Background: Activated adenovirus proteinase (AVP-pVIc) is generated from an inactive form (AVP), but the structural changes that activate AVP are unknown.
Results: The crystal structure of the AVP was determined.
Conclusion: Comparison of AVP with AVP-pVIc reveals why AVP is inactive by changes in one domain.
Significance: Activation of the enzyme may occur along a 62-amino acid pathway by contiguous conformational changes.
Keywords: Adenovirus, Cysteine Protease, Enzyme Catalysis, Enzyme Structure, X-ray Crystallography, Atomic Resolution, Cation-π, Interaction, Cofactors, Enzyme Activation
Abstract
The adenovirus proteinase (AVP), the first member of a new class of cysteine proteinases, is essential for the production of infectious virus, and here we report its structure at 0.98 Å resolution. AVP, initially synthesized as an inactive enzyme, requires two cofactors for maximal activity: pVIc, an 11-amino acid peptide, and the viral DNA. Comparison of the structure of AVP with that of an active form, the AVP-pVIc complex, reveals why AVP is inactive. Both forms have an α + β fold; the major structural differences between them lie in the β-sheet domain. In AVP-pVIc, the general base His-54 Nδ1 is 3.9 Å away from the Cys-122 Sγ, thereby rendering it nucleophilic. In AVP, however, His-54 Nδ1 is 7.0 Å away from Cys-122 Sγ, too far away to be able to abstract the proton from Cys-122. In AVP-pVIc, Tyr-84 forms a cation-π interaction with His-54 that should raise the pKa of His-54 and freeze the imidazole ring in the place optimal for forming an ion pair with Cys-122. In AVP, however, Tyr-84 is more than 11 Å away from its position in AVP-pVIc. Based on the structural differences between AVP and AVP-pVIc, we present a model that postulates that activation of AVP by pVIc occurs via a 62-amino acid-long activation pathway in which the binding of pVIc initiates contiguous conformational changes, analogous to falling dominos. There is a common pathway that branches into a pathway that leads to the repositioning of His-54 and another pathway that leads to the repositioning of Tyr-84.
Introduction
As with many viruses, adenoviruses encode a proteinase whose activity is essential for the production of infectious virus (1). The adenovirus proteinase (AVP),2 whose structure we describe here, is synthesized as an inactive enzyme (2, 3). Late in adenovirus infection, about 50 copies of AVP (4) become activated inside young virions (5) and cleave about 1500 copies (6) of the six different virion precursor proteins used in the assembly of the virion (1). AVP is synthesized in an inactive form because if it were not, it would cleave virion precursor proteins before virion assembly, thereby aborting an infection (7). Two viral cofactors have been discovered that stimulate proteinase activity. One cofactor is pVIc,3 the 11-amino acid peptide from the C terminus of adenovirus precursor protein pVI (2, 7, 8). Its primary sequence is GVQSLKRRRCF. Cys-104 of AVP can form a disulfide bond with Cys-10′ of pVIc in vitro (9, 10) and does so in vivo in the virus particle (11). A second cofactor is the viral DNA (2, 12). The two viral cofactors increase the specificity constant (kcat/Km) for substrate hydrolysis (2, 7, 12, 13). In the presence of human adenovirus serotype 2 (Ad2) DNA, the kcat/Km for AVP increases 110-fold (7, 12, 13); in the presence of pVIc, it increases 1130-fold. With all three components together, AVP, pVIc, and human adenovirus serotype 2 DNA, the kcat/Km increases 15,800-fold.
How AVP acquires pVIc from pVI and how active AVP-pVIc complexes process the virion precursor proteins have presented a conundrum. AVP and pVI are sequence-independent DNA-binding proteins (2, 12, 46). In the tightly packed interior of a young virion, the concentration of viral DNA is 500 g/liter (14); that plus the sieving effect of DNA (15) diminishes the effective three-dimensional diffusion constants of AVP and pVI by more than 1 million-fold (46–48). A model solving the conundrum as to how AVP is activated and how AVP-pVIc complexes cleave the virion precursor proteins has been presented and experimentally verified. AVP binds randomly to DNA and does not slide along the DNA (47). pVI also binds randomly to DNA, but it slides along DNA with a one-dimensional diffusion constant of 1.45 × 106 bp2/s. pVI slides into AVP. AVP, partially activated by being bound to the viral DNA, cleaves pVI first at its N terminus and then at its C terminus. pVIc, released by cleavage of pVI at its C terminus, binds to the AVP that cut it out, and then a disulfide bond is formed between Cys-10′ of pVIc and Cys-104 of AVP, thereby keeping AVP permanently activated. The processing of the virion proteins by AVP-pVIc complexes occurs by the following mechanism (48). Covalent, active AVP-pVIc complexes slide along the viral DNA with a one-dimensional diffusion constant of 21.0 × 106 bp2/s and process the precursor proteins, which are also nonspecifically bound to the viral DNA. Both pVI and AVP-pVIc complexes slide along DNA via one-dimensional diffusion because pVIc is a “molecular sled.”4
An active form of AVP, the AVP-pVIc complex, has been crystallized (16, 17), and its structure has been determined at 2.6 Å resolution (9) and later at 1.6 Å resolution (10). The AVP-pVIc complex is a cysteine proteinase. The peptide cofactor pVIc binds tightly to AVP with extensive contacts. The equilibrium dissociation constant, Kd, for pVIc and AVP is 4.4 μm; in the presence of DNA, the Kd drops to 90 nm (7). Between pVIc and AVP, there are 26 hydrogen bonds, four ion pairs, and a disulfide bond (18). Surprisingly, pVIc, which exerts a powerful control on the rate of catalysis by AVP, binds quite far from the active site residues involved in catalysis; Cys-104 of AVP, which forms the disulfide bond with Cys-10′ of pVIc, is 32 Å away from Cys-122, the active site nucleophile.
The AVP-pVIc complex consists of two domains. Cys-122 resides in one domain, whereas His-54 and Glu-71 reside in the other. pVIc appears to form a “strap” that may help position the two domains in a configuration for optimal catalysis. Comparison of the amino acid residues involved in catalysis by the canonical cysteine proteinase papain with those amino acids in similar positions in the AVP-pVIc complex revealed that they can be superimposed. However, even with these juxtapositions, because the order along the polypeptide chain of these amino acids in AVP and papain is different, AVP is the first member of a new class of cysteine proteinases. The remarkable juxtaposition of catalytic elements strongly suggests that AVP employs the same catalytic mechanism as papain (19).
Although analysis of the structure of the active enzyme, the AVP-pVIc complex, has revealed its catalytic machinery, little information could be discerned as to why in the absence of pVIc, the enzyme is inactive. We crystallized AVP (20) and here present its structure at a resolution of 0.98 Å as determined by x-ray diffraction. With the structure of the inactive form of the enzyme now available, comparison of it with the structure of an active form of the enzyme, the AVP-pVIc complex, revealed at the structural level why AVP is inactive and provided insights as to how the binding of pVIc to AVP activates the enzyme. Furthermore, this high resolution structure revealed novel targets for compounds that may act as antiviral agents.5
EXPERIMENTAL PROCEDURES
AVP
Recombinant AVP was purified from Escherichia coli, as described previously (13, 21). The concentration of AVP was determined using a molar absorbance coefficient at 280 nm of 26,510 calculated according to the method of Gill and von Hippel (22).
Crystallization
Crystals of AVP were obtained by vapor diffusion with microseeding as described previously (20). Prior to data collection, crystals were equilibrated with a cryoprotectant buffer consisting of 0.4 m sodium citrate, pH 5.6, 0.8 m sodium acetate, and 40% (v/v) glycerol. The glycerol concentration in the crystal droplets was increased in 4% increments at 5-min intervals until the glycerol concentration reached 25%. Crystals were then placed in cryoloops and cryo-cooled in the 100 K nitrogen stream. (Oxford Cryosystems, Oxford, UK).
