Abstract
A variety of genome-wide profiling techniques are available to investigate complementary aspects of genome structure and function. Integrative analysis of heterogeneous data sources can reveal higher level interactions that cannot be detected based on individual observations. A standard integration task in cancer studies is to identify altered genomic regions that induce changes in the expression of the associated genes based on joint analysis of genome-wide gene expression and copy number profiling measurements. In this review, we highlight common approaches to genomic data integration and provide a transparent benchmarking procedure to quantitatively compare method performances in cancer gene prioritization. Algorithms, data sets and benchmarking results are available at http://intcomp.r-forge.r-project.org.
Keywords: DNA copy number, gene expression, microarrays, data integration, algorithms, cancer
INTRODUCTION
Genome-wide profiling technologies, in particular microarrays and next-generation sequencing, are used to characterize disease-associated changes at various levels of genome function. Identification of the key players—genes, chromosomal regions or biological processes—is a fundamental step toward mechanistic characterization of the disease and revealing molecular targets for potential therapeutic intervention. Genomic, transcriptomic, epigenomic and proteomic measurements characterize different aspects of genome regulation and function that are particularly relevant for cancer research [1, 2]. Integrative analysis has been used to prioritize disease genes or chromosomal regions for experimental testing, to discover disease subtypes [3, 4] or to predict patient survival or other clinical variables [5]. Co-occurring genomic observations are increasingly available in private and public repositories, such as the Cancer Genome Atlas database [6] and the Leukemia Gene Atlas [7], promoting wide access to data resources. However, the lack of algorithmic implementations forms a bottleneck hampering integrative approaches.
The integration of gene expression (GE) and copy number (CN) data to identify DNA CN alterations that induce changes in the expression levels of the associated genes is a common task in cancer studies [8]. The detection of chromosomal regions with exceptionally high statistical association between CN and GE can pinpoint disease genes and potential cancer mechanisms [9, 10]. First, high-throughput analyses were reported about a decade ago [11–13], evidencing a clear cis-dosage effect of CN alterations on GE levels [14–16]. Although the downstream effect of CN alteration on GE is still a focus of ongoing research [17, 18], a systematic quantitative comparison of alternative approaches for integrating GE/CN data has been missing, as recently highlighted by Huang et al [8]. Hence, we designed a quantitative benchmarking procedure to compare 12 publicly available methods for cancer gene prioritization based on integrative analysis of CN/GE profiling data on two simulated and three real case studies. In the following sections, we give a methodological overview, introduce the analysis pipeline and discuss the benchmarking results.
QUANTIFYING ASSOCIATIONS BETWEEN GE and CN
The available implementations for the integrative analysis of GE and CN can be roughly divided in four main categories. In this section, we provide a general overview of these approaches with further references to individual algorithms.
Two-step approaches
A comparison of GE levels between groups of samples with distinct CN status aims at revealing CN-induced transcriptional responses. Several approaches separately either first assess the alterations in each data set and then compare the results from both or assess alterations in GE in genes or genomic regions previously identified by an assessment of CN alterations to model changes in GE based on the CN signals [16, 19]. This corresponds to the biological intuition concerning the cis-regulatory effect of CN alterations. In the first step, samples and genes are grouped based on estimated CN levels, estimated probabilities of CN alterations [20] or quantiles [21]. In the second step, differential GE is quantified either between such groups or independently (with respect to a reference sample) using standard approaches for GE analysis such as the t-test which assesses the difference between two sample groups based on Gaussian assumptions [13]. Nonparametric [20, 22] and permutation-based alternatives [23, 24, 36] have also been suggested to relax the normality assumptions of the t-test. Cancer-associated changes often affect chromosomal regions with varying sizes, which potentially contain multiple genes. Therefore, some methods have been designed to specifically detect large regions affected by CN alteration rather than prioritize individual genes [19, 24]. Nevertheless, the regional modeling of GE and CN data can help to pinpoint individual driver genes whose expression is most notably affected by a larger chromosomal alteration.
Regression approaches
Another class of tools uses regression models, generally with CN as the predictor and GE as the response variable, again exploiting the biological intuition concerning the cis-regulatory effect of CN alterations. Both linear [12] and nonlinear regression models [25] have been proposed. Univariate linear regression models have been designed to model the associations between individual CN and GE probes [26], as well as multiple and/or multivariate linear regression models that combine statistical power across multiple probes targeting adjacent genes or chromosomal positions [14, 26–28]. Regression models are theoretically related to correlation analysis. For instance, the square of Pearson’s correlation coefficient estimates the proportion of variance in the response variable that is explained by the predictor in a univariate linear regression. In case, variables are standardized beforehand, the regression coefficient of the predictor variable equals Pearson's correlation coefficient.
