Abstract
Various neurodegenerative diseases are associated with aberrant gene expression. We recently identified a novel class of pimelic o-aminobenzamide histone deacetylase (HDAC) inhibitors that show promise as therapeutics in the neurodegenerative diseases Friedreich’s ataxia (FRDA) and Huntington’s disease (HD). Here, we describe the various techniques used in our laboratories to dissect mechanisms of gene silencing in FRDA and HD, and to test our HDAC inhibitors for their ability to reverse changes in gene expression in cellular models.
Keywords: Histone deacetylase, Epigenetics, Friedreich’s ataxia, Huntington’s disease, Gene activation, Pimelic o-aminobenzamide HDAC inhibitor, qRT-PCR, Chromatin immunoprecipitation
1. Introduction
Histone deacetylase (HDAC) inhibitors have received considerable attention as potential therapeutics for cancer (1) and for a variety of neurological and neurodegenerative diseases (2). We recently described a series of pimelic o-aminobenzamide HDAC inhibitors (HDACi) that reverse heterochromatin-mediated silencing of the frataxin (FXN) gene in the neurodegenerative disease Friedreich’s ataxia (FRDA) (3-5) and also show efficacy in a mouse model for Huntington’s disease (HD) (6). FRDA is caused by the expansion of the simple triplet repeat DNA sequence GAA•TTC within intron 1 of the FXN gene, encoding the essential mitochondrial protein frataxin. Repeats over a threshold level of ~70 induce heterochromatin formation (3), and concomitant gene silencing, resulting in decreased levels of frataxin protein in affected individuals. Importantly for therapeutic development, the pimelic o-aminobenzamides cross the blood brain barrier, cause global increases in histone acetylation in cells and in the mouse brain, and show good tolerance in murine models of disease (5, 6). These molecules also directly affect the histone acetylation status of FXN gene chromatin in FRDA patient cells and in the mouse brain, increasing acetylation at particular lysine residues on histones H3 and H4, and increase FXN gene expression in the brain and heart in a mouse model for FRDA (5). Strikingly, gene expression microarray analysis indicates that most of the differentially expressed genes in FRDA mice revert toward wild-type levels on treatment with the pimelic o-aminobenzamide HDAC inhibitor (5). Similar results have been obtained in a mouse model for HD, where one of these compounds ameliorated the disease phenotype and reversed many of the transcriptional abnormalities found in the brain of R6/2 HD mice (6). We find that members of the pimelic o-aminobenzamide HDAC inhibitor family only increase FXN gene expression in FRDA patient cells and in the FRDA mouse model and are without effect on wild-type human or mouse FXN alleles (refs. 3-5 and see below), indicating that these molecule reverse the effects of the GAA•TTC repeat on the transcriptional status of the FXN gene. We reported that the pimelic o-aminobenzamides are inhibitors of class I HDAC enzymes, with a marked preference for HDAC3 (7), and that molecules with selectivity for HDACs 1 and 2 fail to increase FXN gene expression in patient cells (8).
Here, we present the methods used in our laboratories to interrogate the chromatin changes associated with dysregulated genes in cellular models for neurodegenerative diseases, quantitative reverse transcriptase PCR methods for estimation of mRNA levels for these genes and methods for synthesis and characterization of HDAC inhibitors. We focus on Friedreich’s ataxia, but the methods employed can be extended to other neurological and neurodegenerative diseases, where transcriptional dysfunction is at the core of the disease etiology.
2. Materials
2.1. Synthesis of Pimelic o-Aminobenzamide HDACi and Activity Assays
-
Detailed methods for the synthesis pimelic o-aminobenzamide HDACi have been published (3-5, 8) and two such compounds are commercially available from Calbiochem/EMD Biosciences: HDACi 4b (N1-(2-aminophenyl)-N7-phenylheptanediamide, sold as Histone Deacetylase Inhibitor IV) and HDACi 106 (N1-(2-aminophenyl)-N7-p-tolylheptanediamide, sold as Histone Deacetylase Inhibitor VII).
