Abstract
Background
Cia5a is a locus on rat chromosome 10 that regulates disease severity and joint damage in two models of rheumatoid arthritis, collagen- and pristane-induced arthritis (PIA). In this study, we aimed to identify cellular and molecular processes regulated by Cia5a using microarray-based gene expression analysis of synovial tissues from MHC identical DA (severe erosive disease) and DA.F344(Cia5a) congenics (mild non-erosive disease) rats.
Results
Synovial tissues from six DA and eight DA.F344(Cia5a) rats were analyzed 21 days after the induction of PIA using the Illumina RatRef-12 BeadChip (21,922 genes) and selected data confirmed with qPCR. There was a significantly increased expression of pro-inflammatory mediators such as Il1b (5-fold), Il18 (3.9-fold), Cxcl1 (10-fold), Cxcl13 (7.5-fold) and Ccl7 (7.9-fold), and proteases like Mmp3 (23-fold), Mmp9 (32-fold), Mmp14 (4.4-fold) and cathepsins in synovial tissues from DA, with reciprocally reduced levels in congenics. mRNA levels of 47 members of the Spleen Tyrosine Kinase (Syk) pathway were significantly increased in DA synovial tissues compared with DA.F344(Cia5a), and included Syk (5.4-fold), Syk-activating receptors and interacting proteins, and genes regulated by Syk such as NFkB, and NAPDH oxidase complex genes. Nuclear receptors (NR) such as Rxrg, Pparg and Rev-erba were increased in the protected congenics, and so was the anti-inflammatory NR-target gene Scd1 (54-fold increase). Tnn (72-fold decrease) was the gene most significantly increased in DA.
Conclusions
Analyses of gene expression in synovial tissues revealed that the arthritis severity locus Cia5a regulates the expression of key mediators of inflammation and joint damage, as well as the expression of members of the Syk pathway. This expression pattern correlates with disease severity and joint damage and along with the gene accounting for Cia5a could become a useful biomarker to identify patients at increased risk for severe and erosive disease. The identification of the gene accounting for Cia5a has the potential to generate a new and important target for therapy and prognosis.
Keywords: Rheumatoid arthritis, Articular damage, Autoimmune
Background
Rheumatoid arthritis (RA) is a common, chronic and potentially debilitating form of autoimmune erosive arthritis. Advances in the understanding of RA pathogenesis have led to the development of new and better treatments [1-3]. Yet, sustained remission is still rarely achieved [4], and more effective therapies are needed.
The identification of genes implicated in the regulation of arthritis severity and articular damage has the potential to generate new and potentially better targets for therapies aimed at preserving joint architecture and function, and reducing the risk of developing joint deformities. Yet, little is known about those genes [5], and the large cohorts of RA patients used in genome-wide association studies for susceptibility were not designed to address disease severity and articular damage.
We have previously identified several disease severity and articular damage quantitative trait loci (QTL) in rat models of RA [6-10]. Using a combination of positional cloning and functional studies that include transcriptome analyses of synovial cells and synovial tissues we are beginning to understand the molecular processes regulating arthritis severity and joint damage in pristane- and collagen-induced arthritis (PIA and CIA) [10-14]. Similar strategies have been successfully used to identify other autoimmunity genes in rodent models [15,16].
Cia5a is a 20.6Mb QTL on rat chromosome 10 that regulates arthritis severity, cartilage and bone damage, synovial hyperplasia and inflammation in both PIA and CIA [9,10]. In the present study we used synovial tissues from arthritis-protected DA.F344(Cia5a) congenics and from arthritis-susceptible and MHC-identical DA rats in a microarray analysis of gene expression. We determined that the Cia5a locus regulates the expression of several genes central to RA pathogenesis and joint damage, such as cytokines Il1b and Il18, chemokines, proteases, mediators of the synthesis of reactive oxygen species and prostaglandins, and genes involved in Toll-like receptor signaling. Additionally, the expression of 47 members of the Syk kinase pathway genes, including NFκB genes were significantly regulated by the Cia5a locus. Furthermore, the presence of F344 alleles at the Cia5a interval was associated with increased expression of anti-inflammatory genes, including nuclear receptors and Timp3, suggesting that the Cia5a locus contains a gene involved in maintaining an inflammation-free synovial tissue.
Results
DA.F344(Cia5a) congenics develop a mild form of PIA with a distinct pattern of gene expression compared with DA rats
DA.F344(Cia5a) rats developed a significantly milder form of PIA compared with DA rats [median arthritis severity score (25–75 percentiles), DA=26.5 (17–36.9), DA.F344(Cia5a)=5.5 (3.6-7.2); p=0.002, Mann–Whitney test; Figure 1A and B].
Figure 1.
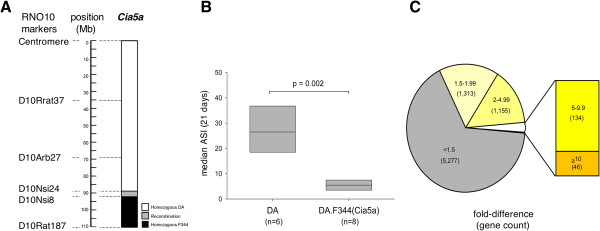
DA and DA.F344(Cia5a) rats differ in arthritis severity and have different synovial gene expression profiles. (A) Map of the Cia5a locus on rat chromosome 10, and the congenic interval boundaries (black=homozygous for F344 alleles; white=homozygous for DA alleles; grey=recombination interval). (B) DA rats had severe disease at 21 days post-induction of PIA; DA.F344(Cia5a) congenics were protected and developed a significantly milder form of arthritis (p=0.002, Mann–Whitney test; boxes show the median and 25%-75% percentiles). (C) 7,925 genes were expressed in all synovial tissues. 2,648 (33.4%) met the 1.5-fold difference and p-value of ≤0.01 (t-test) for significant difference. 134 genes differed by ≥5-fold and less <10-fold, and 46 genes differed by ≥10-fold (inset).
36% (7,925) of the genes in the RatRef-12 BeadChip were consistently expressed in synovial tissues. Nearly one-third of these genes (2,648) met the filtering criteria for differential expression (fold-difference ≥1.5 and p≤0.01). The presence of F344 alleles at the Cia5a interval, as in DA.F344(Cia5a) congenic rats, was associated with increased expression of 1,241 genes and reduced expression of 1,407 genes compared with DA. 134 genes had a ≥5-fold difference between strains (Figure 1C). 46 genes had a ≥10-fold difference in expression, of which 19 were increased and 27 decreased in congenics, compared with DA (Tables 1 and 2).
Table 1.
Genes with ≥10-fold reduction in expression in DA.F344(Cia5a) compared with DA
| Symbol | Name | Entrez Gene ID | Fold reduction | p-value |
|---|---|---|---|---|
|
Tnn |
Tenascin N |
304913 |
71.69 |
8.6x10-13 |
|
Mmp9 |
Matrix metallopeptidase 9 |
81687 |
32.79 |
1.4x10-9 |
|
Cdc2 |
Cell division cycle 2, G1 to S and G2 to M |
54237 |
24.16 |
1.6x10-5 |
|
Mmp3 |
Matrix metallopeptidase 3 |
171045 |
23.94 |
5.0x10-5 |
|
Ccnb2 |
Cyclin B2 |
363088 |
22.95 |
5.0x10-6 |
|
Cthrc1 |
Collagen triple helix repeat containing 1 |
282836 |
17.93 |
3.2x10-9 |
|
Col12a1 |
Collagen, type XII, alpha 1 |
25683 |
16.25 |
7.6x10-8 |
|
Slpi |
Secretory leukocyte peptidase inhibitor |
84386 |
16.16 |
6.4x10-4 |
|
Spc24 |
SPC24, NDC80 kinetochore complex component, homolog |
363028 |
14.72 |
2.5x10-6 |
|
Emilin1 |
Elastin microfibril interfacer 1 |
298845 |
13.55 |
2.4x10-6 |
|
Prc1 |
Protein regulator of cytokinesis 1 |
308761 |
13.08 |
1.6x10-5 |
|
Emb |
Embigin |
114511 |
12.82 |
1.4x10-5 |
|
Nuf2 |
NUF2, NDC80 kinetochore complex component, homolog |
304951 |
12.52 |
2.5x10-6 |
|
Lbp |
Lipopolysaccharide binding protein |
29469 |
11.82 |
8.0x10-5 |
|
Cks2 |
CDC28 protein kinase regulatory subunit 2 |
498709 |
11.62 |
5.9x10-6 |
|
Wisp1 |
WNT1 inducible signaling pathway protein 1 |
65154 |
11.59 |
9.0x10-8 |
|
Cxcl1 |
Chemokine (C-x-C motif) ligand 1 |
81503 |
10.88 |
4.3x10-4 |
|
Steap1 |
Six transmembrane epithelial antigen of the prostate 1 |
297738 |
10.61 |
1.7x10-4 |
| LOC687334 | Similar to cytoskeleton associated protein 2 | 687334 | 10.16 | 9.1x10-6 |
Table 2.
