Background: Conjugative plasmid transfer is the prevalent means for spreading antibiotic resistance genes among bacteria.
Results: Surface exposure of transfer protein TraM from the Gram-positive (G+) plasmid pIP501 was confirmed, and its crystal structure was solved.
Conclusion: Structural relations to type IV secretion (T4S) proteins provide a novel classification scheme.
Significance: The novel classification will help elucidate structure-function relationships in G+ T4S systems.
Keywords: Antibiotic Resistance, Bacterial Conjugation, Bacterial Pathogenesis, Crystal Structure, X-ray Crystallography, Conjugative Plasmid, Enteroccucus, Gram-positive, Type IV Secretion System, pIP501
Abstract
Conjugative plasmid transfer is the most important means of spreading antibiotic resistance and virulence genes among bacteria and therefore presents a serious threat to human health. The process requires direct cell-cell contact made possible by a multiprotein complex that spans cellular membranes and serves as a channel for macromolecular secretion. Thus far, well studied conjugative type IV secretion systems (T4SS) are of Gram-negative (G−) origin. Although many medically relevant pathogens (e.g., enterococci, staphylococci, and streptococci) are Gram-positive (G+), their conjugation systems have received little attention. This study provides structural information for the transfer protein TraM of the G+ broad host range Enterococcus conjugative plasmid pIP501. Immunolocalization demonstrated that the protein localizes to the cell wall. We then used opsonophagocytosis as a novel tool to verify that TraM was exposed on the cell surface. In these assays, antibodies generated to TraM recruited macrophages and enabled killing of pIP501 harboring Enteroccocus faecalis cells. The crystal structure of the C-terminal, surface-exposed domain of TraM was determined to 2.5 Å resolution. The structure, molecular dynamics, and cross-linking studies indicated that a TraM trimer acts as the biological unit. Despite the absence of sequence-based similarity, TraM unexpectedly displayed a fold similar to the T4SS VirB8 proteins from Agrobacterium tumefaciens and Brucella suis (G−) and to the transfer protein TcpC from Clostridium perfringens plasmid pCW3 (G+). Based on the alignments of secondary structure elements of VirB8-like proteins from mobile genetic elements and chromosomally encoded T4SS from G+ and G− bacteria, we propose a new classification scheme of VirB8-like proteins.
Introduction
Bacterial conjugation is the major mechanism of horizontal gene transfer. It is the most prevalent means for the spread of antibiotic resistance and virulence genes (1). During conjugation, plasmid DNA is transported from a donor to a recipient cell. This transport is mediated by a multiprotein complex large enough to span the bacterial cell wall (2, 3). The multiprotein complex is classified as a type IV secretion system (T4SS),4 dedicated to the intercellular transport of proteins or protein-DNA complexes (4–7). The translocation of substrates across the cell envelope is achieved by a mechanism requiring direct contact with a recipient cell (8). The vast majority of information regarding the individual functions, regulation, and interaction of proteins involved in the type IV secretion (T4S) process is available for Gram-negative (G−) bacteria, whereas most knowledge about the equivalent systems of Gram-positive (G+) origin is based on similarity to their counterparts in G− bacteria (9, 10). For the Enterococcus sex pheromone plasmid pCF10, the findings of Chen et al. (11) support a model in which PcfC, the putative coupling protein, initiates substrate transfer through the pCF10 T4S channel by an NTP-dependent mechanism. Li et al. (12) demonstrated for the first time horizontal transfer of a pathogenicity island of G+ origin mediated by a genomic island-type T4SS. They present a hypothetical model for T4S in epidemic Streptococcus suis isolates. Only very recently has structural information on T4SS proteins of G+ origin become available (13, 14).
The multiple antibiotic resistance plasmid pIP501, originally isolated from Streptococcus agalactiae (15), exhibits the broadest known host range for plasmid transfer in G+ bacteria. It is the first plasmid of G+ origin for which stable replication in G− bacteria was shown (16). The transfer region of pIP501 is organized in an operon encoding 15 putative transfer (Tra) proteins. Published and unpublished work in our laboratories has begun to assign structural and functional characteristics to these Tra components. Three of the Tra proteins show significant sequence similarity to the T4SS from Agrobacterium tumefaciens. The ATPase TraE (homolog to VirB4) was shown to interact with itself and with several other potential pIP501 transfer proteins (10) and most likely energizes the conjugation process. The coupling protein TraJ (homolog to VirD4)5 forms hexamers and lacks the transmembrane domain present in other coupling proteins (17). Coupling proteins connect the macromolecular complex of single-stranded plasmid DNA and relaxosome proteins, which is being transported, with the secretory conduit (18). The pIP501 coupling protein TraJ is probably recruited to the cell membrane by TraI (8). The predicted role of the lytic transglycosylase TraG (homolog to VirB1)6 would be to locally punch holes into the peptidoglycan layer of G+ bacteria for the assembly of the conjugative core complex. The relaxase TraA is another component encoded by the pIP501 transfer operon that has been functionally characterized (19, 20). It was shown to bind to the oriT and to autoregulate expression of the T4 transfer genes.
Despite these insights concerning some of the 15 potential transfer proteins, we still lack structural information on the individual molecules. Moreover, the components of the putative T4SS core complex, characterized in structural detail for the pKM101 encoded T4SS of G− origin (3), remain unknown, mainly because of the missing or very low sequence similarities to G− derived T4SS. Potential candidates for the core complex are all Tra proteins for which a transmembrane motif has been predicted, and thus an affinity for the cell envelope is likely, namely TraB, -C, -F, -H, -I, -K, -L, and -M.
Here, we present the biophysical and structural characterization of the TraM C-terminal domain (formerly called ORF13, GenBankTM accession number CAD44393.1; TraM190–322, also referred to as TraMΔ) from the Enteroccocus faecalis conjugative plasmid pIP501. The protein localizes to the cell envelope, and anti-TraMΔ antibodies recruit macrophages to pIP501 harboring E. faecalis cells, suggesting that TraM is a part of the pIP501 transfer system that is accessible from outside of the cell. This is the first time that the opsonophagocytosis assay has been employed to demonstrate the surface accessibility of a putative T4SS protein. TraMΔ forms a trimer in the crystal and reveals structural similarity to the T4SS protein VirB8 from G− bacteria, leading to a novel, secondary structure-based classification of VirB8-like proteins.
EXPERIMENTAL PROCEDURES
Details on purification, biophysical characterization and crystallization will be reported in a separate publication.7
Immunolocalization of TraM
Subcellular fractionation of E. faecalis JH2-2 (pIP501) was performed according to Buttaro et al. (21) with minor modifications. An exponentially growing culture (A600 = 0.5) of E. faecalis JH2-2 (pIP501) was chilled on ice for 15 min, washed twice in an equal volume of potassium phosphate buffer (50 mm, pH 7.0), and resuspended (1:50, v/v) in lysis buffer (50 mm KH2PO4/K2HPO4, pH 7.0, 1 mm EDTA, 1 mm MgCl2, 100 μg·ml−1 DNase, 100 μg·ml−1 RNase). The cells were broken by FastPrep®-24 (MP Biomedicals, Illkirch, France) using lysing matrix E (1.4-mm ceramic spheres, 0.1-mm silica spheres, 4-mm glass beads; MP Biomedicals). Unlysed cells were removed by low speed centrifugation. The cell wall fraction was then harvested by high speed centrifugation at 17,000 × g for 20 min at 4 °C, and the membrane fraction was obtained by ultracentrifugation of the supernatant at 45,000 rpm for 2 h at 4 °C (OTD combi ultracentrifuge; Thermo Fisher Scientific). The remaining supernatant contained the soluble proteins. TraM was detected in the fractions (cell wall, membrane, and cytoplasm) by immunostaining of TraM with primary polyclonal anti-TraMΔ antibody and a secondary horseradish-conjugated anti-rabbit IgG antibody (Promega GmbH, Mannheim, Germany).