Compound dilution: Test compounds at a starting concentration of 9 mM (in DMSO) are first diluted in DMSO at a 1/3 ratio to give 11 concentration points (9 mM, 3 mM, 1 mM, 0.33 mM, 0.11 mM, 0.037 mM, 0.012 mM, 4.11 μM, 1.37 μM, 0.47 μM, 0.15 μM), and a control with no compound, then the whole series (the 12 concentration points in DMSO) is diluted in HDAC buffer at a 1/20 ratio to give 12 new concentrations. 10 μL inhibitor is used per assay, and the assay volume is 25 μL, giving final compound concentrations of 180 μM, 60 μM, 20 μM, 6.7 μM, 2.2 μM, 0.74 μM, 0.25 μM, 82.3 nM, 27.4 nM, 9.1 nM, 3.0 nM, and 0 nM, respectively.
HDAC buffer: 50 mM Tris–Cl, pH 8.0, 137 mM NaCl, 2.7 mM KCl, 1 mM MgCl2.
Fluor-de-Lys™ deacetylase substrate: 50 mM in DMSO (Enzo Life Sciences, Plymouth Meeting, PA).
BSA: Stock (10 mg/mL), prepared in HDAC buffer.
HDAC enzymes: BPS Bioscience, San Diego, CA.
Trypsin: Stock (10 mg/mL), prepared in HDAC buffer.
96-well plates: Greiner Bio-one, black, U-shape.
Lys-C from EMD: 500 μL HDAC buffer is added to Lys-C vial to provide a stock solution.
Equipment: 96-well plate reader, such as Tecan Infinite M200; centrifuge, such as Eppendorf 5810 with rotor adaptor for 96-well plates.
2.2. Growth of Lymphoblastoid Cells in Culture
Cell lines GM15850 and GM15851, from an FRDA affected male and from an unaffected male sibling, respectively (Coriell Cell Repositories, Camden, New Jersey).
Lymphocyte growth medium: RPMI Medium 1640+ GlutaMAX-I supplemented with 15% fetal bovine serum and antimycotic, antibiotic.
Hank’s balanced salt solution (HBSS).
2.3. Isolation of Peripheral Blood Mononuclear Cells
Lymphocyte growth medium.
HBSS.
Ficoll-Paque PLUS (GE Healthcare, Uppsala, Sweden).
2.4. Incubation with HDAC Inhibitors
DMSO and HDACi of appropriate dilutions in DMSO.
CellTiter 96® AQueous Non-Radioactive Cell Proliferation Assay (MTS, Promega, Madison, WI).
2.5. In Cell Deacetylase Assay
Phosphate buffered saline (PBS), pH 7.4.
HDAC buffer: 50 mM Tris–Cl, pH 8.0, 137 mM NaCl, 2.7 mM KCl, 1 mM MgCl2.
Deacetylase lysis buffer: 50 mM Tris–Cl, pH 8.0, 137 mM NaCl, 2.7 mM KCl, 1 mM MgCl2, 1% IGEPAL.
BCA Protein Assay Kit (Thermo Scientific).
96-well, flat-bottom, black opaque microplate.
Fluor-de-Lys™ deacetylase substrate: 50 mM in DMSO.
10 mg/mL BSA, prepared in HDAC buffer.
10 mg/mL trypsin, prepared in HDAC buffer.
Equipment: 96-well plate reader; centrifuge, such as Sorvall RT6000B; Vortex mixer.
2.6. qRT-PCR to Detect FXN mRNA Expression
RNeasy Plus Mini Kit (QIAgen Hilden, Germany).
qScript™One-Step SYBR® Green qRT-PCR Kit (Quanta Biosciences, Gaithersburg, MD).
Primer sequences for the FXN gene, 5′-CAGAGGAAACGC-TGGACTCT-3′ and 5′-AGCCAGA TTTGCTTGTTTGG-3′.
Equipment: MJ Research Chromo4 thermal cycler.
2.7. Western Blotting for Detection of Frataxin and Acetylated Histones
RIPA buffer: 50 mM Tris–HCl pH 7.5, 150 mM NaCl, 1 mM EDTA, 1% Triton X-100, 1% sodium deoxycholate, 0.1% SDS, protease inhibitors.
Equipment: sonicator, such as Branson sonifier 150.
BCA™ Protein Assay Kit (Thermo Scientific, Rockford, IL).
4–12% Bis-tris polyacrylamide gel.
MES running buffer.
Nitrocellulose membrane.
TBS-T: 10 mM Tris–HCl pH 8, 150 mM NaCl, 0.1% Tween.
Dry non-fat milk.
- Antibodies
- Mouse anti-frataxin antibody (Invitrogen, Carlsbad, CA).
