Background: pVIc, an 11-amino acid peptide from the C terminus of adenovirus precursor protein pVI, activates the adenovirus proteinase (AVP).
Results: pVI slides on DNA into AVP, which excises and then covalently binds pVIc thereby rendering AVP fully active.
Conclusion: Activation of AVP requires pVI in cis on DNA.
Significance: These results indicate a new way a protein substrate interacts with a proteinase, via one-dimensional diffusion on DNA.
Keywords: Adenovirus, DNA-binding Protein, DNA Viruses, DNA-Protein Interaction, Protease, Enzyme Activation, One-dimensional Diffusion, Proteinase, Sliding on DNA, Viral Proteins
Abstract
Late in an adenovirus infection, the viral proteinase (AVP) becomes activated to process virion precursor proteins used in virus assembly. AVP is activated by two cofactors, the viral DNA and pVIc, an 11-amino acid peptide originating from the C terminus of the precursor protein pVI. There is a conundrum in the activation of AVP in that AVP and pVI are sequence-independent DNA-binding proteins with nm equilibrium dissociation constants such that in the virus particle, they are predicted to be essentially irreversibly bound to the viral DNA. Here, we resolve that conundrum by showing that activation of AVP takes place on the one-dimensional contour of DNA. In vitro, pVI, a substrate, slides on DNA via one-dimensional diffusion, D1 = 1.45 × 106 bp2/s, until it binds to AVP also on the same DNA molecule. AVP, partially activated by being bound to DNA, excises pVIc, which binds to the AVP molecule that cut it out. pVIc then forms a disulfide bond with AVP forming the fully active AVP-pVIc complex bound to DNA. In vivo, in heat-disrupted immature virus, AVP was also activated by pVI in DNA-dependent reactions. This activation mechanism illustrates a new paradigm for virion maturation and a new way, by sliding on DNA, for bimolecular complexes to form among proteins not involved in DNA metabolism.
Introduction
Human adenovirus, a eukaryotic virus with an ∼36,000-bp, linear DNA genome, encodes the adenovirus proteinase (AVP),5 a 204-amino acid cysteine proteinase (1) whose activity is essential for the synthesis of infectious virus particles (2). One of the functions of the proteinase, after virion assembly, is to cleave six virion precursor proteins to their mature counterparts found in wild-type virions (2). Recombinant AVP exhibited little or no enzymatic activity (3, 4), prompting a search for cofactors. One cofactor is pVIc,6 the 11-amino acid residue peptide (GVQSLKRRRCF) originating from the C terminus of the 250-amino acid adenovirus precursor protein pVI. A second cofactor is the viral DNA (3, 5–7). These cofactors stimulate the macroscopic kinetic constants for substrate hydrolysis by AVP (5, 8–10).
AVP is synthesized as an essentially inactive enzyme, which raises the question how pVIc is cleaved from pVI inside immature particles to activate AVP, i.e. to form AVP-pVIc complexes. Restricting any model for the activation of AVP by pVI in such particles is the inability of AVP and pVI to undergo bimolecular interactions by diffusion in three-dimensional space. Both AVP and pVI are sequence-independent DNA-binding proteins (3, 5, 9, 11). The high concentration of DNA inside the virion (>500 g/liter)) (12) drives both AVP and pVI onto the DNA by mass action. For AVP, the DNA-bound state predominates by at least one hundred thousand-fold over free AVP (5); this, in combination with the sieving action in the dense DNA environment (13), diminishes the effective three-dimensional diffusion constant of AVP by at least one million-fold. The DNA genome cannot move either, thereby enabling two DNA-bound proteins to interact. The pressure exerted by the tightly packed genome on the envelope of the virion creates considerable friction, freezing the DNA in place and rendering DNA-bound proteins likewise immobile. Given this situation inside the virion, it is not clear how a bimolecular interaction between AVP and pVI can occur that leads to cleavage of pVI and activation of the enzyme by released pVIc. Without this occurring, the virus particle cannot become infectious.
Here, we solve this conundrum by presenting evidence that these two proteins can form a bimolecular interaction in the one-dimensional compartment present along the viral DNA by sliding on the DNA via one-dimensional diffusion; they do not diffuse in three-dimensional space to meet. The activation reaction takes place by a novel biochemistry, one-dimensional biochemistry. This mechanism for promoting bimolecular interactions is a new paradigm for how substrates interact with “non-nucleic acid enzymes” and a new paradigm for virion maturation.
EXPERIMENTAL PROCEDURES
Materials
The 11-amino acid peptide pVIc (GVQSLKRRRCF) was purchased from New England Peptide Inc. (Gardner, MA). The 5′-fluorescein-labeled 33-mer ssDNA, 36-mer ssDNA, 60-mer ssDNA, and the strands complementary to these DNAs were purchased from Invitrogen, as was streptavidin Alexa Fluor 546. Annealing of complementary DNAs was done as described previously (9). The 1500-mer dsDNA was obtained by sonicating Cupriavidus metallidurans CH34 genomic DNA. n-Dodecyl-β-d-maltopyranoside (DDM) was purchased from Anatrace (Maumee, OH). Cy3B monomaleimide was purchased from GE Healthcare. 5-Iodoacetamidofluorescein was purchased from Pierce. The complex (pVIc-biotin)·streptavidin was synthesized as described.7 The fluorogenic substrates (Leu-Arg-Gly-Gly-NH)2-rhodamine and (Cbz-Leu-Arg-Gly-Gly-NH)2-rhodamine were synthesized and purified as described previously (3, 14). AVP and the AVP mutant C122A8 were purified using published procedures (3, 8). Coomassie Blue-stained or silver-stained protein gels were scanned on a flatbed scanner, and the bands were quantitated with the GE Healthcare ImageQuant software. Buffer A was 20 mm Hepes (pH 7.0), 0.025% (w/v) DDM, and 0.1 mm DTT. Buffer B was 20 mm Tris-HCl (pH 8.0), 0.025% DDM, and 0.1 mm DTT. pVIc was labeled with Cy3B as described.7
AVP-pVIc Complex Formation
Disulfide-linked AVP-pVIc complexes were prepared by overnight incubation at 4 °C of 75 μm AVP and 75 μm pVIc in 20 mm Tris-HCl (pH 8.0), 250 mm NaCl, 0.1 mm EDTA, and 20 mm β-mercaptoethanol. Under these conditions, Cys-104 of AVP and Cys-10′ of pVIc, the penultimate amino acid residue of pVIc, undergo oxidative condensation (15, 16).
Labeling of Proteins with Fluorescent Dyes
pVI at a concentration of 216 μm in 25 mm Tris-HCl (pH 7.5), 150 mm NaCl, 0.1 mm EDTA, 1 mm DTT, and 0.05% DDM was incubated with freshly prepared 5 mm DTT for 2 h at room temperature. The DTT was removed by passage through a Bio-Spin 6 chromatography column (Bio-Rad) previously equilibrated with 1× PBS containing 0.05% DDM, at pH 7.4. A 3-fold molar excess of either Cy3B monomaleimide or 5-iodoacetamidofluorescein was added., and the solution incubated overnight at 4 °C. The reaction was stopped by an 8-fold molar excess of DTT and centrifuged at room temperature in a Microfuge for 15 min. The unreacted dye was removed by size-exclusion chromatography on a 0.3 × 30-cm Superdex S-200 column equilibrated with 25 mm MES (pH 6.5), 0.05% DDM, and 150 mm NaCl. The dye-conjugated protein peaks were pooled. Protein concentration was measured using a molar extinction coefficient at 280 nm of 30,480 m−1 cm−1 for pVI, at 564 nm of 130,000 m−1 cm−1 for Cy3B conjugated to pVI, and at 492 nm of 68,000 m−1 cm−1 for fluorescein conjugated to pVI. A dye/pVI ratio of 0.4 and 0.6 was calculated for fluorescein- and Cy3B-conjugated pVI, respectively, using a 280 nm correction factor of 0.3 and 0.08, respectively.
To label AVP, we made AVP-pVIc complexes and labeled them using maleimide chemistry. Then, the pVIc in the AVP-pVIc complexes was removed by reduction. By labeling this way, Cys-104 could not be labeled with a dye molecule. Experimentally, disulfide-linked AVP-pVIc complexes were labeled in 25 mm HEPES (pH 7.0), 50 mm NaCl, and 20 mm ethanol by the addition of a 3-fold molar excess of Cy3B maleimide. Labeling reactions were incubated at room temperature in the dark for 2.5 h. Excess reagents were removed from the labeled sample by passage through Bio-Spin 6 chromatography columns (Bio-Rad) equilibrated in the labeling buffer. The degree of labeling was determined using ϵ280 nmAVP = 26,510 m−1 cm−1, ϵ558 nmCy3B = 130,000 m−1 cm−1, and ϵ280 nmCy3B = 10,400 m−1 cm−1. The ratio of labeled AVP-pVIc to total AVP-pVIc was determined to be 0.84. The labeled materials were characterized by MALDI-TOF mass spectrometry. Cy3B-AVP was generated by incubating Cy3B-AVP-pVIc in 20 mm β-mercaptoethanol at room temperature for 2 h. This was followed by removal of pVIc and β-mercaptoethanol by passage through a spin column.
