Abstract
With the development of ultra-high-throughput technologies, the cost of sequencing bacterial genomes has been vastly reduced. As more genomes are sequenced, less time can be spent manually annotating those genomes, resulting in an increased reliance on automatic annotation pipelines. However, automatic pipelines can produce inaccurate genome annotation and their results often require manual curation. Here, we discuss the automatic and manual annotation of bacterial genomes, identify common problems introduced by the current genome annotation process and suggests potential solutions.
Keywords: bacteria, genomics, annotation, automatic, errors
BACKGROUND
Prokaryotic genomics has seen an explosion in the number of genome projects, driven by the advent of next generation sequencing (NGS), resulting in a huge reduction in the time and money investment per project [1]. Microbial genome annotation often consists of running an automatic annotation pipeline followed by manual curation of the results [2]. Most annotation pipelines use homology methods to transfer information from a closely related reference genome to the new sequence. Automatic pipelines can lead to the introduction and propagation of poor annotation and errors, and it is the purpose of the manual curation step to catch and remove these. However, as it is now possible to sequence multiple microbial genomes in a single day at low cost using a single sequencing machine [3], it is no longer feasible to manually curate the annotation of all sequenced genomes. Fully-automatic annotation pipelines, while essential to the modern microbial genomicist, may introduce and propagate inconsistent and incorrect gene annotations.
High-quality annotation goes beyond applying gene prediction software and transferring the annotation from the genome’s closest relative. We have to include features other than coding sites (CDS), such as ribosomal-binding sites (RBSs), termination sites and conserved motifs/domains. Not only do these features give a fuller annotation they actually can rectify errors from earlier parts of the annotation process. For example, predicting RBS and termination sites will give a much clearer idea of a gene’s true location rather than using gene prediction alone. Luckily, there are many software tools for the prediction of these features [4–8].
Transferring annotation purely based on the closest annotated relative does have its limitations. When we consider the reason the new strain has been sequenced, often it will be to identify how this strains differ genetically to its close relatives. This is paradoxical because we are trying to find the differences between these strains but using a similarity based method to annotate it. Potential areas of interest may not be annotated because they are not in the reference genome.
With this surge in sequencing, we will also see an increase in the number of annotated genomes submitted to the public databases. Sequence databases have introduced more stringent requirements for submitters meaning that running an annotation pipeline alone is not enough to ensure acceptance of the genome annotation [9, 10]. There has also been a surge in other next generation techniques such as RNA-seq, incorporating experimental methods gives a better indication of a protein’s role and whether it is functional. These annotations would be more accurate because they are based on actual experiment data rather than homology. Currently genomes can include evidence tags stating how the annotation was assigned, however, they are often omitted from the process. Including evidence qualifiers gives the user an idea of the reliability of the reference genome. The concept of assigning a level of quality to annotation is not novel, but is seldom used [11, 12].
This article discusses some of the current steps for prokaryotic genome annotation and offers a guide to some of the common problems that are encountered during automatic annotation. It goes on to identify the limitations of reference genomes and why choosing the closest relative is not always the best option. We also discuss the rules of the public sequence databases, and go on to suggest possible next steps toward a more accurate, comprehensive annotation with minimal propagation of errors.
Annotation of bacterial genomes
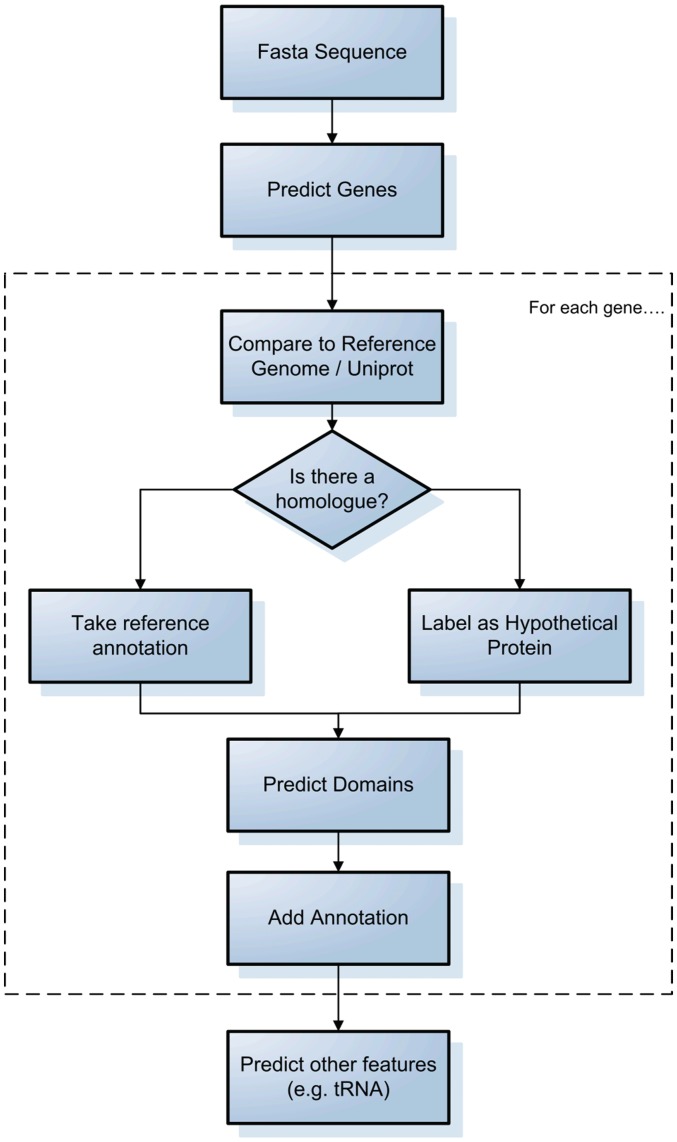
Here we describe a very general process used for bacterial genome annotation (Figure 1). A more thorough review can be found in Stothard and Wishart [2]. In many cases there is a closely related strain/serovar available which has already been sequenced and annotated. Most annotation pipelines employ gene prediction software, the most common of which is GLIMMER [13]. This uses a reference set of sequences to train a model and then utilizes that model to predict coding regions in the genome of interest. Many other ab initio gene prediction algorithms exist and these are reviewed by Do and Choi [14]. Alternatively, gene finding can be performed by extrinsic methods, identifying open reading frames directly from comparisons to protein databases [15, 16].
Figure 1:
A generic process for bacterial genome annotation.