- Rabbit anti-RPL13a antibody (Cell Signaling, Beverly, MA).
- Rabbit anti-histone H3 antibody (Abcam, Cambridge, MA).
- Rabbit anti-acetylated histone H3 antibody (Millipore, Billerica, MA).
- Anti-mouse IgG HRP-linked secondary antibody.
- Anti-rabbit IgG HRP-linked secondary antibody.
HRP chemiluminescent substrate.
2.8. Chromatin Immunoprecipitation
Equipment: sonicator (Branson sonifier 150); MJ Research Chromo4 thermal cycler.
37% formaldehyde.
2 M glycine.
HBSS.
Lysis buffer: 1% SDS, 10 mM EDTA pH 8, 50 mM Tris–Cl pH 8, protease inhibitors.
Dilution buffer: 1% Triton X-100, 150 mM NaCl, 2 mM EDTA, 20 mM Tris–Cl pH 8, protease inhibitors.
Protein A agarose beads.
- Antibodies:
- Rabbit anti-histone H3 (Abcam).
- Rabbit anti-histone H3AcK9 (Millipore).
- Rabbit anti-histone H3AcK14 (Millipore).
- Rabbit anti-histone H4 (Millipore).
- Rabbit anti-histone H4AcK5 (Millipore).
- Rabbit anti-histone H4AcK8 (Millipore).
- Rabbit anti-histone H4AcK12 (Millipore).
- Rabbit anti-histone H4AcK16 (Abcam).
- Normal rabbit IgG.
Low salt wash buffer: 1% Triton X-100, 0.1% SDS, 150 mM NaCl, 2 mM EDTA pH 8, 20 mM Tris–Cl pH 8.
High salt wash buffer: 1% Triton X-100, 0.1% SDS, 500 mM NaCl, 2 mM EDTA pH 8, 20 mM Tris–Cl pH 8.
Elution buffer: 1% SDS, 100 mM NaHCO3.
20 mg/mL proteinase K.
Phenol–chloroform–isoamyl alcohol (25:24:1).
100% ethanol.
3 M sodium acetate pH 5.2.
Glycogen.
75% ethanol.
SYBR® Green PCR mix.
- Primer sequences:
- GAPDH coding sequence: 5′-CACCGTCAAGGCTGAGAACG-3′ and 5′-ATACCCAAGGGAGCCACACC-3′.
- FXN promoter: 5′-CCCCACATACCCAACTGCTG-3′ and 5′-GCCCGCCGCTTCTAAAATTC-3′.
- FXN upstream of the GAA repeats: 5′-GAAACCCAAAGAATGGCTGTG-3′ and 5′-TTCCCTCCTCGTGAAACACC-3′.
- FXN downstream of the GAA repeats: 5′-CTGGAAAAATAGGCAAGTGTGG-3′ and 5′-CAGGGGTGGAAGCCCAA TAC-3′.
3. Methods
3.1. In Vitro HDAC Assays
IC50 protocol:
5 μL of BSA, at 1 mg/mL, is added to 12 wells in a 96-well plate and 5 μL of enzyme at an appropriate dilution is added to the above 12 wells (given by BPS Biosciences for the individual HDAC enzymes).
10 μL of inhibitor at 12 serial (1/3) dilutions in HDAC buffer are added to each well.
The mixture is briefly centrifuged and incubated for 2 h at ambient temperature.
-
5 μL Fluor-de-Lys™ deacetylase substrate (100 μM) is added to the above 12 wells.
The solution is centrifuged briefly and incubated for 1 h at ambient temperature.
25 μL trypsin (1 mg/mL) is added to each well, to a final volume of 50 μL.
The mixture is centrifuged briefly and transferred to a fluorometric plate reader (excitation wavelength 360 nm, emission wavelength 460 nm). Readings are taken at 1-min intervals. A 20-min data set is used to determine the IC50 using GraphPad Prism fitting software.
Ki protocol:
10 μL of BSA (1 mg/mL) is added to eight wells in 96-well plate.
10 μL Fluor-de-Lys™ deacetylase substrate (31.25 μM, 1/1,600 dilution from 50 mM stock solution) is added to each plate.
10 μL of inhibitor at eight serial (1/2) dilutions (10–0 μM) is added to each well.
10 μL Lys-C (1/20 dilution from stock) is added to each well, the solution is centrifuged briefly and incubated for 10 min at ambient temperature.