Location of Cy3B Label
Specific enzymatic digestions followed by matrix-assisted laser desorption ionization time-of-flight (MALDI-TOF) mass spectrometry were used to locate cysteinyl-Cy3B conjugates in AVP. 1.2 μg of labeled AVP was digested by incubation with 0.01 μg of each endoproteinase Glu-C and trypsin at 21 °C in 25 mm Tris-HCl (pH 7.5). At 1, 2, 4, and 22 h, 0.5 μl of the reaction was removed and added to 4.5 μl of a saturated matrix solution (α-cyano-4-hydroxycinnamic acid) in 50% acetonitrile and 0.1% TFA. The matrix-analyte solution was then immediately spotted onto a 100-well stainless-steel sample plate. The sample plate was calibrated using Applied Biosystems peptide calibration mixtures 1 and 2. Mass spectrometric characterization was carried out on a Voyager-DE Biospectrometry work station (Applied Biosystems; Foster City, CA). The m/z peak list generated for each chromatogram was analyzed by the FindPept Tool (17). The Cy3B modification was entered as a post-translational modification with an atomic composition of C37H38N4O7S (molecular weight 682.785). AVP was found to be labeled at Cys-199.
Fluorescence Anisotropy
Steady-state fluorescence anisotropy measurements were performed using an ISS model PC-1 photon counting spectrofluorometer (ISS, Champaign, IL) equipped with polarization accessories. Measurements were made in L-format using a 300-watt xenon arc lamp with 10- and 14-mm Glan-Thompson polarizers in the excitation and emission channels, respectively. For Cy3B dye, the excitation wavelength was 564 nm, with 8-nm slits placed before and after a monochromator. The parallel and vertical emission components were measured through a 580-nm band-pass filter with a full width at half maximum of 10 nm.
Slow kinetic anisotropy measurements were made at room temperature and recorded every 60 s; the excitation shutter was closed in between measurements to minimize photobleaching. Single-point titration experiments involving 5′-fluorescein-labeled DNA were made using an excitation wavelength of 495 nm and a 530-nm long pass emission filter. In both cases, the G factor was measured before the start of each experiment and was always 1.
AVP Activity Assay
The enzymatic activity of AVP-DNA, AVP-pVIc, and AVP-pVIc·DNA complexes was measured using the fluorogenic substrate (Leu-Arg-Gly-Gly-NH)2-rhodamine (3, 14). Reactions in 100 μl of buffer B containing 5 μm (Leu-Arg-Gly-Gly-NH)2-rhodamine were initiated by adding enzyme and cofactors. The solutions were mixed and placed in a Costar 96-well half-area black plate. The fluorescence intensity was monitored continuously for about 500 s in 10-s intervals in a Tecan Safire2 plate reader. The excitation and emission wavelengths were 468 and 523 nm, respectively, each with a 10-nm band pass. The gain was set to 113, and the Z-value was 9320 μm. The rate in relative fluorescence intensity per second was determined from a linear fit of the data.
AVP Activation Assays by pVI and DNA
Activation of AVP by pVI in the presence of DNA was assayed by first reacting 77 nm pVI with 130 nm AVP and varying amounts of DNA in 20 mm Hepes, pH 7, 0.025% DDM, and 0.1 mm DTT. The reaction was allowed to proceed at room temperature (∼21 °C). At various times, a 100-μl aliquot was removed and added to 1 μl of 0.5 mm (Leu-Arg-Gly-Gly-NH)2-rhodamine and 1 μl of Tris base. The latter addition was necessary to raise the pH of the final reaction to 8, the optimal pH for AVP activity.
MALDI-TOF Analysis of the Proteins and Peptides during the Activation of AVP by pVI in the Presence of DNA
Small volumes of activation reaction mixtures were diluted by an equal volume of 0.2% TFA to stop the reactions. The reactions were further diluted with α-cyano-4-hydroxycinnamic acid matrix and placed onto a MALDI-TOF stainless steel 100-well plate. For proteins larger than 10 KDa, sinapinic acid matrix was used. MALDI-TOF data were collected on a Voyager Biospectrometry instrument according to standard protocols.
Sliding Assay Conditions
Flow cells containing λ DNA immobilized at one end were constructed as described previously (18). Labeled proteins were infused at concentrations of 1–2 nm at flow rates of 20–50 ml/h. High flow rates were chosen to drive the longitudinal DNA fluctuation faster than the imaging frame rate (18). The assay buffer consisted of 10 mm MES (pH 6.5), 2–6 mm NaCl, 50 μm EDTA, 20 mm ethanol, 5% glycerol, and where indicated, reducing agent (DTT and mercaptoethanol gave equivalent results).
Fluorescence Imaging
AVP was labeled with Cy3B at Cys-199, pVI at Cys-249, and pVIc at Cys-10′. Individual, fluorescently labeled molecules were imaged by total internal reflection fluorescence microscopy as described previously (18), with the exception that a faster electron-multiplying charge-coupled device camera (Photometrics Cascade:128+) was used for the highest time-resolution measurements. λ DNA was tethered to a glass surface at one end and stretched by a laminar flow of buffer. Single molecules that bound to and diffused along the DNA were illuminated by an evanescent wave via laser beam (532 nm) and imaged with a fluorescence microscope.
Centroid Determination and Analysis of Molecular Trajectories
All AVP and pVI DNA binding events noted were included in the analyses. pVI binding events were identified manually by noting movements consistent with DNA fluctuations in the direction orthogonal to the flow of buffer. These signals were tracked using Gaussian centroid determination in the MATLAB environment. Detection of DNA binding by AVP was found to be more challenging due to the shorter average residence time. For this reason, automated particle-tracking software (Diatrack 3.0, Semasopht, Switzerland), previously used to identify large numbers of individual DNA-bound protein molecules and track their trajectories using Gaussian centroid determination (18), was applied to this purpose. All AVP molecules exhibiting transverse displacements consistent with flow-stretched DNA were included in the analyses. Molecular trajectories were analyzed in MATLAB by methods similar to those previously published (18).
ts-1 Virus and Heat-disrupted ts-1 Virus
Immature virus was obtained by propagating the Ad2 ts-1 mutant in HeLa cells at 39.5 °C as described previously (19). Particles were purified by equilibrium centrifugation in CsCl gradients, desalted on a 10DC column (Bio-Rad), and stored in 20 mm Hepes, pH 7.8, 150 mm NaCl plus 10% glycerol at −70 °C at a final concentration of 1 × 1013 particles/ml. Heat-disrupted ts-1 virus particles were generated by heating ts-1 virus for 10 min at 60 °C (19).
Activation of AVP by pVI in Heat-disrupted Ad2 ts-1 Virions
All reactions were carried out in 10 mm Tris-HCl (pH 8.2), 5 mm MgCl2 buffer. Ad2 ts-1 virions at a concentration of 1.6 × 1012 particles/ml were disrupted by heating at 60 °C for 10 min. When present, reactions contained 0.25 μm AVP. To remove DNA, heat-disrupted virus was incubated with 50 μg/ml DNase I (Sigma D5025) at 37 °C for 24 h. DNase was inactivated by adding 10 mm EDTA. After 30 min, 0.25 μm AVP was added, and the samples were incubated at 37 °C for either 2 h or 24 h. In the indicated cases, purified Ad2 ts-1 DNA was added after DNase inactivation at a final concentration of 50 ng/ml. For DNA isolation, 25 × 1010 Ad2 ts-1 viral particles were treated with proteinase K at a final concentration of 400 μg/ml, and the DNA was extracted by phenol/chloroform precipitation. Proteins were fractionated by SDS-PAGE on an 8–25% gradient PhastGel and visualized by silver staining. An E1-deleted Ad5 vector (Ad5GL) (19) was used as the source for mature, wild-type virus structural proteins.
RESULTS
DNA Is Required for the Activation of AVP by pVI in Vitro
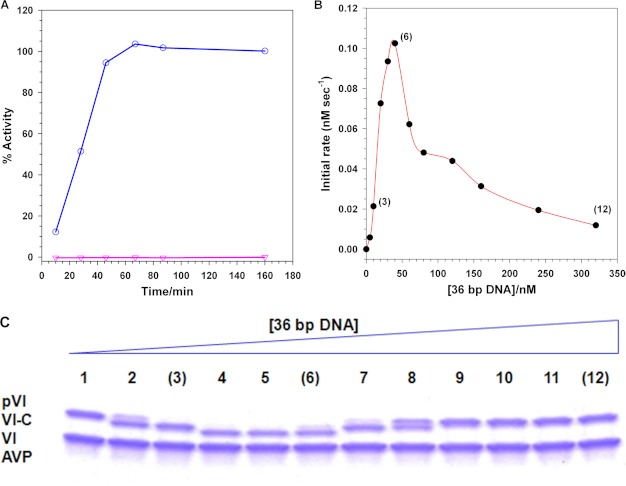
Which components are required for the activation of AVP by pVI to produce the enzymatically active AVP-pVIc complex? If only a simple bimolecular interaction between AVP and pVI were needed, then mixing purified AVP (8) with purified pVI (32) should result in the cleavage of pVI to yield pVIc followed by the formation of active AVP-pVIc complexes. However, when this was done, no enzymatic activity was detected (Fig. 1A). Both AVP and pVI bind to DNA with apparent equilibrium dissociation constants of 63 nm (5) and 46 nm, respectively (32). Is DNA required for the activation of AVP by pVI? We repeated the experiment but in the presence of dsDNA. By 1 h, 100% of the pVI was cleaved and used to form active AVP-pVIc complexes. Thus, activation of AVP to AVP-pVIc complexes by pVI required the presence of DNA.
FIGURE 1.