Data Collection
X-ray diffraction data were collected at Beamline X25 at the National Synchrotron Light Source at Brookhaven National Laboratory. The intensities were recorded using a Brandeis 4k charge-coupled device detector (23, 24). The wavelength was 0.92 Å. High resolution data were collected with 60-s exposures, at a crystal to detector distance of 71.8 mm, with 0.6° oscillations. The low resolution data were obtained at the same distance also with 0.6° oscillations using an attenuated beam and 10-s exposures. The data were integrated and scaled with the software package HKL2000 (25), and Rp.i.m. assessment was determined using SCALA from the CCP4 program suite (26) Analysis of the data indicated that the AVP crystals belonged to the P21 space group with unit cell parameters a = 36.3, b = 54.5, c = 42.4 Å, β = 100.1°.
Structure Refinement
A clear molecular replacement solution was obtained with the starting model the coordinates of AVP from the AVP-pVIc structure (Protein Data Bank (PDB) code 1NLN) (10) using the program AMoRe (27). The refinement was cross-validated by the Rfree index (28), calculated using 5% of all reflections. The refinement was initiated using the program REFMAC (29). The energy function as well as x-ray terms were used as targets in the minimization procedure, and the model was refined using isotropic atomic displacement parameters. Rigid body refinement in the resolution range of 8.0–3.0 Å was performed to compensate for any small differences in unit cell parameters between the 1NLN structure and the current structure. This refinement was followed by positional and overall B factor refinements with the resolution range extended to 1.5 Å. After each round of refinement, visual inspection and model corrections were made using the program Quanta (Molecular Simulations, Inc). B factors were individually refined for all non-hydrogen atoms. All reflections were used in all steps of both the refinement and the map calculations.
After several rounds of refinement, the diffraction data were extended to their highest resolution, 0.98 Å, and refinement continued using the program SHELXL (30). During the first five cycles of refinement, isotropic B factors were refined for all atoms. Later, non-hydrogen atoms were refined using anisotropic displacement parameters. At this stage, hydrogen atoms were introduced into well ordered parts of the structure at stereochemically calculated positions. For all hydrogen atoms included in the refinement, isotropic B factors that were 20% higher than those of the parent atoms (50% higher in the case of methyl hydrogens) were applied. Manual adjustments of the model were performed with the program Quanta. The occupancies of atoms present in double conformations were refined as constrained (x) and (1 − x). Water molecules were classified as either fully occupied or half-occupied on the basis of their electron density and their distance to neighboring atoms. Their occupancies were not refined because refinement of both occupancies and temperature factors at resolutions approaching 1 Å is generally not stable (31). The quality of the geometrical and stereochemical indices was continuously monitored using the program PROCHECK (32) and MOLPROBITY (33).
RESULTS
X-ray Diffraction Data
The unit cell dimensions and mass of the molecular species in the crystal were consistent with there being one 23,087-Da monomer of AVP per asymmetric unit, which gives a Matthew's coefficient of 1.75 Å3/Da (34). This corresponds to a solvent content of 29.2%, which may be why this crystal diffracted to such high resolution. Overall, the model consisted of 1786 non-hydrogen atoms, with 194 residues, 258 waters, and 1 sodium ion. The data collection statistics are listed in Table 1.
TABLE 1.
Data collection and refinement statistics
| Parameter | Value |
|---|---|
| Data collection | |
| Wavelength (Å) | 0.92 |
| Resolution (Å) | 20-0.98 (1.00-0.98) |
| Space Group | P21 |
| Cell Dimensions | |
| a, b, c | a = 36.27 Å, b = 54.54 Å, c = 42.41 Å |
| α, γ, β | α = γ = 90°, β = 100.1° |
| Matthews Coefficient (molecules/asymmetric unit) | 1.75 Å3/Da (1) |
| Redundancy (overall/outer shell) | 4.9 (2.2) |
| I/σI | 25.4 (1.6) |
| Rmergea (overall/outer shell) | 0.057 (0.594) |
| Rp.i.m.b | 0.041 |
| Rmeas.c | 0.144 |
| Completeness (%) (overall/outer shell) | 99.1 (97.1) |
| Refinement statistics | |
| Resolution range (Å) | 20–0.98 |
| No. of reflections | 90549 |
| Completeness (working + test) (%) | 97.3 |
| R factor | 0.1354 |
| Rfree | 0.1678 |
| No. of protein atoms | 1535 |
| No. of sodium ions | 1 |
| No. of water molecules | 258 |
| B values | |
| From Wilson plot (Å2) | 9.93 |
| Mean B value (overall, Å2) | 18.43 |
| Root mean square deviation bonds (Å) | 0.029 |
| Root mean square deviation angles (degrees) | 0.045 |
| Ramachandran plot analysis | |
| Favored region (%) | 98.4 |
| Allowed region (%) | 1.6 |
a Rmerge = ΣhklΣi|Ii(hkl) − 〈I(hkl)〉/ΣhklΣi(hkl), where Ii(hkl) is the integrated intensity of a given reflection and 〈I(hkl)〉 is the mean intensity of multiple corresponding symmetry-related reflections.
b Rp.i.m. = Σhkl[1/N − 1]½Σi|Ii(hkl) − 〈I(hkl)〉/ΣhklΣi(hkl), where Ii(hkl) is the integrated intensity of a given reflection, 〈I(hkl)〉 is the mean intensity of multiple corresponding symmetry-related reflections and N is the multiplicity of a given reflection.
c Rmeas. = Σhkl[N/N − 1]½Σi|Ii(hkl) − 〈I(hkl)〉/ΣhklΣi(hkl), where Ii(hkl) is the ith measurement of the intensity of reflection hkl, 〈I(hkl)〉 is the mean intensity of reflection hkl, and N is the number of observations of intensity I(hkl) (multiplicity).
Quality of the Model
The structure of AVP was solved with molecular replacement. The final model refined against the diffraction data measured from crystals of the native protein gave a crystallographic R factor of 13.5% (Rfree = 16.8%) for the data from 20 to 0.98 Å. An example of the high resolution of the electron density is shown in Fig. 1. Residues 29–32, highly conserved among AVP genes, are depicted with the 2Fo − Fc map contoured at 1.5 σ. Residues 48–51 and 97–104 could not be modeled due to poorly defined or missing electron density in those regions. These two “disordered” regions lie within loops of the AVP-pVIc structure used to initiate the molecular replacement solution. Seven residues were found whose side chains exhibited multiple conformations: Glu-7, Asp-26, Cys-67, Arg-169, Gln-173, Ser-176, and Ser-194. The weights of the stereochemical restraints used during the refinement together with the final deviations of the geometrical parameters from ideal values are shown in Table 1.
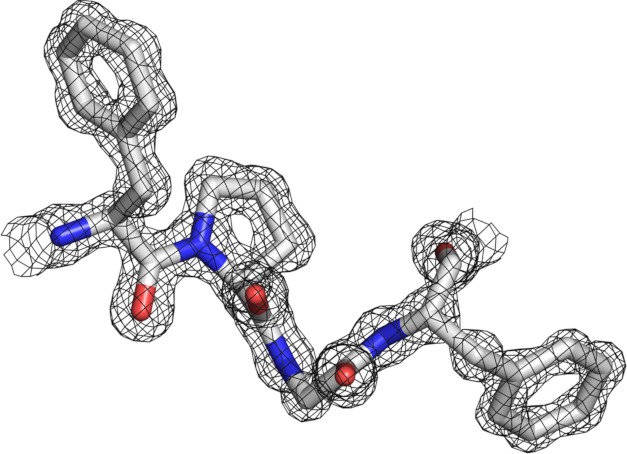
FIGURE 1.
