on immunoprecipitation of the AGO2 (Argonaute 2 protein) component of the RNA-induced silencing complex (RISC), along with its associated microRNAs. Briefly, Daoy cells were transfected with miR-218 plasmid using Lipofectamine 2000 reagent. Daoy cells overexpressing miR-218 and control cells (∼3–5 × 106) were UV-irradiated (Stratalinker 2400) to covalently cross-link RNA-protein complexes that are in direct contact (∼1 Å). UV irradiation induces covalent bonds between proteins and nucleic acids when contact distances are within 1 Å. Following cross-linking, the AGO-RNA complex was co-immunoprecipitated from cells using 2A8 antibody against AGO2, isolated, and radiolabeled (32P) with linker. The labeled AGO-RNA complex was then transferred to nitrocellulose by SDS-PAGE and visualized by autoradiography. AGO-RNA was purified and subjected to two sequential rounds of adapter ligation. The ligated product was reverse-transcribed, amplified by PCR, and bio-analyzed for the microRNA peak around 110 bp. The RNA was then sequenced using Ilumina sequencer with Solexa primers (16, 17). Quality control measures showed successful pull-down of AGO2 bound to RNA (supplemental Fig. S6A), a smear of band around 100–200 bp, and the presence of appropriately sized RNA transcripts for input to sequencing (supplemental Fig. S6B).
RNA Sequencing and Analysis
RNA sequencing was done at the Biochemistry and Molecular Genetics core laboratory (University of Colorado, Denver, CO). RNA sequencing was done using an Illumina GAllx sequencer with Solexa sequencing primer (5′-CTA TGG ATA CTT AGT CAG GGA GGA CGA TGC GG-3′). Two FASTQ files were output from the high throughput sequencing: one containing all reads from the miR-218-treated cells (the experimental file) and one containing all reads from the control cells (the control file). Both files were converted to SCARF format, and any poor quality bases were trimmed from the 3′-ends of the reads prior to alignment. BOWTIE was then used to align both the experimental and control files to the current version of the human genome NCBI V36. Aligned reads were then annotated with the gene name and whether the read occurred in the 3′-UTR or not. Counts for reads occurring in annotated genes in the 3′-UTR and that contained the seed sequence for miR-218 were compared between the experimental data and the control data. Reads with higher counts in the experimental data represent genes with increased binding to the RISC complex and therefore indicate genes that are likely targets of the miR-218. -Fold change was then calculated as the number of reads in miR-218-overexpressing condition/number of reads in control condition. For purpose of this calculation, the number of reads in the control condition was set to 0.5 for any genes that were absent in the control.
Analysis of Biological Enrichment of miR-218 Targets
Functional analysis of genes identified by HITS-CLIP as miR-218 targets was performed with the National Institutes of Health Database for Annotation, Visualization, and Integrated Discovery (DAVID) bioinformatic Web tool, using PANTHER (protein analysis through evolutionary relationships) biological process terms (18–20).
Target Validations
Western Blotting
Protein expression levels were determined by Western blotting as described previously (21). The primary antibodies used in this study are CDK6 (Cell Signaling), RICTOR (Cell Signaling), cathepsin B (Cell Signaling), JARID1A (Bethyl Laboratories), BMI1 (Cell Signaling), MEL-18 (Santa Cruz Biotechnology, Inc., Santa Cruz, CA), β-actin (Millipore), and α-tubulin (Cell Signaling).
Luciferase Reporter Assay
The 3′-UTR of RICTOR and CDK6 or the mutated sequences where three nucleotides in the seed sequence were mutated of the respective genes were PCR-amplified and cloned into the siCHECK2 vector (Promega, catalog no. C8021). Plasmids were confirmed by sequencing. The primer sequences used are given in supplemental Table S1. The seed sequence for RICTOR and CDK6 used in the cloning was the same as the sequence that was pulled down with AGO2 using HITS-CLIP for each of these genes. HEK293T cells were seeded in a 96-well plate 24 h before the transfection (7000 cells/well). siCHECK vector containing the CDK6 or RICTOR 3′-UTR sequence was co-transfected with 100 nm control miR or with miR-218 oligonucleotide. Forty-eight hours after transfection, cells were lysed, and the luciferase activities were measured using the Dual-Glo luciferase reporter assay system (Promega). Renilla luciferase activity was normalized to firefly luciferase activity for each well (n > 3). A p value was calculated using Student's t test.
CDK6 Open Reading Frame Vector
CDK6 cDNA vector clone that entirely lacks the 3′-UTR (Vector EX-M0442-M29) along with the corresponding vector control (EX-EGFP-M03) were purchased from GeneCopoeia (Rockville, MD). These clones were co-transfected with miR-218 plasmid in medulloblastoma cells to study their effect on the cell colony focus assay and on the cell tumor sphere formation assay.
Statistical Analysis
p values were calculated by either Student's t test or analysis of variance or as indicated. All of the error bars represent S.E. For the microRNA microarray profiling results, the p values were calculated as described by Northcott et al. (13).
RESULTS
miR-218 Is Down-regulated in both Medulloblastoma Patient Samples and in Cell Lines
To investigate the role of miR-218 in medulloblastoma, we first measured mature miR-218 expression by quantitative RT-PCR in a small cohort of pediatric medulloblastoma patient samples and normal pediatric cerebellum. miR-218 expression was significantly lower in our patient samples, p < 0.0001 (Fig. 1A). Next we evaluated miR-218 expression in a large cohort of 90 patient samples (13). Recent genomic analysis showed four distinct subgroups of medulloblastoma. The four subgroups are WNT signaling (WNT), Sonic Hedgehog (SHH), Group 3, and Group 4 (12). miR-218 expression was significantly down in the SHH group compared with adult cerebellum (p < 0.014, Mann-Whitney U rank test) and Group 3 compared with adult cerebellum (p < 0.01, Mann-Whitney U rank test) (Fig. 1B). Similar down-regulation of miR-218 expression was observed in well characterized medulloblastoma cell lines (p < 0.05) (Fig. 1C). The expression level in cell lines, although significantly down-regulated, varied from 20 to 0.0001% of normal levels. To evaluate the functional significance of this decreased miR-218, we analyzed the biological role of miR-218 governing medulloblastoma cells with respect to the key tumorigenic properties of cell proliferation, long term survival, cell motility, differentiation, and malignant transformation.