Activation of AVP by pVI requires DNA and both proteins to be on the same DNA molecule. A, DNA is required for the activation of AVP by pVI. Assays contained 3.38 μm pVI, 6.54 μm AVP, and either 3.38 μm 36-bp DNA or no DNA. After the indicated time intervals, aliquots were removed and assayed for enzymatic activity. 100% activity is the enzyme activity of 3.38 μm AVP-pVIc complexes bound to DNA. ○, plus DNA; ▿, minus DNA. B, for activation of AVP by pVI, both proteins must be on the same DNA molecule. Reactions contained 77 nm pVI, 130 nm AVP, and the indicated concentrations of 36-mer dsDNA. After 30 min at 21 °C, the amount of AVP-pVIc formed was assayed. C, SDS-PAGE (15% polyacrylamide gel) analysis of the reactions in B except that the concentrations of pVI, AVP, and DNA were 26-fold higher. The positions of pVI, the intermediate VI-C, and VI are indicated by the lines on the left of the gel. The DNA concentration in the reactions fractionated on the gel increased from left to right. The numbers in parentheses in B are used as visual indicators to the corresponding gel lanes in C.
AVP and pVI Must Both Be on the Same DNA Molecule for Activation to Occur, and Reactions Proceed Only in cis and Not in trans
To see whether both enzyme (AVP) and substrate (pVI) must be on the same molecule of DNA for activation to occur or whether they can interact when bound to different DNA molecules, we incubated increasing concentrations of DNA with a mixture of AVP and pVI, each at sufficient concentrations to drive DNA binding at any concentration of DNA, and then we assayed for enzyme activation (Fig. 1B). At low DNA concentrations, the DNA was saturated with AVP and pVI; the rate of AVP activation was proportional to the DNA concentration. The rate of activation of AVP reached a peak at the concentration of DNA at which all the AVP and pVI were bound to all the DNA molecules. Beyond the peak, the rate of AVP activation progressively decreased. For example, at a DNA concentration of 320 nm, the rate of AVP activation was 12% of the rate exhibited at the DNA concentration at the peak, 50 nm. This is the type of curve expected if AVP and pVI must be on the same DNA molecule for activation to occur. Beyond the peak, as the concentration of DNA was progressively increased, the probability that both an AVP and a pVI molecule would be bound to the same DNA molecule would progressively decrease. If an AVP on one DNA molecule can be activated by a pVI on another DNA molecule, one would predict an initial curve similar to that shown in Fig. 1B, but beyond the peak, the rate of activation would have remained constant. This is because all the AVP and pVI would have been bound to DNA and, therefore, the bound protein concentration would not have changed as the DNA concentration was increased. These conclusions were corroborated by the SDS-PAGE analysis of the proteins present at the various DNA concentrations in Fig. 1C; pVI was not processed at the higher DNA concentrations. Thus, activation only proceeds in cis, i.e. both AVP and pVI must be on the same molecule of DNA.
AVP Does Not Slide Efficaciously on DNA
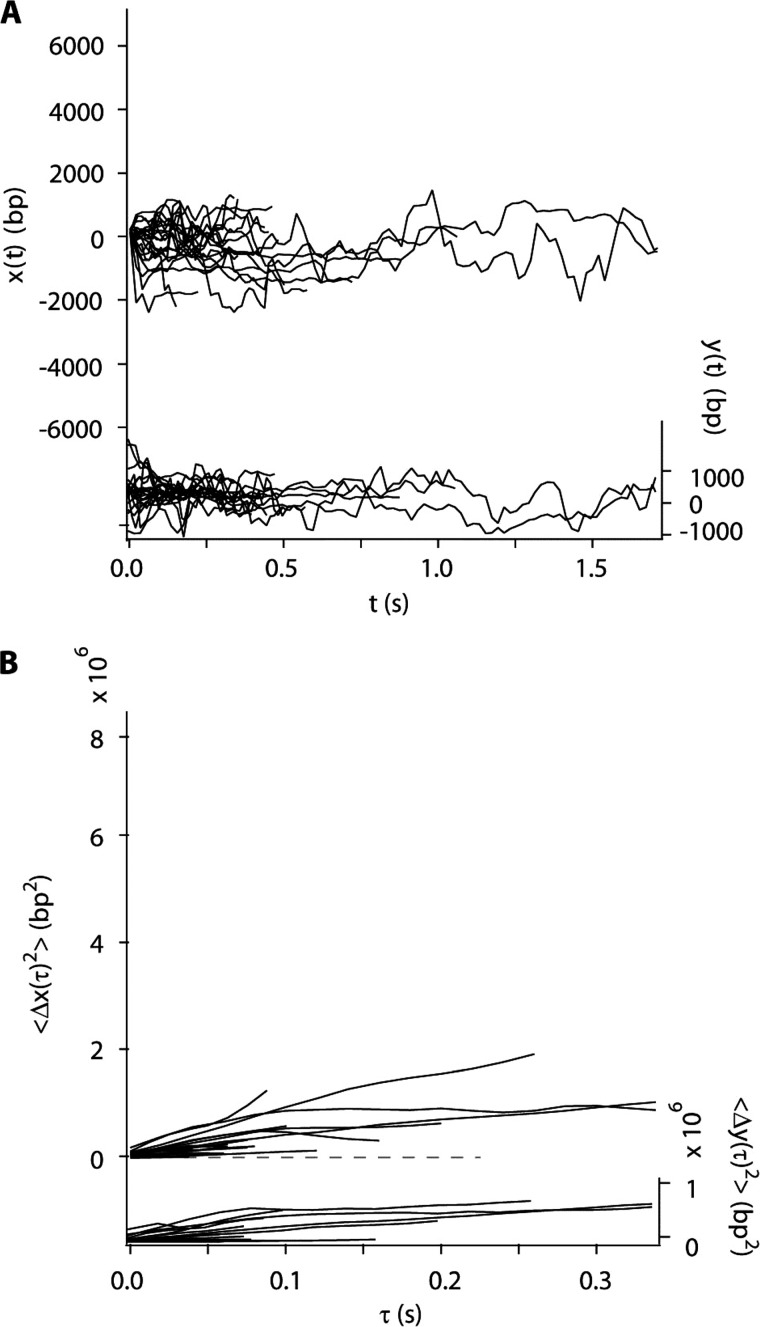
If both AVP and pVI must be on the same molecule of DNA for activation to occur, then presumably, one molecule must slide into the other to promote the bimolecular interaction that leads to the cutting out of pVIc and its binding to AVP. Does AVP slide along DNA? Using total internal reflection fluorescence microscopy as described previously (18, 20, 33), Cy3B-labeled AVP was introduced to flow-stretched DNA. As predicted (5), many molecules were observed to bind to random sites along the DNA.9 The average time AVP stayed bound to DNA was about 0.5 s, and the vast majority of these molecules remained at their initial locations until dissociating from the DNA. Less than 4% of the DNA-bound AVP molecules could be assigned diffusion constants statistically distinguishable from zero. Thus, the diffusion constants appeared to be bimodally distributed, with an effectively immobilized majority cluster and a potentially mobile minority cluster. Data from the minority, mobile fraction are shown in Fig. 2, A and B. The trajectories of 19 AVP molecules are plotted in Fig. 2A. The mean square displacement (MSD) of each molecular trajectory versus diffusion time is shown in Fig. 2B. From the MSD slopes, the mean one-dimensional diffusion constant (D1) of this subpopulation was 1.7 × 106 bp2/s, with S.D. of 1.9 × 106 bp2/s (Table 1). The diffusion constant relevant to searching along DNA is the mean across the entire population of bound AVP molecules. Assigning the immobilized majority cluster a diffusion constant of zero, the whole population mean D1 is very low, 0.02–0.07 × 106 bp2/s.
FIGURE 2.
Only a small minority of AVP molecules slide along flow-stretched dsDNA. A, sliding of a minority of AVP molecules, less than 4%, along the DNA (x(t), left axis, 19 trajectories). B, mean square displacement of the trajectories shown in A along the DNA (〈Δx(τ)2〉, left axis). In A and B, motion transverse to the DNA (y(t) and 〈Δy(τ)2〉, respectively, right axes) is represented on the same scale, as a control.
TABLE 1.
Binding to DNA, Kd, and sliding along DNA via one-dimensional diffusion and D1 values of adenovirus proteins
| Species molecular weights (amino acids in parentheses) | Ligand for Kd(app) analysis | Kd(app) | DNA-binding site length | One-dimensional diffusion constanta |
|---|---|---|---|---|
| nm | bp | bp2/s × 10−6 | ||
| pVI 27,014 (1–250) | 33-mer dsDNA | 46 ± 1.6b | 8 | 1.45 ± 0.13 |
| VI 22,118 (34–239) | 33-mer dsDNA | 307 ± 14b | ||
| pVIcc 1350 (240–250) | 12-mer dsDNA | 264 ± 25 | 7 | 26.0 ± 1.8 |
| AVP 23,087 (1–204) | 12-mer dsDNA | 63 ± 5.8d | 0.02–0.07e | |
| AVP-pVIc 24,435 (215) | 36-mer dsDNA | 4.6 ± 2.2d | 6 | 21.0 ± 1.9f |
| pVIc-biotin streptavidine ∼57,000 | 18-mer dsDNA | 35 ± 5.0c | 2.21 ± 0.21c |
a To convert from bp to nm: 106 bp2/s = 102,400 (nm)2/s.
b Ref. 32.
c Footnote 7.
d Ref. 5.
e Whole population-mean D1 calculated from one population (99–96% of the molecules bound to DNA) have a D1 of zero and another population (1–4% of the molecules bound to DNA) having a D1 of 1.7 × 106 bp2/s, with S.D. of 1.9 × 106 bp2/s.
f Ref. 33.
