Abstract
Background
Glioma is the most common primary malignant central nervous system tumor in adult, and is usually not curable in spite of various therapeutic approaches. Clarification of the oncogenic process in its early stage is important for the diagnosis and effective therapy.
Methods
In the present study, we used the serological identification of antigens by recombinant cDNA expression cloning (SEREX) to explore the subtle changes of the protein expression in low-grade glioma. The levels of serum autoantibodies to the SEREX-identified glioma-related antigens were analyzed by ELISA, and the epitope site was identified using deletion mutants and overlap peptide array. Changes in the serum autoantibody levels were examined in the rat glioma model using C6 and 9 L glioma cell lines.
Results
We identified 31 glioma-related antigens by SEREX. Among them, the serum level of autoantibody to src-homology 3-domain GRB2-like 1 (SH3GL1) was significantly higher in patients with low-grade glioma than healthy volunteers or high-grade gliomas. The 10 amino-acids at the C-terminal were identified as the epitope site by the overlap peptide array and the ELISA using deletion mutants. The tissue expression of SH3GL1 protein increased in proportion to glioma progression. The rat glioma models confirmed the increase of anti-SH3GL1 autoantibody level in the early stage and the suppression in the late stage.
Conclusion
SH3GL1 may be involved in the oncogenic process of gliomas and effectively elicit an autologous antibody response in low-grade gliomas. The immunological reaction to SH3GL1 would contribute to the establishment of a novel diagnostic and therapeutic target for gliomas.
Keywords: Src, SH3GL1, Autoantibody, Glioma, SEREX
Introduction
Glioma is the most common primary malignant central nervous system (CNS) tumor in adults and arises from neuroepithelial cells, mostly astrocytes or oligodendrocytes. Glioma is divided into 4 grades according to World Health Organization (WHO) histological classification, and the prognosis of glioma is still poor [1,2]. Glioblastoma (GB), WHO grade IV, and anaplastic astrocytoma (AA), WHO grade III, are referred to as high-grade glioma, and the median survival time of patients with AA and GB is 2–3 years and only approximately 1.5 years, respectively [2]. In the cases of WHO grade II tumor, the median survival time of patients with diffuse astrocytoma (WHO grade II) is also limited to approximately 5–7 years [3]. In most cases, patients with glioma present large cerebral lesion at diagnosis, which prevents effective removal without neurological deficits, and the remnant tumors relapse even though receiving post-operative treatments with radiotherapy and chemotherapy [4]. The clarification of the oncogenic process especially in the early stage would contribute to its early diagnosis and to new molecular targets.
Serological identification of antigens by recombinant cDNA expression cloning (SEREX) is one of the powerful tools for finding novel cancer antigens [5] and has been applied on a nationwide basis to target many cancers, including gliblastoma [6-8]. However, the specific and crucial changes in the protein expression in low-grade gliomas have not been identified yet. In contrast, it is well known that activation of the receptor tyrosine kinases such as epidermal growth factor receptor (EGFR) is the most frequent molecular aberration found in high-grade gliomas [9]. The receptor tyrosine kinases make the ras pathway activation through a protein-protein interaction of the adaptor protein called GRB2 with Son of Sevenless (Sos) protein through src-homology 3 (SH3) domain [10,11]. The connection of the adaptor protein and Sos is a key step toward activating the ras-mediated oncogenic pathways in the downstream of receptor tyrosine kinases.
In the present study, the authors applied SEREX to glioma to find SH3-domain GRB2-like 1 (SH3GL1) as a novel glioma-related antigen. The levels of serum autoantibodies to SH3GL1 were significantly higher in patients with low-grade gliomas than in healthy donors by ELISA. In contrast, the serum autoantibody level was significantly depressed in high-grade glioma patients compared with low-grade gliomas patients. We identified the epitope site of SH3GL1 by overlap peptide array and an ELISA using deletion mutants. The rat glioma model using C6 and 9 L glioma cells also showed the increases of the anti-SH3GL1 autoantibody level in the early stage and decreases in the late stage. Although low-grade gliomas are not always in an early-stage of the disease, it is usually accepted that gliomas often progress from low-grade tumors to higher-grade tumors as the time proceeds [12]. The present clinical data and the animal models suggested the immunosurveillance can work in low-grade glioma patients and the immune tolerance would occur in those with high-grade gliomas. The present findings would contribute to the knowledge of molecular basis of low-grade gliomas and the establishment of a novel diagnostic and therapeutic target.
