Abstract
Background
Asthma is a complex disease characterized by sex-specific differences in incidence, prevalence and severity, but little is known about the molecular basis of these sex differences. Objective: To investigate the genetic architecture of sex differences in asthma risk, we evaluated i) associations between polymorphisms in the interferon-gamma (IFNG) gene and childhood onset asthma in combined and sex-specific samples, and ii) interactions between polymorphisms and sex on asthma risk.
Methods
Main and sex-interaction effects of IFNG genetic diversity on asthma risk and IFN-γ levels were examined in a birth cohort of children at high risk for asthma and allergic diseases. Replication of the genetic association was assessed in an independent sample of asthma cases.
Results
Significant genotype-by-sex interactions on asthma were observed for two IFNG SNPs, rs2069727 and rs2430561, which were in strong linkage disequilibrium with each other. In contrast, none of the ten IFNG SNPs showed significant main effects on asthma. The observed genotype-by-sex interaction on asthma was characterized by non-additivity, i.e. heterozygote boys had the highest risk for asthma, while heterozygote girls had the lowest risk. The interaction effect was robust to other asthma risk factors but was limited to children who experienced wheezing illnesses with viral infections during the first three years of life. Genotype-by-sex interactions were also observed in IFN-γ response to LPS in the first year of life. Finally, the sex interaction effect was replicated in an independent population of childhood asthma cases.
Conclusions
These results provide insight into the genetic basis of sex differences in asthma and highlight the potential importance of interactions among sex, genotype, and environmental factors in asthma pathogenesis.
Keywords: IFN-γ, asthma, children, sex differences, single nucleotide polymorphism, association study
INTRODUCTION
Asthma is a common, chronic inflammatory disorder of the airways that arises from interactions between environmental stimuli, particularly those in early life, and genetic (and epigenetic) factors. Significant age- and sex-specific differences are observed in patterns asthma prevalence and severity,1, 2 suggesting that the gene-environment interactions are not static, and instead may respond to developmental and physiological changes occurring in each sex.
Sex differences in asthma are expressed as a higher prevalence of wheezing and asthma in boys before puberty, a shift towards girls around and following puberty, and finally by an increased prevalence, incidence, and severity in women during adulthood.1–4 Sex-specific differences have also been reported for numerous asthma-related traits, including bronchial hyperresponsiveness5, allergic sensitization6, serum IgE levels7, and developmental cytokine response profiles.8 The genetic architecture of asthma risk also appears to differ by sex, as evidenced by reports of sex-specific genetic associations with asthma risk or severity for genes such as TSLP, 9 VDR,10 KCNB1,11 and ADRB2.12, 13
Early life developmental cytokine response profiles have been studied as potential indicators of postnatal immune development. Of the cytokines studied, diminished production of IFN-γ by stimulated peripheral blood mononuclear cells (PBMCs) collected in the first year of life has been repeatedly linked to the subsequent development of atopy, wheezing, and/or asthma in children.14–18 The finding that diminished ex vivo IFN-γ responses in peripheral blood cells are associated with subsequent asthma risk suggests that factors influencing IFN-γ production may be important in asthma pathogenesis.
The IFN-γ pathway has also been implicated in asthma susceptibility in some,19–21 but not all, genetic association studies.22, 23 No variation in the coding exons of the IFNG gene has been reported so all associations with asthma were with polymorphisms located in introns or regions adjacent to the gene. For example, a single nucleotide polymorphism (SNP) in intron 3 of IFNG was associated with asthma in a case-control study of adults in India,19 and a (CA)n repeat polymorphism in intron 1 of IFNG was associated with childhood asthma in a family-based study in Taiwan20 and with atopic asthma in a case-control study of Japanese children.21 In contrast, two other studies failed to find a significant association between the (CA)n repeat polymorphism and asthma.22, 23 The heterogeneity of these results suggests that genetic variation in IFNG may be associated with asthma risk in complex ways.
We previously showed that sex-specific differences in IFN-γ response profiles were present during early childhood and that sex modified the association between IFN-γ production and wheezing phenotypes in a prospective birth cohort study of children with a parental history of allergy and/or asthma.8 In the same cohort, it was also observed that reduced IFN-γ neonatal production was associated with increased frequency and severity of viral respiratory illnesses and wheezing.24, 25 To elucidate the genetic basis of variation in IFN-γ production and better understand the relationship between sex-specific patterns of IFN-γ response and sex differences in the occurrence of asthma, we characterized genetic variation at the IFNG locus in the same high-risk birth cohort and tested for main and sex-interaction effects between IFNG variants and asthma diagnosed at age 8.
METHODS
Ethics Statement
Written informed consent was obtained from all participants. The COAST study was approved by approved by the University of Wisconsin Human Subjects Committee and the University of Chicago Institutional Review Board. The Chicago Asthma Genetics study was approved by the University of Chicago Institutional Review Board.
Childhood Origins of ASThma (COAST) study subjects
A total of 289 subjects were enrolled at birth into the Childhood Origins of ASThma study between November 1998 and May 2000, as previously described.26 To be eligible for inclusion, each newborn was required to have at least one parent with respiratory allergies (defined as one or more positive aeroallergen skin test responses) and/or a history of physician-diagnosed asthma.
Clinical definitions
Asthma was diagnosed at eight years of age, as previously described,27, 28 based on the presence of at least one of the following: (1) physician-diagnosed wheezing at an office visit; (2) use of albuterol for coughing or wheezing episodes as prescribed by a physician; (3) use of a daily controller medicine; (4) step-up plan including use of albuterol or short-term use of inhaled corticosteroids during illness, and (5) use of prednisone for asthma exacerbation.
A wheezing respiratory illness in the first three years of life was defined by the occurrence of one or more of the following: (1) physician-diagnosed wheezing at an office visit; (2) an illness for which a child was prescribed short- or long-acting β-agonists and/or controller medicines; (3) an illness given the following diagnoses: bronchiolitis, wheezing illness, reactive airway disease, asthma, and/or asthma exacerbation, as previously described.28, 29
Aeroallergen and food allergen sensitization were evaluated based on the measurement of allergen-specific IgE in children at 3 years of age for dust mite, Alternaria alternata, cat dander, dog, egg white, milk, peanut, and soy by automated fluoroenzyme immunoassay (Unicap 100; Pharmacia and Upjohn Diagnostics, Kalamazoo, MI) as previously described.17 Specific IgE values above the sensitivity for detection (0.35 kU/L) were considered positive. Allergic sensitization was defined as having one or more positive specific IgE values.
