Abstract
Background
Glioblastoma (GBM) is the most lethal and common type of primary brain tumor. Recent evidence suggests that a subpopulation of GBM cells (glioblastoma stem cells [GSCs]) is critical for tumor progression, invasion, and therapeutic resistance. We and others have demonstrated that MET, a receptor tyrosine kinase, positively regulates the stemness phenotype and radioresistance of GSCs. Here, we interrogated the downstream effector pathways of MET signaling in GSCs.
Methods
We have established a series of GSCs and xenograft tumors derived from freshly dissociated specimens from patients with GBM and characterized a subpopulation enriched with MET activation (METhigh/+). Through global expression profiling and subsequent pathways analysis, we identified signaling pathways that are enriched in METhigh/+ populations, one of which is Wnt/β-catenin signaling pathway. To determine molecular interaction and the biological consequences of MET and Wnt/β-catenin signaling, we used pharmacological and shRNA-mediated genetic inhibition and performed various molecular and cellular analyses, including flow cytometry, immunohistochemistry, and clonogenicity assays.
Results
We found that Wnt/β-catenin signaling is highly active in METhigh/+ cells, compared with bulk tumor cells. We also showed that Wnt/β-catenin signaling activities in GBM are directly modulated by the addition of ligand-mediated MET activation or MET inhibition. Furthermore, the ectopic expression of active-β-catenin (S37A and S45Y) rescued the phenotypic effects caused by MET inhibition.
Conclusion
These data suggest that Wnt/β-catenin signaling is a key downstream effector of MET signaling and contributes to the maintenance of GSC and GBM malignancy.
Keywords: glioblastoma, glioblastoma stem cell, hepatocyte growth factor, MET protooncogene, Wnt/β-catenin signaling
Glioblastoma multiforme (GBM) is the most aggressive and common type of primary brain tumor. Despite its maximal therapies consisting of surgical resection followed by concurrent irradiation and chemotherapy, the median survival among patients with GBM is ∼14.6 months, the worst one among all human cancers.1 Indeed, most patients with GBM show a high degree of radio- and chemoresistance and almost all experience tumor relapse. There is thus an urgent need to develop novel therapeutics for this devastating disease.
A number of recent studies support the hypothesis that a relatively small population of GBM has stem-like characteristics and drives tumor propagation and recurrence against multimodal treatments (i.e., glioblastoma stem cells [GSCs]).2–4 We and others have identified several GSC enrichment markers, including CD133, CD15, integrin alpha 6, CD44, and MET.3,5–10 Although many questions regarding the generality of the proposed GSC surface markers, frequency of GSCs, and reversible nature of cell status are insufficiently addressed, the concept of GSC has brought new insights for understanding GBM initiation, progression, and resistance to standard therapy.4,11
The MET receptor tyrosine kinase (RTK) was first identified as the product of a fusion oncogene TPR-MET.12,13 Later, the full-length MET protooncogene was identified, which encodes the cell surface receptor for a specific ligand, hepatocyte growth factor (HGF), also known as scatter factor.14 Ligand-dependent activation of MET occurs via an autocrine loop (intratumoral HGF) or paracrine loops (HGF produced by stromal and/or endothelial cells).15 MET activation leads to efficient induction of downstream signal transduction pathways that include the mitogen-activated protein kinase (MAPK) cascades, the phosphoinositide 3-kinase-AKT (PI3K-AKT) axis, and signal transducer and activator of transcription proteins (STATs).15 All of these pathways positively control MET-dependent cell proliferation, survival, and migration, although a few exceptions exist.
Genetic abnormalities of MET can cause the aberrant activation of HGF/MET signaling, which can in turn cause primary tumor progression and secondary metastasis.16 In patients with GBM, gains of chromosome 7, where MET is located, have been frequently reported.17,18 Furthermore, focal genomic MET amplification is found in 4% of patients with GBM.19 Indeed, MET is one of the most highly amplified RTKs found in GBM, along with EGFR and PDGFR.20 We and others have shown that MET overexpression is associated with the poor prognosis and invasiveness of GBM tumors.21–23 Thus, targeting the MET pathway will be a powerful therapeutic approach that may potentiate the standard treatments for patients with GBM.24–26
Recently, we and others have suggested that MET signaling plays a pivotal role in maintaining the GSC-like characteristics.9,10,27 Li et al. reported that MET signaling regulates the expression levels of stem cell marker genes, such as SOX2, NESTIN, and NANOG, suggesting that MET is critical for maintaining the stem-like phenotype of GSC.9 We have shown that a subpopulation of cells expressing high levels of MET receptor (METhigh/+) cells has the enriched self-renewal potential and capacity of tumor initiation, compared with the bulk GBM cells, and that MET inhibition either by shRNA-mediated knockdown or pharmacological means effectively decreased clonogenicity and tumoriogenicity in xenograft tumor models.10 However, our understanding of the signaling cascades and/or transcriptional regulators that link MET signaling to the stem-like properties of GSC is still limited.
Several developmental signaling pathways, including Notch, bone morphogenetic protein, Wnt/β-catenin, and Sonic hedgehog, have been identified as crucial for the maintenance of stem-like properties and for treatment resistance.28–31 In this study, we have investigated the potential crosstalk between MET and other signaling cascades extending from the initial expression profiling studies. Here, we provide the evidence supporting that Wnt/β-catenin signaling is a key mediator of MET signaling in GSCs.
Materials and Methods
Reagents and Plasmids
The MET inhibitors were purchased from Calbiochem (SU11274, Germany), Tocris (PHA665752, United Kingdom), and Selleckchem (SGX523, USA). The shRNA knockdown plasmid targeting MET was obtained from Sigma (USA) and the pGreenFire™ TCF/LEF lentiviral reporter vector from System Biosciences (USA). The expression vectors for the constitutively active forms of β-catenins (S37A and S45Y) were kindly denoted by Prof. Sung Hee Baek (Seoul National University, Korea).
Expansion and Neurosphere Culture of GBM Patient-Derived Cells
After signed informed consent, tumor samples were previously obtained and GBM patient-derived cells were isolated.6,10,32–34 The GBM cells used in this study and detailed procedures were described in our prior publications.6,10 For the in vivo expansion of the GBM cells, one million of the patient-derived GBM cells were dissociated, resuspended in Hanks’ balanced salt solution (HBSS) medium, mixed with an equal volume of cold Matrigel (BD Bioscience, USA), and then subcutaneously injected into the flanks of nude mice. When the size of the xenograft tumor was already >1000 mm3, the tumor mass was mechanically and enzymatically dissociated into single cells.10,33,35 For short-term in vitro expansion, both the primary and xenograft GBM cells were cultured and passaged in Neurobasal A media (Invitrogen, USA) supplemented with B27 and N2 supplements (0.5X each; Invitrogen, USA) and recombinant bFGF and EGF (20 ng/mL each; R&D Systems, USA).
Neurosphere Forming Limiting Dilution Assay
The cultured GBM cells were enzymatically dissociated into single-cell suspensions, plated into 24 wells of 96-well plates with various seeding densities (2, 5, 10, 20, 50, 100, 200, and 500 cells per well, depending on the experiments) and incubated at 37°C for 2–3 weeks. At the time of quantification, each well was observed under a microscope for the determination of neurosphere formation. For statistical analysis, the numbers of responded events were plotted, and neurosphere frequency was calculated using the Extreme Limiting Dilution Analysis software.36
Lentivirus Production and Transduction of the GBM Cells
To generate recombinant lentivirus, a MET-targeting shRNA vector or a pGreenFire™ TCF/LEF lentivirus reporter vector was cotransfected with lentiviral packaging vectors (VSVG and PAX2) into HEK293FT cells (Invitrogen, USA). Then, media supernatants were collected 3 times (days 2, 3, and 4, respectively), and lentiviruses were concentrated through ultracentifugation. For the transduction of GBM cells, lentiviruses were added into the culture medium in the presence of polybrene (8 μg/mL). After 24 h, puromycin (1 μg/mL) selection was performed to eliminate noninfected GBM cells.