Opsonophagocytosis Killing and Killing Inhibition Assay
Opsonophagocytic assay and opsonophagocytic inhibition assay were performed as described previously (22). In brief, polymorphonuclear neutrophils from healthy volunteers were prepared using heparin-dextran sedimentation and hypotonic lysis and adjusted to 1 × 107 ml−1 in RPMI with 10% FBS. Sera against TraMΔ were produced in rabbits (Biogenes, Berlin, Germany) and used at a dilution of 1:10 in RPMI + FBS. Lyophilized baby rabbit serum (Cedarlane, Burlington, Canada) was reconstituted in RPMI, diluted at 1:15, and preadsorbed at 4 °C for 1 h with the target strain tested subsequently in the assay. Bacteria were grown to midlog phase in TSB medium and adjusted to ∼1 × 107 cfu ml−1 photospectrometrically, and serial dilutions were plated on tryptic soy agar plates to confirm the viable counts. Equal volumes of all four components (bacterial strain, rabbit serum against TraMΔ, baby rabbit serum as complement source, and human neutrophils) were combined, and incubated on a rotor rack for 90 min. At the end of the experiments, serial dilutions were prepared, plated on tryptic soy agar plates, incubated overnight, and enumerated. Opsonic killing was measured as compared with a control containing no PMNs. For the opsonophagocytic inhibition assay, increasing amounts of purified TraMΔ were preincubated with serum and added to PMNs, complement, and bacteria, as described above.
Cross-linking Experiments
Cross-linking experiments were performed as follows: 50-μl sample (9.2 μg of TraMΔ; 300 mm NaCl, 100 mm Bicine, 1 mm DTT, 0/0.001/0.01/0.05/0.1% glutardialdehyde, and distilled H2O) were incubated for 20 min at room temperature. Glycine was added to a final concentration of 140 mm, and the samples were incubated for 5 min at room temperature. 400 μl of acetone (−20 °C) were added, and the samples were precipitated at −20 °C for 2 h, followed by centrifugation for 15 min at 16,100 × g and 4 °C. The pellet was resuspended in 10 μl of H2O and 10 μl of loading buffer for SDS-PAGE. A molecular weight standard was used to assess the size of the cross-linked oligomers (26630, PageRuler unstained broad range protein ladder; Thermo Fisher Scientific).
Mass Spectroscopy of TraMΔ Crystals
Several crystals of TraMΔ were dissolved in 10 μl of pure H2O and investigated by MALDI-TOF analysis (ultrafleXtreme; Bruker, Vienna, Austria). After first standard size evaluation experiments, one of the samples was digested with trypsin and further analyzed via MS/MS to define the N-terminal sequence of TraMΔ.
Structure Refinement
Details on data collection and processing are presented in a separate publication.7 The model was refined in COOT (23) and REFMAC5 (24). The refined x-ray model was validated with MolProbity (25). The secondary structure elements were determined using STRIDE (26). Three-dimensional alignments of the TraMΔ structure with structural homologs were performed with DALI (27) and MATRAS (28). The structural alignment of TraMΔ monomers was conducted with MASS (29). PyMOL (30) was used to prepare structure representations and to calculate the RMSD of TraMΔ monomer alignments. The PDBePISA (31) server was used to calculate the interaction surfaces in TraMΔ trimers. The structural and sequential similarities between TraM214–322 and its close homologs were examined using the pairwise structural alignment feature of MATRAS, as well as the SSM algorithm (32) included in the program Coot.
Sequence-based Comparison and Characterization
The following online services were used to search for transmembrane motifs in the TraM sequence and potential homologous proteins: TMPRED (33), PHDhtm (34), HMMTOP (35), TMHMM (36), SOSUI (37), MemsatSVM (38), Memsat3 (39), and MemBrain (40).
PSIpred (41) was used to predict the secondary structure content of TraM and of homologous proteins, but where known, the secondary structure was derived from the crystal structure. General features of the His-tagged TraMΔ construct were assessed with ProtParam (42). Coiled-coil motif searches were performed with COILS (43), MultiCoil (44), and PairCoil2 (45). A search for other VirB8-like proteins in G− and G+ conjugative plasmids, transposons, integrative conjugative elements (ICEs), and genetic islands was performed by comparing secondary structure and the position of the predicted transmembrane helix to the known structures of VirB8, TcpC, and TraM.
Coiled-coil Motif Molecular Dynamics Simulations
The amino acid sequence of the predicted coiled-coil motif (36 amino acids) was submitted to the HHpred (46) protein similarity detection server via the Bioinformatics Toolkit (47). The best 20 hits were selected for automatic submission to homology model building with Modeler (48). The homology modeling resulted in an all α-helical, slightly curved peptide. PyMOL was used to cut several loose residues on both ends of the helix (final sequence 7–32, QVQLQSVKKESELLEEQIERVKETDI, residue 188–213 of the TraM sequence), as well as to duplicate and align the peptide to a known coiled-coil domain. Because the TraM coiled-coil motif was predicted to consist of three helices, we searched for a suitable reference structure. Eventually, the triple coiled-coil motif of the human surfactant protein D (Protein Data Bank code 3DBZ) served as the template. The resulting TraM coiled-coil model was used in molecular dynamics simulations.
Employing GROMACS and the OPLS All-atom Force Field
The GROMACS 4.5.5 software package (49) was used to perform molecular dynamics simulations and equilibrations. pKa values and protonation states of the titratable amino acids were calculated at pH 7 using TITRA (50) employing the Tanford-Kirkwood sphere model (50). The structure was solvated with water inside a cubic box and minimized and equilibrated for 0.1 ns (ensembles of NPT and NVT, respectively), with position restraint on all heavy atoms followed by five unrestrained simulations of 10 ns with explicit solvent employing OPLS all-atom force field and the TIP3P water model (51). The backbone RMSD was monitored to ensure complete equilibration of the protein model. All of the calculations were carried out with a 2-fs time step, and long range electrostatic interactions were computed using the particle mesh Ewald method (52). All bonds in the system were constrained using the LINCS algorithm (53). The neighbor list search was updated every five steps within a 1.0 nm cut-off. van der Waals interactions were computed with a cut-off of 1.4 nm. The isotropic Parrinello-Rahman (54, 55) protocol was used for pressure (1 bar), and the velocity-rescaling thermostat (56) was used for temperature coupling. The components of the system are separately coupled at 300 K with a coupling constant of 0.1 ps. Periodic boundary conditions were applied in all three dimensions (57). Calculations were performed on 64-bit Linux 48-core nodes of an AMD Magny-Cours cluster. Data analysis and image rendering were carried out with standard tools provided within the GROMACS package (49), VMD (58), YASARA (59), and in-house scripts for post-processing and quality control.
Employing YASARA and the Amber03 Force Field
Simulations were carried out using the program YASARA (59), with the Amber03 force field (60, 61) using a 7.86 Å cut-off. Model structures were energy-minimized to remove bumps and correct the covalent geometry and charges were neutralized by adding counter ions. After removal of conformational stress by a short steepest descent minimization, simulated annealing (time step, 2 fs) was performed until convergence was reached. Periodic boundary simulations were done on cubic cells of extra extension along each axis of the protein of 10 Å. The simulation cell was filled with explicit water to a density of 0.997 g/liter and gradually minimized. Minimization was followed by an equilibration procedure to 298 K. Resulting minimized and equilibrated models were subsequently used for MD simulations. Temperature was kept at 298 K by rescaling the atom velocities every 25 simulation steps. Production simulations were carried out for 10 ns on quad-core 64-bit Linux workstations.