Collection and stimulation of blood samples
Cord blood was collected from the umbilical cord vein using standard techniques, as described previously.30 Peripheral blood samples were collected annually beginning at age 1 year. Mononuclear cells were separated using density centrifugation (LSM Lymphocyte Separation Medium, ICN Biomedicals Inc., Aurora, OH). Aliquots of 106 cells were suspended in RPMI-1640 supplemented with 10% fetal bovine serum (Hyclone, Logan, UT, USA), L-glutamine (2mM), penicillin (100 U/mL), and streptomycin (100 μg/mL). Cells were incubated (24 h, 37°C, 5% CO2) in a 24-well flat bottom cell culture plate (1mL/well, Corning Inc., Corning, NY) with lipopolysaccharide (LPS). At the beginning of the study, LPS was aliquoted into single-use portions that were stored at −80°C until use, ensuring that all cells were treated with identical product.
Cytokine enzyme-linked immunoassay (ELISA)
IFN-γ was measured in culture supernatants by ELISA, according to manufacturer’s instructions (Pharmigen, San Diego, CA), except that sample volume was reduced to 50 μl. The sensitivity of the IFN-γ ELISA was 3.1 pg/mL. Samples were measured in duplicate and the mean of those measurements was used for analysis. IFN-γ was measured in the cord blood cells of 95 children and detectable levels, i.e. values >3.1 pg/mL, were observed in 59 of those samples. IFN-γ was measured in the year 1 peripheral blood cells of 220 children and detectable levels were observed in 60 of those samples. Detectable IFN-γ values were log transformed and regressed against a bivariate measure of the immediacy of processing time (0 for processing time of <12 hours after blood draw, 1 for processing time of >12 hours).30 The residuals of this regression were used for tests of genetic associations.
IFNG Genotyping
SNPs were chosen to capture common variation in IFNG and its flanking region using data from European Americans (CEU) in the International HapMap project31 and the SeattleSNPs database. Specifically, SNPs with a minor allele frequency ≥0.05 were selected to tag known linkage disequilibrium (LD) bins and to evaluate reported functional variation at rs2069727, rs2430561, rs2069705, and rs1861493. Five IFNG SNPs (rs2430561, rs1861493, rs2069705, rs2069718, rs2069727) were genotyped using the SNaPshot® Multiplex System (Applied Biosystems, Carlsbad, CA). Two SNPs (rs2069728, rs2193050) were genotyped using TaqMan® OpenArray® technology (Applied Biosystems, Carlsbad, CA). Three SNPs (rs2069732, rs2069707, rs2193048) were genotyped using TaqMan® Assay-on-Demand technology (Applied Biosystems, Carlsbad, CA). Pairwise linkage disequilibrium between SNPs was calculated and visualized in Haploview 4.2.32
Statistical analysis
Main and sex-interaction effects on asthma status were assessed using contingency table analysis and logistic regression, as implemented in JMP® 8.0.2.2 (SAS Institute Inc., Cary, NC). Main effects were assessed using a genotype test, which assumes no specific underlying genetic model. The association between asthma status and observed genotype counts at each SNP were compared in 2×3 contingency tables using the likelihood ratio chi-square test. To assess sex interaction effects, asthma status was used as the outcome variable in a logistic regression model that included genotype, sex, and a genotype-by-sex interaction term as predictor variables.
Genetic associations were evaluated using the 195 COAST children who were assigned a definitive asthma diagnosis at age 8. To correct for multiple testing of SNPs in IFNG, we calculated the effective number of independent tests given the patterns of pairwise LD between SNPs using Li and Ji’s method,33 as implemented in the matrix spectral decomposition (matSpD) program34 at http://gump.qimr.edu.au/general/daleN/matSpD/. Using Li and Ji’s method, the ten SNPs in IFNG reduced to 6.0 independent variables. P values were corrected for 6.0 tests using the Sidak test for multiple comparisons: Pc=1−(1−P)k, where k equals the number of independent comparisons.35
Genetic associations with LPS-stimulated IFN-γ levels were determined in each sex separately using the non-parametric Van der Waerden test. Genotype-by-sex interactions on LPS-stimulated IFN-γ level were assessed using a standard least square regression using IFNG genotype class (i.e. heterozygote versus homozygote), sex, and a genotype-by-sex interaction term as predictor variables.
Chicago Asthma Genetics study subjects
Participants were recruited through the adult and pediatric asthma clinics at the University of Chicago Medical Center as part of the Chicago Asthma Genetics study.36 Subjects diagnosed with asthma were at least 6 years of age at the time of recruitment and met the following criteria: (1) a physician’s diagnosis of asthma, (2) presence of at least two self-reported symptoms (cough, wheeze, and/or shortness of breath), (3) current use of asthma medications, and (4) either bronchial hyperresponsiveness (defined as a ≥20% decrease in baseline FEV1 after inhalation of ≤25 mg of methacholine per milliliter) or an increase of ≥15% in baseline FEV1 after treatment with a short-acting bronchodialator inhalation or treatment with inhaled corticosteroids.37 Subjects were excluded from study if they smoked the equivalent of ≥3 pack-years, had a birthweight <4.4 pounds, or had a conflicting pulmonary diagnosis. Age of asthma onset was ascertained by questionnaire. Testing for a genotype-by-sex interaction effect was performed in 79 individuals with childhood onset of asthma (≤8 years of age) using a 2×3 contingency table, with the P value representing the likelihood ratio chi-square statistic.
RESULTS
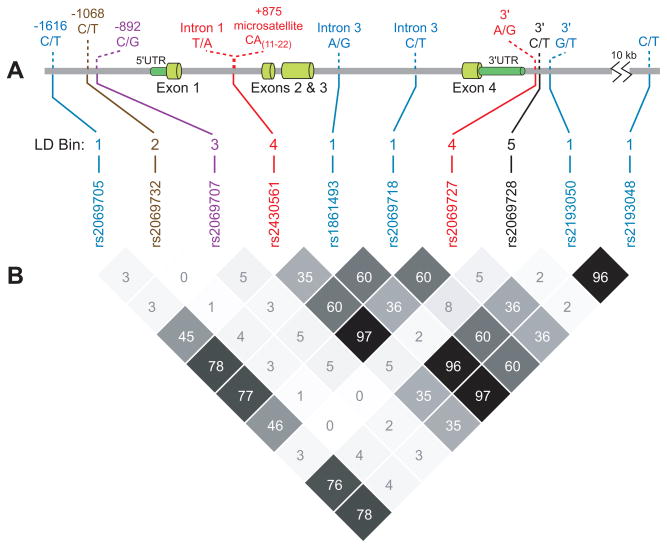
Ten SNPs at the IFNG locus were genotyped in 234 COAST children of European descent (Figure 1, A). Observed genotype counts at all 10 SNPs did not differ from those expected under Hardy-Weinburg equilibrium (P>0.05; data not shown). Pairwise linkage disequilibrium (LD), measured by r2, was observed among a subset of IFNG SNPs (Figure 1, B). Using an r2 cutoff of 0.75, the ten IFNG SNPs fell into five LD bins: five SNPs comprised bin 1 (rs2069705, rs1861493, rs2069718, rs2193050, and rs2193048), two SNPs comprised bin 4 (rs2430561 and rs2069727), and one SNP each comprised bins 2 (rs2069732), 3 (rs2069707), and 5 (rs2069728).
