Abstract
The availability of complete genomes and global gene expression profiling has greatly facilitated analysis of complex genetic regulatory systems. We describe the use of a bioinformatics strategy for analyzing the cis-regulatory design of genes diferentially regulated during viral infection of a target cell. The large-scale transcriptional activity of human embryonic kidney (HEK293) cells to reovirus (serotype 3 Abney) infection was measured using the Affymetrix HU-95Av2 gene array. Comparing the 2000 base pairs of 5’ upstream sequence for the most differentially expressed genes revealed highly preserved sequence regions, which we call “modules”. Higher-order patterns of modules, called “super-modules”, were significantly over-represented in the 5’ upstream regions of transcriptionally responsive genes. These supermodules contain binding sites for multiple transcription factors and tend to define the role of genes in processes associated with reovirus infection. The supermodular design encodes a cis-regulatory logic for transducing upstream signaling for the control of expression of genes involved in similar biological processes. In the case of reovirus infection, these processes recapitulate the integrated response of cells including signal transduction, transcriptional regulation, cell cycle control, and apoptosis. The computational strategies described for analyzing gene expression data to discover cis-regulatory features and associating them with pathological processes represents a novel approach to studying the interaction of a pathogen with its target cells.
Keywords: Cis-regulation, gene arrays, promoters, sequence modules, transcription factors, Gene Ontology, signaling, reovirus, apoptosis
INTRODUCTION
Though the mechanisms of eukaryotic transcriptional regulation are not completely understood, they are thought to be controlled by two main factors. First, cis-regulation involves the combinatorial binding of transcription factors to the 5’ upstream regions of genes, which controls the activation or repression of the basal transcriptional assembly. Second, signal transduction pathways integrate the cellular stimuli that lead to the activation of transcription factors, which is known as trans-regulation. Understanding the mechanisms of cis- and trans-regulation can help decipher the form and function of living systems. The availability of the complete genome and the ability to assay the large-scale transcriptional response of an organism bring us closer to this understanding.
In this manuscript, we utilized a computational methods strategy to analyze cis-regulation from high-throughput gene expression data. Similar approaches have been used to characterize microarray data from yeast [1] and plants [2], however they have not been previously utilized to study the interactions of a pathogen with host target cells. We utilized this approach to investigate reovirus-induced alteration of host gene expression profiles. Mammalian reoviruses are non-enveloped viruses that contain a double-stranded RNA genome. Most mammalian species, including humans, serve as hosts for reovirus infection. Reovirus infection in neonatal mice provides a classic experimental system for studying viral pathogenesis [3, 4]. Similarly, the interaction of reoviruses with a variety of host cells have provided valuable insights into how viruses perturb cellular signaling pathways including those involving transcription factor activation, mitogen-activated protein kinase cascades, and cell death. [5-8].
We test the hypothesis that significant similarity between the 5’ upstream regions of genes is key to understanding their co-regulation and their role in the integrated signaling response of cells. Using genes that were identified as most transcriptionally responsive to reovirus infection and by computationally searching their upstream sequences, we identified putative transcriptional control modules, which are regions with highly similar 5’ upstream sequence. These modules were significantly over-represented in transcribed genes as compared to untranscribed or unchanged genes. We also identified patterns of modules, called supermodules, which were highly specific to transcriptionally active genes. Using this modular cis-regulatory architecture we could resolve the role of downstream genes in different cellular processes, such as signal transduction pathways leading to apoptosis. Our results suggest that the modular architecture of the upstream regions of differentially expressed genes plays an important role in cis-regulation. We believe that this indicates that these modules are candidate target regulatory regions whose activity is modulated by virus-induced perturbations in host-cell signal transduction events. That is, activated sets of transcription factors binding in defined combinations to modules regulate the changes in host cell gene expression induced following infection with a viral pathogen.
A variety of methods have been developed to identify significant differentially expressed genes and to cluster genes based on expression profiles. Computational approaches for identifying cis-regulatory sites and for analyzing gene expression data have been of great interest. Our work contributes to more recent computational approaches for analyzing transcriptional control by combining high-throughput gene array data with whole genomic sequence (a recent review can be found in [9]). We have put our results in the context of: (i) the use of gene array data, (ii) the nature of upstream sequence motifs involved in transcriptional control, (iii) algorithms for motif discovery, and (iv) biological systems for studying transcription control.
Gene array data is used to identify coregulated genes, which narrows the search for sequence motifs to the 5’ upstream regions of a relevant subset the genes. Some approaches use levels of expression to cluster genes [10, 11]. A completely different approach involves the use of the level of expression of a gene as some function of the putative transcription factor binding sites [12, 13]. We treat the gene array data as a coarse measure of biological function by grouping genes into three broad categories: significant differential expression (de), unchanged expression (uc) and no expression (off). We believe that these three functional classes are useful for representing the important biological groups of transcriptional activity and for understanding cis-regulatory logic.
The nature of cis-regulatory motifs is another major attribute differentiating existing methods for analysis of transcription factor binding sites. While earlier methods focused on identifying individual motifs to represent a single transcription factor binding site, recent approaches capture the binding of multiple transcription factors through combinations of motifs. Individual and complex motifs can be further classified either by their use of known binding sites [10, 12, 14-19] or by their discovery of novel ones [20-24]. While it is important to search for novel motifs, as the available information about transcription factor binding sites is limited, it is computationally difficult to discover short regulatory elements accurately. This is because short and variable nucleotide sequence motifs have a high rate of false positive matches. Our approach overcomes these issues by focusing on long motifs (greater than 40 nucleotides). We term these motifs “modules” and higher-order patterns of these motifs “supermodules”. We believe that the putative modules and supermodules in the 5’ upstream regions of differentially expressed genes play a critical role in trans-regulation. That is, activated transcription factors bind combinatorially to the modules; interactions between groups of modules, manifested as supermodules, regulate the expression of their 3’ downstream genes.
A number of methods have been developed for discovering novels motifs in sequence data. We utilize MEME (Motif Expectation-Maximation for Motif Elicitation) [25, 26] for this purpose. The significance of motifs is generally evaluated by calculating the representation of motifs in the 5’ upstream regions of a subset of putatively coregulated genes as compared to random sequence [11, 14, 17, 18, 21-23]. Our approach uses all of the highly regulated genes to identify putative cis-regulatory modules but tests their significance by comparison with genes that are unchanged or off. We also identify longer stretches (typically 15-50 bases) of imperfectly conserved sequence similarity motifs, instead of shorter motifs, corresponding to the core and surrounding nucleotides specific for individual transcription factors. This approach is biologically more plausible and it makes complete use of the information derived from the gene array analysis.
One of the most important factors in analyzing cis-regulatory patterns in the context of gene expression is the utilization of a biological system that is known to strongly involve transcriptional control. We chose reovirus infection of a human cell line in this analysis because it is an important model for studying eukaryotic processes that lead to host cell damage and death [5-8]. The strain used in this study, serotype 3 Abney (T3A), efficiently induces apoptosis in HEK293 cells through the binding of tumor necrosis factor (TNF)-related apoptosis-inducing ligand (TRAIL) to its cell surface death receptors DR4 and DR5 [27]. It has been shown that the activated transcription factors c-Jun and NF-κB play a critical role in these processes [28-33]. Since the expression of new genes is critical to reovirus-induced apoptosis, this is a useful system to study the control of transcription.
Finally, to interpret the biological relevance of our analysis we compared supermodules with the biological roles of genes from the GeneOntology (GO) database [34], using a previously described tool [35]. By analyzing the involvement of supermodules in controlling the transcription of downstream genes and the role of genes in biological processes, we were able to elucidate the pathways involved in the integrated response of mammalian cells to reovirus infection. The advantage conferred by using this particular tool for inferring biological associations for different supermodules is conferred by the fact that the method generates p-value scores for the category frequency counts using a variety of distribution models (e.g. binomial and hypergeometric) and adjusts the p-values for multiple correction comparisons in order to minimize the risk of Type 1 errors.