FIGURE 1.
Differential expression of miR-218 in medulloblastoma. A, relative expression of miR-218 mRNA, normalized to U6b expression, shows significant down-regulation in pediatric medulloblastoma patient samples compared with normal cerebellum by quantitative PCR analysis (***, p < 0.0001). B, miR-218 expression as measured by microRNA microarray analysis in 90 medulloblastoma tumor specimens that are characterized and divided into four subgroups (WNT, SHH, Group 3, and Group 4). miR-218 is significantly down in SHH (*, p < 0.02) and Group 3 (**, p < 0.01) compared with adult cerebellum. C, in all well characterized medulloblastoma cell lines, the miR-218 expression was significantly lower than that of the normal cerebellum by quantitative PCR analysis. p < 0.05 normal versus all medulloblastoma cell lines shown above by analysis of variance. Error bars, S.E.
Expression of miR-218 Decreases Malignant Properties of Medulloblastoma Cells
To evaluate the biological role of miR-218 in the ability of medulloblastoma cells to proliferate and to form colonies, miR-218 was overexpressed in Daoy cells. This cell line was chosen for these studies because its native expression of miR-218 was only 20% of that seen in normal cerebellum. Daoy cells were transfected with three different clones of miR-218-expressing plasmids. Results showed that clone miR-218-1 produced the best increase in miR-218 expression (supplemental Fig. S1), and therefore this clone was used in subsequent experiments. The expression of miR-218 after transfection was 25-fold greater than that of control-transfected Daoy (supplemental Fig. S1), and Daoy cells expressed miR-218 at 20% of the level of normal cerebellum (Fig. 1C), making ectopic expression of miR-218 ∼4–5-fold greater compared with normal cerebellum.
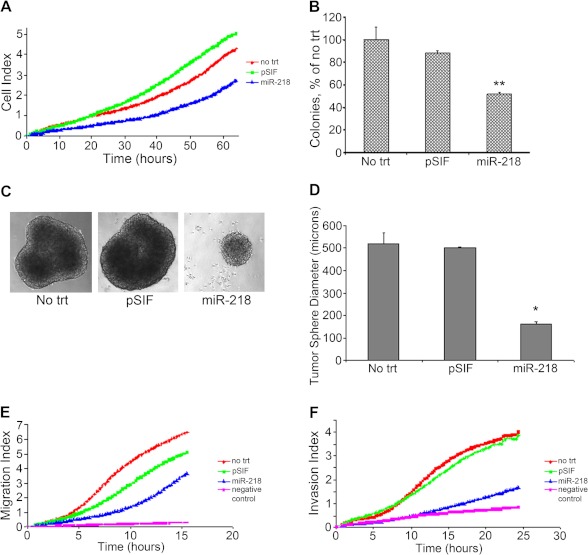
Cell proliferation was measured in real time using the xCELLigence system in Daoy cells. The results demonstrated that increased miR-218 expression induced a decrease in cell proliferation when compared with either cells transfected with empty vector pSIF or non-transfected cells (Fig. 2A). To evaluate the long term effect of increased expression of miR-218, colony focus assays were performed. Approximately 9 days after transfection, the number of colonies formed was significantly less in Daoy cells overexpressing miR-218 compared with that of either control: empty vector pSIF-transfected or non-transfected Daoy cells (Fig. 2B). We tested the effect of miR-218 on cell growth and cell colony formation in an additional medulloblastoma cell line, ONS76, and similar results were obtained (supplemental Fig. S2, A and B). In summary, re-expression of miR-218 in medulloblastoma cells decreased both cell proliferation and colony formation.
FIGURE 2.
Effect of re-expression of miR-218 in the medulloblastoma cell lines. A, Daoy cells transfected with miR-218 showed a significant decrease in cell growth compared with that of both non-transfected (no trt) cells and cells transfected with empty vector pSIF as measured in real time using xCELLigence. By analysis of variance, p < 0.05 for miR-218 versus empty vector pSIF for the indicated time period. B, Daoy cells transfected with miR-218 resulted in a significant inhibition in cell colony-forming ability compared with that of non-transfected and empty vector. **, p < 0.01, miR-218-transfected cells versus pSIF. C and D, Daoy cells transfected with miR-218 resulted in the formation of tumor spheres with smaller diameters compared with that of non-transfected and empty vector. Representative images of tumor spheres formed from each of the treatments, cells treated either with empty vector pSIF or miR-218, or cells with no treatments are shown. *, p < 0.02 miR-218 versus pSIF. E and F, real-time measurement of cell migration and invasion shows that ectopic expression of miR-218 in Daoy cells inhibits cell migration (E) and invasion (F) compared with that of control cells. Error bars, S.E.
The growth of various cancers has been associated with the presence of a subpopulation of cells with stemlike properties (22). The presence of stemlike cells in the Daoy cell population has been previously established (23). Tumor stem cells have the characteristic of forming tumor spheres. Studies from our laboratory (21, 24) and from others (25) have shown that Daoy cells form tumor spheres when grown in serum-free neurobasal medium. Quantitative PCR analysis has shown that these tumor spheres express significantly increased levels of the neural stem cell (NSC) markers SOX2, NANOG, and NESTIN compared with those of cells grown as monolayers in medium containing serum (24). To determine the role of miR-218 in tumor sphere formation, transfected Daoy cells were seeded onto an ultra-low attachment plate in serum-free neurobasal medium containing B27, EGF, and FGF for 7 days. Ectopic expression of miR-218 decreased tumor sphere diameter significantly compared with controls (Fig. 2, C and D). The average diameter of the spheres formed by miR-218-transfected cells was roughly one-third to one-half the average diameter of cells transfected with empty vector pSIF or non-transfected cells. The expression of stem cell markers was significantly increased in medulloblastoma cells grown as tumor spheres (21, 24). NANOG, whose mRNA expression is strongly associated with the capacity of stem cells to self-renew, decreased in tumor spheres overexpressing miR-218 compared with control transfected cells. Conversely, MAP2, whose mRNA expression is a hallmark of neuronal differentiation, was significantly increased in miR-218-expressing tumor spheres compared with controls, as measured by quantitative RT-PCR (supplemental Fig. S3). The expression data from each treatment are normalized to that of no treatment control. These results support our hypothesis that the change in tumor sphere diameter was due to a diminished self-renewal capacity and increased cell differentiation rather than cell growth.