Example of high resolution (0.98 Å) data. A region of the AVP structure is depicted with the 2Fo − Fc map contoured at 1.5 σ. The amino acid residues, Phe-29, Pro-30, Gly-31, and Phe-32, are conserved among AVP genes and lie in the His activation pathway. All figures were generated with PyMOL (49).
The Ramachandran plot (35) shows 92.3% of the non-proline, non-glycine residues in most favored regions, 6.1% in additional allowed regions. The average B value for the main chain atoms was 11.63 Å2, and 18.43 Å2 for side chain atoms. The overall B value from the Wilson plot was 9.9 Å2.
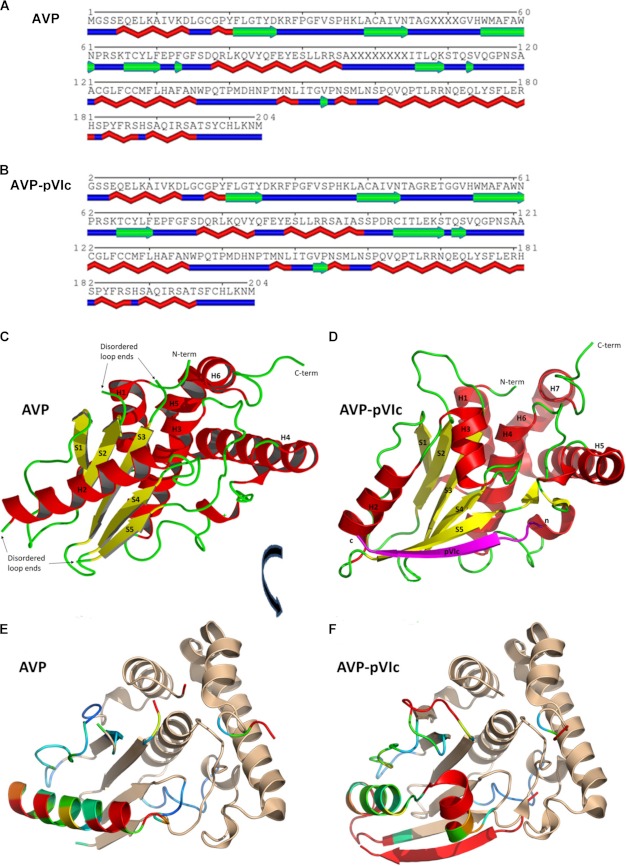
Overall Fold
Secondary structure elements of AVP and of the AVP moiety in the AVP-pVIc complex are listed below the amino acid sequence in Fig. 2, A and B, respectively. AVP has an α + β fold structure, ovoid in shape with dimensions of ∼45 × 35 × 33 Å. The structure contains five β-strands, six α-helices, and two 3-10 helices and is arranged into two domains with the active site situated at the domain interface (Fig. 2C). One domain, the α-helical domain, is composed of four α-helices and both 3-10 helices, encompassing the N-terminal 15 residues and residues 113–204. The other domain, the β-strand domain, contains five β-strands arranged in a β-sheet that is sandwiched between a long α-helix and the helical domain, encompassing residues 16–112. The molecule contains 204 amino acids, 194 of which could be mapped into the electron density. The β-sheet domain contains the two loops whose backbone positions could not be mapped due to a lack of electron density.
FIGURE 2.
Structural comparisons between AVP and the AVP-pVIc complex. A and B, secondary structure elements of AVP are depicted below the amino acid sequence for both AVP (A) and the AVP-pVIc complex (B). The two structures, AVP and the AVP-pVIc complex, were superimposed by least squares fitting. For C and D, helices are colored red, strands are colored yellow, and coils are colored green. In C, the secondary structure representation of AVP is shown. In D, the secondary structure representation of the AVP-pVIc complex is shown with the pVIc peptide depicted in magenta. N-term, N terminus; C-term, C terminus. In E and F, the aligned structures of C and D have been rotated ∼60° on the x axis and 20° on the y axis to highlight the structural changes. In E, the aligned structural graphic of AVP is shown with residues colored by r.m.s.d. using a spectrum from blue, similar in structure, through red, highly different in structure. Those amino acid residues that are essentially identical in structure are colored tan. In F, the alignment of AVP-pVIc with AVP is shown with residues colored as in C.
Overall Structure
The structure of AVP was solved by molecular replacement using as a search model the structure of the proteinase from the AVP-pVIc covalent complex (PDB entry 1NLN) (Fig. 2D). The aligned structures of AVP and of the AVP-pVIc complex are shown in Fig. 2, E and F, with the root mean square differences between each highlighted in color and the similarities between the two colored in beige. Overall, the structures of AVP and the AVP-pVIc complex are very similar, with an r.m.s.d. for 680 backbone atoms of 0.78 Å; for 1024 atoms from all aligned residues, the r.m.s.d. was 0.37 Å. The similarities and differences seem to be domain-specific.
The similarities in the two structures are mostly in the α-helical domain. The backbone in the α-helical domain is practically identical between the two structures; only the last 3 residues at the C terminus of the protein diverge in position between the two structures. The r.m.s.d. for the 404 backbone atoms of the helical domain between the two structures is 0.32 Å. The 2 catalytic residues of this domain, Gln-115 (oxyanion hole) and Cys-122 (active site nucleophile), are in virtually identical positions.
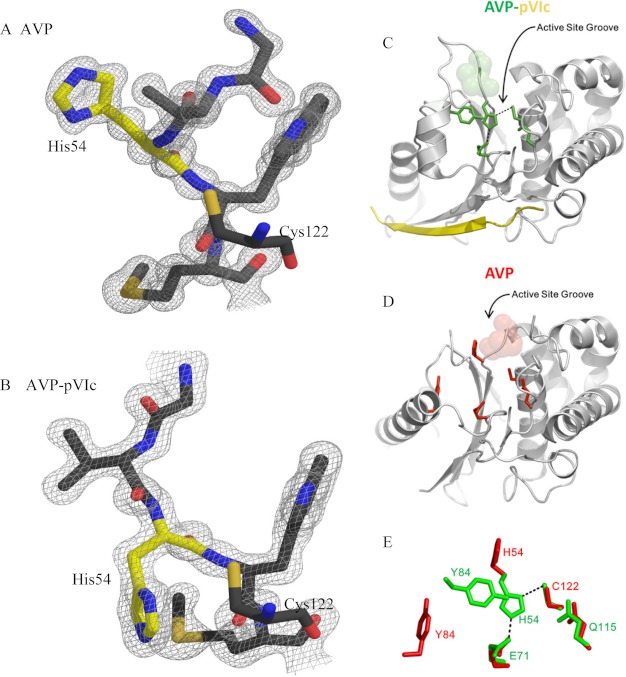
The major differences between the two structures are found in the β-sheet domain. Here, the r.m.s.d. for the 380 backbone atoms whose positions are defined in this domain between the two structures is 3.23 Å. The β-strands at each end of the central β-sheet are altered when compared with the AVP-pVIc structure. Strand S1 is extended by one residue in AVP encompassing residues 21 through 26. Strand S5 is 3 residues shorter, extending from residues 106 through 109 rather than from residues 104 through 110. The region connecting strands S1 and S2 has residues that have undergone a significant backbone rearrangement, resulting in a different arrangement of their side chains. The most significant difference in structure in the β-sheet domain is in the long helix above the β-sheet, extending from residues 78 through 95 in AVP. In the AVP-pVIc structure, a helix-coil-helix motif extends from residue 77 through 98 followed by a small coil from residues 99 through 103. There are two loops whose backbone could not be completely traced in the β-sheet domain. One of these loops extends from residues 45 through 53. There is insufficient electron density to map residues 48 through 51. This loop connects strands S2 and S3. Because one end of this undefined loop lies close to the catalytic His-54, the fit of the model to the density was examined using an average B factor per residue analysis. The residues at the apex of the loop in the AVP-pVIc structure, residues 48–51, have the highest average B factors found in the structure. These are the residues whose positions could not be mapped in the AVP structure. In keeping with the flexible nature of this region of AVP, the average B factors for residues on each end of the undefined region of AVP have the highest average B factors. The analysis supports the altered positions of the residues within this region of the AVP structure whose positions could be refined, as exemplified by the fit of the model to the density (Fig. 3, A and B). The second loop extends from residues 96 through 105. Residues 97 through 104 could not be mapped in the structure. This loop extends from the C-terminal end of the long helix to strand S5.