Transient Transfection of the GBM Cells
Overexpression vectors for the mutated β-catenin (S37A and S45Y) were transfected into GBM cells using Neon™ electroporation system according to the manufacturer's protocol (Invitrogen, USA). In brief, 106 GBM cells were treated with double pulses of 1400 V for 20 ms in a 100 μL electroporation tip. Transfection efficiencies of 464T were about 50%–70%. After 24 h, cells were counted and plated for limiting dilution assays.
FACS Analysis and Sorting
Patient-derived GBM cells were enzymatically dissociated into single cells and then labeled with the following antibodies: anti-HGFR/MET-biotin (BAF358; R&D Systems, USA), streptavidin-APC (#17-4317-82; eBioscience, USA), anti-CD133/2-APC (#130-090-854; Miltenyi Biotec, Germany), anti-β-catenin-PE (#6898S; Cell Signaling, USA), and TCF4 (ab60727; Abcam, United Kingdom). MET and CD133 staining were performed according to the manufacturer's protocols. Intracellular staining of β-catenin and TCF4 was performed using the BD Cytofix/Cytoperm™ buffers (BD Biosciences, USA). The stained cells were analyzed on a FACS Calibur (for analysis) and a FACS Aria (for sorting). The FlowJo™ program was used for the quantification of the fluorescent cells.
RNA Isolation and Real-Time Quantitative Polymerase Chain Reaction (PCR)
The total RNA was isolated with an RNeasy Mini Kit (Qiagen, USA). Then, equal amounts of RNA were subjected to cDNA synthesis using a SuperScript™ III First-Strand cDNA synthesis kit (Invitrogen, USA). The relative amount of mRNA was evaluated using an Applied Biosystems 7900HT Real-Time PCR System (Applied Biosystems, USA) and was calculated following normalization to β-actin mRNA. The primer sequences that were used for the real-time quantitative PCR analyses are available upon request.
Microarray Analysis
The global gene expressions of the METhigh/+ and METlow/− cells were analyzed using Affymetrix GeneChip Human Gene 1.0 ST Array.10 Total RNA was processed and hybridized to the array chip according to the manufacturer's manual (Affymetrix, USA). CEL files were normalized using the MAS5.0 algorithm.
Immunohistochemistry
The isolated xenograft tumors were prefixed in 4% paraformaldehyde overnight, cryoprotected in 25% sucrose in PBS at 4°C, and cryo-embedded in OCT compound at −42°C. The frozen tumor was sectioned in 8 micron thickness at −20°C, using cryostat, and the sections were dehydrated before refreezing at −20°C. The sections were then rehydrated, and the OCT was removed in 0.2% Tween-20 for 10 min at room temperature. Slides were fixed in 4% paraformaldehyde for 10 min and were simultaneously permeablized and blocked in 1% Triton X-100, 0.2% Tween-20, 5% normal goat serum, and 1% normal horse serum in PBS for 2 h at room temperature. The slides were incubated with primary antibodies overnight at 4°C, were subsequently labeled with 4.0 ng/μL Alexa Fluor antibodies in a blocking buffer for 2 h at room temperature, and were counterstained in DAPI (Invitrogen, USA). Primary antibodies were prepared by diluting 2.5 ng/μL anti-phospho-MET rabbit polyclonal IgG (Santa Cruz, USA) and 5 ng/μL anti-active β-catenin mouse monoclonal IgG (Milipore, USA) in a blocking buffer. The stained slides were mounted in a fluorescence mounting medium (DAKO, Denmark), and the signals were revealed using an LSM700 confocal microscope (Carl Zeiss, Germany).
Luciferase Assay
GBM cells from a reporter tumor were treated with MET inhibitors (PHA665752 and SU11274) at various concentrations (1, 2, and 4 μM, respectively). After one day, total cell extracts were prepared with Glo Lysis Buffer, and the luciferase activities were evaluated using the Steady-Glo Luciferase Assay System, according to the manufacturer's protocol (Promega, USA). Each value was normalized to the total protein amount.
Fractionation of Nuclear/Cytoplasmic Proteins and Western Blotting
The patient-derived GBM cells (named as 131) were treated with HGF or PHA665752 for 4 h. For the analysis of the β-catenin translocation, nuclear and cytoplasmic lysates were isolated using NE-PER Nuclear and Cytoplasmic Extraction Reagents according to the manufacturer's protocol (Thermo Fisher Scientific, USA). Each lysate was used for Western blotting analysis with the following antibodies: phospho-MET, MET, and β-catenin (Cell Signaling, USA); active β-catenin and Histone H3 (Millipore, USA); phospho-Stat3 (Cell Signaling, USA); and α-tubulin (Sigma Aldrich, USA).
Results
MET Inhibition Decreases GSC Clonogenicity
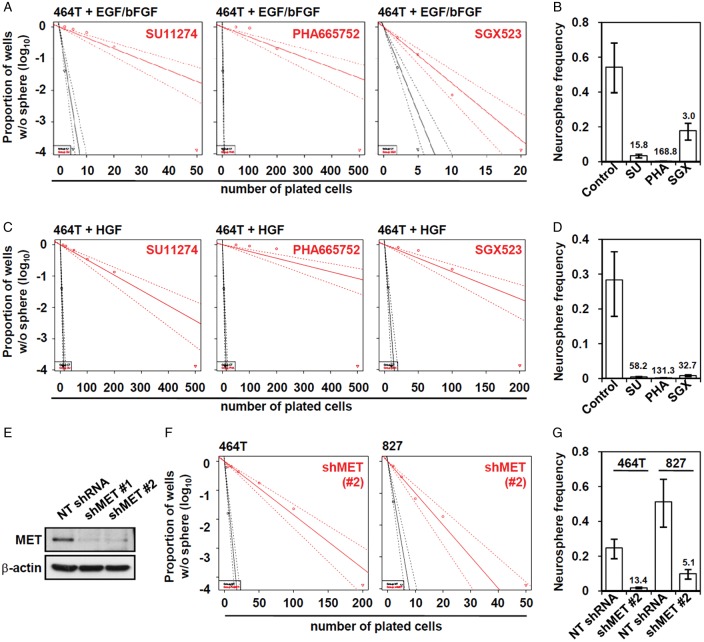
We previously showed that MET signaling is critical for the maintenance of GSC stemness.9,10 To determine whether the inhibition of MET signaling influences the clonogenicity of GBM cells, we treated GBM cells with several small-molecule MET inhibitors, such as SU11274, PHA665752, and SGX523,15,37,38 and evaluated clonogenic potentials of these cells by limiting dilution clonogenic assay. As shown in Fig. 1A and B, the treatments with MET inhibitors significantly suppressed the clonogenicity of GBM cells in the EGF/bFGF-containing culture condition. GBM cells cultured in the presence of HGF without EGF/bFGF are also highly clonogenic, similar to the cells cultured in EGF/bFGF-containing media, suggesting that HGF-MET signaling can sustain clonogenic growth of GBM cells. When the cells were cultured in the presence of HGF without EGF/bFGF, GBM cells responded more sensitively to the MET inhibitors (Fig. 1C and D), and this tendency was similarly observed in other GBM cells (Supplementary material, Fig. S1). To verify this finding, lentiviral shRNA knockdown system was applied to exclude the possibility of potential off-target effect caused by chemical MET inhibitors. Transduction with the lentivirus carrying MET-targeting shRNA in GBM cells strongly decreased basal expression of MET (Fig. 1E) and decreased the clonogenic growth of these GBM cells (Fig. 1F and G).