Once coding regions have been identified, they are aligned either to a reference genome annotation or the entirety of UniProt [17] using fast sequence alignment tools (e.g. FASTA [18] or BLAST [19]), the top hits are accepted as homologs and the annotation is transferred across for genes displaying high similarity. Other features such as tRNAs and rRNAs may then added using other prediction software [20].
A range of automatic bacterial annotation pipelines have been published, including web-based systems such as RAST [21], BASys [22], WeGAS [23] and MaGe/Microscope [24]; and systems to be locally installed, such as AGeS [25], DIYA [26] and PIPA [27]. There is also MICheck [28] which checks annotated sequences for syntactic errors. All of these systems carry out the basic process outlined above, with various additions to check for errors or add additional information. It is worth noting that in order to submit to a genome repository that the annotation needs to be in a compatible format (e.g. .tab or .asn). Some pipelines do not output in this manner as they are designed to either hold the annotation online or for in-house analysis [22, 23]. Further processing may therefore be necessary before submission to a public database.
Other feature types
For acceptance to databases such as GenBank or EMBL, only gene, CDS and structural RNA features need to be added [9, 10]. However, many other features should be added. This section gives a broad overview of some of the other features and how they can be predicted; a comprehensive guide is available [29].
Gene prediction software sometimes assigns the wrong start/termination sites. Glimmer for example assigns the start site as the most upstream start codon [5]. By searching for RBS, one can infer and reassign the start site; RBSFinder does this by looking for motifs such as the Shine-Dalgarno sequence pattern [5]. For termination sites, TransTerm searches for rho-independent transcription terminators to assign the correct termination site [6]. As well as correcting start/termination sites these features should be added to the annotation, using the tags ‘RBS’ and ‘terminator’ respectively.
Regions of conservation within proteins such as motifs and domains should be added to the annotation after the gene finding step. There are many databases which store protein families such as ProSite, PRINTS and Pfam [4, 7, 8]. InterproScan can perform searches against a range of domain/motif databases [30]. Hits to motif/domain databases should be assigned the qualifier ‘db_xref’ within the corresponding CDS feature [9, 10].
Areas of horizontal gene transfer (HGT) such as pathogenicity islands and prophage can be predicted by looking at asymmetries in codon composition and the GC content as these will often differ between areas of HGT and the rest of the genome [31]. They are often associated with the presence of integrases, transposases and IS elements [31]. Software tools exists to predict these [32, 33], and these are reviewed and compared by Langille, et al. [34]. There are clear guidelines for annotating phage, this should be assigned under the ‘source’ feature with the name of the bacteriophage in the ‘organism’ qualifier and the type of sequence in ‘mol_type’ (usually genomic DNA). There is no specific annotation tag for other GIs so these should be annotated as miscellaneous features. The mobile genetic elements themselves use the ‘mobile_element’ tag.
Sequence repeats such as ‘clustered regularly interspaced short palindromic repeats’ (CRISPRs) and other tandem repeats are of biological interest. For example, they can be used to understand the bacterial defense mechanism [35] and to distinguish between closely related strains [36]. Software tools exist [37, 38] and databases such as MICdb store predicted microsatellites as well as offering a prediction tool for user inputted sequence [39].
Identifying a protein’s cellular localization can be indicative of function and this can be used in the identification of drug targets. There are many methods of prediction including homology and keywords [40], amino acid composition [41–43] and a mixture of these [44], Gardy and Brinkman [45] have performed a comprehensive review of the many tools available.
LIMITATIONS OF THE ANNOTATION PROCESS
In an ideal world this would be the end of the annotation process. The fact that homology is the basis for these pipelines means that many genomes currently available may have been annotated using old, out of date genomes as a reference which in turn have been annotated based on even older more out of date genomes. The misannotations and errors may perpetuate throughout each new genome, ultimately propagating into secondary databases such as UniProt [17] and KEGG [46], and domain-specific databases such as PATRIC [47].
The public sequence databases have recognized the need for controlling this replication of errors and provide validation software for checking the standard of one’s annotation prior to submission [9, 10]. This section looks at common errors that are the product of automated annotation and tries to address methods of overcoming these.
Inconsistent annotation
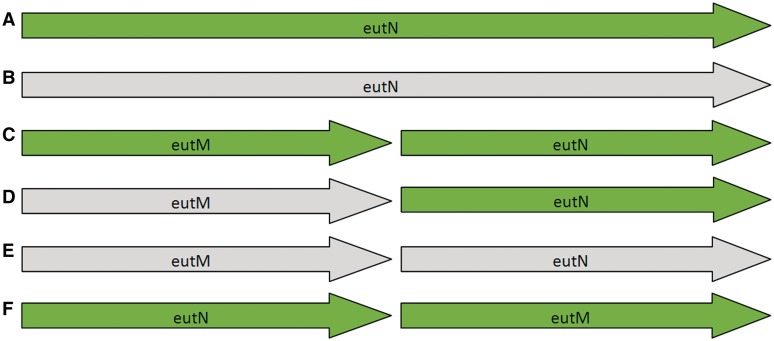
Many bacterial genera now have multiple species and strains with complete genomes, representing a fantastic resource for comparative genomics. However, each genome is annotated separately, by a range of different groups using different protocols, and this introduces inconsistencies. One particular problem is that of split/fused genes and domains; Kummerfield and Teichman [48] found that, of 7116 distinct domain architectures examined across 131 archaeal, bacterial and eukaryotic genomes, 47% showed evidence of gene fusion/fission events. An example of this is the eutM/eutN locus in Salmonella. Figure 2 shows six different models that have been used to annotate this region in the 17 RefSeq records for Salmonella at time of publication. In Salmonella typhi CT18 (NC_003198) and Salmonella typhi Ty2 (NC_004631) there is a single ORF of 690 bp annotated as eutN (Figure 2A). The protein sequence maps to two domains in PFAM, a BMC domain (PF00936) and a EutN_CcmL domain (PF03319). In all other Salmonella genomes in RefSeq, stop codons within this region split the gene, and the domains, in two. In one genome (NC_012125) the region has been annotated as a single long pseudogene of 690 bp (Figure 2B); a further four genomes annotate two intact gene/CDS features, eutM and eutN, each ∼300 bp in length (Figure 2C). A further three genomes are annotated with one pseudogene, a 291 bp ORF equivalent to the eutM gene in Figure 2C, and one intact gene, a 288 bp ORF labeled as eutN (Figure 2D). A further two genomes annotate two ORFs, 291 bp and 300 bp in length respectively, both annotated as pseudogenes (Figure 2E), equivalent to the eutM and eutN genes in Figure2C. Finally, one genome (NC_006511) includes two intact genes, but has reversed the order of eutM and eutN (Figure 2F).