Genes with ≥10-fold increased expression in DA.F344(Cia5a) compared with DA *
| Symbol | Name | Entrez Gene ID | Fold increase | p-value |
|---|---|---|---|---|
|
Scd1 |
Stearoyl-Coenzyme A desaturase 1 |
246074 |
54.62 |
8.8x10-5 |
|
Mpz |
Myelin protein zero |
24564 |
39.80 |
7.7x10-5 |
|
Akr1c19 |
Aldo-keto reductase family 1, member C19 |
307096 |
26.45 |
3.0x10-6 |
|
Nnat |
Neuronatin |
94270 |
22.08 |
5.2x10-7 |
|
Ces3 |
Carboxylesterase 3 |
113902 |
21.06 |
5.4x10-6 |
|
Mup5 |
Major urinary protein 5 |
298107 |
18.41 |
9.5x10-5 |
|
LOC688457 |
Similar to Major urinary protein precursor (MUP) |
688457 |
17.42 |
1.7x10-4 |
|
Abcd2 |
ATP-binding cassette, sub-family D (ALD), member 2 |
84356 |
16.95 |
4.2x10-6 |
|
S100b |
S100 calcium binding protein B |
25742 |
16.81 |
8.9x10-8 |
|
Tshr |
Thyroid stimulating hormone receptor |
25360 |
16.61 |
1.8x10-7 |
|
LOC689147 |
Hypothetical protein LOC689147 |
689147 |
15.84 |
7.9x10-6 |
|
Thrsp |
Thyroid hormone responsive |
25357 |
15.76 |
1.2x10-5 |
|
LOC259244 |
Alpha-2u globulin PGCL3 |
259244 |
15.74 |
1.8x10-4 |
|
Mup4 |
Major urinary protein 4 |
362527 |
15.38 |
9.9x10-5 |
|
Omd |
Osteomodulin |
83717 |
13.82 |
5.8x10-6 |
|
Plekhb1 |
Pleckstrin homology domain containing, family B (evectins) member 1 |
64471 |
13.26 |
2.9x10-5 |
|
Ankrd5 |
Ankyrin repeat domain 5 |
296184 |
13.14 |
4.3x10-5 |
|
Cidea |
Cell death-inducing DNA fragmentation factor, a subunit-like effector A |
291541 |
12.91 |
2.4x10-6 |
|
Atp1a2 |
ATPase, Na+/K+ transporting, alpha 2 polypeptide |
24212 |
12.62 |
6.9x10-5 |
|
Pck1 |
Phosphoenolpyruvate carboxykinase 1 (soluble) |
362282 |
11.95 |
2.4x10-5 |
|
Mrap |
Melanocortin 2 receptor accessory protein |
288271 |
11.80 |
1.8x10-7 |
|
Timp3 |
TIMP metallopeptidase inhibitor 3 |
25358 |
11.48 |
2.9x10-9 |
|
MGC72973 |
Beta-glo |
361619 |
11.38 |
1.4x10-6 |
|
Plp1 |
Proteolipid protein 1 |
24943 |
11.33 |
1.3x10-5 |
|
Plin |
Perilipin |
25629 |
10.99 |
9.1x10-7 |
|
Adipoq |
Adiponectin, C1Q and collagen domain containing |
246253 |
10.96 |
3.3x10-6 |
| Pcbd1 | Pterin-4 a-carbinolamine dehydratase/dimerization cofactor of hepatocyte nuclear factor 1 a |
29700 | 10.74 | 5.8x10-8 |
* Bold= nuclear receptor-inducible gene.
Expression of pro-inflammatory genes, proteases (including matrix metalloproteases, MMPs) and adhesion molecules was significantly increased in DA and decreased in DA.F344(Cia5a)
The 1,407 genes with increased expression in DA and reciprocally decreased expression in DA.F344(Cia5a) congenics included pro-inflammatory cytokines and chemokines implicated in arthritis pathogenesis such as Il1b (5.17-fold on microarray, and 2.46-fold on qPCR), Il18, Mif, Ccl2, Ccl7 and Cxcl13 (Table 3 and Additional file 1: Table S3 and Additional file 2: Table S4). Genes with significantly decreased expression in congenics also included those implicated in the development of cartilage and bone erosions such as MMPs (Mmp3 [24-fold], Mmp9 and Mmp14), and other proteases (cathepsins D, E, K and S) (Table 3 and Figure 2). Interestingly, Syk (see below) has been shown to regulate the expression of differentially expressed MMPs such as Mmp3[17] and Mmp9[18], further suggesting a potential central role for Syk in arthritis and a Syk-regulatory effect of Cia5a. Components of the extracellular matrix (ECM; Cthrc1, Col12a1, Emilin1) also had reduced expression in congenics, and together with the levels of proteases suggested that there was reduced matrix turnover and reduced degradation, compared with arthritic DA rats (Table 3).
Table 3.
Mediators of inflammation and articular damage up-regulated in DA synovium and down-regulation in DA.F344(Cia5a)
| Gene Symbol | Gene Name | Entrez Gene ID | Fold DA/Cia5a | p-value* |
|---|---|---|---|---|
|
Cytokines and chemokines |
|
|
|
|
|
Il1b |
interleukin 1 beta |
24494 |
5.17 |
0.002 |
|
Il18 |
interleukin 18 |
29197 |
3.91 |
0.0002 |
|
Ltb |
lymphotoxin beta |
361795 |
3.77 |
0.0002 |
|
Mif |
macrophage migration inhibitory factor |
81683 |
2.37 |
0.0002 |
|
Aif1 |
allograft inflammatory factor 1 |
29427 |
2.48 |
0.0001 |
|
Ccl2 |
chemokine (C-C motif) ligand 2 |
24770 |
3.95 |
0.01 |
|
Ccl7 |
chemokine (C-C motif) ligand 7 |
287561 |
7.90 |
0.002 |
|
Cxcl1 |
chemokine (C-X-C motif) ligand 1 |
81503 |
10.88 |
0.0004 |
|
Cxcl13 |
chemokine (C-X-C motif) ligand 13 |
498335 |
7.53 |
0.000004 |
|
Proteases |
|
|
|
|
|
Mmp3 |
matrix metallopeptidase 3 |
171045 |
23.94 |
0.0001 |
|
Mmp9 |
matrix metallopeptidase 9 |
81687 |
32.79 |
0.000000001 |
|
Mmp14 |
matrix metallopeptidase 14 |
81707 |
4.46 |
0.000000004 |
|
Mmp19 |
matrix metallopeptidase 19 |
304608 |
5.63 |
0.0000003 |
|
Ctsc |
cathepsin C |
25423 |
2.64 |
0.00007 |
|
Ctsd |
cathepsin D |
171293 |
1.90 |
0.000002 |
|
Ctse |
cathepsin E |
25424 |
2.03 |
0.0002 |
|
Ctsk |
cathepsin K |
29175 |
3.20 |
0.0000003 |
|
Ctss |
cathepsin S |
50654 |
1.68 |
0.002 |
|
Extra-cellular matix |
|
|
|
|
|
Cthrc1 |
collagen triple helix repeat containing 1 |
282836 |
17.93 |
0.000000003 |
|
Col12a1 |
collagen, type XII, alpha 1 |
25683 |
16.25 |
0.00000008 |
|
Emilin1 |
elastin microfibril interfacer 1 |
298845 |
13.55 |
0.000002 |
|
Adhesion molecules |
|
|
|
|
|
Itga5 |
integrin alpha 5 (fibronectin receptor alpha) |
315346 |
3.47 |
0.00000001 |
|
Itgam |
integrin alpha M |
25021 |
3.58 |
0.0001 |
|
Itgav |
integrin alpha V |
296456 |
1.61 |
0.0004 |
|
Itgb2 |
integrin beta 2 |
309684 |
3.44 |
0.0001 |
|
Itgb7 |
integrin, beta 7 |
25713 |
3.86 |
0.002 |
|
Cd44 |
Cd44 molecule |
25406 |
1.83 |
0.005 |
|
Cdh11 |
cadherin 11 |
84407 |
1.87 |
0.003 |
|
Toll-like receptors and regulators of their activity |
||||
|
Cd14 |
CD14 |
60350 |
2.13 |
0.001 |
|
Irak4 |
interleukin-1 receptor-associated kinase 4 |
300177 |
1.65 |
0.0004 |
|
Lpb |
lipopolysaccharide binding protein |
29469 |
4.36 |
0.0003 |
|
Myd88 |
myeloid differentiation primary response gene 88 |
301059 |
1.59 |
0.002 |
|
Pycard |
PYD and CARD domain containing |
282817 |
1.95 |
0.0004 |
|
Tlr2 |
toll-like receptor 2 |
310553 |
4.36 |
0.000 |
|
Tlr6 |
toll-like receptor 6 |
305353 |
1.76 |
0.001 |
|
Prostaglandin and leukotriene synthesis |
||||
|
Ptgs2 |
prostaglandin-endoperoxide synthase 2 |
29527 |
9.73 |
0.0006 |
| Pla2g4a | phospholipase A2, group IVA (cytosolic, calcium-dependent) | 24653 | 2.04 | 0.0004 |
*t-test.
Figure 2.
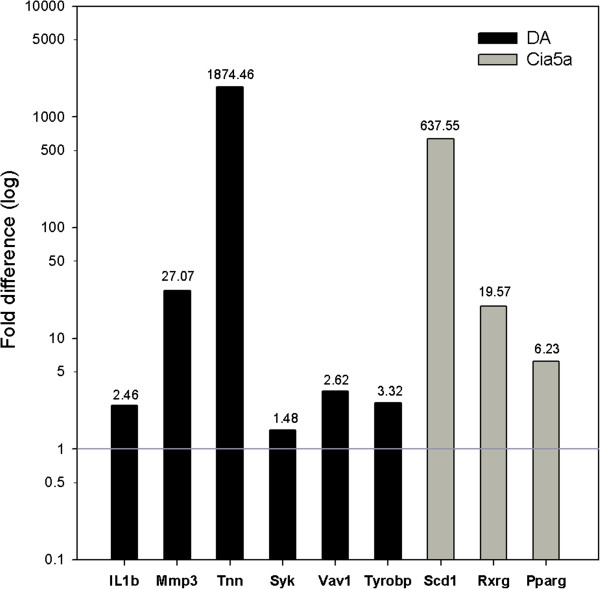
qPCR validation of the microarray results. Genes expressed in increased levels in DA (six genes, black bars) and genes expressed in increased levels in DA.F344(Cia5a) congenics (three genes, grey bars) were selected for qPCR confirmation. Fold-differences were log-transformed. The same RNAs used in the microarray experiments were used for qPCR. ΔCt was used for statistical analyses; all genes had p≤0.04, t-test.
Adhesion molecules required for leukocyte migration into the synovium were increased in DA synovial tissues and decreased in DA.F344(Cia5a), including integrins Itga5, Itgam, Itgb2, Itgb7, and Cd44 (Table 3). Cadherin-11 (Cdh11), a FLS-specific gene required for cell-cell interactions and implicated in FLS invasion and synovial hyperplasia was also decreased in DA.F344(Cia5a) congenics, consistent with the non-hyperplastic synovial tissue previously described in this strain, as opposed to the highly hyperplastic synovial tissue seen in DA [10].