FIGURE 3.
Repositioning of His-54 and comparisons of the active site of AVP-pVIc complexes with the “active” site of AVP. Repositioning of His-54 was determined. In A, residues 52 through 56 of AVP are shown with the 2Fo − Fc electron density map describing the model contoured at 1.5 σ around the residues. In B, residues 52 through 56 of the AVP-pVIc complex are shown with the 2Fo − Fc electron density map describing the model contoured at 1.5 σ around the residues. The nucleophile Cys-122 is shown without electron density to indicate its position relative to His-54 in both A and B. Nitrogen atoms are colored blue, oxygen atoms are colored red, sulfur atoms are colored light orange, and carbon atoms are colored gray except for those on His-54, which are colored yellow. Comparisons of the active site of AVP-pVIc complexes with the “active” site of AVP and repositioning of His-54 are shown. In C, the secondary structure representation of the AVP-pVIc complex is shown with the residues involved in catalysis shown in stick form and colored green. The pVIc peptide is colored yellow. In D, the secondary structure of AVP is shown with the same residues depicted in stick form and colored red. In C and D, the positions of the residues that block the active site groove in AVP are colored semitransparent spheres. In E, the positions of the amino acids involved in catalysis in AVP (red) and in the AVP-pVIc complex (green) are overlaid to reveal the differences in position. Only His-54 and Tyr-84 are in different positions in the two structures. Hydrogen bonds from Glu-71 to His-54 and His-54 to Cys-122 are depicted as dashed lines.
The Active Sites and the Amino Acid Residues Involved in Catalysis
In the AVP-pVIc complex, the active site is located within a 25 Å long bent groove that is ∼8 Å wide (Fig. 3C). Cys-122 and His-54, the active site nucleophile and the general base, respectively, are located in the middle of the groove. These amino acids are conserved among adenovirus serotypes. The distance between atoms Sγ of Cys-122 and Nδ1 of His-54 is 3.87 Å. This is probably a thiolate-imidazolium ion pair, like the nucleophilic Cys-His ion pair in papain (36), because a thiolate anion in AVP can be titrated at pH 5.0 with dithiodipyridine (13). Glu-71, probably the third member of the charge-relay system (37), lies on the other side of the imidazole ring of His-54 from Cys-122. A hydrogen bond is formed between atoms Oϵ2 of Glu-71 and Nϵ2 of His-54. Glu-71 is replaced only by Asp among adenovirus strains. The backbone nitrogen of Cys-122 and side chain nitrogen of Gln-115 form the presumed oxyanion hole (Fig. 3C).
In the AVP structure, the active site is in a similar position to its location in the structure of the AVP-pVIc complex (Fig. 3D). Comparison of the positions of the 4 amino acids involved in catalysis by the AVP-pVIc complex to the positions of those amino acids in AVP reveals why AVP is inactive (Fig. 3E). In AVP, 3 of the catalytic amino acids, Cys-122, Glu-71 and Gln-115, occupy nearly identical positions to the ones they have in the active AVP-pVIc complex. However, the position of His-54 in AVP is different from its position in the AVP-pVIc complex (Fig. 4, A and B). The His-54 Nδ1 has moved from being 3.87 Å away from the Cys-122 Sγ nucleophile in the AVP-pVIc complex to 7.0 Å away from Cys-122 Sγ in AVP. This movement is a consequence of the repositioning of the loop containing residues 46–54 in AVP. In place of the His-54, there is a series of water molecules that form a hydrogen-bond network extending from the surface down toward Glu-71. In the AVP structure, His-54 is the third residue with defined density extending from the C-terminal side of the disordered loop from residues 48–51. The backbone atoms of this portion of the loop have undergone significant φ/ψ rotations that result in the repositioning of the amino acid residues. With His-54, carbonyl and α-carbon atoms sit in the same positions as in the AVP-pVIc structure (Fig. 3, A and B). However, there is an approximate 80° rotation of the ψ angle that results in the repositioning of the histidine side chain away from Cys-122. This repositioning is why AVP has little or no enzymatic activity. Now His-54 and its Nδ1 atom are at a distance and in an orientation by which it can no longer abstract the proton on the Cys-122 Sγ. Thus, a thiolate-imidazolium ion pair cannot form to render Cys-122 nucleophilic. As a consequence, AVP would have little or no catalytic activity.
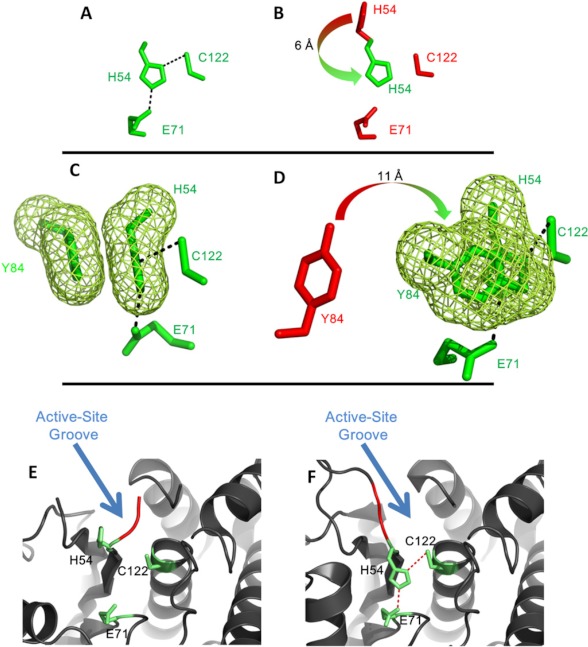
FIGURE 4.
Movements of amino acids in the activation of AVP by pVIc. In A, the orientation of the Cys-His-Glu triad of the AVP-pVIc complex is shown. In B, that triad in AVP is shown along with the position of the His from the AVP-pVIc complex. The arrow indicates the rotation and distance His-54 must move to align with the catalytic Cys. In C, the catalytic triad of AVP-pVIc and Tyr-84 is shown in an orientation to highlight the cation-π interaction of Tyr-84 with His-54 (green mesh). In D, the location of Tyr-84 (red) in AVP, with an arrow showing the direction and distance it must move to form the cation-π interaction in AVP-pVIc, is shown. In E and F, the occlusion of the active site groove in AVP by the loop containing His-54 is shown. In E, the location of the loop relative to the active site residues in AVP is shown. The residues extending across the loop blocking the groove (Gly-52 and Val-53) are colored red, and the active site residues are shown in stick form and colored green. In F, the location of the same residues in the AVP-pVIc complex structure indicating that the groove is no longer blocked is shown. Unblocking the groove also enabled His-54 to move opposite the Cys-122 to render it nucleophilic.
There is a fifth amino acid involved in catalysis by the AVP-pVIc complex. In the AVP-pVIc complex structure, His-54 forms a cation-π interaction with Tyr-84 (Fig. 4C). The preferred mode for such an interaction is usually stacked offset (face-to-face with the rings in a staggered arrangement) (38). In barnase, Tyr-94 interacts more strongly with the protonated form of His-18 (39). This aromatic-histidine interaction stabilizes the protonated form of histidine by 0.8–1 kcal mol−1 relative to the unprotonated form, and thereby, increases its pKa value. This function of the aromatic group would be analogous to that of the aspartic residue in the catalytic triad of the serine proteinase (40), i.e. to stabilize the protonated form of histidine in a transition state of a reaction in which the histidine acts as a proton acceptor. A second function of the cation-π interaction between Tyr-84 and His-54 may be that it prevents the imidazole ring from rotating, thereby freezing its Nδ1 atom in a position that is optimal for the ion pair interaction with Cys-122. In AVP, Tyr-84 is more than 11 Å away from its position in the AVP-pVIc complex (Fig. 4D). Thus, another reason AVP is inactive is because Tyr-84 is too far away from His-54 for a cation-π interaction to take place.