Figure 1. IFNG gene structure and patterns of pairwise LD.
(A) Genetic polymorphisms in the 18.4 kb IFNG region on chromosome 12. SNPs included in this study, and the intron 1 microsatellite, are shown. (B) Patterns of pairwise linkage disequilibrium (r2) observed in the COAST children. SNPs in the same LD bin are shown in the same color.
IFNG SNPs show significant genotype-by-sex interaction effects on asthma
In the combined cohort, only one SNP in LD bin 5, rs2069728, showed significant main effects on asthma status (P=0.023), but this association was not significant after correction for multiple testing (Pc=0.13) (Table I). In the sex-stratified cohorts, the association between SNP rs2069727, in LD bin 4, and asthma in the girls-only cohort approached statistical significance after correcting for multiple tests (Pc=0.076; Table I). In the boys-only cohort, rs2069727 exhibited a non-significant trend in the opposite direction of the girls-only cohort, suggesting possible sex-specific differences in the effects of this variant. The other SNP in LD bin 4, rs2430561, showed sex-specific genetic associations similar to those of rs2069727. These two SNPs are in strong LD (r2=0.97); because genotype data were more complete for rs2069727 and because all results were similar for the two SNPs, we present results only for rs2069727 in subsequent analyses. SNPs in two additional LD bins (1 and 5) exhibited significant associations with asthma in the girls-only cohort before, but not after, correction for multiple tests, rs2069718 (P=0.029; Pc=0.16) and rs2069728 (P=0.048; Pc=0.26).
Table I.
IFNG genotype associations with asthma at age 8 years in the COAST children.
| SNP | Position | LD bin | Minor Allele Freq. | Cohort | Asthma at age 8 years (P values) | |||
|---|---|---|---|---|---|---|---|---|
| Main effect | Main effect (corrected) | Sex interaction | Sex interaction (corrected) | |||||
| rs2069705 (C/T) | 66,841,278 | 1 | 0.36 | Combined (n=193) Girls-only (n=80) Boys-only (n=113) |
0.44 0.18 0.55 |
0.34 | ||
| rs2069732 (C/T) | 66,840,728 | 2 | 0.04 | Combined (n=163) Girls-only (n=68) Boys-only (n=95) |
0.70 0.92 0.81 |
0.92 | ||
| rs2069707 (C/G) | 66,840,555 | 3 | 0.06 | Combined (n=181) Girls-only (n=76) Boys-only (n=105) |
0.78 0.51 0.83 |
0.63 | ||
| rs2430561 (A/T) | 66,838,787 | 4 | 0.44 | Combined (n=182) Girls-only (n=77) Boys-only (n=105) |
0.95 0.026 0.13 |
0.15 | 0.0042 | 0.025 |
| rs1861493 (A/G) | 66,837,463 | 1 | 0.31 | Combined (n=186) Girls-only (n=78) Boys-only (n=108) |
0.24 0.42 0.26 |
0.88 | ||
| rs2069718 (C/T) | 66,836,429 | 1 | 0.43 | Combined (n=185) Girls-only (n=78) Boys-only (n=107) |
0.43 0.029 0.85 |
0.16 | 0.063 | |
| rs2069727 (A/G) | 66,834,490 | 4 | 0.45 | Combined (n=193) Girls-only (n=80) Boys-only (n=113) |
0.95 0.013 0.089 |
0.076 | 0.0014 | 0.0084 |
| rs2069728 (C/T) | 66,834,051 | 5 | 0.06 | Combined (n=172) Girls-only (n=73) Boys-only (n=99) |
0.023 0.048 0.37 |
0.13 0.26 |
0.44 | |
| rs2193050 (G/T) | 66,833,210 | 1 | 0.30 | Combined (n=172) Girls-only (n=73) Boys-only (n=99) |
0.13 0.17 0.12 |
0.50 | ||
| rs2193048 (C/T) | 66,822,891 | 1 | 0.31 | Combined (n=186) Girls-only (n=78) Boys-only (n=108) |
0.11 0.16 0.26 |
0.78 | ||
P values significant before correction for multiple comparisons are shown in boldface.
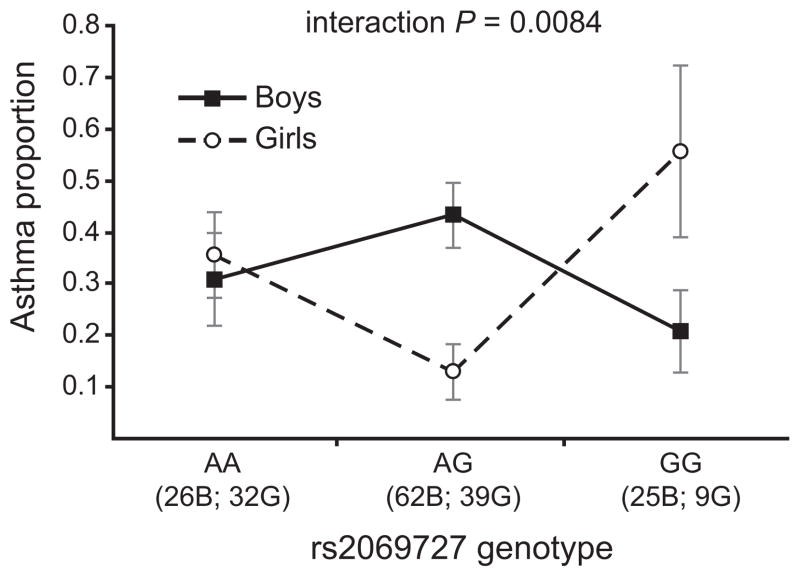
Two SNPs in LD bin 4 (rs2069727 and rs2430561) showed significant sex interaction effects on asthma: interaction Pc=0.0084 for rs2069727 and interaction Pc=0.025 for rs2430561 (Table I). This genotype-by-sex interaction was manifested as an overdominant pattern in which heterozygous girls exhibited lower rates of asthma than homozygous girls, and heterozygous boys exhibited relatively higher rates of asthma than homozygous boys (Figure 2). At rs2069727, only 5 of 39 heterozygote girls were diagnosed with asthma by age 8, compared to 11 of 32 AA homozygotes and 5 of 9 GG homozygotes, resulting in a heterozygote OR = 0.23 (95% CI = 0.074–0.71). In boys, the pattern was reversed: 27 of 62 heterozygotes at rs2069727 were diagnosed with asthma by age 8, compared to only 8 of 26 AA homozygotes and 5 of 25 GG homozygotes, resulting in a heterozygote OR = 2.17 (95% CI = 0.97–4.87).