To study other hallmarks of cancer, we examined the effect of miR-218 on migration and invasion. Daoy cells were transfected with miR-218 or control empty vector, and measurement of migration was performed in real time using the xCELLigence system. The rate of migration over the entire time period was significantly slower in cells overexpressing miR-218 compared with either controls (Fig. 2E).
Next, to investigate whether miR-218 is involved in cell invasion, Daoy cells transfected with miR-218 were allowed to invade in Matrigel-coated E 16 plates, and cell invasion was again measured in real time using the xCELLigence system. Cells overexpressing miR-218 showed a substantially slower rate of invasion compared with that of controls (Fig. 2F). Taken together, re-expression of miR-218 in medulloblastoma cells down-regulated a variety of cellular functions, including proliferation, colony formation, cell motility, cell stemness, and cell invasion.
Expression of miR-218 Decreases Survival, Increases Differentiation, and Blocks Malignant Transformation of C17.2 NSCs
Immortalized C17.2 NSCs are multipotent self-renewing cells. These cells can be transformed using oncogenes, such as REST (26). REST-transformed C17.2 cells form medulloblastoma tumors when implanted orthotopically in murine models (26). miR-218 expression in C17.2 cells was low (only 50%) compared with Daoy cells (supplemental Fig. S4A), and after transfection with miR-218 plasmid, there was a significant increase in miR-218 mRNA level (supplemental Fig. S4B). Due to the inhibitory effect of miR-218 in medulloblastoma tumor sphere size, we hypothesized that miR-218 could play a significant role in changing the phenotype of NSCs. To investigate this, we transfected C17.2 cells with miR-218 and measured cell colony formation. Cells transfected with miR-218 showed significantly decreased colony formation compared with that of the empty vector (Fig. 3A). On the other hand, transfection of C17.2 cells with anti-miR-218 abrogated the inhibition of colony formation seen with miR-218, suggesting that miR-218 regulates NSCs proliferation.
FIGURE 3.
Biological effects of differential expression of miR-218 in murine neural progenitor stem cells. A, C17.2 cells transfected with miR-218 showed significant inhibition in their ability to form colonies compared with empty vector pSIF-transfected and non-transfected cells. #, p < 0.05 miR-218 versus pSIF. B, C17.2 cells form a negligible number of colonies in soft agar. C17.2 cells, when co-transfected with REST and pSIF plasmids, formed a robust number of colonies in soft agar that is partially inhibited by the overexpression of miR-218 with REST (**, p < 0.01). Representative wells from each treatment are shown (left). The colonies were counted using the Qpro software program and graphed (right). C, microRNA 218 promotes differentiation of neural stem cells in vitro. C17.2 cells were transfected with miR-218 or anti-miR-218 or with their corresponding control vectors and grown in differentiation medium. Tuj1 and NESTIN proteins were detected by immunofluorescence. i, representative merged immunofluorescence images of C17.2 cells stained for Tuj1 (green) that include the signal detected from DAPI staining of nuclear cells (blue). ii, representative merged immunofluorescence images of C17.2 cells treated as indicated and stained for NESTIN (green); DNA was fluorescently labeled with DAPI (blue). D, percentage of cells with either Tuj1 or NESTIN positive in DAPI-positive cells were counted and normalized to the total DAPI-stained nuclei in different fields in n = 4 wells. p values were calculated by Student's t test. The increase in miR-218 showed more cells expressing the neuronal differentiation marker Tuj1 and a concomitant decrease in cells expressing the neural progenitor marker NESTIN. #, p < 0.05; **, p < 0.01, comparisons as denoted in the figure. Error bars, S.E.
To further investigate the role of miR-218 in malignant transformation of cells, a soft agar assay was done. REST-transformed C17.2 cells form colonies in soft agar, whereas unmodified C17.2 cells form a negligible number of colonies in soft agar (21). We hypothesized that miR-218 would inhibit the colony formation induced by oncogenic REST. C17.2 cells were co-transfected with both REST and empty vector pSIF or with REST and miR-218. Results showed that miR-218 significantly reduced the number of colonies induced by REST in C17.2 cells compared with that of the empty vector (Fig. 3B).
Next we used C17.2 cells as a model to evaluate whether miR-218 has differentiation-inducing properties. To determine the effect of miR-218 on C17.2 cell fate, C17.2 cells overexpressing miR-218, anti-miR-218, or the respective control vectors were grown in differentiation medium. Immunofluorescence staining of cells using antibodies to NESTIN, a stem cell marker, and to Tuj1, a differentiation marker, was performed after 5–7 days. There was approximately a 2-fold increase in Tuj1/DAPI-positive staining and a concomitant (∼2-fold) decrease in NESTIN/DAPI-positive staining in cells treated with miR-218 compared with that of its control vector pSIF (p < 0.05) (Fig. 3, C and D), indicating that miR-218 increased differentiation of neuronal stem cells. Conversely there was a 3.5-fold increase in NESTIN/DAPI-positive cells and ∼1.5-fold decrease in Tuj1/DAPI-positive cells in anti-miR-218-treated cells compared with miR-218-treated cells, suggesting decreased differentiation and an increased stemlike phenotype. There was no significant change in either NESTIN or Tuj1 protein staining in C17.2 cells treated with anti-miR-218 when compared with its control vector, miRZip. The immunofluorescence images for individual protein staining and for DAPI staining for each treatment are given in supplemental Fig. S5. The data are quantified and are expressed as the percentage of cells that are expressing NESTIN or Tuj1 normalized to the total number of DAPI-positive cells (Fig. 3D). Taken together, our results show that an increase in miR-218 induces stem cell differentiation and a decrease in miR-218 leaves the cells in a less differentiated or more primitive state.