Fig. 1.
Inhibition of GSC clonogenic growth by MET inhibition. The 464T GBM cells were cultured in the EGF/bFGF (A and B) or HGF-containing media (C and D). (A and C) To determine the clonogenicity of GBM cells after MET inhibition, we performed neurosphere-limiting dilution assays with (red line) or without (black line) MET inhibitors. MET inhibitors were added to the media with the following concentrations: SU11274, 2 μM; PHA665752, 1 μM; and SGX523, 10 μM. (B and D) Neurosphere frequencies were calculated using ELDA software and fold inhibition by MET inhibitor was denoted. (E) 827 GBM cells were infected with lentiviruses carrying MET shRNA, and uninfected cells were selected out in the presence of puromycin (1 μg/mL) for 3 days. MET expression was analyzed by Western blotting. Neurosphere-limiting dilution assays were conducted to evaluate the changes of clonogenicities in respond to MET knockdown (F), and neurosphere frequencies were calculated (G).
Next we investigated whether MET inhibition is associated with the stem cell population of GBM cells. When GBM cells were stained with CD133, which is a prototypic GSC marker,5 CD133+ cell populations decreased in response to the MET inhibitor treatment (Supplementary material, Fig. S2). These results suggest that pharmacological or genetic inhibition of MET in GBM cells lead to the loss of clonogenicity and reduction of the GSC population.
Wnt/β-catenin Signaling Activity Correlates with MET Signaling Activity in GSCs
To obtain a clue for the potential signaling cascade that links MET signaling to the stemness of GSC, we have isolated METhigh/+ and METlow/− cells and examined expression profiles of these subpopulations.10 The METhigh/+ and METlow/− cells were sorted on the basis of the surface expression of MET, and cDNA microarray analysis was conducted.10 The MET expression level was high in the METhigh/+ cells, compared with METlow/− cells, as expected. Expression levels of a number of Wnt/β-catenin signaling target genes were significantly high in METhigh/+ cells (Supplementary material, Table S1), compared with their counterpart METlow/− cells. Of note, there was no evident alteration in the expression of Wnt ligands, Frizzled receptors, and cellular mediators of Wnt/β-catenin signaling, such as Axin and GSK3β (data not shown).
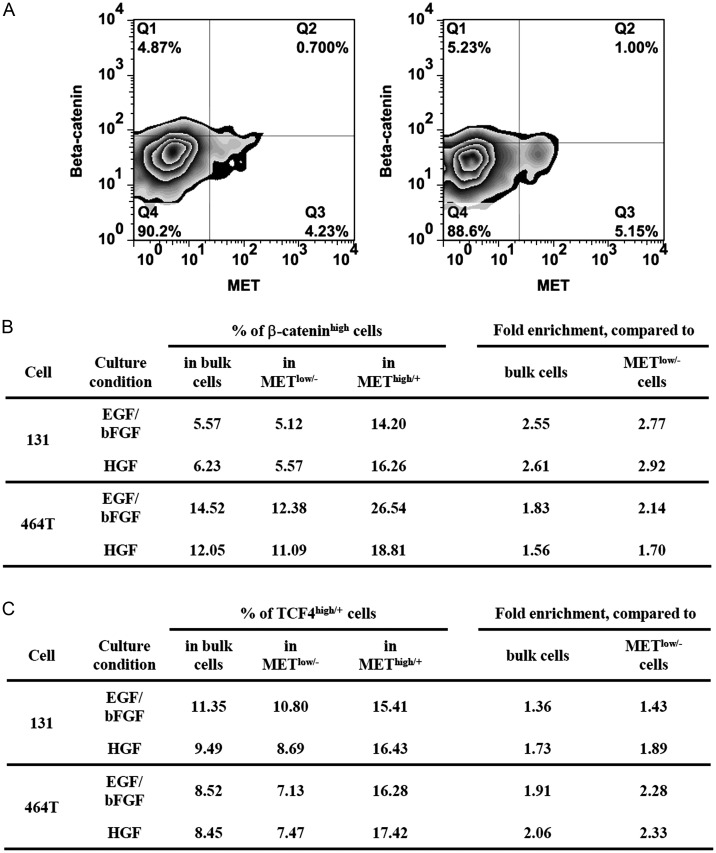
To determine whether MET and Wnt/β-catenin signaling are closely associated at a single cell level, we first performed dual FACS analysis using anti-MET and anti-β-catenin or anti-MET and anti-TCF4 (a transcriptional activator component of Wnt/β-catenin pathway) antibodies in GBM cells.39–41 Under 2 different culture conditions (EGF/bFGF and HGF), METhigh/+ subpopulations expressed the relatively high levels of β-catenin (Fig. 2A and B). Consistent with the β-catenin data, TCF4high/+ cells were enriched in the METhigh/+ subpopulation, compared with the bulk or METlow/− cells (Fig. 2C), suggesting the enrichment of Wnt/β-catenin-active cells in METhigh/+ GBM subpopulations.
Fig. 2.
Enrichment of β-cateninhigh and TCF4high/+ cells in the METhigh/+ cells. (A) After the culture of the patient GBM cells (131) in EGF/bFGF- and HGF-containing growth medium, the cells were costained with β-catenin and MET antibodies by FACS analysis. Representative FACS images were shown. (B) 131 and 464T cells were costained with these antibodies, and the percentage of the β-cateninhigh cells within the whole tumor cells (i.e., bulk cells), the METlow/−, and METhigh/+ subpopulations were calculated. The relative enrichment scores were shown as fold changes compared with the bulk or METlow/− cells. (C) 131 and 464T cells were double stained with TCF4 and MET antibodies, and the enrichment scores were presented.
Wnt/β-Catenin Signaling Activity Correlates with MET Activation in Xenograft Tumor
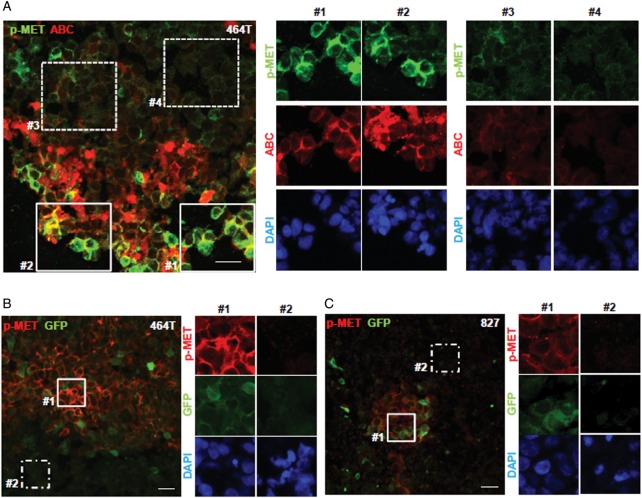
We next investigated whether colocalization of Wnt/β-catenin and MET signaling is also prevalent in a GSC-derived GBM xenograft tumor. Immunofluorescence analysis was performed using phospho-MET and activated β-catenin (ABC, a surrogate marker for the activated Wnt pathway42) antibodies on the sections derived from xenograft tumors. Consistent with the in vitro results, a small population of phospho-MET–positive cells was costained with an ABC antibody, indicating that the cells harboring a high level of phospho-MET (p-MET) are also positive for activated β-catenin signal (Fig. 3A).
Fig. 3.