Figure 2. IFNG genotype-by-sex interaction effects on asthma in COAST children.
Proportion (± SE) of COAST boys (solid line) and girls (dashed line) diagnosed with asthma at age 8 for each IFNG rs2069727 genotype. The numbers of boys (B) and girls (G) of each genotype are indicated.
IFNG genotype-by-sex interaction effect on asthma is robust to risk factors
Previous research on the COAST cohort identified risk factors that contributed to risk of developing asthma at age 6.28 Two risk factors, wheezing illnesses in the first three years of life and aeroallergen sensitization at age 3, were also significantly associated with increased risk of asthma at age 8 (Table II). To determine if these risk factors modified the observed IFNG genotype, they were included as covariates in a reanalysis of IFNG interaction effects. The genotype-by-sex interaction effect on asthma remained significant for both rs2069727 (interaction P=0.0086) and rs2430561 (interaction P=0.0040) when wheezing history and aeroallergen sensitization were included, similar to the results of testing for interaction effects without them (Table I).
Table II.
Risk factors for asthma at age 8 years in COAST children.
| Risk Factors | Children with risk factor (%) | Asthma at age 8 univariate P value | Asthma at age 8 multivariate P value |
|---|---|---|---|
| Wheezing in first 3 years of life | 99/195 (50.8) | <0.0001 | <0.0001 |
| Aeroallergen sensitization at 3 years | 52/200 (26.0) | 0.0051 | 0.024 |
| Older siblings | 109/233 (46.8) | 0.069 | 0.18 |
| Dog in household at birth | 88/233 (37.8) | 0.15 | 0.40 |
| Food sensitization at 3 years | 81/201 (40.3) | 0.31 | 0.86 |
Genotype-by-sex interaction effect is limited to children who experienced wheezing illnesses in the first 3 years of life
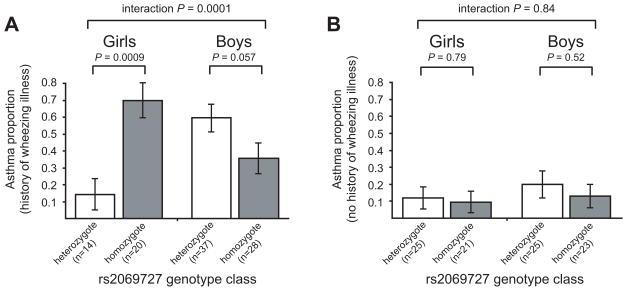
To further examine the relationship between virus-induced wheezing in the first three years of life and the observed IFNG-sex interaction effect on asthma risk, we stratified the COAST cohort by the occurrence of early life wheezing illnesses and reexamined IFNG interaction effects on asthma. The interaction effect between IFNG and asthma was different in children with a history of wheezing illnesses (Figure 3, A) compared to those with no history (Figure 3, B). Specifically, the IFNG genotype-by-sex interaction effect on asthma was highly significant in children who experienced wheezing illnesses in early life (n=99; interaction P=0.0001 for rs2069727) but non-significant in children with no history of wheezing illnesses (n=94; interaction P=0.84 for rs2069727).
Figure 3. Influence of wheezing history on IFNG association with asthma.
Proportion (± SE) of rs2069727 heterozygous and homozygous COAST children diagnosed with asthma by age 8 years, stratified by the occurrence (A) or absence (B) of viral wheezing illnesses in the first three years of life.
IFN-γ protein levels show genotype-by-sex interaction effect
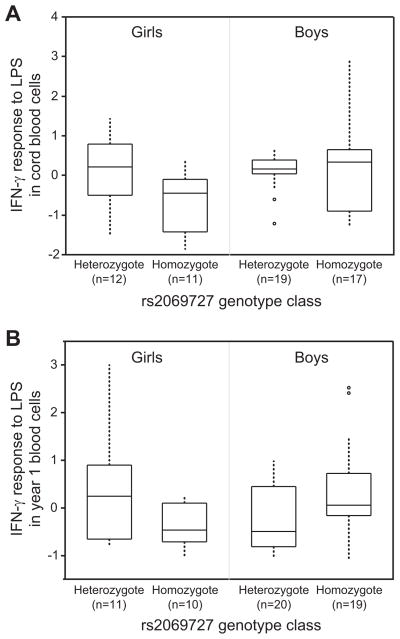
To investigate the potential mechanisms underlying the observed IFNG genotype-by-sex interaction effect on asthma in the COAST children, the association between IFNG genetic variation and the strength of IFN-γ response to LPS during the first year of life was examined. In cord blood cells, IFN-γ response to LPS was significantly higher in girls who were heterozygous for SNP rs2069727 compared to girls who were homozygous (P=0.027) (Figure 4, A). Cord blood IFN-γ response was not significantly different in heterozygote versus homozygote boys (P=0.61). In cells collected at year 1, LPS-stimulated IFN-γ levels were non-significantly higher in heterozygote girls compared to homozygotes (P=0.14), whereas rs2069727 homozygote boys had significantly higher IFN-γ response to LPS than heterozygotes (P=0.041) (Figure 4, B). The observed sex differences in the rs2069727 association with IFN-γ response resulted in significant genotype-by-sex interaction effects on the magnitude of IFN-γ response to LPS in both cord blood (interaction P=0.038) and year 1 (interaction P=0.0092) cells. Similar interactions with IFN-γ response to LPS were observed for the rs2430561 SNP (data not shown).
Figure 4. LPS-induced IFN-γ response in COAST boys and girls by rs2069727 genotype class.
(A) Median IFN-γ response in cord blood mononuclear cells. (B) Median IFN-γ response in PBMCs collected at 1 year of age.
Genotype-by-sex interaction effect on asthma is replicated in Chicago asthma cases
The interaction between IFNG genotype and sex was examined in a separate group of unrelated individuals of European descent who experienced a childhood onset of asthma. In these Chicago subjects with asthma, the rs2069727 genotype frequency distribution differed significantly by sex: 55% of males with asthma were heterozygous, while only 25% of females with asthma were heterozygous (Table III). The observed genotype frequencies resulted in significant genotype-by-sex interaction effects for both SNPs (rs2069727 P=0.016, rs2430561 P=0.0052; Table III). IFNG genotype frequencies in the male and female Chicago asthma cases were similar to those observed in the COAST boys and girls who developed asthma by age 8.