pVI Slides on DNA via One-dimensional Diffusion
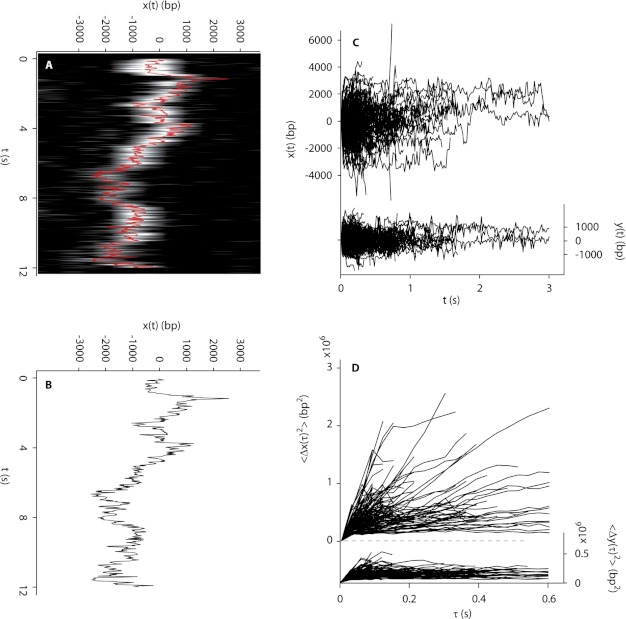
The observation that AVP molecules moved little while bound on DNA suggested the possibility that pVI might move on DNA. We labeled pVI molecules with the fluorophore Cy3B and observed the molecules binding to DNA as monomers (32) at random locations. Most important, the molecules slid rapidly over tens of thousands of base pairs before dissociating from the DNA. For example, the molecule whose motion is shown in the raw image data in Fig. 3A was determined by centroid analysis (Fig. 3B) to have traveled more than 10,000 bp during a 12-s binding event. The trajectories of 126 pVI molecules sliding on DNA are plotted in Fig. 3C; the MSD of each trajectory is shown versus diffusion time in Fig. 3D. The MSD for each molecule is approximately linear with diffusion time, indicating transport dominated by Brownian motion. From the MSD slopes, the mean diffusion constant was calculated to be 1.45 ± 0.13 × 106 bp2/s (Table 1).
FIGURE 3.
Sliding of pVI on flow-stretched DNA. A, rapid motion of a pVI molecule along flow-stretched dsDNA recorded at 284 Hz. Cinemagraph was generated from raw images showing motion along DNA (horizontal axis; each line is a strip of pixels from an image series frame) as a function of time (vertical axis). B, trajectory of the molecule depicted in A produced by Gaussian centroid determination of the signal of the molecule in each of 262 frames. C, pVI molecules diffuse rapidly along DNA (x(t), left axis, 126 trajectories). D, mean square displacement of the trajectories shown in C along the DNA (〈Δx(τ)2〉, left axis). In C and D, motion transverse to the DNA (y(t) and 〈Δy(τ)2〉, respectively, right axes) is represented on the same scale, as a control.
pVI Is Cleaved Sequentially by AVP Bound to DNA
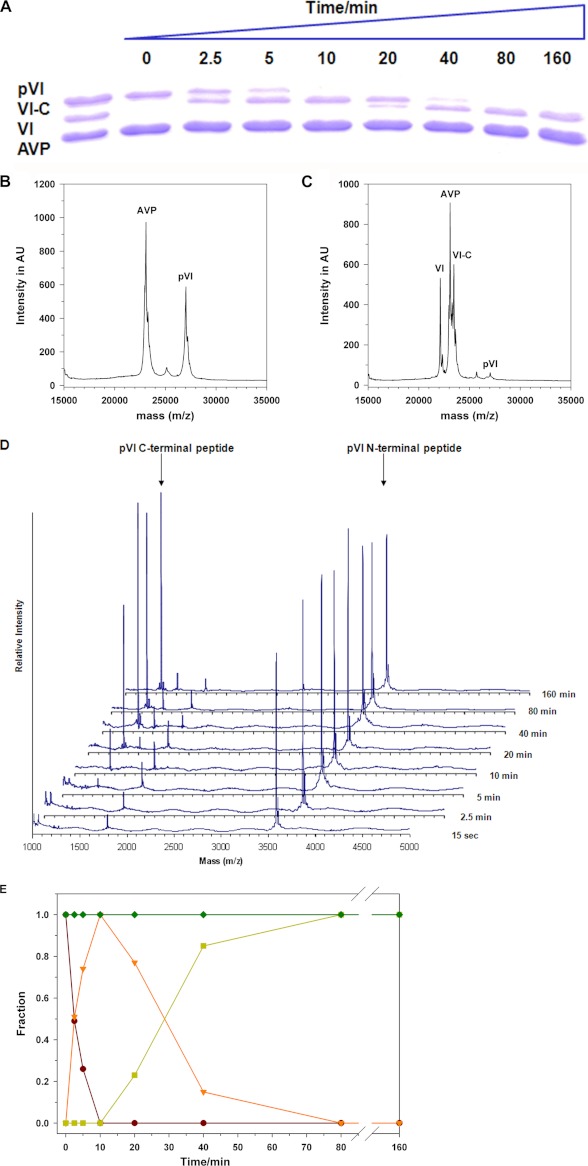
To determine the sequence of events in the activation of AVP by pVI in the presence of DNA, we assayed aliquots of the reaction in Fig. 1A for processing intermediates. SDS-polyacrylamide gel electrophoresis (Fig. 4A) showed that by 2.5 min, an intermediate in the processing of pVI to protein VI appeared. By 10 min, all the pVI had disappeared. At 20 min, protein VI began to appear, and by 40 min, almost all the intermediate had been converted to protein VI. A similar analysis of the reaction in the absence of DNA showed that pVI is not cleaved by AVP.10 MALDI-TOF mass spectroscopic analysis of the reactions in Fig. 1A showed that before the addition of DNA, two masses were present, AVP and pVI (Fig. 4B). At the 20-min time point, the pVI mass had disappeared (Fig. 4C). Masses corresponding to VI-C (pVI from which the N-terminal peptide (pVIn), amino acids 1–33, was cleaved) and VI had appeared. A MALDI-TOF mass spectroscopic analysis of the peptides generated at each time point is shown in Fig. 4D. By 15 s, the N-terminal peptide pVIn of pVI, amino acids 1–33, began to appear. At 5 min, pVIc, the peptide from the C terminus of pVI, amino acids 240–250, began to appear. Thus, the processing of pVI by AVP occurred in two steps, first cleavage at the N terminus of pVI and then at its C terminus.
FIGURE 4.
Sequence of events during the activation of AVP by pVI in the presence of DNA. During the activation of AVP by pVI in the presence of DNA depicted in Fig. 1A, aliquots were removed after various time intervals and assayed. A, SDS-PAGE (15% polyacrylamide gel) analysis. The first lane on the left contains the markers AVP, pVI, and VI. B, MALDI-TOF analysis of the proteins in the reaction in Fig. 1A before the DNA was added; this represents the 0-min time point. AU, arbitrary units. C, the 20-min time point is shown. D, MALDI-TOF analysis of the peptides produced in the reaction mixture in Fig. 1A at each time point. The arrows point to the peaks representing the masses (m/z) of the C-terminal (molecular weight 1,350, amino acids 240–250 from pVI) and N-terminal (molecular weight 3583, amino acids 1–33 from pVI) peptides. E, summary of the sequence of events in the activation of AVP by pVI in the presence of DNA. The gel in A was scanned for protein density, and the data were plotted as the fraction of the initial amount of AVP or pVI versus time. ♦, AVP; ●, pVI; ▾, VI-C; and ■, VI.
The results of the analysis of the activation of AVP by pVI in the presence of DNA shown in Fig. 4, A–D are summarized in Fig. 4E. The plot is based upon scans for protein density of the gel in Fig. 4A. The data were plotted as the fraction of the initial amount of AVP or pVI versus time. Initially, pVI was cleaved; half of it was processed by 2.5 min. Concomitantly, VI-C (pVI from which the N-terminal peptide, amino acids 1–33, was cleaved) appeared; half of its maximum appeared by 2.5 min. At 12 min, all of the pVI had disappeared, and there was a maximum amount of VI-C. Then, VI-C began to decline. After 30 min, half the maximum amount of VI-C was present with the concomitant appearance of half the maximal amount of VI. By 80 min, all the pVI and VI-C had disappeared, and the maximal amount of protein VI had appeared. N-VI (pVI from which the C-terminal peptide, amino acids 240–250, was cleaved) was not observed, indicating that the processing was sequential. The amount of AVP did not change during the time course, indicating that aliquots put on the gel all contained the same amount of protein.