MATERIALS AND METHODS
Cells and Virus
Human embryonic kidney (HEK293) cells (ATCC CRL1573) were plated in T75 plated flasks and incubated for 24 hours. When cells were 70% confluent they were infected with reovirus strain T3A at a multiplicity of infection (MOI) of 100 PFU per cell in a volume of 2 ml at 37 °C for 1 hour. A high MOI was used to ensure that all susceptible cells were infected. Cells used for control infections were inoculated with a virus-free cell lysate control. Cells were harvested at 12 hours post infection and washed with phosphate-buffered saline. cRNA was prepared in accordance with protocols recommended by Affymetrix and each sample was prepared in duplicate.
Gene Expression Analysis
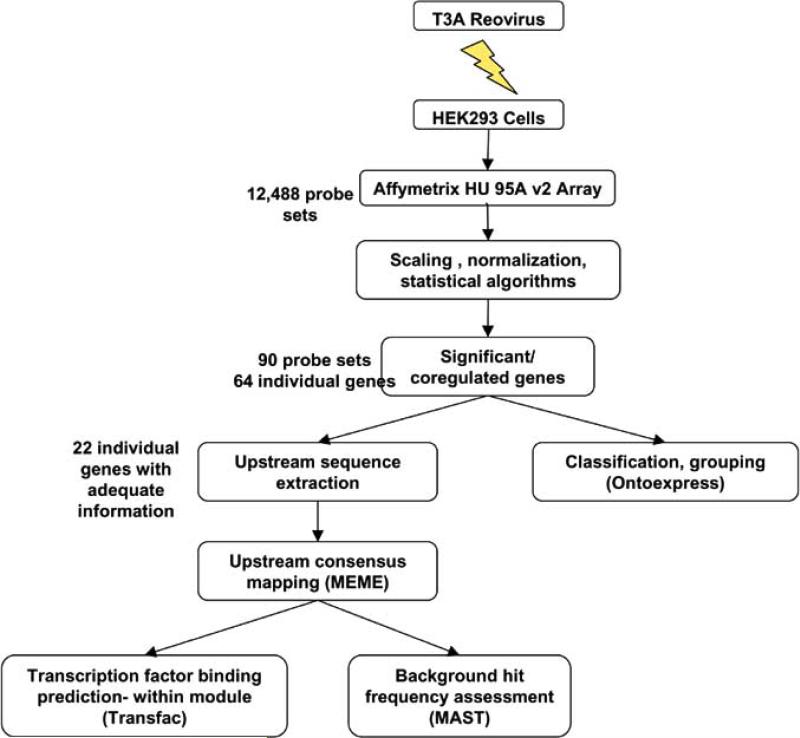
We used Affymetrix U95A version 2 arrays to assay expression levels of genes in infected and control samples. Gene array data was read with Agilent GeneArray. Using Affymetrix algorithms in GeneChip 5 software, transcripts was classified as present, marginally present or absent. Present versus absent calls were used later to group genes that were absent across all samples into the set of untranscribed genes. A median filter was used to filter genes that did not show a significant variation across all samples. The data was analyzed using GeneSpring suite (from Silicon Genetics). Data was normalized in order to facilitate cross array comparison and to account for variations. Linear regression was performed between replicates in order to filter out genes that showed poor consistency between replicates and genes that lay within the 5% confidence interval were retained for further analysis. Genes that were both well-replicated and present were subjected to parametric (t-test) and nonparametric (Wilcoxon signed rank) tests with the False Discovery Rate (FDR) correction. The FDR threshold was varied between 0.08 and 0.15 and the resulting gene list was chosen based on the minimum percentage of false positives produced for a given number of genes. This yielded 90 probe set identifiers corresponding to 64 distinct genes showing statistically significant differential expression, while 4,300 genes showing insignificant differential expression, and 6,000 genes were found to be transcriptionally inactive. Out of the 64 initially identified differentially expressed genes, 22 genes were available for upstream characterization (see Fig. (1)). The remaining 42 genes could not be analyzed in this fashion for a variety of reasons including: (1) no upstream sequences were available, (2) short upstream sequences were available for certain genes having large spans of N's, (3) genes were not represented/included within the genomic assembly; this included genes that could not be mapped to any of the assembled human contigs used for sequence retrieval.
Fig. (1).
Flow chart showing the overall experimental design for analysis of reovirus-induced changes in gene expression. HEK293 cells were infected with reovirus T3A. 24 hrs post-infection cRNA was prepared from infected cell lysates. Gene expression was analyzed using Affymetrix U95A (version 2) arrays. 90 probe sets corresponding to 64 distinct genes were found to be differentially expressed following reovirus infection. 22 genes had sufficient information available for extraction of 2000 bp of 5'-sequence upstream of the first start codon. A bioinformatics program for motif discovery (MEME, http://meme.sdsc.edu </exchweb/bin/redir.asp?URL=http://meme.sdsc.edu/>) was used to identify conserved sequence motifs of 15-50 bp size (“modules”) within the 5'-upstream region of differentially expressed genes. Transcription factor binding arrangement within modules was predicted using the TRANSFAC database. Differentially expressed genes were also classified and grouped using Ontoexpress, an online analysis tool based on the Gene Ontology annotation database (See Materials and Methods and text for further details).
Upstream Genomic Sequence Analysis
The complete human genome was obtained from GenBank and the first 2,000 base pairs of the 5’ upstream regions for each gene in the Affymetrix U95A version 2 gene array were extracted from it. We found upstream regions for 7,022 out of the total 10,390 genes including, 22 significantly differentially expressed genes, 4,300 expressed genes, and 2,700 untranscribed genes. The similarity between the upstream genomic sequences was computed using MEME (Motif Expectation-Maximization for Motif Elicitation) [25, 26], which finds conserved sequence motifs in a set of biomolecular sequences. Sequence modules of length between 15 and 50 nucleotides were identified in the significant differentially transcribed. These conserved modules were then searched in the genes whose expression was unchanged by reovirus infection and the genes that remained transcriptionally inactive using MAST (Motif Alignment and Search Tool) [36]. MAST is a tool for searching the biological sequence databases for sequences that contain one or more of a group of known motifs. It takes as input a file containing the descriptions of one or more motifs and searches a sequence database that the user selects that matches the motifs. The motif file can be the output of the MEME motif discovery tool or any file in the appropriate format.
Significance of Modules
To assess the significance of the conserved modules identified by MEME the upstream regions of all available genes in the unchanged and untranscribed groups were searched using MAST (using p < 0.0001 for each match). The observed counts of modules in each expression group were compared with the expected frequency according to a simple random model. Assuming module occurrences are equally likely in all expression groups, the expected frequency of hits for one module or supermodule in any category, denoted as Ni, is proportional to the fraction of genes in each group, given by fi. Since the number of genes in each group and the total number of genes are fixed, we have , , and . The expected number of observations of a module in a given expression group is the product of the fraction of genes in that group and the total number of observed hits for a module or supermodule, No. Hence, . To test the independence between the observed and expected counts of modules we employed the Fisher exact test (instead of the χ2 test due to some low expected and observed counts).
Over-Representation and Under-Representation of Modules
We calculated the likelihood ratio of the observed and the expected frequencies of occurrence of each module and supermodule in the 5’ upstream regions of genes in the different expression categories. This ratio is denoted as , where α = (de; uc; off) and β = (module; supermodule) and calculated as shown in equation 1 below:
| (1) |
There are three main cases for the value of this ratio. First, when a particular module or supermodule occurs more frequently in an expression group than expected by chance then and . Second, when a particular module or supermodule occurs at the expected frequency then and . Third, when the frequency of the module or supermodule is lower than what would be expected by chance then and . The interpretation of the likelihood ratio is summarized in equation 2 below:
| (2) |
Gene Ontology Annotation
Gene functional annotations were obtained by querying the latest release of the Gene Ontology (GO) database [34]. The score of the molecular functions and biological processes was calculated as the frequency of occurrence for each GO identifier in the differentially expressed genes with the total number of genes with the same GO identifiers for all genes on the Affymetrix HU-95v2 gene array. This percentage used each GO node and all of its children [35]. The p-values associated to each category were calculated using the hypergeometric distribution model and adjusted using FDR to compensate for Type I errors introduced by multiple comparisons. Onto-express [35] was utilized to correlate expression profiles with biochemical and molecular functions, biological properties, and cellular roles of proteins encoded by differentially expressed genes.
