Background: The adenovirus proteinase and its precursor protein substrates are all sequence independent DNA binding proteins.
Results: The proteinase slides along DNA to locate and process its substrates.
Conclusion: Processing of precursor proteins by the adenovirus proteinase occurs on DNA.
Significance: This is a new way an enzyme not involved in nucleic acid metabolism interacts with its substrates: sliding on DNA via one-dimensional diffusion.
Keywords: Adenovirus, Cysteine Protease, DNA-Protein Interaction, Protein Domains, Viral Protease, Virus Assembly, One-dimensional Diffusion, Proteinase, Sliding on DNA, Viral Proteins
Abstract
Precursor proteins used in the assembly of adenovirus virions must be processed by the virally encoded adenovirus proteinase (AVP) before the virus particle becomes infectious. An activated adenovirus proteinase, the AVP-pVIc complex, was shown to slide along viral DNA with an extremely fast one-dimensional diffusion constant, 21.0 ± 1.9 × 106 bp2/s. In principle, one-dimensional diffusion can provide a means for DNA-bound proteinases to locate and process DNA-bound substrates. Here, we show that this is correct. In vitro, AVP-pVIc complexes processed a purified virion precursor protein in a DNA-dependent reaction; in a quasi in vivo environment, heat-disrupted ts-1 virions, AVP-pVIc complexes processed five different precursor proteins in DNA-dependent reactions. Sliding of AVP-pVIc complexes along DNA illustrates a new biochemical mechanism by which a proteinase can locate its substrates, represents a new paradigm for virion maturation, and reveals a new way of exploiting the surface of DNA.
Introduction
The adenovirus proteinase (1) (AVP),5 a 23-kDa cysteine proteinase (see Fig. 1A), is required for the synthesis of infectious virus. It is synthesized in an inactive form that when packaged inside a nascent virion becomes activated to process multiple copies of the six different virion precursor proteins (2) used in the assembly of the virus particle; only then is the virus particle infectious (2). Maximal activation of AVP in vitro requires two cofactors (3, 4). One cofactor is pVIc, the 11-amino acid peptide (GVQSLKRRRCF) from the C terminus of virion precursor protein pVI (see Fig. 1A). The other cofactor is the entire viral DNA genome of 36,000 bp of linear DNA (3, 5, 6). Together, in an AVP-pVIc-DNA complex,6 the viral cofactors dramatically increase the relative kcat/Km for substrate hydrolysis by more than 15,000-fold (5, 7, 8).
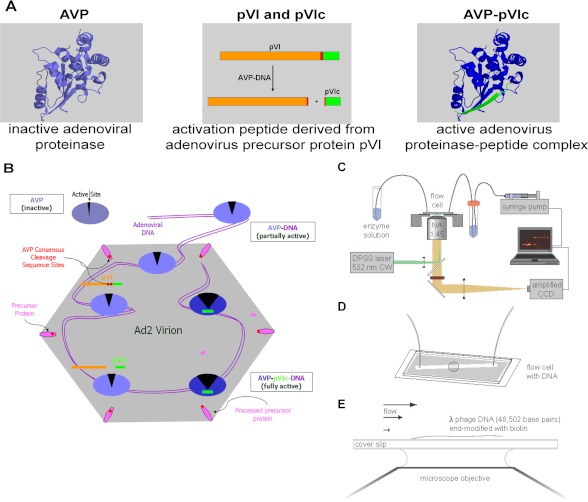
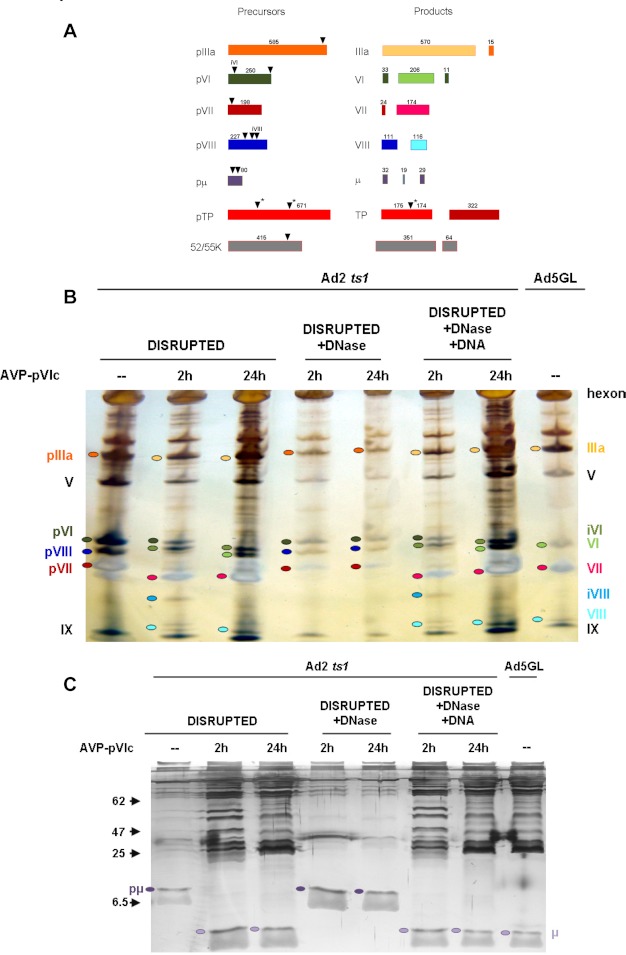
FIGURE 1.
Components of the adenovirus proteinase system. A model for the activation of AVP and cleavage of precursor proteins (3) and a method to assay proteins sliding along DNA in vitro (25) are presented. A, the AVP is inactive. Partially activated by being bound to the viral DNA, AVP cleaves pVI to liberate the 11-amino acid peptide (pVIc), in green, which then binds to AVP, forming an active AVP-pVIc complex. B, AVP-pVIc complexes slide along the viral DNA processing virion precursor proteins also bound to the DNA (3). C and D, single-molecule DNA sliding assay using an inverted microscope fitted for total internal reflection fluorescence imaging (C) with a mounted flow cell to which λDNA molecules (48,502 bp) had been attached at one end (25–27, 47, 48) (D). E, AVP-pVIc complexes, each labeled at Cys-199 with one molecule of the fluorescent dye Cy3B, were continuously flowed over the surface of the coverslip. Single protein molecules bound to DNA or sliding along DNA were illuminated by evanescent laser excitation and tracked using a high speed charge-coupled device camera (25, 29). DPSS, diode-pumped solid state.
Recently, we showed how AVP can be activated by pVI to form the AVP-pVIc complex in the core of the immature virion (50). AVP binds nonspecifically to the viral DNA and does not move. pVI also binds nonspecifically to the viral DNA; however, it slides along the DNA via one-dimensional diffusion, eventually sliding into AVP. AVP, partially activated by being bound to its cofactor DNA (3, 5, 6, 9), cleaves DNA-bound pVI twice, once at its N terminus and then at its C terminus. Cleavage at the C terminus liberates pVIc, the 11-amino acid peptide cofactor. pVIc then preferentially binds to the AVP molecule that cut it out. Finally, pVIc forms a disulfide bond with AVP, keeping the now fully activated enzyme, the AVP-pVIc-DNA complex, permanently activated (8, 10, 11).
There is a conundrum in the processing of the virion precursor proteins by an AVP-pVIc complex (see Fig. 1A). How can about 50 molecules (12) of the fully active proteinase (both cofactors bound) cleave multiple copies (about 1500) of six different virion precursor proteins at about 1900 processing sites (13–15) within the tightly packed interior of a nascent particle under conditions in which almost no three-dimensional diffusion can occur? Like AVP and the AVP-pVIc complex (3, 5, 8), the adenoviral precursor proteins pVI, preterminal protein (pTP), pVII, pIIIa, and pμ are sequence-independent DNA-binding proteins (16–19). The high concentration of DNA inside the virion (178–500 g/liter) (20) drives by mass action all these proteins onto the DNA. For AVP-pVIc complexes, the DNA-bound state predominates by at least one hundred thousand-fold over free AVP (5); this, in combination with the sieving action of the densely packaged DNA (21), diminishes the effective three-dimensional diffusion constant of AVP-pVIc by at least 1 million-fold. Given these circumstances, by what mechanism can crucial bimolecular associations occur when both enzymes and substrates are essentially irreversibly bound to a fixed matrix, the viral DNA?
