Background: PEP-19 modulates the kinetics of Ca2+ binding to CaM.
Results: An acidic region in PEP-19 binds Ca2+ and is essential for both modulating Ca2+ binding to CaM and sensitizing cells to ATP-induced Ca2+ release.
Conclusion: Simply binding to CaM is not sufficient to account for the biological activities of PEP-19.
Significance: Regulating ligand-induced Ca2+ release gives PEP-19 the potential to broadly affect cell signaling.
Keywords: Calcium Signaling, Calmodulin, Intrinsically Disordered Proteins, NMR, Purinergic Receptor, IQ Motif, PEP-19, Ligand-induced Calcium Release
Abstract
PEP-19 is a small, intrinsically disordered protein that binds to the C-domain of calmodulin (CaM) via an IQ motif and tunes its Ca2+ binding properties via an acidic sequence. We show here that the acidic sequence of PEP-19 has intrinsic Ca2+ binding activity, which may modulate Ca2+ binding to CaM by stabilizing an initial Ca2+-CaM complex or by electrostatically steering Ca2+ to and from CaM. Because PEP-19 is expressed in cells that exhibit highly active Ca2+ dynamics, we tested the hypothesis that it influences ligand-dependent Ca2+ release. We show that PEP-19 increases the sensitivity of HeLa cells to ATP-induced Ca2+ release to greatly increase the percentage of cells responding to sub-saturating doses of ATP and increases the frequency of Ca2+ oscillations. Mutations in the acidic sequence of PEP-19 that inhibit or prevent it from modulating Ca2+ binding to CaM greatly inhibit its effect on ATP-induced Ca2+ release. Thus, this cellular effect of PEP-19 does not depend simply on binding to CaM via the IQ motif but requires its acidic metal binding domain. Tuning the activities of Ca2+ mobilization pathways places PEP-19 at the top of CaM signaling cascades, with great potential to exert broad effects on downstream CaM targets, thus expanding the biological significance of this small regulator of CaM signaling.
Introduction
PEP-19 (Purkinje cell protein 4, Pcp4) is a small protein (62 amino acids) with no known intrinsic activity other than binding to CaM2 in the presence or absence of Ca2+. Although it was originally identified in the central nervous system, PEP-19 mRNA is also found in human bladder, kidney, prostate, uterus, thyroid, and adrenal tissues (1). Changes in expression levels suggest biological roles for PEP-19 in both normal and pathological conditions. For example, PEP-19 mRNA levels are significantly reduced in a mouse model for Parkinson disease (2) and in the prefrontal cortex of alcoholics (3), but its levels are increased in anergic B cells (4) and in human uterine leiomyomas (5).
Animal and cellular model systems have demonstrated effects of PEP-19 on diverse cellular processes. PEP-19 null mice show a dramatic reduction in long term plasticity at synapses between granule cell parallel fibers and Purkinje cells (6). Overexpression of PEP-19 in PC12 cells increases neurite outgrowth (7), and premature neuronal differentiation is seen in transgenic mice with three copies of the PEP-19 gene (Pcp4) (8). The latter suggests a role for PEP-19 in Down syndrome because the human PEP-19 gene (PCP4) is present on chromosome 21. In addition, PEP-19 has anti-apoptotic activity when expressed in PC12 and HEK293T cells (9, 10), and it provides protection against Ca2+ overload in cortical neurons (10). These experimental observations are consistent with a proposed neuroprotective role for PEP-19 based on expression patterns in neuronal tissues that are susceptible to Huntington and Alzheimer diseases (11).
The above studies emphasize the need to understand the mechanism of action of PEP-19. Two models for PEP-19 have been proposed based on studies using peptides and the homologous proteins neurogranin (Ng) and neuromodulin (12–14). The first, or camstatin model, proposes that PEP-19 competitively inhibits activation of CaM target proteins. The second, or calpacitin model, proposes that PEP-19 binds with high affinity to apo-CaM to retard its release from PEP-19, thereby affecting the temporal profile of available CaM during a Ca2+ pulse. We proposed an alternative or additional mechanism for PEP-19 based on its ability to modulate the Ca2+ binding properties of CaM. Specifically, PEP-19 increases both the Ca2+ kon and koff rates at the C-domain of CaM up to 40-fold with little effect on the KCa (15). We also showed that an acidic sequence located adjacent to the IQ motif is required to modulate Ca2+ binding to the C-domain of CaM, even though it has no apparent intrinsic affinity for CaM (16). Thus, the acidic/IQ motif of PEP-19 has the potential to modulate the rate-limiting kinetics of Ca2+ binding to CaM.
This study investigates the molecular mechanism by which PEP-19 modulates Ca2+ binding to CaM, and it tests the hypothesis that the biological activities of PEP-19 rely on synergy between the biochemical properties of its acidic and IQ sequences. Our results show that the acidic sequence in PEP-19 has intrinsic metal binding properties that play a role in increasing the rates of Ca2+ binding to CaM, at least in part, by electrostatically steering Ca2+ to and from Ca2+ binding sites III and/or IV. We also show that PEP-19 sensitizes HeLa cells to ATP-dependent Ca2+ release and that this effect is greatly reduced or eliminated by mutations in PEP-19 that inhibit or eliminate its ability to modulate Ca2+ binding to CaM. Tuning the activities of Ca2+ mobilization pathways by PEP-19 greatly expands the biological significance of this small regulator of CaM signaling.
EXPERIMENTAL PROCEDURES
Mutagenesis and Protein Purification
QuikChange II XL site-directed mutagenesis kit (Stratagene) was used to generate a panel of PEP-19 mutants. CaM and C-CaM (isolated C domain of calmodulin) were decalcified by addition of 5 mm EDTA and 0.1 mm BAPTA as a UV marker and then desalting on a Bio-Gel P2 column (Bio-Rad) in 10 mm NH4HCO3 that had been decalcified using a Ca2+ sponge column (Molecular Probes). Decalcified proteins were then lyophilized and resuspended in desired buffers. Protein concentrations were estimated using an extinction coefficient of ϵ276 nm = 0.18 ml−1 mg−1 for C-CaM and ϵ215 nm = 0.59 ml−1 mg−1 for PEP-19.
Ca2+ Binding Measurements
The rate of Ca2+ dissociation (koff) from CaM or C-CaM in the presence or absence of PEP-19 derivatives was determined using stopped-flow fluorescence and the Ca2+-sensitive dye Quin-2 as described previously (15). Typically, solutions of 2–5 μm CaM or C-CaM in 20 mm MOPS, pH 7.5, 100 mm KCl, 30 μm CaCl2 were rapidly mixed with 20 mm MOPS, pH 7.5, 300 μm Quin-2. Excess free Ca2+ and Ca2+ that is rapidly released from the N-domain of CaM bind to Quin-2 in the 1.7-ms dead time of the stopped-flow instrument. The subsequent increase in Quin-2 fluorescence is due to binding Ca2+ released slowly from the C-domain. Experiments were performed at 23 °C using an Applied Photophysics Ltd. (Leatherhead, UK) model SX20 MV sequential stopped-flow spectrofluorimeter with a 150 watt Xe/Hg lamp.
Equilibrium Ca2+ binding constants for CaM in the presence or absence of PEP-19 derivatives were determined using tyrosine fluorescence at 23 °C as described previously (17). Data were collected with a QuantaMaster fluorimeter (Photo Technology International). Intrinsic Tyr emission spectra were recorded from 290 to 320 nm with the excitation wavelength of 276 nm. Solutions contained 20 mm MOPS, pH 7.5, 0 or 100 mm KCl, 1 mm EGTA, 1 mm HEDTA, 1 mm nitrilo-2,2′,2′-triacetic acid, 5 μm CaM or C-CaM with or without PEP-19 or its derivatives. Calcium was added from a concentrated stock prepared in the same buffer with CaM, PEP-19, and chelators, so that only the concentration of Ca2+ changes during the titration even though the volume increases. The concentration of total Ca2+ needed to achieve a desired free Ca2+ concentration was determined using the on-line calculator MaxChelator. Control titrations were performed using Br2BAPTA as an indicator instead of CaM or C-CaM to confirm that the calculated free Ca2+ was accurate at high and low ionic strength. The KCa for Br2BAPTA is 1.59 μm at 100 mm KCl and 0.15 μm at 10 mm KCl (18).