What Happens to pVIc Once It Is Cleaved from the VI-C Intermediate
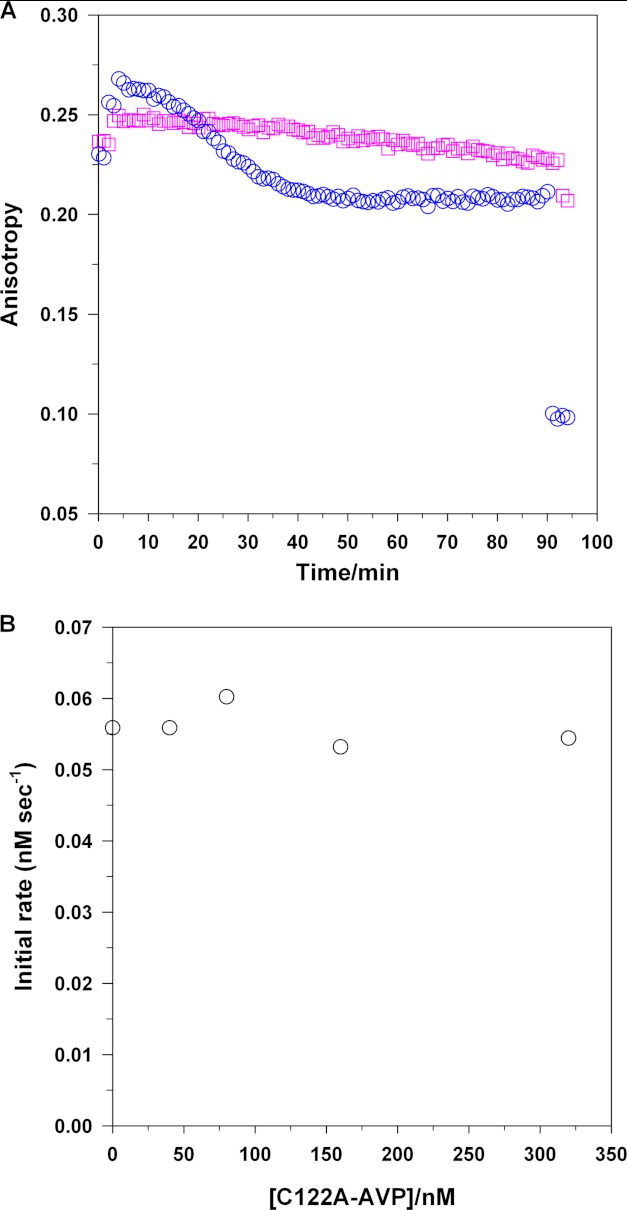
After pVIc is cut out from VI-C on DNA, does the resultant pVIc dissociate from the DNA, or does it stay associated with the DNA, bound or not bound to AVP? The experimental design to answer these questions is complicated by the knowledge that pVIc not only binds to AVP reversibly (9) and irreversibly (16) but, by itself, binds to DNA7 (5). To determine where pVIc resides after being cut out of pVI by AVP bound to DNA, we used fluorescence anisotropy. This is a rapid, sensitive, and quantitative technique used to measure protein-protein and protein-DNA interactions in solution. Fluorescence anisotropy is a measure of the depolarization of emitted fluorescence intensity obtained after excitation by a polarized light source. It depends directly on the relative rate of fluorescence emission versus the rate of tumbling in solution. We labeled pVI at Cys-249 with Cy3B. Cys-249 of pVI is Cys-10′ of the released pVIc, so the fluorescent label follows released pVIc, not protein VI, after cleavage of pVI by AVP bound to DNA. We used fluorescence anisotropy to monitor the tumbling of the Cy3B. The degree of tumbling is directly proportional to the size of the object the reporter is bound to. Thus, when it is attached to pVIc bound to AVP and or when Cy3B is attached to pVI bound to DNA, the tumbling is low as compared with when it is attached to pVIc in solution, and this is reflected by an increase in anisotropy.
Experimentally, we measured the fluorescence anisotropy of labeled pVI for 2 min (Fig. 5A). Then, we added 36-mer dsDNA. The anisotropy increased from 0.23 to 0.25, due to the binding of pVI to the DNA. After 4 min, AVP was added; the anisotropy increased further, to 0.27, due to the binding of AVP to the DNA. Over the next 40 min, the anisotropy decreased, from 0.27 to 0.21. Concomitant with the decrease in anisotropy was the formation of AVP-pVIc complexes as deduced by activity assays.10 The decrease in anisotropy was due to the release of VI from the DNA because its Kd(app), 307 nm, is much higher than the maximal concentration of VI that can be generated in the experiment, 77 nm (Table 1). After 50 min, the change in anisotropy decreased no more, stopping at 0.21. The experiment indicated after 50 min that all the pVI had been converted to pVIc and that the pVIc remained bound to the DNA by itself or associated with AVP.
FIGURE 5.
Released pVIc remains associated with the DNA, after excision from pVI, and binds to the AVP molecule that cut it out. A, pVIc remains associated with the DNA. The steady-state fluorescence anisotropy of 10 nm Cy3B-pVI and 67 nm unlabeled pVI in buffer A with 10 mm NaCl at 21 °C was measured for 2 min. Next, 67 nm 36-bp DNA (○) or 201 nm 12-bp DNA (□) was added. After an additional 2 min, 130 nm AVP was added, and the change in anisotropy was monitored for 80 min. Lastly, 399 nm histone H1 was added, and the anisotropy was monitored for 7 min. B, pVIc binds to the AVP molecule that cut it out. Increasing amounts of the AVP mutant C122A were added to reaction mixtures in buffer A containing 130 nm pVI, 130 nm AVP, and 2.8 nm 1500-mer dsDNA. After a 5-min incubation at 21 °C, the formation of active AVP-pVIc complexes was assayed.
These conclusions are based upon the following arguments. The anisotropy of free pVI is 0.23, and the anisotropy of free pVIc is 0.08. The addition of a high concentration of histone H1 (21), a nonspecific DNA-binding protein, to the reaction displaced all the other proteins bound to DNA, and this caused the anisotropy to drop to 0.08, the anisotropy of free pVIc (Fig. 5A). Thus, all the pVI had been processed to pVIc. Cy3B-labeled pVIc cannot form a covalent complex with AVP because its Cys-10′ contains the Cy3B fluorophore. Because the anisotropy of pVIc remained constant and high (0.21) for 50 min, we concluded that during the entire activation reaction and after, most of the pVIc remained associated with the DNA, bound to either AVP or DNA. Most of the labeled pVIc was probably bound to AVP bound to DNA because the apparent Kd for labeled pVIc binding to DNA is 264 nm,7 whereas the Kd for pVIc binding to AVP bound to DNA is 90 nm (9). The experiment was repeated to determine whether activation of AVP by pVI could occur in trans, i.e. with one molecule of AVP bound to DNA interacting with a molecule of pVI bound to another molecule of DNA. We chose 12-mer dsDNA because it was too short for an AVP and a pVI molecule to bind to the same molecule of DNA. Experimentally, no change in anisotropy occurred by 8 min after the addition of AVP to a solution of DNA and labeled pVI. This indicated that AVP and pVI did not bind to the same molecule of DNA. The addition of histone H1 after 95 min indicated that no pVIc had formed as the anisotropy remained at about 0.25. If pVIc had formed, the anisotropy should have decreased to 0.08. These results are also consistent with the conclusion that AVP cannot be activated by pVI unless both molecules reside on the same DNA molecule.
Does pVIc Bind to the AVP Molecule That Cut It Out?
To answer this question, we incubated AVP, pVI, and DNA with increasing concentrations of the C122A mutant of AVP and then assayed for AVP-pVIc complex activity (Fig. 5B). This mutant of AVP lacks enzymatic activity as the nucleophilic cysteine has been substituted; however, the mutant binds pVIc and binds to DNA like wild-type AVP.8 No decrease in AVP-pVIc complex activity was observed regardless of the mutant AVP concentration. That enzyme activity remained constant indicates that the newly cleaved pVIc did not bind to any AVP molecule present; it bound to the active AVP molecule that cut it out. If newly cleaved pVIc could bind to any AVP molecule, as the concentration of C122A was increased, the pVIc would have a greater probability to bind to the inactive mutant, and the amount of enzyme activity observed would decrease. Thus, pVIc binds to the AVP molecule that cut it out from pVI.
Does a Disulfide Bond Form between pVIc and the AVP Molecule That Cut It Out from pVI?
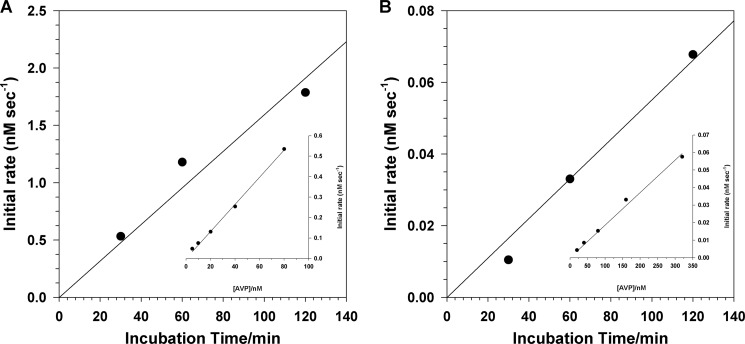
To determine whether a disulfide bond forms between pVIc and the AVP molecule that cut it out from pVI, we set up an activation reaction with AVP, pVI, and DNA at pH 8 and, as a function of time, measured: 1) the total concentration of reversible AVP-pVIc complexes plus disulfide-linked AVP-pVIc complexes and 2) the concentration of disulfide-linked AVP-pVIc complexes. If a disulfide bond is formed between pVIc and the AVP molecule that cut it out, then the total concentration of reversible AVP-pVIc complexes plus disulfide-linked AVP-pVIc complexes should equal the concentration of disulfide-linked AVP-pVIc complexes. In the experiment, we incubated 0.77 μm pVI, 1.3 μm AVP, and 0.67 μm 36-mer dsDNA. After various time intervals, two aliquots were removed. One aliquot was diluted with an equal volume of buffer containing the fluorogenic substrate (Leu-Arg-Gly-Gly-NH)2-rhodamine. The other aliquot was diluted into the same solution, but it also contained 0.5 m NaCl. Enzyme activity in the absence of NaCl should be reflective of all AVP-pVIc complexes formed, both reversible and disulfide-linked. This is because the highest possible concentration of pVIc generated is 3.5-fold higher than the Kd of pVIc for AVP in the presence of DNA, 90 nm (9). Thus, reversible complexes bound to DNA should remain intact, as would disulfide-linked complexes. Each type of complex, reversible and disulfide-linked, has the same specific activity (16). Enzyme activity in the presence of NaCl should be reflective only of those AVP-pVIc complexes formed that contained a disulfide bond between Cys-10′ of pVIc and Cys-104 of AVP. At that concentration of salt and those protein concentrations, AVP, pVIc, and all AVP-pVIc complexes cannot bind to DNA (5). Not being bound to DNA, the reversible AVP-pVIc complexes will dissociate to AVP and pVIc because the highest possible concentration of pVIc that could be generated is 11-fold lower than the Kd of pVIc for AVP in the absence of DNA, 4.4 μm (9). Because AVP alone has no detectable enzyme activity, the only enzymatic activity observed will be from disulfide-linked AVP-pVIc complexes.