Correlation-based approaches
DR-Correlate [21] and a modified version of Ortiz-Estevez algorithm [16] use correlation-based analysis to scan over the genome and detect loci with exceptionally high associations between CN/GE. To address potential shortcomings with respect to a biologically inadequate reflection of CN and GE abnormalities by ordinary correlation analysis, Schäfer et al. [29] substitute sample means by the reference medians, and Lipson et al. [30] use quantile-based analysis to obtain improved correlation coefficients. Furthermore, canonical correlation analysis (CCA) has been suggested to identify general linear associations between CN and GE data through flexible detection of weighted combinations of probes, which reveal maximal correlations between the two data sources. This is expected to more efficiently distinguish the relevant shared variation of the GE/CN data from the data set-specific effects [34]. Various modifications for dimensionality reduction and model regularization have also been proposed based on principal component analysis [31] and penalized approaches based on LASSO, elastic net or other constraints to obtain sparse or regularized versions of CCA [5, 32–34]. Although regularization may reduce overfitting and sparsity can simplify interpretation of the results, setting the appropriate regularization parameters may be a challenging task.
Latent variable models
Latent variable approaches are used to model directly the data-generating processes. For instance, the pint/simcca algorithm [34] decomposes GE and CN data sets into shared and independent Gaussian components based on regularized probabilistic CCA. A comparison of the shared and data set-specific signals is used to pinpoint chromosomal regions with exceptionally high levels of dependence between the GE/CN observations. Related matrix decomposition models and iterative, dependence-seeking projections have been suggested based on generalized singular value decomposition [3] and independent component analysis [35]. The advantage of latent variable models in comparison with the two-step-, correlation- or regression-based approaches is that they explicitly model both the signal and noise in the data, and take into account the uncertainty in the model by integrating over the unknown latent variables. These properties help distinguishing signal from noise in a robust manner, but often come at an increased computational cost.
BENCHMARKING THE ALGORITHMS
Manual literature search in PubMed and Google Scholar using combinations of the keywords ‘gene expression’, ‘copy number’, ‘integration’ and inspection of the Bioconductor repository (http://www.bioconductor.org) were performed to identify available implementations, yielding 12 algorithms that were applicable for cancer gene prioritization based on integrative analysis of GE/CN data (Table 1). The source code for Ortiz-Estevez [16] was obtained from the authors. An automated benchmarking pipeline was created to compare method performance on two simulated data sets and three real case studies (http://intcomp.r-forge.r-project.org).
Table 1:
Summary of the comparison algorithms
| Implementation | CN preprocessing | Methodology | Significance scoring | Reference |
|---|---|---|---|---|
| CNAmet (R) | Called | Custom statistic; | PPT; aberrant regions | [24] |
| Two step | [36] | |||
| DR-Correlate/t-test (BC) | Raw/segmented | Two step | PPT; P-values | [21] |
| DR-Correlate (BC) | Raw/segmented | COR | PPT; P-values | [21] |
| edira (R) | Raw/segmented | Custom statistic; | NT; P-values | [29] |
| COR | ||||
| intCNGEan (R) | cghCall object | Custom statistic; | PNT; P-values | [20] |
| Two step | ||||
| Ortiz-Estevez (R) | Raw/segmented | Two step | PNT; P-values | [16] |
| PMA (CRAN) | Raw/segmented | LV; COR | PLV; P-values | [56] |
| PREDA/SODEGIR (BC) | Raw/segmented | Custom statistic; | PPT; aberrant regions/ | [19] |
| Two step | q-values | [48] | ||
| pint/simcca | Raw/segmented | LV; COR | PLV; P-values | [34] |
| SIM (BC) | Raw/segmented | REG | PT; P-values | [26] |
The implementations are available through Bioconductor (BC); CRAN or R source code (R). The CN preprocessing methods required by each algorithm are listed. COR, correlation analysis; REG, regression analysis; LV, latent variables analysis; PT, parametric test; NT, nonparametric test; PNT, permutation test based on statistic of nonparametric test; PPT, permutation test based on statistic of parametric test; PLV, permutation test based on latent variable score.
Each method was used to prioritize candidate cancer genes, followed by a comparison with a golden standard list of known cancer genes, and ranking of the methods based on receiver operating characteristic (ROC) analysis of the prioritized gene lists and running times. Investigating the true positive rate among the top findings complemented the standard area under curve (ROC/AUC) analysis, which considers the overall prioritized gene list. Default parameters for each method were used where possible. The following exceptions were made to apply the algorithms to cancer gene prioritization. In DR-Correlate [21], empirical P-values from 1000 random gene permutations were used to rank the genes. The DR-Correlate t-test option was not applicable on the Ferrari simulations due to the low number of replicate samples. CNAmet [24, 36] requires called CN values and provides separate lists for amplifications and deletions; thus, the two lists were pooled and ranked based on the P-values. Moreover, to enable an unbiased AUC comparison of CNAmet with all other methods (that prioritize all genes), random ranks were assigned to genes labeled by CNAmet with no P-value (nonsignificant genes). With intCNGEan [20], the weighted Mann–Whitney test with univariate analysis was used with an effective P-value threshold of 0.1. In pint/simcca [34], segmented CN data were used only when the resolution of the CN platform was higher than the resolution of the GE microarray. In PREDA/SODEGIR, we used ‘spline’ for smoothing, 1000 random gene orderings of the output regions and the median AUC as an unbiased output for gene prioritization.