Substrate Binding Site in AVP
In AVP, a substrate cannot bind in the active site (Fig. 4E). In the AVP-pVIc complex, the active site lies with a long deep curved groove at the domain interface that extends across one face of the structure (Fig. 4F). Near the middle, at the bend of the curve, is Cys-122. A salt bridge between Glu-5 and Arg-48, located ∼13 Å away from the Cys-122 Sγ, effectively seals one end of the groove. This salt bridge forms an end wall of the groove and forms part of a pocket deep enough to accommodate the P4 residue (Leu, Ile, or Met) of substrates containing AVP consensus cleavage sites. In the AVP structure, that salt bridge is absent. Furthermore, part of the loop containing His-54 that connects β-strands S2 and S3 is repositioned. In AVP, residues 52 through 54 extend across the active site groove, effectively blocking it at Cys-122 (Fig. 4E). This leaves Cys-122 at the base of this new wall. The repositioned portion of the His-54 loop that lies across the active site groove also shortens the groove by about 11 Å. These changes render the active site unable to bind an AVP substrate.
pVIc Binding Site on AVP
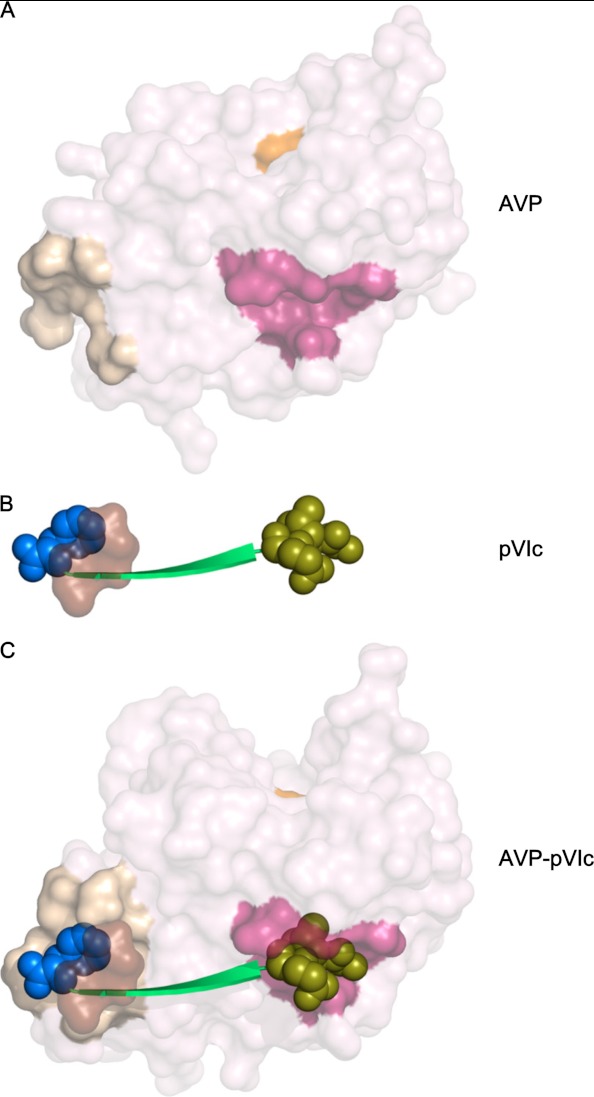
In the AVP-pVIc structure, pVIc appears to function as a strap holding together one domain containing Cys-122 with the other domain containing His-54 and Glu-71 in a configuration for optimal catalysis (Fig. 3A). The N terminus of pVIc (Gly-1′, Val-2′, and Gln-3′) binds in a pocket, the “NT-pocket,” which is an invagination within the helical domain of AVP (Fig. 5A). Binding displaces a well ordered sodium atom in the NT-pocket. That this pocket is structurally conserved between AVP (Fig. 5A) and AVP-pVIc (Fig. 5C) implies that perhaps the first step in the interaction of pVIc (Fig. 5B) with AVP is the binding of the N terminus of pVIc in this pocket. The binding of the next 3 amino acids of pVIc (Ser-4′, Leu-5′, and Lys-6′) also does not alter the structure of AVP; only surface side chain movements are necessary to accommodate these residues binding as an extended β-strand. It is at Arg-7′ and beyond that the binding of pVIc begins to induce significant rearrangements in AVP. The net results of these changes are formation of a disulfide bond between Cys-10′ of pVIc and Cys-104 of AVP and the formation of a new pocket in AVP, the “CT-pocket,” into which Phe-11′ of pVIc binds (Fig. 5C). Because in AVP, the location of Cys-104 could not be defined, the extension of strand S5 and the formation of the disulfide bond between Cys-104 and Cys-10′ can only happen as the binding of pVIc induces the formation of the CT-pocket.
FIGURE 5.
The NT- and CT-pockets on AVP and the AVP-pVIc complex into which the two termini of pVIc bind. In A, the accessible surface of AVP is shown with the active site cysteine colored orange, residues that form the NT-pocket in the AVP-pVIc complex colored dark pink, and those residues whose positions could be determined in the structure that aid in forming the CT binding pocket colored tan. In B, pVIc is shown in green with the residues that interact in the binding pockets shown as van der Waals spheres; the NT-pocket residues are in olive, and the CT-pocket residues are in blue. Those residues of pVIc that aid in forming the CT-pocket are shown as a light brown surface. In C, the AVP-pVIc complex is shown with the NT- and CT-pockets colored as in A and the pVIc colored as in B. Three residues that aid in forming the CT-pocket are undefined in the AVP structure.
DISCUSSION
We have solved the structure of AVP to 0.98 Å resolution. Comparison of the similarities and differences between this structure and that of the AVP-pVIc complex revealed why AVP is inactive and provided insights into the mechanisms of activation of AVP by the binding of pVIc. In AVP and in AVP-pVIc complexes, most of their α-helical domains are almost identical in structure. The orientation of the nucleophile and the oxyanion hole is maintained, as is the region of the molecules that interact with the N terminus of pVIc. There are, however, major differences in structure within the β-sheet domain between the active and inactive forms of the enzymes. Loop flexibility, in particular, plays a significant role in the change from the inactive to the active state.
The observation that the region of AVP quite far from the active site where the disulfide bond forms between Cys-104 and Cys-10′ of pVIc has an altered structure in the AVP-pVIc complex suggests this region of the molecule as a potential initiation point for the conformational changes that result in the activation of the enzyme. The changes from a single long helix to a helix-coil-helix and the rearrangements of the two disordered loops have two major consequences. Firstly, His-54 moves to a position in the structure where it can interact with the nucleophile, Cys-122. Secondly, Tyr-84 moves to a position over His-54 such that a cation-π interaction occurs; this undoubtedly adds to the nucleophilicity of the active site and a rigidification of the position of His-54 relative to Cys-122.
pVIc, which exerts powerful control on the rate of catalysis by AVP, binds quite far from the active site residues involved in catalysis; Cys-104 of AVP, which forms the disulfide bond with Cys-10′ of pVIc, is 32 Å away from Cys-122, the active site nucleophile. One reason for this is that pVIc is a molecular sled that slides the AVP-pVIc complex along the viral DNA via one-dimensional diffusion to process the virion precursor proteins also bound to the viral DNA (48).4 If the active site were too close to pVIc, and therefore, close to the DNA, it might be difficult for the active site to interact with the precursor protein substrates. However, this then raises the question as to how the binding of pVIc far from the active site influences the active site residues involved in catalysis.