Association of Wnt/β-catenin and MET signaling activities in the xenograft tumor. (A) Frozen sections of a xenograft tumor derived from 464T GBM cells were costained with phospho-MET (p-MET; green) and active β-catenin (ABC; red) antibodies to represent the MET and Wnt/β-catenin signaling activities (scale bar = 20 μm). The middle (#1 and #2) and right (#3 and #4) panels showed the areas with high and low signal intensities, respectively. (B and C) Lentiviruses harboring a pGreenFire™ vector with TCF/LEF1-binding sites were used to generate Wnt/β-catenin reporter tumor. The intensity of the green fluorescence reflected Wnt/β-catenin signaling activities. The xenograft tumor sections from the (B) 464T and (C) 827 reporter tumors were stained with a phospho-MET antibody (p-MET; red) and were visualized using confocal microscopy. The representative images with high or low signal intensities of MET and Wnt/β-catenin signaling were shown (scale bar = 20 μm). DAPI was used to visualize the nuclei.
To further explore the association between MET and Wnt/β-catenin signaling in GBM, we generated the xenograft tumor transduced with Wnt/β-catenin reporter. These tumors expressed a GFP/luciferase reporter vector driven by a minimal CMV promoter and TCF/LEF binding sites (the transcriptional target sequence for Wnt/β-catenin pathway).43 To validate the fidelity of a TCF/LEF reporter expressed in the xenograft tumor cells, xenograft sections were stained with an active β-catenin antibody (ABC). The cells that were positive for ABC had a high GFP-intensity (Supplementary material, Fig. S3). In addition, we isolated the tumor cells from a GBM tumor having TCF/LEF reporter and performed Wnt reporter assays with the addition of a Wnt ligand, Wnt3a. The reporter activity was increased in proportion to the concentration of the added Wnt3a (Supplementary material, Fig. S4). Having confirmed that Wnt/β-catenin reporter is a reliable readout for the Wnt/β-catenin activity, we next performed immunohistochemistry analysis on the sections from these reporter tumors using phospho-MET antibody. A majority of the stained cells with a phospho-MET antibody revealed strong GFP-positivity (ie, active Wnt/β-catenin signaling) (Fig. 3B and C). Together, these data support the notion that MET and Wnt/β-catenin signaling activities are colocalized within a small subset of the GBM xenograft tumor.
MET Inhibition Decreases Wnt/β-catenin Signaling Through Suppression of β-catenin Nuclear Translocation
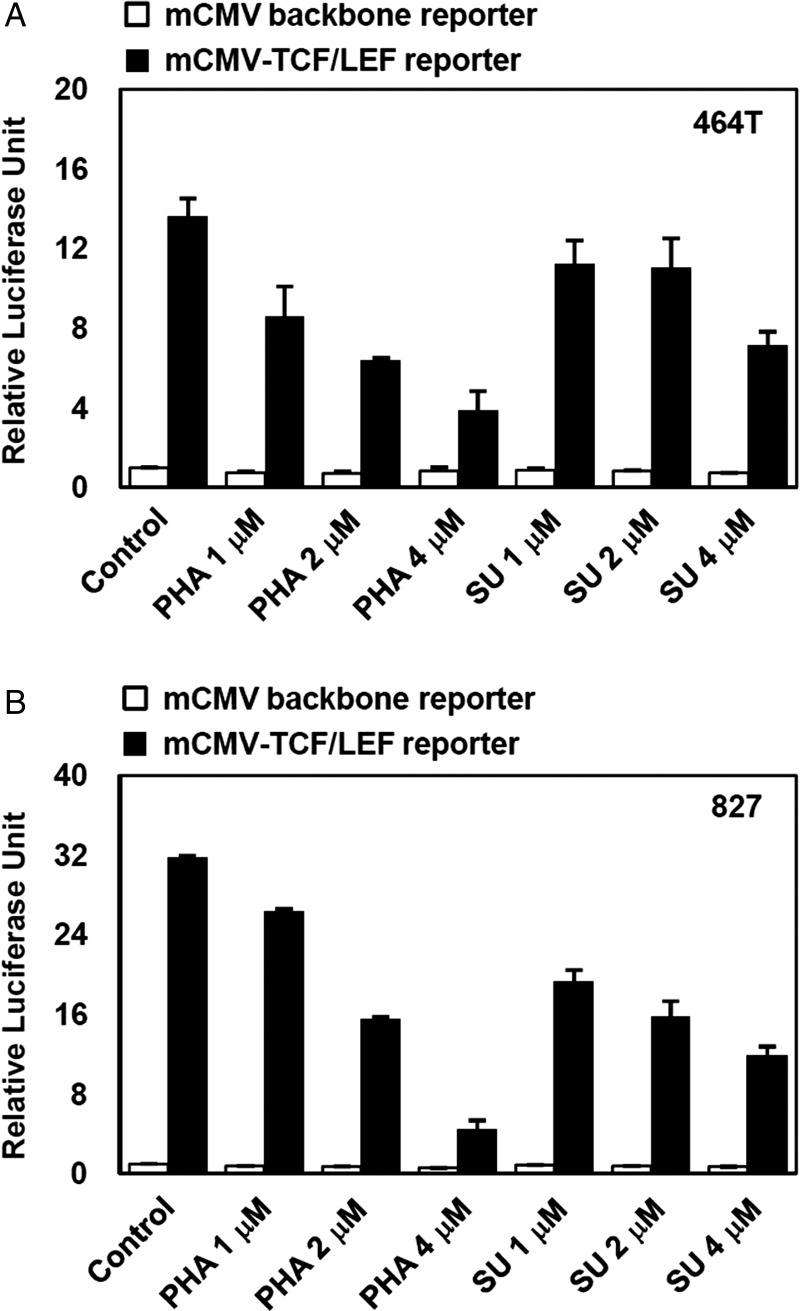
To determine whether MET signaling directly affects Wnt/β-catenin signaling activity, we evaluated the TCF/LEF-mediated transcriptional activity of GBM cells after MET inhibition. Treatment with the MET inhibitors SU11274 and PHA665752 significantly attenuated the TCF/LEF reporter activity of GBM cells in a concentration-dependent manner, suggesting that Wnt/β-catenin activity in GBM is concomitantly decreased by MET inhibition (Fig. 4A and B).
Fig. 4.
Decrease of TCF/LEF promoter activity by MET inhibition in GBM cells. GBM cells dissociated from Wnt/β-catenin reporter-expressing tumors (derived from 464T cells [A] and 827 cells [B]) were treated with MET inhibitors (PHA665752 and SU11274, concentrations of 1, 2, and 4 μM) for one day, protein lysates harvested, and then the relative luciferase activities were measured. Experiments were performed in triplicate, and error bar represents standard deviation (SD).
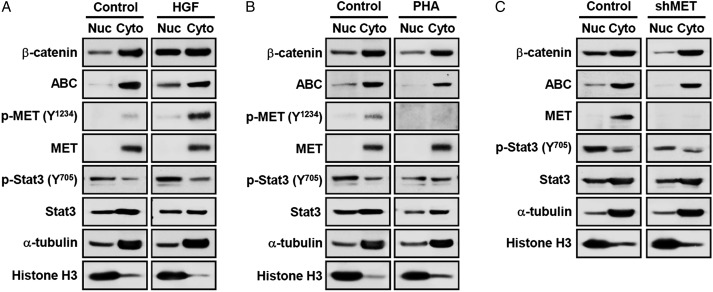
To elucidate whether β-catenin is involved in this process, we determined the status of nuclear translocation of β-catenin in response to activation or inhibition of MET signaling. Phospho-MET was sharply increased after the addition of HGF (Fig. 5A) and decreased after MET inhibition (Fig. 5B and C). Similarly, an activated form of Stat3 (phospho-Stat3) was clearly accumulated in the nucleus by the HGF treatment and was decreased by MET inhibition. Expression levels of both total β-catenin and ABC in nucleus were significantly increased by HGF treatment (Fig. 5A), whereas MET inhibition either by PHA665752 or MET knockdown suppressed nuclear translocation of β-catenin (Fig. 5B and C). Taken together, these data demonstrate that MET signaling directly influences Wnt/β-catenin signaling activity through regulation of the active β-catenin and its nuclear translocation.