For all methods, GE and CN probes were matched by selecting for each GE probe the closest CN probe within the same chromosomal arm. One-to-one matching between the GE and CN data was required in the real case studies [34, 37]; in simulation experiments, the original simulation procedures [19, 29] were followed as described below. The preprocessing of CN data depends partially on the platform resolution. On the latest high-density SNP arrays, for instance, segmentation strategies are essential for estimating the CN for individual genes [8]. Various approaches consider to investigate only certain genomic regions at a time, e.g. to avoid bias, and propose different strategies to select the size of the chromosomal region, including fixed windows in terms of consecutive probes or base pairs [28, 30, 34], chromosome arms or minimal common regions [26] or performing kernel regression [19], where the probe signals are modeled with a smoothing function which accounts for the nonuniform distribution of the genes along the genome.
Simulated data
Two simulated data sets were generated by roughly following Schäfer et al. ([29]; 'Schäfer' data) and Bicciato et al. ([19]; 'Ferrari' data). The simulations are based on general assumptions regarding the associations between the (altered) CN and GE signals in genome-wide profiling studies, as detailed in the original publications. For the ‘Schäfer’ data set, CN and GE values are drawn from a normal mixture where two components represent aberrations of different extent for each locus; 100 samples were created for each input with mixing proportions of either 10% or 90% for the affected and normal regions. Varying noise levels were imposed using multiple variance parameters (0.25, 0.5, 1, 2 and 4 times an adjusted median absolute deviation of the data). The data points are organized in 16 equally sized blocks to mimic affected regions. The ‘Ferrari’ data with six samples was created by manipulating a renal cell carcinoma data set through permutation of loci and adding or subtracting constants to both CN and GE values within 10 blocks of 10 Mbp. Normal control data was generated by subtracting the median across the samples [19].
Real case studies
We investigated two publicly available breast cancer data sets [12, 13] and a leukemia study [38]. Expert-curated lists of known breast cancer genes [39] and leukemia genes from the Cancer Gene Census [40] were used as the ground truth for the benchmarking experiments, respectively. The preprocessed ‘Hyman’ data set [13] contains 14 breast cancer cell lines, 7489 genes and 48 known breast cancer genes. The preprocessed 'Pollack' data set [12] contains 41 breast cancer samples, 4287 genes and 38 known breast cancer genes. The preprocessed ‘Mullighan’ data set consists of 171 acute lymphoblastic leukemia (ALL) samples divided into 9 subtypes [38, 41], 2162 genes in the matched CN/GE data and 39 known leukemia genes. A combination of standard algorithms was used to preprocess the 500 K Affymetrix CN data [42–44] and the Affymetrix GE data [45–47] for the Mullighan data set. The CN data (Affymetrix Human Mapping 500 K) was downloaded from ftp://ftp.studje.org and normalized with CRMA v2 [42]. The log-additive model from the CRMA v1 algorithm [43] was used for probe summarization. Data values from the Nsp and Sty array of the 500 K set were combined and segmented with CBS [44]. GE profiles of the same ALL specimens, measured with the Affymetrix HG-U133A platform, were obtained from GEO (GSE12995; [45]) and preprocessed with the RPA algorithm [46] and EntrezID-based custom chip definition file (v13; [47]). The reference for GE and CN data was defined as the median normalized log ratios across all samples. In all data sets, probes with no EntrezID or location information and probes mapping to multiple locations or in sex chromosomes were excluded. Missing values were imputed by Gaussian random samples using the mean and variance of the data.
RESULTS
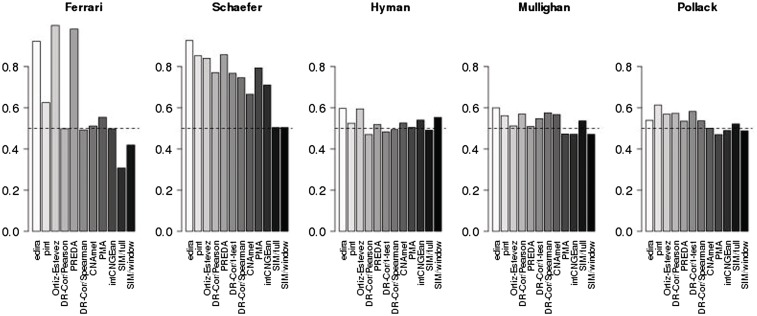
The cancer gene prioritization performance of the comparison methods as quantified by the AUC analysis is summarized in Figure 1 (for the ROC curves, see Supplementary Figure S1). The highest median ranking across the five benchmarking data sets was obtained by edira (1), followed by Ortiz-Estevez (4) and pint/simcca (4). Each of these three methods outperformed the others on at least one data set. Note that the performance of edira with the ‘Schäfer’ data set and of PREDA/SODEGIR with the ‘Ferrari’ data set needs to be carefully interpreted, since these simulations were originally constructed to follow the particular modeling assumptions of these algorithms in the original publications [19, 29]. The complete benchmarking results are available at the project website.