Materials and methods
Sera and glioma tissue
Sera were obtained from patients with various types of glioma and from healthy volunteers after they had provided written informed consent. Patients with glioma underwent surgery and the tumor was histologically diagnosed as grade II–IV glioma at Chiba University Hospital in 1998–2008; healthy donors were confirmed to have no cerebral diseases using radiological imaging such as computed tomography or magnetic resonance imaging. No patient received steroid therapy at the time of blood sampling. Each sample was centrifuged at 3 000 × g for 5 min and then frozen at –80°C until use. Glioma tissue was collected from the tumor tissue during surgical treatment. Normal brain tissue, which did not show any glioma cell infiltration under microscopic examination, was isolated from the circumference of the glioma specimen and from non-neoplastic CNS tissues that were obtained during a lesionectomy from a patient with intractable epilepsy or during a lobectomy from patients with benign CNS tumors, such as meningioma. The Local Ethical Review Board of the Graduate School of Medicine, Chiba University approved the studies in this issue, and we obtained written informed consent from the patients and healthy volunteers concerning the use of material for scientific research.
Phage cDNA library
A total RNA was prepared from the human glioblastoma cell-line U-87 MG (ATCC, HTB-14) using the acid guanidium thiocyanate-phenol-chloroform method with an mRNA purification kit (AquaPure RNA isolation kit, BioRad, Hercules, CA) used in accordance with the manufacturer’s instructions. Double-stranded cDNA was synthesized through conventional procedures and ligated into the EcoRI-XhoI site of λZAP II phage. The library size was over 1.0 × 106 PFU/ml.
Immunological screening using SEREX
E. coli XL1-Blue MRF’ was infected with λZAP II phages containing a cDNA library and the expression of cDNA was induced by blotting on nitrocellulose membranes, pretreated with 10 mM isopropyl-β-D-thiogalactoside (IPTG; Wako Pure Chemicals, Osaka, Japan). After washing and blocking, the membranes were exposed in 1:2000-diluted serum for 1 h. The membranes were treated with 1:5000-diluted alkalinephosphatase-conjugated goat anti-human IgG (Jackson ImmunoResearch Laboratories, West Grove, PA). After incubation in a color development solution containing 0.3 mg/ml of nitroblue tetrazolium chloride (Wako Pure Chemicals) and 0.15 mg/ml of 5-bromo-4-chloro-3-indolylphosphate (Wako Pure Chemicals), positive reactions were detected. Positive clones were re-cloned twice to obtain monoclonality.
Sequence analysis of identified clones
Monoclonalized phage cDNA clones were converted to pBluescript phagemids through in vivo excision using ExAssist helper phage (Stratagene, La Jolla, CA). Plasmid DNA was obtained from an E. coli SOLR strain transformed by the phagemid. The inserted cDNAs were sequenced using the dideoxy chain termination method and the sequences were analyzed for homology with a public database provided by the National Center for Biotechnology Information (NCBI).
Production of glutathione S-transferase (GST) fusion proteins
cDNA inserts of these clones incorporated in pBluescript were cleaved by EcoRI and XhoI generally and cloned into the EcoRI-XhoI site of pGEX-4 T-3, pGEX-4 T-2, and pGEX-4 T-1 vectors (Amersham Bioscience, Piscataway, NJ) that express recombinant GST fusion proteins. E. coli JM109 cells containing pGEX clones (A600 = 0.3–0.5) were cultured in 200 ml of Luria broth (LB), and lysed through sonication. The lysate was then centrifuged and the GST-fusion proteins in the supernatants were purified by glutathione-Sepharose. These samples were centrifuged and affinity-purified with glutathione-Sepharose.
ELISA
Purified recombinant proteins diluted at 10 μg protein/ml in PBS were added to each well of 96-well plates and incubated at room temperature overnight. As a control, the same amount of GST was applied. Sera diluted at 1:100 in PBS with 10% FBS were added to the wells and incubated for 1 h. The wells were exposed to 1:2 000-diluted horseradish peroxidase-conjugated goat anti-human IgG antibody (Jackson ImmunoResearch Laboratories, West Grove, PA). Then, 100 μl of a peroxidase substrate (o-phenylenediamine, 0.4 mg/ml) containing 0.02% (v/v) H2O2 were added. Absorbance at 490 nm was determined using a microplate reader (Emax, Molecular Devices, Sunnyvale, CA).