Fig. 5.
Regulation of β-catenin nuclear translocation by MET signaling. 131 GBM cells were grown in the presence and absence of a growth factor overnight and were treated with (A) HGF (50 ng/mL) and (B) PHA665752 (5 μM) for 4 h. The nuclear and cytoplasmic portions of each sample were fractionated and submitted for Western blot analysis. (C) 131 GBM cells were infected with lentiviruses carrying MET shRNA, and the nuclear/cytoplasmic portions were fractionated for Western blot analysis. Abbreviation: ABC, active β-catenin.
Restoration of Wnt/β-Catenin Signaling Rescues MET Inhibition-Mediated Loss of Clonogenicity of GBM Cells
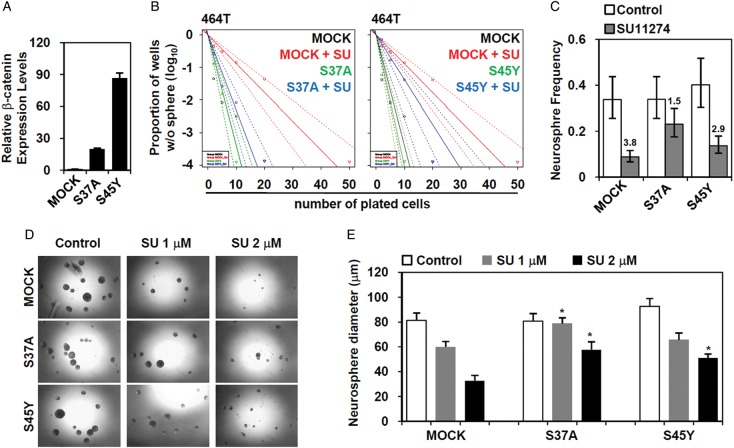
The above data indicate that MET inhibition decreases the clonogenic growth of GBM cells and that Wnt/β-catenin signaling is a downstream effector of MET signaling in GBM. To interrogate the significance of Wnt/β-catenin signaling activity in MET-dependent GSC self-renewal, we performed a functional rescue experiment. We hypothesized that the restoration of Wnt/β-catenin signaling might recover GSC clonogenicity caused by MET inhibition. To test this hypothesis, we overexpressed 2 mutated constructs of β-catenin (S37A and S45Y) (Fig. 6A). These β-catenin mutants could not be phosphorylated by GSK3β, thereby escaping from proteasomal degradation and resulting in the constitutive activation of the downstream WNT target genes.44,45 Cells expressing either of these mutants remained highly clonogenic, despite the fact that they were treated with MET inhibitor (Fig. 6B and C), determined by limiting dilution assay. The average size of neurospheres in the mutant-expressing cells was much bigger than in that of MET inhibitor-treated cells (Fig. 6D and E). These results further support that Wnt/β-catenin signaling positively regulates the clonogenicity of GBM cells, at least in part, as a downstream mediator of MET.
Fig. 6.
Recovery of GBM clonogenicity by restoration of Wnt/β-catenin signaling. (A) The 464T GBM cells were transfected with overexpression vector of mutated β-catenin constructs (S37A and S45Y). (A) After 2 days, the overexpression of each construct was confirmed by real-time quantitative PCR. (B) The changes in clonogenicity were addressed via neurosphere-limiting dilution assay in the presence of the MET inhibitor, SU11274 (2 μM). (C) The changes in the neurosphere frequency were calculated using ELDA software, and the fold inhibition in response to MET inhibition was denoted. (D) After the treatment with the MET for 10 days (SU11274, 1 and 2 μM), microscopic images were taken, and the representative images obtained were shown here. (E) The average neurosphere size was calculated. P < .01 compared with MOCK transfected cells.
Discussion
Wnt/β-catenin signaling plays important roles in maintaining cancer stem cell stemness in various types of cancer, such as colon, breast, and lung cancer, and hepatocellular carcinoma.46–49 In GBM, β-catenin expression is correlated with tumor malignancy, and it has been proposed as a prognostic marker in GBM.50–52 Of more importance, the accumulated evidence suggests that Wnt/β-catenin signaling contributes to the maintenance of stem-like cell properties, inhibition of differentiation, the acquired radioresistance, and the development of an invasive phenotype.30,53,54
Here, we have unraveled a crosstalk between MET and Wnt/β-catenin signaling as a critical molecular network operating in GSC. We demonstrated that inhibition of MET signaling concurrently suppressed Wnt/β-catenin activity (Figs 4 and 5). Moreover, we showed that ectopic expression of a constitutively active form of β-catenin rescued the loss of clonogenicity of GBM cells through MET inhibition (Fig. 6). In support of this contention, we found that a small population of GSCs residing in the tumor possessed high levels of both MET and Wnt/β-catenin activities (Figs 2 and 3). As previously elucidated, MET-positive cells are dominantly positioned in the GSC niche and have higher expression levels of stem cell–related markers, such as CD133, Nestin, and Sox2.9,10 Our data presented here support the notion that Wnt/β-catenin signaling contributes to sustaining the GSC stemness in conjunction with MET signaling.
MET and Wnt/β-catenin signaling pathways appear to share similar functions in tumor growth and metastasis.55,56 For example, the association of MET and Wnt/β-catenin signaling has been identified as a critical factor for the aggressive phenotype of breast cancer cell lines leading to bone metastasis56 and for the poor outcome in patients with breast cancer.55 Moreover, it was reported that WNT/β-catenin signaling blockade by a dominant-negative form of TCF4 diminished HGF-induced gene expression profiles and the resulting phenotypic changes.57 However, MET signaling leads to the transformation of colon epithelial cells without an aberrant Wnt/β-catenin activity,58 suggesting that MET signaling affect various cellular processes in both Wnt/β-catenin-dependent and independent manner. Here, we demonstrate that Wnt/β-catenin signaling is responsible for connecting MET signaling to GSC biology, and further studies are in process to reveal additional and more detailed molecular network of MET and Wnt/β-catenin signaling. In this regard, we postulated that β-catenin formed a complex with MET and dissociates from MET after HGF treatment.59–61 In preliminary studies, we also found that MET inhibition attenuated the phosphorylation level of low-density lipoprotein receptor-related protein 6 (LRP6),62,63 which is a co-receptor of Wnt/β-catenin signaling (data not shown).
There are several non-mutually exclusive mechanisms that link MET signaling to Wnt/β-catenin signaling, including (1) the increased expression of Wnt/β-catenin signaling components, such as Wnt7b and LEF1;64,65 (2) the activation of cellular signaling pathways, including GSK3 and Src;66,67 and/or (3) the direct interaction between MET and β-catenin.59–61 In addition, EGFR signaling can promote the Wnt/β-catenin signaling through the stabilization and subsequent nuclear accumulation of β-catenin, which depends on several mediators, such as caveolin-1, ERK, ERK/LRP6, and pyruvate kinase M2.68–71 MET is known to interact with EGFR and acts as a compensatory pathway for EGFR signaling, suggesting that they have substantially overlapped downstream mediators.72–75 Thus, it is also possible that regulation of Wnt/β-catenin signaling by MET inhibition might be caused by the similar mechanisms in the downstream of EGFR, at least in part. Combinations of several effectors are likely to be involved and cellular context will be one of major determinants in the MET-WNT signaling axis.