A model proposing a solution to the conundrum was published in 1993 (3). That model (see Fig. 1B) postulated that AVP-pVIc complexes slide along the viral DNA to locate and process the virion precursor proteins. We know that in infectious wild-type virus, pVIc is covalently attached to AVP (11), indicating that the form of AVP that processes the virion precursor proteins is the AVP-pVIc complex. There are two key predictions from the model: 1) AVP-pVIc complexes slide along the viral DNA via one-dimensional diffusion and 2) AVP-pVIc complexes bound to DNA cleave virion precursor proteins also bound to DNA. Here, we test these predictions directly. Not only did we observe AVP-pVIc complexes sliding along DNA via one-dimensional diffusion, but we also saw that AVP-pVIc complexes processed the virion precursor proteins only in DNA-dependent reactions both in vitro and in vivo. There is no precedent for a proteinase sliding along DNA to locate and process its substrates.
EXPERIMENTAL PROCEDURES
Materials
The gene for AVP was expressed in Escherichia coli, and the resultant protein was purified as described previously (3, 7). pVI was purified as described (49). AVP concentrations were determined using a calculated (22) extinction coefficient of 26,510 m−1cm−1 at 280 nm. pVIc (GVQSLKRRRCF), the 5′-fluorescein-labeled 12-mer DNA (Fl-GACGACTAGGAT), and 5′-fluorescein-labeled 18-mer DNA (Fl-CAGGAAACAGCTATGACC) were purchased from Invitrogen. Fluorescent DNAs were annealed to their complimentary strands according to standard protocols. Cy3B-maleimide was purchased from GE Healthcare. pVIc concentrations were determined by titration of the cysteine residue with Ellman's reagent (23) using an extinction coefficient of 14,150 m−1cm−1 at 412 nm for released thionitrobenzoate. Octylglucoside (Fisher Scientific) and endoproteinase Glu-C (Sigma) were both obtained from commercial sources. Where indicated, buffer A was 20 mm Hepes (pH 7.0), 10 mm NaCl, 0.025% n-dodecyl-β-d-maltopyranoside, and 0.1 mm DTT, and buffer B was 20 mm Tris-HCl (pH 8.0), 0.025% n-dodecyl-β-d-maltopyranoside, 10 mm NaCl, and 0.1 mm DTT. n-Dodecyl-β-d-maltopyranoside was purchased from Anatrace (Maumee, OH). pVIc was labeled with Cy3B as described previously.7
AVP-pVIc Complex Formation
Disulfide-linked AVP-pVIc complexes were prepared by overnight incubation at 4 °C of 75 μm AVP and 75 μm pVIc in 20 mm Tris-HCl (pH 8.0), 250 mm NaCl, 0.1 mm EDTA, and 20 mm β-mercaptoethanol. Under these conditions, Cys-104 of AVP and Cys-10′ of pVIc undergo oxidative condensation (10, 11).
Fluorescent Labeling
Disulfide-linked AVP-pVIc complexes, 75 μm, were labeled in 25 mm HEPES (pH 7.0), 50 mm NaCl, and 20 mm ethanol by the addition of Cy3B maleimide to 225 μm. Labeling reactions were incubated at room temperature in the dark for 2.5 h. Excess reagents were removed from the labeled sample by passage through Bio-Spin 6 chromatography columns (Bio-Rad) equilibrated in the labeling buffer. The degree of labeling was determined using ϵ280 nmAVP = 26,510 m−1cm−1, ϵ558 nmCy3B = 130,000 m−1cm−1, and ϵ280 nmCy3B = 10,400 m−1cm−1. The ratio of labeled AVP-pVIc to total AVP-pVIc was determined to be 0.84. The labeled materials were characterized by matrix-assisted laser desorption ionization time-of-flight mass spectrometry (MALDI-TOF).
Location of Cy3B Label on AVP
Specific enzymatic digestions followed by MALDI-TOF mass spectrometry were used to locate cysteinyl-Cy3B conjugates in AVP-pVIc complexes. 1.2 μg of labeled AVP-pVIc complexes were digested by incubation with 0.01 μg of each endoproteinase Glu-C or trypsin at 21 °C in 25 mm Tris-HCl (pH 7.5). At 1, 2, 4, and 22 h, 0.5 μl of each reaction were removed and added to 4.5 μl of a saturated matrix solution (α-cyano-4-hydroxycinnamic acid) in 50% acetonitrile and 0.1% TFA. The matrix-analyte solution was then immediately spotted onto a 100-well stainless steel sample plate. The sample plate was calibrated using Applied Biosystems peptide calibration mixtures 1 and 2. Mass spectrometric characterization was carried out on a Voyager-DE Biospectrometry work station (Applied Biosystems; Foster City, CA). The m/z peak list generated for each chromatogram was analyzed by the FindPept tool (24). The Cy3B modification was entered as a post-translational modification with an atomic composition of C37H38N4O7S (molecular weight 682.785). AVP-pVIc complexes were found to be labeled at Cys-199.8
Fluorescence Imaging
AVP-pVIc complexes were labeled with Cy3B at Cys-199. Individual, fluorescently labeled molecules were imaged by total internal reflection fluorescence microscopy as described previously (25), with the exception that a faster electron-multiplying charge-coupled device camera (Photometrics Cascade:128+) was used for the highest time-resolution measurements. λDNA was tethered to a glass surface at one end and stretched by a laminar flow of buffer (see Fig. 1, C–E). The flow rate in the experiments was chosen such that the timescale of the DNA fluctuations was fast enough to negate any impact of the DNA motion on the measured diffusion constants. The concentration of fluorophore was 1 nm or less, to achieve a low background fluorescence. Because the concentration of AVP-pVIc complexes in the assays was about 1 nm and because the binding of AVP-pVIc complexes to DNA decreases with increasing ionic strength (5), to see the interaction of AVP-pVIc complexes with DNA, the ionic strength of the assay had to be low. Single molecules that bound to and diffused along the DNA were illuminated by a laser beam (532 nm) and imaged with a fluorescence microscope.
Centroid Determination and Analysis of Molecular Trajectories
Due to the speed and duration of sliding by AVP-pVIc complexes, these events were readily identified manually. All AVP-pVIc complex DNA binding events noted were included in the sliding analyses. Once events had been identified, molecular trajectories were tracked and analyzed using Gaussian centroid determination in the MATLAB environment as described previously (25). Primary trajectory data were converted to the mean square displacement (MSD) of the distance traveled versus time, and a plot of the MSD versus time yielded a straight line whose slope is the one-dimensional diffusion constant (25, 27, 51).
RESULTS
Sliding of AVP-pVIc Complexes along DNA
To determine whether AVP-pVIc complexes slide on DNA, we used single-molecule fluorescence microscopy (Fig. 1C) with flow-stretched DNA (Fig. 1, D and E) (25, 28, 29). In the DNA sliding assay (25), AVP-pVIc molecules labeled with one molecule of Cy3B at Cys-199 were observed to bind DNA at random locations, as predicted (3, 5), and furthermore, to diffuse rapidly over tens of thousands of base pairs before dissociating from the DNA. For example, the molecule whose motion is shown in the raw image data in Fig. 2A traveled more than 20,000 bp during a 0.9-s binding event. A trajectory of the molecule depicted in Fig. 2A produced by Gaussian centroid determination of the signal of the molecule in each of 262 frames is shown in Fig. 2B.