The gene with the most significantly increased expression in DA versus DA.F344(Cia5a) was Tnn (Tenascin N; Table 1 and Figure 2). Tnn has been implicated in osteogenesis and angiogenesis but not in arthritis or inflammation.
These results demonstrate that DA rats with PIA have increased synovial expression of many genes implicated in RA pathogenesis, further validating the molecular similarities between PIA and RA, and underscoring the potential relevance of both Cia5a in arthritis pathogenesis and the present study in discovering new key genes and pathways regulating arthritis.
Increased expression of members of the Syk (spleen tyrosine kinase) pathway in DA synovial tissues, and reciprocally decreased expression in DA.F344(Cia5a)
47 members of the Syk pathway were expressed in significantly increased levels in DA, and in reduced levels in congenics (Table 4, and Figures 2 and 3). These included: a) Syk-activating receptors such as Fcgr2a, Fcer1g, integrins (Itga5, Itgb2, Itgam), c-lectin receptors (Clec4a3, Clec7a [Dectin 1], Clec11a), Trem2 and Dap12 (Tyrobp), b) Syk (5.4-fold) itself, c) Syk-interacting and downstream signaling genes including Vav1, Lcp2 (Slp76), Ptk2b (Pyk2), Lat, Rac2, and Ezr (Vil2), and d) genes belonging to pathways activated by Syk and implicated in arthritis pathogenesis and synovial hyperplasia and pannus formation such as NFκB pathway genes (Fadd, Ikbkb, Nfkb1, Nfkb2), cytokines (Il1b, Ltb, Mif), genes implicated in cell proliferation (Ccnb2, Cdc2, Cks2, Nuf2), cytoskeleton regulation (Actr3, Arpc4, Coro1b, Ezr/Vil2, Myo9b, Parva, Tubb5), and NAPDH oxidase complex genes implicated in the production of reactive oxygen species (ROS) (Ncf1, Ncf2, Ncf4, Cyba) (Table 4).
Table 4.
Members of the Syk kinase pathway up-regulated in DA synovium compared with down-regulation in DA.F344(Cia5a)
| Gene Symbol | Gene Name | Entrez Gene ID | Fold DA/Cia5a | p-value* |
|---|---|---|---|---|
|
Activating receptors |
|
|
|
|
|
Fcer1g |
Fc fragment of IgE, high affinity I, receptor for; gamma polypeptide |
25441 |
2.50 |
0.0004 |
|
Fcgr2a |
Fc fragment of IgG, low affinity IIa, receptor (CD32) |
116591 |
3.08 |
0.005 |
|
Tcrg |
T cell receptor gamma locus |
24821 |
3.32 |
0.0001 |
|
Trem2 |
triggering receptor expressed on myeloid cells 2 |
301227 |
2.69 |
0.0006 |
|
Tyrobp |
Tyro protein tyrosine kinase binding protein |
361537 |
4.80 |
0.0001 |
|
Integrins |
|
|
|
|
|
Itgam |
integrin alpha M |
25021 |
3.58 |
0.0001 |
|
Itgav |
integrin alpha V |
296456 |
1.61 |
0.0004 |
|
Itga5 |
integrin alpha 5 (fibronectin receptor alpha) |
315346 |
3.47 |
0.00000001 |
|
Itgb2 |
integrin beta 2 |
309684 |
3.44 |
0.0001 |
|
Itgb7 |
integrin, beta 7 |
25713 |
3.86 |
0.002 |
|
c-type lectin receptors |
|
|
|
|
|
Clec4a1 |
C-type lectin domain family 4, member a1, Dcir4 |
362430 |
2.38 |
0.001 |
|
Clec4a3 |
C-type lectin domain family 4, member a3, Dcir3 |
362431 |
3.00 |
0.0004 |
|
Clec7a |
C-type lectin domain family 7, member a, Dectin 1 |
502902 |
8.48 |
0.001 |
|
Clec11a |
C-type lectin domain family 11, member a, Scgf |
29313 |
4.24 |
0.00000008 |
|
Syk and Syk-binding and intermediate partners |
||||
|
Ezr** |
ezrin |
54319 |
2.42 |
0.0001 |
|
Lat |
linker for activation of T cells |
81511 |
3.83 |
0.000001 |
|
Lcp2 |
lymphocyte cytosolic protein 2 |
155918 |
2.02 |
0.007 |
|
Ptk2b |
PTK2B protein tyrosine kinase 2 beta |
50646 |
2.54 |
0.0000002 |
|
RhoG |
ras homolog gene family, member G (rho G) |
308875 |
1.54 |
0.0008 |
|
RhoH |
ras homolog gene family, member H |
305341 |
2.25 |
0.001 |
|
Syk |
spleen tyrosine kinase |
25155 |
5.43 |
0.00004 |
|
Vav1 |
vav 1 guanine nucleotide exchange factor |
25156 |
4.60 |
0.0001 |
|
NFκB genes and pathway |
|
|
|
|
|
Ikbkb |
inhibitor of kappa light polypeptide gene enhancer in B-cells, kinase beta |
84351 |
1.60 |
0.0006 |
|
Ikbke |
inhibitor of kappa light polypeptide gene enhancer in B-cells, kinase epsilon |
363984 |
1.64 |
0.0001 |
|
Nfkb1 |
nuclear factor of kappa light polypeptide gene enhancer in B-cells 1 |
81736 |
1.88 |
0.002 |
|
Nfkb2 |
nuclear factor of kappa light polypeptide gene enhancer in B-cells 2, p49/p100 |
309452 |
1.98 |
0.00003 |
|
Cytokine and chemokine transcription§ |
||||
|
Ccl2 |
chemokine (C-C motif) ligand 2 |
24770 |
3.95 |
0.01 |
|
Ccl7 |
chemokine (C-C motif) ligand 7 |
287561 |
7.90 |
0.002 |
|
Il1b |
interleukin 1 beta |
24494 |
5.17 |
0.002 |
|
Ltb |
lymphotoxin beta (TNF superfamily, member 3) |
361795 |
3.77 |
0.0002 |
|
Cell Proliferation |
|
|
|
|
|
Cdc2 |
cell division cycle 2, G1 to S and G2 to M |
54237 |
24.16 |
0.00002 |
|
Ccnb2 |
cyclin B2 |
363088 |
22.95 |
0.000005 |
|
Cks2 |
CDC28 protein kinase regulatory subunit 2 |
498709 |
11.62 |
0.000006 |
|
Prc1 |
protein regulator of cytokinesis 1 |
308761 |
13.08 |
0.00002 |
|
Spc24 |
SPC24, NDC80 kinetochore complex component, homolog (S. cerevisiae) |
363028 |
14.72 |
0.000002 |
|
Cytoskeletal changes |
|
|
|
|
|
Actr3 |
ARP3 actin-related protein 3 homolog (yeast) |
81732 |
1.88 |
0.0001 |
|
Arpc4 |
actin related protein 2/3 complex, subunit 4 |
297518 |
1.61 |
0.00005 |
|
Capzb |
capping protein (actin filament) muscle Z-line, beta |
298584 |
1.64 |
0.0003 |
|
Coro1b |
coronin, actin-binding protein, 1B |
29474 |
1.68 |
0.0001 |
|
Myo9b |
myosin IXb |
25486 |
1.93 |
0.00003 |
|
Tiam2 |
T-cell lymphoma invasion and metastasis 2 |
308142 |
2.61 |
0.00003 |
|
Parva |
parvin, alpha |
57341 |
2.19 |
0.00005 |
|
Reactive oxygen species production |
||||
|
Cyba |
cytochrome b-245, alpha polypeptide |
79129 |
2.72 |
0.00001 |
|
Ncf1 |
neutrophil cytosolic factor 1 |
114553 |
6.00 |
0.0001 |
|
Ncf2 |
neutrophil cytosolic factor 2 |
364018 |
4.75 |
0.0003 |
|
Ncf4 |
neutrophil cytosolic factor 4 |
500904 |
2.46 |
0.004 |
| Rac2¶ | ras-related C3 botulinum toxin substrate 2 (small GTP binding protein Rac2) | 366957 | 2.32 | 0.0007 |
Syk gene is underlined. * t-test;
§=All four chemokine and cytokine listed genes are NFkB-inducible targets.
** Ezrin is also involved in cytoskeleton regulation.
¶ Rac also regulates NFkB activity, cytoskeleton and cell proliferation.
Figure 3.
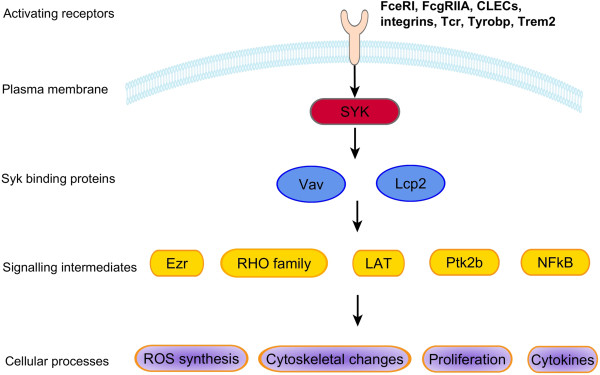
Syk pathway genes and Syk-regulated cellular processes. Genes, gene families and cellular processes with members expressed in increased levels in DA synovial tissues, and significantly reduced levels in DA.F344(Cia5a) congenics.
Interestingly, genes that neutralize ROS (Cat, Sod1, Gss) went on the opposite direction with increased expression in congenics (Table 5).
Table 5.