Tyrosine fluorescence intensity was plotted against the free Ca2+ concentration and fit to the following form of the Hill Equation 1,
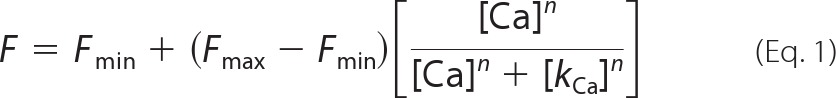 |
where [Ca2+] is the free Ca2+ concentration; F is the fluorescence intensity at a given free Ca2+ concentration; Fmin is the initial fluorescence intensity in the absence of added Ca2+; Fmax is the fluorescence at maximal Ca2+; KCa is the concentration of Ca2+ at which the change in fluorescence is half-maximal, and n is the Hill coefficient.
NMR Methodology
NMR experiments were performed on a Bruker DRX 600 MHz spectrometer equipped with a 5-mm triple resonance cryoprobe at 298 K. Protein samples were dissolved in buffer containing 10 mm imidazole, 5% D2O (v/v), pH 6.3, 100 mm KCl. 1H,15N HSQC spectra were used to determine residues in PEP-19 that are affected by binding to C-CaM. Briefly, 1H,15N HSQC spectra were collected during titration of 15N-labeled PEP-19 with C-CaM in the presence or absence of Ca2+. Characteristics of fast exchange were seen at saturating Ca2+, so backbone amides could be assigned by following cross-peaks during the titration. Slow exchange was seen in the apo-state, so assignments in the bound state were made using HNCO, HNCA, HN(CO)CA, HNCACB, CBCA(CO)NH, 15N HSQC-TOCSY, and 15N-edited NOESY-HSQC experiments. All NMR spectra were processed and analyzed using Topspin 2.0 (Bruker) and FELIX 2004 (MSI, San Diego). 1H chemical shifts were referenced to 2,2-dimethyl-2-silapentane-5-sulfonate, and 15N/13C chemical shifts were referenced indirectly using their respective gyromagnetic ratios. The average amide chemical shift change was calculated using Equation 2,
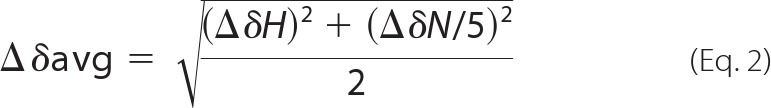 |
where ΔδH is the change in 1H chemical shift and ΔδN is the change in 15N chemical shift.
Calcium Imaging
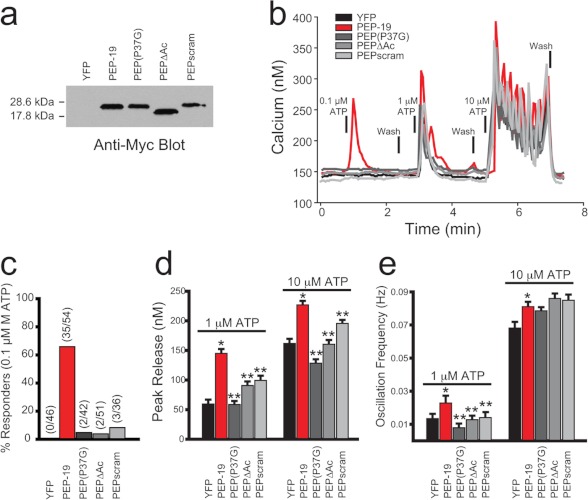
Calcium imaging was performed exactly as described previously (19). HeLa cells were transfected with yellow fluorescent protein (YFP) only (control) or co-transfected with YFP and PEP-19 constructs at a DNA ratio of 1:4 using Lipofectamine 2000. Twenty four hours after transfection, single cell calcium responses evoked by NaATP were recorded from all YFP-positive cells in a given field. All experiments were repeated at least three times, and the data were pooled for statistical analysis. The actual number of single cell records averaged for each condition is indicated above the bars in Fig. 6c.
FIGURE 6.
Effect of PEP-19 and its derivatives on ATP-induced intracellular Ca2+ release. Western blots in a show the relative level of expression of Myc-tagged PEP-19 and mutant proteins in HeLa cells that were transiently transfected with the corresponding expression plasmids. b shows intracellular Ca2+ levels in single cells in response to increasing concentrations of ATP. c shows the percentage of cells from each group that showed increased intracellular Ca2+ in response to 0.1 μm ATP. d and e show the effect of PEP-19 and mutated derivatives on peak Ca2+ levels and Ca2+ oscillation frequency, respectively, in response to 1 μm and 10 μm ATP. Data in d and e are represented as mean ± S.E. Statistical significance was determined with a Student's t test with * indicating p < 0.05 versus YFP and ** indicating p < 0.05 versus PEP-19.