HITS-CLIP Identifies 618 Genes as Likely Targets of miR-218 in Medulloblastoma Cells
Several databases based on various algorithms, like TargetScan, miRanda, and PicTar, are available for predicting the targets of selected microRNAs (27–29). These algorithms have proved to be a major advancement in the field of microRNA biology. However, the problem with current computer algorithms is that they suggest, but cannot accurately identify, which genes are targeted by a given microRNA in a given cellular context (16, 30, 31). Therefore, unbiased experimental approaches are needed to identify and validate the direct targets of microRNAs. One novel method is the HITS-CLIP technique. This method provides a robust unbiased means to directly identify functional protein-RNA interactions (16, 32).
We performed HITS-CLIP to identify direct targets of miR-218 by following the protocol reported earlier with minimum modifications (16, 32). HITS-CLIP was performed with Daoy cells transfected with miR-218 or control cells. Argonaute-bound microRNA-mRNA complexes were successfully pulled down (supplemental Fig. S6, A and B) and comprehensively sequenced. We obtained 23,639,136 reads for the control and 20,117,446 reads for miR-218-overexpressing cells. The target mRNAs were aligned to the human genome, and the reads that contained a Watson-Crick match to the miR-218 seed sequence and occurred in the 3′-UTR of aligned genes were tabulated by their -fold changes in the miR-218-overexpressing library compared with the control library. Genes that showed a higher number of reads in the miR-218-overexpressing condition than in the control condition (i.e. -fold change >1) were considered targets of miR-218. Using the above criteria, 692 targets in 618 genes were identified using HITS-CLIP (supplemental Table S2). The list of targets generated by HITS-CLIP was compared with the targets predicted by TargetScan for miR-218. The number of targets predicted by TargetScan was 713 targets in 616 genes. Although both methods identified approximately the same number of targets, only 66 were common to both methods (Fig. 4). This represents a large discrepancy between targets identified using the unbiased experimental approach of HITS-CLIP compared with those predicted computationally.
FIGURE 4.
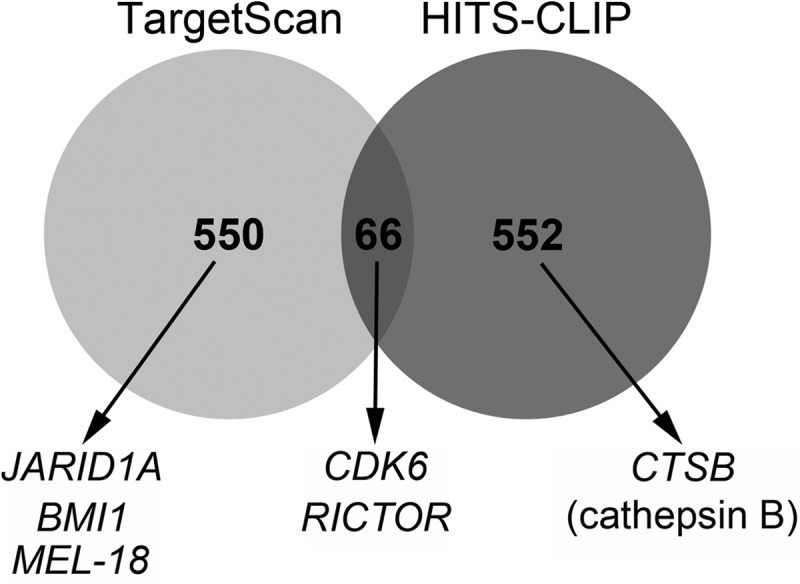
Venn diagram showing the number of overlapping genes targeted by miR-218 that were identified by the HITS-CLIP method and predicted by TargetScan. Shown below are the genes that were selected for validation in this study.
To examine whether the miR-218 targets identified by HITS-CLIP were associated with particular biological processes, the target list of 638 unique genes was input into DAVID. This analysis showed that miR-218 targets are enriched among multiple processes relating to cell cycle, transcription, and cell motility (supplemental Table S3). Multiple functions associated with protein synthesis and modifications are also enriched.
To check the accuracy of the HITS-CLIP method, we validated selected targets from each of the following categories: 1) both found in HITS-CLIP and predicted by TargetScan and validated by previous studies as an miR-218 target (RICTOR; rapamycin-independent companion of mTOR); 2) found both in HITS-CLIP and predicted by TargetScan but not previously validated (CDK6; cyclin-dependent kinase 6); 3) high -fold change in HITS-CLIP but not predicted by TargetScan (CTSB; cathepsin B); and 4) not identified by HITS-CLIP but predicted by TargetScan (BMI1, JARID1A, and MEL-18). The -fold changes in HITS-CLIP and the TargetScan scores of each of these six genes are given in supplemental Table S2. CDK6 and RICTOR had a 2-fold increase, whereas CTSB had a 20-fold enrichment in the 218 transfected cells compared with control. BMI1, JARID1A, and MEL-18 did not have any enrichment in the HITS-CLIP data (supplemental Table S2).
For the validation of these targets, Daoy cells were transfected with miR-218 or empty vector, and protein lysates were collected after 48 h and run on a Western blot. CDK6, RICTOR, and CTSB proteins were significantly knocked down in cells ectopically expressing miR-218, confirming that these genes are among the true targets of miR-218 (Fig. 5A). This validation was repeated in another medulloblastoma cell line, ONS76. Overexpression of miR-218 decreased the protein levels of targets CDK6, RICTOR, and CTSB as similarly seen in Daoy cells (Fig. 5A). CDK6 and RICTOR were consistently up-regulated in low miR-218-expressing medulloblastoma cell lines, whereas CTSB was more variably up-regulated in different medulloblastoma cell lines compared with normal cerebellum (Fig. 5B).