Figure 1:
AUC values in ROC analysis quantify cancer gene prioritization performance of the methods for the five benchmarking data sets. High values indicate high true-positive versus false-positive ratio among the top findings; the dashed line indicates the expected AUC value for a random gene list (AUC = 0.5). The methods have been ordered by their median rank across all data sets. For the ROC curves, see Supplementary Figure S1.
Considering the true-positive rate among the top 200 genes of each algorithm, pint/simcca had the highest median ranking (1), followed by edira, Ortiz-Estevez and PREDA/SODEGIR (3; Supplementary Figure S2). These methods had systematically the highest median rankings with multiple thresholds (20, 50 and 100 top genes). Notably, although edira and PREDA/SODEGIR had the highest AUC scores on the Schäfer data, most of other algorithms outperformed these methods with respect to known true positives among the top findings in this data set.
Differences regarding the running times were considerable (Supplementary Table S1). Specifically, edira and PMA were the fastest methods with less than 1 min running time in all data sets, closely followed by Ortiz-Estevez with a maximum running time of <3 min. The number of permutations in significance testing affects remarkably the running times of CNAmet, DR-Correlate, intCNGEan and PREDA/SODEGIR, although in the latest version of PREDA/SODEGIR a parallelized version has been implemented to reduce computation time [48].
DISCUSSION
Prioritization of disease genes is a key-modeling task in functional genomics [49–52]. This review provides an overview and quantitative benchmarking of publicly available algorithms for detecting associations between GE and CN alterations. Our work complements the recent review by Huang et al. [8], who pointed out the lack of quantitative comparisons of the available methods. The ‘intcomp’ benchmarking package applied in this review is freely available at R-forge (http://intcomp.r-forge.r-project.org) to facilitate transparent comparisons and the addition of new algorithms, benchmarking procedures and validation data sets.
The comparison of 12 algorithms with respect to their cancer gene prioritization performance revealed systematic differences across independent data sets, preprocessing scenarios and sample sizes. Interestingly, while no systematic differences between the four main categories of GE/CN integration approaches were seen, systematic differences between individual methods were evident. In particular, edira, Ortiz-Estevez and pint/simcca consistently outperformed the other methods. Considering both relative performance and running time, edira and Ortiz-Estevez seem to offer an optimal trade-off, although all methods have acceptable running times for practical applications. While none of the methods outperformed the others in all data sets, identification of the few best-performing implementations provides quantitative guidance for the selection of analysis tools and has therefore direct practical relevance for cancer studies.
Benchmarking the algorithms on real data is crucial since simulation studies are unlikely to capture all complexities present in real data. However, the availability of suitable benchmarking data sets is limited. We selected publicly available data sets in which both GE and CN data from the same samples are available and independent lists of known cancer genes obtained from the literature. The model performance is in general better in the simulation studies, compared to the real cancer data sets, suggesting that manually curated cancer gene lists may be only coarse approximations of the ground truth in the real case studies and that simulations may have lower noise levels. On the other hand, simulation procedures are only rough approximations of the biological reality and the simulation schema can remarkably affect model performance. For instance, variants of DR-Correlate and CNAmet performed well with ‘Schäfer’ simulated data, but their performance dropped close to random expectation in the ‘Ferrari’ data set. The ‘Ferrari’ simulations assume that the CN effect is visible in all tumor samples, which can be particularly disadvantageous for DR-Correlate and other methods that rely on variations between the aberration profiles across the samples. The ‘Ferrari’ and ‘Schäfer’ simulated data sets were originally designed to evaluate the performances of PREDA/SODEGIR and edira methods, and this aspect potentially causes positive bias on these methods in the respective data sets. Moreover, certain methods, such as CNAmet [36], Ortiz-Estevez [16] or PREDA/SODEGIR [19], have originally been designed to prioritize altered chromosomal regions rather than individual genes. Our benchmarking procedure is based on the prioritization of individual genes since this is the most prevalent objective shared by the available GE/CN integration algorithms.
Since chromosomal CN alterations represent a key feature of cancer, well-performing GE/CN analysis methods are expected to have a good prioritization performance of known cancer genes. However, certain cancer genes may be overlooked by integrative approaches that focus only on simultaneous changes in both GE and CN levels since gene activity is also affected by cellular mechanisms other than GE/CN alterations. For such reason, it was not un-expected that 33–73% of the known cancer genes were not included among the first 200 prioritized genes by any comparison method in the five benchmarking data sets. The relatively low number (0–8) of the known cancer genes among the first 200 findings in the real case studies highlights the need for efficient approaches to identify key mutations and genes that drive cancer development and progression [23]. Moreover, although any algorithm detected certain cancer genes, none of the known cancer genes was detected by all methods in any benchmarking data set among the first 200 findings. Since different methods emphasize different aspects of the GE/CN data, efficient joint analysis of the results from multiple independent methodologies might outperform individual methods. One could, for instance, consider mean or median ranks across the prioritized lists, or weight the different lists according to certain criteria. Related approaches have been suggested elsewhere [49], but have not been investigated in the context of GE/CN analysis yet. In our experiments, straightforward ranking of the genes based on their mean or median rank across the different methods did not outperform the best-performing methods in any benchmarking data set.