RESULTS
PEP-19 Proteins Generated for Study
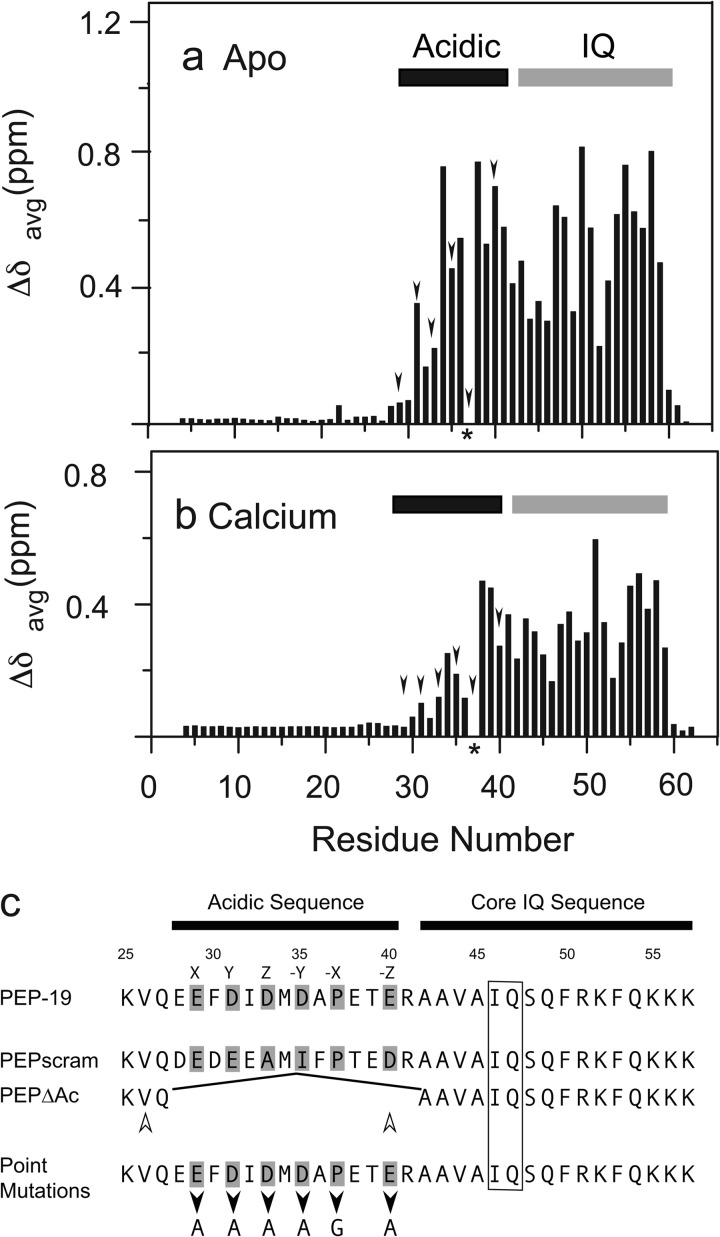
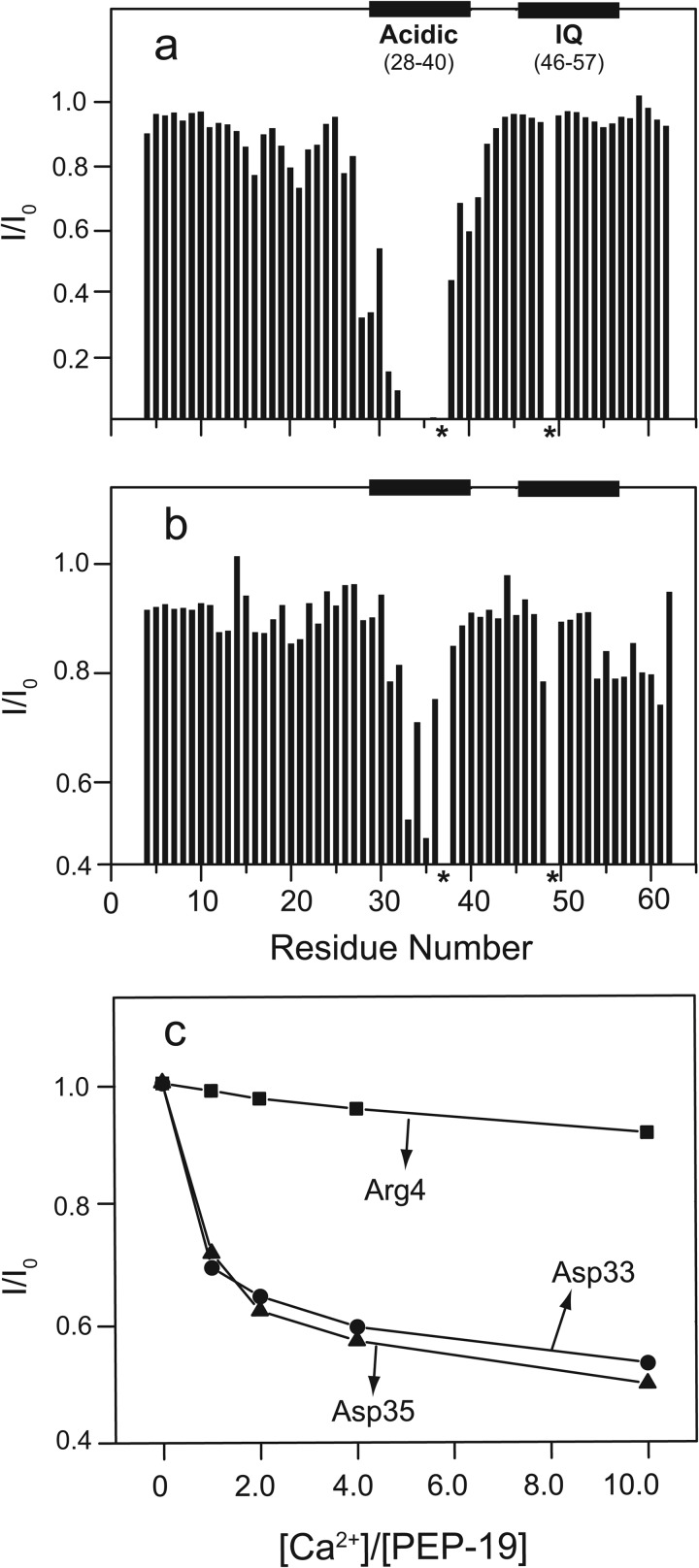
We used amide chemical shift perturbation to identify residues in PEP-19 that experience significant structural transitions upon binding to the C-domain of CaM because these residues will likely play key roles in regulating Ca2+ binding to CaM. C-CaM, which encodes residues 76–148 of CaM, was used for these experiments because we showed previously that PEP-19 binds to C-CaM and had the same effects on its Ca2+-binding proteins as seen for full-length CaM (20). Fig. 1, a and b, shows that backbone amide chemical shifts for residues 1–30 in PEP-19 are unchanged upon binding to C-CaM in the absence or presence of Ca2+. Because free PEP-19 is intrinsically disordered (21), these data show that residues 1–30 remain disordered when bound to C-CaM. Amide chemical shift perturbations are restricted to residues in the acidic/IQ region of PEP-19 upon binding to either apo- or Ca2+-C-CaM.
FIGURE 1.
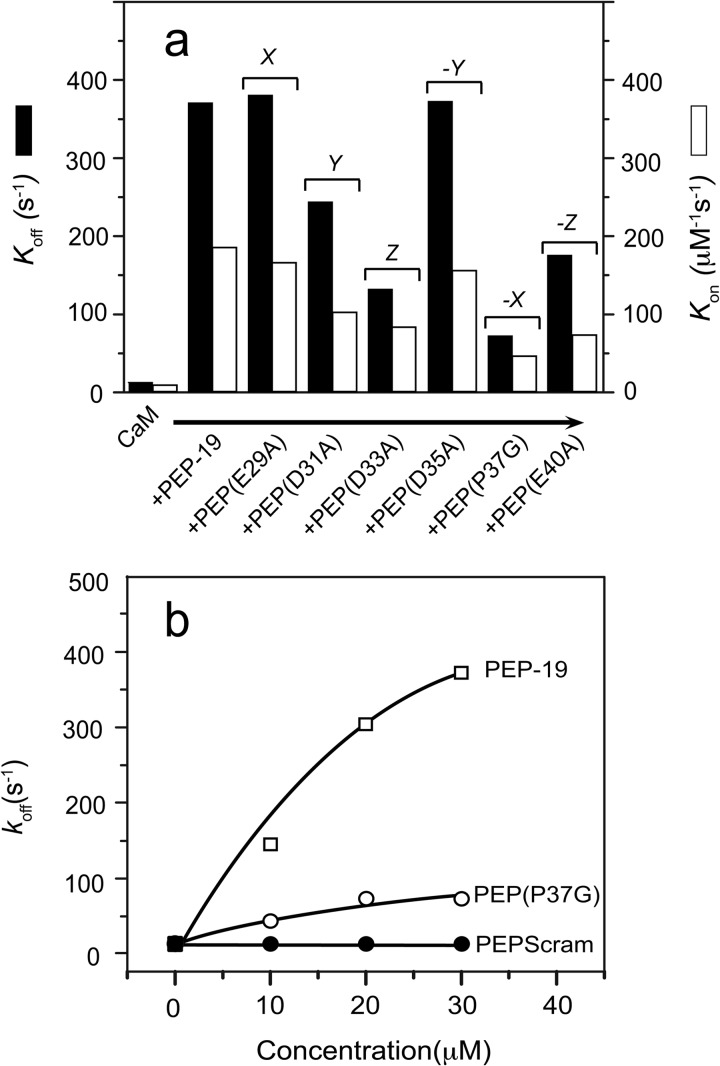
Amide chemical shift changes in PEP-19 upon binding to C-CaM are restricted to the acidic/IQ region. Average amide chemical shift perturbations were determined for residues in 15N-labeled PEP-19 caused by binding to either apo (a) or Ca2+-bound (b) C-CaM as described under “Experimental Procedures.” The arrowheads indicate acidic residues that could potentially bind Ca2+. The asterisk indicates Pro-37, which does not have a backbone amide. The core IQ sequence shown in c is based on comparison of multiple IQ motif proteins. Residues denoted by X, Y, Z, and −Y, −X, and −Z in native PEP-19 correspond to potential Ca2+ ligand positions based on a canonical EF-hand Ca2+-binding motif. PEPΔAc was generated by deletion of residues Glu-28 to Arg-41. The open arrowheads indicate that Val-26 is shifted to the position of Glu-40 in PEPΔAc. Residues 28–40 were randomly scrambled in PEPscram while retaining Pro-37. Black arrowheads indicate positions of point mutations.
Based on the above chemical shift perturbations, two sets of proteins were generated to test the biochemical and functional significance of the acidic sequence in PEP-19 (see Fig. 1c). The acidic sequence is deleted in PEPΔAc such that Val-26 effectively substitutes for Glu-40 of native PEP-19. We anticipated that a hydrophobic residue at this position would promote association of PEPΔAc with both the N- and C-domains of CaM because a Phe residue at the homologous position in the CaV1.2 channel anchors its IQ region to the N-domain (22). Residues in the acidic sequence of PEPscram are randomized to determine whether the native sequence is important for modulating Ca2+ binding to CaM or whether a cluster of negative charges is sufficient.