Anti-inflammatory genes and nuclear receptors up-regulated in DA.F344(Cia5a) congenics
| Gene Symbol | Gene Name | Entrez Gene ID | Fold difference DA/Cia5a | p-value* |
|---|---|---|---|---|
|
Anti-inflammatory and regulators of immune responses |
||||
|
Scd1 |
stearoyl-Coenzyme A desaturase 1 |
246074 |
54.62 |
0.0001 |
|
Timp3 |
TIMP metallopeptidase inhibitor 3 |
25358 |
11.48 |
0.000000003 |
|
Adipoq |
adiponectin, C1Q and collagen domain containing |
246253 |
10.96 |
0.000003 |
|
Ptpn11 |
protein tyrosine phosphatase, non-receptor type 11 |
25622 |
4.18 |
0.00002 |
|
Cyp2j3 |
cytochrome P450, family 2, subfamily j, polypeptide 3 |
313375 |
3.38 |
0.002 |
|
Ptgis |
prostaglandin I2 (prostacyclin) synthase |
25527 |
2.63 |
0.0003 |
|
Cat |
catalase |
24248 |
1.91 |
0.002 |
|
Gss |
glutathione synthetase |
25458 |
1.56 |
0.001 |
|
Sod1 |
superoxide dismutase 1, soluble |
24786 |
1.53 |
0.00002 |
|
Nuclear receptors and an interacting protein |
||||
|
Rxrg |
retinoid X receptor gamma, Nr2b3 |
83574 |
5.39 |
0.0001 |
|
Pparg |
peroxisome proliferator-activated receptor gamma, Nr1c3 |
25664 |
5.32 |
0.00005 |
|
Rev-erba |
Nr1d1 |
252917 |
3.41 |
0.004 |
|
Arp1 |
Nr2f2 |
113984 |
2.45 |
0.0002 |
|
Nrip1 |
nuclear receptor interacting protein 1 |
304157 |
2.35 |
0.00004 |
|
Thrb |
thyroid hormone receptor beta, Nr1a2 |
24831 |
2.07 |
0.002 |
|
Thra |
thyroid hormone receptor alpha, Nr1a1 |
81812 |
1.97 |
0.001 |
|
Ncor1 |
nuclear receptor co-repressor 1 |
54299 |
1.89 |
0.003 |
|
Rora |
RAR-related orphan receptor alpha, Nr1f1 |
300807 |
1.88 |
0.0001 |
| Lxra | Liver X receptor alpha, Nr1h3 | 58852 | 1.52 | 0.0006 |
* t-test.
Additionally, Syk and Vav1 expression levels correlated with the cumulative arthritis severity score (Pearson’s correlation coefficient of 0.8 and p=0.0006 for both genes). Taken together, these observations suggest that the Cia5a QTL contains an arthritis gene that directly or indirectly regulates the expression of Syk pathway genes, providing a possible mechanistic explanation for this locus’ effect on the regulation of disease severity.
DA.F344(Cia5a) congenics have reduced synovial expression of innate immune response-activating genes, including members of the inflammasome
Expression levels of genes implicated in innate immune responses were significantly increased in DA, and decreased in DA.F344(Cia5a). In addition to the Syk pathway, and mediators of ROS synthesis, and regulators of cytokine transcription such as members of the NFκB pathways discussed above, DA.F344(Cia5a) congenics also had reduced expression of AP-1 genes (Fos and JunB), Il1b and other members of the inflammasome (Card11, Nalp3 [both detected only in DA], and Pycard). Pattern recognition receptors such as Zbp1 and Lgp2 (both detected only in DA), and components of the toll-like receptor (TLR) pathway (Cd14, Ikbke, Irak4, Lbp, MyD88, Tlr2, Tlr6, Ticam1; Table 3) were expressed in increased levels in DA and decreased in congenics. Interestingly, and in line with these observations, the expression levels of negative regulators of TLR signaling such as Ptpn11 and Pparg was conversely increased in congenics, suggesting that the arthritis gene located within the Cia5a QTL might mediate the balance between activating and inhibitory signals implicated in TLR signaling.
Genes implicated in the synthesis of prostaglandins and leukotrienes (Pla2g4a, Ptgs2/Cox2, Ptges) were also increased in DA (Table 3), while genes that counteract eicosanoid-mediated inflammation (Ptgis, Cyp2j3) were increased in congenics (Table 5).
Increased expression of anti-inflammatory genes, including nuclear receptors (NRs), in synovial tissues from DA.F344(Cia5a) Congenics
Several genes with known anti-inflammatory and cytokine-suppressing properties were expressed in increased levels in DA.F344(Cia5a) synovial tissues, and reduced in DA. These included the NRs Lxra, Pparg, Rev-erba, Rora, Thra, and Thrb (Table 5).
Scd1 was the gene with the most significantly increased expression in DA.F344(Cia5a) congenics with a 55-fold difference compared with DA (Tables 2 and 5). Scd1 has been shown to reduce cytokine levels and to have anti-inflammatory activity [19,20]. We have previously reported that Scd1 is expressed in significantly reduced levels in synovial tissues from rats with severe arthritis, and increased in the synovial tissues of yet another arthritis-protected congenic strain [14].
Adipoq and Timp3, which is an inhibitor of the TNFα converting enzyme (TACE), were two additional anti-inflammatory genes expressed in significantly increased levels (>10-fold) in DA.F344(Cia5a) (Table 5).
Scd1 and some of the other genes up-regulated in DA.F344(Cia5a) synovial tissues such as Adipoq, Cidea, Cd36, Fabp4, Gpd1, Lpl, Lpin1, Mgst1, Plin, Pck1, Slc2a4 and Srebf1, are known to be inducible by NRs (Table 2 and Table 5). These observations suggest that NRs were not only expressed in increased levels but also had increased activity in synovial tissues from DA.F344(Cia5a) compared with DA.
Genes located within the Cia5a interval have significantly different expression levels
75 of the 7,925 genes expressed by all samples were located within the Cia5a interval. 21 of these 75 had increased expression in DA synovial tissues, and 11 were increased in the congenics. 14 of these 32 differentially expressed genes had ≥2-fold-difference. Sphk1 and Sectm1b were the genes contained within the Cia5a interval with the most significantly increased expression in DA (7.58 and 7.61-fold, respectively), while Itgb4 and DIgr1 and were those with the most significantly increased expression in DA.F344(Cia5a) congenics (2.77 and 2.85-fold, respectively) (Table 6). Additionally, four genes located within the Cia5a interval were expressed only or predominantly in DA synovium, while two other genes were expressed predominantly in DA.F344(Cia5a) (Table 6). It is conceivable that these differences in expression levels of genes located within the Cia5a interval could be explained at least in part by a polymorphism in the 5’ untranslated region (UTR) that affects a transcription factor binding site in cis, thus affecting transcription efficiency, or a 3’ UTR polymorphism affecting mRNA stability.
Table 6.
Differentially expressed candidate genes located within the Cia5a interval on rat chromosome 10*
| Gene Symbol | Gene name | Entrez Gene ID | Difference | p-value§ |
|---|---|---|---|---|
|
Increased in DA |
|
|
Fold DA/Cia5a |
|
|
Igsf7 |
immunoglobulin superfamily, member 7 |
287813 |
4.42 |
0.00008 |
|
Lgals3bp |
lectin, galactoside-binding, soluble, 3 binding protein |
245955 |
3.85 |
0.000001 |
|
RGD1309310 |
similar to mKIAA0195 protein |
303677 |
3.89 |
0.00001 |
|
Sectm1b |
secreted and transmembrane 1B |
287884 |
7.61 |
0.00001 |
|
Slc16a3 |
solute carrier family 16, member 3 |
80878 |
5.36 |
0.000005 |
|
Sphk1 |
sphingosine kinase 1 |
170897 |
7.58 |
0.0000003 |
|
Increased in DA.F344(Cia5a) |
Fold Cia5a/DA |
|
||
|
Itgb4 |
integrin beta 4 |
25724 |
2.85 |
0.0004 |
|
RGD1311422 |
similar to CG8841-PA |
287822 |
2.57 |
0.002 |
|
RGD1561778 |
similar to dendritic cell-derived immunoglobulin(Ig)-like receptor 1, DIgR1 |
303666 |
2.77 |
0.002 |
|
Slc25a10 |
solute carrier family 25, member 10 |
170943 |
2.15 |
0.004 |
|
Expressed only or predominantly in DA |
Frequency DA:Cia5a |
|
||
|
Cd300lf |
CD300 molecule-like family, member f |
287818 |
6:0 |
0.0003 |
|
Fdxr |
ferredoxin reductase |
79122 |
6:1 |
0.0047 |
|
Cd7 |
CD7 molecule |
303747 |
6:0 |
0.0003 |
|
Sectm1a |
secreted and transmembrane 1A |
287885 |
6:1 |
0.0047 |
|
Expressed only or predominantly in DA.F344(Cia5a) | ||||
|
Aanat |
arylalkylamine N-acetyltransferase |
25120 |
0:7 |
0.0047 |
| Hrnbp3 | hexaribonucleotide binding protein 3 | 287847 | 1:6 | 0.1♯ |
*List contains the most signficantly differentially expressed genes; § t-test was used to compare means for fold-difference calculations and Fisher's Exact test to compare frequencies. #not statistically significant.
Gene targets of microRNAs (miRNA) contained within the Cia5a interval were not differentially expressed
The Cia5a interval contains six predicted miRNAs. We considered the possibility that polymorphisms in one of those six miRNAs could account for the Cia5a effect on gene expression and arthritis severity. In that case, such polymorphism would affect the miRNA activity on the transcription of its target genes. Therefore, we look for the differential expression of targets of all six predicted miRNAs located within the Cia5a interval. A list of target and non-target genes was generated for each of the six miRNAs, but no significant over-representation of targets was detected (Figure 4), suggesting that polymorphisms affecting the expression or function of the miRNAs contained within the Cia5a interval are less likely to explain the differences in gene expression identified in this study.
Figure 4.
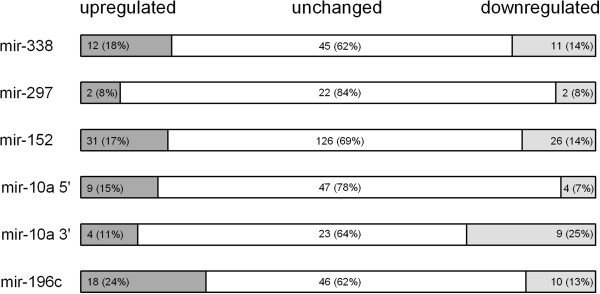
Differentially expressed predicted targets of the miRNAs located within the Cia5a interval. Six miRNAs map to the Cia5a interval. The number of predicted target genes of each miRNA that were up-regulated and down-regulated in DA and DA.F344(Cia5a) congenics was not statistically significant (Chi-square test with the Yates correction). Numbers (percentages intra-parenthesis) of genes expressed in increased levels (dark grey), reduced levels (light grey) or unchanged (white) in DA compared with DA.F344(Cia5a) are shown.