RESULTS
TraM Localizes to the Cell Envelope
To localize the TraM protein in vivo, an exponentially growing culture of E. faecalis JH2-2 (pIP501) was fractionated into cell wall, membrane, and cytoplasmic fraction as described by Buttaro et al. (21). TraM was exclusively found in the cell envelope fractions (cell wall and membrane; Fig. 1A), whereas the jointly expressed TraN protein, predicted by PSORTb to localize to the cytoplasm, was exclusively found in the cytoplasmic fraction, consistent with a sound separation of cytoplasmic and cell envelope proteins.
FIGURE 1.
TraM localization and characterization. A, TraM localizes to the cell envelope of pIP501 harboring E. faecalis JH2-2 cells. The localization of TraM in the cell fractions was detected by Western blot with anti-TraMΔ antibodies. CW, cell wall; M, membrane; CP, cytoplasm. B, cross-linking assay of TraMΔ. Lane M, molecular mass standard; lane 1, control (no glutardialdehyde, no treatment); lanes 2–6, TraMΔ with 0/0.001/0.01/0.05/0.1% glutardialdehyde. C, opsonophagocytic killing and inhibition of killing assays using anti-TraMΔ antisera.
Characterizing the TraMΔ Protein
Attempts to overexpress and purify full-length TraM (37.5 kDa) failed because of solubility problems. Consequently, a stable truncation derivative, TraMΔ (18.6 kDa), was constructed and purified. It lacks the N-terminal domain and the putative transmembrane domain, which is positioned between the N- and C-terminal domains. Details of the purification and crystallization procedure will be reported in a separate publication.7 The size of TraMΔ in solution has been evaluated via gel filtration, showing an apparent molecular mass of 24.4 kDa, which indicates a homogenous monomeric protein. Additional biophysical experiments were performed to confirm the oligomerization state of TraMΔ in solution. DLS and SAXS experiments yielded a monomer under the conditions tested. In contrast, in vitro cross-linking studies showed that TraMΔ is able to form multimers in solution (Fig. 1B). Dimer formation was already visible at the lowest glutardialdehyde concentration. A trimeric form of TraMΔ was detected at the highest cross-linker concentration.
Anti-TraMΔ Antibodies Recruit Macrophages to pIP501 Harboring E. faecalis Cells
The opsonophagocytic killing assay showed effective killing of 75.5% of E. faecalis JH2-2 (pIP501) cells with sera raised against TraMΔ at a serum dilution of 1:10, whereas no killing was observed for E. faecalis JH2-2 without plasmid pIP501 (Fig. 1C). Complete inhibition of killing was obtained when 20 or 100 μg ml−1 of purified TraMΔ were incubated with the sera, whereas lower concentrations of TraMΔ (i.e., 0.8 and 4 μg ml−1) resulted in a dose-dependent killing.
The TraMΔ Crystal Structure
Because of the lack of structures with significant sequence similarity with TraMΔ, selenomethionine-containing TraMΔ crystals were used for structure solution by single-wavelength anomalous dispersion. A single selenomethionine crystal showed a nontwinned pattern and diffracted to 2.5 Å resolution at the synchrotron. The crystal belonged to space group P1, with unit cell parameters a = 39.21, b = 54.98, c = 93.47 Å, α = 89.91°, β = 86.44°, γ = 78.63°, and six molecules/asymmetric unit. Details on crystallization, data collection, and processing are part of a separate publication.7 Table 1 provides an overview of the refinement statistics.
TABLE 1.
Refinement statistics
A single crystal was used for the data collection.
| Space group | P1 |
|---|---|
| Unit cell dimensions (Å, °) | a = 39.21, b = 54.98, c = 93.47, α = 89.91, β = 86.44, γ = 78.63 |
| Refinement | |
| Resolution (Å) | 46.89-2.5 |
| No. reflections | 24,589 |
| Rwork/Rfree | 0.2074/0.2627 |
| No. atoms | |
| Protein | 5412 |
| Water | 399 |
| B-factors | |
| Protein | 28.62 |
| Water | 31.33 |
| RMSDs | |
| Bond lengths (Å) | 0.014 |
| Bond angles (°) | 1.762 |
| MolProbity validation | |
| Ramachandran outliers | 1.25% (8 of 624) |
| Ramachandran favored | 95.17% |
| Cβ deviations > 0.25 Å | 3 of 624 |
| Residues with bad bonds | 0.0% |
| Residues with bad angles | 0.31% |
| MolProbity score | 59th percentile |
Six molecules were found in the asymmetric unit. The N-terminal ends of the monomers, namely residues 194–213, appeared to be flexible and were not observed in the electron density map. The final coordinates and structure factor amplitudes have been deposited in the Protein Data Bank (code 4EC6).
The crystal structure of TraMΔ consists of two anti-parallel α-helices (h1 and h2) at the N terminus and an anti-parallel, highly curved β-sheet, made up of five β-strands (s1–s4′) in the C-terminal part of the protein (Fig. 2A). The β-sheet is wrapped around helix 1. A twist in β-strand 1 is located at residue Tyr269 (Fig. 2B). Because of this distortion, the backbone oxygen of Tyr269 is positioned in a way to form a intermolecular hydrogen bond at the trimerization interface (Fig. 2C). The overall alignment of the six TraMΔ monomers showed a mean RMSD value of 0.41 Å. RMSDs for individual pair-wise alignments of the monomers are shown in supplemental Table S1.
FIGURE 2.

The structure of TraM214–322. A, cartoon representation of TraM214–322 with view onto the twisted β-sheet and 90° turned about the 3-fold axis. Secondary structure elements are highlighted (helices in cyan and strands in purple). B, detailed view of the twist in strand 1. C, trimerization interface between β-strands 1 and 2 of chain F and α-helix 1 of chain D. D, one TraM trimer shown in cartoon representation. The monomers are colored green (chain D), red (chain E), and cyan (chain F), respectively. The monomer-monomer contact region is indicated. IS, interaction site. E, surface representation of the interaction area between monomers D and F. F, the C-terminal end of monomer F (stick representation) nestled in a hydrophobic cleft of monomer D (surface representation).
The six molecules of the asymmetric unit build up two independent trimers, in which the monomers are related by a noncrystallographic 3-fold axis. Each trimer forms a triangular pyramid (Fig. 2D). The primary interaction surface between TraMΔ monomers (Fig. 2E) is formed by residues of the C terminus (Phe319–Asn322) and of the strands s1, s2, and s3 (Ser265, Asn267–Ser271/Glu281, Leu283–Asn285/Met294, and Lys296) in the first molecule and residues in the α-helices (Ser214, Ser216, Lys217, Thr220, Phe221, Arg223, Tyr224/Thr245, and Tyr246), as well as the loop between β-strands s3 and s4 (Thr304 and Asn306–Leu309) in the second molecule. Intermolecular hydrogen bonds are formed between residues Lys217–Tyr269, Thr220–Tyr268, and Tyr224–Asn285. In addition, several van der Waals interactions are formed between the complementary surfaces, resulting in a monomer-monomer interface area of 492 Å2. Consequently, the total buried area of one monomer amounts to 984 Å2 or ∼14% of the calculated solvent-accessible surface area of one isolated monomer. The size of the interaction surface within the trimers suggests that trimer formation in solution is disfavored (62), which is consistent with our experimental data.