10 μL enzyme (1/1,000 dilution from stock) is added, the solution is centrifuged briefly and the plate transferred to a fluorometric plate reader. Data are taken from 0 to 90 min to determine the Ki using the GraphPad Prism fitting software.
3.2. Growth of Lymphoblastoid Cells in Culture
Lymphoblastoid cells are grown in suspension culture in lymphocyte growth medium at 37°C in a CO2 incubator. Cells are split 1:3 every 48 h (see Note 1). To split, spin cells for 5 min at 200 × g, and resuspend them in HBSS. Spin again for 5 min at 200 × g and resuspend in lymphocyte growth medium.
3.3. Isolation of Peripheral Blood Mononuclear Cells
50 mL of donor human blood is collected in heparin tubes or EDTA tubes (see Note 2).
Distribute blood in four 50 mL tubes and add 12.5 mL of HBSS to each tube.
Underlay the diluted blood with 12.5 mL of Ficoll-Paque PLUS. Care should be taken not to mix the two solutions.
Spin at 400 × g for 40 min (see Note 3).
After centrifugation, three layers should be visible: the upper layer of plasma, the middle layer of ficoll, and the lower layer of granulocytes and erythrocytes. At the interface of ficoll and plasma, a thin white layer is also visible. This is the lymphocyte layer or “buffy coat.” Aspirate the lymphocyte layer with a plastic pipette (ensure that the whole layer is removed by aspirating a minimum amount of the ficoll layer) and transfer to a new tube.
Add at least three volumes of HBSS and centrifuge at 200 × g for 10 min.
Remove the supernatant and resuspend in 20 mL of lymphocyte growth medium.
Incubate cells at 37°C in a CO2 incubator for 4–16 h.
3.4. Incubation with HDAC Inhibitors
Dilute cells at a concentration of 1–2 × 106 cells/mL for primary lymphocytes or 3–4 × 105 cells/mL for lymphoblastoid cells. Distribute the cells in a 6-well plate, 4 mL per well.
Make four dilutions of HDACi in DMSO: 10, 5, 2.5, and 1 mM (see Note 4).
Add 4 μL of each HDAC inhibitor dilution to each well and 4 μL of DMSO to the vehicle control sample.
Incubate at 37°C in a CO2 incubator and collect cells for in cell deacetylase assay after 4 h, or RNA extraction, Western blot or chromatin immunoprecipitation (ChIP) after 24–48 h.
Use a cytotoxicity assay to determine the EC50 of each compound (see Note 5). Remove 100 μL and transfer to a 96-well plate. Use the CellTiter 96® AQueous Non-Radioactive Cell Proliferation Assay, following manufacturer’s instructions (see Note 6).
3.5. In-Cell Deacetylase Assay
Collect 2 × 106 cells by centrifugation at 400 × g for 5 min and aspirate supernatant.
Resuspend cell pellet in 5 mL PBS, centrifuge at 200 × g for 5 min, and aspirate supernatant.
Add 150 μL deacetylase lysis buffer to the pellet and mix by vortex until lysate appears homogeneous.
Centrifuge samples at 200 × g for 5 min to pellet insoluble material.
Transfer 120 μL of each supernatant to a 2 mL microcentrifuge tube and place on ice.
Remove 5 μL from each cell lysate and determine protein concentration using BCA protein assay kit following the manufacturer’s instructions.
Dilute all samples to the lowest protein concentration using cell lysis buffer (see Note 7).
Transfer 100 μL of each cell lysate to one well of a 96-well black opaque microplate.
Dilute Fluor-de-Lys™ Substrate 1:100 in HDAC buffer to make a working concentration of 500 μM.
Transfer 50 μL HDAC buffer to one well of the assay plate for a “substrate only” control.
Add 10 μL of diluted Fluor-de-Lys™ Substrate to 100 μL of each sample. Add 5 μL of diluted Fluor-de-Lys™ Substrate to the “substrate only” and control well.
Transfer 55 μL of each sample from one well of the assay plate to a second well of the plate to generate duplicate assay wells.
Incubate the assay plate at room temperature (20–23°C) for 30 min.
Dilute trypsin solution to 1 mg/mL with HDAC buffer, add 50 μL to each well of the assay plate.
Read plate in fluorometric plate reader (excitation wavelength of 360 nm and emission wavelength of 460 nm) at 2-min intervals for a total of 30 min.