Construction of SH3GL1 deletion mutants
Some deletion constructs of SH3GL1 were obtained through digestion with restriction enzymes or the inverse PCR method. The SEREX-identified phage clone was containing a full-length coding sequence of SH3GL1 (1–368 amino acids), that comprised Bin-Amphiphysin-Rvs (BAR) domain (amino acid positions between 5 and 242) in the N-terminal portion, coiled-coil (CC) domain (amino acid proteins between 180 and 250) at the middle, and the SH3 domain (amino acid positions between 309 and 364) in the C-terminal portion. The region of SH3GL1 cDNA corresponding to amino acids between 260 and 368 was cleaved by SmaI and XhoI and subcloned into the pGEX 4 T-3 vector at the SmaI-XhoI digestion sites (SH3GL1 mut-1). Amino acids 316–368, 260–289 and 354–368 were deleted through the inverse PCR method with the KOD–Plus Mutagenesis Kit (Toyobo, Osaka, Japan) using the SH3GL1 mut-1 cDNA as a template (SH3GL1 mut-2, 3 and 4, respectively). The primers for SH3GL1 mut-2 were forward 5′-CCAGTCTTCCGACAAGCCCATC-3′, reverse 5′-TGGGGATCCACGCGGAACCAG-3′; for SH3GL1 mut-3 were forward 5′-TCGAGCGGCCGCATCGTGAC-3′, reverse 5′-GCCCGACTGGCCGTCCAGCATG-3′; and for SH3GL1 mut-4 were; forward 5′-TCGAGCGGCCGCATCGTGAC-3′, reverse 5′-GCCCGACTGGCCGTCCAGCATG-3′.
Overlap peptide array
Peptides spanning amino acid residues 1–368 of SH3GL1 were synthesized on cellulose membranes as a series of peptides with the overlapping by 12 amino acids using F-moc amino acids according to the manufacturer’s protocol (Auto spot robot ASP222; ABIMED Analysen-Technik GmbH, Langenfeld, Germany) as previously described [13]. Membranes were incubated with the sera of patients at 1:200 dilutions for more than 12 h. Then, the antigen-antibody complexes were detected with FITC-conjugated goat anti-human IgG (109-095-098; Jackson ImmunoResearch, West Grove, PA) at 1:10000 dilutions. The fluorescence of the peptide spots were detected using Typhoon 9400 (Amersham Biosciences, Stockholm, Sweden) with a 488 nm/520 nm filter. The scanned image was also analyzed with CS analyzer ver. 3.0 (Atto & Rise Corporation, Tokyo, Japan) and fluorescent intensity of each spot was calculated.
Immunohistochemical staining for SH3GL1 protein
Immunohistochemistry with the polyclonal antibody against SH3GL1 (sc-25495; Santa Cruz) was performed using commercially available reagents, Histofine (Nichirei Bioscience Inc, Tokyo, Japan), and according to the manufacturer’s recommendations. This antibody was confirmed to be cross-reactive for human, mouse, and rat SH3GL1. Sections were counterstained with hematoxylin, then dehydrated and mounted.
Staining of tissue specimens was observed in 100 × fields with approximately all fields presenting glioma cells. The staining intensity in cytosole was classified into 5 groups, absent (−), light partial staining (±), homogeneous light staining (+), partly strong positive staining (++) and homogeneous strong positive staining (+++).
Brain Tumor Model, Monitoring of Tumor Size, and Serum Sampling
Rat C6 glioma cells and 9 L gliosarcoma cells were originally obtained from ATCC and maintained in Dulbecco's modified Eagle medium (D-MEM) supplemented with 10% fetal calf serum in a humidified atmosphere of 5% CO2. Male Wister rats for C6 cells and Fisher rats for 9 L cells, weighing between 200 and 240 g (7–8 weeks old) were used. The animals were anesthetized and placed in a stereotaxic apparatus. A burr hole was made at 4 mm posterior to bregma and 3 mm right to midline. A 25-gauge needle was inserted to the point of 3 mm ventral from dura where 1 × 105 syngeneic C6 or 9 L tumor cells in 10 μl medium were slowly injected. To estimate i.c. tumor volume sequentially, all the animals were examined with a 7 tesla MRI every 7 days started on day 7 after the tumor inoculation. The sera were obtained from tail vein every 7 days. The animal experimentation was reviewed and approved by the Institutional Animal Care and Use Committee of National Institute of Radiological Science.