Analyses of cell type specific genes suggests synovial tissue cellularity differences between DA and DA.F344(Cia5a) Congenics
13 genes known to be specifically expressed by the cell types of interest to this study were used to compare DA and DA.F344(Cia5a) synovial tissues (Additional file 3: Table S2). The expression levels of those genes suggested increased numbers of FLS (consistent with synovial hyperplasia), macrophages, dendritic cells (DC), neutrophils and T cells in the synovial tissues of DA, compared with congenics (Additional file 3: Table S2). No gene specific for B cells, NK cells or Tregs were among those differentially expressed between the two strains, suggesting that the number of these cells in the synovial tissues of these two strains was not significantly different.
Discussion
Disease severity and articular damage are associated with increased risk for disability, joint deformities and reduced life expectancy in patients with RA [21-23]. Yet, little is known about the genes implicated in the regulation of disease severity and articular damage genes in RA, and these genes could be the most relevant targets for new therapies aimed at preserving the joint architecture and function.
We have previously identified Cia5a, a 20.6 Mb arthritis severity and joint damage regulatory locus, on rat chromosome 10 [10]. Cia5a co-localizes with other arthritis severity loci identified in other rodent models of arthritis such as oil-induced arthritis (Oia3) [24], and CIA in a DAxACI intercross (Cia27) [25]. There have been no genome-wide association or linkage studies of disease severity and joint damage in RA, and therefore, it is unknown whether the Cia5a syntenic region on human chromosome 17q22-q25 harbors a severity or joint damage arthritis regulatory gene. However, the human 17q22-q25 region contains a locus previously linked with RA susceptibility [26]. In the present study we analyzed synovial tissues from DA rats, which develop severe arthritis (PIA) with pronounced synovial hyperplasia and cartilage and bone destruction, and synovial tissues from the DA.F344(Cia5a) congenics, which develop mild and non-erosive disease. These two strains share the same MHC and are genetically identical except for the presence of F344 alleles at the Cia5a interval, underscoring the magnitude of the effect of this single locus on clinical disease, on histologic joint damage [10] and on gene expression (present study). DA.F344(Cia5a) congenics had significantly reduced expression of genes previously implicated in RA pathogenesis, RA severity and articular damage, including Il1b, Il18, Mif, Mmp3 and Mmp14. These and other similarities between DA rats and RA synovial tissues’ gene expression, such as increased expression of chemokines, matrix proteins, adhesion molecules, mediators of innate immune responses, and others, underscore and further validate the potential clinical relevance of our model and discovery strategy.
We identified a new role for Cia5a on the regulation of the expression of members of the Syk pathway, where forty-seven genes directly or indirectly related to Syk activation were expressed in increased levels in DA, and significantly reduced levels in DA.F344(Cia5a) congenics. Syk is a tyrosine kinase that phosphorylates ITAM motifs in trans-membrane receptors or adaptors, and interacts with partners like Vav, PI3K and Slp76[27]. Syk activation mediates signaling through several cell surface receptors, including those with significantly different levels in this study such C-lectin type receptors, Fcer1g, Fcgr2a, Trem2, Tyrobp, integrins, and the T-cell receptor (TCR) (Figure 3). Resident and infiltrating inflammatory cells in the RA synovial pannus, such as mast cells, macrophages, B and T cells, express these Syk-activating receptors. These resident cells and infiltrating cells have been implicated in arthritis pathogenesis and joint damage, raising the possibility that part of their effect may be mediated by Syk-activating receptors.
Analyses of cell-specific genes suggested reduced numbers of macrophages, dendritic cells, neutrophils and T cells in the synovial tissues of congenics compared with DA, which is in agreement with our previous histologic analyses and might explain part, but not all of the differences in expression of Syk genes. Additionally, DA.F344(Cia5a) congenics had significantly lower levels of the FLS-specific gene Cdh11, compatible with the reduced synovial hyperplasia that we have previously described.
Syk pathway members regulate several cellular processes implicated in arthritis pathogenesis and articular damage, ranging from the production of reactive oxygen species, NFκB activation and the transcription of pro-inflammatory mediators such as Il1b and Ccl2, to the cell proliferation required for the development of synovial hyperplasia, and actin cytoskeleton rearrangements [27]. NFκB activity is regulated by Syk and by several other pathways including TLRs and cytokine receptors [28]. The NFκB pathway has a central role in the production of pro-inflammatory cytokines such as IL-1β, IL-6 and TNFα, in the development of synovial hyperplasia and in disease severity [29-31]. Actin cytoskeleton rearrangements are also regulated by the Syk pathway [27], and are required for the migration of inflammatory cells into the synovial tissue, and for synovial cells and synovial tissue invasion and destruction of cartilage [13,32]. Therefore, our observations suggest that a gene located within the Cia5a interval is a new regulator of the expression of Syk pathway genes implicated in key processes in arthritis pathogenesis.
The precise mechanisms whereby Cia5a regulates the expression of Syk genes remain unclear, and might reflect differences in tissue cellularity, multiple cell-activating processes, or a polymorphism in transcription factor located within the Cia5a interval that affects transcription. Studies by our group of synovial tissues obtained from four different congenic strains yielded different results in gene expression (Brenner et al., manuscript in preparation) [12,14,33], suggesting that the Syk-regulatory effect of Cia5a is a specific observation, and not simply related cellularity differences or inflammation.
Syk has been recently implicated in arthritis pathogenesis and joint damage, and Syk-deficient mice are protected from autoantibody-induced erosive arthritis [34], and treatment with a SYK inhibitor significantly reduced disease severity and joint erosions and damage in collagen-induced arthritis [35]. Both the total and phosphorylated forms of SYK are expressed in increased levels in RA synovial tissues compared with osteoarthritis, and SYK inhibition reduced the expression of IL-6 and MMP-3 [17]. More importantly, the use of a SYK inhibitor significantly reduced disease activity in patients with RA [36], with 67%, 43% and 28% of patients achieving ACR20, ACR50 and ACR70, respectively, in a 3-month double-blind and placebo-controlled study [37]. Therefore, it is conceivable that the Syk pathway genes differentially expressed in this study could help identify patients more likely to benefit from therapy with SYK inhibitors. Additionally, Syk is critical to TNFα-induced responses [38], raising the possibility that the Syk pathway 47-gene signature could be used to predict increased TNFα activity prior to choosing a biologic therapeutic agent. Additionally, the increased expression of Syk pathway genes could identify patients at increased risk to develop erosive disease and could become a prognostic tool. Lastly, the Cia5a gene itself has the potential to become a new target for therapies aimed at reducing articular damage via inhibition of Syk pathway genes, including processes downstream from Syk such as NFκB.
While several genes with pro-inflammatory, proteolytic, innate immunity and inflamasome-related activity were expressed in reduced levels in DA.F344(Cia5a) congenics, groups of genes with known anti-inflammatory properties were expressed in increased levels in congenics. These genes included Timp3, Ptpn11, antagonists of reactive oxygen species (Cat, Gss, Sod1) and nuclear receptors. Nuclear receptors such as Lxra, Pparg and Rora have been shown to interfere with NFκB and AP-1 activation [39-41], and to have anti-inflammatory and arthritis-suppressive properties [42-45]. Rxrg was another nuclear receptor expressed in significantly increased levels in DA.F344(Cia5a) congenics. While Rxrg itself has not been studied in the context of arthritis, it dimerizes with Lxra, Pparg, and with Vdr, and is required for their anti-inflammatory activity. Additionally, several nuclear receptor-inducible genes, including the inflammation-suppressor Scd1[20] were expressed in increased levels in the synovial tissues of the congenics. These observations suggest that not only nuclear receptor levels were increased, but also their activity. We have recently identified a similar nuclear receptor expression signature in another arthritis-protective congenic strain, DA.ACI(Cia25) [14], suggesting that this effect is not specific to the Cia5a locus, but more broadly correlates with preservation of both a normal synovial environment and articular architecture.
The gene with the most significantly increased expression in DA compared with congenics was Tnn (Tenascin N). While little is known about this secreted extra-cellular matrix glycoprotein, it has been implicated in cancer-associated angiogenesis [46], and in integrin-dependent cancer motility [47]. Another member of the tenascin family, Tenascin C (Tnc), was recently shown to be an endogenous activator of TLR4, an inducer of IL-6 and TNFα, and was required for joint damage in arthritic mice [48], suggesting that Tnn could have a function similar to Tnc in arthritis.
Lastly, we considered the possibility that a polymorphism in the 5’ UTR or 3’UTR region of the gene accounting for Cia5a could interfere with its transcription and/or mRNA stability, respectively, leading to increased or reduced gene-specific mRNA levels. We looked for differentially expressed genes and genes preferentially expressed by only one of the strains and located within Cia5a as a clue to the above possibility. Thirty-eight genes met these criteria, and particularly the most significant sixteen genes are interesting candidates that will be studied in detail (Table 6).
Conclusion
In conclusion, in the present study we identified a pattern of gene expression regulated by Cia5a, which included several inflammatory mediators and 47 members of the Syk pathway. Levels of several mediators of arthritis pathogenesis, synovial hyperplasia and articular damage were also reduced in DA.F344(Cia5a) congenics, underscoring the importance of the gene accounting for this locus. Increased expression of nuclear receptors correlated with joint preservation, and a new potential mediator of inflammation, Tnn, was identified for the first time in synovial tissues. Our observations suggest that the gene accounting for Cia5a has the potential to become an important new target for therapies aimed at preserving joint architecture free of damage, and reducing inflammation.