Figure 2:
The six different models present across 17 RefSeq entries for Salmonella species for the eutM/eutN locus. Green indicates normal gene/CDS features, lighter grey indicates gene features annotated as pseudogenes. (A) A single intact gene of 690 bp; (B) a single pseudogene of 690 bp; (C) two short intact genes ∼300 bp in length; (D) one pseudogene and one intact gene, each ∼300 bp in length; (E) two pseudogenes, each 300 bp in length; and (F) two intact genes with the order reversed.
The various ways in which the eutN and eutM genes have been annotated represents a problem for further genome annotation. We cannot know, simply from the genome sequences alone, whether this locus represents a single long gene that has been split in two, or two shorter genes that have become fused. All six models represent different interpretations of a locus that is highly conserved at the nucleotide level across Salmonella species, and any novel genome that is compared to just one of those models will have annotation heavily influenced by that model. For example, if a novel genome is compared only to genomes represented by Figure 2B (two short ORFs annotated as a single long pseudogene) the interpretation will be very different than if the genome were compared to Figure 2C (two short ORFs annotated as two separate intact genes).
Predicting domains directly, rather than genes, using tools such as PfamAlyzer [49], may help in regions with split genes. In the case of eutM/eutN in Salmonella, a domain search would identify two intact domains in all cases; however, the question of whether or not those domains come from the same or separate genes would remain unresolved. We are left with two different versions of the eutN gene from Salmonella in the public databases, one of 690 bp containing two domains, and one of ∼290 bp with one domain.
The only way to annotate this region correctly in silico would be to compare any new genome to each of the six different models. It is difficult to imagine a set of rules that could be given to an automatic annotation pipeline to interpret correctly the evolution of this region and apply that interpretation to a newly sequenced genome. To truly get the full story we would need to look at experimental data (such as RNA-Seq data) to see what the patterns of expression are.
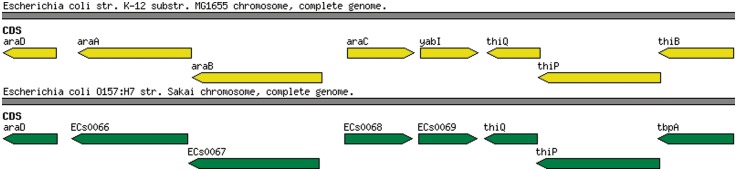
In the eutN/eutM example above, we see a case where genes of vastly differing lengths have been given the same gene name in different genomes; in contrast to this, it is also possible for orthologous genes to be assigned different gene names. Figure 3 shows a syntenic block of genes annotated in Escherichia coli K12 MG1655 (NC_000913) and E. coli O157:H7 Sakai (NC_002695). These two regions are more than 97% identical at the nucleotide level; however, the annotation differs considerably. While E. coli K12 MG1655 contains features with gene names araA, araB and araC, the equivalent features in E. coli 0157:H7 Sakai do not have those gene names and have been assigned uninformative locus tags. Further information is available for the features with only locus tags, including their involvement in arabinose metabolism, however, the gene names remain absent. At the far right of the gene block, two orthologous features exist, both with gene names, however, this time the problem is that they are different: thiB in K12 MG1655 and tbpA in 0157:H7 Sakai. A simple search of the NCBI gene database (search term ‘thiB AND Escherichia coli [Organism]’ versus search term ‘tbpA AND Escherichia coli [Organism]’) reveals that both features code for a thiamin(e) transporter subunit, but the gene is given the gene name tbpA in over 30 E. coli species, whereas it is given the name thiB in only one. Luckily, the thiB feature in K12 MG1655 lists tbpA as a ‘synonym’. Finally, in the centre of the image, K12 MG1655 contains a feature with the gene name yabI, whereas its ortholog in 0157:H7 Sakai only has a locus tag. This is an example of a y-gene, which we discuss in greater detail in the ‘Hypothetical proteins’ section.
Figure 3:
A syntenic block of genes showing inconsistent gene name annotations in E.coli K12 MG1655 and E. coli 0157:H7 Sakai.
The major issue here is that not only do different genomes annotate orthologous genes differently, and provide inconsistent information; they also contain differing amounts of information. This means that, when annotating a new genome, it is essential to choose a reference genome that contains the most accurate and up-to-date information, and that it is also preferable to compare any new genome to multiple references such that inconsistent annotations can be identified and resolved.
Spelling mistakes
There are 128 proteins in UniProt that contain the word ‘syntase’, an incorrect spelling of the word ‘synthase’. To put this into context, the RefSeq entry for Rhizobium etli CFN 42 (accession NC_007761) assigns the function ‘dihydrofolate syntase’ to gene folC. This has propagated into other databases such as UniProt (accession: Q2KE79), KEGG (accession: RHE_CH00024), and xBASE (accession: RHE_CH00024). If a user was to visit any of these databases and search for ‘dihydrofolate synthase’ the misspelled entries would be omitted from the search results. Large scale detection and correction of spelling mistakes in public databases is a difficult task, and so there is a reliance on the submitter to correct these. Automatic annotation pipelines simply copy and propagate what is there already. Spelling mistakes may be highlighted by the validation software provided by the public databases during submission, however, an alternative correct spelling isn’t offered, making it difficult to amend the mistakes without manual intervention.
This can be solved by writing rules to find spelling mistakes [16]. However, this approach is limited to spelling mistakes which are explicitly written in the code. A solution may exist beyond biological science. The search engine Google upon receiving the input ‘syntase’ automatically states ‘Did you mean: synthase’. There are programming languages which have classes or plugins to produce such ‘did you mean’ results [50, 51].
‘Same gene name, different product name’
This issue occurs when two features, either within or between genomes, are assigned the same short gene name yet different product names. The NCBI validation software specifically highlights when this occurs intra-genomically with the description ‘Same gene name, different product name’ [9, 10]. In the current set of 2696 microbial genome and plasmid sequences in RefSeq, we detected 23,843 genes with at least two different product names (see http://www.ark-genomics.org/genomeannotation.html for the full list). The most extreme example of this is gene ‘tnp’ which has 151 different product names (‘tnpA’ has a further 97). A more manageable example can be seen in Table 1. The ‘int’ gene has a total of 12 different product names across 17 Salmonella RefSeq entries. These product names contain huge variation in terms of information content. When using an automatic annotation pipeline, there is a danger that if the top hit is to an entry labeled ‘Hypothetical protein’, then you will capture far less information than if your top hit is to ‘phage integrase family site specific recombinase’. In order to correctly annotate this gene in a new genome, it is necessary to take into account all of these product names in the annotation process. It is difficult to imagine a set of text-mining rules that could efficiently interpret the range of annotations and assign the most suitable one to a new gene.
