Abstract
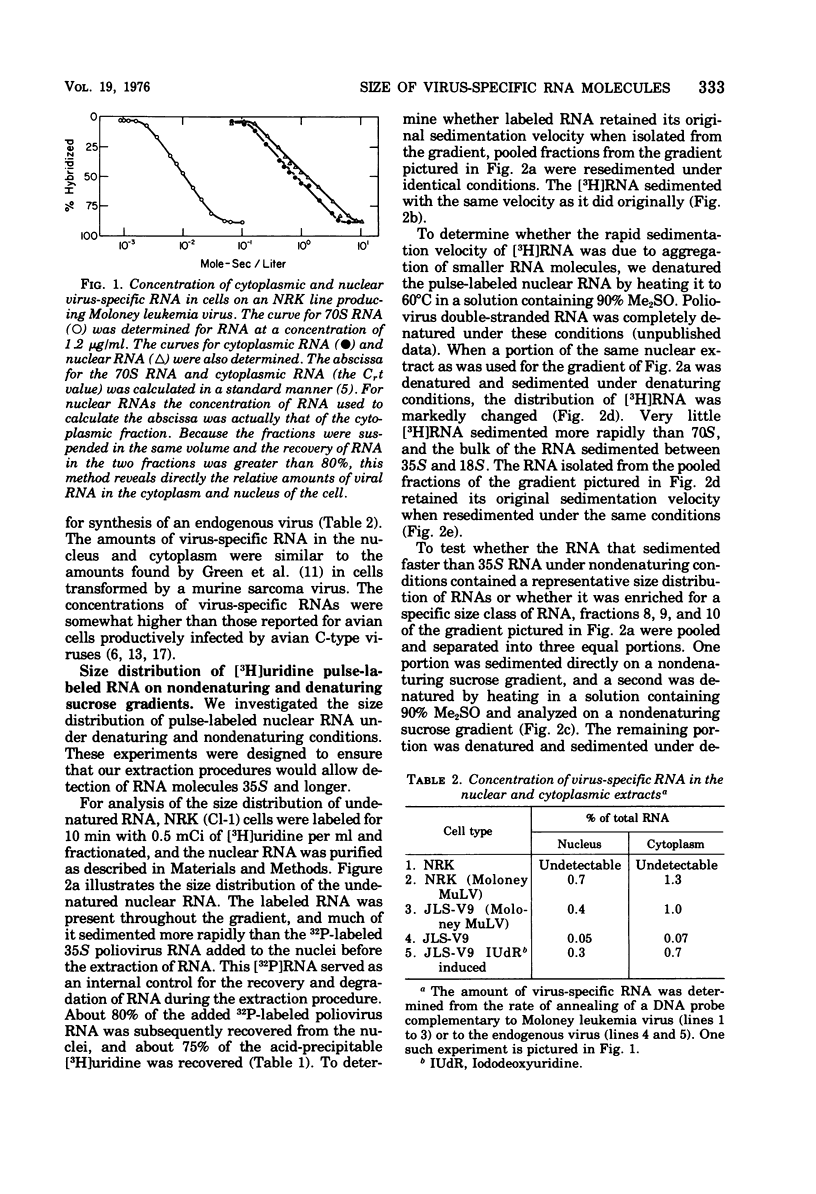
About 1% of the total RNA of cell lines producing murine leukemia virus is virus-specific RNA. About one-third of the virus-specific RNA is located within the nucleus. The size distribution of virus-specific RNA was determined before and after denaturation. Before denaturation, virus-specific RNA sequences sedimented as a heterogeneous population of RNA molecules, some of which sedimented very rapidly. After denaturation, most of the virus-specific RNA had a sedimentation coefficient of 35S or lower, but a small fraction of the nuclear virus-specific RNA sedimented more rapidly than 35S RNA even after denaturation.