BMI1, MEL-18, and JARID1A were predicted by TargetScan alone. Because of their TargetScan scores and their intrinsic association with cell proliferation, cell stemness, and differentiation, these genes were chosen for validation. However, BMI1, MEL-18, and JARID1A showed no down-regulation in their protein levels after re-expression of miR-218 in Daoy cells (Fig. 5C). Thus, these genes are not true targets of miR-218 in medulloblastoma. This is consistent with the HITS-CLIP data, which did not identify any -fold change in the mRNA of these genes, although they were predicted by TargetScan. Overall, these results emphasize the need for actual experimental validation of computationally predicted targets and show that HITS-CLIP is a valuable technique for accurate identification of microRNA targets.
To further determine whether miR-218 directly regulates our validated targets, we cloned the HITS-CLIP-identified target sequence of CDK6 and RICTOR into luciferase reporter constructs. The sequence alignments are given in Fig. 5D. The overexpression of miR-218 in HEK293T cells significantly decreased luciferase activity in CDK6 and RICTOR target sequence-containing vectors. This shows that miR-218 directly binds to CDK6 and RICTOR target sequences identified by HITS-CLIP (Fig. 5D). The mutated sequence of both CDK6 and RICTOR showed no regulation.
CDK6 Rescues Tumor Suppressor Properties of miR-218
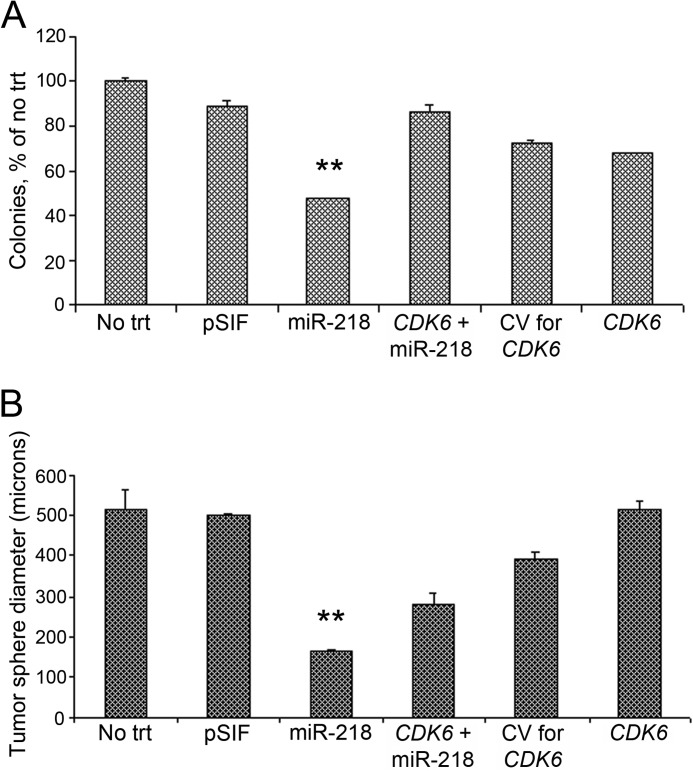
CDK6, which regulates cell cycle progression, was identified as an adverse prognostic marker in medulloblastoma (33). Pierson et al. (10) have shown that CDK6 is regulated by miR-124 among other microRNAs. To obtain direct evidence that CDK6 is one of the targets of miR-218 and to evaluate whether CDK6 suppression is a functional component of the miR-218 growth arrest phenotype, we investigated whether CDK6 would rescue miR-218-induced clonogenic arrest in medulloblastoma cells. A miR-218-expressing plasmid was co-transfected with either a CDK6 open reading frame vector that lacks the 3′-UTR (and therefore would not be regulated by miR-218) or with corresponding control vector. The colony focus assay showed that miR-218-mediated inhibition of the cell colony formation was rescued by CDK6 (Fig. 6A). This suggests that CDK6 may regulate cell colony formation and survival.
FIGURE 6.
Rescue effect of CDK6 on colony formation and tumor sphere size in Daoy cells overexpressing miR-218. Co-transfection with miR-218 and CDK6 that lacks the 3′-UTR rescued the cells from the inhibitory effect of miR-218 alone on the cell colony-forming ability (A) and on the tumor sphere diameter (B). Cells co-transfected with CDK6 that lacks the 3′-UTR and miR-218 increased the number of colonies and increased the tumor sphere size significantly (**, p < 0.01 miR-218 versus CDK6 + miR-218). CV, control vector for CDK6 that lacks the 3′-UTR. Error bars, S.E.
To further investigate whether CDK6 repression would rescue miR-218-induced tumor sphere size reduction, a tumor sphere assay was carried out with CDK6 that lacks the 3′-UTR co-transfected with miR-218 vector along with the appropriate controls. Co-expression of CDK6 rescued the miR-218-induced reduction in tumor sphere size (Fig. 6B) but provided less rescue effect compared with that expected based on its effect on cell colony formation. These data suggest that CDK6 is a functional target of miR-218 in medulloblastoma tumor cells. It is possible that expression of cell cycle drivers in general overcome the miR-218 effect, and these studies will have to be further pursued.
DISCUSSION
Understanding the biological underpinnings of aggressive brain tumors, such as medulloblastoma, is key for advances in therapy. Aberrant expression of microRNAs is found in many cancers. Studies of the functional role of microRNAs as a tumor suppressor or as an oncogene can identify new pathways in cancer and provide insight for the development of new or adjuvant therapies using these small non-coding RNAs. Because each microRNA can target multiple genes, they can provide attractive targets for treatment.
In this study, we show that miR-218 expression is down in medulloblastoma patient samples and is significantly down in SHH and Group 3 subgroups of medulloblastoma tumor specimens and in tumor cell lines. Poor expression of miR-218 is also found in other types of cancer, including gastric, colon, prostate, pancreatic, and cervical cancers (34, 35). Reduced expression of miR-218 was also found in glioma cells (36).
We found that miR-218 functions as a tumor suppressor in medulloblastoma by decreasing cell proliferation, cell migration, and invasion. One could argue that the reduction in the miR-218-overexpressing Daoy cells' migration and invasion may be due to a decrease in cell proliferation. The doubling time for Daoy cells is ∼28 h. The changes in the cell invasion and cell migration were noted before the cells' mitotic division. Therefore, miR-218 has a cumulative quadruple positive effect; i.e. it reduces medulloblastoma cell proliferation, cell migration, cell invasion, and cellular self-renewal capacity (Fig. 7). Additionally, forced expression of miR-218 in neural stem cells increases cell differentiation and decreases cell malignant transformation in the presence of oncogenic REST.