Methods
Rats
DA/BklArbNsi (DA) rats were originally purchased from Bantin & Kingman (Freemont, CA), maintained at the Arthritis and Rheumatism Branch, National Institute of Arthritis and Musculoskeletal and Skin Disease, National Institutes of Health, and then transferred to the Feinstein Institute for Medical Research (FIMR; formerly North Shore-LIJ Research Institute, Nsi). DA.F344(Cia5a) congenic rats were generated as previously described [9]. Briefly, a 20.6Mb interval from chromosome 10 from the arthritis-resistant F344 strain was introgressed into arthritis-susceptible DA rats through genotype-guided breeding (Figure 1A). This strategy selected for F344 alleles at the Cia5a interval while excluding donor genome contamination at other loci known to regulate arthritis [10,49]. Experiments were done with 8–12 week-old male rats homozygous at the congenic interval. All experiments were conducted under an Institutional Animal Care and Use Committee (IACUC)-approved protocol.
PIA and tissue collection
Male DA (n=6) and DA.F344(Cia5a) (n=8) congenic rats were anesthetized and injected intradermally with 150 μl of pristane (MP Bio, Solon, OH) divided into two injection sites at the base of the tail (day 0) [50]. Arthritis severity was assessed with a previously described 80-point scoring system [51]. Ankle synovial tissues were collected 21 days post-induction of arthritis.
RNA extraction
Total RNA was extracted from synovial tissues using the RNeasy Mini Kit (Qiagen, Valencia, CA) according to the manufacturer's instructions and including a DNase treatment step. RNA was quantified and assessed for purity using the NanoDrop spectrophotometer (Rockland, DE). RNA integrity was verified with the BioAnalyzer 2100 (Agilent, Palo Alto, CA).
Microarray
All reagents and procedures were previously optimized for use with the Illumina Whole-Genome Expression platform [12]. Briefly, total RNA (200 ng) was amplified and biotinylated using the TotalPrep labeling kit (Ambion, Austin, TX). Each individual sample was hybridized to one individual array in the RatRef-12 Expression BeadChip (Illumina, San Diego, CA), which contains 22,522 probes covering 21,922 rat genes selected primarily from the NCBI RefSeq database (Release 16). Hybridization was done in Illumina IntelliHyb chambers, followed by washing and staining with Cy3-streptavidin. The BeadChip was scanned on a high-resolution Illumina BeadArray reader using a two-channel 0.8 μm resolution confocal laser scanner.
cDNA synthesis and quantitative real-time PCR (qPCR) expression analysis
Differences in the expression of selected genes from the microarray analyses were validated with qPCR. The qPCR conditions have been described elsewhere [12]. Briefly, total RNA (200 ng) from each sample was used for cDNA synthesis using Superscript III (Invitrogen). Primers and qPCR probes were designed to target the same exons as the corresponding Illumina RatRef-12 Expression BeadChip probes (Additional file 4: Table S1). We used Universal ProbeLibrary (Roche, Indianapolis, IN) and Taqman (ABI, Applied Biosystems, Foster City, CA) probes labeled with FAM at the 5' end and TAMRA at 3' end. Reactions were prepared in duplicates with Eurogentec qPCR MasterMix (San Diego, CA), and run on an ABI Prism 7700 thermocycler using SDS software version 1.9.1 (ABI). Ct (threshold cycle) values were adjusted for Gapdh in each sample (ΔCt). Expression levels (ΔCt) were compared using the t-test and a p-value ≤0.01 was considered significant. Fold-differences were calculated with the 2-ΔΔCt method [52].
MicroRNAs (miRNA)
We considered the possibility that polymorphisms in a miRNA located within the Cia5a interval could account for the Cia5a effect on gene expression and arthritis severity. In that case, such polymorphism would affect the miRNA activity on the transcription of its target genes. Therefore, we looked for miRNAs mapping to the Cia5a interval using the miRBase [53]. Target genes for the miRNAs contained within the Cia5a interval were predicted with TargetScan [54]. Enrichment for differentially expressed predicted targets of miRNAs located within Cia5a was calculated using the Chi-square test with the Yates correction.
Cellular subset gene expression signatures
Differences in tissue resident and infiltrating cell populations can affect the interpretation of gene expression analyses. We looked for cell-specific genes using the GNF Mouse GeneAtlas V3 (Affymetrix MOE430, GEO code GSE10246), a database containing gene expression information for 96 resting and stimulated mouse cell types and tissues [55,56], as well as the BioGPS website (http://www.biogps.org, Scripps Research Institute). The GNF Mouse GeneAtlas V3 did not include fibroblast-like synoviocytes (FLS) or regulatory T cells (Tregs). Therefore, additional non-redundant cell signature genes were obtained from the literature to represent FLS and Tregs [57-65]. We generated a list of genes specific for B cells, T cells, Treg cells, NK cells, FLS, dendritic cells, mast cells, macrophages and neutrophils. We next looked for those cell-specific genes within the list of genes differentially expressed between DA and DA.F344(Cia5a) congenics, as well as in the list of genes preferentially expressed, or only expressed in one strain and not in the other in order to gain insight into differences in cell populations.
Microarray analysis and statistics
Microarray fluorescence intensities were extracted using BeadStudio 2.0 (Illumina). Fluorescent intensities were background-subtracted and then normalized using the cubic spline algorithm. Normalized data were log2-transformed prior to all analyses. Probes consistently expressed in all arrays were included in the analyses. Genes with ≥1.5-fold difference in intensity between DA and DA.F344(Cia5a) and a t-test p-value ≤ 0.01 were considered differentially expressed and selected for pathway detection analyses using IPA 5.5.1 (Ingenuity Systems, Redwood City, CA), as well as public online databases (Ensembl, Genecards, Oncomine, BioGPS, Rat Genome Database) and literature search (Pubmed).
Strain-specific (genes only in one of the strains), or preferential strain (genes expressed in a higher percentage of rats of one strain, and in lower percentage of rats of the other strain) gene expression was determined with the Fisher’s exact test.
Enrichment for biological functions and disease groups was determined with the IPA software and calculated using the Fisher’s exact test with the Benjamini-Hochberg correction and a cutoff p-value of ≤0.05. Enrichment for differentially expressed genes within specific cell subsets, or genes located within the Cia5a interval was calculated using the Fisher’s exact test. Non-normally distributed arthritis severity scores were compared using the Mann–Whitney test.
Funded by a Postdoctoral Fellowship Award from the New Jersey Chapter of the Arthritis Foundation to Dr. M. Brenner, and by the National Institutes of Health grants R01-AR46213, R01-AR052439 (NIAMS) and R01-AI54348 (NIAID) to Dr. P. Gulko.
Competing interests
The authors have no competing financial interests to declare. The results presented in this manuscript are the basis for a recently submitted patent application.
Authors’ contributions
MB carried out the work with rats, including induction of arthritis, tissue dissection and all the steps in the microarray experiments, including a significant role in the analyses, interpretation of the results and manuscript writing. PSG conceived and designed the study and did the statistical and pathway analyses analysis, as well as the manuscript writing. Both authors read and approved the final manuscript.
Supplementary Material
Table S3. Functional categories related to angiogenesis and extra-cellular matrix turnover that were significantly down-regulated in DA.F344(Cia5a) synovium.
Table S4. Functional categories related to pro-inflammatory signals, chemotaxis, and activation of myeloid cells that were significantly down-regulated in DA.F344(Cia5a) synovium*.
Table S2. Detection frequency and expression values of cell subset specific genes in DA and DA.F344(Cia5a) synovial tissues.
Table S1. Primers and probes used for qPCR and the exons they targeted.
Contributor Information
Max Brenner, Email: mbrenner@nshs.edu.
Pércio S Gulko, Email: pgulko@nshs.edu.