Statistical analysis
The significance of differences among healthy donors, patients with low-grade glioma, and patients with high-grade glioma was calculated using the Kruskal Wallis H-test and the Mann–Whitney U-test with Bonferroni correction. Differences were considered significant only if p < 0.05. The overall survivals from the date of initial diagnosis were estimated using Kaplan-Meier methodology and compared by the Log rank test to estimate the clinical significance of production of autoantibody for SH3GL1.
Results
Serological screening of cDNA library
The phage expression library was constructed using mRNA derived from the U-87 MG glioblastoma cell-line. To identify glioma-associated antigens, a total of 5 × 106 cDNA clones were screened using sera from 48 patients with glioma and 57 reacting clones were isolated from 19 of 48 sera. DNA sequence analysis and a search for homologous sequences in an NCBI-accessible database indicated that these isolated clones comprised 31 independent genes (Table 1).
Table 1.
Genes identified by SEREX
| Gene name | Symbol | NCBI accession no. | Coding sequence | cDNA inserts of recombinant protein† |
|---|---|---|---|---|
| amplified in breast cancer 1 |
ABC1 |
NM_022070 |
18.3563 |
|
| anillin, actin binding protein (scraps homolog, Drosophilia) |
ANLN |
NM_018685 |
205.3579 |
|
| ATP synthase, H + transporting, mitochondrial F1complex, beta polypeptide, nuclear gene encoding mitochondrial protein |
ATP5B |
NM_001686 |
106.1695 |
|
| catenin (cadherin-associated protein), alpha-like 1 |
CTNNAL1 |
NM_003798 |
22.2248 |
|
| CDV3 homolog (mouse) |
CDV3 |
NM_017548 |
316.1092 |
|
| centromere protein F, 350/400 ka (mitosin) |
CENPF |
NM_016343 |
175.9519 |
3553.4866 |
| chromosome 14 open reading frame 145 |
C14orf145 |
NM_152446 |
172.3456 |
|
| coagulation factor III (thromboplastin, tissue factor) |
F3 |
NM_001993 |
124.1011 |
|
| coiled-coil domain containing 86 |
CCDC86 |
NM_024098 |
56.1138 |
|
| cyclin G1, transcript variant 2 |
CCNG1 |
NM_199246 |
135.1022 |
|
| eukaryotic translation elongation factor 1 alpha 1 |
EEF1A1 |
NM_001402 |
64.1452 |
|
| ferritin, heavy polypeptide 1 |
FTH1 |
NM_002032 |
236.787 |
|
| ferritin, light polypeptide |
FTL |
NM_000146 |
200.727 |
|
| heterogeneous nuclear ribonucleoprotein C (C1/C2), transcript variant 4 |
HNRPC |
NM_001077443 |
219.1100 |
|
| homeobox B2 |
HOXB2 |
NM_002145 |
121.1191 |
|
| Homo sapiens mRNA for KIAA0146 gene, partial cds. |
KIAA0146 |
NM_001080394 |
1.3218 |
|
| macrophage migration inhibitory factor |
MIF |
NM_002415 |
98.445 |
23.561 |
| myosin phosphatase-Rho interacting protein, transcript variant 1 |
M-RIP |
NM_015134 |
57.3173 |
2194.3856 |
| nucleolar protein 8 |
NOL8 |
NM_017948 |
304.3807 |
|
| oral-facial-digital syndrome 1 |
OFD1 |
NM_003611 |
312.3350 |
|
| postmeiotic segregation increased 1(S.cerevisiae) |
PMS1 |
NM_000534 |
231.3029 |
|
| retinoblastoma binding protein 8, transcript variant 1 |
RBBP8 |
NM_002894 |
332.3025 |
473.1274 |
| ribosomal protein, large, P0, transcript variant 1 |
RPLP0 |
NM_001002 |
179.1131 |
433.1217 |
| RNA export 1 homolog (S.pombe), transcript variant 1 |
RAE1 |
NM_003610 |
342.1448 |
|
| serine/threonine kinase 3(STE20 homolog, yeast) |
STK3 |
NM_006281 |
142.1617 |
|
| SH3-domain GRB2-like 1 |
SH3GL1 |
NM_003025 |
107.1213 |
43.1615 |
| synaptonemal complex protein SC65 |
SC65 |
NM_006455 |
289.1598 |
|
| TAF7 RNA polymerase II, TATA box binding protein (TBP)-associated factor, 55 kDa |
TAF7 |
NM_005642 |
741.1790 |
1578.2310 |
| talin 1 |
TLN1 |
NM_006289 |
91.7716 |
5712…8187 |
| transforming growth factor, beta-induced, 68 kDa |
TGFBI |
NM_000358 |
48.2099 |
1371…2691 |
| unc-45 homolog A (C.elegans), transcript variant 2 or 3 | UNC45A | NM_001039675 | 836.3625 | 1924.3471 |
† cDNA inserts of positive clones were successfully expressed into proteins followed by ELISA.