FIGURE 7.
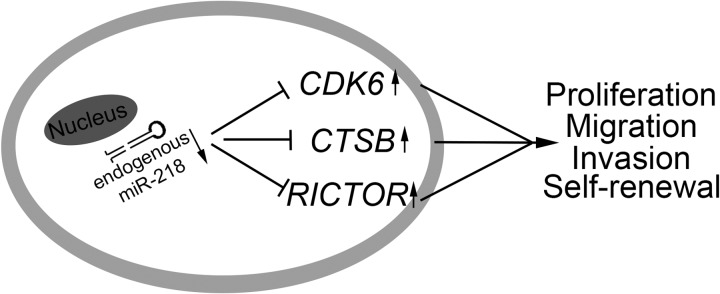
A working model of miR-218 interactions with its target genes in medulloblastoma. Down-regulation of miR-218 contributes to increased expression of CDK6, CTSB, and RICTOR, which promotes a malignant phenotype in medulloblastoma cells.
The HITS-CLIP technique was used to identify the direct true targets of miR-218. Although there are many bioinformatic tools based on computer algorithms available to predict microRNA targets, HITS-CLIP resolved several problems associated with predicting targets computationally. Using HITS-CLIP, 618 genes were identified as probable targets of miR-218. Functional analysis of targets identified by HITS-CLIP shows that genes targeted by miR-218 control multiple functional pathways, including cell cycle, cell metabolism, and cell motility.
It has been shown that miR-218 acts as a tumor suppressor in oral cancer by targeting the mTOR component RICTOR in a manner associated with inhibition of Akt phosphorylation (37). Consistent with that report, in our study, HITS-CLIP identified RICTOR as a direct target of miR-218. Another target found by HITS-CLIP that has shown potential importance in medulloblastoma is CDK6. CDK6 has been identified as an adverse prognostic marker in medulloblastoma (33) that up-regulates cell cycle progression and blocks cell differentiation. These effects were blocked by expression of miR-218 in our study, suggesting that miR-218 might exert its function in these areas, at least in part, through CDK6.
Another target of miR-218 identified by HITS-CLIP and validated was CTSB (cathepsin B). It is interesting to note that CTSB is known to regulate cell migration and cell invasion, suggesting that miR-218-mediated inhibition of CTSB may, in part, account for the observed effects on cell invasion and migration in this study. Further investigations are clearly needed to study the role of CTSB.
One key factor is that our data did not establish a strong inverse correlation between miR-218 expression (Fig. 1C) and protein level of its target in the medulloblastoma cell lines (Fig. 5B). This suggests that the target protein expression is regulated by other mechanisms besides miR-218 regulation and also that the regulatory activity of microRNAs depends on various factors besides their expression level (38).
Most significantly, HITS-CLIP can be more effective in identifying valid microRNA targets than computational algorithms. Targets predicted only by TargetScan and not by HITS-CLIP did not validate in further assays. Conversely, CDK6 and RICTOR, which were present in both HITS-CLIP and TargetScan lists, were validated. Therefore, the computer algorithms are useful tools, but they can be associated with an increased probability in finding false targets compared with the experimental HITS-CLIP method (16). These facts provide a strong rationale to evaluate more systematically the targets of microRNAs using experimental methods. NF-κB, ROBO1, LASP1, and PXN (paxillin) are the other predicted genes that have also been reported earlier as targets of miR-218 (39–42). Among these reported targets of miR-218, we found ROBO1 (4-fold increase) and LASP1 (8-fold increase) in our HITS-CLIP target list. There was no up-regulation in the expression of the other two targets, NF-κB and PXN, in miR-218-restored medulloblastoma cells. It is possible that these targets (NF-κB and PXN) may not be direct targets of miR-218 but rather indirect targets. Another possibility is that these targets are found in different cancer cell lines, like gastric and lung, and therefore could be organ-specific targets.
This current report also provides compelling evidence that suggests the need for experimental methods to identify direct targets of microRNAs. In summary, we believe that screening for miR-218 and induction of miR-218 in medulloblastoma patients will be clinically relevant and can lead to new therapeutic strategies. In fact, we have now validated CDK6 as a therapeutic target using small molecule inhibitors in medulloblastoma (43). Further experiments are needed to unravel the exact mechanisms leading to the observed effects.
Acknowledgments
We thank Dr. Jay Hesselberth for help with sequencing. We also thank Dr. Manoj Pillai for technical assistance with the HITS-CLIP method. We thank Dr. Robert Darnell for valuable input on the HITS-CLIP method. We thank Janet Parrish for help with the luciferase assay. Imaging experiments were performed in the University of Colorado Anschutz Medical Campus Advance Light Microscopy Core, supported in part by National Institutes of Health, NCRR, Colorado Clinical and Translational Services Institute Grant UL1 RR025780. Plasmid sequencing was done in the University of Colorado, Denver, Diabetes and Endocrinology Research Center Molecular Biology Core, supported by National Institutes of Health Grant P30 DK57516.
This work was supported, in whole or in part, by National Institutes of Health, NINDS, Grant K08NS059790 (to R. V.). This work was also supported by Morgan Adams foundation grants (to R. V. and N. K. F.).

This article contains supplemental Tables S1–S3 and Figs. S1–S6.
- miR
- microRNA
- HITS-CLIP
- high throughput sequencing of RNA isolated by cross-linking immunoprecipitation
- RISC
- RNA-induced silencing complex
- NSC
- neural stem cell.