Analyze fluorescent intensity at 30 min. Inspect fluorescence vs. time curves for possible anomalies, such as nonlinear behavior.
3.6. qRT-PCR to Detect FXN mRNA Expression
Isolate total RNA from 1–2 × 106 cells for primary lymphocytes or 2.5–5 × 105 cells for lymphoblastoid cells using RNeasy Plus Mini Kit, following manufacturer’s instructions (see Note 8) and measure its concentration with a spectrophotometer (see Note 9).
qRT-PCR is performed using qScript™One-Step SYBR® Green qRT-PCR Kit. Each RNA sample is analyzed in triplicate using FXN primer set (see Note 10).
Each 20 μL reaction contains: 20 ng of total RNA, 10 μL of Quanta qScript™One-Step SYBR® Green mix, 0.1 μL of reverse transcriptase, and 10 pmol of each primer.
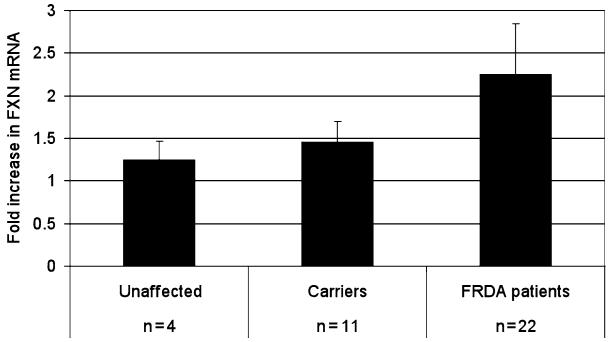
Use the following conditions for the thermocycler (see Note 11): 55°C for 10 min, 95°C for 5 min, 40 cycles of 95°C for 10 s, 55°C for 10 s, and 72°C for 30 s. After cycling is complete set the PCR machine to calculate dissociation curves from 70 to 96°C (see Note 12). Visualize the amplification curve in a logarithmic scale and set the baseline from cycle 3 to cycle 9. Set the threshold in the linear phase of amplification and record the threshold cycle (Ct) for each well. Average the Ct values for the three replicates and calculate the ΔCt for each sample (ΔCt = Ct (HDACi-treated sample) – Ct (DMSO sample)). The relative amount of FXN mRNA compared to the DMSO-treated sample is 2−ΔCt. Figure 1 provides an example of results with compound 4b (5 μM) in primary lymphocytes from normal donors, nonaffected carriers (individuals who are heterozygous for the expanded GAA repeats in FXN) and FRDA patients, after a 24 h incubation of these cells in culture. No effects on FXN mRNA levels are noted in peripheral blood mononuclear cells (PBMCs) from healthy donors, an intermediate effect is seen in PBMCs from carriers and a clear ~2- to 2.5-fold increase in FXN mRNA is noted in patient cells, showing that this compound only affects transcription of pathogenic FXN alleles and has no affect on the wild-type gene.
Fig. 1.
qRT-PCR determination of FXN mRNA levels in primary lymphocytes from unaffected individuals, carriers and Friedreich’s ataxia patients, after the treatment with HDACi 4b at 5 μM, for 24 h. The fold-increase in FXN mRNA compared to the DMSO control is shown, and the number of patient samples is shown for each group. HDACi treatment fails to show a statistically significant increase in FXN mRNA levels in PBMCs from unaffected individuals, while significant increases in FXN mRNA levels in PBMCs from FRDA patients are observed (p = 0.0036). FXN mRNA levels in this experiment were normalized to GAPDH mRNA, which did not change significantly from the DMSO control. We thank Dr. Ryan Burnett for these data (see ref. 3).
3.7. Western Blotting for Detection of Frataxin and Acetylated Histones
Collect 2 × 106 cells for primary lymphocytes or 1 × 106 cells for lymphoblastoid cells by centrifugation and wash them in HBSS.
Resuspend cells in 100 μL of RIPA buffer.
Sonicate for 5 s with a Branson sonifier at 4 W output to reduce the viscosity of the solution.
Determine protein concentration with the BCA™ Protein Assay Kit, following manufacturer’s instructions.
Separate 30 μg of total protein extract for frataxin and RPL13a detection or 5 μg of total protein extract for H3 and acetylated H3 detection, onto a 4–12% bis-tris polyacrylamide gel in MES buffer and transfer to a nitrocellulose membrane following manufacturer’s instructions.