RESULTS
Upstream Sequence Analysis of Differentially Expressed Genes
We used Affymetrix U95A version 2 arrays to measure the large-scale transcriptional profile of human embryonic kidney (HEK293) cells in response to reovirus (T3A) infection. By applying stringent statistical filters (see methods) we divided the genes on the array into the following groups: significant differential expression, unchanged expression, and transcriptionally inactive. The 2 kb 5’ upstream regions of the genes on the array were extracted from the human genome using GenBank. This resulted in 22 genes in the differentially expressed group, 4,300 genes in the unchanged group and 2,700 untranscribed genes. The 5’ upstream regions of the 22 differentially expressed genes were analyzed using MEME to find the most similar sequence regions.
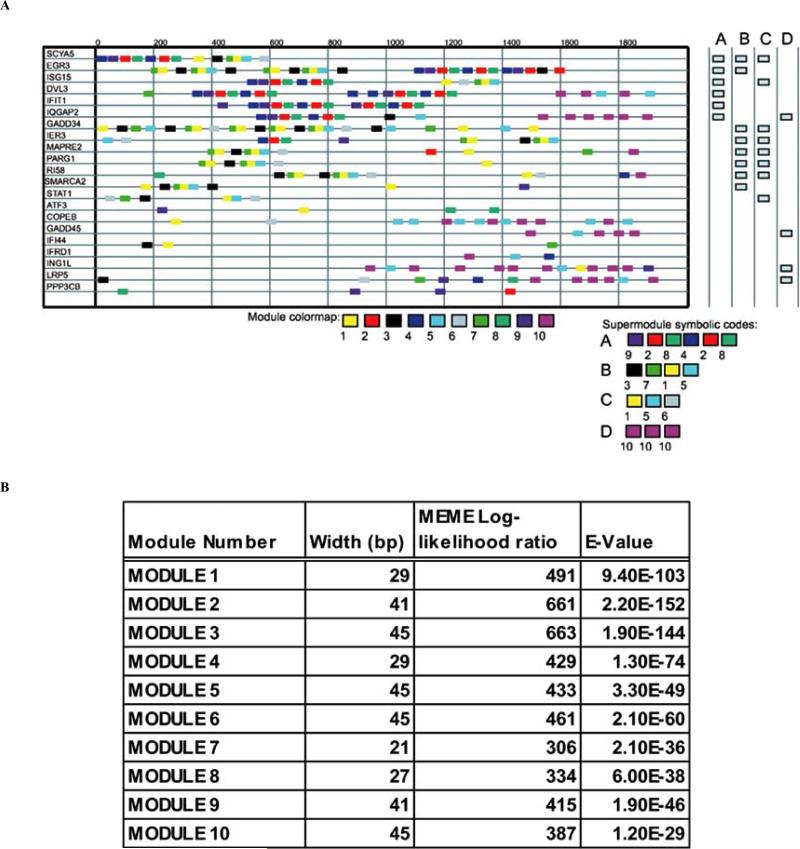
We termed similar sequence regions in the 5’ upstream regions of genes “modules.” Modules are longer (15-50 bp in length) than the short regulatory element consensus sequences, which are generally used in analyzing upstream regions of genes. A schematic of the ten most significant modules in the 22 differentially expressed genes is shown in Fig. (2a). This visualization shows the organization of the upstream region of individual genes, where each module is represented by a uniquely colored rectangle. For example, SCYA5 (the first gene from the top) has the following arrangement for the first five modules starting 2000 bp upstream from the start codon: module 9, module 2, module 8, module 4 and module 2. The figure also displays the overall architecture of the cis-regulatory regions of the differentially expressed genes. The visualization shows that number of other genes have modules in common with SCYA5 including EGR3, ISG15, DVL3 and IFIT1, to mention a few. For instance, EGR3 has two occurrences of module 9; ISG15 has a single occurrence of module 9; both DVL3 and IFIT1 have multiple occurrences of module 9. Since the occurrence of these modules in these sequences is highly statistically significant, and the expression of these genes is highly regulated, it is plausible that these modules play a role in transcriptional control.
Fig. (2).
(a) Schematic showing the 10 most significant modules in the 22 analyzed genes that were differentially expressed during reovirus infection. Each module is represented by a different colored block, and the sequential arrangement of these modules within the 2000 bp's 5'-upstream of the first start codon of each differentially expressed gene is shown. Supermodules are composed of sets of modules in a particular arrangement. The arrangement of modules characterizing four supermodules (A-D) is shown at the bottom of the figure, and the presence of each of these supermodules within the 22 genes is shown at the right of the figure. (b) The size of each module shown in (a) is indicated along with the log-likehood ratio and E-value for its occurrence.
The size, log-likelihood ratios and corresponding E-values associated to each individual module are shown in Fig. (2b). The size of each identified modular element varies between 21-45 base-pairs. Using this type of approach allows identification of rather long stretches of closely related sequence elements, which is extremely useful given the fact that individual transcription factors identify degenerate oligonucleotides sequences as binding sites. Each individual module captures the spatial relationship between putative individual transcription factor binding sites both in terms of the order of the hits and the distance between them. This represents an important difference between our approach and more commonly employed methods which rely heavily on the identification of individual transcription factor binding sites as the starting material for regulatory element discovery.
Modular Organization and Supermodules
In addition to the individual modules, we also found a unique “supermodular” organization in the upstream regions in 16 out of the 22 differentially expressed genes. A supermodule is a sequence of modules in which the order and relative location of the modules is preserved in the upstream regions of different genes. For example, the upstream region of SCYA5 had the following sequence of modules: 9, 2, 8, 4, 2, 8. We call this sequence, (9 2 8 4 2 8), a supermodule because it is also preserved in the upstream regions of the EGR3, ISG15, DVL3, IFIT1, and IQGAP2. The occurrences of supermodules in the upstream regions of these 22 genes are summarized on the right hand side of Fig. (2). We identified four supermodules, A, B, C and D, which were defined by the sequences of modules given by (9 2 8 4 2 8), (3 7 1 5), (1 5 6) and (10 10 10), respectively.
Significance of Modules and Supermodules
To assess the biological relevance of modules and supermodules in transcriptional control we evaluated their significance across the complete dataset in two ways. First, we compared the observed versus expected frequency of occurrence of modules in the three categories of transcriptional response: differentially expressed (de), unchanged (uc), and untranscribed (off). For each of these categories we computed the occurrence of modules and supermodules in the upstream regions of all 7022 genes. For instance, module 1 was found in 14 out of 22 of the significantly differentially expressed genes; in 1759 of the genes that were unchanged; and 948 out of the transcriptionally inactive genes. Using a random model, we calculated the expected number of occurrences of module 1 as 8, 1666, 1046 in the de, uc and off categories, respectively. Using the Fisher exact test of independence we found the observed and expected frequencies of occurrence for module 1 to be highly statistically different across the expression groups (p < 0.001). Applying this procedure to each of the modules and supermodules showed that the observed and expected frequency of modules in the upstream regions of transcriptionally active and inactive genes was statistically different (Table 1). Hence, the distribution of modules and supermodules in the upstream regions of genes is significantly different from what would be expected by chance.
Table 1.