Model for the Activation of AVP by pVIc and the Activation Pathways
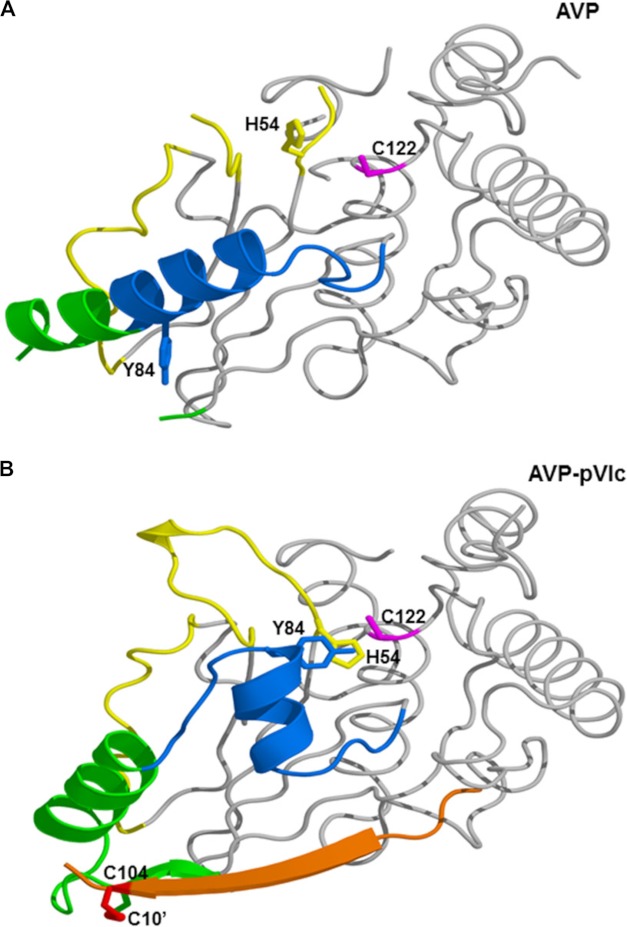
We propose a model for the activation of AVP upon the binding of pVIc that is consistent with the structural differences between AVP and AVP-pVIc complexes. The structural changes that occur upon the binding of pVIc to AVP are localized to more than half of the β-strand domain and appear to involve a path over 62 amino acids long (Fig. 6). This implies that there may be an activation pathway in which contiguous conformation changes occur, analogous to falling dominos. The secondary structure changes seen in the AVP-pVIc structure relative to this AVP structure (Fig. 6A) imply that pVIc binding initiates the extension of the S5 β strand and the loop rearrangements involving residues 97–104 necessary for the CT-pocket to form. Our working model is as follows. Upon the binding of pVIc to AVP, a series of structural transitions occurs in AVP beginning with the induction of the CT-pocket (Fig. 5C). There is a common pathway, green in Fig. 6B, that then bifurcates into pathways that lead to the repositioning of His-54, blue in Fig. 6B, and to a repositioning of Tyr-84, yellow in Fig. 6B. His-54 and Tyr-84 are the 2 amino acids in AVP that must be reoriented in order for the AVP-pVIc complex to become active.
FIGURE 6.
Structural transition in the activation of AVP by pVIc via the activation pathways. A top view of the aligned structures is shown in graphic form. In A, AVP is shown with the residues involved in the common pathway colored green, the His-54 pathway colored yellow, and the Tyr-84 pathway colored blue. His-54 (pink) and Tyr-84 (tan) are shown in stick form. In B, the orientations of those residues are depicted in the AVP-pVIc structure. The cation-π interaction of His-54 and Tyr-84 is shown by the overlapping side chains, and pVIc is colored red.
Common Activation Pathway
The activation pathway is triggered when the 3 N-terminal amino acids of pVIc bind in a preformed, hydrophobic pocket, the NT-pocket, on AVP. Beginning with Leu-5′, the remaining amino acids of pVIc lay down upon AVP as an extended β-strand. Cys-10′ of pVIc forms a disulfide bond with Cys-104 of AVP. The C-terminal amino acid, Phe-11′, binds in an induced, hydrophobic pocket. The differences in the structure of AVP and the AVP-pVIc complex indicate that pVIc binding causes an extension in the S5 β-strand of the β-sheet by 3 amino acids: Cys-104, Ile-105, and Ser-110. The CT-pocket formation is coincident with the formation of the tight turn involving residues 100–103; the C terminus of the long helix is extended from Ser-95 to Ser-99. Then, this portion of the helix rotates ∼20° from the long helix axis and changes its pitch by a similar amount. This is the common activation pathway, colored green (Fig. 6).
His-54 Activation Pathway
The extension of the lower end of the helix by a full turn and its movement alter the positions of its side chains and their interactions with the residues in the coil connecting strands S1 and S2 (colored yellow in Fig. 6, A and B). This results in repositioning of the backbone between amino acid residues 26 and 33 as indicated by the extensive φ and ψ angle differences observed between the two structures (Table 2). With these changes in the AVP structure, different portions of the coil interact with the repositioned helix, and other residues are now in positions to interact with the region of the undefined loop between residues 47 and 52 in AVP; this change allows hydrogen bonding to occur between residues 26 and 28 and residues within the undefined loop such that it now becomes much less flexible. A further consequence of this rearrangement is that the backbone moves, allowing the φ/ψ rotation of the His-54 backbone, which would enable its side chain to drop down to a position where it forms a hydrogen bond to Glu-71 and thus be in a more optimal position for interacting with Cys-122.
TABLE 2.
Phi and Psi angles and alignment r.m.s.d. in residues between β-strands S1 and S2 in AVP versus AVP-pVIc
r.m.s.d. values are calculated with AVP as the reference molecule. The largest deviations are between residues 28 through 32.
| AVP |
r.m.s.d. | AVP-pVIc |
|||
|---|---|---|---|---|---|
| φ | ψ | φ | ψ | ||
| Å | |||||
| Tyr-25 | −128 | 148 | 0.534 | −137 | 163 |
| Asp-26 | −99 | 107 | 1.488 | −89 | −172 |
| Lys-27 | 68 | −49 | 1.120 | −60 | −13 |
| Arg-28 | −89 | 159 | 2.595 | −85 | 9 |
| Phe-29 | −72 | 164 | 2.109 | −46 | 122 |
| Pro-30 | −55 | 157 | 2.907 | −66 | −30 |
| Gly-31 | −57 | −43 | 3.655 | 120 | −176 |
| Phe-32 | −156 | 160 | 2.889 | −98 | 131 |
| Val-33 | −123 | 153 | 2.513 | −118 | 79 |
| Ser-34 | −105 | 146 | 0.926 | −158 | 66 |
| Pro-35 | −71 | −4 | 0.967 | −62 | −23 |
| His-36 | −113 | −2 | 0.324 | −129 | 15 |
Tyr-84 Activation Pathway
Triggering of the common activation pathway also initiates changes in the Tyr-84 branch of the activation pathway (Fig. 6, A and B, colored blue). At Tyr-88, the long helix breaks, extending into a coil through Tyr-84. The extension of this portion of the long helix into a coil, along with the anchoring of its lower end due to the disulfide bond formation with pVIc and the “tethering” of the N-terminal portion of the helix to the central strand of the β sheet, makes the N-terminal portion of the long helix rotate roughly 105°, generating the helix-coil-helix motif of AVP-pVIc. This movement also completes the formation of the active site groove across the domain interface. These events allow Tyr-84 to move almost 11 Å so that it can now form a cation-π interaction with His-54.
How Does AVP Cut Out pVIc from pVI?