We have elucidated the biological effects of MET activation via ether by the addition of HGF or overexpression of a constitutively active form of MET (TPR-MET76) on Wnt/β-catenin signaling. In contrast to HGF treatment (Fig. 5A), overexpression of TPR-MET in GBM has a marginal effect on nuclear accumulation of total and activated β-catenin (data not shown). These observations could be attributable to differential signal transduction mechanisms between the HGF-MET ligand-receptor signaling and ligand-independent TPR-MET signaling. Alternatively, a high level of MET receptor in GBM cells can potentially mask the effect of TPR-MET.
Because the profound impact of MET signaling in GSC has only just emerged, a more complete understanding of MET signaling will provide novel insights about GBM growth and recurrence, which will eventually lead to the development of effective therapeutic options and diagnostic markers. Here, we provide evidence that Wnt/β-catenin signaling connects MET signaling to the maintenance of GSC stemness, which might be a crucial hallmark to understand the molecular mechanism maintaining the GSC characteristics.
Supplementary Material
Funding
This work was supported by the National Research Foundation of Korea Grant funded by the Korean Government, Ministry of Education, Science and Technology (NRF-M1AXA002-2011-0028414).
Supplementary Material
Acknowledgment
The authors thank Sung Hee Baek (Seoul National University) for providing the S37A and S45Y β-catenin plasmid DNA.
Conflict of interest statement. None declared.
References
- 1.Stupp R, Mason WP, van den Bent MJ, et al. Radiotherapy plus concomitant and adjuvant temozolomide for glioblastoma. N Engl J Med. 2005;352(10):987–996. doi: 10.1056/NEJMoa043330. [DOI] [PubMed] [Google Scholar]
- 2.Liu G, Yuan X, Zeng Z, et al. Analysis of gene expression and chemoresistance of CD133+ cancer stem cells in glioblastoma. Mol Cancer. 2006;5:67. doi: 10.1186/1476-4598-5-67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bao S, Wu Q, McLendon RE, et al. Glioma stem cells promote radioresistance by preferential activation of the DNA damage response. Nature. 2006;444(7120):756–760. doi: 10.1038/nature05236. [DOI] [PubMed] [Google Scholar]
- 4.Chen J, Li Y, Yu TS, et al. A restricted cell population propagates glioblastoma growth after chemotherapy. Nature. 2012;488(7412):522–526. doi: 10.1038/nature11287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Singh SK, Hawkins C, Clarke ID, et al. Identification of human brain tumour initiating cells. Nature. 2004;432(7015):396–401. doi: 10.1038/nature03128. [DOI] [PubMed] [Google Scholar]
- 6.Son MJ, Woolard K, Nam DH, Lee J, Fine HA. SSEA-1 is an enrichment marker for tumor-initiating cells in human glioblastoma. Cell Stem Cell. 2009;4(5):440–452. doi: 10.1016/j.stem.2009.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lathia JD, Gallagher J, Heddleston JM, et al. Integrin alpha 6 regulates glioblastoma stem cells. Cell Stem Cell. 2010;6(5):421–432. doi: 10.1016/j.stem.2010.02.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.He J, Liu Y, Xie X, et al. Identification of cell surface glycoprotein markers for glioblastoma-derived stem-like cells using a lectin microarray and LC-MS/MS approach. J Proteome Res. 2010;9(5):2565–2572. doi: 10.1021/pr100012p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Li Y, Li A, Glas M, et al. c-Met signaling induces a reprogramming network and supports the glioblastoma stem-like phenotype. Proc Natl Acad Sci U S A. 2011;108(24):9951–9956. doi: 10.1073/pnas.1016912108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Joo KM, Jin J, Kim E, et al. MET signaling regulates glioblastoma stem cells. Cancer Res. 2012;72(15):3828–3838. doi: 10.1158/0008-5472.CAN-11-3760. [DOI] [PubMed] [Google Scholar]
- 11.Singh SK, Clarke ID, Terasaki M, et al. Identification of a cancer stem cell in human brain tumors. Cancer Res. 2003;63(18):5821–5828. [PubMed] [Google Scholar]
- 12.Cooper CS, Park M, Blair DG, et al. Molecular cloning of a new transforming gene from a chemically transformed human cell line. Nature. 1984;311(5981):29–33. doi: 10.1038/311029a0. [DOI] [PubMed] [Google Scholar]
- 13.Park M, Dean M, Cooper CS, et al. Mechanism of met oncogene activation. Cell. 1986;45(6):895–904. doi: 10.1016/0092-8674(86)90564-7. [DOI] [PubMed] [Google Scholar]
- 14.Bottaro DP, Rubin JS, Faletto DL, et al. Identification of the hepatocyte growth factor receptor as the c-met proto-oncogene product. Science. 1991;251(4995):802–804. doi: 10.1126/science.1846706. [DOI] [PubMed] [Google Scholar]
- 15.Buchanan SG, Hendle J, Lee PS, et al. SGX523 is an exquisitely selective, ATP-competitive inhibitor of the MET receptor tyrosine kinase with antitumor activity in vivo. Mol Cancer Ther. 2009;8(12):3181–3190. doi: 10.1158/1535-7163.MCT-09-0477. [DOI] [PubMed] [Google Scholar]
- 16.Lesko E, Majka M. The biological role of HGF-MET axis in tumor growth and development of metastasis. Front Biosci. 2008;13:1271–1280. doi: 10.2741/2760. [DOI] [PubMed] [Google Scholar]
- 17.Lopez-Gines C, Cerda-Nicolas M, Gil-Benso R, et al. Association of chromosome 7, chromosome 10 and EGFR gene amplification in glioblastoma multiforme. Clin Neuropathol. 2005;24(5):209–218. [PubMed] [Google Scholar]
- 18.Fischer U, Wullich B, Sattler HP, Gottert E, Zang KD, Meese E. Coamplification on chromosomes 7p12-13 and 9q12-13 identified by reverse chromosome painting in a glioblastoma multiforme. Hum Genet. 1994;93(3):331–334. doi: 10.1007/BF00212033. [DOI] [PubMed] [Google Scholar]
- 19.Network CGAR. Comprehensive genomic characterization defines human glioblastoma genes and core pathways. Nature. 2008;455(7216):1061–1068. doi: 10.1038/nature07385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Snuderl M, Fazlollahi L, Le LP, et al. Mosaic amplification of multiple receptor tyrosine kinase genes in glioblastoma. Cancer Cell. 2011;20(6):810–817. doi: 10.1016/j.ccr.2011.11.005. [DOI] [PubMed] [Google Scholar]
- 21.Koochekpour S, Jeffers M, Rulong S, et al. Met and hepatocyte growth factor/scatter factor expression in human gliomas. Cancer Res. 1997;57(23):5391–5398. [PubMed] [Google Scholar]
- 22.Kong DS, Song SY, Kim DH, et al. Prognostic significance of c-Met expression in glioblastomas. Cancer. 2009;115(1):140–148. doi: 10.1002/cncr.23972. [DOI] [PubMed] [Google Scholar]
- 23.Nabeshima K, Shimao Y, Sato S, et al. Expression of c-Met correlates with grade of malignancy in human astrocytic tumours: an immunohistochemical study. Histopathology. 1997;31(5):436–443. doi: 10.1046/j.1365-2559.1997.3010889.x. [DOI] [PubMed] [Google Scholar]