Table 1:
Different product names assigned to features with the gene name ‘int’ across 17 different RefSeq entries for Salmonella species
| Gene name | Product name | Accession |
|---|---|---|
| int | bacteriophage integrase | NC_003198, NC_004631, NC_015761 |
| int | Gifsy-1 prophage Int | NC_006905 |
| int | hypothetical protein | NC_006905 |
| int | Integrase | NC_003198, NC_004631, NC_006511, NC_012125 |
| int | integrase (fragment) | NC_003198 |
| int | phage integrase family site specific recombinase | NC_006905 |
| int | putative cytoplasmic protein | NC_006905 |
| Int | Putative integrase | NC_003384 |
| int | putative integrase protein | NC_006905 |
| int | putative P4-type integrase | NC_006905 |
| int | putative phage integrase protein | NC_006905 |
| int | site-specific recombinase, phage integrase family | NC_012125 |
Hypothetical proteins
The term ‘hypothetical protein’ often refers to a gene that has been predicted by software but which finds no homolog of known function in the databases, and which has no known functional domain. There are currently 53 035 proteins whose product name contains both words in UniProt (search term: ‘name:hypothetical AND name:protein’) and there are a further 5 178 212 proteins in UniProt that contain the words ‘uncharacterized’ and ‘protein’ (search term: ‘name:uncharacterized AND name:protein’). These may be real genes with no known function or they may be artifacts of the gene prediction process.
Many bacterial genes of unknown function are assigned y-gene names based on their orthologous location in E. coli K-12 [52]. The letters denote the location in terms of minutes around a circular genome. This gene annotation has propagated throughout many strains and species of bacteria, losing the relevance and context of its name as the genes are not all in the same relative location to the original annotation in E. coli K-12. For example the yabF gene has a known function, ‘glutathione-regulated potassium-eflux system ancillary protein’. The gene name yabF is completely meaningless in all genomes other than the original and actually has a synonym kefF. With that in mind annotators should use more informative gene names as a preference, choosing alternative gene names over the original y-gene annotation.
Often there are features which are only orthologous to other hypothetical features and do not contain any domains. These could either be regions with no functionality, a relic of the feature prediction software or the domains present have not been discovered yet. Whether or not to include them is often a decision made by the annotation team and varies between groups. Thus, many artifactual ‘hypothetical proteins’ may be annotated, published and disseminated into the public databases, reinforcing the annotator’s belief that their new gene predictions do indeed have homologs in other species. It would be more informative to actually state in the annotation a score for each feature. This will allow users to make informed assessments of the features and programmers to easily parse genomes to handle hypothetical proteins based on their quality of hits. Gilks, et al. [12] discuss the possibility of assigning scores based on the source of annotation.
There are arguments for and against keeping these proteins in the annotation. If they are indeed a misannotation by the gene prediction software they should be removed as they will perpetuate through secondary and tertiary databases as a recognized protein awaiting functional discovery. Searching for conserved domains or motifs in databases such as Pfam or InterPro can give an indication of whether a hypothetical protein is functional but this has pitfalls too. The fact that a protein has a domain hit doesn’t necessarily convey its function. Pfam [8], for example, contains over 3000 ‘domains of unknown function’, or DUFs, representing over 20% of known families [53] and as more novel genomes are sequenced the number of new DUFs will increase. A hit to a DUF does not inform us of a feature’s function, but as they are areas of high conservation they indicate a potential region of biological interest.
Through computational methods alone there are no means to conclusively determine whether a genomic region is functional. With that in mind conserved features of unknown function should be kept because in the future they may be recognized as a true region of interest; however, they should be annotated differently to discriminate them from features with stronger evidence. Evidence tags are available but they are often not present, and are not a prerequisite for submission to GenBank or Embl. Evidence qualifiers such as how the feature was predicted (e.g. glimmer, blast, homology) and what entries it hits in a given database provide a clear audit trail for anyone who wants to assess the quality of a particular annotation. The type of data source used, that is, whether it is manually curated or automated should be stated, providing the user with a clear method of judging the annotation. As experimental data becomes more ubiquitous evidence tags should play a larger role in annotation.
Experimental methods such as RNA-Seq [54] and Signature Tagged Mutagenesis (STM) [55] may help to identify regions of functionality. RNA-Seq data can help delineate and quantify areas of transcription, and overlaying this expression data on the genome may help biologists to identify pseudogenes and the true locations of features. STM can help identify the function of genes by monitoring the phenotype of single-gene mutants.
The most important point is that one’s annotation is only ever as good as the reference data sources. In terms of publicly available genome sequences the quality is varied. It is worth actually looking at the annotation and assessing the quality. Choosing a genome because it is the closest relative will give the most homologous features but might not give the best quality annotation.
Combining additional data with the original annotation gives scientists a new way of viewing the genome. Experimental data could be able to solve the eutM/eutN problem described above; for example, RNA-Seq data would show which areas of the genome are actively transcribed and STM may indicate whether knocking out either of the genes alters the phenotype of the mutant.