References
- Kremer JM, Westhovens R, Leon M, Di Giorgio E, Alten R, Steinfeld S, Russell A, Dougados M, Emery P, Nuamah IF. et al. Treatment of rheumatoid arthritis by selective inhibition of T-cell activation with fusion protein CTLA4Ig. N Engl J Med. 2003;349(20):1907–1915. doi: 10.1056/NEJMoa035075. [DOI] [PubMed] [Google Scholar]
- Weinblatt ME, Kremer JM, Bankhurst AD, Bulpitt KJ, Fleischmann RM, Fox RI, Jackson CG, Lange M, Burge DJ. A trial of etanercept, a recombinant tumor necrosis factor receptor:Fc fusion protein, in patients with rheumatoid arthritis receiving methotrexate. N Engl J Med. 1999;340(4):253–259. doi: 10.1056/NEJM199901283400401. [DOI] [PubMed] [Google Scholar]
- Nishimoto N, Yoshizaki K, Miyasaka N, Yamamoto K, Kawai S, Takeuchi T, Hashimoto J, Azuma J, Kishimoto T. Treatment of rheumatoid arthritis with humanized anti-interleukin-6 receptor antibody: a multicenter, double-blind, placebo-controlled trial. Arthritis Rheum. 2004;50(6):1761–1769. doi: 10.1002/art.20303. [DOI] [PubMed] [Google Scholar]
- Wolfe F, Rasker JJ, Boers M, Wells GA, Michaud K. Minimal disease activity, remission, and the long-term outcomes of rheumatoid arthritis. Arthritis Rheum. 2007;57(6):935–942. doi: 10.1002/art.22895. [DOI] [PubMed] [Google Scholar]
- Marinou I, Maxwell JR, Wilson AG. Genetic influences modulating the radiological severity of rheumatoid arthritis. Ann Rheum Dis. 2010;69(3):476–482. doi: 10.1136/ard.2009.117721. [DOI] [PubMed] [Google Scholar]
- Gulko PS, Kawahito Y, Remmers EF, Reese VR, Wang J, Dracheva SV, Ge L, Longman RE, Shepard JS, Cannon GW. et al. Identification of a new non-major histocompatibility complex genetic locus on chromosome 2 that controls disease severity in collagen- induced arthritis in rats. Arthrititis Rheum. 1998;41(12):2122–2131. doi: 10.1002/1529-0131(199812)41:12<2122::AID-ART7>3.0.CO;2-#. [DOI] [PubMed] [Google Scholar]
- Kawahito Y, Cannon G, Gulko P, Remmers E, Longman R, Reese V, Wang J, Griffiths M, Wilder R. Localization of quantitative trait loci regulating adjuvant induced arthritis in rats: evidence for genetic factors common to multiple autoimmune diseases. J Immunol. 1998;161(8):4411–4419. [PubMed] [Google Scholar]
- Remmers EF, Joe B, Griffiths MM, Dobbins DE, Dracheva SV, Hashiramoto A, Furuya T, Salstrom JL, Wang J, Gulko PS. et al. Modulation of multiple experimental arthritis models by collagen-induced arthritis quantitative trait loci isolated in congenic rat lines: different effects of non-major histocompatibility complex quantitative trait loci in males and females. Arthritis Rheum. 2002;46(8):2225–2234. doi: 10.1002/art.10439. [DOI] [PubMed] [Google Scholar]
- Joe B, Remmers EF, Dobbins DE, Salstrom JL, Furuya T, Dracheva S, Gulko PS, Cannon GW, Griffiths MM, Wilder RL. Genetic dissection of collagen-induced arthritis in chromosome 10 quantitative trait locus speed congenic rats: evidence for more than one regulatory locus and sex influences. Immunogenetics. 2000;51(11):930–944. doi: 10.1007/s002510000231. [DOI] [PubMed] [Google Scholar]
- Brenner M, Meng HC, Yarlett NC, Joe B, Griffiths MM, Remmers EF, Wilder RL, Gulko PS. The Non-MHC quantitative trait locus Cia5 contains three major arthritis genes that differentially regulate disease severity, pannus formation, and joint damage in collagen- and pristane-induced arthritis. J Immunol. 2005;174(12):7894–7903. doi: 10.4049/jimmunol.174.12.7894. [DOI] [PubMed] [Google Scholar]
- Laragione T, Brenner M, Mello A, Symons M, Gulko PS. The arthritis severity locus Cia5d is a novel genetic regulator of the invasive properties of synovial fibroblasts. Arthritis Rheum. 2008;58(8):2296–2306. doi: 10.1002/art.23610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laragione T, Brenner M, Li W, Gulko PS. Cia5d Regulates a new fibroblast-like synoviocyte invasion-associated gene expression signature. Arthritis Res Ther. 2008;10(4):R92. doi: 10.1186/ar2476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laragione T, Gulko PS. MTOR regulates the invasive properties of synovial fibroblasts in rheumatoid arthritis. Mol Med. 2010;16(9–10):352–358. doi: 10.2119/molmed.2010.00049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brenner M, Linge CP, Li W, Gulko PS. Increased synovial expression of nuclear receptors correlates with protection in pristane-induced arthritis: a possible novel genetically regulated homeostatic mechanism. Arthritis Rheum. 2011;63(10):2918–2929. doi: 10.1002/art.30507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rozzo SJ, Allard JD, Choubey D, Vyse TJ, Izui S, Peltz G, Kotzin BL. Evidence for an interferon-inducible gene, Ifi202, in the susceptibility to systemic lupus. Immunity. 2001;15(3):435–443. doi: 10.1016/S1074-7613(01)00196-0. [DOI] [PubMed] [Google Scholar]
- Karp CL, Grupe A, Schadt E, Ewart SL, Keane-Moore M, Cuomo PJ, Kohl J, Wahl L, Kuperman D, Germer S. et al. Identification of complement factor 5 as a susceptibility locus for experimental allergic asthma. Nat Immunol. 2000;1(3):221–226. doi: 10.1038/79759. [DOI] [PubMed] [Google Scholar]
- Cha HS, Boyle DL, Inoue T, Schoot R, Tak PP, Pine P, Firestein GS. A novel spleen tyrosine kinase inhibitor blocks c-Jun N-terminal kinase-mediated gene expression in synoviocytes. J Pharmacol Exp Ther. 2006;317(2):571–578. doi: 10.1124/jpet.105.097436. [DOI] [PubMed] [Google Scholar]
- Letellier E, Kumar S, Sancho-Martinez I, Krauth S, Funke-Kaiser A, Laudenklos S, Konecki K, Klussmann S, Corsini NS, Kleber S. et al. CD95-Ligand on peripheral myeloid cells activates Syk kinase to trigger their recruitment to the inflammatory site. Immunity. 2010;32(2):240–252. doi: 10.1016/j.immuni.2010.01.011. [DOI] [PubMed] [Google Scholar]
- Chen C, Shah YM, Morimura K, Krausz KW, Miyazaki M, Richardson TA, Morgan ET, Ntambi JM, Idle JR, Gonzalez FJ. Metabolomics reveals that hepatic stearoyl-CoA desaturase 1 downregulation exacerbates inflammation and acute colitis. Cell metabolism. 2008;7(2):135–147. doi: 10.1016/j.cmet.2007.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacDonald ML, van Eck M, Hildebrand RB, Wong BW, Bissada N, Ruddle P, Kontush A, Hussein H, Pouladi MA, Chapman MJ. et al. Despite antiatherogenic metabolic characteristics, SCD1-deficient mice have increased inflammation and atherosclerosis. Arterioscler Thromb Vasc Biol. 2009;29(3):341–347. doi: 10.1161/ATVBAHA.108.181099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Zeben D, Breedveld FC. Prognostic factors in rheumatoid arthritis. J Rheumatol Suppl. 1996;44:31–33. [PubMed] [Google Scholar]
- Gossec L, Dougados M, Goupille P, Cantagrel A, Sibilia J, Meyer O, Sany J, Daures JP, Combe B. Prognostic factors for remission in early rheumatoid arthritis: a multiparameter prospective study. Ann Rheum Dis. 2004;63(6):675–680. doi: 10.1136/ard.2003.010611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolfe F, Mitchell DM, Sibley JT, Fries JF, Bloch DA, Williams CA, Spitz PW, Haga M, Kleinheksel SM, Cathey MA. The mortality of rheumatoid arthritis. Arthritis Rheum. 1994;37(4):481–494. doi: 10.1002/art.1780370408. [DOI] [PubMed] [Google Scholar]
- Holm BC, Wei Xu H, Jacobsson L, Larsson A, Luthman H, Lorentzen JC. Rats made congenic for Oia3 on chromosome 10 become susceptible to squalene-induced arthritis. Hum Mol Genet. 2001;10(6):565–572. doi: 10.1093/hmg/10.6.565. [DOI] [PubMed] [Google Scholar]
- Brenner M, Laragione T, Yarlett NC, Li W, Mello A, Gulko P. Cia27 Is a novel non-MHC arthritis severity locus on rat chromosome 10 syntenic to the rheumatoid arthritis 17q22-q25 locus. Genes Immun. 2006;7(5):335–341. doi: 10.1038/sj.gene.6364304. [DOI] [PubMed] [Google Scholar]
- Barton A, Eyre S, Myerscough A, Brintnell B, Ward D, Ollier WE, Lorentzen JC, Klareskog L, Silman A, John S. et al. High resolution linkage and association mapping identifies a novel rheumatoid arthritis susceptibility locus homologous to one linked to two rat models of inflammatory arthritis. Hum Mol Genet. 2001;10(18):1901–1906. doi: 10.1093/hmg/10.18.1901. [DOI] [PubMed] [Google Scholar]
- Mocsai A, Ruland J, Tybulewicz VL. The SYK tyrosine kinase: a crucial player in diverse biological functions. Nat Rev Immunol. 2010;10(6):387–402. doi: 10.1038/nri2765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oeckinghaus A, Hayden MS, Ghosh S. Crosstalk in NF-kappaB signaling pathways. Nat Immunol. 2011;12(8):695–708. doi: 10.1038/ni.2065. [DOI] [PubMed] [Google Scholar]
- Li X, Makarov SS. An essential role of NF-kappaB in the "tumor-like" phenotype of arthritic synoviocytes. Proc Natl Acad Sci U S A. 2006;103(46):17432–17437. doi: 10.1073/pnas.0607939103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jimi E, Aoki K, Saito H, D'Acquisto F, May MJ, Nakamura I, Sudo T, Kojima T, Okamoto F, Fukushima H. et al. Selective inhibition of NF-kappa B blocks osteoclastogenesis and prevents inflammatory bone destruction in vivo. Nat Med. 2004;10(6):617–624. doi: 10.1038/nm1054. [DOI] [PubMed] [Google Scholar]
- Miagkov AV, Kovalenko DV, Brown CE, Didsbury JR, Cogswell JP, Stimpson SA, Baldwin AS, Makarov SS. NF-kappaB activation provides the potential link between inflammation and hyperplasia in the arthritic joint. Proc Natl Acad Sci U S A. 1998;95(23):13859–13864. doi: 10.1073/pnas.95.23.13859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan A, Akhtar M, Brenner M, Zheng Y, Gulko PS, Symons M. The GTPase Rac regulates the proliferation and invasion of fibroblast-like synoviocytes from rheumatoid arthritis patients. Mol Med. 2007;13(5–6):297–304. doi: 10.2119/2007-00025.Chan. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jenkins E, Brenner M, Laragione T, Gulko PS. Synovial expression of Th17-related and cancer-associated genes is regulated by the arthritis severity locus Cia10. Genes Immun. 2012;13(3):221–231. doi: 10.1038/gene.2011.73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jakus Z, Simon E, Balazs B, Mocsai A. Genetic deficiency of Syk protects mice from autoantibody-induced arthritis. Arthritis Rheum. 2010;62(7):1899–1910. doi: 10.1002/art.27438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pine PR, Chang B, Schoettler N, Banquerigo ML, Wang S, Lau A, Zhao F, Grossbard EB, Payan DG, Brahn E. Inflammation and bone erosion are suppressed in models of rheumatoid arthritis following treatment with a novel Syk inhibitor. Clin Immunol. 2007;124(3):244–257. doi: 10.1016/j.clim.2007.03.543. [DOI] [PubMed] [Google Scholar]