- 24.Lal B, Xia S, Abounader R, Laterra J. Targeting the c-Met pathway potentiates glioblastoma responses to gamma-radiation. Clin Cancer Res. 2005;11(12):4479–4486. doi: 10.1158/1078-0432.CCR-05-0166. [DOI] [PubMed] [Google Scholar]
- 25.Wen PY. American Society of Clinical Oncology 2010: report of selected studies from the CNS tumors section. Expert Rev Anticancer Ther. 2010;10(9):1367–1369. doi: 10.1586/era.10.117. [DOI] [PubMed] [Google Scholar]
- 26.Zhang Y, Guessous F, Kofman A, Schiff D, Abounader R. XL-184, a MET, VEGFR-2 and RET kinase inhibitor for the treatment of thyroid cancer, glioblastoma multiforme and NSCLC. IDrugs. 2010;13(2):112–121. [PMC free article] [PubMed] [Google Scholar]
- 27.De Bacco F, Casanova E, Medico E, et al. The MET oncogene is a functional marker of a glioblastoma stem cell subtype. Cancer Res. 2012;72(17):4537–4550. doi: 10.1158/0008-5472.CAN-11-3490. [DOI] [PubMed] [Google Scholar]
- 28.Bar EE, Chaudhry A, Lin A, et al. Cyclopamine-mediated hedgehog pathway inhibition depletes stem-like cancer cells in glioblastoma. Stem Cells. 2007;25(10):2524–2533. doi: 10.1634/stemcells.2007-0166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Fan X, Matsui W, Khaki L, et al. Notch pathway inhibition depletes stem-like cells and blocks engraftment in embryonal brain tumors. Cancer Res. 2006;66(15):7445–7452. doi: 10.1158/0008-5472.CAN-06-0858. [DOI] [PubMed] [Google Scholar]
- 30.Kim Y, Kim KH, Lee J, et al. Wnt activation is implicated in glioblastoma radioresistance. Lab Invest. 2012;92(3):466–473. doi: 10.1038/labinvest.2011.161. [DOI] [PubMed] [Google Scholar]
- 31.Piccirillo SG, Reynolds BA, Zanetti N, et al. Bone morphogenetic proteins inhibit the tumorigenic potential of human brain tumour-initiating cells. Nature. 2006;444(7120):761–765. doi: 10.1038/nature05349. [DOI] [PubMed] [Google Scholar]
- 32.Joo KM, Nam DH. Prospective identification of cancer stem cells with the surface antigen CD133. Methods Mol Biol. 2009;568:57–71. doi: 10.1007/978-1-59745-280-9_5. [DOI] [PubMed] [Google Scholar]
- 33.Lee J, Kotliarova S, Kotliarov Y, et al. Tumor stem cells derived from glioblastomas cultured in bFGF and EGF more closely mirror the phenotype and genotype of primary tumors than do serum-cultured cell lines. Cancer Cell. 2006;9(5):391–403. doi: 10.1016/j.ccr.2006.03.030. [DOI] [PubMed] [Google Scholar]
- 34.Joo KM, Kim SY, Jin X, et al. Clinical and biological implications of CD133-positive and CD133-negative cells in glioblastomas. Lab Invest. 2008;88(8):808–815. doi: 10.1038/labinvest.2008.57. [DOI] [PubMed] [Google Scholar]
- 35.Hemmati HD, Nakano I, Lazareff JA, et al. Cancerous stem cells can arise from pediatric brain tumors. Proc Natl Acad Sci U S A. 2003;100(25):15178–15183. doi: 10.1073/pnas.2036535100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hu Y, Smyth GK. ELDA: extreme limiting dilution analysis for comparing depleted and enriched populations in stem cell and other assays. J Immunol Methods. 2009;347(1–2):70–78. doi: 10.1016/j.jim.2009.06.008. [DOI] [PubMed] [Google Scholar]
- 37.Sattler M, Pride YB, Ma P, et al. A novel small molecule met inhibitor induces apoptosis in cells transformed by the oncogenic TPR-MET tyrosine kinase. Cancer Res. 2003;63(17):5462–5469. [PubMed] [Google Scholar]
- 38.Christensen JG, Schreck R, Burrows J, et al. A selective small molecule inhibitor of c-Met kinase inhibits c-Met-dependent phenotypes in vitro and exhibits cytoreductive antitumor activity in vivo. Cancer Res. 2003;63(21):7345–7355. [PubMed] [Google Scholar]
- 39.Wang W, Pan H, Murray K, Jefferson BS, Li Y. Matrix metalloproteinase-1 promotes muscle cell migration and differentiation. Am J Pathol. 2009;174(2):541–549. doi: 10.2353/ajpath.2009.080509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Schenkel JM, Zloza A, Li W, Narasipura SD, Al-Harthi L. Beta-catenin signaling mediates CD4 expression on mature CD8+ T cells. J Immunol. 2010;185(4):2013–2019. doi: 10.4049/jimmunol.0902572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zhang J, Huang K, Shi Z, et al. High beta-catenin/Tcf-4 activity confers glioma progression via direct regulation of AKT2 gene expression. Neuro Oncol. 2011;13(6):600–609. doi: 10.1093/neuonc/nor034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.van Noort M, Meeldijk J, van der Zee R, Destree O, Clevers H. Wnt signaling controls the phosphorylation status of beta-catenin. J Biol Chem. 2002;277(20):17901–17905. doi: 10.1074/jbc.M111635200. [DOI] [PubMed] [Google Scholar]
- 43.Conley SJ, Gheordunescu E, Kakarala P, et al. Antiangiogenic agents increase breast cancer stem cells via the generation of tumor hypoxia. Proc Natl Acad Sci U S A. 2012;109(8):2784–2789. doi: 10.1073/pnas.1018866109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Easwaran V, Song V, Polakis P, Byers S. The ubiquitin-proteasome pathway and serine kinase activity modulate adenomatous polyposis coli protein-mediated regulation of beta-catenin-lymphocyte enhancer-binding factor signaling. J Biol Chem. 1999;274(23):16641–16645. doi: 10.1074/jbc.274.23.16641. [DOI] [PubMed] [Google Scholar]
- 45.Porfiri E, Rubinfeld B, Albert I, Hovanes K, Waterman M, Polakis P. Induction of a beta-catenin-LEF-1 complex by wnt-1 and transforming mutants of beta-catenin. Oncogene. 1997;15(23):2833–2839. doi: 10.1038/sj.onc.1201462. [DOI] [PubMed] [Google Scholar]
- 46.Vermeulen L, De Sousa EMF, van der Heijden M, et al. Wnt activity defines colon cancer stem cells and is regulated by the microenvironment. Nat Cell Biol. 2010;12(5):468–476. doi: 10.1038/ncb2048. [DOI] [PubMed] [Google Scholar]
- 47.King TD, Suto MJ, Li Y. The Wnt/beta-catenin signaling pathway: a potential therapeutic target in the treatment of triple negative breast cancer. J Cell Biochem. 2012;113(1):13–18. doi: 10.1002/jcb.23350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Teng Y, Wang X, Wang Y, Ma D. Wnt/beta-catenin signaling regulates cancer stem cells in lung cancer A549 cells. Biochem Biophys Res Commun. 2010;392(3):373–379. doi: 10.1016/j.bbrc.2010.01.028. [DOI] [PubMed] [Google Scholar]
- 49.Yamashita T, Ji J, Budhu A, et al. EpCAM-positive hepatocellular carcinoma cells are tumor-initiating cells with stem/progenitor cell features. Gastroenterology. 2009;136(3):1012–1024. doi: 10.1053/j.gastro.2008.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Liu X, Wang L, Zhao S, Ji X, Luo Y, Ling F. beta-Catenin overexpression in malignant glioma and its role in proliferation and apoptosis in glioblastma cells. Med Oncol. 2011;28(2):608–614. doi: 10.1007/s12032-010-9476-5. [DOI] [PubMed] [Google Scholar]