Trimerization of TraM Is Facilitated by the N-terminal Coiled-coil Motif
The N-terminal ends of TraMΔ point toward the tip of the triangular pyramids (Fig. 2D) in the crystals. Thus, the flexible N-terminal ends of the TraMΔ constructs, as well as the predicted TM helix of the full-length protein, would be perfectly positioned to form a triple-helix coiled-coil structure. To clarify this possibility, the TraM amino acid sequence was analyzed in detail. An extended transmembrane area (residues 166 to ∼190) was consistently found by all used trans-membrane helix search algorithms. The N-terminal domain of TraM was predicted to be cytoplasmic, whereas the C-terminal domain would face toward the outside of the bacterial cell. The sequence between the trans-membrane helix and the C-terminal domain was indicative of a coiled-coil motif with a high probability of trimer formation (residues 184–213). The lack of electron density for the coiled-coil in the crystal structure could be explained by imperfect trimerization, because of missing residues (supplemental Fig. S1). We think that these residues (184–193) are crucial for the formation of a stable coiled-coil interface. To test our hypothesis, a model of the putative coiled-coil trimer was used in molecular dynamics simulations to evaluate the stability of the trimerization motif. The RMSD of the model backbone after alignment to the starting model converges to a value of ∼3 Å (using the Amber03 force field) and 4.5 Å (using GROMACS), respectively (Fig. 3A). The solvent-accessible hydrophobic surface of the trimer remains constant over the time of the simulation (10 ns), with a value of 2600 Å2 (Amber03) and 2800 Å2 (GROMACS) (Fig. 3B). The final models are very similar (Amber03: RMSD 1.23 Å). Their hydrophobic residues face toward the center of the trimer (Fig. 3, C and D), thus stabilizing the triple coiled-coil motif. From these results we conclude that the complete coiled-coil motif would favor the formation of a stable TraM trimer in solution.
FIGURE 3.

The potential TraM coiled-coil motif. A, RMSD of the resulting backbone after least square fit to the starting model backbone. The results from GROMACS simulations (black lines) and Amber simulations (gray lines) are shown. The graphs are represented as progressive means of 25 data points. B, hydrophobic fraction of the solvent-accessible surface area per residue. The area remains stable in Amber03 (left panel) and GROMACS (right panel) simulations (three each). The data graphs are represented as progressive means of 50 data points. C, alignment of the three final Amber03 coiled-coil models in cartoon representation; the view is along the coiled-coil axis and 90° turned. D, highlighting on the hydrophobic residues (stick representation) facing toward the center of the triple coiled-coil model.
TraMΔ Is a Structural Homolog of VirB8 and TcpC
A structural similarity search revealed that TraM214–322 is structurally related to members of the nuclear transport factor-2 (NTF-2) like superfamily (supplemental Table S2) with a high similarity to the NTF-2 protein from Rattus norvegicus (Protein Data Bank code 1OUN; Fig. 4A). Consequently, TraM214–322 can be considered a protein of the NTF-2 like family. A closer examination of the list of structural homologs revealed a relation to the published structures of the periplasmic region of VirB8 from B. suis (Protein Data Bank code 2BHM) (63) and A. tumefaciens (Protein Data Bank code 2CC3) (64). Although VirB8 was described as a member of the T4SS from A. tumefaciens (65) and seems to interact with many T4SS components, it was not found in the structurally characterized core complex of the T4SS of plasmid pKM101 (3). VirB8 was proposed to play a role as an assembly factor or scaffolding protein for the correct complex formation and localization (66, 67). Furthermore we found a close similarity to the recently published structure of TcpC from the C. perfringens conjugative model plasmid pCW3 (Protein Data Bank code 3UB1 (13)). Again, the homologous protein interacts with potential members of the pCW3 encoded T4SS. Thus, a function similar to that of VirB8 was postulated for TcpC.
FIGURE 4.

Structural comparison of TraM214–322 to related proteins. A, cartoon representation of TraMΔ, NTF2 (R. norvegicus, Protein Data Bank code 1OUN), the C-terminal and the central domain of TcpC (C. perfringens, Protein Data Bank code 3UB1), and the periplasmic domain of VirB8 from A. tumefaciens (Protein Data Bank code 2CC3) and B. suis (Protein Data Bank code 2BHM), respectively; secondary structure elements are highlighted (helices in cyan and strands in purple). B, comparison of the TraMΔ trimer to the TcpC trimer, formed by the central domains of TcpC monomers.
In contrast to VirB8, TcpC consists of two NTF2-like domains. TraM214–322 shares a higher sequence identity, as well as structural similarity with these TcpC domains than with NTF2 or any VirB8 protein. Detailed information on the individual alignments is depicted in Table 2 (parts A and B). Despite low sequence identity (supplemental Fig. S2), the overall fold of all aligned molecules (Fig. 4A) is quite conserved. The two NTF2-like domains of TcpC are connected by a seven-amino acid linker, which is nestled in a cleft formed by two loops of the central domain. Similarly, the TraMΔ C-terminal end is nestled in a cleft formed by the loop between β-strands s3 and s4 and the C-terminal end of α-helix h2 (Fig. 2F). Variable intermolecular hydrogen bonds are formed, depending on the side chain positions of the C-terminal residues. In addition, the side chains of Phe319 and Phe321 occupy a hydrophobic pocket of the adjacent monomer, stabilizing the interaction between TraMΔ monomers via van der Waals interactions. Furthermore, TcpC forms trimers in the crystal, with the central domains aligned as a triangular pyramid around the molecular 3-fold axis (13). The two independent TraM194–322 trimers adopt a very similar structural organization (Fig. 4B).
TABLE 2.
Validating the structural similarity of TraMΔ to related structures

A New Classification of VirB8-like Proteins
We further evaluated the differences between the VirB8 proteins from G− A. tumefaciens and B. suis and the G+ homologs TcpC and TraM. Based on their domain composition, we categorized the homologous proteins into three distinct classes (Fig. 5). We performed an extended search for VirB8-like proteins in a broad spectrum of conjugative plasmids, transposons, ICEs, and genetic islands from G− and G+ bacteria. The candidates were sorted according to their similarity to either of the VirB8-like classes, based on secondary structure prediction. The results for the prediction-based comparison and sequence alignments of potential NTF2-like domains can be found in supplemental Table S3 and in detail in supplemental Fig. S3.
FIGURE 5.
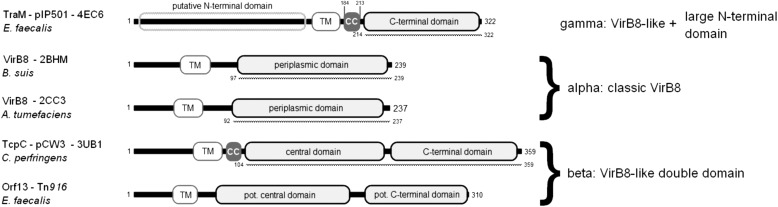
Comparison of the domain arrangement of TraM and its structurally related proteins from G+ and G− putative T4SS. The amino acid sequence contained in the available structures is indicated by a dotted line below the individual representations. Because the structure of Orf13 from Tn916 is not available, the two potential domains are assigned according to secondary structure predictions with PsiPred. Transmembrane helices have been predicted for all proteins as described above. The potential coiled-coiled motifs of TraM and TcpC are highlighted as gray boxes (CC).