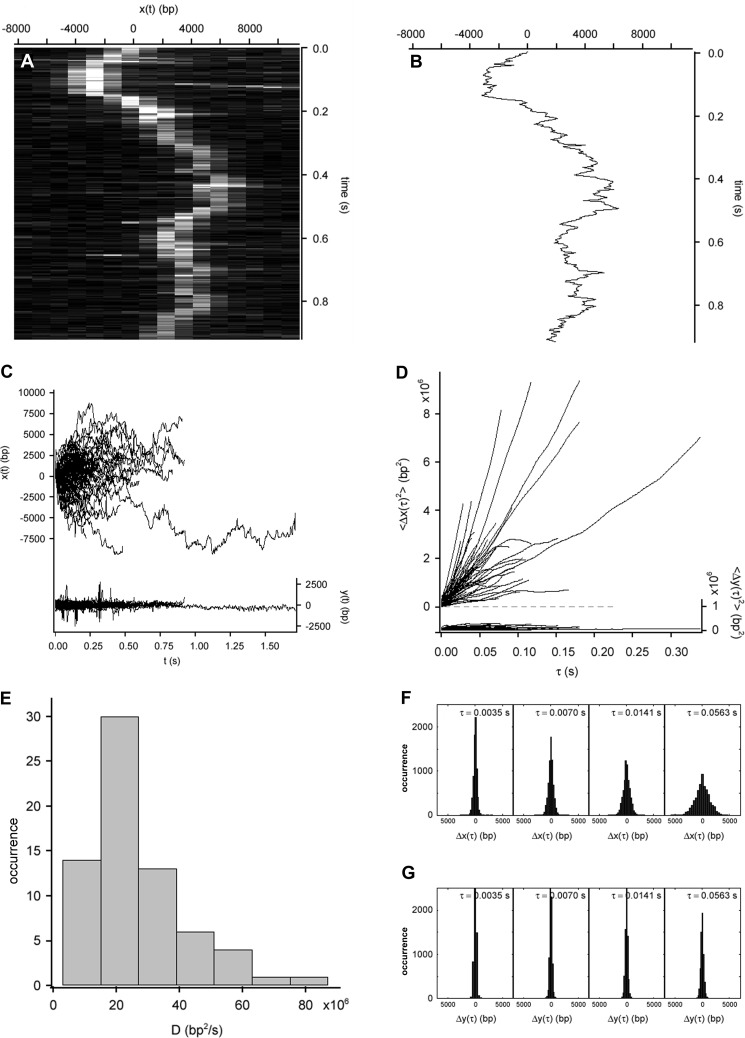
FIGURE 2.
Rapid diffusion of AVP-pVIc complexes along flow-stretched dsDNA. A, rapid motion of an AVP-pVIc molecule sliding along flow-stretched dsDNA recorded at 284 Hz. Cinegraph was generated from the overlay of 262 raw images showing motion along DNA (horizontal axis; each line is a strip of pixels from an image series frame) as a function of time (vertical axis). B, trajectory of the molecule depicted in A produced by Gaussian centroid determination of the signal of the molecule in each of 262 frames. C, AVP-pVIc complexes diffuse rapidly along DNA (x(t), left axis, 72 trajectories). D, mean square displacement of the trajectories shown in C along the DNA (〈Δx(τ)2〉, left axis). E, histogram of the measured diffusion constants for AVP-pVIc diffusing along dsDNA with the mean equal to 21 × 106 bp2/s. F, AVP-pVIc complex displacements along DNA grew as a function of time (Δx(τ), upper panels). G, displacements of AVP-pVIc complexes transverse to DNA (Δy(τ), lower panels) did not grow as the protein was confined to the DNA. In C and D, motion transverse to the DNA (y(t) and 〈Δy(τ)2〉, respectively, right axes) is represented on the same scale as a control.
One-dimensional Diffusion Constant
The quality of the data was sufficient to be able to obtain one-dimensional diffusion constants for individual molecules. The trajectories of 72 AVP-pVIc complexes sliding on DNA are plotted in Fig. 2C. The MSD of each molecular trajectory in Fig. 2C is plotted versus diffusion time in Fig. 2D. The MSD for each molecule was approximately linear with diffusion time, indicating transport dominated by Brownian motion. From the MSD slopes, one-dimensional diffusion constants (D1) were calculated according to D1 = 〈Δx2〉/2Δτ and are displayed in the histogram in Fig. 2E. Another indication the one-dimensional diffusion was due to Brownian motion was that the AVP-pVIc complex displacements along DNA increased as a function of time (Fig. 2F), whereas the displacements transverse to the DNA did not (Fig. 2G), as expected for particles confined to diffuse in one dimension only, i.e. along the DNA. AVP-pVIc diffusion did not seem to be biased by the direction of flow; during longer intervals, the mean displacements along DNA were indistinguishable from zero. The mean diffusion constant was 21.0 ± 1.9 × 106 bp2/s with the variation among D1 measured for individual AVP-pVIc complexes yielding an S.D. of 15.6 × 106 bp2/s (Table 1). This one-dimensional diffusion constant and that of proliferating cell nuclear antigen (30) are among the fastest yet reported for anything sliding on DNA.
TABLE 1.
One-dimensional diffusion constants on λDNA, equilibrium dissociation constants (Kd) on DNA, and size of DNA binding site
Kd(app) is the apparent equilibrium dissociation constant.
| Proteina,b (molecular mass in kDa) | Mean D1c ± S.E. | S.D. D1 | Kd(app) | Size of DNA binding site |
|---|---|---|---|---|
| bp2/s × 10−6 | bp2/s × 10−6 | nm | bp | |
| AVP (23.1) | 0.02–0.97d | 63 ± 5.8e | ||
| pVIcf (1.35) | 26.1 ± 1.8 | 11 | 264 ± 25c | 7 |
| AVP-pVIc (24.4) | 21.0 ± 1.9 | 15.6 | 4.6 ± 2.2e | 6 |
| AVP-pVIc (24.4) (high salt) | 17.1 ± 3.5 | 16.2 | ||
| pIIIag (64.3) | 19.4 ± 3.9h | 33 | ||
| pVI (27.0) | 1.45 ± 0.13d | 1.61 | 46 ± 1.6i,j |
a AVP and AVP-pVIc complexes were labeled with Cy3B at Cys-199. pVIc was labeled at Cys-10′ with Cy3B. pVI was labeled at Cys-249 with Cy3B.
b The NaCl concentration in assay buffer (see “Experimental Procedures”) was 2–6 mm NaCl; in high salt assay buffer, 20–25 mm NaCl was added to assay buffer.
c To convert from bp to nm: 106 bp2/s = 102,400 nm2/s.
d Ref. 50.
e Ref. 5.
f Footnote 7.
g V. Graziano and W. F. Mangel, unpublished observations.
h Determined in 50 mm NaCl.
i Determined in 1 mm MgCl2.
j Ref. 49.
Mechanism of Translation along DNA
The high one-dimensional diffusion constant of AVP-pVIc complexes caused us to consider the possibility that processes other than persistent sliding along DNA might be occurring (31). Sliding consists of one-dimensional translocation along the DNA with the protein in continuous contact with the DNA; it closely approximates idealized one-dimensional diffusion (31, 32). Hopping, also called microdiffusion, is a process wherein the protein repeatedly dissociates from the DNA and rebinds at new locations on the DNA. The excursions from the DNA involved in hopping can give rise to apparent one-dimensional diffusion (33, 34). In principle, hopping can lead to much faster apparent one-dimensional translocation of a protein molecule along DNA than sliding due to decreased friction of the protein with the solvent and the DNA (32, 51).