Distribution of Modules and Supermodules
| Module | No | P-value | ||||||
|---|---|---|---|---|---|---|---|---|
| 1 | 14 | 8.52 | 1759 | 1666.23 | 948 | 1046.24 | 2721 | 1.29 × 10-4 |
| 2 | 10 | 9.61 | 2003 | 1878.11 | 1054 | 1179.28 | 3067 | 2.01 × 10-5 |
| 3 | 12 | 7.75 | 1647 | 1514.37 | 814 | 950.88 | 2473 | 4.92 × 10-8 |
| 4 | 12 | 10.33 | 2128 | 2018.34 | 1156 | 1267.33 | 3296 | 3.34 × 10-4 |
| 5 | 19 | 13.38 | 2739 | 2616.01 | 1514 | 1642.61 | 4272 | 1.11 × 10-4 |
| 6 | 12 | 6.2 | 1240 | 1211.86 | 727 | 760.94 | 1979 | 2.25 × 10-2 |
| 7 | 13 | 7.56 | 1596 | 1477.02 | 803 | 927.43 | 2412 | 2.77 × 10-7 |
| 8 | 12 | 8.21 | 1761 | 1605 | 848 | 1007.79 | 2621 | 6.70 × 10-10 |
| 9 | 13 | 11.58 | 2363 | 2262.67 | 1319 | 1420.75 | 3695 | 2.59 × 10-3 |
| 10 | 11 | 13.03 | 2721 | 2546.2 | 1426 | 1598.78 | 4158 | 1.87 × 10-7 |
| A: (3 7 1 5) | 9 | 1.1 | 230 | 214.94 | 112 | 134.96 | 351 | 3.96 × 10-14 |
| B: (1 5 6) | 8 | 1.3 | 269 | 253.52 | 137 | 159.19 | 414 | 3.99 × 10-9 |
| C: (9 2 8 4 2 8) | 7 | 0.9 | 198 | 175.75 | 82 | 110.35 | 287 | 6.57 × 10-12 |
| D: (10 10 10) | 5 | 6.72 | 1447 | 1313.51 | 693 | 824.77 | 2145 | 2.44 × 10-8 |
This table summarizes the observed and expected counts for each module and supermodule in the 5’ upstream regions of the genes in each of three expression categories: differentially expressed (de), unchanged (uc) and not expressed (off). The first column in the table from left to right is Module, which is the label of the module or sequence of supermodules. Columns two to seven contain the different frequency counts: , and α= (de; uc; off) and c = (o = observed; e = expected). Column eight shows the total number of observed occurrences of a module or supermodule in the upstream region of all 7022 genes. Finally, column nine shows the p-value computed using the Fisher exact test for the independence between the observed and expected counts. From top to bottom, the first 10 rows summarize the occurrence of the ten modules in the upstream regions of all genes in the three expression categories. The last 4 rows summarize the statistics on the occurrence of supermodules. Please see the text for additional information.
Though the occurrence of modules and supermodules is statistically different across the expression categories, additional information is necessary to infer their role in transcriptional control. For this purpose, we calculated the likelihood ratio, denoted as Rα,β, of the observed and the expected frequencies of occurrence of each module and supermodule in the 5’ upstream regions of genes in the different expression categories. We calculated Rα,β and their descriptive statistics for the ten modules and the supermodules across the three expression categories as shown in Table 2. The likelihood ratios between categories were found to be statistically different using a one-way ANOVA (p = 0.006). The average over-representation of the modules and supermodules in the differentially expressed genes and under-representation in the transcriptionally inactive genes is summarized in the row labeled R̄ (see Table 2). The mean value of the likelihood ratio was greater than unity in the differentially expressed genes . In the unchanged and the inactive genes the ratios were and , respectively. Using a one-sided t-test to compare means, p < 0.05 (p-value adjusted for multiple testing by the false discovery rate correction). Therefore we can be confident that modules and supermodules are generally significantly over-represented in the 5’ upstream regions of differentially expressed genes, under-represented in the 5’ upstream transcriptionally inactive genes and close to randomly distributed in the 5’ upstream regions of unchanged genes. Comparing the values of the ratio for the modules and supermodules in Table 2 also highlights an important result: the supermodules occur up to eight times more frequently upstream of differentially expressed genes than the unresponsive genes (p < 0.05).
Table 2.
Representation of Modules and Supermodules in Different Expression Categories
| Module | Rde | Ruc | Roff |
|---|---|---|---|
| 1 | 1.64 | 1.06 | 0.91 |
| 2 | 1.04 | 1.07 | 0.89 |
| 3 | 1.55 | 1.09 | 0.86 |
| 4 | 1.16 | 1.05 | 0.91 |
| 5 | 1.42 | 1.05 | 0.92 |
| 6 | 1.94 | 1.02 | 0.96 |
| 7 | 1.72 | 1.08 | 0.87 |
| 8 | 1.46 | 1.1 | 0.84 |
| 9 | 1.12 | 1.04 | 0.93 |
| 10 | 0.84 | 1.07 | 0.89 |
| A: (3 7 1 5) | 8.18 | 1.07 | 0.83 |
| B: (1 5 6) | 6.17 | 1.06 | 0.86 |
| C: (9 2 8 4 2 8) | 7.78 | 1.13 | 0.74 |
| D: (10 10 10) | 0.74 | 1.1 | 0.84 |
| R̄ | 2.77 | 1.07 | 0.88 |
| σ R | 2.68 | 0.03 | 0.05 |
| Rmax | 8.18 | 1.13 | 0.96 |
| Rmin | 0.84 | 1.02 | 0.74 |
The table summarizes the likelihood ratios of the observed and expected counts for each module and supermodule in the 5’ upstream regions of the genes in each of the three expression categories: differentially expressed (de), unchanged (uc) and not expressed (off). The first column in the table from left to right is Module, which is the label of the module or sequence of supermodules. Columns two to four show the ratios of the observed and expected counts for each module and supermodule in the three expression categories, denoted by Rα where α = (de;uc;off). The ratios are computed using data from Table 1.
It is important to note that the likelihood ratios for module 10 and supermodule D do not follow the general trend. Specifically, module 10 and supermodule D were under-represented in the differentially expressed genes. On the other hand, supermodule D was slightly over-represented in genes that were not differentially expressed. We discuss possible reasons for these observations later.
The Anatomy of a Module
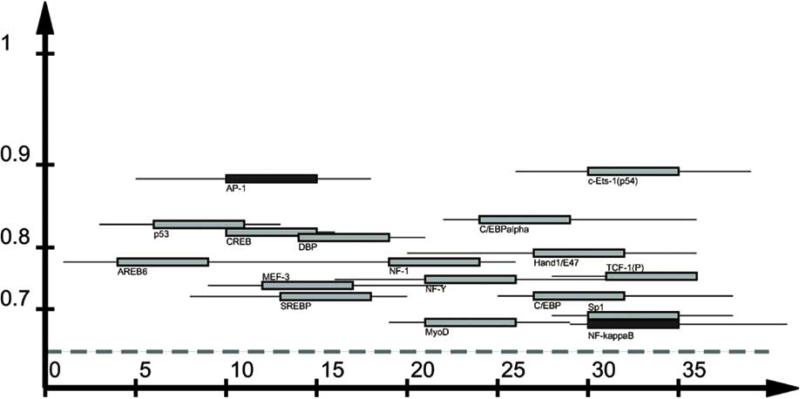
We explored the potential combinatorial interactions of the modules with transcription factors by searching the consensus sequences of the modules [37] against the known regulatory elements in the TransFac database [38]. For example, Fig. (3) shows a visualization of the high-scoring putative regulatory elements that were found in module 2. We found a number of potential binding sites for different transcription factors including, c-Jun (activating protein 1, AP-1) and NF-κB. In addition, we also observed predicted matches for c-Jun binding in modules 3, 5 and 6. This is consistent with our prior studies and those of others showing that both NF-κB and c-Jun are activated following reovirus infection [28-33].
Fig. (3).
A schematic illustrating the pattern of putative regulatory elements found in module 2. The two elements (AP-1 and NF-κB) shown in dark blocks are known to be regulated during reovirus infection of cells.