Table III.
Genotype frequencies for the rs2069727 and rs2430561 SNPs in Chicago asthma cases.
| SNP | Sex of case | Genotype counts (frequency) | Sex interaction P-value* | ||
|---|---|---|---|---|---|
| G/G | G/A | A/A | |||
| rs2069727 | Male | 10 (0.20) | 28 (0.55) | 13 (0.25) | 0.016 |
| Female | 13 (0.46) | 7 (0.25) | 8 (0.29) | ||
| rs2430561 | Male | 10 (0.20) | 28 (0.55) | 13 (0.25) | 0.0052 |
| Female | 14 (0.50) | 6 (0.21) | 8 (0.29) | ||
Significance was determined from χ2 analyses in 2×3 contingency tables.
DISCUSSION
Identification of the genetic and environmental factors that contribute to the developmental and sex-specific patterns of asthma prevalence is complicated by potential interactions between them.38, 39 Here we provide evidence that interactions between sex and IFNG polymorphisms contribute to the risk of developing childhood asthma. Heterozygosity at two IFNG SNPs (rs2069727 and rs2430561) was protective in girls, but associated with increased asthma risk in boys. This genotype-by-sex interaction was itself modified by the occurrence of viral wheezing illnesses in the first three years of life: the interaction effect was highly significant in children who wheezed and absent in children who did not wheeze. Nonetheless, the IFNG genotype-by-sex interaction effect proved to be robust to other significant asthma risk factors in the COAST cohort, and reproducible in an independent population of subjects with a childhood onset of asthma.
IFN-γ production by PBMCs has been studied as a potential biomarker of the developmental immune maturation processes associated with the subsequent development of asthma, allergy, or impaired lung function.14–17, 40 Generally, diminished IFN-γ in the first year of life has been associated with subsequent disease. Our results support this hypothesis, but in a sex-specific pattern: girls who are homozygous at the IFNG rs2069727 or rs2430561 SNPs exhibit lower levels of cord blood IFN-γ after LPS stimulation and a higher risk of asthma at age 8 compared to girls who are heterozygous. Conversely, boys who are heterozygous for those IFNG SNPs exhibit lower levels of year 1 IFN-γ after LPS stimulation and a corresponding higher risk of asthma at age 8 compared to boys who are homozygous. This study does not address whether variation in IFN-γ expression directly contributes to asthma pathogenesis or if it is simply a marker for a more global immune maturation phenotype, such as an imbalance between TH2 and TH1 responses. Stern et al15 proposed a model by which impaired early life IFN-γ production contributes to increased susceptibility to viral illnesses, which then results in childhood wheezing and a higher subsequent risk of developing asthma. Our data is consistent with this model in that the IFNG genotype-by-sex interaction is limited to children who wheezed after viral infections in their first three years of life (Figure 3). Thus, the IFN-γ pathway may be involved in the network of immune processes that predispose individuals who wheeze to future asthma risk.
The IFNG genotype-by-sex interaction effect on childhood asthma observed here for rs2069727 and rs2430561 has not been reported previously. However, others have reported associations between these IFNG SNPs and adult asthma. For example, the rs2069727 SNP was associated with asthma in adults living in India, although the most significant association in that study was with the intron 3 SNP, rs186149419. In contrast, we found no evidence that SNP rs1861493, which is in nearly perfect LD with rs1861494 in LD bin 1, was associated with asthma in the COAST cohort. In a separate study, the rs2430561 (LD bin 2) SNP was associated with asthma in Chinese adults.41 Finally, the (CA)n microsatellite polymorphism in intron 1, which is in strong LD with both the rs2430561 and rs2069727 SNPs,42, 43 differed significantly between individuals with asthma and controls in at least two studies,20, 21 but not in two others.22, 23 In all of these studies, the authors did not report results for sex-stratified samples or for genotype-by-sex interactions so it is not possible to determine if their results confirm or refute the sex-specific heterozygote effects observed in this study.
The two IFNG SNPs (rs2069727 and rs2430561) involved in the genotype-by-sex interaction on asthma in this study were also associated with sex-specific differences in IFN-γ levels in PBMCs stimulated with LPS (Figure 4). Several previous studies have reported an association between the rs2430561 SNP and IFN-γ levels in vitro in response to LPS, PHA, and other stimulants.44–48 However, it remains unclear if variation at rs2430561 is directly responsible for the functional effects, perhaps through differential NF-κB binding to the site,42 or if the rs2430561 SNP simply tags other functional variation on the haplotype. It is also unclear what role sex hormones play in the sex-specific IFNG associations and genotype-by-sex interactions observed here. Estrogen, for example, is known to interact with the IFNG locus and promote IFN-γ production,49–51 and thus may contribute to IFNG-mediated sex differences in asthma risk.
Single locus overdominance, where heterozygotes show a phenotype beyond the range of the homozygotes, has been documented for many diseases and phenotypes.52, 53 Several recent studies have reported sex-specific patterns of overdominance similar to those observed here. For example, sex-specific overdominance was observed between the IL-1β gene and asthma,54 between the β2-adrenergic receptor gene and hypertension,55 and between the insulin-degrading enzyme (IDE) gene and human lifespan, plasma insulin levels, and spliceform distribution.56 Thus, there is precedent for the type of interaction observed in this study.
In conclusion, we report a genotype-by-sex interaction effect on childhood asthma risk. These results provide further evidence that asthma is a complex disease resulting from the effects of multiple diverse, yet interacting, factors. To accurately elucidate the genetic architecture of asthma and other complex diseases future studies should be sensitive to the contribution of genetic interactions with age, sex, and environment.
Key messages.
IFNG polymorphisms are associated with the occurrence of childhood asthma in a sex-specific manner.
IFNG polymorphisms are associated with in vitro IFN-γ levels in the first year of life.
IFNG association with asthma was limited to children who experienced early life wheezing illnesses.