The structure of AVP reported here revealed why it is inactive. An obvious question is because AVP is inactive, how does it cut out pVIc from pVI, thereby enabling AVP-pVIc complexes to form? We have shown that for this to happen, both AVP and pVI must be bound to DNA (47). The kcat/Km for substrate hydrolysis by AVP is stimulated 200-fold by AVP being bound to DNA such that when pVI slides into AVP, it is active enough to cut out pVIc.
The Four Amino Acids Involved in catalysis in AVP and AVP-like Proteins
Upon activation by pVIc, only one of the 4 amino acids involved in catalysis, His-54, changes position. This is brought about by structural changes in one of the two domains of AVP in a region in that domain encompassing 62 amino acids. The 4 amino acids involved in catalysis by AVP-pVIc complexes are in a similar position as in papain; however, the folds of AVP-pVIc complexes and papain are totally different, an example of convergent evolution (9). Recently, other proteinases have been added to the AVP family. Among viruses, this includes vaccinia virus (41) and African swine fever virus (42). Chlamydia trachomatis has a gene similar to that of AVP (43). In yeast, Ulp1 is a member of a family of proteinases involved in desumoylation (44). YopJ from Yersinia pestis is an acetyltransferase involved in the inhibition of mitogen-activated protein kinase and nuclear factor κB signaling in animal cells and in the induction of localized cell death in plants (45). The high resolution crystal structure reported here should facilitate the identification of compounds that prevent AVP from being active and therefore act as antiviral agents.
Acknowledgments
We thank Zbigniew Dauter for advice during data collection and analysis. We also thank Michael Becker and Annie Heroux for assistance in data collection. Data for this study were measured at beam line X-25 of the National Synchrotron Light Source. Financial support comes principally from the Offices of Biological and Environmental Research and of Basic Energy Sciences of the United States Department of Energy, and from the National Center for Research Resources (Grant P41RR012408) and the NIGMS (Grant P41GM103473) of the National Institutes of Health. The National Synchrotron Light Source, Brookhaven National Laboratory, is supported by the United States Department of Energy, Office of Science, Office of Basic Energy Sciences, under Contract Number DE-AC02-98CH10886.
This work was supported, in whole or in part, by National Institutes of Health Grant AI R0141599 (to W. F. M.). This work was also supported by the Office of Biological and Environmental Research of the United States Department of Energy under Prime Contract DE-AC0298CH10886 with Brookhaven National Laboratory.
The atomic coordinates and structure factors (code 4EKF) have been deposited in the Protein Data Bank (http://wwpdb.org/).
The following designations are used in this study: pVIc, 11-amino acid cofactor (GVQSLKRRRCF) originating from the C-terminus of virion precursor protein pVI; AVP-pVIc, noncovalent or covalently linked heterodimer of AVP and pVIc.
P. C. Blainey, V. Graziano, W. J. McGrath, G. Luo, X. S. Xie, and W. F. Mangel, submitted for publication.
W. J. McGrath, V. Graziano, and W. F. Mangel, submitted for publication.
- AVP
- adenovirus proteinase
- pVI
- the precursor to adenovirus protein VI
- r.m.s.d.
- root mean square deviation.
REFERENCES
- 1. Weber J. (1976) Genetic analysis of adenovirus type 2 III. Temperature sensitivity of processing viral proteins. J. Virol. 17, 462–471 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Mangel W. F., McGrath W. J., Toledo D. L., Anderson C. W. (1993) Viral DNA and a viral peptide can act as cofactors of adenovirus virion proteinase activity. Nature 361, 274–275 [DOI] [PubMed] [Google Scholar]
- 3. Webster A., Hay R. T., Kemp G. (1993) The adenovirus protease is activated by a virus-coded disulphide-linked peptide. Cell 72, 97–104 [DOI] [PubMed] [Google Scholar]
- 4. Brown M. T., McGrath W. J., Toledo D. L., Mangel W. F. (1996) Different modes of inhibition of human adenovirus proteinase, probably a cysteine proteinase, by bovine pancreatic trypsin inhibitor. FEBS Lett. 388, 233–237 [DOI] [PubMed] [Google Scholar]
- 5. Edvardsson B., Everitt E., Jörnvall H., Prage L., Philipson L. (1976) Intermediates in adenovirus assembly. J. Virol. 19, 533–547 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Pérez-Berná A. J., Marabini R., Scheres S. H. W., Menéndez-Conejero R., Dmitriev I. P., Curiel D. T., Mangel W. F., Flint S. J., San Martín C. (2009) Structure and uncoating of immature adenovirus. J. Mol. Biol. 392, 547–557 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Baniecki M. L., McGrath W. J., McWhirter S. M., Li C., Toledo D. L., Pellicena P., Barnard D. L., Thorn K. S., Mangel W. F. (2001) Interaction of the human adenovirus proteinase with its 11-amino acid cofactor pVIc. Biochemistry 40, 12349–12356 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Webster A., Kemp G. (1993) The active adenovirus protease is the intact L3 23K protein. J. Gen. Virol. 74, 1415–1420 [DOI] [PubMed] [Google Scholar]
- 9. Ding J., McGrath W. J., Sweet R. M., Mangel W. F. (1996) Crystal structure of the human adenovirus proteinase with its 11 amino acid cofactor. EMBO J. 15, 1778–1783 [PMC free article] [PubMed] [Google Scholar]
- 10. McGrath W. J., Ding J., Didwania A., Sweet R. M., Mangel W. F. (2003) Crystallographic structure at 1.6-Ä resolution of the human adenovirus proteinase in a covalent complex with its 11-amino-acid peptide cofactor: insights on a new fold. Biochim. Biophys. Acta 1648, 1–11 [DOI] [PubMed] [Google Scholar]
- 11. McGrath W. J., Aherne K. S., Mangel W. F. (2002) In the virion, the 11-amino-acid peptide cofactor pVIc is covalently linked to the adenovirus proteinase. Virology 296, 234–240 [DOI] [PubMed] [Google Scholar]
- 12. McGrath W. J., Baniecki M. L., Li C., McWhirter S. M., Brown M. T., Toledo D. L., Mangel W. F. (2001) Human adenovirus proteinase: DNA binding and stimulation of proteinase activity by DNA. Biochemistry 40, 13237–13245 [DOI] [PubMed] [Google Scholar]
- 13. Mangel W. F., Toledo D. L., Brown M. T., Martin J. H., McGrath W. J. (1996) Characterization of three components of human adenovirus proteinase activity in vitro. J. Biol. Chem. 271, 536–543 [DOI] [PubMed] [Google Scholar]
- 14. Casjens S. (1997) Principles of virion structure, function and assembly. In: Structural Biology of Viruses (Chiu W., Burnett R. M., Garcea R. L., eds) pp. 3–37, Oxford University Press, Oxford [Google Scholar]
- 15. Mangenot S., Keller S., Rädler J. (2003) Transport of nucleosome core particles in semidilute DNA solutions. Biophys. J. 85, 1817–1825 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Keefe L. J., Ginell S. L., Westbrook E. M., Anderson C. W. (1995) Crystallization and preliminary X-ray diffraction studies of the human adenovirus serotype 2 proteinase with peptide cofactor. Protein Sci. 4, 1658–1660 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. McGrath W. J., Ding J., Sweet R. M., Mangel W. F. (1996) Preparation and crystallization of a complex between human adenovirus serotype 2 proteinase and its 11-amino-acid cofactor pVIc. J. Struct. Biol. 117, 77–79 [DOI] [PubMed] [Google Scholar]