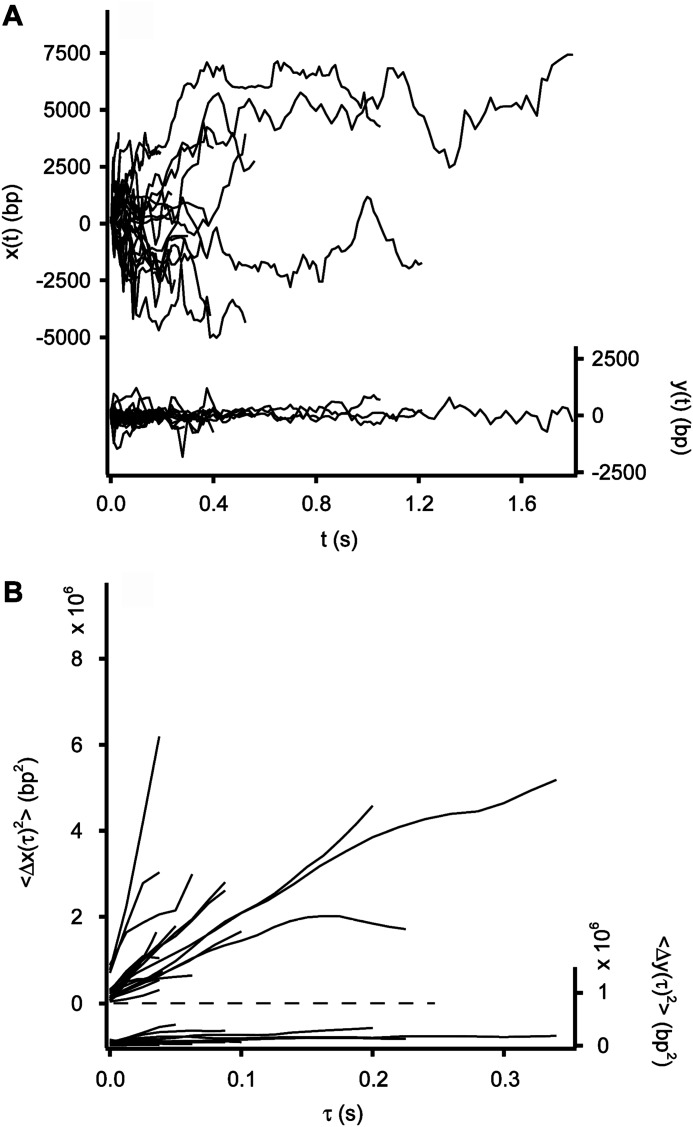
Formally, any protein, including AVP-pVIc complexes, capable of entering a dynamic binding equilibrium with DNA can undergo hopping. For those proteins whose nonspecific DNA binding has an electrostatic component, such as the AVP-pVIc complex (5), hopping and sliding can be distinguished experimentally with respect to observed one-dimensional diffusion by measuring the observed one-dimensional diffusion constant at varying ionic strengths. Were the observed protein motions to occur by hopping rather than by sliding, increasing the salt concentration would decrease the residence time of the protein on DNA. This is because stronger electrostatic screening lowers the nonspecific binding affinity, thereby increasing the fraction of time that the enzyme is unbound and mobile. This would result in an apparent increase in the one-dimensional diffusion constant. For these reasons, the sliding activities of AVP-pVIc complexes were assayed in solutions with higher salt concentrations, 20–25 mm NaCl. Twenty-seven trajectories are shown in Fig. 3A, and the MSDs versus time for 22 of those trajectories are shown in Fig. 3B. 〈D1〉 was found to be 17.1 ± 3.5 × 106 bp2/s. Because this is not a higher value than the 21.0 ± 1.9 × 106 bp2/s we observed in low salt, 2–6 mm NaCl (Table 1), it appears that the observed transport is dominated by sliding of AVP-pVIc complexes in contact with the DNA and not by hopping.
FIGURE 3.
Diffusion of the AVP-pVIc complex along flow-stretched dsDNA in 0.020–0.025 m NaCl. A, AVP-pVIc diffuses rapidly along DNA (x(t), left axis, 27 trajectories). Motion transverse to the DNA (y(t), right axis) is represented on the same scale as a control. B, mean square displacement of 22 AVP-pVIc complexes sliding along DNA (〈Δx(τ)2〉, left axis) and transverse to the DNA (〈Δy(τ)2〉, right axis.
Indications for Single Molecule Sliding along Viral DNA
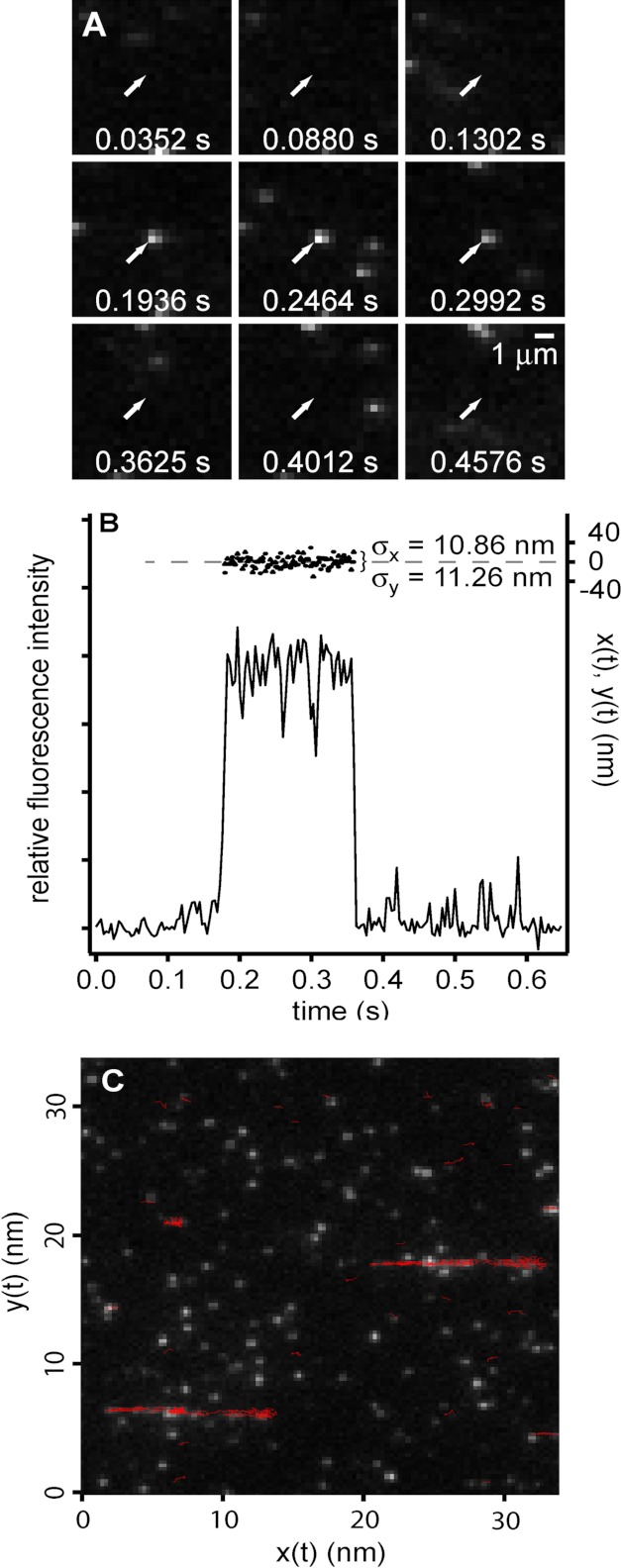
There are several indications that these data represent one-dimensional diffusion of individual protein molecules along a single DNA molecule. Performing assays at low protein and DNA concentrations ensured, given the image resolution, that the observation of two like molecules in the same diffraction-limited volume would be exceedingly rare. An example of a typical fluorescent image is shown in Fig. 4, A and B. A molecule adsorbed to the glass slide at 0.1936 s; its fluorescence remained constant before disappearing at 0.4012 s (Fig. 4, A and B). This abrupt, one-step photo bleaching of a molecule irreversibly bound to the glass slide is characteristic of the visualization of an individual protein molecule labeled with a single dye molecule. Were the fluorescence from more than one dye molecule on a protein or were more than one labeled protein present, upon bleaching, the fluorescence would have diminished in multiple steps as the several colocalized dye molecules would not likely bleach simultaneously. Thus, the images we observed in the sliding assays corresponded to single protein molecules. To show that the labeled protein molecules that moved during sliding assays were interacting with the flow-stretched phage DNA, we overlaid the trajectories we observed in the course of a diffusion experiment onto the first frame of the fluorescent image of the image series. The trajectories outlined the DNA strands (Fig. 4C) whose positions were confirmed after the sliding assay by staining the DNA with an intercalating dye. These data indicate that the binding and movement we observed were occurring along the DNA and not at random locations on the surface of the coverslip.