- 51.Liu C, Tu Y, Sun X, et al. Wnt/beta-Catenin pathway in human glioma: expression pattern and clinical/prognostic correlations. Clin Exp Med. 2011;11(2):105–112. doi: 10.1007/s10238-010-0110-9. [DOI] [PubMed] [Google Scholar]
- 52.Rossi M, Magnoni L, Miracco C, et al. beta-catenin and Gli1 are prognostic markers in glioblastoma. Cancer Biol Ther. 2011;11(8):753–761. doi: 10.4161/cbt.11.8.14894. [DOI] [PubMed] [Google Scholar]
- 53.Zheng H, Ying H, Wiedemeyer R, et al. PLAGL2 regulates Wnt signaling to impede differentiation in neural stem cells and gliomas. Cancer Cell. 2010;17(5):497–509. doi: 10.1016/j.ccr.2010.03.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Jin X, Jeon HY, Joo KM, et al. Frizzled 4 regulates stemness and invasiveness of migrating glioma cells established by serial intracranial transplantation. Cancer Res. 2011;71(8):3066–3075. doi: 10.1158/0008-5472.CAN-10-1495. [DOI] [PubMed] [Google Scholar]
- 55.Ponzo MG, Lesurf R, Petkiewicz S, et al. Met induces mammary tumors with diverse histologies and is associated with poor outcome and human basal breast cancer. Proc Natl Acad Sci U S A. 2009;106(31):12903–12908. doi: 10.1073/pnas.0810402106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Previdi S, Maroni P, Matteucci E, Broggini M, Bendinelli P, Desiderio MA. Interaction between human-breast cancer metastasis and bone microenvironment through activated hepatocyte growth factor/Met and beta-catenin/Wnt pathways. Eur J Cancer. 2010;46(9):1679–1691. doi: 10.1016/j.ejca.2010.02.036. [DOI] [PubMed] [Google Scholar]
- 57.Pan FY, Zhang SZ, Xu N, et al. Beta-catenin signaling involves HGF-enhanced HepG2 scattering through activating MMP-7 transcription. Histochem Cell Biol. 2010;134(3):285–295. doi: 10.1007/s00418-010-0729-3. [DOI] [PubMed] [Google Scholar]
- 58.Boon EM, Kovarikova M, Derksen PW, van der Neut R. MET signalling in primary colon epithelial cells leads to increased transformation irrespective of aberrant Wnt signalling. Br J Cancer. 2005;92(6):1078–1083. doi: 10.1038/sj.bjc.6602405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Apte U, Zeng G, Muller P, et al. Activation of Wnt/beta-catenin pathway during hepatocyte growth factor-induced hepatomegaly in mice. Hepatology. 2006;44(4):992–1002. doi: 10.1002/hep.21317. [DOI] [PubMed] [Google Scholar]
- 60.Herynk MH, Tsan R, Radinsky R, Gallick GE. Activation of c-Met in colorectal carcinoma cells leads to constitutive association of tyrosine-phosphorylated beta-catenin. Clin Exp Metastasis. 2003;20(4):291–300. doi: 10.1023/a:1024024218529. [DOI] [PubMed] [Google Scholar]
- 61.Monga SP, Mars WM, Pediaditakis P, et al. Hepatocyte growth factor induces Wnt-independent nuclear translocation of beta-catenin after Met-beta-catenin dissociation in hepatocytes. Cancer Res. 2002;62(7):2064–2071. [PubMed] [Google Scholar]
- 62.Tamai K, Semenov M, Kato Y, et al. LDL-receptor-related proteins in Wnt signal transduction. Nature. 2000;407(6803):530–535. doi: 10.1038/35035117. [DOI] [PubMed] [Google Scholar]
- 63.Bilic J, Huang YL, Davidson G, et al. Wnt induces LRP6 signalosomes and promotes dishevelled-dependent LRP6 phosphorylation. Science. 2007;316(5831):1619–1622. doi: 10.1126/science.1137065. [DOI] [PubMed] [Google Scholar]
- 64.Huang FI, Chen YL, Chang CL, Yuan RH, Jeng YM. Hepatocyte growth factor activates Wnt pathway by transcriptional activation of LEF1 to facilitate tumor invasion. Carcinogenesis. 2012;33(6):1142–1148. doi: 10.1093/carcin/bgs131. [DOI] [PubMed] [Google Scholar]
- 65.Liu Y, Chattopadhyay N, Qin S, et al. Coordinate integrin and c-Met signaling regulate Wnt gene expression during epithelial morphogenesis. Development. 2009;136(5):843–853. doi: 10.1242/dev.027805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Bertotti A, Bracco C, Girolami F, et al. Inhibition of Src impairs the growth of met-addicted gastric tumors. Clin Cancer Res. 2010;16(15):3933–3943. doi: 10.1158/1078-0432.CCR-10-0106. [DOI] [PubMed] [Google Scholar]
- 67.Papkoff J, Aikawa M. WNT-1 and HGF regulate GSK3 beta activity and beta-catenin signaling in mammary epithelial cells. Biochem Biophys Res Commun. 1998;247(3):851–858. doi: 10.1006/bbrc.1998.8888. [DOI] [PubMed] [Google Scholar]
- 68.Ji H, Wang J, Nika H, et al. EGF-induced ERK activation promotes CK2-mediated disassociation of alpha-Catenin from beta-Catenin and transactivation of beta-Catenin. Mol Cell. 2009;36(4):547–559. doi: 10.1016/j.molcel.2009.09.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Yang W, Xia Y, Ji H, et al. Nuclear PKM2 regulates beta-catenin transactivation upon EGFR activation. Nature. 2011;480(7375):118–122. doi: 10.1038/nature10598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Krejci P, Aklian A, Kaucka M, et al. Receptor tyrosine kinases activate canonical WNT/beta-catenin signaling via MAP kinase/LRP6 pathway and direct beta-catenin phosphorylation. PLoS One. 2012;7(4):e35826. doi: 10.1371/journal.pone.0035826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Lu Z, Ghosh S, Wang Z, Hunter T. Downregulation of caveolin-1 function by EGF leads to the loss of E-cadherin, increased transcriptional activity of beta-catenin, and enhanced tumor cell invasion. Cancer Cell. 2003;4(6):499–515. doi: 10.1016/s1535-6108(03)00304-0. [DOI] [PubMed] [Google Scholar]
- 72.Jo M, Stolz DB, Esplen JE, Dorko K, Michalopoulos GK, Strom SC. Cross-talk between epidermal growth factor receptor and c-Met signal pathways in transformed cells. J Biol Chem. 2000;275(12):8806–8811. doi: 10.1074/jbc.275.12.8806. [DOI] [PubMed] [Google Scholar]
- 73.Karamouzis MV, Konstantinopoulos PA, Papavassiliou AG. Targeting MET as a strategy to overcome crosstalk-related resistance to EGFR inhibitors. Lancet Oncol. 2009;10(7):709–717. doi: 10.1016/S1470-2045(09)70137-8. [DOI] [PubMed] [Google Scholar]
- 74.Kawaguchi K, Murakami H, Taniguchi T, et al. Combined inhibition of MET and EGFR suppresses proliferation of malignant mesothelioma cells. Carcinogenesis. 2009;30(7):1097–1105. doi: 10.1093/carcin/bgp097. [DOI] [PubMed] [Google Scholar]
- 75.Stommel JM, Kimmelman AC, Ying H, et al. Coactivation of receptor tyrosine kinases affects the response of tumor cells to targeted therapies. Science. 2007;318(5848):287–290. doi: 10.1126/science.1142946. [DOI] [PubMed] [Google Scholar]
- 76.Gupta PB, Kuperwasser C, Brunet JP, et al. The melanocyte differentiation program predisposes to metastasis after neoplastic transformation. Nat Genet. 2005;37(10):1047–1054. doi: 10.1038/ng1634. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.