- 18. McGrath W. J., Baniecki M. L., Peters E., Green D. T., Mangel W. F. (2001) Roles of two conserved cysteine residues in the activation of human adenovirus proteinase. Biochemistry 40, 14468–14474 [DOI] [PubMed] [Google Scholar]
- 19. Polgár L. (1974) Mercaptide-imidazolium ion-pair: the reactive nucleophile in papain catalysis. FEBS Lett. 47, 15–18 [DOI] [PubMed] [Google Scholar]
- 20. Baniecki M. L., McGrath W. J., Dauter Z., Mangel W. F. (2002) Adenovirus proteinase: crystallization and preliminary X-ray diffraction studies to atomic resolution. Acta Crystallogr. D Biol Crystallogr. 58, 1462–1464 [DOI] [PubMed] [Google Scholar]
- 21. Anderson C. W. (1993) Expression and purification of the adenovirus proteinase polypeptide and of a synthetic proteinase substrate. Protein Expr. Purif. 4, 8–15 [DOI] [PubMed] [Google Scholar]
- 22. Gill S. C., von Hippel P. H. (1989) Calculation of protein extinction coefficients from amino acid sequence data. Anal. Biochem. 182, 319–326 [DOI] [PubMed] [Google Scholar]
- 23. Phillips W. C., Stanton M., Stewart A., Qiian H., Ingersoll C., Sweet R. M. (2000) Multiple CCD detector for macromolecular X-ray crystallography. J. Appl. Crystallogr. 33, 243–251 [Google Scholar]
- 24. Strauss M. G., Westbrook E. M., Naday I., Coleman T. A., Westbrook M. L., Travis D. J., Sweet R. M., Pflugrath J. W., Stanton M. (1990) A CCD-based detector for protein crystallography. Nucl. Instrum. Methods A297, 275–295 [Google Scholar]
- 25. Otwinowski Z., Minor W. (1997) Processing of x-ray diffraction data collected in oscillation mode. Methods Enzymol. 276, 307–326 [DOI] [PubMed] [Google Scholar]
- 26. Winn M. D., Ballard C. C., Cowtan K. D., Dodson E. J., Emsley P., Evans P. R., Keegan R. M., Krissinel E. B., Leslie A. G., McCoy A., McNicholas S. J., Murshudov G. N., Pannu N. S., Potterton E. A., Powell H. R., Read R. J., Vagin A., Wilson K. S. (2011) Overview of the CCP4 suite and current developments. Acta Crystallogr. D Biol Crystallogr. 67, 235–242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Navaza J., Saludijan P. (1997) AMoRe: an automated molecular replacement program package. Methods Enzymol. 276, 581–594 [DOI] [PubMed] [Google Scholar]
- 28. Brünger A. T. (1992) Free R value: a novel statistical quantity for assessing the accuracy of crystal structures. Nature 355, 472–475 [DOI] [PubMed] [Google Scholar]
- 29. Murshudov G. N., Vagin A. A., Dodson E. J. (1997) Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr. D Biol Crystallogr. 53, 240–255 [DOI] [PubMed] [Google Scholar]
- 30. Sheldrick G. M., Schneider T. R. (1997) SHELXL: high-resolution refinement. Methods Enzymol. 277, 319–343 [PubMed] [Google Scholar]
- 31. Sevcik J., Dauter Z., Lamzin V. S., Wilson K. S. (1996) Ribonuclease from Streptomyces aureofaciens at atomic resolution. Acta Crystallogr. D Biol Crystallogr. 52, 327–344 [DOI] [PubMed] [Google Scholar]
- 32. Laskowski R. A., MacArthur M. W., Moss D. S., Thornton J. M. (1993) PROCHECK: a program to check the stereochemical quality of protein structures. J. Appl. Crystallogr. 26, 283–291 [Google Scholar]
- 33. Davis I. W., Leaver-Fay A., Chen V. B., Block J. N., Kapral G. J., Wang X., Murray L. W., Arendall W. B., 3rd, Snoeyink J., Richardson J. S., Richardson D. C. (2007) MolProbity: all-atom contacts and structure validation for proteins and nucleic acids. Nucleic Acids Res. 35, W375–W383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Kantardjieff K. A., Rupp B. (2003) Matthews coefficient probabilities: Improved estimates for unit cell contents of proteins, DNA, and protein-nucleic acid complex crystals. Protein Sci. 12, 1865–1871 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Ramakrishnan C., Ramachandran G. N. (1965) Stereochemical criteria for polypeptide and protein chain conformations. II. Allowed conformations for a pair of peptide units. Biophys. J. 5, 909–933 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Drenth J., Jansonius J. N., Koekoek R., Wolthers B. G., eds (1971) in The Enzymes, Vol. 3, pp. 485–499, Academic Press, New York [Google Scholar]
- 37. Blow D. M., Birktoft J. J., Hartley B. S. (1969) Role of a buried acid group in the mechanism of action of chymotrypsin. Nature 221, 337–340 [DOI] [PubMed] [Google Scholar]
- 38. Samanta U., Pal D., Chakrabarti P. (1999) Packing of aromatic rings against tryptophan residues in proteins. Acta Crystallogr. D Biol Crystallogr. 55, 1421–1427 [DOI] [PubMed] [Google Scholar]
- 39. Loewenthal R., Sancho J., Fersht A. R. (1992) Histidine-aromatic interactions in barnase. Elevation of histidine pKa and contribution to protein stability. J. Mol. Biol. 224, 759–770 [DOI] [PubMed] [Google Scholar]
- 40. Blow D. M. (1976) Structure and mechanism of chymotrypsin. Acc. Chem. Res. 9, 145–152 [Google Scholar]
- 41. Ansarah-Sobrinho C., Moss B. (2004) Role of the I7 protein in proteolytic processing of vaccinia virus membrane and core components. J. Virol. 78, 6335–6343 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Andrés G., Alejo A., Simón-Mateo C., Salas M. L. (2001) African swine fever virus protease, a new viral member of the SUMO-1-specific protease family. J. Biol. Chem. 276, 780–787 [DOI] [PubMed] [Google Scholar]
- 43. Stephens R. S., Kalman S., Lammel C., Fan J., Marathe R., Aravind L., Mitchell W., Olinger L., Tatusov R. L., Zhao Q., Koonin E. V., Davis R. W. (1998) Genome sequence of an obligate intracellular pathogen of humans: Chlamydia trachomatis. Science 282, 754–759 [DOI] [PubMed] [Google Scholar]
- 44. Li S.-J., Hochstrasser M. (1999) A new protease required for cell-cycle progression in yeast. Nature 398, 246–251 [DOI] [PubMed] [Google Scholar]
- 45. Mukherjee S., Keitany G., Li Y., Wang Y., Ball H. L., Goldsmith E. J., Orth K. (2006) Yersinia YopJ acetylates and inhibits kinase activation by blocking phosphorylation. Science 312, 1211–1214 [DOI] [PubMed] [Google Scholar]
- 46. Graziano V., McGrath W. J., Suomalainen M., Greber U.F., Freimuth P., Blainey P.C., Luo G., Xie X.S., Mangel W.F. (2012) Regulation of a viral proteinase by a peptide and DNA in one-dimensional space. I. Binding to DNA and to hexon of the precursor to protein VI, pVI, of human adenovirus. J. Biol. Chem. 287, 2059–2067 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Graziano V., Luo G., Blainey P.C., Pérez-Berná A. J., McGrath W. J., Flint S. J., San Martín C., Xie X.S., Mangel W.F. (2012) Regulation of a viral proteinase by a peptide and DNA in one-dimensional space. II. Adenovirus proteinase is activated in an unusual one-dimensional biochemical reaction. J. Biol. Chem. 287, 2068–2080 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Blainey P. C., Graziano V., Pérez-Berná A. J., McGrath W. J., Flint S. J., San Martín C., Xie X. S., Mangel. W. F. (2012) Regulation of a viral proteinase by a peptide and DNA in one-dimensional space. IV. Viral proteinase slides along DNA to locate and process its substrates. J. Biol. Chem. 287, 2092–2102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. DeLano W. L. (2010) The PyMOL Molecular Graphics System, version 1.3r1, Schrödinger, LLC, New York [Google Scholar]