FIGURE 4.
Observation and localization of individual AVP-pVIc complexes. A, single molecule bound to cover slip. Fluorescence images of a single AVP-pVIc molecule (arrow) immobilized on a coverslip (1.2 kW/cm2, 0.00352 s/frame) are shown. B, fluorescence intensity (left axis) and position estimate (right axis) of the molecule indicated in A. Circles represent measured positions in the direction parallel to flow (x), and triangles represent measured positions in the direction perpendicular to flow (y) versus time. C, sliding on DNA. Shown is an overlay of trajectories (red) on a grayscale fluorescence image from the recording showing the position of flow-stretched λDNA molecules; the position of the DNA was confirmed by staining with intercalating dye after the sliding experiment.
Does Processing of a Virion Precursor Protein in Vitro Require DNA?
If sliding by AVP-pVIc complexes on DNA is required for the processing of the virion precursor proteins, then one would predict that processing of virion precursor proteins would occur only in the presence of DNA. Previously, we cloned, expressed, and purified the precursor to protein VI, pVI (49). We showed that pVI binds tightly to DNA independent of DNA sequence, Kd(app) = 46 nm (Table 1) (where Kd(app) is the apparent equilibrium dissociation constant). pVI contains 250 amino acids and must be cleaved twice, liberating amino acids 1–33 from the N terminus and amino acids 239–250 from the C terminus, to become protein VI (13, 14). To see whether the processing of pVI to protein VI by AVP-pVIc complexes requires DNA, we incubated AVP-pVIc complexes with pVI in the presence of DNA. The reactions contained low salt to increase the duration of DNA binding events and excess DNA over proteins to slow the rate of processing so that processing intermediates could be observed. At various time intervals, we withdrew aliquots of the reactions and assayed them for the presence of protein VI using SDS-polyacrylamide gel electrophoresis and MALDI-TOF mass spectrometry. In the presence of 36-mer dsDNA, after 1 min, an intermediate in the processing of pVI to VI appeared (Fig. 5B). By 4 min, most of the pVI had disappeared. At 2 min, protein VI began to appear, and by 64, min there was more protein VI than intermediate. In the absence of DNA, no conversion of pVI to VI was observed, even after 256 min (Fig. 5A). Thus, for processing of pVI to protein VI to occur via AVP-pVIc complexes in vitro, DNA is required.
FIGURE 5.
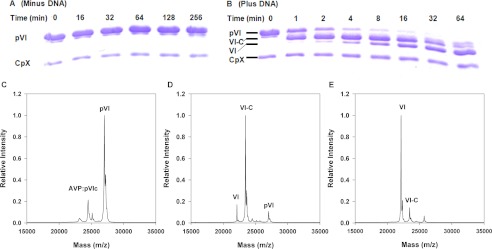
DNA-dependent processing of pVI by AVP-pVIc complexes in vitro. DNA-dependent cleavage of pVI by AVP-pVIc complexes is shown. A and B, SDS-PAGE analysis of the cleavage of pVI by AVP-pVIc complexes in the absence (A) or presence (B) of DNA. A 100-μl reaction in buffer A contained 6.7 μm pVI, 1.4 μm AVP-pVIc, and either 3.4 μm 36-bp DNA or no DNA. After the indicated time intervals, aliquots were removed, and the proteins were fractionated on a 15% polyacrylamide gel by SDS-PAGE. The first lane on the left contains the markers pVI and AVP-pVIc complexes. MALDI-TOF analysis of the DNA-dependent processing of pVI by AVP-pVIc complexes is shown. C, the zero time point before DNA was added. D, 2 min after DNA was added. E, 4 min after DNA was added. The intermediate processing product VI-C (pVI fragment amino acids 34–250; 23,450 m/z) is observed, but the other intermediate product N-VI (pVI fragment amino acids 1–239; 25,683 m/z) is not observed.
Processing Intermediate
To identify the specific intermediates during the processing reaction depicted in the gel in Fig. 5B, we performed MALDI-TOF mass spectroscopic analysis. The concentrations of components in the assay were chosen not to optimize the processing rate but to reveal potential processing intermediates. Before the addition of DNA, two masses were present, AVP-pVIc complexes and pVI (Fig. 5C). At the 2-min time point (Fig. 5D), much of the pVI mass had disappeared. Masses corresponding to VI-C (pVI from which the N-terminal peptide, amino acids 1–33, was cleaved) and VI had appeared. At 4 min (Fig. 5E), no pVI was observed, and there was more protein VI than VI-C. Thus, the processing of pVI by AVP-pVIc complexes occurred sequentially, in two steps: first cleavage at the N terminus of pVI and then cleavage at its C terminus.
Processing of Virion Precursor Proteins in Vivo
To determine whether AVP-pVIc complexes can process virion precursor proteins in vivo, we used extracts from ts-1 virus as a source of virion precursor proteins. ts-1 virus is a temperature-sensitive mutant of adenovirus that when grown at the nonpermissive temperature produces virions devoid of AVP; as such, all the virion precursor proteins are intact (2). The precursor proteins and their mature forms in wild-type virus are shown in the graphic in Fig. 6A. In one experiment, AVP-pVIc complexes were incubated with heat-disrupted ts-1 virus particles for 2 and 24 h before fractionating the proteins on an SDS-polyacrylamide gel (Fig. 6B). Some pVI was processed within 2 h, and all of it was processed within 24 h. On the other hand, most of the pVII was processed by 2 h, and all of it was processed by 24 h. Processing of pIIIa is difficult to resolve as the change in mass is the loss of only 15 out of 585 amino acids. As for precursor to adenovirus protein VIII (pVIII), at 2 h, a band consistent with an ∼17-kDa processing intermediate is visible; the band disappears at 24 h, leaving only one band corresponding to the large (12-kDa) fragment of mature VIII (35). The processing of pμ was observed with a 24% polyacrylamide gel (Fig. 6C). pμ was processed by AVP-pVIc complexes within 2 h.
FIGURE 6.
DNA-dependent processing of the precursor proteins by AVP-pVIc complexes in heat-disrupted ts-1 virus particles. A, virion precursor proteins and their processed forms in wild-type virus. The precursor proteins, their cleavage sites (▾), and the processed precursor proteins are color-coded. ▾* indicates AVP consensus cleavage sites where processing has not been confirmed experimentally because there are only two copies of pTP per virion. B, DNA-dependent cleavage of the precursor proteins by AVP-pVIc complexes in heat-disrupted ts-1 virus. Reaction conditions are listed under “Experimental Procedures.” Proteins were fractionated on PhastGel 8–25% gradient gels. Lane 1 contained heat-disrupted ts-1 virus incubated for 24 h. Lanes 2 and 3 contained heat-disrupted ts-1 virus incubated with AVP-pVIc complexes for 2 or 24 h, respectively. Lanes 4 and 5 contained heat-disrupted ts-1 virus treated with DNase and then incubated with AVP-pVIc complexes for 2 or 24 h, respectively. Lanes 6 and 7 contained heat-disrupted ts-1 virus treated with DNase, with the DNase inactivated, the DNA returned, and then the reactions incubated with AVP-pVIc complexes for 2 or 24 h, respectively. Lane 8 contained mature, wild-type adenovirus serotype 5 virus. C, the proteins in the reactions in B were fractionated on a 24% polyacrylamide gel to be able to visualize the processing of pμ, which consists of 80 amino acids and is cleaved twice. The positions of molecular mass standards are indicated with arrowheads on the left.