The GST-fusion recombinant proteins were successfully produced using pGEX-4 T vectors in 10 of 31 antigens—centromere protein F, 350/400 ka (CENPF); macrophage migration inhibitory factor (MIF); myosin phosphatase-Rho interacting protein (M-RIP); retinoblastoma binding protein 8 (RBBP8); ribosomal protein, large, P0 (RPLP0); SH3GL1, TAF7 RNA polymerase II, TATA box binding protein-associated factor, 55 kDa (TAF7); talin 1 (TLN1); transforming growth factor beta-induced 68 kDa (TGFBI), and unc-45 homolog A (UNC45A) (Figures 1 and 2).
Figure 1.
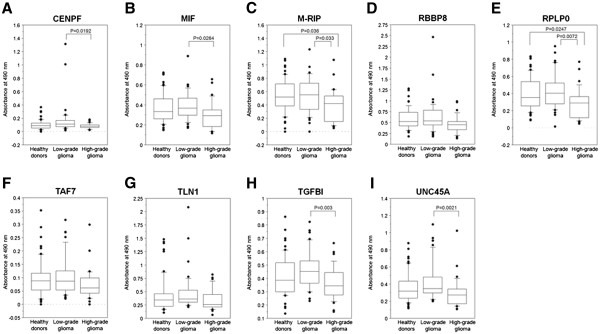
Serum antibody levels of glioma SEREX antigens. cDNA inserts of identified clones were recombined in-frame into pGEX vectors that express recombinant GST fusion proteins. Using the fusion proteins as antigens, the levels of antibodies were examined by the ELISA and shown by the ordinate, as (A) CENPF, (B) MIF, (C) M-RIP, (D) RBBP8, (E) RPLP0, (F) TAF7, (G) TLN1, (H) TGFBI, (I) UNC45A. The significance of differences among healthy donors, patients with low-grade glioma and with high-grade glioma was calculated using Kruskal Wallis H-test and Mann–Whitney U-test with Bonferroni correction. The box-and-whisker plots display the 10th, 25th, 50th, 75th and 90th percentiles.
Figure 2.
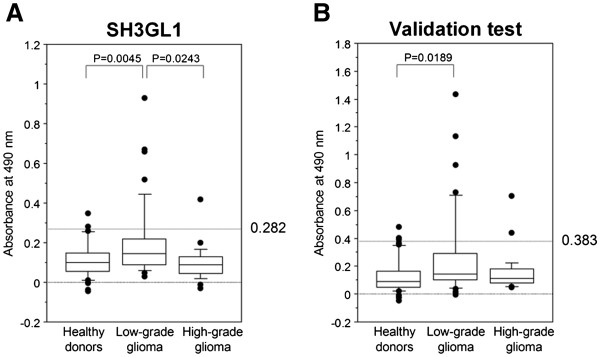
The increasing levels of antibodies to SH3GL1 in sera of the patients with low-grade glioma. Serum antibody level to SH3GL1 was examined by the ELISA as described in the legends of Figure 1. First screening test (A) and the individual validation test (B), revealed the significant higher levels of autologous antibody against SH3GL1 in low-grade glioma patients, than healthy donors (P = 0.045 and 0.0189).
ELISA to detect serum antibodies
Using a recombinant antigen protein, ELISA was performed on sera from 32 patients with high-grade glioma, 40 with low-grade glioma and 56 healthy volunteers, which were collected between 1998 and 2005 in Chiba University Hospital. The serum used for SEREX screening was excluded. The characteristics of the sera are shown in Table 2 (left).