Supermodules and Signaling
In order to understand the biological importance of the supermodules we used the GeneOntology [34] and OntoExpress [35] to analyze the biological processes in which the differentially expressed genes were involved. Table 3 summarizes the involvement of the genes 3’ downstream of supermodules in different biological processes. These processes include apoptosis, cell cycle arrest, signal transduction, inflammation and viral response pathways. For example, all of the genes downstream from supermodules A, B, C, and D were involved in signal transduction, but only supermodule D appeared to be upstream of genes involved in DNA repair. The genes putatively controlled by supermodules B and C shared most of their biological process annotations, whereas genes regulated by supermodule A shared just a subset of these processes. Supermodule D controlled genes that showed a marked difference in terms of their molecular process annotations: they were involved in five distinct processes including, DNA repair, lipid metabolism, positive regulation of cell proliferation, protein biosynthesis and regulation of CDK activity. In terms of just their molecular functions, genes downstream of supermodules A and D shared a distinct subset of functional annotations than did supermodules B and C. The association of supermodules with different biological processes is highly informative because it aids in grouping genes based on cis-regulatory information as opposed to their level of expression alone.
Table 3.
Supermodules and GO Processes
| GO Biological Processes | A | Adj p-value | B | Adj p-value | C | Adj p-value | D | Adj p-value |
|---|---|---|---|---|---|---|---|---|
| Anti -apoptosis | □ | ■ | 2.75E-04 | ■ | 2.02E-04 | □ | ||
| Apoptosis | □ | ■ | 5.44E-05 | ■ | 4.28E-05 | □ | ||
| Calcium ion homeostasis | ■ | 2.54E-05 | ■ | 5.05E-05 | ■ | 4.14E-05 | □ | |
| Cell cycle arrest | □ | ■ | 1.72E-04 | ■ | 1.28E-04 | ■ | 4.51E-05 | |
| Cell growth and/or maintenance | □ | ■ | 0.0017627 | ■ | 0.0013941 | □ | ||
| Cell proliferation | □ | ■ | 0.002006 | ■ | 0.0016282 | □ | ||
| Cellular defense response | ■ | 1.91E-04 | ■ | 7.28E-05 | ■ | 4.00E-05 | □ | |
| Circadian rhythm | ■ | 1.51E-05 | ■ | 3.74E-05 | □ | □ | ||
| DNA repair | □ | □ | ■ | 1.06E-04 | ||||
| Embryogenesis and morphogenesis | ■ | 2.66E-04 | ■ | 6.27E-04 | ■ | 4.51E-04 | □ | |
| Immune response | ■ | 9.17E-05 | ■ | 0.0024494 | ■ | 1.38E-04 | □ | |
| Inflammatory response | ■ | 2.98E-04 | ■ | 7.62E-04 | ■ | 5.47E-04 | □ | |
| Intracellular signaling cascade | □ | ■ | 0.00237 | ■ | 0.0019023 | □ | ||
| Lipid metabolism | □ | □ | □ | ■ | 9.66E-05 | |||
| Positive regulation of cell proliferation | □ | □ | □ | ■ | 1.10E-04 | |||
| Protein biosynthesis | □ | □ | □ | ■ | 1.30E-04 | |||
| Regulation of CDK activity | □ | □ | □ | ■ | 3.01E-05 | |||
| Regulation of transcription DNA-dependent | ■ | 0.0057307 | ■ | 0.0019621 | □ | ■ | 0.0027668 | |
| Regulation of transcription from Pol II promoter | □ | ■ | 0.0011795 | □ | □ | |||
| Response to DNA damage stimulus | □ | ■ | 2.81E-05 | ■ | 2.46E-05 | □ | ||
| Response to oxidative stress | ■ | 3.78E-05 | ■ | 6.04E-05 | ■ | 5.05E-05 | □ | |
| Response to viruses | ■ | 3.01E-05 | ■ | 5.60E-05 | ■ | 4.90E-05 | □ | |
| Rho protein signal transduction | □ | ■ | 5.38E-05 | ■ | 4.59E-05 | □ | ||
| Signal transduction | ■ | 9.32E-05 | ■ | 0.0022916 | ■ | 0.0017359 | ■ | 1.49E-05 |
| Small GTPase mediated signal transduction | ■ | 2.88E-04 | □ | □ | ■ | 1.16E-04 |
The table summarizes GeneOntology biological process annotation for differentially expressed genes containing a supermodule in the 5’ upstream region. The ■ symbol denotes participation of the supermodule regulated gene in a biological process, and the □ symbol shows the absence of annotation for the regulated gene in a process. The adjusted p-values associated to the occurrence frequencies for each category/module are displayed on the right side adjacent of the corresponding module.
We used the four supermodules and their associated genes to elucidate the signal transduction processes in which they may be involved. Supermodule A was shared by interferon signaling genes (ISG15, IFIT1), transcriptional control genes EGR3 (involved in FasL apoptosis signaling) and signal transduction. Supermodule B was shared among a different subset of differentially expressed genes involving the above categories and there were two genes that had supermodules A and B (EGR3, SCYA5). The genes involved in cell signaling, interferon and cell proliferation control (GADD34, MAPRE2) were downstream of supermodule C. Thus, supermodule C could be a common control circuit for the genes linked to signaling and early cellular response to viral infection. The presence of multiple supermodules in the upstream regions of these genes suggests that the transcripts related to cell signaling and interferon response are under the control of multiple pathways. These findings point to several mechanisms that are involved in the onset of reovirus-mediated apoptosis. For instance, DNA damage response is modulated via p53-dependent mechanisms. Interferon and other chemokine-mediated signaling indicates the activation of a wider network of signal transduction pathways including G-proteins; Wnt and growth receptor signaling; p38 / JNK pathway via MEKK4 and MAPK8; and the activation of the apoptotic process via TRADD and FasL. Transcriptional modulators are also a key component of these processes including, nuclear matrix binding proteins, transcription factors and core promoter binding proteins. In combination, these processes represent the effector arm of the cell signaling machinery.
DISCUSSION
In this manuscript, we present a computational approach to analyze cis-regulation induced by viral infection of a target cell from high-throughput gene expression data. Our analysis of the 5’ upstream regions of genes differentially expressed in response to reovirus (T3A) infection of HEK293 cells shows a distinct modular cis-regulatory organization. The significant over-representation of modules and supermodules in the upstream regions of differentially expressed genes, strongly suggests their involvement in transcriptional control. Statistical analysis of the functional annotations for the genes downstream from the same supermodules reveals their involvement in similar biological processes including apoptosis, cell cycle regulation, antiviral defense and DNA repair. Our analysis of the consensus sequence for each module against a large set of known transcription factor binding sites yielded matches with NF-κB and c-Jun, which are known to be activated during reovirus infection [28-33]. These results suggest that modular organization is preserved to ensure an effective cellular response to external stimuli through signaling pathways.
Our results strongly suggest a role for modules and supermodules in transcriptional control. Analysis of the 5’ upstream regions of a large set of genes in differing transcriptional states suggests that modules are preserved because they are the loci for gene regulation through signaling; and that higher-order patterns of modules, or supermodules, encode some of the logic necessary for controlling the transcriptional state of their 3’ downstream genes. In general, we found a significant over-representation of supermodules in highly differentially expressed genes and an under-representation in transcriptionally inactive genes. Further analysis of each module revealed potential binding sites for transcription factors that are known to be involved in reovirus-induced apoptosis.
The annotations for molecular functions and for biological processes of transcriptionally active genes suggest an association between upstream supermodules and known pathways. However, we did not always identify a direct correspondence between supermodules and pathways. Some supermodules appear to regulate genes involved in identical pathways; others regulate multiple overlapping pathways. There are several interpretations for overlapping control of pathways by supermodules. First, the similarity between supermodules, measured by shared modular components, could be responsible for their regulation of similar processes. Second, the continuity of cellular responses to changes in homeostasis could be affected by a core group of modules in combination with a divergent set of modules to fine-tune the response to different stimuli. Third, overlapping could be a manifestation of redundancy in the control of gene regulation. Fourth, it is also possible that cis-regulation is more complicated than can be explained by a modular representation of only the 2000bp in the 5’ upstream regions of genes. We are further analyzing the relationship between modular organization and signaling pathways to understand their biologically relevance.