Does Processing of Virion Precursor Proteins by AVP-pVIc Complexes in Vivo Require DNA?
In vivo, inside immature virions, the viral DNA is compacted and decorated with tightly bound proteins such as pVII and protein V (36). Is processing of the precursor proteins in heat-disrupted ts-1 by AVP-pVIc complexes virus DNA-dependent? Heat disruption of purified adenovirus virions renders the viral DNA accessible to the action of DNase and other proteins (37). If, before adding AVP-pVIc complexes, the heat-disrupted ts-1 virus particles were incubated with DNase, no processing of pVI or the other precursor proteins was observed 2 or 24 h after adding AVP-pVIc complexes (Fig. 6B). Most convincing was the experiment in which heat-disrupted ts-1 virus particles were incubated with DNase, the DNase was inactivated, and ts-1 viral DNA was added back. Here, upon adding AVP-pVIc complexes and incubating for 24 h, the processing of pVI and the other precursor proteins was identical to that observed with heat-disrupted ts-1 virus particles incubated with AVP-pVIc complexes for 24 h. The remaining precursor protein, pTP, was not visualized in these experiments because it is present at only two copies per virion.
DISCUSSION
Our results present a solution to the enigma of how bimolecular associations can occur between enzymes and substrates inside an immature virus particle. The solution, sliding of an enzyme along DNA to locate and process its substrates, is sufficient to enable all the precursor proteins to be processed. Sliding of AVP-pVIc complexes along DNA via facilitated diffusion represents a new biochemical mechanism by which a proteinase can locate its substrates. By sliding along DNA, AVP-pVIc complexes can find their substrates, although both the enzymes and the protein substrates remain tightly bound to the DNA matrix. Although the lac repressor, the canonical example of facilitated diffusion, locates its operator in E. coli via a combination of sliding along DNA and jumping among segments of DNA (38, 39), sliding alone may be sufficient for AVP to locate quickly all the precursor proteins bound to the viral DNA. Searching along the DNA via one-dimensional diffusion rather than hopping in three-dimensional space is particularly effective under conditions when the three-dimensional diffusion constant is low and when many targets are present on a relatively short DNA segment, i.e. short relative to the number of base pairs covered during an average slide. The adenovirus genome contains 36,000 bp. Each of the 50 molecules of AVP present in the virion (12) needs to interrogate only about 700-bp segments of viral DNA for the entire genome to be scanned for bound precursor protein molecules. Many of the AVP-pVIc and pVI (50) sliding events we observed covered tens of thousands of base pairs, a distance much longer than the target spacing.
The quality of the AVP-pVIc single-molecule sliding data we obtained was exceptionally good. This was because single AVP-Cy3B and AVP-pVIc-Cy3B conjugates emitted photons at unusually high rates (∼350,000 photons detected per second), giving signals several times brighter than those from free Cy3B molecules or other Cy3B-protein conjugates. For example, these complexes were nearly five times brighter than co-imaged Cy3B-labeled human oxoguanine DNA glycosylase 1 conjugates (25). Steric constraint of Cy3 by conjugation to macromolecules has been shown to increase the fluorescence emission relative to that from an equimolar solution of the free dye (40). Cy3B is a modified form of Cy3 that contains a rigid backbone preventing conformational changes that facilitate transitions to dark states (41). This rigidification results in an 8-fold increase in the fluorescence quantum yield when compared with Cy3. In our AVP conjugates, we showed that Cy3B is attached to Cys-199, which has adjacent to it Phe-198. It is possible that interactions of the dye with Phe-198 lower the rates of transition to dark states or accelerate the decays from dark states, enabling the molecule to emit photons even more productively. Minimal sticking of AVP-pVIc to the flow cell was observed despite the fact that blocking agents, e.g. BSA, were excluded from the flow solution.
The increased brightness allowed exceptional spatiotemporal resolution of protein displacements. Using centroid determination to estimate the position of the molecule in each frame, we were able to construct the most probable trajectory of a molecule along the DNA with 3.5-ms time resolution (Fig. 2B). The data (Fig. 2, A and B) demonstrated 11 nm resolution of the position of AVP-pVIc in images collected continuously at 284 Hz under typical experimental conditions. Also, in experiments determining the DNA fluctuation time scale using images of flow-stretched λDNA molecules stained with an intercalating dye,9 the data indicated that at the flow rate used for the highest time resolution measurements, 50 ml/h, the longitudinal DNA fluctuations are well averaged over 3.52 ms and thus do not contribute significantly to fluctuations in the measured position of DNA-bound molecules imaged at this high frame rate.
Can sliding along DNA occur in the core of the immature adenovirus virion? It is likely because the concentration and protein decoration of highly compacted DNA in the core of the adenovirus virion are not unlike that of the DNA inside a bacterium (42) or in the nucleus of a eukaryotic cell (43), where facilitated diffusion has been shown to take place (38, 39). We calculate the double-stranded DNA concentration inside adenovirus virions to be 178–500 g/liter, which is similar to the concentration of DNA in double-stranded DNA bacteriophages (44). The concentration of double-stranded DNA in the nucleoid regions of E. coli is estimated at ∼50–100 g/liter (45, 46). In a living E. coli cell, labeled lac repressor molecules have been observed binding to chromosomal lac operator sites (38, 39). In searching for operator sequences, the lac repressor spends ∼90% of its time nonspecifically bound to and diffusing along DNA. Given that the density of DNA inside the E. coli (42) nucleoid is not all that different from the density of DNA inside an adenovirus virion, that the density of proteins decorating DNA in E. coli is not that dissimilar to that of those decorating adenovirus DNA inside a virion, and that sliding along DNA via one-dimensional diffusion occurs in a live E. coli cell (39), there is no reason why sliding along DNA via one-dimensional diffusion cannot occur in the core of an immature adenovirus virion.
A model for the role of AVP in maturation of the virus particle, based upon recent, extensive evidence including data presented here, proposes the following. 1) AVP is synthesized in a catalytically inactive form (3, 4). If AVP were synthesized as an active enzyme, it could cleave virion precursor protein before virion assembly, and this would abort the infection (8). 2) Binding of AVP to DNA inside immature virions partially activates the enzyme and restricts its access to substrates by constraining its diffusion (3, 5). 3) pVI slides via one-dimensional diffusion on DNA into AVP, and 4) the partially activated AVP cuts out pVIc from pVI (49). 5) The released pVIc binds to and forms a disulfide bond with the AVP that cut it out, forming the covalent AVP-pVIc complex. 6) AVP-pVIc complexes bind tightly to DNA (5), and the resultant ternary complex is the most active form of the enzyme (5, 7, 8). 7) Although AVP binds to but does not slide on DNA (50), the fully active proteinase, the AVP-pVIc complex bound to DNA, does slide along the DNA via one-dimensional diffusion as it searches for binds to and processes its substrates, the DNA-bound precursor proteins. In a different study, we address the mechanism of sliding by pVI and AVP-pVIc complexes, showing that pVIc is a “molecular sled,” capable of sliding by itself or carrying heterologous cargos such as protein VI and AVP.7
This solution to the conundrum establishes a new paradigm for virion maturation. The surprising discovery of the robust sliding activity of AVP-pVIc complexes illustrates how sliding, a conspicuous feature of cellular proteins with functions related to nucleic acid metabolism, can operate in a completely different context: to facilitate bimolecular interactions between enzymes and substrates that are forced by thermodynamic imperatives to bind tightly on a fixed matrix, in this case a viral DNA genome. Given this novel exploitation of the “sliding potential” of the DNA contour by AVP described here, it is not difficult to imagine that other proteins and peptides will be found that make noncanonical use of facilitated diffusion along DNA.