Table 2.
Characteristics of screening serum sets
| |
1st sampling test |
Validation test |
|---|---|---|
| Sampling periods | 1998-2005 | 2005-2008 |
|
No. of patients |
128 |
115 |
| Healthy donors, n (%) |
56 (43.8) |
48 (41.7) |
| Age (mean ± SD), range |
48.1 ± 18.3, 16-76 |
51.3 ± 15.2, 16-76 |
| Low-grade glioma, n(%) |
40 (31.3) |
42 (36.5) |
| Age (mean ± SD), range |
45.8 ± 14.8, 20-74 |
44.2 ± 14.1, 22-78 |
| Pilocytic astrocytoma, n (%) |
2 (5.0) |
4 (9.5) |
| Diffuse astrocytoma, n (%) |
18 (45.0) |
15 (35.7) |
| Oligodendroglioma, n (%) |
16 (40.0) |
19 (45.2) |
| Oligoastrocytoma, n (%) |
3 (7.5) |
1 (2.4) |
| Ependymoma, n (%) |
1 (2.5) |
|
| Ganglioglioma, n (%) |
|
3 (7.1) |
| High-grade glioma, n (%) |
32 (25.0) |
25 (21.7) |
| Age (mean ± SD), range |
49.7 ± 18.3, 8-78 |
49.8 ± 15.5, 28-78 |
| Glioblastoma, n (%) |
24 (75.0) |
17 (68.0) |
| Anaplastic astrocytoma, n (%) |
5 (15.6) |
3 (12.0) |
| Anaplastic oligodendroglioma, n (%) |
2 (6.3) |
2 (8.0) |
| Anaplastic oligoastrocytoma, n (%) |
1 (3.1) |
1 (4.0) |
| Anaplastic ependymoma, n (%) |
|
1 (4.0) |
| Choroid plexus carcinoma, n (%) | 1 (4.0) |
The levels of serum antibodies of CENPF, MIF, M-RIP, RPLP0, TGFBI and UNC45A were significantly lower in patients with high-grade glioma than in those with low-grade glioma (Figure 1A-C, E, H and I) and, moreover, the levels of anti-M-RIP and anti-RPLP0 antibodies in patients with high-grade glioma were also significantly lower than in healthy volunteers (Figure 1C and E).
The levels of serum antibodies to SH3GL1 were significantly higher in patients with low-grade glioma than those with high-grade glioma (P = 0.0243) and healthy volunteers (P = 0.0045) (Figure 2A). When the antibody levels were divided into 2 groups with a cut-off value of 0.383 corresponding to the mean + 2 standard deviations (SD) of SH3GL1 antibodies in healthy volunteers, the positive rate of patients with low-grade glioma was 62.5% (25 of 40), whereas those of patients with high-grade glioma and healthy volunteers were 8.9% (5 of 56) and 15.6% (5 of 32), respectively.
Independent validation test for the levels of antibodies to SH3GL1
To verify the generality of low-grade glioma-specific increase in serum antibodies to SH3GL1, an independent validation test was carried out using other serum set. In validations, consecutive serum samples that were collected in 2005–2008 after the first serum sampling, were enrolled, and no apparent differences in the characteristics were observed between the 2 serum sets (Table 2). The results of the ELISA based on the newly collected serum set showed that the levels of serum autoantibodies to SH3GL1 were significantly higher than those of healthy donors (P = 0.0189) (Figure 2B). Although there was no statistical significance in the levels of antigens between patients with low- and high-grade glioma, similar distribution was recognized. In the combined population of the first sampling test and the validation test, there was a significant difference between low-grade gliomas and high-grade gliomas (p = 0.0351).
The same results of both ELISA based on the independent serum sets support the possibility that SH3GL1 is aberrantly expressed and efficiently elicits a systemic immune response in low-grade glioma patients. The level of anti-SH3GL1 autoantibody could be a novel low-grade glioma-specific serum marker. In contrast, the lower serum autoantibody levels against these determined SEREX-antigens in patients with high-grade glioma as opposed to those with low-grade glioma and healthy volunteers suggest that the existence of some immunosuppressive mechanisms in high-grade gliomas.