The second set of proteins was designed to test the functional significance of sequence similarity between the acidic region of PEP-19 and the consensus EF-hand Ca2+-binding site where alternating residues provide oxygens to coordinate Ca2+ at X, Y, Z and −Y,−X, and −Z positions (see Fig. 1c). Thus, Ala was substituted individually for Glu-29, Asp-31, Asp-33, Asp-35, or Glu-40. In addition, Pro-37 was changed to Gly to test the hypothesis that backbone constraints imposed by the cyclized Pro side chain dictates the relative positions of adjacent acidic residues when PEP-19 is bound to CaM, thereby affecting Ca2+ binding.
Deletion of the Acidic Sequence Prevents Modulation of Ca2+ Binding to CaM
Calcium-dependent Tyr fluorescence was used to measure the KCa of the C-domain of CaM in the presence or absence of native and mutated PEP-19. Table 1 shows that neither native PEP-19 nor its mutated derivatives have large effects on KCa, although most decreased the cooperativity of Ca2+ binding.
TABLE 1.
Effect of mutants on Ca2+ binding to CaM
Apparent KCa values were determined by monitoring Tyr fluorescence during titration with Ca2+ and fitting the data to the Hill equation as described under “Experimental Procedures.” ND indicates not determined. Calcium dissociation rates (koff) were derived by fitting Ca2+ dissociation curves to mono- or diexponential equations. Values for koff are the average of 3–5 determinations in the presence or absence of 20 μm PEP-19 proteins. Stoichiometry of Ca2+ release was determined by calibrating Quin-2 fluorescence. Rates of Ca2+ association (kon) were calculated from kon = koff/KCa.
| PEP-19 protein | Equilibrium Ca2+ binding |
Binding kinetics |
Stoichiometry |
|||
|---|---|---|---|---|---|---|
| KCa | Hill coefficient | koff, 1 | koff, 2 | kon | Ca2+/protein | |
| μm | s−1 | s−1 | s−1 μm−1 | |||
| None | 1.6 ± 0.1 | 1.8 ± 0.1 | 10.4 ± 0.2 | 6.5 | 2.0 | |
| PEP-19 | 2.0 ± 0.1 | 1.1 ± 0.01 | 298 ± 10 | 149 | 1.8 | |
| mycPEP-19 | 2.0 ± 0.2 | 1.3 ± 0.04 | 303 ± 15 | 151 | 1.8 | |
| PEPΔAc | ND | ND | 11.7 ± 0.6 | 36 ± 4 | ND | 1.7 |
| PEPscram | 1.9 ± 0.2 | 1.6 ± 0.05 | 10.6 ± 0.4 | 5.7 | 2.0 | |
| PEP(E29A) | 2.3 ± 0.1 | 1.1 ± 0.03 | 276 ± 11 | 120 | 1.8 | |
| PEP(D31A) | 2.4 ± 0.1 | 1.1 ± 0.01 | 157 ± 5 | 65 | 1.7 | |
| PEP(D33A) | 1.6 ± 0.1 | 1.4 ± 0.04 | 94 ± 6 | 59 | 1.8 | |
| PEP(D35A) | 2.4 ± 0.1 | 1.1 ± 0.1 | 288 ± 12 | 120 | 1.8 | |
| PEP(P37G) | 1.6 ± 0.2 | 1.5 ± 0.02 | 69 ± 5 | 43 | 1.9 | |
| PEP(E40A) | 2.4 ± 0.2 | 1.1 ± 0.03 | 159 ± 8 | 66 | 1.7 | |
The relatively slow Ca2+ koff rate of 10.4 s−1 for free CaM in Table 1 is due to dissociation of 2 Ca2+ from the C-domain because dissociation of Ca2+ from the N-domain is very rapid and occurs in the dead-time (1.7 ms) of the stopped-flow fluorimeter. PEP-19 greatly increases the rate of Ca2+ dissociation to about 300 s−1, but the stoichiometry remains 2 Ca2+ released per CaM. Table 1 shows that deletion of the acidic sequence in PEPΔAc prevents the increase in Ca2+ koff. Thus, the acidic sequence of PEP-19 is required for modulation of Ca2+ binding to CaM.
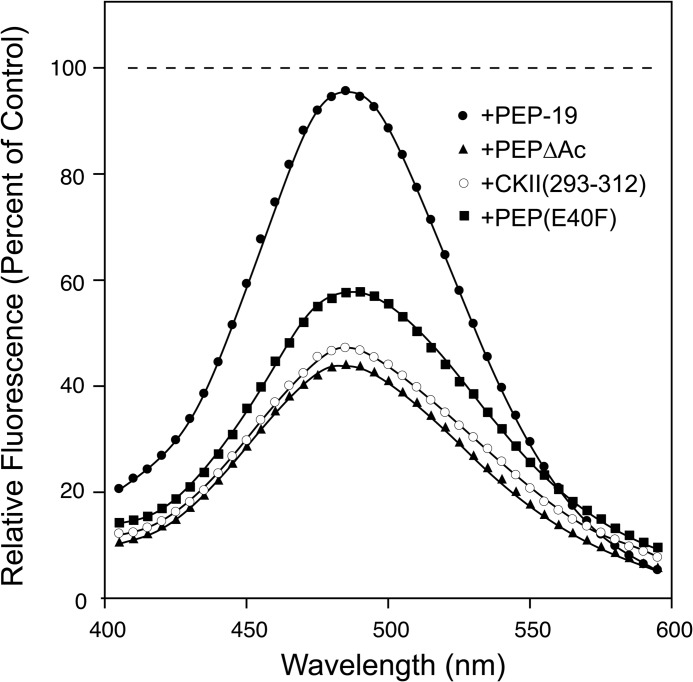
Interestingly, the stoichiometry of Ca2+ release in the presence of PEPΔAc is 4 Ca2+/mol of CaM instead of 2 seen the presence of all other PEP-19 proteins. This is consistent with the above prediction that PEPΔAc binds to both the N- and C-domains of CaM, thereby slowing the rate of release of Ca2+ from the N-domain as is seen for other CaM-binding proteins and peptides (23). We confirmed this mode of binding using a donor- and acceptor-labeled CaM (CaM(DA)) (24), which gives a large decrease in fluorescence due to FRET when CaM adopts a compact structure upon binding both domains to one peptide. Fig. 2 shows that fluorescence from CaM(DA) is not greatly affected by native PEP-19 because it binds preferentially to the C-domain of CaM, but a large decrease in fluorescence is seen upon binding to either PEPΔAc or a CaM-binding peptide from CaM kinase II, CKII(293–312), which is known to bind to both domains of CaM (25). As a further test, we generated PEP(E40F), with Phe at the homologous position to the Phe that anchors the IQ motif of the CaV1.2 channel to the N-domain of CaM (22). Fig. 2 shows that PEP(E40F) also causes a large decrease in fluorescence upon binding to CaM(DA). These results show that the absence of an appropriately positioned hydrophobic group in the acidic region of PEP-19 allows preferential binding to the C-domain of CaM.
FIGURE 2.
PEPΔAc binds to both domains of CaM to induce a compact structure. CaM(DA) has Thr-38 (N-domain) and Thr-110 (C-domain) converted to Cys and is labeled with a sulfhydryl-specific fluorescent donor (5-((((2-iodoacetyl)amino)ethyl)amino)naphthalene-1-sulfonic acid) and the nonfluorescent acceptor (N-(4-dimethylamino-3,5-dinitrophenyl)maleimide). The dashed line indicates the fluorescence maximum if no FRET effect was observed.