Distinguishing orthologs from paralogs
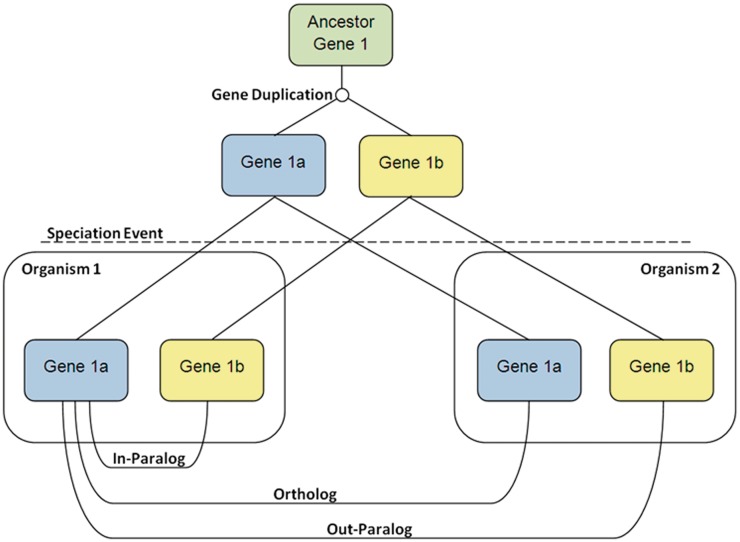
The definition of orthologous and paralogous genes is of great importance when annotating novel genomes. Whereas ‘homology’ refers to genes that simply share a common origin, ‘orthology’ refers to genes that arise by speciation and ‘paralogy’ refers to genes that arise by duplication. Figure 4 shows some of the processes that can lead to, and define, orthologs and paralogs. Beginning with a single ancestral, a gene duplication event occurs to create two paralogous genes. After a speciation event, there are two different organisms that both contain the paralogous genes from the gene duplication event. Gene 1a in Organism 1 has three homologs after the speciation event. Gene 1a in Organism 1 and Gene 1a in Organism 2 are orthologs as they have only been separated by the speciation event. Gene 1a in Organism 1 and Gene 1b in Organism 1 are in-paralogs, as they have only been separated by the gene duplication event. Finally, Gene 1a in Organism 1 and Gene 1b in Organism 2 are out-paralogs, as they have been separated by the gene duplication and the speciation event.
Figure 4:
A diagram displaying the processes that can lead to, and define, orthologs and paralogs. Gene duplication and speciation events create complex evolutionary relationships between genes.
These processes are not only crucial in defining evolutionary relationships, but also functional relationships, as orthologs tend to retain similar functions, whereas paralogs tend to diverge over time to perform different functions (reviewed in ref. [56]). Therefore, when transferring functional annotation from a sequenced genome to a novel genome, it is essential that orthologs are accurately defined. There are several computational approaches which can be used to accurately define orthologs (reviewed in ref. [57]). Phylogenetic tree-based approaches attempt to reconstruct the evolutionary relationship between gene sequences and thus define orthologs and paralogs; however, it may be impractical to construct a phylogenetic tree for every gene in a newly sequence genome. An alternative is the ‘bidirectional’ or ‘reciprocal’ best-hit approach [58], usually determined by comparing the top-ranking matches found by a search algorithm such as BLAST or FASTA [18, 19]. Gene Synteny, the conservation of local gene order, can also help distinguish orthologs from paralogs in closely related genomes. However, it is important to note that a number of processes can lead to the breakdown of absolute gene synteny, resulting in genuine orthologs having a different gene order. These processes include gene duplication or fusion events, local rearrangements (insertions/deletions) and translocations. It is important that we model these processes to allow the correct identification of orthologs in complex cases, and the MaGe [24] system attempts to do this. Finally, it has been observed that orthologs exhibit a greater level of protein domain architecture conservation than paralogs [59]. In practice, it may be essential to use a combination of approaches, and several software applications exist [57].
THE RULES OF THE SEQUENCING DATABASE
Many scientists go through the process of annotation with the final aim of submitting to a genome database such as GenBank or EMBL. In order to realize this goal there are many rules which need to be followed [9, 10] and often validation software is provided to verify one’s annotation. These rules are imposed to ensure a better standard of genome annotation, however, they do mean that often the output of an automatic annotation pipeline must be manually checked and altered prior to publication. Many of the issues described in the ‘Limitations of the Annotation Process’ section may be identified as potential problems and the submitter is provided with long lists of features that represent these. They must be checked, and either altered or justified. In addition to those mentioned above, there are others described below.
CDS nomenclature
There are many words which may be unacceptable in protein names, such as ‘binding’, ‘domain’, ‘like’, ‘motif’, ‘gene’ and ‘homolog’. Submitters may be encouraged to change these: for example ‘bacteriophage replication gene’ can be changed to ‘bacteriophage replication protein’ and ‘peptidyl-tRNA hydrolase domain protein’ can be changed to ‘peptidyl-tRNA hydrolase protein’; a note may be added to state that the feature contains the aforementioned domain. These rules add complications if the submitter wants to fully automate the process of annotation. As a rule of thumb, if a predicted coding region has homologs in SwissProt these are the best protein names to transfer across and running the validation software after using SwissProt initially can greatly reduce the number of suspect names. As an aside, ‘probable’ and ‘predicted’ are not flagged up by the validation software but ‘putative’ is the preferred alternative.
Some CDSs have the same protein name as the protein next to them, which can be the sign of either a disrupted gene or a valid gene duplication event. It can also be because the protein name is very general such as ‘hypothetical protein’ or ‘inner membrane protein’. These features may be flagged up by the validation software and, if they are not pseudogenes, need a note stating that they overlap a CDS with the same protein name.
CDS gene names that appear more than once in a genome and have different proteins names to one another (e.g. Table 1) may also be identified as potential errors. These may be brought to the submitter’s attention who often has to use their discretion and knowledge to assign gene names correctly. This can be as simple as performing a similarity search and seeing which gene names are associated with the hits.
Problems with coding regions
The NCBI validation software flags up all instances where a coding region completely contains another coding region on the opposite strand. The submitter is asked to check these coding regions and decide whether these are true features. If the coding region only hits hypothetical proteins and doesn’t contain any domains, it may be either removed or demoted to a miscellaneous feature.
FUTURE
Gold standard genomes
RefSeq is one attempt to standardize and improve the quality of genome annotation; however, as we have shown, problems persist. With the implementation of stricter rules for submission we should see an increase in annotation quality. While genomes of varying quality are available there should be a means for scientists to see the quality of any given annotation. Evidence qualifiers such as how the feature was predicted and what entries in a given database the feature sequence hit, including the database version and date, would provide a clear audit trail for anyone who wants to assess the quality of a particular annotation. The type of data source used, that is, whether it is manually curated or automatically generated should also be stated, providing the user with a clear method of judging the annotation.
Out of the 1851 publicly available completed bacterial genomes 102 have a version number of 0.2 or higher [60]. This means that the submitting group have revisited the original sequence and changed it. The fact that the sequences have been changed is indicative of a higher quality sequence. This, however, does not reflect the quality of the annotation. It is possible to look at the revision history of genomes within GenBank, this will give users an idea of changes on a genome by genome basis, no small feat when there are 1851 genomes available. In the literature there have been several papers which have revisited and reannotated genomes, these include strains of E. coli, Campylobacter jejuni and Mycobacterium tuberculosis [61–63]. In terms of what is currently available these genomes are likely to be the closest to realizing ‘gold standard genome annotation’.