Acknowledgments
Declaration of all sources of funding: This work was supported by the National Institutes of Health grants R01 HL61879, P01 HL70831, M01 RR03186, R01 HL085197, and M01 RR00055. D. Loisel was supported by the National Institutes of Health grants F32 HL095268 and T32 HL007605. J.E. Gern receives grant support from AstraZeneca and GlaxoSmithKline; is a consultant for Biota, Centocor, 3V Biosciences, and Boehringer Ingelheim; and has stock options in 3V Biosciences and EraGen Biosciences. R. F. Lemanske, Jr, is a speaker for Merck, AstraZeneca, Doembecher Children’s Hospital, Washington University, the Medicus Group, the Park Nicolet Institute, the American College of Allergy, Asthma & Immunology, the Los Angeles Allergy Society, the Michigan Allergy/Asthma Society, the Medical College of Wisconsin, the Fund for Medical Research and Education (Detroit), the Children’s Hospital of Minnesota, the Toronto Allergy Society, the American Academy of Allergy, Asthma & Immunology, Beaumont Hospital (Detroit), the University of Illinois, the Canadian Society of Allergy and Clinical Immunology, New York Presbyterian, Med Media Educational Group, Onpointe Medical Communication, Medical University of South Carolina, Health Matters Communication, Bishop McCann, Donohue, Purohit, Miller, Inc, Center for Health Care Education, University of California-San Francisco, American Thoracic Society, University of Iowa, Indiana University, American Lung Association of Upper Midwest, Vanderbilt University, and Rochester Children’s Hospital; is a consultant for AstraZeneca, Smith Research, Inc, the Merck Childhood Asthma Network, Novartis, Quintiles/Innovax, RC Horowitz & Co, Inc, International Meetings and Science, Scienomics, Scientific Therapeutics, Cognimed, Inc, Map Pharmaceuticals, and Gray Consulting; is an author for Up-to-Date; is a textbook editor for Elsevier, Inc; and receives grant support from the National Heart, Lung, and Blood Institute. C. Ober receives research support from the National Institutes of Health.
The authors thank Dan Nicolae for helpful discussions, Gaixin Du for data management and Kristen Patterson for technical support. We want to acknowledge the many clinical coordinators, fellows, research associates, support staff, and students who have contributed immeasurably to sample procurement, data acquisition, analyses, and presentation. We also want to thank the COAST families for their continued interest and support of this project.
Abbreviations used
- COAST
Childhood Origins of Asthma study
- ELISA
Enzyme-linked immunosorbent serological assay
- IFN-γ
Interferon-gamma
- IFNG
Genes encoding interferon-gamma
- LD
Linkage disequilibrium
- LPS
Lipopolysaccharide
- OR
Odds ratio
- PBMC
Peripheral blood mononuclear cell
- SNP
Single nucleotide polymorphism
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Almqvist C, Worm M, Leynaert B. Impact of gender on asthma in childhood and adolescence: a GA2LEN review. Allergy. 2008;63:47–57. doi: 10.1111/j.1398-9995.2007.01524.x. [DOI] [PubMed] [Google Scholar]
- 2.Kynyk JA, Mastronarde JG, McCallister JW. Asthma, the sex difference. Curr Opin Pulm Med. 17:6–11. doi: 10.1097/MCP.0b013e3283410038. [DOI] [PubMed] [Google Scholar]
- 3.Sears MR, Greene JM, Willan AR, Wiecek EM, Taylor DR, Flannery EM, et al. A longitudinal, population-based, cohort study of childhood asthma followed to adulthood. N Engl J Med. 2003;349:1414–22. doi: 10.1056/NEJMoa022363. [DOI] [PubMed] [Google Scholar]
- 4.Vink NM, Postma DS, Schouten JP, Rosmalen JG, Boezen HM. Gender differences in asthma development and remission during transition through puberty: the TRacking Adolescents’ Individual Lives Survey (TRAILS) study. J Allergy Clin Immunol. 2010;126:498–504. e1–6. doi: 10.1016/j.jaci.2010.06.018. [DOI] [PubMed] [Google Scholar]
- 5.Leynaert B, Bousquet J, Henry C, Liard R, Neukirch F. Is bronchial hyperresponsiveness more frequent in women than in men? A population-based study. Am J Respir Crit Care Med. 1997;156:1413–20. doi: 10.1164/ajrccm.156.5.9701060. [DOI] [PubMed] [Google Scholar]
- 6.Xuan W, Marks GB, Toelle BG, Belousova E, Peat JK, Berry G, et al. Risk factors for onset and remission of atopy, wheeze, and airway hyperresponsiveness. Thorax. 2002;57:104–9. doi: 10.1136/thorax.57.2.104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kulig M, Tacke U, Forster J, Edenharter G, Bergmann R, Lau S, et al. Serum IgE levels during the first 6 years of life. J Pediatr. 1999;134:453–8. doi: 10.1016/s0022-3476(99)70203-9. [DOI] [PubMed] [Google Scholar]
- 8.Uekert SJ, Akan G, Evans MD, Li ZH, Roberg K, Tisler C, et al. Sex-related differences in immune development and the expression of atopy in early childhood. J Allergy Clin Immunol. 2006;118:1375–81. doi: 10.1016/j.jaci.2006.09.008. [DOI] [PubMed] [Google Scholar]
- 9.Hunninghake GM, Soto-Quiros ME, Avila L, Kim HP, Lasky-Su J, Rafaels N, et al. TSLP polymorphisms are associated with asthma in a sex-specific fashion. Allergy. 2010;65:1566–75. doi: 10.1111/j.1398-9995.2010.02415.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Raby BA, Lazarus R, Silverman EK, Lake S, Lange C, Wjst M, et al. Association of vitamin D receptor gene polymorphisms with childhood and adult asthma. Am J Respir Crit Care Med. 2004;170:1057–65. doi: 10.1164/rccm.200404-447OC. [DOI] [PubMed] [Google Scholar]
- 11.Seibold MA, Wang B, Eng C, Kumar G, Beckman KB, Sen S, et al. An African-specific functional polymorphism in KCNMB1 shows sex-specific association with asthma severity. Hum Mol Genet. 2008;17:2681–90. doi: 10.1093/hmg/ddn168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Santillan AA, Camargo CA, Ramirez-Rivera A, Delgado-Enciso I, Rojas-Martinez A, Cantu-Diaz F, et al. Association between beta(2)-adrenoceptor polymorphisms and asthma diagnosis among Mexican adults. J Allergy Clin Immunol. 2003;112:1095–100. doi: 10.1016/j.jaci.2003.09.029. [DOI] [PubMed] [Google Scholar]