All of the analyzed VirB8-like proteins found in putative T4SS of G− origin (17 plasmids and 6 T4SS located on the chromosome, including ICEs, a gonococcal genetic island and a pathogenicity island) belong to class ALPHA. Four proteins from plasmids encoded by Salmonella typhi, Pseudomonas aeruginosa, Enterobacter aerogenes, and Bordetella pertussis exhibit a slightly different composition with a longer N-terminal tail compared with the classical fold of VirB8 from A. tumefaciens. With the exception of the VirB8-like protein from S. typhi, the amino acid sequences of the NTF2-like domains for these proteins are nearly identical. We found five proteins of G+ origin that share the class ALPHA fold (two plasmids, two ICEs, and one transposon). Proteins that belong to class BETA could be further divided into three subgroups, with minor differences in their domain composition. The first group consists of candidates strictly similar to TcpC that are exclusively found in conjugative plasmids of C. perfringens (4). The second group contains proteins encoded on transposons (10) from diverse G+ genera, like streptococci, enterococci, and clostridiae. The third group contains more distantly related proteins from two ICEs of Streptococcus gallolyticus and Bacillus subtilis. Class GAMMA-like proteins were exclusively found in E. faecalis conjugative plasmids (4). A sequence alignment of all analyzed NTF2-like sequences showed that class BETA and GAMMA proteins were closer related to each other than to class ALPHA proteins (supplemental Fig. S4). Additionally, neither TraM (class GAMMA) nor TcpC (class BETA) possesses the recently described binding pocket of VirB8-like class ALPHA proteins, recognized by VirB8 interaction inhibitors (69, 68). It is worth noting that the E. faecalis plasmid pCF10 harbors two VirB8-like proteins from different classes: PrgD resembles the GAMMA class composition, and PrgL is very similar to ALPHA class proteins.
DISCUSSION
Conjugative transfer greatly increases prokaryotic genome plasticity and has enormous importance in human health care as a major means of antibiotic resistance spread among pathogens and commensal bacteria (7). Consequently, the research field has attracted rising attention over the last decades.
The VirB/D4 T4SS from A. tumefaciens is the most investigated model system and has been studied since the late 1970s (70). Starting from this prototype of T-DNA transfer from A. tumefaciens to plant cells, much effort has been spent on the elucidation of other T4SS, originated from G− bacteria. Structural information has been obtained for individual transfer proteins, like TrwB (R388, VirD4 homolog; Protein Data Bank code 1E9S) (71) or TraC (pKM101, VirB5 homolog; Protein Data Bank code 1R8I) (72) from Escherichia coli, as well as VirB11 from B. suis (Protein Data Bank code 2GZA) (73) and Helicobactor pylori (Cag pathogenicity island; Protein Data Bank code 1NLZ) (74). In particular, the structure elucidation of the T4SS core complex of the pKM101 T4SS from E. coli by electron microscopy and crystallography (3) contributed to the understanding of the assembly and partial architecture of the conjugative transfer apparatus. Very recently, structural information has become available for the transfer proteins TcpC from C. perfringens (13) and VirB4 from Thermoanaerobacter pseudethanolicus (14). However, advances equal to those made with the G− T4SS have not been achieved for systems originating from G+ bacteria. Thus, the molecular mechanisms of DNA transfer in G+ bacteria remain largely unknown. The lack of knowledge is a particular matter of concern, because many severe human pathogens belong to this group of prokaryotes (75).
In this study, structural and biophysical approaches were used to characterize TraM, a putative transfer protein from the E. faecalis conjugative model plasmid pIP501. This task was especially demanding, because no sequence similarities to T4SS of G− origin have been detected. We showed that TraM is a membrane-associated protein (Fig. 1A). Surface accessibility of TraM was further confirmed, because we could demonstrate that only enterococci expressing the TraM protein were killed in opsonophagocytosis assays using anti-TraMΔ antibodies, whereas the isogenic strain not encoding TraM was completely resistant to killing. The specificity of the TraMΔ-mediated killing was confirmed, because increasing amounts of purified TraMΔ were able to inhibit the killing activity in a concentration-dependent manner. We conclude that these results identify TraM as an important part of the secretion apparatus, accessible from the cell exterior. This defines TraM as a highly interesting target for structure/function studies. In addition, the opsonophagocytosis assay may prove a valuable tool for all potential transfer proteins, for which no sequence similarities are found, and biochemical characterization does not provide evidence of their specific function in the respective T4SS. The use of opsonophagocytosis assays is a simple and elegant means to gain additional information on the surface exposure of target proteins, as well as a strong evidence for their actual involvement in the build-up of the T4S machinery.
The surprising structural similarities of TraM to transfer proteins originating from G− and G+ bacteria were revealed. The crystallized NTF2-like C-terminal domain of TraM is structurally similar to VirB8 from B. suis (Protein Data Bank code 2BHM) (63) and A. tumefaciens (2CC3) (64). In addition, we found a similar structure in the recently published transfer protein TcpC from C. perfringens plasmid pCW3 (13). Despite the very low sequence similarity, the overall structural features of the NTF2-like domains appear to be conserved. The structural similarities reinforce the prediction that TraM performs a key role in the secretion process, which is underlined by its surface accessibility. The data are particularly interesting in that, although VirB8 co-purifies with core complex components, the protein is not present in the actual core structure (3), and its function is still a matter of debate.
To obtain more information about the putative localization and function of the VirB8-like proteins, we compared the individual domain composition of available VirB8-like structures from G− and G+ bacteria; as displayed in Fig. 5, VirB8 of A. tumefaciens and B. suis consists of a small N-terminal domain, followed by a TM helix and the C-terminal NTF2-like domain. TcpC from C. perfringens exhibits a similar composition but features a second NTF2-like domain, separated from the central domain by a short linker region. Secondary structure prediction indicated that the putative transfer protein Orf13 from E. faecalis transposon Tn916 shares this arrangement. However, the overall composition of TraM differs significantly from other VirB8-like proteins. Unlike any known transfer protein that contains a NTF2-like domain, it possesses a large N-terminal domain, for which no potentially homologous structures were found. This domain is followed by the predicted transmembrane motif and the NTF2-like segment. The findings suggest that despite sharing a common domain architecture, the VirB8-like proteins possess a variable, modular domain composition. Based on this observation, we propose a new classification of VirB8-like proteins (Fig. 5); classic VirB8-like proteins contain only one NTF2-like domain (class ALPHA). Proteins with two succeeding NTF2-like domains, such as TcpC from C. perfringens, belong to class BETA. Class GAMMA proteins consist of a single, C-terminal NTF-like domain plus a large cytoplasmic N-terminal domain of yet unknown fold.
We propose that the significant variation between VirB8-like proteins of G− and G+ bacteria is due to the diverse composition of the cell envelopes. G− cell walls are comprised of two membranes, separated by a thin peptidoglycan layer. In contrast, G+ bacteria possess only a single membrane, coated by a thick peptidoglycan layer, which itself may be covered by a protein or glycoprotein outer layer (76). This difference in thickness and composition may have led either to the gradual adaptation of an ancestral set of proteins involved in conjugative transfer, as proposed by Gillespie et al. (77) for the Rickettsiales Vir homolog (rvh) T4SS, or to the co-evolutional development of proteins with a similar fold that serve an equivalent function, as suggested by Feldman et al. (78) for the Legionella pathogenesis system. Very recently the co-evolutional development of conjugative plasmid transfer was proposed in a broader sense by Harrison and Brockhurst (79). Another explanation for the distinction of the three protein classes may be the functional adaptation for the conjugation apparatus of G− and G+ bacteria, respectively.
TraMΔ and the central domain of TcpC both crystallized as trimers. The N-terminal helices in both structures are positioned in a way to allow a smooth transition into a triple coiled-coil motif, which was predicted for TraM (Fig. 4), as well as for TcpC between the TM helix and the C-terminal domain. MD simulations of the triple-helix motif indicate that the coiled-coil structure is stable under aqueous conditions. In contrast, we found that the TraMΔ construct, which contains only a part of the trimerization motif, behaves as a monomer in solution. In case of TcpC, the N-terminal deletion construct was also found to be a monomer in solution (13). This contradicts previous data by Parsons et al. (80) and Steen et al. (81), who suggested that the N-terminal part of TcpC serves as an oligomerization domain. We propose trimerization for both full-length proteins in vivo, based on the putative coiled-coil motifs.