The choice of preprocessing and model parameters can have a remarkable effect on the results. The key decisions in the context of GE/CN data are associated with selecting the CN preprocessing approach [53], size of the investigated chromosomal regions and the matching approach for the integrated data sets. These and related issues are extensively discussed in the recent review by Huang et al. [8]. It is also possible to utilize class information of the samples, for instance, by including both tumor and reference samples [21]. However, in many cases, the references are included as a pooled control for two-color microarray experiments but not as a separate group, as with the Hyman and Pollack data sets. Moreover, genomic aberrations often affect only a subset of the cancer patients, and multiple cancer subtypes may be present, as in the Mullighan data set. The matching approach for GE/CN data may also affect the results. In the current pipeline, each GE probe is matched to the closest CN probe or segment. Requiring one-to-one matching of the GE/CN data may lead to exclusion of many GE probes in particular on high-density arrays such as in the Mullighan data set. The publicly available benchmarking pipeline will allow further experimentation with alternative preprocessing scenarios. All data presented in this study come from microarray studies, where several matched GE/CN data sets are available from public sources, but the approach should be in principle applicable also to high-throughput sequencing data. Since the underlying biological phenomena remain unaltered, and methodological approaches proposed for GE/CN integration are based on relatively general modeling assumptions, it can be expected that the proposed methods are applicable also in the context of next-generation sequencing after appropriate data preprocessing.
Further integrative tasks in GE/CN analysis would include modeling of trans-regulatory effects of CN aberrations on genes outside the affected region [54, 55], disease subtype discovery [4], prediction of patient survival or of clinical covariates [56] and integrative analysis of other data sources, such as methylation [57], microRNA [58–59] or protein expression [60]. However, fewer implementations for such tasks are currently available. Availability of reference implementations would facilitate benchmarking and optimizing new algorithms. The benchmarking pipeline introduced in this review can be adjusted to incorporate additional algorithms and data sets as they become available.
CONCLUSION
A variety of methods is available for the integrative analysis of GE and CN data. The algorithms can be classified as two-step, regression, correlation-based and latent variable approaches. Implementation quality, running time and accuracy of the algorithm, as well as preprocessing, sample size and availability of control samples need to be considered when selecting the appropriate method. The benchmarking pipeline reveals systematic differences in cancer gene prioritization performance of available implementations across five case studies.
SUPPLEMENTARY DATA
Supplementary Data are available online at http://bib.oxfordjournals.org/.
Key Points.
Integrative analysis algorithms for GE and CN data include two-step, regression, correlation-based and latent variable approaches.
The benchmarking pipeline reveals systematic differences in cancer gene prioritization performance of currently available implementations.
Implementation quality, running time and accuracy of the algorithm, as well as data preprocessing, sample size and availability of control samples need to be considered when selecting the analysis approach.
FUNDING
This work was supported by EuGESMA COST Action BM0801 (European Genomics and Epigenomics Study on MDS and AML). L.L. has been supported by Helsinki Institute for Information Technology HIIT and Finnish Center of Excellence on Adaptive Informatics Research (AIRC). M.D. is supported by the European Leukemia Network of Excellence (LSHC-CT-2004); Deutsche Kinderkrebsstiftung (grant number DKS 2010.21) and Carreras Foundation (grant number DJCLS 09/04). M.S. is supported by the Deutsche Forschungsgemeinschaft (Research Training Group Statistical Modeling). S.B. is supported from AIRC Special Program Molecular Clinical Oncology ‘5 per mille’.
Supplementary Material
Acknowledgement
We would like to thank Francesco Ferrari for providing the simulated data set for this study.
Biographies
Leo Lahti is a postdoctoral researcher in Wageningen University, Netherlands, focusing on integrative high-throughput data analysis, human genomics and microbial ecology.
Martin Schäfer is a PhD student at the Department of Statistics, TU Dortmund University. His main interest is in developing statistical methods to understand diseases on a molecular level.
Hans-Ulrich Klein is a computational scientist working at the Institute of Medical Informatics, University of Münster. His main interest is the development and application of methods for the analysis of high-dimensional molecular data for a better understanding of cancer.
Silvio Bicciato is an Assistant Professor at the Center for Genome Research of the University of Modena and Reggio Emilia. His research interests include bioinformatics and analysis of high-throughput genomics data.
Martin Dugas is Professor and Managing Director of the Institute of Medical Informatics at the University of Münster. His research activity focuses on biomedical, genomics, and health informatics.