- 13.Guerra S, Graves PE, Morgan WJ, Sherrill DL, Holberg CJ, Wright AL, et al. Relation of beta(2)-adrenoceptor polymorphisms at codons 16 and 27 to persistence of asthma symptoms after the onset of puberty. Chest. 2005;128:609–17. doi: 10.1378/chest.128.2.609. [DOI] [PubMed] [Google Scholar]
- 14.Guerra S, Lohman IC, Halonen M, Martinez FD, Wright AL. Reduced interferon gamma production and soluble CD14 levels in early life predict recurrent wheezing by 1 year of age. Am J Respir Crit Care Med. 2004;169:70–6. doi: 10.1164/rccm.200304-499OC. [DOI] [PubMed] [Google Scholar]
- 15.Stern DA, Guerra S, Halonen M, Wright AL, Martinez FD. Low IFN-gamma production in the first year of life as a predictor of wheeze during childhood. J Allergy Clin Immunol. 2007;120:835–41. doi: 10.1016/j.jaci.2007.05.050. [DOI] [PubMed] [Google Scholar]
- 16.Macaubas C, de Klerk NH, Holt BJ, Wee C, Kendall G, Firth M, et al. Association between antenatal cytokine production and the development of atopy and asthma at age 6 years. Lancet. 2003;362:1192–7. doi: 10.1016/s0140-6736(03)14542-4. [DOI] [PubMed] [Google Scholar]
- 17.Neaville WA, Tisler C, Bhattacharya A, Anklam K, Gilbertson-White S, Hamilton R, et al. Developmental cytokine response profiles and the clinical and immunologic expression of atopy during the first year of life. J Allergy Clin Immunol. 2003;112:740–6. doi: 10.1016/s0091-6749(03)01868-2. [DOI] [PubMed] [Google Scholar]
- 18.Vuillermin PJ, Ponsonby AL, Saffery R, Tang ML, Ellis JA, Sly P, et al. Microbial exposure, interferon gamma gene demethylation in naive T-cells, and the risk of allergic disease. Allergy. 2009;64:348–53. doi: 10.1111/j.1398-9995.2009.01970.x. [DOI] [PubMed] [Google Scholar]
- 19.Kumar A, Ghosh B. A single nucleotide polymorphism (A -> G) in intron 3 of IFN gamma gene is associated with asthma. Genes and Immunity. 2008;9:294–301. doi: 10.1038/gene.2008.17. [DOI] [PubMed] [Google Scholar]
- 20.Wang TN, Chu YT, Chen WY, Feng WW, Shih NH, Hsiang CH, et al. Association of interferon-gamma and interferon regulatory factor 1 polymorphisms with asthma in a family-based association study in Taiwan. Clin Exp Allergy. 2006;36:1147–52. doi: 10.1111/j.1365-2222.2006.02551.x. [DOI] [PubMed] [Google Scholar]
- 21.Nakao F, Ihara K, Kusuhara K, Sasaki Y, Kinukawa N, Takabayashi A, et al. Association of IFN-gamma and IFN regulatory factor 1 polymorphisms with childhood atopic asthma. J Allergy Clin Immunol. 2001;107:499–504. doi: 10.1067/mai.2001.113051. [DOI] [PubMed] [Google Scholar]
- 22.Shao CC, Suzuki Y, Kamada F, Kanno K, Tamari M, Hasegawa K, et al. Linkage and association of childhood asthma with the chromosome 12 genes. J Hum Genet. 2004;49:115–22. doi: 10.1007/s10038-003-0118-z. [DOI] [PubMed] [Google Scholar]
- 23.Barnes KC, Freidhoff LR, Nickel R, Chiu YF, Juo SH, Hizawa N, et al. Dense mapping of chromosome 12q13.12-q23.3 and linkage to asthma and atopy. J Allergy Clin Immunol. 1999;104:485–91. doi: 10.1016/s0091-6749(99)70398-2. [DOI] [PubMed] [Google Scholar]
- 24.Gern JE, Brooks GD, Meyer P, Chang A, Shen KL, Evans MD, et al. Bidirectional interactions between viral respiratory illnesses and cytokine responses in the first year of life. J Allergy Clin Immunol. 2006;117:72–8. doi: 10.1016/j.jaci.2005.10.002. [DOI] [PubMed] [Google Scholar]
- 25.Copenhaver CC, Gern JE, Li ZH, Shult PA, Rosenthal LA, Mikus LD, et al. Cytokine response patterns, exposure to viruses, and respiratory infections in the first year of life. Am J Respir Crit Care Med. 2004;170:175–80. doi: 10.1164/rccm.200312-1647OC. [DOI] [PubMed] [Google Scholar]
- 26.Lemanske RF. The Childhood Origins of Asthma (COAST) study. Pediatr Allergy Immunol. 2002;13:38–43. doi: 10.1034/j.1399-3038.13.s.15.8.x. [DOI] [PubMed] [Google Scholar]
- 27.Jackson DJ, Virnig CM, Gangnon RE, Evans MD, Roberg KA, Anderson EL, et al. Fractional exhaled nitric oxide measurements are most closely associated with allergic sensitization in school-age children. J Allergy Clin Immunol. 2009;124:949–53. doi: 10.1016/j.jaci.2009.07.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jackson DJ, Gangnon RE, Evans MD, Roberg KA, Anderson EL, Pappas TE, et al. Wheezing rhinovirus illnesses in early life predict asthma development in high-risk children. Am J Respir Crit Care Med. 2008;178:667–72. doi: 10.1164/rccm.200802-309OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lemanske RF, Jackson DJ, Gangnon RE, Evans MD, Li ZH, Shult PA, et al. Rhinovirus illnesses during infancy predict subsequent childhood wheezing. J Allergy Clin Immunol. 2005;116:571–7. doi: 10.1016/j.jaci.2005.06.024. [DOI] [PubMed] [Google Scholar]
- 30.Sullivan Dillie KT, Tisler CJ, Dasilva DF, Pappas TE, Roberg KA, Carlson-Dakes KT, et al. The influence of processing factors and non-atopy-related maternal and neonate characteristics on yield and cytokine responses of cord blood mononuclear cells. Clin Exp Allergy. 2008;38:298–304. doi: 10.1111/j.1365-2222.2007.02891.x. [DOI] [PubMed] [Google Scholar]
- 31.Gibbs RA, Belmont JW, Hardenbol P, Willis TD, Yu FL, Yang HM, et al. The International HapMap Project. Nature. 2003;426:789–96. doi: 10.1038/nature02168. [DOI] [PubMed] [Google Scholar]
- 32.Barrett JC, Fry B, Maller J, Daly MJ. Haploview: analysis and visualization of LD and haplotype maps. Bioinformatics. 2005;21:263–5. doi: 10.1093/bioinformatics/bth457. [DOI] [PubMed] [Google Scholar]
- 33.Li J, Ji L. Adjusting multiple testing in multilocus analyses using the eigenvalues of a correlation matrix. Heredity. 2005;95:221–7. doi: 10.1038/sj.hdy.6800717. [DOI] [PubMed] [Google Scholar]