The data (Fig. 6) indicated that all of the AVP-pVIc complexes that formed during the activation reaction became disulfide-linked. For the first 100 min of the activation reaction, the rates of formation of AVP-pVIc complexes, reversible and irreversible (Fig. 6A) or irreversible (Fig. 6B), were linear. Assays of the reaction mixture in low salt (bound to DNA) gave a much higher rate of substrate hydrolysis than that in high salt (not bound to DNA). This was to be expected. The kcat/Km ratio for substrate hydrolysis by AVP-pVIc complexes bound to DNA is 16-fold higher than by AVP-pVIc complexes in the absence of DNA (5, 9). Using standard curves relating rates of substrate hydrolysis versus the concentrations of AVP-pVIc complexes bound and not bound to DNA (Fig. 6, A and B, insets), we calculated that at the 30-min time point, 128 nm AVP-pVIc complexes, reversible and irreversible, had formed and that 130 nm of them were disulfide-linked. Thus, it appears as if all the pVIc generated formed a disulfide bond with the AVP molecule that cut it out.
FIGURE 6.
pVIc forms a disulfide bond with the AVP molecule that cut it out. A reaction mixture was set up in buffer B containing 0.77 μm pVI, 1.3 μm AVP, and 0.67 μm 36-mer dsDNA. After the indicated time intervals, 55-μl aliquots were removed and added to 55 μl of either 5 μm (Cbz-Leu-Arg-Gly-Gly-NH)2-rhodamine in buffer B containing 0.67 μm 36-mer dsDNA (A) or 5 μm (Cbz-Leu-Arg-Gly-Gly-NH)2-rhodamine in buffer B but with 0.5 m NaCl (B). The rates of substrate hydrolysis were measured over a 30-min period. The insets are standard curves relating the initial rate as a function of time versus molar concentration of AVP-pVIc complexes bound to DNA (A) and AVP-pVIc complexes not bound to DNA (B). In the inset for A, the reactions in buffer B contained the indicated concentrations of AVP plus 20 μm pVIc, 0.67 μm 36-mer dsDNA, and 5 μm (Cbz-Leu-Arg-Gly-Gly-NH)2-rhodamine. In the inset for B, the reaction in buffer B with 250 mm NaCl contained 5 μm (Cbz-Leu-Arg-Gly-Gly-NH)2-rhodamine.
Is DNA Required for the Activation of AVP by pVI in Vivo?
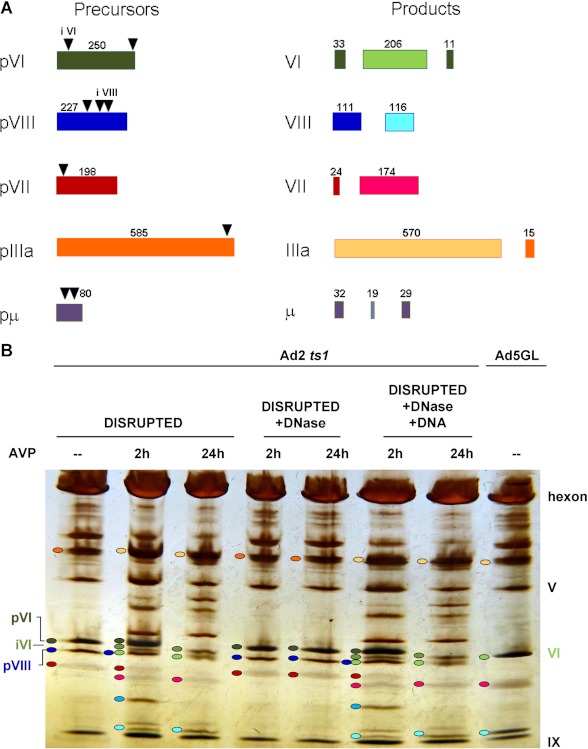
Is DNA required for the activation of AVP in vivo, inside immature particles where the viral DNA is decorated with tightly bound proteins such as pVII and protein V (22)? There are two observations that indicate it is. First, sliding does occur in vivo, at a DNA concentration not unlike that inside the adenovirus virion, by the lac repressor inside Escherichia coli (23). Secondly, we observe DNA-dependent activation of AVP in a quasi in vivo situation, using heat-disrupted ts-1 virus particles (19). ts-1 virus is a temperature-sensitive mutant of adenovirus that when grown at the nonpermissive temperature produces noninfectious virions devoid of AVP; as such, all the virion precursor proteins are intact (2). The graphic in Fig. 7A depicts the precursor proteins, the processing sites, and the mature proteins. In one experiment, AVP was incubated with heat-disrupted ts-1 virus for 2 and 24 h before fractionating the proteins on an SDS-polyacrylamide gel. The results (Fig. 7B) indicated that some pVI was processed within 2 h, and all of it was processed within 24 h as indicated by the disappearance of the pVI band. During the processing of pVI, other precursor proteins (pIIIa, pVII, and pVIII) were observed to be processed, presumably by the newly formed AVP-pVIc complexes (33). If, before adding AVP, heat-disrupted ts-1 virus was incubated with DNase, no processing of pVI or the other precursor proteins was observed, 2 or 24 h after adding AVP. Most convincing was the experiment in which heat-disrupted ts-1 virus was incubated with DNase, the DNase was inactivated, and ts-1 viral DNA was added back. Here, upon adding AVP and incubating for 2 or 24 h, the processing pattern of pVI and the other precursor proteins was identical to that observed with heat-disrupted ts-1 virus particles just incubated with AVP. Thus, in heat-disrupted virus, DNA is required for the activation of AVP.
FIGURE 7.
In heat-disrupted ts-1 virus particles, activation of AVP by pVI requires DNA. A, the precursor proteins, their cleavage sites (▾), and the processed precursor proteins are labeled and color-coded. B, in heat-disrupted ts-1 virus particles, activation of AVP requires DNA. Proteins were fractionated on PhastGel 8–25% gradient gels. pVI is cleaved to protein VI via an intermediate iVI. pVIII, the precursor to protein VIII, migrates like protein VI. Lane 1 contained heat-disrupted ts-1 virus incubated for 24 h. Lanes 2 and 3 contained heat-disrupted ts-1 virus incubated with AVP for 2 or 24 h, respectively. Lanes 4 and 5 contained heat-disrupted ts-1 virus treated with DNase and then incubated with AVP for 2 or 24 h, respectively. Lanes 6 and 7 contained heat-disrupted ts-1 virus treated with DNase, with the DNase inactivated, the DNA returned, and then the reactions incubated with AVP for 2 or 24 h, respectively. Lane 8 contained wild-type virus (Ad5GL). Ovals next to bands refer to the specific precursor proteins and their products that are color-coded as in B.
DISCUSSION
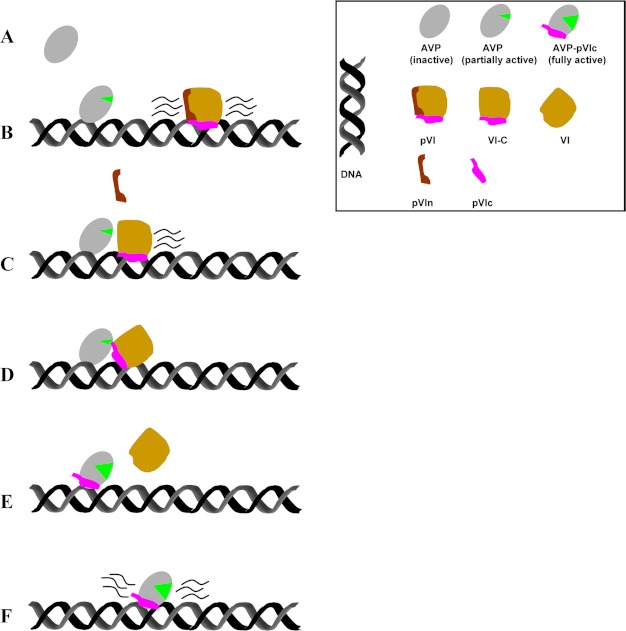
We have shown how AVP can be activated by pVI in the core of the immature virion, i.e. how an AVP-pVIc complex can form in a dense DNA environment. AVP, synthesized as an inactive proteinase to prevent premature processing of viral precursor proteins, binds to the viral DNA and does not move. pVI binds to the viral DNA and slides via one-dimensional diffusion into AVP. AVP, partially activated by being bound to its cofactor DNA (3, 5–7), cleaves DNA-bound pVI twice, once at its N terminus and then at its C terminus. Because pVI probably binds to DNA via its pVIc moiety7 (32, 33), to stay on the DNA for both cleavages, pVI must first be cleaved at its N terminus to generate the processing intermediate VI-C and liberate a 33-amino acid peptide. Then, presumably, a rate-limiting, conformational change occurs, making the C-terminal cleavage site accessible. Secondly, AVP cleaves VI-C near its C terminus, generating protein VI and liberating pVIc, the 11-amino acid peptide cofactor. pVIc then preferentially binds to the AVP molecule that cut it out. Finally, pVIc forms a disulfide bond with AVP, keeping the now fully activated enzyme, the AVP-pVIc·DNA complex, permanently activated (9, 15, 16). These data are summarized in the model in Fig. 8.