Block membrane with 5% nonfat milk for 1 h at room temperature. For frataxin and RPL13a detection, incubate the blot overnight at 4°C with the antibody in 5% nonfat milk (1:1,000 dilution for frataxin and 1:2,000 dilution for RPL13). For H3 and acetylated H3 detection, incubate the blot 1 h at room temperature with the antibody in 5% nonfat milk (1:10,000 dilution for H3 and 1:5,000 dilution for acetylated H3).
Wash the blot four times for 5 min with TBS-T at ambient temperature.
Incubate for 1 h at room temperature with HRP-conjugated anti-mouse (for frataxin detection) or anti-rabbit (for RPL13a, H3 and acetylated H3 detection) secondary antibody.
Wash with TBS-T four times for 5 min at room temperature.
Incubate for 5 min at room temperature with HRP chemiluminescent substrate and expose to X-ray films.
Scan the film and quantify band intensity using image analysis software, such as Imagequant or imageJ.
3.8. Chromatin Immunoprecipitation
Add formaldehyde to cell media to a final concentration of 1% and incubate 10 min at ambient temperature with shaking.
Add glycine to a final concentration of 0.125 M and incubate for 5 min at ambient temperature with shaking.
Spin cells for 5 min at 200 × g and wash with HBSS.
Resuspend at 1 × 107 cell/mL in lysis buffer and incubate for 15 min on ice.
Sonicate four times for 15 s using a Brason sonifier at 9 W output, incubating the samples on ice for 1 min after each round (see Note 13).
Spin at 12,000 × g for 10 min at 4°C and collect the supernatant. This is the whole cell extract.
Preclear the cell extract by incubating with 60 μL of protein A-agarose for 1 h at 4°C.
Spin the sample for 30 s at 800 × g and transfer the supernatant to a new tube.
Use 100 μL of whole cell extract per immunoprecipitation (IP, include a sample to be incubated with normal rabbit IgG, as a negative control) and save 10 μL (label this sample as INPUT).
Dilute the 100 μL of extract with 0.9 mL of dilution buffer and add 2–4 μg of the desired histone antibody or rabbit normal IgG.
Incubate at 4°C overnight on a rotator.
The next day add 60 μL of protein A-agarose and incubate at 4°C for 2 h on a rotator.
Spin for 30 s at 800 × g to pellet the beads and remove the supernatant (see Note 14).
Add 1 mL of low salt wash buffer to the beads and rotate for 5 min at 4°C. Spin as above and remove the supernatant. Repeat two times, for a total of three low salt washes.
Perform a final wash with 1 mL of high salt wash buffer, spin as above, and remove the supernatant.
Add 150 μL of elution buffer and incubate at 65°C for 10 min to elute protein-DNA complexes. Spin as above and collect supernatant.
Repeat the elution step with 150 μL of elution buffer and combine the two supernatants.
Add 300 μL of elution buffer to the 10 μL of INPUT previously set aside.
Add 3 μL of 20 mg/mL proteinase K to the eluates and the INPUT samples and incubate at 37°C for 30 min.
Reverse crosslink from 6 h to overnight at 65°C.
DNA purification: Add 300 μL of phenol–chloroform–isoamyl alcohol, vortex for 30 s and spin in a microcentrifuge at 12,000 × g for 5 min. Collect the upper phase and transfer in a new tube.
Precipitate DNA by adding 600 μL of 100% ethanol, 30 μL of 3 M sodium acetate pH 5.2 and 20 μg of glycogen.
Place on dry ice for 30 min and centrifuge at 12,000 × g at 4°C for 30 min. Remove the supernatant and wash the pellet with 500 μL of 75% ethanol.
Spin for 10 min at 4°C, remove the ethanol and air-dry the pellet for 15 min at ambient temperature. Resuspend in 100 μL of water.
Prepare the following dilution of the INPUT samples: 1:3, 1:9, 1:27, 1:81, 1:243, and dilute each IP sample 1:4.
Each 20 μL qPCR reaction contains: 4 μL of DNA (INPUT or IP), 10 μL of 2× SYBR® Green PCR mix and 10 pmol of each primer. For each primer set, generate a standard curve using the undiluted INPUT sample and its dilutions and analyze each diluted IP sample in triplicate. Use the following conditions for the thermocycler (see Note 11): 95°C for 5 min, 40 cycles of 95°C for 10 s, 55°C for 10 s, and 72°C for 30 s.