Janssen, et al. [11] calculated the number of publications per gene for all completed genome to calculate a Species Knowledge Index (SKI) for each genome. They showed that, in bacteria, there is a pronounced bias toward certain organisms namely E. coli, Pseudomona aeruginosa and Bacillus subtilis. With this in mind perhaps there should be a focus to annotate genomes with a high SKI to the highest level possible as there is such an abundance of experimental data available. These can then be used as gold standard genomes for annotations of other species.
As we learn more about genes and protein function it becomes clear that a simple protein name is inadequate. Some proteins are multi-functional, performing different tasks depending on the context it is expressed in. We can say that a protein has a one-to-many relationship with function, meaning that assigning a protein name based on the first function associated with it can be misleading and inaccurate. The Gene Ontology (GO) may provide a more flexible way of describing a range of functions explicitly and concisely, and GO annotations natively include evidence qualifiers. However, GO terms are not frequently included as part of the initial annotation of bacterial genomes. The EBI offer UniProtKB-GOA Proteome Sets [64], GO annotations for all completely sequenced genomes in the public domain, however, these are not included with or clearly linked to the original genome submission. The development and use of GO annotations is encouraged and these should be included in genome annotation efforts.
Improving automated annotation
The pipelines currently on offer do not take many of the pitfalls outlined above into account, meaning that a lot of manual effort is required to correct errors and inconsistencies. It is easy to imagine adjustments to current pipelines that take into account certain aspects (e.g. common spelling mistakes) but not others (e.g. correctly interpreting pseudogenes). Realistically, completely removing the manual stage of annotation would be imprudent, however, improving current automated pipelines may greatly reduce the time spent manually checking the annotation.
New data types
There have been a flood of new genome-wide data types in the post-genomic era, for example microarray and RNA-Seq data, many of which can assist with genome annotation. However, these are often large, unwieldy, come in a variety of different formats and can be hard to integrate with one another. Allowing scientists to visualize this data alongside genome annotation can be hugely powerful [65]; however, genome annotation is often kept in specific flat file formats where integrating non-text data is virtually impossible. Secondary and tertiary databases may include additional data alongside the original genome annotation [20], but these ‘data warehouse’ approaches employ copies of the original data which can become out-of-date and out-of-synch with the original data. The advent of bioinformatics web services [66] may allow new systems that query data live over the internet, ensuring the latest data is displayed.
CONCLUSION
Advances in sequencing technologies are allowing researchers to sequence microbial genomes at a huge rate. It is becoming harder to devote time to manually annotate these genomes, leading to a rise in automatic annotation pipelines. However, due to a range of problems, the output of these automatic annotation pipelines is unsuitable for publication. Some changes can be made to improve this output; however, it is difficult to envisage an end to manual checking and curation.
Additional data from post-genomics experiments can help improve genome annotation; however, a line has to be drawn regarding what data should be included in the annotation and what should be in separate databases. Tools and services need to be developed which offer scientists a means of viewing genome annotation augmented with other experimental data. This will empower the user to make meaningful judgments on the quality of annotation and the relevance of a particular region to their research.
For the foreseeable future bacterial annotation requires both automated and manual steps. Offering users a measure of quality for the whole genome and individual genes will allow user to make an informed choice regarding reference genomes and transferring annotation between genomes. Using GO terms would improve protein description and reduce syntactic errors.
Key Points.
Advances in sequencing technology now allow modern researchers to rapidly sequence multiple bacterial genomes.
Automatic annotation pipelines that work via comparison to a reference database can introduce and propagate errors.
Manual checking and curation of annotation is essential to maintain a high quality.
Additional data-sources from post-genomic experiments can assist in the annotation process.
FUNDING
This work was funded by an Institute Strategic Programme grant awarded to The Roslin Institute by the Biotechnology and Biological Sciences Research Council (BBSRC), and by a studentship funded by the Institute for Animal Health.
Acknowledgements
We would like to acknowledge the help, assistance and advice provided by staff at the NCBI and EBI during genome submission.
Biographies
Emily Richardson is a PhD student at The Roslin Institute, University of Edinburgh. Her project focuses on the use of next-generation bioinformatic and informatic tools for the multi-dimensional annotation of bacterial genomes.
Mick Watson is Director of ARK-Genomics, a genomics facility at the The Roslin Institute, University of Edinburgh. His research interests are the bioinformatics and functional genomics of farmed animal species and their pathogens.