Native Sequence of the Acidic Region Is Necessary to Modulate Ca2+ Binding to CaM
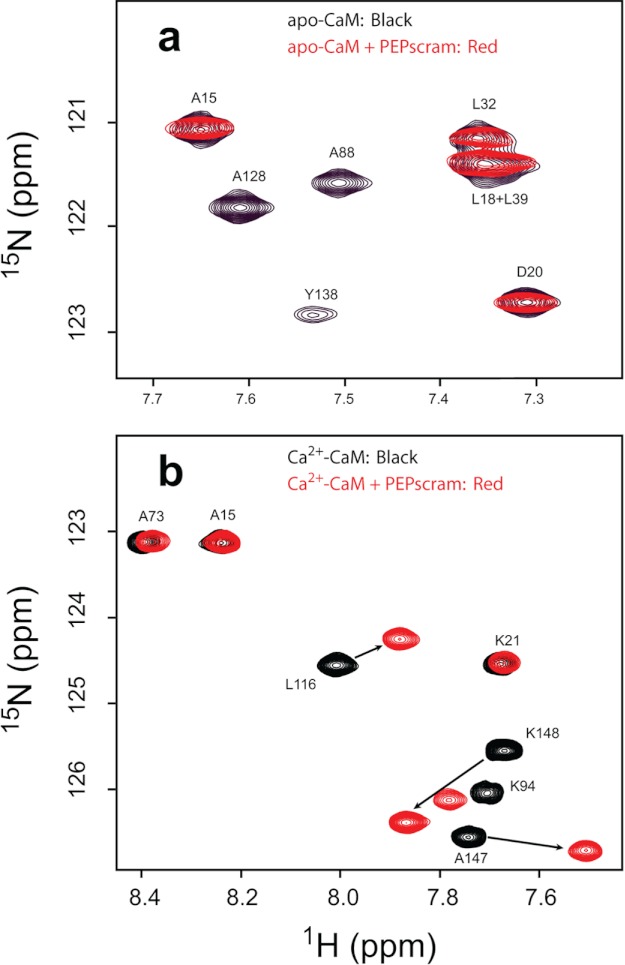
Table 1 shows that PEPscram has essentially no effect on KCa, koff, kon, or the stoichiometry of Ca2+ binding to CaM. This lack of effect was so striking that we used NMR to determine whether PEPscram binds to CaM with the same domain specificity and exchange properties as native PEP-19. We showed previously that native PEP-19 binds to apo-CaM and Ca2+-CaM with characteristics of slow and fast exchange, respectively, on the NMR time scale (21). Fig. 3 shows that PEPscram retains these properties. Specifically, Fig. 3a shows that PEPscram binds to apo-CaM with slow to intermediate exchange on the NMR time scale, causing severe broadening of backbone amide cross-peaks for residues in the C-domain, but it has little effect on amides in the N-domain (full spectra are supplied as supplemental material). Fig. 3b shows that PEPscram also selectively binds to the C-domain of Ca2+-CaM, but with characteristics of fast exchange. Thus, both PEPscram and PEP-19 bind to the C-domain of apo- or Ca2+-CaM, and with similar exchange characteristics, but PEPscram is incapable of modulating the Ca2+ binding properties of CaM.
FIGURE 3.
1H,15N HSQC NMR spectra indicate binding of PEPscram to apo- and Ca2+-CaM. a shows a selected region of the overlaid 1H,15N HSQC spectra of apo-CaM (black) and apo-CaM in complex with PEPscram (red); b shows a selected region of the overlaid 1H,15N HSQC spectra of Ca2+-CaM (black) and Ca2+-CaM in complex with PEPscram (red). PEPscram binding causes amide resonances in Ca2+-CaM undergoing fast exchange on the NMR time scale. The arrows in b highlight the movements of amide resonances in Ca2+-CaM due to PEPscram binding.
None of the PEP-19 point mutations had significant effects on KCa of CaM, but Table 1 and Fig. 4a show that they have varying effects on koff and kon. Conversion of Glu-29 to Ala at the putative X coordination position had no effect. Mutation of Asp-31, Asp-33, or Glu-40 to Ala inhibited the ability of PEP-19 to increase koff but to different extents. The properties of PEP(D35A) are very similar to native PEP-19, even though Asp-35 at the putative −Y coordination position is centered between residues 31, 33, and 40. This could be explained by the fact that the −Y position is highly variable in canonical EF-hand Ca2+-binding loops because the backbone carbonyl oxygen, not the side chain, of this residue coordinates Ca2+.
FIGURE 4.
Contribution of acidic residues in PEP-19 to modulating Ca2+ binding to the C-domain of CaM. a shows the effects of specific acidic residues in PEP-19 on the rates of Ca2+ dissociation (black bars) and association (white bars) at the C-domain of CaM. Dissociation of Ca2+ was measured in the presence of 30 μm PEP-19 or the indicated mutant PEP-19. Dissociation rates are the average of 4–5 determinations. Association rates were calculated from koff and KCa by kon = koff/KCa (see Table 1). b shows the effect of increasing concentrations of PEP-19, PEP(P37G), and PEPscram on the rate of Ca2+ dissociation.
Fig. 4b shows that conversion of Pro-37 to Gly significantly decreased the ability of PEP-19 to modulate Ca2+ binding to CaM, although not to the extent seen for PEPscram. This suggests that backbone constraints imposed by the imide side chain of Pro-37 positions acidic residues in PEP-19 such that they can properly modulate Ca2+ binding to CaM. Therefore, mutation of Pro would be equivalent to mutating multiple acidic residues. This is consistent with the fact that PEPscram is incapable of modulating Ca2+ binding to CaM because it effectively has multiple acidic mutations.
Acidic Region of PEP-19 Binds Ca2+
The distribution of acidic residues in PEP-19 led us to determine whether the acidic sequence has intrinsic Ca2+ binding activity. Its similar ionic radii and metal coordination geometries to Ca2+ make paramagnetic Tb3+ a sensitive probe for identifying Ca2+-binding sites (26). Fig. 5a shows that Tb3+ broadens backbone amide chemical shifts for residues in the acidic sequence of PEP-19, especially residues 31–36, which are severely broadened at a Tb3+/PEP-19 ratio of 1:50. Amides for Asp-33 and Asp-35 are most affected and are broadened beyond detection at a Tb3+/PEP-19 ratio of 1:100. These spectral perturbations indicate that Tb3+ binds to the acidic region in PEP-19.
FIGURE 5.
Interactions between PEP-19 and metal ions monitored by 1H,15N HSQC spectra. I/I0 is the intensity ratio for backbone amide cross-peaks in the 1H,15N HSQC spectra of 0.5 mm PEP-19 collected in the presence (I) and absence (I0) of 10 μm Tb3+ (a) or 5 mm Ca2+ (b) in the absence of KCl. Panel c shows the effect of increasing Ca2+ on I/I0 for Asp-33 and Asp-35 relative to Arg-4, which is unaffected by specific binding of Ca2+. * indicate the absence of backbone amides for Pro-37 and Gln-49, which could not be assigned.
Although Ca2+ is not paramagnetic, we reasoned that it might affect specific amide resonance intensities due to exchange broadening if Ca2+ binds to PEP-19. Indeed, Fig. 5b shows that addition of Ca2+ to PEP-19 causes exchange broadening of amide resonances in the acidic sequence relative to other regions in PEP-19. Similar to the effect of Tb3+, Asp-33 and Asp-35 are most affected by addition of Ca2+ and show maximal broadening at a Ca2+:PEP-19 molar ratio between 1 and 2 as shown in Fig. 5c with Arg-4 in the N-domain of CaM as a control. These spectral perturbations indicate that Ca2+ binds weakly to the acidic sequence of PEP-19.
Effect of Electrostatics on Ca2+ Binding
We reasoned that the acidic sequence of PEP-19 with intrinsic Ca2+ binding properties may increase the Ca2+ kon if positioned near site III and/or IV of CaM by attracting or electrostatically steering Ca2+ to these binding sites. Because the contribution of electrostatic interactions would be decreased by monovalent cations, we predicted that decreasing the KCl concentration would increase the kon for Ca2+ binding to the C-domain of CaM in the presence or absence of PEP-19. Table 2 shows the KCa, koff, and kon values for Ca2+ binding to the C-domain of CaM with or without 100 mm KCl and with or without 30 μm PEP-19. The Ca2+ binding affinity is increased about 13-fold at low ionic strength due primarily to a large increase in kon. The effect of KCl on kon can be explained by electrostatic shielding of acidic side chains on CaM that coordinate or attract Ca2+. PEP-19 increases Ca2+ kon by 27- and 45-fold at 100 and 0 mm KCl, respectively. This effect of PEP-19 can be attributed, at least in part, to electrostatic steering of Ca2+ ions via weak Ca2+ binding activity of the acidic sequence in PEP-19.
TABLE 2.