FIGURE 8.
A model, based upon the data in this study, of the activation of AVP by pVI on DNA. A, AVP is inactive, no green mouth. B, AVP bound to DNA is slightly active, green mouth. pVI moves back and forth along the DNA via one-dimensional diffusion. C, pVI slides into AVP. AVP, slightly active, cleaves pVI at its N terminus, releasing the 33-amino acid peptide pVIn. D, a conformational change occurs so that the active site of AVP is at the C terminus of VI-C. E, AVP then cleaves VI-C at its C terminus, releasing the 11-amino acid peptide pVIc that binds to the AVP molecule that cut it out. The fully active AVP-pVIc complex bound to DNA is formed. Protein VI dissociates from the DNA. F, the fully active AVP-pVIc complex bound to DNA slides along DNA via one-dimensional diffusion to locate and process virion proteins also bound to DNA.
That activation of AVP by pVI requires DNA in vitro is consistent with the quasi in vivo data on the activation of AVP using ts-1 virus particles disrupted by heat. We showed that when heat-disrupted ts-1 virus particles are incubated with AVP, by 2 h after the addition of AVP, some pVI is processed concomitant with the beginnings of processing of some of the precursor proteins. If the heat-disrupted ts-1 virus particles are treated with DNase and then AVP is added, no processing of pVI or any of the other virion precursor proteins was observed, even after a 24-h incubation. Most significant, if after DNase treatment, the DNase is inactivated and DNA is added back, by 2 h after the addition of AVP, some pVI is processed concomitant with the beginnings of processing of some of the precursor proteins. By 24 h, all the pVI had been processed. These and other data (3, 8, 33) imply that in the immature virus particle, AVP is activated by pVI while both are bound to the viral DNA.
Binding to and sliding along DNA are required for the activation of AVP by pVI. Binding of AVP to DNA partially activates the enzyme, allowing it to cleave pVI also bound to DNA. Also, binding of AVP and pVI to DNA must orient them such that a productive collision occurs when the substrate-binding site near the N terminus of pVI slides into the active site of AVP. This is a novel type of biochemistry, one-dimensional biochemistry.7 Sliding on DNA is required for the activation of AVP by pVI. In the absence of DNA, AVP will not even bind to pVI.10 That both AVP and pVI must be on the same DNA molecule for formation of AVP-pVIc complexes is consistent with a requirement for sliding. Because AVP binds to DNA and does not slide, the only way AVP could become activated via pVI is for pVI to slide on DNA into AVP. We recently showed that molecules sliding along DNA, including AVP-pVIc complexes, diffuse along a helical path defined by the double helix and rotate to keep the DNA-binding face of the protein in contact with DNA (20, 24).
The very small fraction of AVP molecules that may slide on DNA is not relevant biologically, i.e. in vivo AVP probably does not participate in the cleavage of the precursor proteins by sliding along the viral DNA. Overall, the sliding rate of AVP is 300–1,000-fold lower than that for AVP-pVIc complexes. Because there are 50 molecules of AVP per virion, only 1–3 molecules per virion could be actively sliding. Also, because the relative kcat/Km of AVP-DNA complexes is more than 300-fold lower than that of AVP-pVIc·DNA complexes, it seems unlikely that AVP-DNA complexes would be active enough to be able to process virion precursor proteins efficiently (5, 8, 9). In wild-type, infectious virions, all the AVP molecules present are in the form of covalent AVP-pVIc complexes (15).
This activation mechanism for AVP ensures that AVP will not be prematurely activated. If AVP and pVI collide in the cytoplasm or nucleus of the host cell, AVP will not become activated because both must be bound to DNA. Secondly, pVI may not even be able to bind to DNA before it gets into a nascent virion. In the cytoplasm, pVI has been shown to bind tightly to hexon, the major structural protein of the adenovirus capsid, and to escort hexon into the nucleus (25, 26). Three molecules of pVI bind to one hexon trimer (32). The two regions on pVI for hexon binding are at amino acids 48–74 and 233–239 (27). The nuclear location signal, at the C terminus of pVI (KRRR), is within the pVIc sequence (amino acids 240–250). Premature activation of AVP to AVP-pVIc complexes can interfere with virus replication. We have shown that if pVIc is added along with adenovirus to cells in culture, there is a large reduction in the production of infectious virus, a 99.8% reduction 28 h after infection (9). The mechanism of virion assembly is not known. It is not known how the viral DNA enters a nascent virion nor, once inside, how the viral DNA is situated. Thus, it is difficult to speculate how this process prevents AVP from being prematurely activated by pVI.
This novel mechanism for the activation of AVP should be sufficient to allow for the activation of all the 50 molecules of AVP in the virion (28). There are 360 molecules of pVI (29, 30), and given that individual molecules can slide over as many as 20,000 bp before dissociating from the DNA, they should be able to efficiently locate all the AVP molecules bound along the 36,000-bp adenovirus genome. The remaining pVI molecules are processed by AVP-pVIc complexes; we have shown that AVP-pVIc complexes bound to DNA slide along the DNA via one-dimensional diffusion, processing the virion precursor proteins, including pVI, also bound to the viral DNA (33).
DNA performs several, crucial functions during the activation of AVP to AVP-pVIc complexes by pVI. Both AVP (5) and pVI bind to DNA. DNA, as a cofactor, partially activates AVP (3, 5) so that it can cleave pVI twice. We had previously shown in vitro that AVP bound to DNA is partially active, i.e. it can cleave synthetic substrates (3, 5, 9). pVI slides on DNA, and AVP does not. Binding to DNA must orient AVP and pVI such that a productive collision occurs when the substrate-binding site at the N terminus of pVI slides into the active site of AVP; this is a new type of one-dimensional biochemistry.7 Also, if AVP and pVI are bound to different DNA molecules, they do not interact, implying that the active site and the substrate-binding site are both not readily accessible in trans. DNA lowers the equilibrium dissociation constant of pVIc for AVP 50-fold, from 4.4 μm in the absence of DNA to 90 nm in its presence (9). This helps to ensure that pVIc, when liberated from pVI, binds to the AVP molecule that cut it out.
A conundrum about the assembly of adenovirus virions arises when it is realized that AVP and the six virion precursor proteins are essentially irreversibly bound to the viral DNA inside the immature virion. However, under these conditions, somehow AVP becomes activated by pVIc and the virion precursor proteins become processed by AVP-pVIc complexes. Here, we addressed the part of the conundrum as to how AVP becomes activated by pVI under these conditions. The solution is an unprecedented series of reactions between a proteinase and its substrate. In a different study, we address the remaining enigma of how fully activated AVP-pVIc complexes process the virion precursor proteins (33). AVP-pVIc complexes slide along the DNA via one-dimensional diffusion processing the virion precursor proteins also bound to the DNA. Finally, we address the issue of how pVI and AVP-pVIc complexes slide along the DNA. The element they both have in common is pVIc, and we show that pVIc is a “molecular sled” that can slide by itself or slide with different adenovirus cargos attached to it such as protein VI or AVP or even slide with heterologous cargos attached to it.7
Although many proteins, including viral proteins (31), have been shown to slide on DNA, all the examples to date have been of “nucleic acid” proteins and enzymes with functions relevant to specific loci or features in the genome. pVI, by contrast, is an adenovirus structural protein tasked with no known functions at particular genomic loci. Not only is there no precedent for a “non-nucleic acid protein” sliding on DNA, there is no precedent for a substrate sliding along DNA into an enzyme that will cleave it. Given this novel exploitation by pVIc of the ability of DNA to present a sliding surface, following this new precedent, other proteins/peptides may also be discovered that make noncanonical use of facilitated diffusion along DNA.
Acknowledgments
We thank Eileen Kasmarcik in the F. William Studier laboratory for cloning pVI and Michelle Louie for cloning protein VI. We are grateful to María López (CNB-CSIC) and Wenying Huang (Princeton University) for technical assistance and to Elena Molina and Rodrigo Jiménez-Saiz (Instituto de Investigacion en Ciencias de la Alimentacion - Consejo Superior de Investigaciones Cientificas) for access to the PhastGel electrophoresis system.
This work was supported, in whole or in part, by National Institutes of Health Grants R01AI41599 (to W. F. M.) and GM037705 (to S. J. F.) and the National Institutes of Health Director's Pioneer Award and a National Science Foundation grant (to X. S. X.). This work was also supported by Grants BFU2010-16382 and FIS2010-10552-E from the Ministerio de Ciencia e Innovación of Spain (to C. S. M.)
The following designations are used in this study: AVP-pVIc, noncovalent or covalently linked heterodimer of AVP and pVIc; N-VI, pVI from which the C-terminal peptide, amino acids 240–250, was cleaved; pVIc, 11-amino acid cofactor (GVQSLKRRRCF) originating from the C terminus of virion precursor protein pVI; pVIn, the 33 amino acid peptide originating from the N terminus of virion precursor protein pVI; VI-C, the cleavage product of virion precursor protein pVI from which the N-terminal 33 amino acids have been removed. Cbz, carboxybenzyl; pVII, the precursor to adenovirus protein VII; pIIIa, the precursor to adenovirus protein IIIa; pμ, the precursor to adenovirus protein μ.