References
- 1.Chin L, Gray J. Translating insights from the cancer genome into clinical practice. Nature. 2008;452:553–63. doi: 10.1038/nature06914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hawkins R, Hon G, Ren B. Next-generation genomics: an integrative approach. Nat Rev Genet. 2010;11:476–86. doi: 10.1038/nrg2795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Berger J, Hautaniemi S, Mitra S, et al. Jointly analyzing gene expression and copy number data in breast cancer using data reduction models. IEEE/ACM Tr Comp Biol Bioinf. 2006;3:2–16. doi: 10.1109/TCBB.2006.10. [DOI] [PubMed] [Google Scholar]
- 4.Shen R, Olshen A, Ladanyi M. Integrative clustering of multiple genomic data types using a joint latent variable model with application to breast and lung cancer subtype analysis. Bioinformatics. 2009;25:2906–12. doi: 10.1093/bioinformatics/btp543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Witten D, Tibshirani R. Extensions of sparse canonical correlation analysis, with applications to genomic data. Stat Appl Genet Mol Biol. 2009;8:28. doi: 10.2202/1544-6115.1470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.The Cancer Genome Atlas Research Network. Comprehensive genomic characterization defines human glioblastoma genes and core pathways. Nature. 2008;455:1061–68. doi: 10.1038/nature07385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Leukemia Gene Atlas. www.leukemia-gene-atlas.org (15 December 2011, date last accessed) [Google Scholar]
- 8.Huang N, Shah PK, Li C. Lessons from a decade of integrating cancer copy number alterations with gene expression profiles. Brief Bioinformatics. doi: 10.1093/bib/bbr056. Advance Access published on 23 September 2011, doi:10.1093/bib/bbr056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.De S, Babu M. Genomic neighborhood and the regulation of gene expression. Curr Opin Cell Biol. 2010;22:7. doi: 10.1016/j.ceb.2010.04.004. [DOI] [PubMed] [Google Scholar]
- 10.Lee H, Kong S, Park P. Integrative analysis reveals the direct and indirect interactions between DNA copy number aberrations and gene expression changes. Bioinformatics. 2008;24:889–96. doi: 10.1093/bioinformatics/btn034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Phillips J, Hayward S, Wang Y, et al. The Consequences of chromosomal aneuploidy on gene expression profiles in a Cell line model for prostate carcinogenesis. Cancer Res. 2001;61:8143–9. [PubMed] [Google Scholar]
- 12.Pollack J, Sorlie T, Perou C, et al. Microarray analysis reveals a major direct role of DNA copy number alteration in the transcriptional program of human breast tumors. Proc Natl Acad Sci. 2001;99:12963–8. doi: 10.1073/pnas.162471999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hyman E, Kauraniemi P, Hautaniemi S, et al. Impact of DNA amplification on gene expression patterns in breast cancer. Cancer Res. 2002;62:6240–5. [PubMed] [Google Scholar]
- 14.Gu W, Choi H, Ghosh D. Global associations between copy number and transcript mRNA microarray data: an empirical study. Cancer Informatics. 2008;6:17–23. doi: 10.4137/cin.s342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Myllykangas S, Junnila S, Kokkola A, et al. Integrated gene copy number and expression microarray analysis of gastric cancer highlights potential target genes. Int J Cancer. 2008;123:817–25. doi: 10.1002/ijc.23574. [DOI] [PubMed] [Google Scholar]
- 16.Ortiz-Estevez M, De Las Rivas J, Fontanillo C, et al. Segmentation of genomic and transcriptomic microarrays data reveals major correlation between DNA copy number aberrations and gene-loci expression. Genomics. 2011;97:86–93. doi: 10.1016/j.ygeno.2010.10.008. [DOI] [PubMed] [Google Scholar]
- 17.Harvey R, Mullighan C, Wang X, et al. Identification of novel cluster groups in pediatric high-risk B-precursor acute lymphoblastic leukemia with gene expression profiling: correlation with genome-wide DNA copy number alterations, clinical characteristics, and outcome. Blood. 2010;116:4874–84. doi: 10.1182/blood-2009-08-239681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yuan Y, Rueda O, Curtis C, et al. Penalized regression elucidates aberration hotspots mediating subtype-specific transcriptional responses in breast cancer. Bioinformatics. 2011;27:2679–85. doi: 10.1093/bioinformatics/btr450. [DOI] [PubMed] [Google Scholar]
- 19.Bicciato S, Spinelli R, Zampieri M, et al. A computational procedure to identify significant overlap of differentially expressed and genomic imbalanced regions in cancer datasets. Nucleic Acids Res. 2009;37:5057–70. doi: 10.1093/nar/gkp520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.van Wieringen W, van de Wiel M. Nonparametric testing for DNA copy number induced differential mRNA gene expression. Biometrics. 2009;65:19–29. doi: 10.1111/j.1541-0420.2008.01052.x. [DOI] [PubMed] [Google Scholar]