Effect of ionic strength on Ca2+ binding to CaM
Apparent KCa, Hill coefficients, and koff rates were determined as described under “Experimental Procedures.” Values are the average ± S.D. of at least three experiments. The kon was calculated from kon = koff/KCa. The buffer is 20 mm MOPS, pH 7.5, with the indicated concentration of KCl. An ionic strength of 13 mm is contributed by 20 mm MOPS at this pH. PEP-19 was added to a final concentration of 30 μm.
| KCl | PEP-19 | KCa | Hill coefficient | koff | kon |
|---|---|---|---|---|---|
| mm | μm | s−1 | μm−1 s−1 | ||
| 100 | − | 1.6 ± 0.1 | 1.8 ± 0.1 | 12.5 ± 0.1 | 7.8 |
| 0 | − | 0.12 ± 0.01 | 1.8 ± 0.04 | 7.0 ± 0.2 | 58 |
| 100 | + | 2.0 ± 0.1 | 1.1 ± 0.01 | 428 ± 40 | 214 |
| 0 | + | 0.14 ± 0.01 | 1.0 ± 0.05 | 363 ± 24 | 2592 |
PEP-19 Sensitizes HeLa Cells to ATP-induced Ca2+ Release
ATP-induced Ca2+ release in HeLa cells was selected as an initial model system to determine whether PEP-19 can impact a Ca2+ release pathway because this pathway involves multiple potential points of regulation by CaM, including P2Y G-protein-coupled receptors, phospholipase C, and the IP3 receptor. PEPscram, PEP(P37G), and PEPΔAc were selected to test the biological significance of the acidic sequence because they all bind to CaM but have little or no effect on its Ca2+ binding properties. Expression plasmids for native and mutated PEP-19 were engineered with N-terminal Myc tags to readily determine relative expression levels in transfected cells. We anticipated that the Myc tag would not affect interactions between PEP-19 and CaM because residues 1–23 in PEP-19 are disordered when bound to CaM. As shown in Table 1, PEP-19 and mycPEP-19 have essentially identical effects on Ca2+ binding to CaM.
HeLa cells were transfected with a control YFP plasmid or cotransfected with YFP and PEP-19 plasmids at a 1:4 ratio. YFP-positive cells from different coverslips were selected for analysis. The Western blot in Fig. 6a shows comparable levels of expression of PEP-19 proteins in transfected cells, and it confirms that the apparent molecular mass of PEPΔAc is smaller than the other proteins due to deletion of the acidic sequence. Fig. 6b shows intracellular Ca2+ in response to stimulation with 0.1, 1, and 10 μm ATP. The most striking observation is that only cells expressing PEP-19 showed a robust increase in intracellular Ca2+ in response to 0.1 μm ATP. As summarized in Fig. 6c, control cells were unresponsive to 0.1 μm ATP, but 65% (35/54) of cells expressing PEP-19 responded with a significant increase in intracellular Ca2+ levels. Fig. 6d shows that peak intracellular Ca2+ release stimulated by 1 and 10 μm ATP was also significantly higher in cells expressing PEP-19 relative to control cells. Finally, Fig. 6e shows that PEP-19 increases the frequency of Ca2+ oscillations induced by 1 μm ATP relative to control cells.
In contrast to native PEP-19, Fig. 6c shows that only 5% of all cells expressing PEP(P37G), PEPscram, or PEPΔAc responded to 0.1 μm ATP with an increase in Ca2+. Fig. 6d shows that peak Ca2+ levels induced by 1 and 10 μm ATP are also significantly lower in cells expressing mutant PEP(P37G), PEPscram, or PEPΔAc relative to PEP-19. Moreover, Fig. 6d shows the mutant PEP-19 proteins do not mimic the effect of native PEP-19 on Ca2+ oscillation frequency at 1 mm ATP. These data demonstrate that simply binding CaM is not sufficient and that the native acidic sequence in PEP-19 is required to sensitize HeLa cells to ATP-dependent Ca2+ release.
DISCUSSION
Cell signaling pathways must be regulated at multiple levels to control the amplitude and temporal characteristics of cellular responses and to prevent chaotic signaling that can lead to cell damage or death. Calmodulin is primarily regulated by intracellular Ca2+, which is in turn controlled by cell-specific arrays of Ca2+ channels, pores, and pumps (27). A poorly understood regulatory mechanism involves the actions of dedicated regulators of CaM signaling, which have no known intrinsic activity other than binding to CaM. For example, the small neuronal phosphoprotein called ARPP-21, or regulator of calmodulin signaling, binds to Ca2+-CaM to competitively inhibit activation of calcineurin and block suppression of L-type Ca2+ currents (28). PEP-19 is also a small protein with the potential to broadly affect CaM signaling by binding to apo- or Ca2+-CaM via its IQ motif.
An obvious potential mechanism for PEP-19 is to competitively inhibit activation of CaM targets as proposed in the camstatin model (12). A caveat to this is that enzymes such as CaM kinase II bind CaM with 10,000-fold greater affinity than does PEP-19. Nevertheless, CaM binds to many proteins with low affinity, and PEP-19 would be particularly effective as an antagonist of proteins that bind preferentially to the apo- or Ca2+-bound C-domain of CaM. Another mechanism, the calpacitin model (14), proposes that higher affinity binding of PEP-19 to apo-CaM relative to Ca2+-CaM retards its release during a Ca2+ pulse thereby affecting the temporal profile of available CaM and decreasing the overall rate of association of CaM with Ca2+-dependent target proteins, especially at low Ca2+ levels. This model stems from early studies showing that the homologous protein, neuromodulin, binds preferentially to apo-CaM (29). However, this selectivity is only observed at low salt, and neuromodulin binds with equal affinity to apo- and Ca2+-CaM in buffers containing 150 mm KCl (30). PEP-19 also has little selectivity for apo- versus Ca2+-CaM at physiologically relevant concentrations of salt (12, 16).
These caveats to proposed mechanisms for PEP-19 led us to explore alternatives. We first showed that PEP-19 increased kon and koff rates for Ca2+ binding to the C-domain of CaM by 30–40-fold without greatly affecting KCa (15). Importantly, an acidic sequence located adjacent to the IQ motif is required for PEP-19 to modulate Ca2+ binding to CaM (16). Thus, PEP-19 has the potential to modulate the rate-limiting kinetics of Ca2+ binding to CaM and to provide a regulatory mechanism that is analogous to regulators of G-protein signaling, or RGS proteins, that modulates nucleotide hydrolysis (31).
The first goal of this study was to investigate the molecular mechanism of action of PEP-19. Our results show the following: 1) the native sequence of the acidic region as well as backbone constraints imposed by Pro-37 are required for PEP-19 to modulate Ca2+ binding to CaM; 2) the acidic sequence has weak Ca2+ binding properties. Interestingly, mutations that compromise the ability of PEP-19 to modulate Ca2+ binding to CaM have proportional effects on both kon and koff (see Table 1), which suggests that a similar mechanism is responsible, at least in part, for modulating both parameters. A role for acidic residues in tuning the Ca2+ kon but not koff for binding to Ca2+ EF-hand proteins was demonstrated by Martin et al. (32), who showed that neutralizing three acidic surface residues near EF loop I in calbindin D9k decreased the kon up to 50-fold. By analogy, the acidic sequence of PEP-19 may mimic an increase in negative surface charge near site III and/or IV of CaM, thereby increasing the Ca2+ kon CaM by stabilizing a Ca2+-CaM initiation complex or by electrostatically steering Ca2+ to sites III and/or IV. PEP-19 may increase the Ca2+ koff of CaM by providing a low affinity transition Ca2+-binding site that shuttles Ca2+ to the solvent rather than allowing it to rebind to the EF-hands of CaM. The inability of PEP(P37G) and PEPscram to modulate Ca2+ kon and koff may be due to repositioning the acidic residues relative to the EF-hand Ca2+-binding loops in CaM and/or compromising Ca2+ binding to PEP-19.