The GO annotations of the genes and the relevant biomedical literature on reovirus-induced apoptosis present a congruent view of the association between supermodules, the genes that are transcriptionally controlled by them, and biological pathways in which these genes are involved. Supermodules B and C were most closely associated with apoptosis, which is supported by the available literature on the genes controlled by these modules. The propagation of apoptotic and antiapoptotic pathways in mammalian cells through signal transduction pathways is well-described. In reovirus-induced cell death, genes involved in signaling can be resolved on the basis of the modules and supermodules in their 5’ upstream regions. For example, the genes associated with interferon signaling including, ISG15, IFIT1, IFI44 and IFRD1 appear to be controlled supermodule A, but IFI44 and IFRD1 by supermodule D, and RI58 by supermodule B. Since all of these genes are associated with interferon-related signaling, their upstream modular architecture hints at the existence of alternative pathways for regulating their transcriptional activity. Interferon signaling pathways have previously been shown to play a key role in reovirus-induced myocardial injury and inflammatory responses [39-43]. These genes also share similar consensus control elements with genes involved in different processes, creating a highly interconnected network for controlling their transcriptional modulation.
We summarize the association of supermodules with biological processes in Table 3 and Table 4 in the context of a transcription control network as follows: Genes controlled by supermodules B and C are associated with signal transduction, antiviral defense and cell cycle regulation. Supermodules B and C act as a potential “switchboard” for transcriptional regulation. Similarly, supermodule A is associated with a subset of signal transduction and antiviral defense pathways but it is also associated with distinct processes like transcriptional regulation and cytoskeletal rearrangement. Module D stands apart because its regulatory targets are involved in distinct processes including, DNA repair, lipid metabolism, cell proliferation and protein biosynthesis. The involvement of some supermodule-regulated genes in opposing processes like proapoptosis and antiapoptosis is not discordant. For example, genes involved in sensing DNA damage and initiating repair mechanisms after irradiation can also function later as triggers for cell death if the repair mechanisms are unsuccessful [44]. Therefore analyzing modules and supermodules from gene expression data can aid in deciphering the complex genetic regulatory network of a living systems in a novel way.
Table 4.
Supermodules and GO Functions
| GO Molecular Function | A | Adj p-value | B | Adj p-value | C | Adj p-value | D | Adj p-value |
|---|---|---|---|---|---|---|---|---|
| Actin binding | ■ | 2.93E-04 | □ | □ | ■ | 1.16E-04 | ||
| Antiviral response protein activity | ■ | 2.85E-05 | ■ | 4.72E-05 | ■ | 4.14E-05 | □ | |
| Apoptosis inhibitor activity | □ | ■ | 1.18E-04 | ■ | 9.58E-05 | □ | ||
| Calmodulin binding | ■ | 2.00E-04 | □ | □ | ■ | 1.02E-04 | ||
| Chemokine activity | ■ | 5.51E-05 | ■ | 9.11E-05 | ■ | 7.48E-05 | □ | |
| GTPase inhibitor activity | ■ | 2.54E-05 | □ | □ | ■ | 6.78E-06 | ||
| Helicase activity | □ | ■ | 5.72E-05 | □ | □ | |||
| Microtubule binding | □ | ■ | 4.30E-05 | ■ | 2.82E-05 | □ | ||
| Protein binding | ■ | 0.00170446 | □ | ■ | 2.58E-03 | □ | ||
| Ras GTPase activator activity | ■ | 1.27E-05 | □ | □ | ■ | 3.39E-06 | ||
| Rho GTPase activator activity | □ | ■ | ■ | 5.17E-05 | □ | |||
| Structural constituent of ribosome | □ | □ | □ | ■ | 9.72E-05 | |||
| Transcription co-activator activity | □ | ■ | 9.39E-04 | □ | □ | |||
| Transcription factor activity | ■ | 0.0039627 | ■ | 0.00771924 | □ | □ |
The table summarizes GeneOntology molecular function annotation for differentially expressed genes containing a supermodule in the 5’ upstream region. The ■ symbol denotes the existence of annotation for a supermodule regulated gene with a molecular function, and the □ symbol shows the absence of annotation for a regulated gene with that function. The adjusted p-values associated to the occurrence frequencies for each category/module are displayed on the right side adjacent of the corresponding module
It is important to emphasize that all modules and supermodules may not be biologically relevant in transcriptional control alone. Module 10 and supermodule D, which are both statistically under-represented in the differentially expressed genes, are a possible example of this. On the other hand, their slight over-representation in the unchanged genes and their associated GO processes may suggest their role in regulating “housekeeping” functions. Further analysis of modules, their organization and their putative regulatory elements with novel algorithms will aid in elucidating the finer details of cis-regulatory control.
A preliminary analysis of the predicted transcription factor binding sites the consensus sequence for each module suggests the presence of both NF-κB and c-Jun controlled apoptosis. Modules 2, 4, 5 and 10 have multiple matches against the NF-κB consensus-binding site. The genes downstream from supermodules B and C, which contain the above modules, are closely linked to apoptosis, inflammation, viral infection response, and cell cycle arrest [45, 46]. The involvement of both c-Jun and NF-κB mediated signaling is known to be critical in the onset of reovirus-induced apoptosis: The targeted disruption of NF-κB activation results in the inhibition of programmed cell death. The potential binding sites for c-Jun (in modules 2, 3, 5 and 6) suggest a subtle difference in the distribution of NF-κB and c-Jun binding sites among the supermodules. This may be important because the differential activation of these two transcription factors in response to reovirus infection is associated with differences in cell fate. For example, the transient activation of NF-κB and sustained AP-1 activation is associated with apoptosis in hepatocytes, whereas prolonged NF-κB activation and a lack of AP-1 activation results in proliferation [47]. Furthermore, TNFR1 mediated signaling during hepatitis virus C infection increases the transcript levels of NF-κB and AP-1 through the activation of IκB kinase and JNK [48]. The balance between these two mechanisms during the response to reovirus infection can determine shifts in the cellular commitment for apoptosis or for survival. Therefore further analysis of the distribution of potential transcription factor binding sites in modules could help elucidate the control of gene expression by signaling pathways at a finer level.
The results in Table 1 and in Table 2 indicate that modules and supermodules are found in differentially expressed genes as well as genes that are unchanged in expression or transcriptionally inactive. A possible explanation is that genes sharing common regulatory modules can be subject to different transcriptional enhancement or repression mechanisms. For example, the mechanisms of enhancement and repression can be controlled by variable transcription factor binding sites within the modules or outside the modules. This is plausible, since individual transcription factors can recognize variable sequence motifs, and the differential binding of these proteins to their target sites can alter the expression of their downstream genes. Another explanation emerges from the use of stringent statistical criteria for identifying differential expression, and the limited sensitivity of Affymetrix gene array technology for identifying subtle transcriptional changes. Both factors could account for incorrect categorization of many biologically relevant genes as being unchanged in expression. These are important issues, which we are addressing in ongoing research.
The complex combinatorial nature of transcription regulation events is likely to represent the norm in higher order eukaryotic organisms, especially when addressing complex cellular events such as the response to pathogen infection. This stems in part from the nature of the protein complexes involved in the regulation process and that are involved in the execution of the instructions within the genetic regulatory logic apparatus. Rather than focusing on individual transcription factors we explored the occurrence patterns and frequencies of the identified individual predicted regulatory elements and their higher organization. The organization of individual modular elements into supermodules follows the same spatial constraint rule, i.e. the modules occur in the same order within a given supermodule. Also, the distance between individual modules across the genes that share it is conserved, with the distance variation between supermodules being very small. We believe that it is the specific patterns of transcription factors binding in the modules and supermodules that ultimately determine the pattern of gene expression induced in a cell in response to a stimulus such as a viral infection. However, it is likely that the spatial arrangement of modules and their orientation and location in relation to the transcription start site also play a significant role.