Acknowledgments
We thank Gregory L. Verdine and Sofia Johansson for discussions. The cloning of pVI and pIIIa was done by Eileen Kasmarcik in the F. William Studier laboratory (Brookhaven National Laboratory). We are grateful to María López (CNB-CSIC) and Wenying Huang (Princeton University) for technical assistance and to Elena Molina and Rodrigo Jiménez-Saiz (Instituto de Investigacion en Ciencias de la Alimentacion - Consejo Superior de Investigaciones Cientificas) for access to the PhastGel electrophoresis system.
This work was supported, in whole or in part, by National Institutes of Health Grants R01AI41599 (to W. F. M.) and GM037705 (to S. J. F.) and the National Institutes of Health Director's Pioneer Award and a National Science Foundation (to X. S. X.) This work was also supported by Grants BFU2010-16382 and FIS2010-10552-E from the Ministerio de Ciencia e Innovación of Spain (to C. S. M.).
The following designations are used in this study: pVIc, 11-amino acid cofactor (GVQSLKRRRCF) originating from the C-terminus of virion precursor protein pVI; AVP-pVIc, noncovalent or covalently linked heterodimer of AVP and pVIc; N-VI, pVI fragment amino acids 1–239; 25,683 m/z; VI-C, pVI from which the N-terminal peptide, amino acids 1–33, was cleaved. pVII, the precursor to adenovirus protein VII; pIIIa, the precursor to adenovirus protein IIIa; pμ, the precursor to adenovirus protein μ.
P. C. Blainey, V. Graziano, W. J. McGrath, G. Luo, X. S. Xie, and W. F. Mangel, submitted for publication.
V. Graziano, W. J. McGrath and W. F. Mangel, unpublished observations.
- AVP
- adenovirus proteinase
- pVI
- the precursor to adenovirus protein VI
- pTP
- preterminal protein
- MSD
- mean square displacement.
REFERENCES
- 1. Ding J., McGrath W. J., Sweet R. M., Mangel W. F. (1996) Crystal structure of the human adenovirus proteinase with its 11 amino acid cofactor. EMBO J. 15, 1778–1783 [PMC free article] [PubMed] [Google Scholar]
- 2. Weber J. (1976) Genetic analysis of adenovirus type 2 III. Temperature sensitivity of processing viral proteins. J. Virol. 17, 462–471 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Mangel W. F., McGrath W. J., Toledo D. L., Anderson C. W. (1993) Viral DNA and a viral peptide can act as cofactors of adenovirus virion proteinase activity. Nature 361, 274–275 [DOI] [PubMed] [Google Scholar]
- 4. Webster A., Hay R. T., Kemp G. (1993) The adenovirus protease is activated by a virus-coded disulphide-linked peptide. Cell 72, 97–104 [DOI] [PubMed] [Google Scholar]
- 5. McGrath W. J., Baniecki M. L., Li C., McWhirter S. M., Brown M. T., Toledo D. L., Mangel W. F. (2001) Human adenovirus proteinase: DNA binding and stimulation of proteinase activity by DNA. Biochemistry 40, 13237–13245 [DOI] [PubMed] [Google Scholar]
- 6. Bajpayee N. S., McGrath W. J., Mangel W. F. (2005) Interaction of the adenovirus proteinase with protein cofactors with high negative charge densities. Biochemistry 44, 8721–8729 [DOI] [PubMed] [Google Scholar]
- 7. Mangel W. F., Toledo D. L., Brown M. T., Martin J. H., McGrath W. J. (1996) Characterization of three components of human adenovirus proteinase activity in vitro. J. Biol. Chem. 271, 536–543 [DOI] [PubMed] [Google Scholar]
- 8. Baniecki M. L., McGrath W. J., McWhirter S. M., Li C., Toledo D. L., Pellicena P., Barnard D. L., Thorn K. S., Mangel W. F. (2001) Interaction of the human adenovirus proteinase with its 11-amino acid cofactor pVIc. Biochemistry 40, 12349–12356 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Gupta S., Mangel W. F., McGrath W. J., Perek J. L., Lee D. W., Takamoto K., Chance M. R. (2004) DNA binding provides a molecular strap activating the adenovirus proteinase. Mol. Cell. Proteomics 3, 950–959 [DOI] [PubMed] [Google Scholar]
- 10. McGrath W. J., Baniecki M. L., Peters E., Green D. T., Mangel W. F. (2001) Roles of two conserved cysteine residues in the activation of human adenovirus proteinase. Biochemistry 40, 14468–14474 [DOI] [PubMed] [Google Scholar]
- 11. McGrath W. J., Aherne K. S., Mangel W. F. (2002) In the virion, the 11-amino-acid peptide cofactor pVIc is covalently linked to the adenovirus proteinase. Virology 296, 234–240 [DOI] [PubMed] [Google Scholar]
- 12. Brown M. T., McGrath W. J., Toledo D. L., Mangel W. F. (1996) Different modes of inhibition of human adenovirus proteinase, probably a cysteine proteinase, by bovine pancreatic trypsin inhibitor. FEBS Lett. 388, 233–237 [DOI] [PubMed] [Google Scholar]
- 13. van Oostrum J., Burnett R. M. (1985) Molecular composition of the adenovirus type 2 virion. J. Virol. 56, 439–448 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Lehmberg E., Traina J. A., Chakel J. A., Chang R. J., Parkman M., McCaman M. T., Murakami P. K., Lahidji V., Nelson J. W., Hancock W. S., Nestaas E., Pungor E., Jr. (1999) Reversed-phase high-performance liquid chromatographic assay for the adenovirus type 5 proteome. J. Chromatogr. B Biomed. Sci. Appl. 732, 411–423 [DOI] [PubMed] [Google Scholar]
- 15. Pérez-Berná A. J., Marabini R., Scheres S. H. W., Menéndez-Conejero R., Dmitriev I. P., Curiel D. T., Mangel W. F., Flint S. J., San Martín C. (2009) Structure and uncoating of immature adenovirus. J. Mol. Biol. 392, 547–557 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Russell W. C., Precious B. (1982) Nucleic acid-binding properties of adenovirus structural polypeptides. J. Gen. Virol. 63, 69–79 [DOI] [PubMed] [Google Scholar]
- 17. Chatterjee P. K., Vayda M. E., Flint S. J. (1986) Identification of proteins and protein domains that contact DNA within adenovirus nucleoprotein cores by ultraviolet light crosslinking of oligonucleotides 32P-labelled in vivo. J. Mol. Biol. 188, 23–37 [DOI] [PubMed] [Google Scholar]
- 18. Webster A., Leith I. R., Hay R. T. (1994) Activation of adenovirus-coded protease and processing of preterminal protein. J. Virol. 68, 7292–7300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Greber U. F. (1998) Virus assembly and disassembly: the adenovirus cysteine protease as a trigger factor. Rev. Med. Virol. 8, 213–222 [DOI] [PubMed] [Google Scholar]
- 20. Casjens S. (1997) Principles of virion structure, function and assembly. In: Structural Biology of Viruses (Chiu W., Burnett R. M., Garcea R. L., eds) pp. 3–37, Oxford University Press, Oxford [Google Scholar]
- 21. Mangenot S., Keller S., Rädler J. (2003) Transport of nucleosome core particles in semidilute DNA solutions. Biophys. J. 85, 1817–1825 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Gill S. C., von Hippel P. H. (1989) Calculation of protein extinction coefficients from amino acid sequence data. Anal. Biochem. 182, 319–326 [DOI] [PubMed] [Google Scholar]
- 23. Riddles P. W., Blakeley R. L., Zerner B. (1983) Reassessment of Ellman's reagent. Methods Enzymol. 91, 49–60 [DOI] [PubMed] [Google Scholar]