The second goal of this study was to determine whether PEP-19 modulates CaM-dependent signaling pathways that affect intercellular Ca2+ homeostasis. We selected purinergic ATP-induced Ca2+ release as a model system because this pathway involves multiple potential points of regulation by CaM. The data in Fig. 6 show that PEP-19 sensitizes HeLa cells to ATP-dependent Ca2+ release and also alters the frequency of Ca2+ oscillations. Importantly, these biological effects require an intact acidic sequence, not simply binding of PEP-19 to CaM. Additional studies will be necessary to identify the level at which PEP-19 impacts Ca2+ release, but these effects reinforce the idea that both PEP-19 and Ng play roles in intercellular Ca2+ homeostasis (33). Such a role would be consistent with expression of PEP-19 in neuroendocrine and neuronal cells such as Purkinje cells (34) that have highly active Ca2+ signaling dynamics with robust and prolonged trains of action potentials (35). Ng knock-out mice show multiple effects on Ca2+ dynamics, including increased base-line Ca2+ levels and blunted Ca2+ transients induced by synaptic activity or glutamate receptor agonists (36). We anticipate PEP-19 and Ng will influence distinct sets of Ca2+ mobilization proteins and/or have different effects on the same proteins because PEP-19 increases both kon and koff of Ca2+ binding to the C-domain (15), whereas Ng increases only Ca2+ koff leading to decreased Ca2+ binding affinity (37). Different cellular effects of PEP-19 and Ng are also suggested by different patterns of expression and because PEP-19 has anti-apoptotic effects (9, 10), whereas RC3 is reported to have pro-apoptotic activity (38, 39).
Calmodulin regulates numerous proteins involved in Ca2+ mobilization that could be tuned by PEP-19. With respect to ATP-dependent Ca2+ release, CaM directly and indirectly impacts phospholipase C activity (40), and it also modulates the activity of the IP3 receptor (41) and store-operated Ca2+ entry channels (42) subsequent to IP3 generation. Other CaM-dependent channels and extrusion proteins include the ryanodine receptor (43), plasma membrane Ca2+ pumps (44), and the Na+/Ca2+ exchanger (45). Interestingly, the modes of interaction between CaM and several key Ca2+ mobilization proteins may make them particularly susceptible to PEP-19 because it binds selectively to the C-domain of CaM. For example, voltage-operated Ca2+ channels (46) and the IP3 receptor (47) rely on selective, sequential, or stepwise interactions with the C-domain of CaM in its apo- or Ca2+-bound forms.
In summary, this study reveals new mechanisms of action for PEP-19 and demonstrates novel effects on ATP-dependent Ca2+ release that do not depend solely on binding PEP-19 to CaM, but it also requires its ability to modulate Ca2+ binding to CaM. Tuning the activities of Ca2+ mobilization pathways would place PEP-19 at the top of CaM signaling cascades, with great potential to exert broad effects on downstream CaM targets, thus expanding the biological significance of this small regulator of CaM signaling.
This work was supported, in whole or in part, by National Institutes of Health Grants GM081685 and GM081685-03S1 (to D. B.) and GM06961109 (to J. A. P.). This work was also supported by Fellowships R90 DK071504 from the W. M. Keck Center for Interdisciplinary Biosciences, 10POST3110010 from the American Heart Association (to X. W.), and GRNT2280427 from the American Heart Association (to J. A. P.).

This article contains supplemental Fig. S1.
- CaM
- calmodulin
- C-CaM
- isolated C domain of CaM
- Ng
- neurogranin
- BAPTA
- 1,2-bis(2-aminophenoxy)ethane-N,N,N′,N′-tetraacetic acid
- IP3
- inositol 1,4,5-trisphosphate
- CaM(DA)
- donor- and acceptor-labeled CaM.
REFERENCES
- 1. Ge X., Yamamoto S., Tsutsumi S., Midorikawa Y., Ihara S., Wang S. M., Aburatani H. (2005) Interpreting expression profiles of cancers by genome-wide survey of breadth of expression in normal tissues. Genomics 86, 127–141 [DOI] [PubMed] [Google Scholar]
- 2. Sköld K., Svensson M., Nilsson A., Zhang X., Nydahl K., Caprioli R. M., Svenningsson P., Andrén P. E. (2006) Decreased striatal levels of PEP-19 following MPTP lesion in the mouse. J. Proteome Res. 5, 262–269 [DOI] [PubMed] [Google Scholar]
- 3. Iwamoto K., Bundo M., Yamamoto M., Ozawa H., Saito T., Kato T. (2004) Decreased expression of NEFH and PCP4/PEP-19 in the prefrontal cortex of alcoholics. Neurosci. Res. 49, 379–385 [DOI] [PubMed] [Google Scholar]
- 4. Glynne R., Ghandour G., Rayner J., Mack D. H., Goodnow C. C. (2000) B-lymphocyte quiescence, tolerance, and activation as viewed by global gene expression profiling on microarrays. Immunol. Rev. 176, 216–246 [DOI] [PubMed] [Google Scholar]
- 5. Kanamori T., Takakura K., Mandai M., Kariya M., Fukuhara K., Kusakari T., Momma C., Shime H., Yagi H., Konishi M., Suzuki A., Matsumura N., Nanbu K., Fujita J., Fujii S. (2003) PEP-19 overexpression in human uterine leiomyoma. Mol. Hum. Reprod. 9, 709–717 [DOI] [PubMed] [Google Scholar]
- 6. Wei P., Blundon J. A., Rong Y., Zakharenko S. S., Morgan J. I. (2011) Impaired locomotor learning and altered cerebellar synaptic plasticity in pep-19/PCP4-null mice. Mol. Cell. Biol. 31, 2838–2844 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Harashima S., Wang Y., Horiuchi T., Seino Y., Inagaki N. (2011) Purkinje cell protein 4 positively regulates neurite outgrowth and neurotransmitter release. J. Neurosci. Res. 89, 1519–1530 [DOI] [PubMed] [Google Scholar]
- 8. Mouton-Liger F., Thomas S., Rattenbach R., Magnol L., Larigaldie V., Ledru A., Herault Y., Verney C., Créau N. (2011) PCP4 (PEP-19) overexpression induces premature neuronal differentiation associated with Ca2+/calmodulin-dependent kinase II-δ activation in mouse models of Down syndrome. J. Comp. Neurol. 519, 2779–2802 [DOI] [PubMed] [Google Scholar]
- 9. Erhardt J. A., Legos J. J., Johanson R. A., Slemmon J. R., Wang X. (2000) Expression of PEP-19 inhibits apoptosis in PC12 cells. Neuroreport 11, 3719–3723 [DOI] [PubMed] [Google Scholar]
- 10. Kanazawa Y., Makino M., Morishima Y., Yamada K., Nabeshima T., Shirasaki Y. (2008) Degradation of PEP-19, a calmodulin-binding protein, by calpain is implicated in neuronal cell death induced by intracellular Ca2+ overload. Neuroscience 154, 473–481 [DOI] [PubMed] [Google Scholar]
- 11. Utal A. K., Stopka A. L., Roy M., Coleman P. D. (1998) PEP-19 immunohistochemistry defines the basal ganglia and associated structures in the adult human brain and is dramatically reduced in Huntington's disease. Neuroscience 86, 1055–1063 [DOI] [PubMed] [Google Scholar]
- 12. Slemmon J. R., Morgan J. I., Fullerton S. M., Danho W., Hilbush B. S., Wengenack T. M. (1996) Camstatins are peptide antagonists of calmodulin based upon a conserved structural motif in PEP-19, neurogranin, and neuromodulin. J. Biol. Chem. 271, 15911–15917 [DOI] [PubMed] [Google Scholar]
- 13. Gerendasy D. D., Herron S. R., Watson J. B., Sutcliffe J. G. (1994) Mutational and biophysical studies suggest RC3/neurogranin regulates calmodulin availability. J. Biol. Chem. 269, 22420–22426 [PubMed] [Google Scholar]
- 14. Gerendasy D. D., Sutcliffe J. G. (1997) RC3/neurogranin, a postsynaptic calpacitin for setting the response threshold to calcium influxes. Mol. Neurobiol. 15, 131–163 [DOI] [PubMed] [Google Scholar]