P. C. Blainey, V. Graziano, W. J. McGrath, G. Luo, X. S. Xie, and W. F. Mangel, submitted.
W. J. McGrath and W. F. Mangel, unpublished observations.
P. C. Blainey, W. J. McGrath, and W. F. Mangel, unpublished observations.
V. Graziano and W. F. Mangel, unpublished observations.
- AVP
- adenovirus proteinase
- Ad2
- human adenovirus serotype 2
- DDM
- n-dodecyl-β-d-maltopyranoside
- pVI
- the precursor to adenovirus protein VI
- MSD
- mean square displacement.
REFERENCES
- 1. Ding J., McGrath W. J., Sweet R. M., Mangel W. F. (1996) Crystal structure of the human adenovirus proteinase with its 11 amino acid cofactor. EMBO J. 15, 1778–1783 [PMC free article] [PubMed] [Google Scholar]
- 2. Weber J. (1976) Genetic analysis of adenovirus type 2 III. Temperature sensitivity of processing viral proteins. J. Virol. 17, 462–471 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Mangel W. F., McGrath W. J., Toledo D. L., Anderson C. W. (1993) Viral DNA and a viral peptide can act as cofactors of adenovirus virion proteinase activity. Nature 361, 274–275 [DOI] [PubMed] [Google Scholar]
- 4. Webster A., Hay R. T., Kemp G. (1993) The adenovirus protease is activated by a virus-coded disulphide-linked peptide. Cell 72, 97–104 [DOI] [PubMed] [Google Scholar]
- 5. McGrath W. J., Baniecki M. L., Li C., McWhirter S. M., Brown M. T., Toledo D. L., Mangel W. F. (2001) Human adenovirus proteinase: DNA binding and stimulation of proteinase activity by DNA. Biochemistry 40, 13237–13245 [DOI] [PubMed] [Google Scholar]
- 6. Bajpayee N. S., McGrath W. J., Mangel W. F. (2005) Interaction of the adenovirus proteinase with protein cofactors with high negative charge densities. Biochemistry 44, 8721–8729 [DOI] [PubMed] [Google Scholar]
- 7. Gupta S., Mangel W. F., McGrath W. J., Perek J. L., Lee D. W., Takamoto K., Chance M. R. (2004) DNA binding provides a molecular strap activating the adenovirus proteinase. Mol. Cell. Proteomics 3, 950–959 [DOI] [PubMed] [Google Scholar]
- 8. Mangel W. F., Toledo D. L., Brown M. T., Martin J. H., McGrath W. J. (1996) Characterization of three components of human adenovirus proteinase activity in vitro. J. Biol. Chem. 271, 536–543 [DOI] [PubMed] [Google Scholar]
- 9. Baniecki M. L., McGrath W. J., McWhirter S. M., Li C., Toledo D. L., Pellicena P., Barnard D. L., Thorn K. S., Mangel W. F. (2001) Interaction of the human adenovirus proteinase with its 11-amino acid cofactor pVIc. Biochemistry 40, 12349–12356 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Baniecki M. L., McGrath W. J., McWhirter S. M., Li C., Toledo D. L., Pellicena P., Barnard D. L., Thorn K. S., Mangel W. F. (2002) Interaction of the human adenovirus proteinase with its 11-amino acid cofactor pVIc. Biochemistry 41, 430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Russell W. C., Precious B. (1982) Nucleic acid-binding properties of adenovirus structural polypeptides. J. Gen. Virol. 63, 69–79 [DOI] [PubMed] [Google Scholar]
- 12. Casjens S. (1997) Principles of virion structure, function and assembly. In: Structural Biology of Viruses (Chiu W., Burnett R. M., Garcea R. L., eds) pp. 3–37, Oxford University Press, Oxford [Google Scholar]
- 13. Mangenot S., Keller S., Rädler J. (2003) Transport of nucleosome core particles in semidilute DNA solutions. Biophys. J. 85, 1817–1825 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. McGrath W. J., Abola A. P., Toledo D. L., Brown M. T., Mangel W. F. (1996) Characterization of human adenovirus proteinase activity in disrupted virus particles. Virology 217, 131–138 [DOI] [PubMed] [Google Scholar]
- 15. McGrath W. J., Aherne K. S., Mangel W. F. (2002) In the virion, the 11-amino-acid peptide cofactor pVIc is covalently linked to the adenovirus proteinase. Virology 296, 234–240 [DOI] [PubMed] [Google Scholar]
- 16. McGrath W. J., Baniecki M. L., Peters E., Green D. T., Mangel W. F. (2001) Roles of two conserved cysteine residues in the activation of human adenovirus proteinase. Biochemistry 40, 14468–14474 [DOI] [PubMed] [Google Scholar]
- 17. Gasteiger E., Gattiker A., Hoogland C., Ivanyi I., Appel R. D., Bairoch A. (2003) ExPASy: The proteomics server for in-depth protein knowledge and analysis. Nucleic Acids Res. 31, 3784–3788 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Blainey P. C., van Oijen A. M., Banerjee A., Verdine G. L., Xie X. S. (2006) A base-excision DNA-repair protein finds intrahelical lesion bases by fast sliding in contact with DNA. Proc. Natl. Acad. Sci. U.S.A. 103, 5752–5757 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Pérez-Berná A. J., Marabini R., Scheres S. H. W., Menéndez-Conejero R., Dmitriev I. P., Curiel D. T., Mangel W. F., Flint S. J., San Martín C. (2009) Structure and uncoating of immature adenovirus. J. Mol. Biol. 392, 547–557 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Blainey P. C., Luo G., Kou S. C., Mangel W. F., Verdine G. L., Bagchi B., Xie X. S. (2009) Nonspecifically bound proteins spin while diffusing along DNA. Nat. Struct. Mol. Biol. 16, 1224–1229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Ramakrishnan V. (1997) Histone H1 and chromatin higher-order structure. Crit. Rev. Eukaryot. Gene Expr. 7, 215–230 [DOI] [PubMed] [Google Scholar]
- 22. Flint S. J., Enquist L. W., Racaniello V. R., Skalka A. M. (2009) Principles of Virology, Third Ed., ASM Press [Google Scholar]
- 23. Elf J., Li G.-W., Xie X. S. (2007) Probing transcription factor dynamics at the single-molecule level in a living cell. Science 316, 1191–1194 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Bagchi B., Blainey P. C., Xie X. S. (2008) Diffusion constant of a nonspecifically bound protein undergoing curvilinear motion along DNA. J. Phys. Chem. B 112, 6282–6284 [DOI] [PubMed] [Google Scholar]
- 25. Matthews D. A., Russell W. C. (1995) Adenovirus protein-protein interactions: molecular parameters governing the binding of protein VI to hexon and the activation of the adenovirus 23K protease. J. Gen. Virol. 76, 1959–1969 [DOI] [PubMed] [Google Scholar]
- 26. Wodrich H., Guan T., Cingolani G., Von Seggern D., Nemerow G., Gerace L. (2003) Switch from capsid protein import to adenovirus assembly by cleavage of nuclear transport signals. EMBO J. 22, 6245–66255 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Matthews D. A., Russell W. C. (1994) Adenovirus protein-protein interactions: hexon and protein VI. J. Gen. Virol. 75, 3365–3374 [DOI] [PubMed] [Google Scholar]
- 28. Brown M. T., McGrath W. J., Toledo D. L., Mangel W. F. (1996) Different modes of inhibition of human adenovirus proteinase, probably a cysteine proteinase, by bovine pancreatic trypsin inhibitor. FEBS Lett. 388, 233–237 [DOI] [PubMed] [Google Scholar]
- 29. van Oostrum J., Burnett R. M. (1985) Molecular composition of the adenovirus type 2 virion. J. Virol. 56, 439–448 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Lehmberg E., Traina J. A., Chakel J. A., Chang R. J., Parkman M., McCaman M. T., Murakami P. K., Lahidji V., Nelson J. W., Hancock W. S., Nestaas E., Pungor E., Jr. (1999) Reversed-phase high-performance liquid chromatographic assay for the adenovirus type 5 proteome. J. Chromatogr. B Biomed. Sci. Appl. 732, 411–423 [DOI] [PubMed] [Google Scholar]
- 31. Flint S. J., Enquist L. W., Krug R. M., Racaniello V. R., Skalka A. M. (2000) Principles of Virology: Molecular Biology, Pathogenesis, and Control, ASM Press, Washington, D. C [Google Scholar]
- 32. Graziano V., McGrath W. J., Suomalainen M., Greber U. F., Freimuth P., Blainey P. C., Luo G., Xie X. S., Mangel W. F. Regulation of a viral proteinase by a peptide and DNA in one-dimensional space. I. Binding to DNA and to hexon of the precursor to protein VI, pVI, of human adenovirus. (2012) J. Biol. Chem. 287, 2059–2067 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Blainey P. C., Graziano V., Pérez-Berná A. J., McGrath W. J., Flint S. J., San Martín C., Xie X. S., Mangel. W. F. Regulation of a viral proteinase by a peptide and DNA in one-dimensional. space. IV. Viral proteinase slides along DNA to locate and process its substrates. (2012) J. Biol. Chem. 287, 2092–2102 [DOI] [PMC free article] [PubMed] [Google Scholar]