After cycling is complete, set the PCR machine to calculate dissociation curves from 70 to 96°C (see Note 12).
Visualize the amplification curve in a logarithmic scale and set the baseline from cycle 3 to cycle 9. Set the threshold in the linear phase of amplification and record the Ct value for each well. Average the Ct values for the three replicates of the IP samples and calculate the ΔCt for each IP sample (normalized to the INPUT sample), using the standard curve generated with the Ct values of INPUT sample. Next, calculate the ΔΔCt (ΔΔCt = ΔCt (GAPDH) – ΔCt (target sequence)). For each IP, the relative recovery of the target sequence normalized to the relative recovery of the GAPDH coding region is 2−ΔΔCt.
Footnotes
Cells should not be seeded at a density lower than 2 × 105 cells/mL and should not be grown above 106 cells/mL. Count cells with a hemocytometer before and after splitting.
Extreme care should be taken while handling blood samples. Wear protective equipment (lab coat, goggles, and gloves) throughout the whole procedure and decontaminate hood and pipettes with 25% bleach. Refer to your institution’s health and safety manual for practices when handling blood samples.
Very slow acceleration and deceleration should be applied to the centrifuge run. At the end of the run carefully remove the tubes from the centrifuge without disturbing the lymphocyte layer.
Dilute HDACi in DMSO so that the final concentration of DMSO in cell culture medium does not exceed 0.4%.
Use HDACi at a concentration at or below their EC50. Using HDACi at a concentration above their EC50 can cause an increase of FXN transcript, as a stress response.
If an accurate determination of EC50 is necessary, more dilutions of HDACi are required to produce a more detailed dose–response curve.
The protein concentration should be between 0.1 and 0.5 μg/μL, as 5–25 μg protein is recommended per assay replicate. Samples with protein concentrations under 0.1 μg should be excluded from analysis.
When handling RNA samples, always wear gloves, use clean pipettes and nuclease-free tubes, tips, and reagents.
Measure absorbance at 230, 260, and 280 nm. A good RNA prep will have a 260/280 and a 260/230 ratio close to 2.
Expression of most genes normally used as recovery standards in qRT-PCR changes upon HDACi treatment, hence these genes are not suitable for the quantification of relative FXN mRNA. We recommend normalizing to cell number, which is more important for lymphoblastoid cells than for primary lymphocytes, as we observe a dose-dependent decrease in cell number when lymphoblastoid cells are treated with HDACi, likely due to the cytostatic effect of HDACi on actively dividing cells. When we normalized to total RNA with these cells, there is a dramatic dose-dependent decrease in GAPDH expression, with the GAPDH level decreasing approximately 70–80% at the higher HDACi concentrations. When we normalize to cell number at the time of cell collection, the subsequent decrease in GAPDH is much less severe, with at most a 20% decrease in the expression level. In contrast to lymphoblastoid cells, HDACi concentration does not appear to decrease cell numbers of nondividing cells, such as primary lymphocytes. Even though primary lymphocyte cell numbers are not affected by HDACi treatment, when total RNA normalization is compared with cell number normalization an improvement in the stability of GAPDH expression level is observed. We routinely count cells after the treatment with HDACi just before collecting the cells for RNA isolation and still have not observed any dose-dependent changes in cell numbers. However, adjust the volume collected from each treatment to ensure that an equal number of cells are collected for each treatment condition.
Cycling conditions depend on the thermocycler used, refer to your instrument’s manual for optimal cycling conditions.
The dissociation plot for every primer pair should be a single sharp peak.
Optimal shearing conditions must be determined for different sonicators. The average fragment length of genomic DNA determines the resolution of the assay. To determine the average size of sheared DNA, remove 20 μL of sheared cell extract and add 300 μL of elution buffer. Add proteinase K to a final concentration of 0.2 mg/mL and incubate at 37°C for 30 min and then from 6 h to overnight at 65°C to reverse crosslinking. Add 300 μL of phenol–chloroform–isoamyl alcohol, vortex for 30 s, and spin in a microcentrifuge at 12,000 × g for 5 min. Collect the upper phase and transfer in a new tube. Precipitate DNA by adding 600 μL of 100% ethanol, 30 μL of 3 M sodium acetate pH 5.2, and 20 μg of glycogen. Place on dry ice for 30 min and centrifuge at 12,000 × g at 4°C for 30 min. Remove the supernatant and wash the pellet with 500 μL of 75% ethanol. Spin for 10 min at 4°C, remove the ethanol and air-dry the pellet for 15 min at room temperature. Resuspend in 10 μL of water and load onto a 1% agarose gel to check the fragment size. An average fragment size of 400–500 bp is recommended.