REFERENCES
- 1. Provias J. P., Becker L. E. (1996) Cellular and molecular pathology of medulloblastoma. J. Neurooncol. 29, 35–43 [DOI] [PubMed] [Google Scholar]
- 2. Gilbertson R. J., Ellison D. W. (2008) The origins of medulloblastoma subtypes. Annu. Rev. Pathol. 3, 341–365 [DOI] [PubMed] [Google Scholar]
- 3. Ribi K., Relly C., Landolt M. A., Alber F. D., Boltshauser E., Grotzer M. A. (2005) Outcome of medulloblastoma in children. Long-term complications and quality of life. Neuropediatrics 36, 357–365 [DOI] [PubMed] [Google Scholar]
- 4. Reinhart B. J., Slack F. J., Basson M., Pasquinelli A. E., Bettinger J. C., Rougvie A. E., Horvitz H. R., Ruvkun G. (2000) The 21-nucleotide let-7 RNA regulates developmental timing in Caenorhabditis elegans. Nature 403, 901–906 [DOI] [PubMed] [Google Scholar]
- 5. Pasquinelli A. E., Ruvkun G. (2002) Control of developmental timing by micrornas and their targets. Annu. Rev. Cell Dev. Biol. 18, 495–513 [DOI] [PubMed] [Google Scholar]
- 6. Slack F. J., Basson M., Liu Z., Ambros V., Horvitz H. R., Ruvkun G. (2000) The lin-41 RBCC gene acts in the C. elegans heterochronic pathway between the let-7 regulatory RNA and the LIN-29 transcription factor. Mol. Cell 5, 659–669 [DOI] [PubMed] [Google Scholar]
- 7. Esquela-Kerscher A., Slack F. J. (2006) Oncomirs. MicroRNAs with a role in cancer. Nat. Rev. Cancer 6, 259–269 [DOI] [PubMed] [Google Scholar]
- 8. Calin G. A., Croce C. M. (2006) MicroRNA signatures in human cancers. Nat. Rev. Cancer 6, 857–866 [DOI] [PubMed] [Google Scholar]
- 9. Cho W. C. (2007) OncomiRs. The discovery and progress of microRNAs in cancers. Mol. Cancer 6, 60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Pierson J., Hostager B., Fan R., Vibhakar R. (2008) Regulation of cyclin dependent kinase 6 by microRNA 124 in medulloblastoma. J. Neurooncol. 90, 1–7 [DOI] [PubMed] [Google Scholar]
- 11. Venkataraman S., Alimova I., Fan R., Harris P., Foreman N., Vibhakar R. MicroRNA 128a increases intracellular ROS level by targeting Bmi-1 and inhibits medulloblastoma cancer cell growth by promoting senescence. PLoS One 5, e10748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Northcott P. A., Korshunov A., Witt H., Hielscher T., Eberhart C. G., Mack S., Bouffet E., Clifford S. C., Hawkins C. E., French P., Rutka J. T., Pfister S., Taylor M. D. Medulloblastoma comprises four distinct molecular variants. J. Clin. Oncol. 29, 1408–1414 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Northcott P. A., Fernandez-L A., Hagan J. P., Ellison D. W., Grajkowska W., Gillespie Y., Grundy R., Van Meter T., Rutka J. T., Croce C. M., Kenney A. M., Taylor M. D. (2009) The miR-17/92 polycistron is up-regulated in sonic hedgehog-driven medulloblastomas and induced by N-myc in sonic hedgehog-treated cerebellar neural precursors. Cancer Res. 69, 3249–3255 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Xing J. Z., Zhu L., Jackson J. A., Gabos S., Sun X. J., Wang X. B., Xu X. (2005) Dynamic monitoring of cytotoxicity on microelectronic sensors. Chem. Res. Toxicol. 18, 154–161 [DOI] [PubMed] [Google Scholar]
- 15. Parker M. A., Anderson J. K., Corliss D. A., Abraria V. E., Sidman R. L., Park K. I., Teng Y. D., Cotanche D. A., Snyder E. Y. (2005) Expression profile of an operationally defined neural stem cell clone. Exp. Neurol. 194, 320–332 [DOI] [PubMed] [Google Scholar]
- 16. Chi S. W., Zang J. B., Mele A., Darnell R. B. (2009) Argonaute HITS-CLIP decodes microRNA-mRNA interaction maps. Nature 460, 479–486 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Wilbert M. L., Yeo G. W. (2011) Genome-wide approaches in the study of microRNA biology. Wiley Interdiscip. Rev. Syst. Biol. Med. 3, 491–512 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Dennis G., Jr., Sherman B. T., Hosack D. A., Yang J., Gao W., Lane H. C., Lempicki R. A. (2003) DAVID. Database for Annotation, Visualization, and Integrated Discovery. Genome Biol. 4, P3. [PubMed] [Google Scholar]
- 19. Huang da W., Sherman B. T., Lempicki R. A. (2009) Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protoc. 4, 44–57 [DOI] [PubMed] [Google Scholar]
- 20. Thomas P. D., Kejariwal A., Campbell M. J., Mi H., Diemer K., Guo N., Ladunga I., Ulitsky-Lazareva B., Muruganujan A., Rabkin S., Vandergriff J. A., Doremieux O. (2003) PANTHER. A browsable database of gene products organized by biological function, using curated protein family and subfamily classification. Nucleic Acids Res. 31, 334–341 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Alimova I., Venkataraman S., Harris P., Marquez V. E., Northcott P. A., Dubuc A., Taylor M. D., Foreman N. K., Vibhakar R. (2012) Targeting the enhancer of zeste homologue 2 in medulloblastoma. Int. J. Cancer 131, 1800–1809 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Sell S. (2004) Stem cell origin of cancer and differentiation therapy. Crit. Rev. Oncol. Hematol. 51, 1–28 [DOI] [PubMed] [Google Scholar]
- 23. Srivastava V. K., Nalbantoglu J. (2008) Flow cytometric characterization of the DAOY medulloblastoma cell line for the cancer stem-like phenotype. Cytometry A 73, 940–948 [DOI] [PubMed] [Google Scholar]