- 24. Gasteiger E., Gattiker A., Hoogland C., Ivanyi I., Appel R. D., Bairoch A. (2003) ExPASy: The proteomics server for in-depth protein knowledge and analysis. Nucleic Acids Res. 31, 3784–3788 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Blainey P. C., van Oijen A. M., Banerjee A., Verdine G. L., Xie X. S. (2006) A base-excision DNA-repair protein finds intrahelical lesion bases by fast sliding in contact with DNA. Proc. Natl. Acad. Sci. U.S.A. 103, 5752–5757 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. van Oijen A. M., Blainey P. C., Crampton D. J., Richardson C. C., Ellenberger T., Xie X. S. (2003) Single-molecule kinetics of λ-exonuclease reveal base dependence and dynamic disorder. Science 301, 1235–1238 [DOI] [PubMed] [Google Scholar]
- 27. Kim S., Blainey P. C., Schroeder C. M., Xie X. S. (2007) Multiplexed single-molecule assay for enzymatic activity on flow-stretched DNA. Nat. Methods 4, 397–399 [DOI] [PubMed] [Google Scholar]
- 28. Kabata H., Kurosawa O., Arai I., Washizu M., Margarson S. A., Glass R. E., Shimamoto N. (1993) Visualization of single molecules of RNA polymerase sliding along DNA. Science 262, 1561–1563 [DOI] [PubMed] [Google Scholar]
- 29. Harada Y., Funatsu T., Murakami K., Nonoyama Y., Ishihama A., Yanagida T. (1999) Single-molecule imaging of RNA polymerase-DNA interactions in real time. Biophys. J. 76, 709–715 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Kochaniak A. B., Habuchi S., Loparo J. J., Chang D. J., Cimprich K. A., Walter J. C., van Oijen A. M. (2009) Proliferating cell nuclear antigen uses two distinct modes to move along DNA. J. Biol. Chem. 284, 17700–17710 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. von Hippel P. H., Berg O. G. (1989) Facilitated target location in biological systems. J. Biol. Chem. 264, 675–678 [PubMed] [Google Scholar]
- 32. Bagchi B., Blainey P. C., Xie X. S. (2008) Diffusion constant of a nonspecifically bound protein undergoing curvilinear motion along DNA. J. Phys. Chem. B 112, 6282–6284 [DOI] [PubMed] [Google Scholar]
- 33. Komazin-Meredith G., Mirchev R., Golan D. E., van Oijen A. M., Coen D. M. (2008) Hopping of a processivity factor on DNA revealed by single-molecule assays of diffusion. Proc. Natl. Acad. Sci. U.S.A. 105, 10721–10726 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Gowers D. M., Wilson G. G., Halford S. E. (2005) Measurement of the contributions of 1D and 3D pathways to the translocation of a protein along DNA. Proc. Natl. Acad. Sci. U.S.A. 102, 15883–15888 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Liu H., Jin L., Koh S. B., Atanasov I., Schein S., Wu L., Zhou Z. H. (2010) Atomic structure of human adenovirus by cryo-EM reveals interactions among protein networks. Science 329, 1038–1043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Flint S. J., Enquist L. W., Racaniello V. R., Skalka A. M. (2009) Principles of Virology, Third Ed., ASM Press, Washington, D. C [Google Scholar]
- 37. Russell W. C., Valentine R. C., Pereira H. G. (1967) The effect of heat on the anatomy of the adenovirus. J. Gen. Virol. 1, 509–522 [DOI] [PubMed] [Google Scholar]
- 38. Hammar P., Leroy P., Mahmutovic A., Marklund E. G., Berg O. G., Elf J. (2012) The lac repressor displays facilitated diffusion in living cells. Science 336, 1595–1598 [DOI] [PubMed] [Google Scholar]
- 39. Elf J., Li G.-W., Xie X. S. (2007) Probing transcription factor dynamics at the single-molecule level in a living cell. Science 316, 1191–1194 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Gruber H. J., Hahn C. D., Kada G., Riener C. K., Harms G. S., Ahrer W., Dax T. G., Knaus H. G. (2000) Anomalous fluorescence enhancement of Cy3 and cy3.5 versus anomalous fluorescence loss of Cy5 and Cy7 upon covalent linking to IgG and noncovalent binding to avidin. Bioconjug. Chem. 11, 696–704 [DOI] [PubMed] [Google Scholar]
- 41. Cooper M., Ebner A., Briggs M., Burrows M., Gardner N., Richardson R., West R. (2004) Cy3B: improving the performance of cyanine dyes. J. Fluoresc. 14, 145–150 [DOI] [PubMed] [Google Scholar]
- 42. Thanbichler M., Shapiro L. (2006) Chromosome organization and segregation in bacteria. J. Struct. Biol. 156, 292–303 [DOI] [PubMed] [Google Scholar]
- 43. Lieberman-Aiden E., van Berkum N. L., Williams L., Imakaev M., Ragoczy T., Telling A., Amit I., Lajoie B. R., Sabo P. J., Dorschner M. O., Sandstrom R., Bernstein B., Bender M. A., Groudine M., Gnirke A., Stamatoyannopoulos J., Mirny L. A., Lander E. S., Dekker J. (2009) Comprehensive mapping of long-range interactions reveals folding principles of the human genome. Science 326, 289–293 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Earnshaw W. C., Casjens S. R. (1980) DNA packaging by the double-stranded DNA bacteriophages. Cell 21, 319–331 [DOI] [PubMed] [Google Scholar]
- 45. Zimmerman S. B. (2006) Shape and compaction of Escherichia coli nucleoids. J. Struct. Biol. 156, 255–261 [DOI] [PubMed] [Google Scholar]
- 46. Murphy L. D., Zimmerman S. B. (1995) Condensation and cohesion of λDNA in cell extracts and other media: implications for the structure and function of DNA in prokaryotes. Biophys. Chem. 57, 71–92 [DOI] [PubMed] [Google Scholar]
- 47. Doyle P. S., Ladoux B., Viovy J. L. (2000) Dynamics of a tethered polymer in shear flow. Phys Rev. Lett. 84, 4769–4772 [DOI] [PubMed] [Google Scholar]
- 48. Schroeder C. M., Blainey P. C., Kim S., Xie X. S. (2008) Hydrodynamic Flow-stretching Assay for Single-Molecule Studies of Nucleic Acid–Protein Interactions. In: Single-Molecule Techniques: A Laboratory Manual (Ha T., Selvin P. R., eds) pp. 461–492, Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY [Google Scholar]
- 49. Graziano V., McGrath W. J., Suomalainen M., Greber U. F., Freimuth P., Blainey P. C., Luo G., Xie X. S., Mangel W. F. (2012) Regulation of a viral proteinase by a peptide and DNA in one-dimensional space. I. Binding to DNA and to hexon of the precursor to protein VI, pVI, of human adenovirus. J. Biol. Chem. 287, 2059–2067 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Graziano V., Luo G., Blainey P.C., Pérez-Berná A. J., McGrath W. J., Flint S. J., San Martín C., Xie X. S., Mangel W. F. (2012) Regulation of a viral proteinase by a peptide and DNA in one-dimensional space. II. Adenovirus proteinase is activated in an unusual one-dimensional biochemical reaction. J. Biol. Chem. 287, 2068–2080 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Blainey P. C., Luo G., Kou S. C., Mangel W. F., Verdine G. L., Bagchi B., Xie X. S. (2009) Nonspecifically bound proteins spin while diffusing along DNA. Nat. Struct. Mol. Biol. 16, 1224–1229 [DOI] [PMC free article] [PubMed] [Google Scholar]