Patients survival
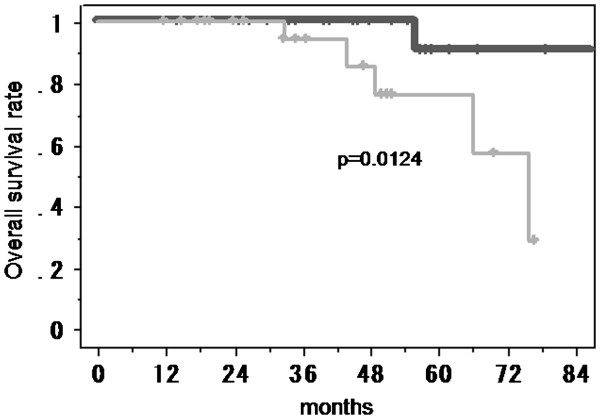
Overall survival of the patients with low-grade gliomas according to the serum level of anti-SH3GL1 autoantibody was analyzed by Kaplan-Meier analysis. The patients included in the test set and the validation set were divided into 2 groups with a cut-off value of the mean + 1 SD of anti-SH3GL1 antibodies in healthy volunteers. The patients with higher serum level of anti-SH3GL1 autoantibody survived significantly longer than those with lower levels (p = 0.0124) (Figure 3).
Figure 3.
Kaplan-Meier analysis for the overall survival of the patients with low-grade gliomas according to the serum level of anti-SH3GL1 autoantibody. The patients with higher serum level of anti-SH3GL1 autoantibody (solid line) survived significantly longer than those with lower levels (gray line) (p = 0.0124).
Search for epitope sites of SH3GL1
To determine the accurate immuno-reactive site, an ELISA using 4 deletion mutants of SH3GL1 cDNA was performed. The BAR domain deletion mutant, identified as SH3GL1 mut-1, was obtained first, and the N-terminal and C-terminal deletion mutants of SH3GL1 mut-1 were produced, as SH3 mut-2 and 3, respectively (Figure 4A). The serum antibody levels to SH3GL1 mut-1 and mut-3 in the patients with low-grade glioma were still significantly higher than those in other groups (Figures 4B and D), while the levels of anti-SH3GL1 mut-2 showed no difference among the groups (Figure 4C). Although these results indicated that the C-terminal of SH3GL1 contributed to the immune-response, the differences were disappeared in SH3GL1 mut-4, deleting only 15 amino acids at the 3′ end of SH3GL1 mut-1 (Figure 4D). These results were suitable for that of overlap peptide array, and approximately the 15 amino acids in the C-terminal of SH3GL1 are indispensable as the epitope recognized by serum antibodies in the patient with low-grade glioma.
Figure 4.
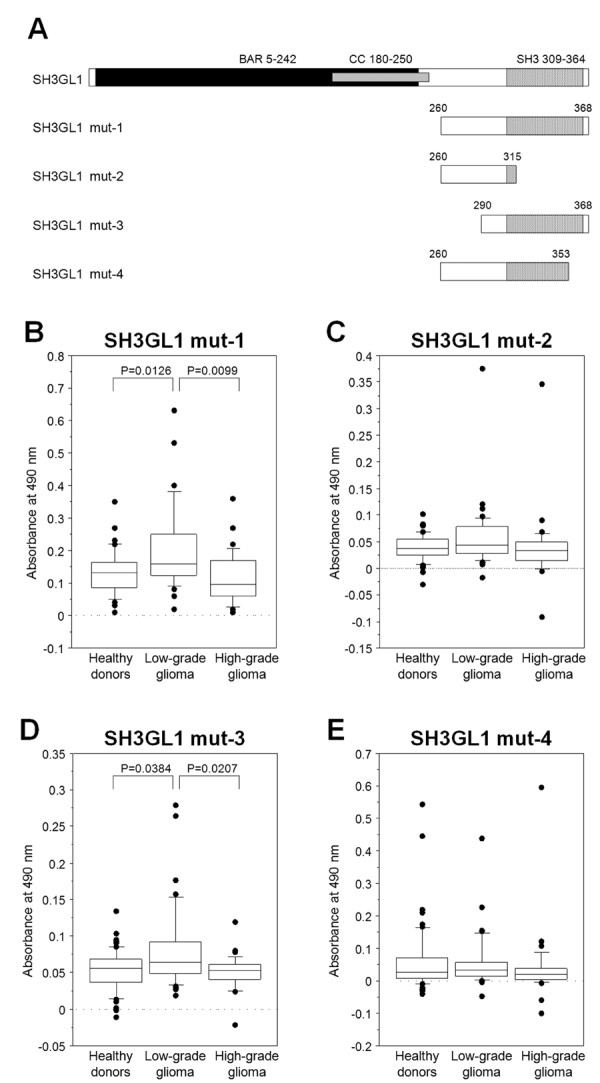
Comparison of serum antibody levels among deletion mutants of SH3GL1. To confirm the epitope site, some SH3GL1 deletion mutants (A) were synthesized. Serum antibody levels were examined by ELISA with SH3GL1 muta-1 (B), mut-2 (C), mut-3 (D) and mut-4 (E), and the 10–20 amino acids at the C-terminal end were indicated as the epitope site.