The striking structural similarities between TraM, the VirB8 proteins of G− origin, and especially TcpC from C. perfringens suggest a similar function. A role as scaffolding factor for the assembly of the conjugative core complex has been proposed for VirB8 (66, 63–65) and TcpC (13). This suggestion was based on the interaction of the protein with other T4SS components. In the case of TcpC, the interactions were observed in bacterial two-hybrid studies (81) (to TcpA, TcpG, and TcpH). In the case of VirB8 from G− bacteria, mutational analyses and binding experiments (82, 83) (to VirB3 and VirB10, respectively), ELISA (67) (to VirB9 and VirB10) and cross-linking, pull-down, and FRET-based experiments (84) (to VirB5 and VirB6) were conducted. However, similar interactions with components of the pIP501 transfer system could not be detected for TraM in yeast two-hybrid and pull-down assays (10). Alternatively, because of its surface accessibility, TraM might provide an attachment site for the recipient cell during the conjugation process. As a third possibility, TraM might be involved in the morphogenesis of the actual T4SS core complex, although VirB8 was not found in the structure of the core complex of the pKM101 encoded T4SS (3). Nevertheless, the striking differences in the cell envelope of G− and G+ bacteria might explain the differences in the composition of VirB8-like proteins. Additionally, they could result in a distinct functional role ofVirB8-like proteins in G+ bacteria.
Despite the growing structural and functional information on T4SSs in general, further efforts are needed to confirm the function of VirB8-like proteins. We propose that the classification of VirB8-like proteins will help to elucidate structure-function relationships in T4SSs of G+ bacteria, by providing insights in the ubiquity and structural adaptation among conjugative transfer proteins.
Acknowledgments
We gratefully acknowledge the staff at the SLS Synchrotron X06DA Beamline and the DESY X33 SAXS Beamline for support during data collection. We thank Ellen Zechner for critical reading of the manuscript and for valuable comments.
This work was supported by Austrian Science Fund Projects P19794 and F4604.

This article contains supplemental Tables S1–S3 and Figs. S1–S4.
E.-K. Çelik, W. Keller, and E. Grohmann, unpublished data.
K. Arends, W. Keller, and E. Grohmann, unpublished data.
N. Goessweiner-Mohr, L. Grumet, T. Pavkov-Keller, R. Birner-Gruenberger, E. Grohmann, and W. Keller, submitted for publication.
- T4SS
- type IV secretion system
- G+
- Gram-positive
- G−
- Gram-negative
- Bicine
- N,N-bis(2-hydroxyethyl)glycine
- ICE
- integrative conjugative element
- RMSD
- root mean square deviation
- NTF
- nuclear transport factor.
REFERENCES
- 1. Williams J. J., Hergenrother P. J. (2008) Exposing plasmids as the Achilles' heel of drug-resistant bacteria. Curr. Opin. Chem. Biol. 12, 389–399 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Llosa M., Gomis-Rüth F. X., Coll M., de la Cruz F. (2002) Bacterial conjugation. A two-step mechanism for DNA transport. Mol. Microbiol. 45, 1–8 [DOI] [PubMed] [Google Scholar]
- 3. Chandran V., Fronzes R., Duquerroy S., Cronin N., Navaza J., Waksman G. (2009) Structure of the outer membrane complex of a type IV secretion system. Nature 462, 1011–1015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Smillie C., Garcillán-Barcia M. P., Francia M. V., Rocha E. P., de la Cruz F. (2010) Mobility of plasmids. Microbiol. Mol. Biol. Rev. 74, 434–452 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Wallden K., Rivera-Calzada A., Waksman G. (2010) Microreview. Type IV secretion systems. Versatility and diversity in function. Cell Microbiol. 12, 1203–1212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Thanassi D. G., Bliska J. B., Christie P. J. (2012) Surface organelles assembled by secretion systems of Gram-negative bacteria. Diversity in structure and function. FEMS Microbiol. Rev. 36, 1046–1082 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Zechner E. L., Lang S., Schildbach J. F. (2012) Assembly and mechanisms of bacterial type IV secretion machines. Philos. Trans. R. Soc. Lond. B Biol. Sci. 367, 1073–1087 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Alvarez-Martinez C. E., Christie P. J. (2009) Biological diversity of prokaryotic type IV secretion systems. Microbiol. Mol. Biol. Rev. 73, 775–808 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Grohmann E., Muth G., Espinosa M. (2003) Conjugative plasmid transfer in Gram-positive bacteria. Microbiol. Mol. Biol. Rev. 67, 277–301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Abajy M. Y., Kopeć J., Schiwon K., Burzynski M., Döring M., Bohn C., Grohmann E. (2007) A type IV-secretion-like system is required for conjugative DNA transport of broad-host-range plasmid pIP501 in Gram-positive bacteria. J. Bacteriol. 189, 2487–2496 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Chen Y., Zhang X., Manias D., Yeo H.-J., Dunny G. M., Christie P. J. (2008) Enterococcus faecalis PcfC, a spatially localized substrate receptor for type IV secretion of the pCF10 transfer intermediate. J. Bacteriol. 190, 3632–3645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Li M., Shen X., Yan J., Han H., Zheng B., Liu D., Cheng H., Zhao Y., Rao X., Wang C., Tang J., Hu F., Gao G. F. (2011) GI-type T4SS-mediated horizontal transfer of the 89K pathogenicity island in epidemic Streptococcus suis serotype 2. Mol. Microbiol. 79, 1670–1683 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Porter C. J., Bantwal R., Bannam T. L., Rosado C. J., Pearce M. C., Adams V., Lyras D., Whisstock J. C., Rood J. I. (2012) The conjugation protein TcpC from Clostridium perfringens is structurally related to the type IV secretion system protein VirB8 from Gram-negative bacteria. Mol. Microbiol. 83, 275–288 [DOI] [PubMed] [Google Scholar]
- 14. Walldén K., Williams R., Yan J., Lian P. W., Wang L., Thalassinos K., Orlova E. V., Waksman G. (2012) Structure of the VirB4 ATPase, alone and bound to the core complex of a type IV secretion system. Proc. Natl. Acad. Sci. U.S.A. 109, 11348–11353 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Horodniceanu T., Bougueleret L., El-Solh N., Bouanchaud D. H., Chabbert Y. A. (1979) Conjugative R plasmids in Streptococcus agalactiae (group B). Plasmid 2, 197–206 [DOI] [PubMed] [Google Scholar]
- 16. Kurenbach B., Bohn C., Prabhu J., Abudukerim M., Szewzyk U., Grohmann E. (2003) Intergeneric transfer of the Enterococcus faecalis plasmid pIP501 to Escherichia coli and Streptomyces lividans and sequence analysis of its tra region. Plasmid. 50, 86–93 [DOI] [PubMed] [Google Scholar]
- 17. Atmakuri K., Cascales E., Christie P. J. (2004) Energetic components VirD4, VirB11 and VirB4 mediate early DNA transfer reactions required for bacterial type IV secretion. Mol. Microbiol. 54, 1199–1211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Gomis-Rüth F. X., Solà M., de la Cruz F., Coll M. (2004) Coupling factors in macromolecular type IV secretion machineries. Curr. Pharm. Des. 10, 1551–1565 [DOI] [PubMed] [Google Scholar]