- Weinblatt ME, Kavanaugh A, Burgos-Vargas R, Dikranian AH, Medrano-Ramirez G, Morales-Torres JL, Murphy FT, Musser TK, Straniero N, Vicente-Gonzales AV. et al. Treatment of rheumatoid arthritis with a Syk kinase inhibitor: a twelve-week, randomized, placebo-controlled trial. Arthritis Rheum. 2008;58(11):3309–3318. doi: 10.1002/art.23992. [DOI] [PubMed] [Google Scholar]
- Weinblatt ME, Kavanaugh A, Genovese MC, Musser TK, Grossbard EB, Magilavy DB. An oral spleen tyrosine kinase (Syk) inhibitor for rheumatoid arthritis. N Engl J Med. 2010;363(14):1303–1312. doi: 10.1056/NEJMoa1000500. [DOI] [PubMed] [Google Scholar]
- Takada Y, Aggarwal BB. TNF activates Syk protein tyrosine kinase leading to TNF-induced MAPK activation, NF-kappaB activation, and apoptosis. J Immunol. 2004;173(2):1066–1077. doi: 10.4049/jimmunol.173.2.1066. [DOI] [PubMed] [Google Scholar]
- Ogawa S, Lozach J, Benner C, Pascual G, Tangirala RK, Westin S, Hoffmann A, Subramaniam S, David M, Rosenfeld MG. et al. Molecular determinants of crosstalk between nuclear receptors and toll-like receptors. Cell. 2005;122(5):707–721. doi: 10.1016/j.cell.2005.06.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogawa D, Stone JF, Takata Y, Blaschke F, Chu VH, Towler DA, Law RE, Hsueh WA, Bruemmer D. Liver x receptor agonists inhibit cytokine-induced osteopontin expression in macrophages through interference with activator protein-1 signaling pathways. Circ Res. 2005;96(7):e59–e67. doi: 10.1161/01.RES.0000163630.86796.17. [DOI] [PubMed] [Google Scholar]
- Konstantinopoulos PA, Vandoros GP, Sotiropoulou-Bonikou G, Kominea A, Papavassiliou AG. NF-kappaB/PPAR gamma and/or AP-1/PPAR gamma 'on/off' switches and induction of CBP in colon adenocarcinomas: correlation with COX-2 expression. Int J Colorectal Dis. 2007;22(1):57–68. doi: 10.1007/s00384-006-0112-y. [DOI] [PubMed] [Google Scholar]
- Chintalacharuvu SR, Sandusky GE, Burris TP, Burmer GC, Nagpal S. Liver X receptor is a therapeutic target in collagen-induced arthritis. Arthritis Rheum. 2007;56(4):1365–1367. doi: 10.1002/art.22528. [DOI] [PubMed] [Google Scholar]
- Park MC, Kwon YJ, Chung SJ, Park YB, Lee SK. Liver X receptor agonist prevents the evolution of collagen-induced arthritis in mice. Rheumatology (Oxford) 2010;49(5):882–890. doi: 10.1093/rheumatology/keq007. [DOI] [PubMed] [Google Scholar]
- Cuzzocrea S, Mazzon E, Dugo L, Patel NS, Serraino I, Di Paola R, Genovese T, Britti D, De Maio M, Caputi AP. et al. Reduction in the evolution of murine type II collagen-induced arthritis by treatment with rosiglitazone, a ligand of the peroxisome proliferator-activated receptor gamma. Arthritis Rheum. 2003;48(12):3544–3556. doi: 10.1002/art.11351. [DOI] [PubMed] [Google Scholar]
- Wiesenberg I, Chiesi M, Missbach M, Spanka C, Pignat W, Carlberg C. Specific activation of the nuclear receptors PPARgamma and RORA by the antidiabetic thiazolidinedione BRL 49653 and the antiarthritic thiazolidinedione derivative CGP 52608. Mol Pharmacol. 1998;53(6):1131–1138. [PubMed] [Google Scholar]
- Martina E, Degen M, Ruegg C, Merlo A, Lino MM, Chiquet-Ehrismann R, Brellier F. Tenascin-W is a specific marker of glioma-associated blood vessels and stimulates angiogenesis in vitro. FASEB J. 2009;24(3):778–787. doi: 10.1096/fj.09-140491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scherberich A, Tucker RP, Degen M, Brown-Luedi M, Andres AC, Chiquet-Ehrismann R. Tenascin-W is found in malignant mammary tumors, promotes alpha8 integrin-dependent motility and requires p38MAPK activity for BMP-2 and TNF-alpha induced expression in vitro. Oncogene. 2005;24(9):1525–1532. doi: 10.1038/sj.onc.1208342. [DOI] [PubMed] [Google Scholar]
- Midwood K, Sacre S, Piccinini AM, Inglis J, Trebaul A, Chan E, Drexler S, Sofat N, Kashiwagi M, Orend G. et al. Tenascin-C is an endogenous activator of toll-like receptor 4 that is essential for maintaining inflammation in arthritic joint disease. Nat Med. 2009;15(7):774–780. doi: 10.1038/nm.1987. [DOI] [PubMed] [Google Scholar]
- Remmers EF, Longman RE, Du Y, O'Hare A, Cannon GW, Griffiths MM, Wilder RL. A genome scan localizes five non-MHC loci controlling collagen-induced arthritis in rats. Nat Genet. 1996;14(1):82–85. doi: 10.1038/ng0996-82. [DOI] [PubMed] [Google Scholar]
- Vingsbo C, Sahlstrand P, Brun JG, Jonsson R, Saxne T, Holmdahl R. Pristane-induced arthritis in rats: a new model for rheumatoid arthritis with a chronic disease course influenced by both major histocompatibility complex and non-major histocompatibility complex genes. Am J Pathol. 1996;149(5):1675–1683. [PMC free article] [PubMed] [Google Scholar]
- Brenner M, Meng H, Yarlett N, Griffiths M, Remmers E, Wilder R, Gulko P. The non-MHC quantitative trait locus Cia10 contains a major arthritis gene and regulates disease severity, pannus formation and joint damage. Arthritis Rheum. 2005;52(1):322–332. doi: 10.1002/art.20782. [DOI] [PubMed] [Google Scholar]
- Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(−delta delta C(T)) method. Methods. 2001;25(4):402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- Griffiths-Jones S, Grocock RJ, van Dongen S, Bateman A, Enright AJ. MiRBase: microRNA sequences, targets and gene nomenclature. Nucleic Acids Res. 2006;34(Database issue)):D140–D144. doi: 10.1093/nar/gkj112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis BP, Burge CB, Bartel DP. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell. 2005;120(1):15–20. doi: 10.1016/j.cell.2004.12.035. [DOI] [PubMed] [Google Scholar]
- Lattin JE, Schroder K, Su AI, Walker JR, Zhang J, Wiltshire T, Saijo K, Glass CK, Hume DA, Kellie S. et al. Expression analysis of G protein-coupled receptors in mouse macrophages. Immunome Res. 2008;4(1):5. doi: 10.1186/1745-7580-4-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu C, Orozco C, Boyer J, Leglise M, Goodale J, Batalov S, Hodge CL, Haase J, Janes J, Huss JW 3rd. et al. BioGPS: an extensible and customizable portal for querying and organizing gene annotation resources. Genome Biol. 2009;10(11):R130. doi: 10.1186/gb-2009-10-11-r130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feuerer M, Hill JA, Mathis D, Benoist C. Foxp3+ Regulatory T cells: differentiation, specification, subphenotypes. Nat Immunol. 2009;10(7):689–695. doi: 10.1038/ni.1760. [DOI] [PubMed] [Google Scholar]
- Hill JA, Feuerer M, Tash K, Haxhinasto S, Perez J, Melamed R, Mathis D, Benoist C. Foxp3 Transcription-factor-dependent and -independent regulation of the regulatory T cell transcriptional signature. Immunity. 2007;27(5):786–800. doi: 10.1016/j.immuni.2007.09.010. [DOI] [PubMed] [Google Scholar]
- Galligan CL, Baig E, Bykerk V, Keystone EC, Fish EN. Distinctive gene expression signatures in rheumatoid arthritis synovial tissue fibroblast cells: correlates with disease activity. Genes Immun. 2007;8(6):480–491. doi: 10.1038/sj.gene.6364400. [DOI] [PubMed] [Google Scholar]
- Palmer C, Diehn M, Alizadeh AA, Brown PO. Cell-type specific gene expression profiles of leukocytes in human peripheral blood. BMC Genomics. 2006;7:115. doi: 10.1186/1471-2164-7-115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fontenot JD, Rasmussen JP, Gavin MA, Rudensky AY. A function for interleukin 2 in Foxp3-expressing regulatory T cells. Nat Immunol. 2005;6(11):1142–1151. doi: 10.1038/ni1263. [DOI] [PubMed] [Google Scholar]
- Fontenot JD, Rasmussen JP, Williams LM, Dooley JL, Farr AG, Rudensky AY. Regulatory T cell lineage specification by the forkhead transcription factor foxp3. Immunity. 2005;22(3):329–341. doi: 10.1016/j.immuni.2005.01.016. [DOI] [PubMed] [Google Scholar]
- Abbas AR, Baldwin D, Ma Y, Ouyang W, Gurney A, Martin F, Fong S, van Lookeren Campagne M, Godowski P, Williams PM. et al. Immune response in silico (IRIS): immune-specific genes identified from a compendium of microarray expression data. Genes Immun. 2005;6(4):319–331. doi: 10.1038/sj.gene.6364173. [DOI] [PubMed] [Google Scholar]
- Hashimoto S, Nagai S, Sese J, Suzuki T, Obata A, Sato T, Toyoda N, Dong HY, Kurachi M, Nagahata T. et al. Gene expression profile in human leukocytes. Blood. 2003;101(9):3509–3513. doi: 10.1182/blood-2002-06-1866. [DOI] [PubMed] [Google Scholar]
- Obata-Onai A, Hashimoto S, Onai N, Kurachi M, Nagai S, Shizuno K, Nagahata T, Matsushima K. Comprehensive gene expression analysis of human NK cells and CD8(+) T lymphocytes. Int Immunol. 2002;14(10):1085–1098. doi: 10.1093/intimm/dxf086. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S3. Functional categories related to angiogenesis and extra-cellular matrix turnover that were significantly down-regulated in DA.F344(Cia5a) synovium.
Table S4. Functional categories related to pro-inflammatory signals, chemotaxis, and activation of myeloid cells that were significantly down-regulated in DA.F344(Cia5a) synovium*.
Table S2. Detection frequency and expression values of cell subset specific genes in DA and DA.F344(Cia5a) synovial tissues.
Table S1. Primers and probes used for qPCR and the exons they targeted.