- 21.Salari K, Tibshirani R, Pollack J. DR-Integrator: a new analytic tool for integrating DNA copy number and gene expression data. Bioinformatics. 2010;26:414–6. doi: 10.1093/bioinformatics/btp702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.van Wieringen W, Belien J, Vosse S, et al. ACE-it: a tool for genome-wide integration of gene dosage and RNA expression data. Bioinformatics. 2006;22:1919–20. doi: 10.1093/bioinformatics/btl269. [DOI] [PubMed] [Google Scholar]
- 23.Akavia U, Litvin O, Kim J. An integrated approach to uncover drivers of cancer. Cell. 2010;143:1005–17. doi: 10.1016/j.cell.2010.11.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hautaniemi S, Ringnér M, Kauraniemi P, et al. A strategy for identifying putative causes of gene expression variation in human cancers. J Franklin Institute. 2004;341:77–88. [Google Scholar]
- 25.Solvang H, Lingjaerde O, Frigessi A, et al. Linear and non-linear dependencies between copy number aberrations and mRNA expression reveal distinct molecular pathways in breast cancer. BMC Bioinformatics. 2011;12:197. doi: 10.1186/1471-2105-12-197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Menezes R, Boetzer M, Sieswerda M, et al. Integrated analysis of DNA copy number and gene expression microarray analysis using gene sets. BMC Bioinformatics. 2009;10:203. doi: 10.1186/1471-2105-10-203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Peng J, Zhu J, Bergamaschi A, et al. Regularized multivariate regression for identifying master predictors with application to integrative genomics study of breast cancer. Ann App Stat. 2010;4:53–77. doi: 10.1214/09-AOAS271SUPP. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Stranger B, Forrest M, Dunning M, et al. Relative impact of nucleotide and copy number variation on gene expression phenotypes. Science. 2007;315:848–53. doi: 10.1126/science.1136678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Schäfer M, Schwender H, Merk S, et al. Integrated analysis of copy number alterations and gene expression: a bivariate assessment of equally directed abnormalities. Bioinformatics. 2009;25:3228–35. doi: 10.1093/bioinformatics/btp592. [DOI] [PubMed] [Google Scholar]
- 30.Lipson D, Ben-Dor A, Dehan E, et al. Joint analysis of DNA copy numbers and gene expression levels. In: Jonassen I, Kim J, editors. Proc Algorithms in Bioinformatics: 4th International Workshop WABI 2004. Germany: Springer, 2004. [Google Scholar]
- 31.Soneson C, Lilljebjorn H, Fioretos T, et al. Integrative analysis of gene expression and copy number alterations using canonical correlation analysis. BMC Bioinformatics. 2010;11:191. doi: 10.1186/1471-2105-11-191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Parkhomenko E, Tritchler D, Beyene J. Sparse canonical correlation analysis with application to genomic data integration. Stat App Genet Mol Biol. 2009;8:1. doi: 10.2202/1544-6115.1406. [DOI] [PubMed] [Google Scholar]
- 33.Waaijenborg C, Verselewel de Witt Hamer PC, Zwinderman AH. Quantifying the association between gene expressions and DNA-markers by penalized canonical correlation analysis. Stat Appl Genet Mol Biol. 2008;7:3. doi: 10.2202/1544-6115.1329. [DOI] [PubMed] [Google Scholar]
- 34.Lahti L, Myllykangas S, Knuutila S, et al. Proc MLSP'09 IEEE International Workshop on Machine Learning for Signal Processing XIX. Piscataway, NJ, USA: IEEE; 2009. Dependency detection with similarity constraints; pp. 89–94. [Google Scholar]
- 35.Sheng J, Deng H, Calhoun V, et al. Integrated analysis of gene expression and copy number data on gene shaving using independent component analysis. IEEE Tr Comp Biol Bioinform. 2011;8:1568–79. doi: 10.1109/TCBB.2011.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Louhimo R, Hautaniemi S. CNAmet: an R package for integrating copy number, methylation and expression data. Bioinformatics. 2011;27:887–8. doi: 10.1093/bioinformatics/btr019. [DOI] [PubMed] [Google Scholar]
- 37.Haverty P, Fridlyand J, Li L, et al. High-resolution genomic and expression analyses of copy number alterations in breast tumors. Genes Chromosomes Cancer. 2008;47:530–42. doi: 10.1002/gcc.20558. [DOI] [PubMed] [Google Scholar]
- 38.Mullighan C, Goorha S, Radtke I, et al. Genome-wide analysis of genetic alterations in acute lymphoblastic leukaemia. Nature. 2007;446:758–64. doi: 10.1038/nature05690. [DOI] [PubMed] [Google Scholar]
- 39.Baasiri R, Glasser S, Steffen D, et al. The breast cancer gene database: a collaborative information resource. Oncogene. 1999;18:7958–65. doi: 10.1038/sj.onc.1203335. [DOI] [PubMed] [Google Scholar]