- 15. Putkey J. A., Kleerekoper Q., Gaertner T. R., Waxham M. N. (2003) A new role for IQ motif proteins in regulating calmodulin function. J. Biol. Chem. 278, 49667–49670 [DOI] [PubMed] [Google Scholar]
- 16. Putkey J. A., Waxham M. N., Gaertner T. R., Brewer K. J., Goldsmith M., Kubota Y., Kleerekoper Q. K. (2008) Acidic/IQ motif regulator of calmodulin. J. Biol. Chem. 283, 1401–1410 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Xiong L. W., Kleerekoper Q. K., Wang X., Putkey J. A. (2010) Intra- and interdomain effects due to mutation of calcium-binding sites in calmodulin. J. Biol. Chem. 285, 8094–8103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Linse S., Helmersson A., Forsén S. (1991) Calcium binding to calmodulin and its globular domains. J. Biol. Chem. 266, 8050–8054 [PubMed] [Google Scholar]
- 19. Stieren E., Werchan W. P., El Ayadi A., Li F., Boehning D. (2010) FAD mutations in amyloid precursor protein do not directly perturb intracellular calcium homeostasis. PLoS ONE 5, e11992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Wang X., Kleerekoper Q. K., Xiong L. W., Putkey J. A. (2010) Intrinsically disordered PEP-19 confers unique dynamic properties to apo and calcium calmodulin. Biochemistry 49, 10287–10297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Kleerekoper Q. K., Putkey J. A. (2009) PEP-19, an intrinsically disordered regulator of calmodulin signaling. J. Biol. Chem. 284, 7455–7464 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Fallon J. L., Halling D. B., Hamilton S. L., Quiocho F. A. (2005) Structure of calmodulin bound to the hydrophobic IQ domain of the cardiac Ca(v)1.2 calcium channel. Structure 13, 1881–1886 [DOI] [PubMed] [Google Scholar]
- 23. Persechini A., White H. D., Gansz K. J. (1996) Different mechanisms for Ca2+ dissociation from complexes of calmodulin with nitric oxide synthase or myosin light chain kinase. J. Biol. Chem. 271, 62–67 [DOI] [PubMed] [Google Scholar]
- 24. Xiong L., Kleerekoper Q. K., He R., Putkey J. A., Hamilton S. L. (2005) Sites on calmodulin that interact with the C-terminal tail of Cav1.2 channel. J. Biol. Chem. 280, 7070–7079 [DOI] [PubMed] [Google Scholar]
- 25. Meador W. E., Means A. R., Quiocho F. A. (1993) Modulation of calmodulin plasticity in molecular recognition on the basis of x-ray structures. Science 262, 1718–1721 [DOI] [PubMed] [Google Scholar]
- 26. Yang W., Jones L. M., Isley L., Ye Y., Lee H. W., Wilkins A., Liu Z. R., Hellinga H. W., Malchow R., Ghazi M., Yang J. J. (2003) Rational design of a calcium-binding protein. J. Am. Chem. Soc. 125, 6165–6171 [DOI] [PubMed] [Google Scholar]
- 27. Berridge M. J., Bootman M. D., Roderick H. L. (2003) Calcium signaling. Dynamics, homeostasis, and remodeling. Nat. Rev. Mol. Cell Biol. 4, 517–529 [DOI] [PubMed] [Google Scholar]
- 28. Rakhilin S. V., Olson P. A., Nishi A., Starkova N. N., Fienberg A. A., Nairn A. C., Surmeier D. J., Greengard P. (2004) A network of control mediated by regulator of calcium/calmodulin-dependent signaling. Science 306, 698–701 [DOI] [PubMed] [Google Scholar]
- 29. Andreasen T. J., Luetje C. W., Heideman W., Storm D. R. (1983) Purification of a novel calmodulin binding protein from bovine cerebral cortex membranes. Biochemistry 22, 4615–4618 [DOI] [PubMed] [Google Scholar]
- 30. Alexander K. A., Cimler B. M., Meier K. E., Storm D. R. (1987) Regulation of calmodulin binding to P-57. A neurospecific calmodulin-binding protein. J. Biol. Chem. 262, 6108–6113 [PubMed] [Google Scholar]
- 31. Xie G. X., Palmer P. P. (2007) How regulators of G protein signaling achieve selective regulation. J. Mol. Biol. 366, 349–365 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Martin S. R., Linse S., Johansson C., Bayley P. M., Forsén S. (1990) Protein surface charges and Ca2+ binding to individual sites in calbindin D9k. Stopped-flow studies. Biochemistry 29, 4188–4193 [DOI] [PubMed] [Google Scholar]
- 33. Slemmon J. R., Feng B., Erhardt J. A. (2000) Small proteins that modulate calmodulin-dependent signal transduction: effects of PEP-19, neuromodulin, and neurogranin on enzyme activation and cellular homeostasis. Mol. Neurobiol. 22, 99–113 [DOI] [PubMed] [Google Scholar]
- 34. Ziai M. R., Sangameswaran L., Hempstead J. L., Danho W., Morgan J. I. (1988) An immunochemical analysis of the distribution of a brain-specific polypeptide, PEP-19. J. Neurochem. 51, 1771–1776 [DOI] [PubMed] [Google Scholar]
- 35. Hartmann J., Konnerth A. (2005) Determinants of postsynaptic Ca2+ signaling in Purkinje neurons. Cell Calcium 37, 459–466 [DOI] [PubMed] [Google Scholar]
- 36. van Dalen J. J., Gerendasy D. D., de Graan P. N., Schrama L. H., Gruol D. L. (2003) Calcium dynamics are altered in cortical neurons lacking the calmodulin-binding protein RC3. Eur. J. Neurosci. 18, 13–22 [DOI] [PubMed] [Google Scholar]
- 37. Gaertner T. R., Putkey J. A., Waxham M. N. (2004) RC3/neurogranin and Ca2+/calmodulin-dependent protein kinase II produce opposing effects on the affinity of calmodulin for calcium. J. Biol. Chem. 279, 39374–39382 [DOI] [PubMed] [Google Scholar]
- 38. Devireddy L. R., Green M. R. (2003) Transcriptional program of apoptosis induction following interleukin 2 deprivation. Identification of RC3, a calcium/calmodulin-binding protein, as a novel proapoptotic factor. Mol. Cell. Biol. 23, 4532–4541 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Gui J., Song Y., Han N. L., Sheu F. S. (2007) Characterization of transcriptional regulation of neurogranin by nitric oxide and the role of neurogranin in SNP-induced cell death. Implication of neurogranin in an increased neuronal susceptibility to oxidative stress. Int. J. Biol. Sci. 3, 212–224 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. McCullar J. S., Larsen S. A., Millimaki R. A., Filtz T. M. (2003) Calmodulin is a phospholipase C-β interacting protein. J. Biol. Chem. 278, 33708–33713 [DOI] [PubMed] [Google Scholar]
- 41. Taylor C. W., Laude A. J. (2002) IP3 receptors and their regulation by calmodulin and cytosolic Ca2+. Cell Calcium 32, 321–334 [DOI] [PubMed] [Google Scholar]
- 42. Zhu M. X. (2005) Multiple roles of calmodulin and other Ca2+-binding proteins in the functional regulation of TRP channels. Pflugers Arch. 451, 105–115 [DOI] [PubMed] [Google Scholar]
- 43. Rodney G. G., Williams B. Y., Strasburg G. M., Beckingham K., Hamilton S. L. (2000) Regulation of RYR1 activity by Ca2+ and calmodulin. Biochemistry 39, 7807–7812 [DOI] [PubMed] [Google Scholar]
- 44. Di Leva F., Domi T., Fedrizzi L., Lim D., Carafoli E. (2008) The plasma membrane Ca2+-ATPase of animal cells. Structure, function and regulation. Arch. Biochem. Biophys. 476, 65–74 [DOI] [PubMed] [Google Scholar]
- 45. Carafoli E., Longoni S. (1987) The plasma membrane in the control of the signaling function of calcium. Soc. Gen. Physiol. Ser. 42, 21–29 [PubMed] [Google Scholar]
- 46. Liang H., DeMaria C. D., Erickson M. G., Mori M. X., Alseikhan B. A., Yue D. T. (2003) Unified mechanisms of Ca2+ regulation across the Ca2+ channel family. Neuron 39, 951–960 [DOI] [PubMed] [Google Scholar]
- 47. Kang S., Kwon H., Wen H., Song Y., Frueh D., Ahn H. C., Yoo S. H., Wagner G., Park S. (2011) Global dynamic conformational changes in the suppressor domain of IP3 receptor by stepwise binding of the two lobes of calmodulin. FASEB J. 25, 840–850 [DOI] [PubMed] [Google Scholar]