- 19. Kopec J., Bergmann A., Fritz G., Grohmann E., Keller W. (2005) TraA and its N-terminal relaxase domain of the Gram-positive plasmid pIP501 show specific oriT binding and behave as dimers in solution. Biochem. J. 387, 401–409 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Kurenbach B., Kopeć J., Mägdefrau M., Andreas K., Keller W., Bohn C., Abajy M. Y., Grohmann E. (2006) The TraA relaxase autoregulates the putative type IV secretion-like system encoded by the broad-host-range Streptococcus agalactiae plasmid pIP501. Microbiology 152, 637–645 [DOI] [PubMed] [Google Scholar]
- 21. Buttaro B. A., Antiporta M. H., Dunny G. M. (2000) Cell-associated pheromone peptide (cCF10) production and pheromone inhibition in E. faecalis. J. Bacteriol. 182, 4926–4933 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Theilacker C., Kropec A., Hammer F., Sava I., Wobser D., Sakinc T., Codée J. D., Hogendorf W. F., van der Marel G. A., Huebner J. (2012) Protection against Staphylococcus aureus by antibody to the polyglycerolphosphate backbone of heterologous lipoteichoic acid. J. Infect. Dis. 205, 1076–1085 [DOI] [PubMed] [Google Scholar]
- 23. Emsley P., Lohkamp B., Scott W. G., Cowtan K. (2010) Features and development of Coot. Acta Crystallogr. D Biol. Crystallogr. 66, 486–501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Murshudov G. N., Vagin A. A., Dodson E. J. (1997) Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr. D Biol. Crystallogr. 53, 240–255 [DOI] [PubMed] [Google Scholar]
- 25. Chen V. B., Arendall W. B., 3rd, Headd J. J., Keedy D. A., Immormino R. M., Kapral G. J., Murray L. W., Richardson J. S., Richardson D. C. (2010) MolProbity. Acta Crystallogr. D Biol. Crystallogr. 66, 12–21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Heinig M., Frishman D. (2004) STRIDE. A web server for secondary structure assignment from known atomic coordinates of proteins. Nucleic Acids Res. 32, W500-W502 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Holm L., Rosenström P. (2010) Dali server. Conservation mapping in 3D. Nucleic Acids Res. 38, W545–W549 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Kawabata T. (2003) MATRAS. A program for protein 3D structure comparison. Nucleic Acids Res. 31, 3367–3369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Dror O., Benyamini H., Nussinov R., Wolfson H. (2003) MASS. Multiple structural alignment by secondary structures. Bioinformatics. 19, i95–i104 [DOI] [PubMed] [Google Scholar]
- 30. Schrödinger L. L. (2011) The PyMOL Molecular Graphics System, version 1.3 [Google Scholar]
- 31. Krissinel E., Henrick K. (2007) Inference of macromolecular assemblies from crystalline state. J. Mol. Biol. 372, 774–797 [DOI] [PubMed] [Google Scholar]
- 32. Krissinel E., Henrick K. (2004) Secondary-structure matching (SSM), a new tool for fast protein structure alignment in three dimensions. Acta Crystallogr. D Biol. Crystallogr. 60, 2256–2268 [DOI] [PubMed] [Google Scholar]
- 33. Hofmann K., Stoffel W. (1993) TMBASE. A database of membrane spanning protein segments. Biol. Chem. Hoppe-Seyler 374, 166 [Google Scholar]
- 34. Rost B., Sander C. (1994) Combining evolutionary information and neural networks to predict protein secondary structure. Proteins. 19, 55–72 [DOI] [PubMed] [Google Scholar]
- 35. Tusnády G. E., Simon I. (2001) The HMMTOP transmembrane topology prediction server. Bioinformatics 17, 849–850 [DOI] [PubMed] [Google Scholar]
- 36. Krogh A., Larsson B., von Heijne G., Sonnhammer E. L. (2001) Predicting transmembrane protein topology with a hidden markov model. Application to complete genomes. J. Mol. Biol. 305, 567–580 [DOI] [PubMed] [Google Scholar]
- 37. Hirokawa T., Boon-Chieng S., Mitaku S. (1998) SOSUI. Classification and secondary structure prediction system for membrane proteins. Bioinformatics. 14, 378–379 [DOI] [PubMed] [Google Scholar]
- 38. Nugent T., Jones D. T. (2009) Transmembrane protein topology prediction using support vector machines. BMC Bioinformatics 10, 159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Jones D. T., Taylor W. R., Thornton J. M. (1994) A model recognition approach to the prediction of all-helical membrane protein structure and topology. Biochemistry 33, 3038–3049 [DOI] [PubMed] [Google Scholar]
- 40. Shen H., Chou J. J. (2008) MemBrain. Improving the accuracy of predicting transmembrane helices. PLoS ONE. 3, e2399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Jones D. T. (1999) Protein secondary structure prediction based on position-specific scoring matrices1. J. Mol. Biol. 292, 195–202 [DOI] [PubMed] [Google Scholar]
- 42. Gasteiger E., Gattiker A., Hoogland C., Ivanyi I., Appel R. D., Bairoch A. (2003) ExPASy. The proteomics server for in-depth protein knowledge and analysis. Nucleic Acids Res. 31, 3784–3788 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Lupas A., van Dyke M., Stock J. (1991) Predicting coiled coils from protein sequences. Science 252, 1162–1164 [DOI] [PubMed] [Google Scholar]
- 44. Wolf E., Kim P. S., Berger B. (1997) MultiCoil. A program for predicting two- and three-stranded coiled coils. Protein Sci. 6, 1179–1189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. McDonnell A. V., Jiang T., Keating A. E., Berger B. (2006) Paircoil2. Improved prediction of coiled coils from sequence. Bioinformatics. 22, 356–358 [DOI] [PubMed] [Google Scholar]
- 46. Söding J., Biegert A., Lupas A. N. (2005) The HHpred interactive server for protein homology detection and structure prediction. Nucleic Acids Res. 33, W244–W248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Biegert A., Mayer C., Remmert M., Söding J., Lupas A. N. (2006) The MPI Bioinformatics Toolkit for protein sequence analysis. Nucleic Acids Res. 34, W335–W339 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Eswar N., Webb B., Marti-Renom M. A., Madhusudhan M., Eramian D., Shen M.-Y., Pieper U., Sali A. (2006) UNIT 2.9 Comparative Protein Structure Modeling Using MODELLER Wiley, Weinheim, Germany: [DOI] [PubMed] [Google Scholar]
- 49. Hess B., Kutzner C., van der Spoel D., Lindahl E. (2008) GROMACS 4. Algorithms for highly efficient, load-balanced, and scalable molecular simulation. J. Chem. Theory Comput. 4, 435–447 [DOI] [PubMed] [Google Scholar]
- 50. Petersen M. T., Martel P., Petersen E. I., Drabløs F., Petersen S. B. (1997) Surface and electrostatics of cutinases. Methods Enzymol. 284, 130–154 [DOI] [PubMed] [Google Scholar]
- 51. Price D. J., Brooks C. L. (2004) A modified TIP3P water potential for simulation with Ewald summation. J. Chem. Phys. 121, 10096–10103 [DOI] [PubMed] [Google Scholar]
- 52. Darden T., York D., Pedersen L. (1993) Particle mesh Ewald. An N-log(N) method for Ewald sums in large systems. J. Chem. Phys. 98, 10089–10092 [Google Scholar]
- 53. Hess B., Bekker H., Berendsen H. J., Fraaije J. G. (1997) LINCS. A linear constraint solver for molecular simulations. J. Comput. Chem. 18, 1463–1472 [Google Scholar]