References
- 1.MacLean D, Jones JD, Studholme DJ. Application of ‘next-generation' sequencing technologies to microbial genetics. Nat Rev Microbiol. 2009;7:287–96. doi: 10.1038/nrmicro2122. [DOI] [PubMed] [Google Scholar]
- 2.Stothard P, Wishart DS. Automated bacterial genome analysis and annotation. Curr Opin Microbiol. 2006;9:505–10. doi: 10.1016/j.mib.2006.08.002. [DOI] [PubMed] [Google Scholar]
- 3.Metzker ML. Sequencing technologies - the next generation. Nat Rev Genet. 2010;11:31–46. doi: 10.1038/nrg2626. [DOI] [PubMed] [Google Scholar]
- 4.Attwood TK, Bradley P, Flower DR, et al. PRINTS and its automatic supplement, prePRINTS. Nucleic Acids Res. 2003;31:400–2. doi: 10.1093/nar/gkg030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Suzek BE, Ermolaeva MD, Schreiber M, et al. A probabilistic method for identifying start codons in bacterial genomes. Bioinformatics. 2001;17:1123–30. doi: 10.1093/bioinformatics/17.12.1123. [DOI] [PubMed] [Google Scholar]
- 6.Ermolaeva MD, Khalak HG, White O, et al. Prediction of transcription terminators in bacterial genomes. J Mol Biol. 2000;301:27–33. doi: 10.1006/jmbi.2000.3836. [DOI] [PubMed] [Google Scholar]
- 7.Sigrist CJ, Cerutti L, de Castro E, et al. PROSITE, a protein domain database for functional characterization and annotation. Nucleic Acids Res. 2010;38:D161–166. doi: 10.1093/nar/gkp885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Finn RD, Mistry J, Tate J, et al. The Pfam protein families database. Nucleic Acids Res. 2010;38:D211–22. doi: 10.1093/nar/gkp985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.The Bacterial Genome Submission Guide. http://www.ncbi.nlm.nih.gov/genbank/genomesubmit.html (25 November 2011, date last accessed) [Google Scholar]
- 10.Genome Project Submission Account guidelines. http://www.ebi.ac.uk/embl/Submission/genomes.html (25 November 2011, date last accessed) [Google Scholar]
- 11.Janssen P, Goldovsky L, Kunin V, et al. Genome coverage, literally speaking. The challenge of annotating 200 genomes with 4 million publications. EMBO Rep. 2005;6:397–9. doi: 10.1038/sj.embor.7400412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gilks WR, Audit B, de Angelis D, et al. Percolation of annotation errors through hierarchically structured protein sequence databases. Math Biosci. 2005;193:223–34. doi: 10.1016/j.mbs.2004.08.001. [DOI] [PubMed] [Google Scholar]
- 13.Delcher AL, Harmon D, Kasif S, et al. Improved microbial gene identification with GLIMMER. Nucleic Acids Res. 1999;27:4636–41. doi: 10.1093/nar/27.23.4636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Do JH, Choi DK. Computational approaches to gene prediction. J Microbiol. 2006;44:137–44. [PubMed] [Google Scholar]
- 15.Frishman D, Mironov A, Mewes HW, et al. Combining diverse evidence for gene recognition in completely sequenced bacterial genomes. Nucleic Acids Res. 1998;26:2941–7. doi: 10.1093/nar/26.12.2941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Badger JH, Olsen GJ. CRITICA: coding region identification tool invoking comparative analysis. Mol Biol Evol. 1999;16:512–24. doi: 10.1093/oxfordjournals.molbev.a026133. [DOI] [PubMed] [Google Scholar]
- 17.Ongoing and future developments at the Universal Protein Resource. Nucleic Acids Res. 2011;39:D214–9. doi: 10.1093/nar/gkq1020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pearson WR. Rapid and sensitive sequence comparison with FASTP and FASTA. Methods Enzymol. 1990;183:63–98. doi: 10.1016/0076-6879(90)83007-v. [DOI] [PubMed] [Google Scholar]
- 19.Altschul SF, Gish W, Miller W, et al. Basic local alignment search tool. J Mol Biol. 1990;215:403–10. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 20.Lowe TM, Eddy SR. tRNAscan-SE: a program for improved detection of transfer RNA genes in genomic sequence. Nucleic Acids Res. 1997;25:955–64. doi: 10.1093/nar/25.5.955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Aziz RK, Bartels D, Best AA, et al. The RAST Server: rapid annotations using subsystems technology. BMC Genomics. 2008;9:75. doi: 10.1186/1471-2164-9-75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Van Domselaar GH, Stothard P, Shrivastava S, et al. BASys: a web server for automated bacterial genome annotation. Nucleic Acids Res. 2005;33:W455–9. doi: 10.1093/nar/gki593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lee D, Seo H, Park C, et al. WeGAS: a web-based microbial genome annotation system. Biosci Biotechnol Biochem. 2009;73:213–6. doi: 10.1271/bbb.80567. [DOI] [PubMed] [Google Scholar]
- 24.Vallenet D, Labarre L, Rouy Z, et al. MaGe: a microbial genome annotation system supported by synteny results. Nucleic Acids Res. 2006;34:53–65. doi: 10.1093/nar/gkj406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kumar K, Desai V, Cheng L, et al. AGeS: a software system for microbial genome sequence annotation. PLoS One. 2011;6:e17469. doi: 10.1371/journal.pone.0017469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Stewart AC, Osborne B, Read TD. DIYA: a bacterial annotation pipeline for any genomics lab. Bioinform. 2009;25:962–3. doi: 10.1093/bioinformatics/btp097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yu C, Zavaljevski N, Desai V, et al. The development of PIPA: an integrated and automated pipeline for genome-wide protein function annotation. BMC Bioinform. 2008;9:52. doi: 10.1186/1471-2105-9-52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Cruveiller S, Le Saux J, Vallenet D, et al. MICheck: a web tool for fast checking of syntactic annotations of bacterial genomes. Nucleic Acids Res. 2005;33:W471–9. doi: 10.1093/nar/gki498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Webin EMBL-EBI annotation features and qualifiers. http://www.ebi.ac.uk/ena/WebFeat/ (25 November 2011, date last accessed) [Google Scholar]
- 30.Mulder N, Apweiler R. InterPro and InterProScan: tools for protein sequence classification and comparison. Methods Mol Biol. 2007;396:59–70. doi: 10.1007/978-1-59745-515-2_5. [DOI] [PubMed] [Google Scholar]
- 31.Hacker J, Blum-Oehler G, Mühldorfer I, et al. Pathogenicity islands of virulent bacteria: structure, function and impact on microbial evolution. Mol Microbiol. 1997;23:1089–97. doi: 10.1046/j.1365-2958.1997.3101672.x. [DOI] [PubMed] [Google Scholar]
- 32.Hsiao W, Wan I, Jones SJ, et al. IslandPath: aiding detection of genomic islands in prokaryotes. Bioinform. 2003;19:418–20. doi: 10.1093/bioinformatics/btg004. [DOI] [PubMed] [Google Scholar]
- 33.Waack S, Keller O, Asper R, et al. Score-based prediction of genomic islands in prokaryotic genomes using hidden Markov models. BMC Bioinform. 2006;7:142. doi: 10.1186/1471-2105-7-142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Langille M, Hsiao W, Brinkman F. Evaluation of genomic island predictors using a comparative genomics approach. BMC Bioinform. 2008;9:329. doi: 10.1186/1471-2105-9-329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Barrangou R, Fremaux C, Deveau H, et al. CRISPR provides acquired resistance against viruses in prokaryotes. Science. 2007;315:1709–12. doi: 10.1126/science.1138140. [DOI] [PubMed] [Google Scholar]