Expression analysis using gene arrays suggests that reovirus infection induces alterations at the transcriptional level on a limited set of genes known to participate in cell cycle regulation, interferon-related response and apoptosis, among other processes. To resolve the transcriptional response in terms of signaling pathways, however, the cis-regulatory analysis of modules and supermodules proves to be very useful. Further analysis of the regulatory elements within modules can be used to understand the putative transcription factor binding sites and to identify possible mechanisms for signal transduction pathways controlling gene expression. The pathways identified in this work are consistent with results from previous studies indicating the involvement of the G2/M cell cycle arrest mechanisms and signal transduction via FasL and TRADD [49]. The role of interferon-mediated signaling has been previously associated with reovirus infection events [39-43], possibly via an autocrine feedback loop. Our work independently elucidates the known processes involved in reovirus infection and also suggests the possibility of a broader transcription control network involving signaling pathways and gene regulation through supermodules.
We conclude that the integrated analysis of gene expression and the 5’ upstream genomic regions in terms of modules is a powerful approach for elucidating signal transduction-mediated activity in the response of cells to extracellular stimuli. The approach presented in this paper can be useful for the analysis of gene regulation from other large-scale expression datasets.
ACKNOWLEDGEMENTS
Research support included NIH grants R01 NS050139 (KLT), R01 NS051403 (KLT), 5U01AA013524 (IS), NIAA U01-INIA-BIOINFORMATICS (RL), GM30324 (GLJ), and a MERIT Grant from the Department of Veterans Affairs (KLT). The UCHSC Cancer Center Genomics Core provided additional technical and analytic assistance with GeneChips.
REFERENCES
- 1.Pipel Y, Sudarsanam P, Church GM. Identifying regulatory networks by combinatorial analysis of promoter elements. Nat. Genet. 2001;29:153–9. doi: 10.1038/ng724. [DOI] [PubMed] [Google Scholar]
- 2.Tatematsu K, Ward S, Leyser O, Kamiya Y, Nambara E. Identification of cis-elements that regulate gene expression during initiation of axillary bud outgrowth in Arabidopsis. Plant Physio. 2005;138:757–66. doi: 10.1104/pp.104.057984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Virgin HW, IVth., Tyler KL, Dermody TS. Reovirus. In: Nathanson N, editor. Viral Pathogenesis. Lippincott Raven; Philadelphia: 1997. pp. 669–699. [Google Scholar]
- 4.Tyler KL. Mammalian Reoviruses. In: Knipe DM, Howley PM, editors. Fields Virology. 4th edition Lippincott-Williams & Wilkins; Philadelphia: 2001. pp. 1729–1745. [Google Scholar]
- 5.Clarke P, Richardson-Burns SM, DeBiasi RL, Tyler KL. Mechanisms of apoptosis during reovirus infection. Current. Topics Microbiol. Immunol. 2005;289:1–24. doi: 10.1007/3-540-27320-4_1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Clarke P, DeBiasi RL, Goody R, Hoyt CC, Richardson-Burns S, Tyler KL. Mechanisms of reovirus-induced cell death and tissue injury: Role of apoptosis and virus-induced perturbation of host-cell signaling and transcription factor activation. Viral Immunol. 2005;18:89–116. doi: 10.1089/vim.2005.18.89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Clarke P, Tyler KL. Reovirus-induced apoptosis: A minireview. Apoptosis. 2003;8:141–150. doi: 10.1023/a:1022966508671. [DOI] [PubMed] [Google Scholar]
- 8.Forrest JC, Dermody TS. Reovirus receptors and pathogenesis. J. Virol. 2003;77:9109–15. doi: 10.1128/JVI.77.17.9109-9115.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Qiu P. Recent advances in computational promoter analysis in understanding the transcriptional regulatory network. Biochem. Biophys. Res. Commun. 2003;309:495–501. doi: 10.1016/j.bbrc.2003.08.052. [DOI] [PubMed] [Google Scholar]
- 10.Aerts S, Thijs G, Coessens B, Staes M, Moreau Y, De Moor B. Toucan: deciphering the cis-regulatory logic of coregulated genes. Nucleic Acids Res. 2003;31:1753–64. doi: 10.1093/nar/gkg268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Vilo J, Brazma A, Jonassen I, Robinson A, Ukkonen E. Mining for putative regulatory elements in the yeast genome using gene expression data. Proc. Int. Conf. Intell. Syst. Mol. Biol. 2000;8:384–94. [PubMed] [Google Scholar]
- 12.Birnbaum K, Benfey PN, Shasha DE. cis element/transcription factor analysis (cis/TF): a method for discovering transcription factor/cis element relationships. Genome Res. 2001;11:1567–73. doi: 10.1101/gr.158301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bussemaker HJ, Li H, Siggia ED. Regulatory element detection using correlation with expression. Nat. Genet. 2001;27:167–71. doi: 10.1038/84792. [DOI] [PubMed] [Google Scholar]
- 14.Ellrott K, Yang C, Sladek FM, Jiang T. Identifying transcription factor binding sites through Markov chain optimization. Bioinformatics. 2002;18(Suppl 2):S100–S109. doi: 10.1093/bioinformatics/18.suppl_2.s100. [DOI] [PubMed] [Google Scholar]
- 15.Halfon MS, Grad Y, Church GM, Michelson AM. Computation-based discovery of related transcriptional regulatory modules and motifs using an experimentally validated combinatorial model. Genome Res. 2002;12:1019–28. doi: 10.1101/gr.228902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hannenhalli S, Levy S. Predicting transcription factor synergism. Nucleic Acids Res. 2002;30:4278–84. doi: 10.1093/nar/gkf535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Qiu P, Ding W, Jiang Y, Greene JR, Wang L. Computational analysis of composite regulatory elements. Mamm. Genome. 2002;13:327–32. doi: 10.1007/s00335-001-2141-8. [DOI] [PubMed] [Google Scholar]
- 18.Rebeiz M, Reeves NL, Posakony JW. SCORE: a computational approach to the identification of cis-regulatory modules and target genes in whole-genome sequence data. Site clustering over random expectation. Proc. Natl. Acad. Sci. USA. 2002;99:9888–93. doi: 10.1073/pnas.152320899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Werner T. Cluster analysis and promoter modeling as bioinformatics tools for the identification of target genes from expression array data. Pharmacogenomics. 2001;2:25–36. doi: 10.1517/14622416.2.1.25. [DOI] [PubMed] [Google Scholar]
- 20.Berman BP, Nibu Y, Pfeiffer BD, Tomancak P, Celniker SE, Levine M, Rubin GM, Eisen MB. Exploiting transcription factor binding site clustering to identify cis-regulatory modules involved in pattern formation in the Drosophila genome. Proc. Natl. Acad. Sci. USA. 2002;99:757–62. doi: 10.1073/pnas.231608898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hughes JD, Estep PW, Tavazoie S, Church GM. Computational identification of cis-regulatory elements associated with groups of functionally related genes in Saccharomyces cerevisiae. J. Mol. Biol. 2000;296:1205–14. doi: 10.1006/jmbi.2000.3519. [DOI] [PubMed] [Google Scholar]
- 22.Qin ZS, McCue LA, Thompson W, Mayerhofer L, Lawrence CE, Liu JS. Identification of co-regulated genes through Bayesian clustering of predicted regulatory binding sites. Nat. Biotechnol. 2003;21:435–9. doi: 10.1038/nbt802. [DOI] [PubMed] [Google Scholar]