- 54. Parrinello M. (1981) Polymorphic transitions in single crystals. A new molecular dynamics method. J. Appl. Phys. 52, 7182 [Google Scholar]
- 55. Nosé S., Klein M. (1983) Constant pressure molecular dynamics for molecular systems. Mol. Phys. 50, 1055–1076 [Google Scholar]
- 56. Bussi G., Donadio D., Parrinello M. (2007) Canonical sampling through velocity rescaling. J. Chem. Phys. 126, 14101–14107 [DOI] [PubMed] [Google Scholar]
- 57. Anézo C., Vries A. H., de, Höltje H.-D., Tieleman D. P., Marrink S.-J. (2003) Methodological Issues in Lipid Bilayer Simulations. J. Phys. Chem. B. 107, 9424–9433 [Google Scholar]
- 58. Humphrey W., Dalke A., Schulten K. (1996) VMD. Visual molecular dynamics. J. Mol. Graph. 14, 33–38 [DOI] [PubMed] [Google Scholar]
- 59. Krieger E., Koraimann G., Vriend G. (2002) Increasing the precision of comparative models with YASARA NOVA: A self-parameterizing force field. Proteins. 47, 393–402 [DOI] [PubMed] [Google Scholar]
- 60. Duan Y., Wu C., Chowdhury S., Lee M. C., Xiong G., Zhang W., Yang R., Cieplak P., Luo R., Lee T., Caldwell J., Wang J., Kollman P. (2003) A point-charge force field for molecular mechanics simulations of proteins based on condensed-phase quantum mechanical calculations. J. Comput. Chem. 24, 1999–2012 [DOI] [PubMed] [Google Scholar]
- 61. Sorin E. J., Pande V. S. (2005) Exploring the helix-coil transition via all-atom equilibrium ensemble simulations. Biophys. J. 88, 2472–2493 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Krissinel E. (2010) Crystal contacts as nature's docking solutions. J. Comput. Chem. 31, 133–143 [DOI] [PubMed] [Google Scholar]
- 63. Terradot L., Bayliss R., Oomen C., Leonard G. A., Baron C., Waksman G. (2005) Structures of two core subunits of the bacterial type IV secretion system, VirB8 from Brucella suis and ComB10 from Helicobacter pylori. Proc. Natl. Acad. Sci. U.S.A. 102, 4596–4601 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Bailey S., Ward D., Middleton R., Grossmann J. G., Zambryski P. C. (2006) Agrobacterium tumefaciens VirB8 structure reveals potential protein-protein interaction sites. Proc. Natl. Acad. Sci. U.S.A. 103, 2582–2587 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Baron C. (2006) VirB8. A conserved type IV secretion system assembly factor and drug target. Biochem. Cell Biol. 84, 890–899 [DOI] [PubMed] [Google Scholar]
- 66. Kumar R. B., Xie Y.-H., Das A. (2000) Subcellular localization of the Agrobacterium tumefaciens T-DNA transport pore proteins. VirB8 is essential for the assembly of the transport pore. Mol. Microbiol. 36, 608–617 [DOI] [PubMed] [Google Scholar]
- 67. Sivanesan D., Hancock M. A., Villamil Giraldo A. M., Baron C. (2010) Quantitative analysis of VirB8-VirB9-VirB10 interactions provides a dynamic model of type IV secretion system core complex assembly. Biochemistry 49, 4483–4493 [DOI] [PubMed] [Google Scholar]
- 68. Cameron T. A., Zambryski P. C. (2012) Disarming bacterial type IV secretion. Chem. Biol. 19, 934–936 [DOI] [PubMed] [Google Scholar]
- 69. Smith M. A., Coinçon M., Paschos A., Jolicoeur B., Lavallée P., Sygusch J., Baron C. (2012) Identification of the binding site of Brucella VirB8 interaction inhibitors. Chem. Biol. 19, 1041–1048 [DOI] [PubMed] [Google Scholar]
- 70. Gurley W. B., Kemp J. D., Albert M. J., Sutton D. W., Callis J. (1979) Transcription of TI plasmid-derived sequences in three octopine-type crown gall tumor lines. Proc. Natl. Acad. Sci. U.S.A. 76, 2828–2832 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Gomis-Rüth F. X., Moncalián G., Pérez-Luque R., González A., Cabezón E., de la Cruz F., Coll M. (2001) The bacterial conjugation protein TrwB resembles ring helicases and F1-ATPase. Nature 409, 637–641 [DOI] [PubMed] [Google Scholar]
- 72. Yeo H.-J., Yuan Q., Beck M. R., Baron C., Waksman G. (2003) Structural and functional characterization of the VirB5 protein from the type IV secretion system encoded by the conjugative plasmid pKM101. Proc. Natl. Acad. Sci. U.S.A. 100, 15947–15952 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Hare S., Bayliss R., Baron C., Waksman G. (2006) A large domain swap in the VirB11 ATPase of Brucella suis leaves the hexameric assembly intact. J. Mol. Biol. 360, 56–66 [DOI] [PubMed] [Google Scholar]
- 74. Savvides S. N., Yeo H. J., Beck M. R., Blaesing F., Lurz R., Lanka E., Buhrdorf R., Fischer W., Haas R., Waksman G. (2003) VirB11 ATPases are dynamic hexameric assemblies. New insights into bacterial type IV secretion. EMBO J. 22, 1969–1980 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Burns D. (2003) Type IV transporters of pathogenic bacteria. Curr. Opin. Microbiol. 6, 29–34 [DOI] [PubMed] [Google Scholar]
- 76. Vollmer W., Seligman S. J. (2010) Architecture of peptidoglycan. More data and more models. Trends Microbiol. 18, 59–66 [DOI] [PubMed] [Google Scholar]
- 77. Gillespie J. J., Brayton K. A., Williams K. P., Diaz M. A., Brown W. C., Azad A. F., Sobral B. W. (2010) Phylogenomics reveals a diverse Rickettsiales type IV secretion system. Infect. Immun. 78, 1809–1823 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Feldman M., Zusman T., Hagag S., Segal G. (2005) Coevolution between nonhomologous but functionally similar proteins and their conserved partners in the Legionella pathogenesis system. Proc. Natl. Acad. Sci. U.S.A. 102, 12206–12211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Harrison E., Brockhurst M. A. (2012) Plasmid-mediated horizontal gene transfer is a coevolutionary process. Trends Microbiol. 20, 262–267 [DOI] [PubMed] [Google Scholar]
- 80. Parsons J. A., Bannam T. L., Devenish R. J., Rood J. I. (2007) TcpA, an FtsK/SpoIIIE homolog, is essential for transfer of the conjugative plasmid pCW3 in Clostridium perfringens. J. Bacteriol. 189, 7782–7790 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Steen J. A., Bannam T. L., Teng W. L., Devenish R. J., Rood J. I. (2009) The putative coupling protein TcpA interacts with other pCW3-encoded proteins to form an essential part of the conjugation complex. J. Bacteriol. 191, 2926–2933 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Mossey P., Hudacek A., Das A. (2010) Agrobacterium tumefaciens type IV secretion protein VirB3 is an inner membrane protein and requires VirB4, VirB7, and VirB8 for stabilization. J. Bacteriol. 192, 2830–2838 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Andrieux L., Bourg G., Pirone A., O'Callaghan D., Patey G. (2011) A single amino acid change in the transmembrane domain of the VirB8 protein affects dimerization, interaction with VirB10 and Brucella suis virulence. FEBS Lett. 585, 2431–2436 [DOI] [PubMed] [Google Scholar]
- 84. Villamil Giraldo A. M., Sivanesan D., Carle A., Paschos A., Smith M. A., Plesa M., Coulton J., Baron C. (2012) Type IV secretion system core component VirB8 from Brucella binds to the globular domain of VirB5 and to a periplasmic domain of VirB6. Biochemistry 51, 3881–3890 [DOI] [PubMed] [Google Scholar]