- 36.Kassai-Jáger E, Ortutay C, Tóth G, et al. Distribution and evolution of short tandem repeats in closely related bacterial genomes. Gene. 2008;410:18–25. doi: 10.1016/j.gene.2007.11.006. [DOI] [PubMed] [Google Scholar]
- 37.Bland C, Ramsey TL, Sabree F, et al. CRISPR recognition tool (CRT): a tool for automatic detection of clustered regularly interspaced palindromic repeats. BMC Bioinform. 2007;8:209. doi: 10.1186/1471-2105-8-209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Grissa I, Vergnaud G, Pourcel C, et al. CRISPRFinder: a web tool to identify clustered regularly interspaced short palindromic repeats. Nucleic Acids Res. 2007;35:W52–7. doi: 10.1093/nar/gkm360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sreenu VB, Alevoor V, Nagaraju J, et al. MICdb: database of prokaryotic microsatellites. Nucleic Acids Res. 2003;31:106–8. doi: 10.1093/nar/gkg002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lu Z, Szafron D, Greiner R, et al. Predicting subcellular localization of proteins using machine-learned classifiers. Bioinformatics. 2004;20:547–56. doi: 10.1093/bioinformatics/btg447. [DOI] [PubMed] [Google Scholar]
- 41.Hua S, Sun Z. Support vector machine approach for protein subcellular localization prediction. Bioinform. 2001;17:721–8. doi: 10.1093/bioinformatics/17.8.721. [DOI] [PubMed] [Google Scholar]
- 42.Yu CS, Lin CJ, Hwang JK. Predicting subcellular localization of proteins for Gram-negative bacteria by support vector machines based on n-peptide compositions. Protein Sci. 2004;13:1402–6. doi: 10.1110/ps.03479604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wang J, Sung W-K, Krishnan A, et al. Protein subcellular localization prediction for Gram-negative bacteria using amino acid subalphabets and a combination of multiple support vector machines. BMC Bioinform. 2005;6:174. doi: 10.1186/1471-2105-6-174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Gardy JL, Laird MR, Chen F, et al. PSORTb v.2.0: Expanded prediction of bacterial protein subcellular localization and insights gained from comparative proteome analysis. Bioinformatics. 2005;21:617–23. doi: 10.1093/bioinformatics/bti057. [DOI] [PubMed] [Google Scholar]
- 45.Gardy JL, Brinkman FSL. Methods for predicting bacterial protein subcellular localization. Nat Rev Micro. 2006;4:741–51. doi: 10.1038/nrmicro1494. [DOI] [PubMed] [Google Scholar]
- 46.Kanehisa M, Goto S. KEGG: kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 2000;28:27–30. doi: 10.1093/nar/28.1.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Snyder EE, Kampanya N, Lu J, et al. PATRIC: the VBI PathoSystems Resource Integration Center. Nucleic Acids Res. 2007;35:D401–6. doi: 10.1093/nar/gkl858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kummerfeld SK, Teichmann SA. Relative rates of gene fusion and fission in multi-domain proteins. Trends Genet. 2005;21:25–30. doi: 10.1016/j.tig.2004.11.007. [DOI] [PubMed] [Google Scholar]
- 49.Hollich V, Sonnhammer EL. PfamAlyzer: domain-centric homology search. Bioinformatics. 2007;23:3382–3. doi: 10.1093/bioinformatics/btm521. [DOI] [PubMed] [Google Scholar]
- 50.Lucene – java based search engine. http://lucene.apache.org/java/docs/index.html (25 November 2011, date last accessed) [Google Scholar]
- 51.PHP class – ‘did you mean?'. http://www.phpclasses.org/package/4569-PHP-Get-spelling-correction-suggestions-from-Google.html (25 November 2011, date last accessed) [Google Scholar]
- 52.Rudd KE. Linkage map of Escherichia coli K-12, edition 10: the physical map. Microbiol Mol Biol Rev. 1998;62:985–1019. doi: 10.1128/mmbr.62.3.985-1019.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Bateman A, Coggill P, Finn RD. DUFs: families in search of function. Acta Crystallogr Sect F Struct Biol Cryst Commun. 2010;66:1148–52. doi: 10.1107/S1744309110001685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wang Z, Gerstein M, Snyder M. RNA-Seq: a revolutionary tool for transcriptomics. Nat Rev Genet. 2009;10:57–63. doi: 10.1038/nrg2484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Saenz HL, Dehio C. Signature-tagged mutagenesis: technical advances in a negative selection method for virulence gene identification. Curr Opin Microbiol. 2005;8:612–9. doi: 10.1016/j.mib.2005.08.013. [DOI] [PubMed] [Google Scholar]
- 56.Koonin EV. Orthologs, paralogs, and evolutionary genomics. Annu Rev Genet. 2005;39:309–38. doi: 10.1146/annurev.genet.39.073003.114725. [DOI] [PubMed] [Google Scholar]
- 57.Kristensen DM, Wolf YI, Mushegian AR, et al. Computational methods for Gene Orthology inference. Brief Bioinform. 2011;12:379–91. doi: 10.1093/bib/bbr030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Overbeek R, Fonstein M, D'Souza M, et al. The use of gene clusters to infer functional coupling. Proc Natl Acad Sci USA. 1999;96:2896–901. doi: 10.1073/pnas.96.6.2896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Forslund K, Pekkari I, Sonnhammer EL. Domain architecture conservation in orthologs. BMC Bioinform. 2011;12:326. doi: 10.1186/1471-2105-12-326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.NCBI Complete Microbial Genomes. www.ncbi.nlm.nih.gov/genomes/lproks.cgi (25 November 2011, date last accessed) [Google Scholar]
- 61.Luo C, Hu GQ, Zhu H. Genome reannotation of Escherichia coli CFT073 with new insights into virulence. BMC Genomics. 2009;10:552. doi: 10.1186/1471-2164-10-552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Gundogdu O, Bentley SD, Holden MT, et al. Re-annotation and re-analysis of the Campylobacter jejuni NCTC11168 genome sequence. BMC Genomics. 2007;8:162. doi: 10.1186/1471-2164-8-162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Camus JC, Pryor MJ, Medigue C, et al. Re-annotation of the genome sequence of Mycobacterium tuberculosis H37Rv. Microbiology. 2002;148:2967–73. doi: 10.1099/00221287-148-10-2967. [DOI] [PubMed] [Google Scholar]
- 64.Barrell D, Dimmer E, Huntley RP, et al. The GOA database in 2009–an integrated Gene Ontology Annotation resource. Nucleic Acids Res. 2009;37:D396–403. doi: 10.1093/nar/gkn803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Watson M. ProGenExpress: visualization of quantitative data on prokaryotic genomes. BMC Bioinform. 2005;6:98. doi: 10.1186/1471-2105-6-98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Bhagat J, Tanoh F, Nzuobontane E, et al. BioCatalogue: a universal catalogue of web services for the life sciences. Nucleic Acids Res. 2010;38:W689–94. doi: 10.1093/nar/gkq394. [DOI] [PMC free article] [PubMed] [Google Scholar]