- 23.GuhaThakurta D, Palomar L, Stormo GD, Tedesco P, Johnson TE, Walker DW, Lithgow G, Kim S, Link CD. Identification of a novel cis-regulatory element involved in the heat shock response in Caenorhabditis elegans using microarray gene expression and computational methods. Genome Res. 2002;12:701–12. doi: 10.1101/gr.228902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sinha S, Tompa M. Discovery of novel transcription factor binding sites by statistical overrepresentation. Nucleic Acids Res. 2002;30:5549–60. doi: 10.1093/nar/gkf669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bailey TL, Baker ME, Elkan CP. An artificial intelligence approach to motif discovery in protein sequences: application to steriod dehydrogenases. J. Steroid Biochem. Mol. Biol. 1997;62:29–44. doi: 10.1016/s0960-0760(97)00013-7. [DOI] [PubMed] [Google Scholar]
- 26.Bailey TL, Elkan CP. The value of prior knowledge in discovering motifs with MEME. Proc. Int. Conf. Intell. Syst. Mol. Biol. 1995;3:21–9. [PubMed] [Google Scholar]
- 27.Clarke P, Meintzer SM, Gibson C, Widmann TP, Garrington GL, Johnson, Tyler KL. Reovirus-induced apoptosis is mediated by TRAIL. J. Virol. 2000;74:8135–9. doi: 10.1128/jvi.74.17.8135-8139.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Connolly JL, Rodgers SE, Clarke P, Ballard DW, Kerr LD, Tyler KL, Dermody TS. Reovirus-induced apoptosis requires activation of transcription factor NF-kB. J. Virol. 2000;74:2981–2989. doi: 10.1128/jvi.74.7.2981-2989.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Clarke P, Meintzer SM, Moffitt LA, Tyler KL. Two distinct phases of virus-induced nuclear factor kappa B regulation enhance tumor necrosis factor-related apoptosis-inducing ligand-mediated apoptosis in virus-infected cells. J. Biol. Chem. 2003;278:18092–100. doi: 10.1074/jbc.M300265200. [DOI] [PubMed] [Google Scholar]
- 30.Clarke P, DeBiasi RL, Meintzer SM, Robinson BA, Tyler KL. Inhibition of NF-κB activity and cFLIP expression contribute to viral-induced apoptosis. Apoptosis. 2005;10:513–524. doi: 10.1007/s10495-005-1881-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.O'Donnell SM, Hansberger MW, Connolly JL, Chappell JD, Watson MJ, Pierce JM, Wetzel JD, Han W, Barton ES, Forrest JC, Valyi-Nagy T, Yull FE, Blackwell TS, Rottman JN, Sherry B, Dermody TS. Organ-specific roles for transcription factor NF-kappaB in reovirus-induced apoptosis and disease. J. Clin. Invest. 2005;115:2341–2350. doi: 10.1172/JCI22428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Clarke P, Meintzer SM, Widmann C, Johnson GL, Tyler KL. Reovirus infection activates JNK and the JNK-dependent transcription factor c-Jun. J. Virol. 2001;75:11275–83. doi: 10.1128/JVI.75.23.11275-11283.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Clarke P, Meintzer SM, Wang Y, Moffitt LA, Richardson-Burns SM, Johnson GL, Tyler KL. JNK regulates the release of proapoptotic mitochondrial factors in reovirus-infected cells. J. Virol. 2004;78:13132–8. doi: 10.1128/JVI.78.23.13132-13138.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM, Davis AP, Dolinski K, Dwight SS, Eppig JT, Harris MA, Hill DP, Issel-Tarver L, Kasarskis A, Lewis S, Matese JC, Richardson JE, Ringwald M, Rubin GM, Sherlock G. Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nat. Genet. 2000;25:25–9. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Draghici S, Khatri, Bhavsar P, Shah A, Krawetz SA, Tainsky MA. Onto-Tools, the toolkit of the modern biologist: Onto-Express, Onto-Compare, Onto-Design and Onto-Translate. Nucleic Acids Res. 2003;31:3775–81. doi: 10.1093/nar/gkg624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bailey TL, Gribskov M. Combining evidence using p-values: application to sequence homology searches. Bioinformatics. 1998;14:48–54. doi: 10.1093/bioinformatics/14.1.48. 1998. [DOI] [PubMed] [Google Scholar]
- 37.Kel AE, Gossling E, Reuter I, Cheremushkin E, Kel-Margoulis OV, Wingender E. MATCH: A tool for searching transcription factor binding sites in DNA sequences. Nucleic Acids Res. 2003;31:3576–9. doi: 10.1093/nar/gkg585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Matys V, Fricke E, Geffers R, Gossling E, Haubrock M, Hehl R, Hornischer K, Karas D, Kel AE, Kel-Margoulis OV, Kloos DU, Land S, Lewicki-Potapov B, Michael H, Munch R, Reuter I, Rotert S, Saxel H, Scheer M, Thiele S, Wingender E. TRANSFAC: transcriptional regulation, from patterns to profiles. Nucleic Acids Res. 2003;31:374–8. doi: 10.1093/nar/gkg108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Azzam-Smoak K, Noah DL, Stewart MJ, Blum MA, Sherry B. Interferon regulatory factor-1, interferon-beta, and reovirus-induced myocarditis. Virology. 2002;298:20–9. doi: 10.1006/viro.2002.1470. [DOI] [PubMed] [Google Scholar]
- 40.Sherry B. The role of interferon regulatory factors in the cardiac response to viral infection. Viral Immunol. 2002;15:17–28. doi: 10.1089/088282402317340206. [DOI] [PubMed] [Google Scholar]
- 41.Hamamdzic D, Phillips-Dorsett T, Altman-Hamamdzic S, London SD, London L. Reovirus triggers cell type-specific proinflammatory responses dependent on the autocrine action of IFN-beta. Am. J. Physiol. Lung Cell Mol. Physiol. 2001;280:L18–29. doi: 10.1152/ajplung.2001.280.1.L18. [DOI] [PubMed] [Google Scholar]
- 42.Stewart MJ, Smoak K, Blum MA, Sherry B. Basal and reovirus-induced beta interferon (IFN-beta) and IFN-beta-stimulated gene expression are cell type specific in the cardiac protective response. J. Virol. 2005;79:79–87. doi: 10.1128/JVI.79.5.2979-2987.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Noah DL, Blum MA, Sherry B. Interferon regulatory factor 3 is required for viral induction of beta interferon in primary cardiac myocyte cultures. J. Virol. 1999;73:10208–13. doi: 10.1128/jvi.73.12.10208-10213.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hagan MP, Yacoub A, Dent P. The switch from DNA repair to apoptosis: discovery of a refractory period for radiation-induced EGFR-MAPK signaling following irradiation. Int. J. Radiat. Oncol. Biol. Phys. 2003;57(2 Suppl):S294–5. [Google Scholar]
- 45.DeBiasi RL, Clarke P, Meintzer S, Jotte R, Kleinschmidt-Demasters BK, Johnson GL, Tyler KL. Reovirus-induced alteration in expression of apoptosis and DNA repair genes with potential roles in viral pathogenesis. J. Virol. 2003;77:8934–47. doi: 10.1128/JVI.77.16.8934-8947.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Liu R, McEachin RC, States DJ. Computationally Identifying Novel NF-kappa B-Regulated Immune Genes in the Human Genome. Genome Res. 2003;13:654–61. doi: 10.1101/gr.911803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ahmed-Choudhury J, Russell CL, Randhawa S, Young LS, Adams DH, Afford SC, Choudhury JA. Differential induction of nuclear factor-Kappa B and activator protein-1 activity after CD40 ligation is associated with primary human hepatocyte apoptosis or intrahepatic endothelial cell proliferation. Mol. Biol. Cell. 2003;14:1334–45. doi: 10.1091/mbc.E02-07-0378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Park KJ, Choi SH, Koh MS, Kim DJ, Yie SW, Lee SY, Hwang SB. Hepatitis C virus core protein potentiates c-Jun N-terminal kinase activation through a signaling complex involving TRADD and TRAF2. Virus Res. 2001;74:89–98. doi: 10.1016/s0168-1702(00)00251-3. [DOI] [PubMed] [Google Scholar]
- 49.Tyler KL, Squier MK, Brown AL, Pike P, Willis D, Oberhaus SM, Dermody TS, Cohen JJ. Linkage between reovirus-induced apoptosis and inhibition of cellular DNA synthesis: role of the S1 and M2 genes. J. Virol. 1996;70:7984–91. doi: 10.1128/jvi.70.11.7984-7991.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]