Full text
PDF





Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Acheson N. H., Buetti E., Scherrer K., Weil R. Transcription of the polyoma virus genome: synthesis and cleavage of giant late polyoma-specific RNA. Proc Natl Acad Sci U S A. 1971 Sep;68(9):2231–2235. doi: 10.1073/pnas.68.9.2231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Axel R., Schlom J., Spiegelman S. Evidence for translation of viral-specific RNA in cells of a mouse mammary carcinoma. Proc Natl Acad Sci U S A. 1972 Mar;69(3):535–538. doi: 10.1073/pnas.69.3.535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bramwell M. E. A comparison of gel electrophoresis and density gradient centrifugation of heterogeneous nuclear RNA. Biochim Biophys Acta. 1972 Oct 27;281(3):329–337. doi: 10.1016/0005-2787(72)90446-7. [DOI] [PubMed] [Google Scholar]
- Britten R. J., Kohne D. E. Repeated sequences in DNA. Hundreds of thousands of copies of DNA sequences have been incorporated into the genomes of higher organisms. Science. 1968 Aug 9;161(3841):529–540. doi: 10.1126/science.161.3841.529. [DOI] [PubMed] [Google Scholar]
- Coffin J. M., Temin H. M. Hybridization of Rous sarcoma virus deoxyribonucleic acid polymerase product and ribonucleic acids from chicken and rat cells infected with Rous sarcoma virus. J Virol. 1972 May;9(5):766–775. doi: 10.1128/jvi.9.5.766-775.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Kloet S. R., Mayo V. S., Andrean B. A. Disaggregation of giant "DNA-like RNA" of yeast by denaturation in the presence of formaldehyde. Biochem Biophys Res Commun. 1970 Jul 27;40(2):454–460. doi: 10.1016/0006-291x(70)91030-2. [DOI] [PubMed] [Google Scholar]
- Fan H., Baltimore D. RNA metabolism of murine leukemia virus: detection of virus-specific RNA sequences in infected and uninfected cells and identification of virus-specific messenger RNA. J Mol Biol. 1973 Oct 15;80(1):93–117. doi: 10.1016/0022-2836(73)90235-0. [DOI] [PubMed] [Google Scholar]
- Georgieff M., Bachenheimer S., Darnell J. E. An examination of the nuclear RNA of adenovirus-transformed cells. Cold Spring Harb Symp Quant Biol. 1975;39(Pt 1):475–482. doi: 10.1101/sqb.1974.039.01.059. [DOI] [PubMed] [Google Scholar]
- Gielkens A. L., Salden M. H., Bloemendal H. Virus-specific messenger RNA on free and membrane-bound polyribosomes from cells infected with Rauscher leukemia virus. Proc Natl Acad Sci U S A. 1974 Apr;71(4):1093–1097. doi: 10.1073/pnas.71.4.1093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green M., Rokutanda H., Rokutanda M. Virus specific RNA in cells transformed by RNA tumour viruses. Nat New Biol. 1971 Apr 21;230(16):229–232. doi: 10.1038/newbio230229a0. [DOI] [PubMed] [Google Scholar]
- Holmes D. S., Bonner J. Preparation, molecular weight, base composition, and secondary structure of giant nuclear ribonucleic acid. Biochemistry. 1973 Jun 5;12(12):2330–2338. doi: 10.1021/bi00736a023. [DOI] [PubMed] [Google Scholar]
- Leong J. A., Garapin A. C., Jackson N., Fanshier L., Levinson W., Bishop J. M. Virus-specific ribonucleic acid in cells producing rous sarcoma virus: detection and characterization. J Virol. 1972 Jun;9(6):891–902. doi: 10.1128/jvi.9.6.891-902.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parsons J. T., Coffin J. M., Haroz R. K., Bromley P. A., Weissmann C. Quantitative determination and location of newly synthesized virus-specific ribonucleic acid in chicken cells infected with Rous sarcoma virus. J Virol. 1973 May;11(5):761–774. doi: 10.1128/jvi.11.5.761-774.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peltz R. The integrity of "giant" nuclear RNA. Biochim Biophys Acta. 1973 Apr 21;308(7):148–153. doi: 10.1016/0005-2787(73)90131-7. [DOI] [PubMed] [Google Scholar]
- Penman S. RNA metabolism in the HeLa cell nucleus. J Mol Biol. 1966 May;17(1):117–130. doi: 10.1016/s0022-2836(66)80098-0. [DOI] [PubMed] [Google Scholar]
- Schincariol A. L., Joklik W. K. Early synthesis of virus-specific RNA and DNA in cells rapidly transformed with Rous sarcoma virus. Virology. 1973 Dec;56(2):532–548. doi: 10.1016/0042-6822(73)90056-1. [DOI] [PubMed] [Google Scholar]
- Scolnick E. M., Parks W. P. Harvey sarcoma virus: a second murine type C sarcoma virus with rat genetic information. J Virol. 1974 Jun;13(6):1211–1219. doi: 10.1128/jvi.13.6.1211-1219.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Temin H. M., Baltimore D. RNA-directed DNA synthesis and RNA tumor viruses. Adv Virus Res. 1972;17:129–186. doi: 10.1016/s0065-3527(08)60749-6. [DOI] [PubMed] [Google Scholar]
- Tsuchida N., Gilden R. V., Hatanaka M. Size of virus-specific RNA in B-34, a hamster tumor cell producing nucleic acids of type C viruses from three species. J Virol. 1975 Oct;16(4):832–837. doi: 10.1128/jvi.16.4.832-837.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsuchida N., Robin M. S., Green M. Viral RNA subunits in cells transformed by RNA tumor viruses. Science. 1972 Jun 30;176(4042):1418–1420. doi: 10.1126/science.176.4042.1418. [DOI] [PubMed] [Google Scholar]