- 34.Nyholt DR. A simple correction for multiple testing for single-nucleotide polymorphisms in linkage disequilibrium with each other. Am J Hum Genet. 2004;74:765–9. doi: 10.1086/383251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sidak Z. Rectangular Confidence Regions for Means of Multivariate Normal Distributions. Journal of the American Statistical Association. 1967;62:626. [Google Scholar]
- 36.Haller G, Torgerson DG, Ober C, Thompson EE. Sequencing the IL4 locus in African Americans implicates rare noncoding variants in asthma susceptibility. J Allergy Clin Immunol. 2009;124:1204–9. doi: 10.1016/j.jaci.2009.09.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lester LA, Rich SS, Blumenthal MN, Togias A, Murphy S, Malveaux F, et al. Ethnic differences in asthma and associated phenotypes: Collaborative Study on the Genetics of Asthma. J Allergy Clin Immunol. 2001;108:357–62. doi: 10.1067/mai.2001.117796. [DOI] [PubMed] [Google Scholar]
- 38.Ober C, Vercelli D. Gene-environment interactions in human disease: nuisance or opportunity? Trends Genet. 2011;27:107–15. doi: 10.1016/j.tig.2010.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ober C, Loisel DA, Gilad Y. Sex-specific genetic architecture of human disease. Nature Reviews Genetics. 2008;9:911–22. doi: 10.1038/nrg2415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yao WG, Barbe-Tuana FM, Llapur CJ, Jones MH, Tiller C, Kimmel R, et al. Evaluation of airway reactivity and immune characteristics as risk factors for wheezing early in life. J Allergy Clin Immunol. 2010;126:483–8. doi: 10.1016/j.jaci.2010.06.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Li YF, Wu B, Xiong HY, Zhu CZ, Zhang L. Polymorphisms of STAT-6, STAT-4 and IFN-gamma genes and the risk of asthmain Chinese population. Respir Med. 2007;101:1977–81. doi: 10.1016/j.rmed.2007.04.006. [DOI] [PubMed] [Google Scholar]
- 42.Pravica V, Perrey C, Stevens A, Lee JH, Hutchinson IV. A single nucleotide polymorphism in the first intron of the human IFN-gamma gene: Absolute correlation with a polymorphic CA microsatellite marker of high IFN-gamma production. Hum Immunol. 2000;61:863–6. doi: 10.1016/s0198-8859(00)00167-1. [DOI] [PubMed] [Google Scholar]
- 43.Tso HW, Ip WK, Chong WP, Tam CM, Chiang AKS, Lau YL. Association of interferon gamma and interleukin 10 genes with tuberculosis in Hong Kong Chinese. Genes and Immunity. 2005;6:358–63. doi: 10.1038/sj.gene.6364189. [DOI] [PubMed] [Google Scholar]
- 44.Vandenbroeck K, Goris A. Cytokine gene polymorphisms in multifactorial diseases: gateways to novel targets for immunotherapy? Trends Pharmacol Sci. 2003;24:284–9. doi: 10.1016/S0165-6147(03)00131-7. [DOI] [PubMed] [Google Scholar]
- 45.Hutchings A, Guay-Woodford L, Thomas JM, Young CJ, Purcell WM, Pravica V, et al. Association of cytokine single nucleotide polymorphisms with B7 costimulatory molecules in kidney allograft recipients. Pediatr Transplant. 2002;6:69–77. doi: 10.1034/j.1399-3046.2002.1o444.x. [DOI] [PubMed] [Google Scholar]
- 46.Vollmer-Conna U, Piraino BF, Cameron B, Davenport T, Hickie I, Wakefield D, et al. Cytokine Polymorphisms Have a Synergistic Effect on Severity of the Acute Sickness Response to Infection. Clin Infect Dis. 2008;47:1418–25. doi: 10.1086/592967. [DOI] [PubMed] [Google Scholar]
- 47.Matos GI, Covas CDF, Bittar RD, Gomes-Silva A, Marques F, Maniero VC, et al. IFNG +874T/A polymorphism is not associated with American tegumentary leishmaniasis susceptibility but can influence Leishmania induced IFN-gamma production. BMC Infectious Diseases. 2007:7. doi: 10.1186/1471-2334-7-33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lopez-Maderuelo D, Arnalich F, Serantes R, Gonzalez A, Codoceo R, Madero R, et al. Interferon-gamma and interleukin-10 gene polymorphisms in pulmonary tuberculosis. Am J Respir Crit Care Med. 2003;167:970–5. doi: 10.1164/rccm.200205-438BC. [DOI] [PubMed] [Google Scholar]
- 49.Gourdy P, Araujo LM, Zhu R, Garmy-Susini B, Diem S, Laurell H, et al. Relevance of sexual dimorphism to regulatory T cells: estradiol promotes IFN-gamma production by invariant natural killer T cells. Blood. 2005;105:2415–20. doi: 10.1182/blood-2004-07-2819. [DOI] [PubMed] [Google Scholar]
- 50.Dai RJ, Phillips RA, Zhang Y, Khan D, Crasta O, Ahmed SA. Suppression of LPS-induced Interferon-gamma and nitric oxide in splenic lymphocytes by select estrogen-regulated microRNAs: a novel mechanism of immune modulation. Blood. 2008;112:4591–7. doi: 10.1182/blood-2008-04-152488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Fox HS, Bond BL, Parslow TG. Estrogen Regulates the IFN-Gamma Promoter. J Immunol. 1991;146:4362–7. [PubMed] [Google Scholar]
- 52.Comings DE, MacMurray JP. Molecular heterosis: A review. Mol Genet Metab. 2000;71:19–31. doi: 10.1006/mgme.2000.3015. [DOI] [PubMed] [Google Scholar]
- 53.Hill AVS. The immunogenetics of human infectious diseases. Annu Rev Immunol. 1998;16:593–617. doi: 10.1146/annurev.immunol.16.1.593. [DOI] [PubMed] [Google Scholar]
- 54.Karjalainen J, Nieminen MM, Aromaa A, Klaukka T, Hurme M. The IL-1 beta genotype carries asthma susceptibility only in men. J Allergy Clin Immunol. 2002;109:514–6. doi: 10.1067/mai.2002.121948. [DOI] [PubMed] [Google Scholar]
- 55.Filigheddu F, Reid JE, Troffa C, PinnaParpaglia P, Argiolas G, Testa A, et al. Genetic polymorphisms of the beta-adrenergic system: association with essential hypertension and response to beta-blockade. Pharmacogenomics Journal. 2004;4:154–60. doi: 10.1038/sj.tpj.6500247. [DOI] [PubMed] [Google Scholar]
- 56.Hong MG, Reynolds C, Gatz M, Johansson B, Palmer JC, Gu HF, et al. Evidence that the gene encoding insulin degrading enzyme influences human lifespan. Hum Mol Genet. 2008;17:2370–8. doi: 10.1093/hmg/ddn137. [DOI] [PMC free article] [PubMed] [Google Scholar]