- 24. Harris P. S., Venkataraman S., Alimova I., Birks D. K., Donson A. M., Knipstein J., Dubuc A., Taylor M. D., Handler M. H., Foreman N. K., Vibhakar R. (2012) Polo-like kinase 1 (PLK1) inhibition suppresses cell growth and enhances radiation sensitivity in medulloblastoma cells. BMC Cancer 12, 80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Wang X., Venugopal C., Manoranjan B., McFarlane N., O'Farrell E., Nolte S., Gunnarsson T., Hollenberg R., Kwiecien J., Northcott P., Taylor M. D., Hawkins C., Singh S. K. (2012) Sonic hedgehog regulates Bmi1 in human medulloblastoma brain tumor-initiating cells. Oncogene 31, 187–199 [DOI] [PubMed] [Google Scholar]
- 26. Su X., Gopalakrishnan V., Stearns D., Aldape K., Lang F. F., Fuller G., Snyder E., Eberhart C. G., Majumder S. (2006) Abnormal expression of REST/NRSF and Myc in neural stem/progenitor cells causes cerebellar tumors by blocking neuronal differentiation. Mol. Cell Biol. 26, 1666–1678 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Lewis B. P., Shih I. H., Jones-Rhoades M. W., Bartel D. P., Burge C. B. (2003) Prediction of mammalian microRNA targets. Cell 115, 787–798 [DOI] [PubMed] [Google Scholar]
- 28. John B., Enright A. J., Aravin A., Tuschl T., Sander C., Marks D. S. (2004) Human MicroRNA targets. PLoS Biol. 2, e363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Krek A., Grün D., Poy M. N., Wolf R., Rosenberg L., Epstein E. J., MacMenamin P., da Piedade I., Gunsalus K. C., Stoffel M., Rajewsky N. (2005) Combinatorial microRNA target predictions. Nat. Genet. 37, 495–500 [DOI] [PubMed] [Google Scholar]
- 30. Grimson A., Farh K. K., Johnston W. K., Garrett-Engele P., Lim L. P., Bartel D. P. (2007) MicroRNA targeting specificity in mammals. Determinants beyond seed pairing. Mol. Cell 27, 91–105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Hendrickson D. G., Hogan D. J., Herschlag D., Ferrell J. E., Brown P. O. (2008) Systematic identification of mRNAs recruited to argonaute 2 by specific microRNAs and corresponding changes in transcript abundance. PLoS One 3, e2126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Licatalosi D. D., Mele A., Fak J. J., Ule J., Kayikci M., Chi S. W., Clark T. A., Schweitzer A. C., Blume J. E., Wang X., Darnell J. C., Darnell R. B. (2008) HITS-CLIP yields genome-wide insights into brain alternative RNA processing. Nature 456, 464–469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Mendrzyk F., Radlwimmer B., Joos S., Kokocinski F., Benner A., Stange D. E., Neben K., Fiegler H., Carter N. P., Reifenberger G., Korshunov A., Lichter P. (2005) Genomic and protein expression profiling identifies CDK6 as novel independent prognostic marker in medulloblastoma. J. Clin. Oncol. 23, 8853–8862 [DOI] [PubMed] [Google Scholar]
- 34. Volinia S., Calin G. A., Liu C. G., Ambs S., Cimmino A., Petrocca F., Visone R., Iorio M., Roldo C., Ferracin M., Prueitt R. L., Yanaihara N., Lanza G., Scarpa A., Vecchione A., Negrini M., Harris C. C., Croce C. M. (2006) A microRNA expression signature of human solid tumors defines cancer gene targets. Proc. Natl. Acad. Sci. U.S.A. 103, 2257–2261 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Martinez I., Gardiner A. S., Board K. F., Monzon F. A., Edwards R. P., Khan S. A. (2008) Human papillomavirus type 16 reduces the expression of microRNA-218 in cervical carcinoma cells. Oncogene 27, 2575–2582 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Song L., Huang Q., Chen K., Liu L., Lin C., Dai T., Yu C., Wu Z., Li J. (2010) miR-218 inhibits the invasive ability of glioma cells by direct downregulation of IKK-β. Biochem. Biophys. Res. Commun. 402, 135–140 [DOI] [PubMed] [Google Scholar]
- 37. Uesugi A., Kozaki K., Tsuruta T., Furuta M., Morita K., Imoto I., Omura K., Inazawa J. (2011) The tumor suppressive microRNA miR-218 targets the mTOR component Rictor and inhibits AKT phosphorylation in oral cancer. Cancer Res. 71, 5765–5778 [DOI] [PubMed] [Google Scholar]
- 38. Liang Z., Zhou H., Zheng H., Wu J. (2011) Expression levels of microRNAs are not associated with their regulatory activities. Biol. Direct. 6, 43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Gao C., Zhang Z., Liu W., Xiao S., Gu W., Lu H. (2010) Reduced microRNA-218 expression is associated with high nuclear factor κB activation in gastric cancer. Cancer 116, 41–49 [DOI] [PubMed] [Google Scholar]
- 40. Chiyomaru T., Enokida H., Kawakami K., Tatarano S., Uchida Y., Kawahara K., Nishiyama K., Seki N., Nakagawa M. (2012) Functional role of LASP1 in cell viability and its regulation by microRNAs in bladder cancer. Urol. Oncol. 30, 434–443 [DOI] [PubMed] [Google Scholar]
- 41. Wu D. W., Cheng Y. W., Wang J., Chen C. Y., Lee H. (2010) Paxillin predicts survival and relapse in non-small cell lung cancer by microRNA-218 targeting. Cancer Res. 70, 10392–10401 [DOI] [PubMed] [Google Scholar]
- 42. Tie J., Pan Y., Zhao L., Wu K., Liu J., Sun S., Guo X., Wang B., Gang Y., Zhang Y., Li Q., Qiao T., Zhao Q., Nie Y., Fan D. (2010) MiR-218 inhibits invasion and metastasis of gastric cancer by targeting the Robo1 receptor. PLoS Genet. 6, e1000879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Whiteway S. L., Harris P. S., Venkataraman S., Alimova I., Birks D. K., Donson A. M., Foreman N. K., Vibhakar R. (2012) Inhibition of cyclin-dependent kinase 6 suppresses cell proliferation and enhances radiation sensitivity in medulloblastoma cells. J Neurooncol. doi: 10.1007/s11060-012-1000-7 [DOI] [PMC free article] [PubMed] [Google Scholar]