Remove as much supernatant as possible without aspirating the agarose beads. The use of 0.4 mm flat tips helps minimizing the amount of beads aspirated with the supernatant.
Contributor Information
Elisabetta Soragni, The Scripps Research Institute 10550 North Torrey Pines Road, MB27 La Jolla, CA 92037 858-784-8924 (Tel).
Chunping Xu, The Scripps Research Institute 10550 North Torrey Pines Road, MB27 La Jolla, CA 92037 858-784-8924 (Tel).
Andrew Cooper, Repligen Corporation 41 Seyon Street Building #1, Suite 100 Waltham, MA 02453 Phone: 781.250.0111.
Heather L. Plasterer, Repligen Corporation 41 Seyon Street Building #1, Suite 100 Waltham, MA 02453 Phone: 781.250.0111
James R. Rusche, Repligen Corporation 41 Seyon Street Building #1, Suite 100 Waltham, MA 02453 Phone: 781.250.0111
Joel M. Gottesfeld, The Scripps Research Institute 10550 North Torrey Pines Road, MB27 La Jolla, CA 92037 858-784-8924 (Tel)
References
- 1.Marks P, Breslow R. Dimethyl sulfoxide to vorinostat: development of this histone deacetylase inhibitor as an anticancer drug. Nat. Biotechnol. 2007;25:84–90. doi: 10.1038/nbt1272. [DOI] [PubMed] [Google Scholar]
- 2.Kazantsev AG, Thompson LM. Therapeutic application of histone deacetylase inhibitors for central nervous system disorders. Nat. Rev. Drug Discov. 2008;7:854–868. doi: 10.1038/nrd2681. [DOI] [PubMed] [Google Scholar]
- 3.Herman D, Jenssen K, Burnett R, Soragni E, Perlman SL, Gottesfeld JM. Histone deacetylase inhibitors reverse gene silencing in Friedreich’s ataxia. Nature Chem. Biol. 2006;2:551–558. doi: 10.1038/nchembio815. [DOI] [PubMed] [Google Scholar]
- 4.Rai M, Soragni E, Chou CJ, Barnes G, Jones S, Rusche JR, Gottesfeld JM, Pandolfo M. Two new pimelic diphenylamide HDAC inhibitors induce sustained frataxin upregulation in cells from Friedreich’s ataxia patients and in a mouse model. PLoS One. 2010;5:e8825. doi: 10.1371/journal.pone.0008825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Rai M, Soragni E, Jenssen K, Burnett R, Herman D, Gottesfeld JM, Pandolfo M. HDAC inhibitors correct frataxin deficiency in a Friedreich ataxia mouse model. PLoS ONE. 2008;3:e1958. doi: 10.1371/journal.pone.0001958. doi:1910.1371/journal. pone.0001958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Thomas EA, Coppola G, Desplats PA, Tang B, Soragni E, Burnett R, Gao F, Fitzgerald KM, Borok JF, Herman D, Geschwind DH, Gottesfeld JM. The HDAC inhibitor 4b ameliorates the disease phenotype and transcriptional abnormalities in Huntington’s disease transgenic mice. Proc. Natl. Acad. Sci. USA. 2008;105:15564–15569. doi: 10.1073/pnas.0804249105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chou CJ, Herman D, Gottesfeld JM. Pimelic diphenylamide 106 is a slow, tight-binding inhibitor of class I histone deacetylases. J. Biol. Chem. 2008;283:35402–35409. doi: 10.1074/jbc.M807045200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Xu C, Soragni E, Chou CJ, Herman D, Plasterer HL, Rusche JR, Gottesfeld JM. Chemical probes identify a role for histone deacetylase 3 in Friedreich’s ataxia gene silencing. Chem. Biol. 2009;16:980–989. doi: 10.1016/j.chembiol.2009.07.010. [DOI] [PMC free article] [PubMed] [Google Scholar]