- 40.Futreal P, Coin L, Marshall M, et al. A census of human cancer genes. Nat Rev Cancer. 2004;4:177–83. doi: 10.1038/nrc1299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Mullighan C, Miller C, Radtke I, et al. BCR-ABL1 lymphoblastic leukaemia is characterized by the deletion of Ikaros. Nature. 2008;453:110–4. doi: 10.1038/nature06866. [DOI] [PubMed] [Google Scholar]
- 42.Bengtsson H, Wirapati P, Speed TP. A single-array preprocessing method for estimating full-resolution raw copy numbers from all Affymetrix genotyping arrays including GenomeWideSNP 5 & 6. Bioinformatics. 2009;25:2149–56. doi: 10.1093/bioinformatics/btp371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bengtsson H, Irizarry R, Carvalho B, et al. Estimation and assessment of raw copy numbers at the single locus level. Bioinformatics. 2008;24:759–67. doi: 10.1093/bioinformatics/btn016. [DOI] [PubMed] [Google Scholar]
- 44.Olshen A, Venkatraman E, Lucito R, et al. Circular binary segmentation for the analysis of array-based DNA copy number data. Biostatistics. 2004;5:557–72. doi: 10.1093/biostatistics/kxh008. [DOI] [PubMed] [Google Scholar]
- 45.Mullighan C, Su X, Zhang J, et al. Deletion of IKZF1 and prognosis in acute lymphoblastic leukemia. N Engl J Med. 2009;360:470–80. doi: 10.1056/NEJMoa0808253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lahti L, Elo LL, Aittokallio T, et al. Probabilistic analysis of probe reliability in differential gene expression studies with short oligonucleotide arrays. IEEE/ACM Tr Comp Biol Bioinform. 2011;8:217–25. doi: 10.1109/TCBB.2009.38. [DOI] [PubMed] [Google Scholar]
- 47.Dai M, Wang P, Boyd A, et al. Evolving gene/transcript definitions significantly alter the interpretation of GeneChip data. Nucleic Acids Res. 2005;33:e175. doi: 10.1093/nar/gni179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ferrari F, Solari A, Battaglia C, et al. PREDA: an R-package to identify regional variations in genomic data. Bioinformatics. 2011;27:2446–7. doi: 10.1093/bioinformatics/btr404. [DOI] [PubMed] [Google Scholar]
- 49.Aerts S, Lambrechts D, Maity S. Gene prioritization through genomic data fusion. Nat Biotech. 2006;24:538–44. doi: 10.1038/nbt1203. [DOI] [PubMed] [Google Scholar]
- 50.de Bie T, Tranchevent L-C, van Oeffelen L, et al. Kernel-based data fusion for gene prioritization. Bioinformatics. 2007;23:i125–32. doi: 10.1093/bioinformatics/btm187. [DOI] [PubMed] [Google Scholar]
- 51.Kao C-F, Fang Y-S, Zhao Z, et al. Prioritization and evaluation of depression candidate genes by combining multidimensional data resources. PLoS One. 2011;6:e18696. doi: 10.1371/journal.pone.0018696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Tranchevent L-C, Capdevila F, Nitsch D, et al. A guide to web tools to prioritize candidate genes. Brief Bioinform. 2010;12:22–32. doi: 10.1093/bib/bbq007. [DOI] [PubMed] [Google Scholar]
- 53.van Wieringen W, van de Wiel M, Ylstra B. Normalized, segmented or called aCGH data? Cancer Inform. 2007;3:321–7. [PMC free article] [PubMed] [Google Scholar]
- 54.Lê Cao K-A, González I, Dèjean S. integrOmics: an R package to unravel relationships between two omics datasets. Bioinformatics. 2009;25:2855–6. doi: 10.1093/bioinformatics/btp515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Vaske C, Benz S, Sanborn J, et al. Inference of patient-specific pathway activities from multidimensional cancer genomics data using PARADIGM. Bioinformatics. 2010;26:i237–45. doi: 10.1093/bioinformatics/btq182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Witten D, Tibshirani R, Hastie T. A penalized matrix decomposition, with applications to sparse principal components and canonical correlation analysis. Biostatistics. 2009;10:515–34. doi: 10.1093/biostatistics/kxp008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Andrews J, Kennette W, Pilon J. Multi-platform whole-genome microarray analyses refine the epigenetic signature of breast cancer metastasis with gene expression and copy number. PLoS One. 2010;5:e8665. doi: 10.1371/journal.pone.0008665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Gaire R, Bailey J, Bearfoot J, et al. MIRAGAA—a methodology for finding coordinated effects of microRNA expression changes and genome aberrations in cancer. Bioinformatics. 2010;26:161–7. doi: 10.1093/bioinformatics/btp654. [DOI] [PubMed] [Google Scholar]
- 59.Qin L-X. An integrative analysis of microrna and mrna expression—a case study. Cancer Inform. 2008;6:369–79. doi: 10.4137/cin.s633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Johnson N, Speirs V, Curtin N, et al. A comparative study of genome-wide SNP, CGH microrray and protein expression analysis to explore genotypic and phenotypic mechanisms of acquired antiestrogen resistance in breast cancer. Breast Cancer Res Treat. 2008;111:55–63. doi: 10.1007/s10549-007-9758-6. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.