To confirm the epitope site in the deletion mutant ELISA, overlap peptide array, which is a much useful analysis based on the SPOT-synthesis technique, was applied. In the present analysis, series of peptides of 14 amino acid residues, composed of full-length SH3GL1, were synthesized with overlapping by 12 amino acids, and were blotted in nitrocellulose membranes using F-moc amino acids (Figure 5A). Three representative higher immune-reactive sera of the patients with low-grade glioma, two of the normal volunteers and PBS without serum as background control, were applied in the peptide array (Figure 5B-C). All of three sera of patients showed the fine specific reaction in two consecutive blots, spot 177 and 178, indicating the C-terminal-end of SH3GL1, comparing with the sera from normal volunteers. The calculated fluorescence intensity normalized by background control (Figure 5E) revealed that the common sequence in 2 reactive blots, FPLSYVEVLVPL, was suggested as a minimum epitope site.
Figure 5.
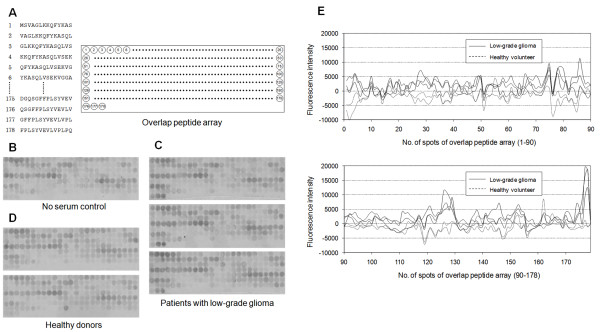
The detection of epitope site by overlapped peptide array. Series of peptides of 14 amino acid residues, composed of SH3GL1, were synthesized with overlapping by 12 amino acids, and were blotted in nitrocellulose membranes using F-moc amino acids (A). Three sera of the patients with low-grade glioma indicated the fine reaction in spot 177 and 178 (C), compared to two normal volunteers (D) and no serum control (B). The calculated fluorescence intensity, normalized by background control, revealed that these spots were suggested as a minimum epitope site (E).
Immunohistochemical staining for SH3GL1 protein
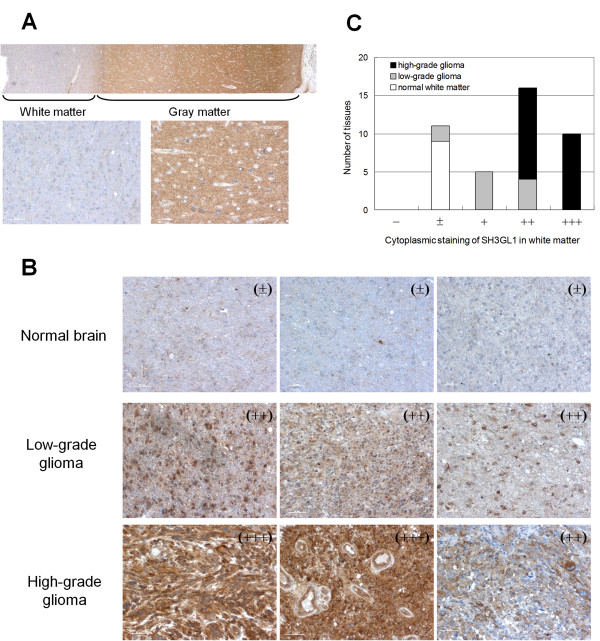
To verify the SH3GL1 expression in glioma tissues directly, immunohistochemical stains for SH3GL1 was obtained in normal brain, low-grade glioma and high-grade glioma. In the normal brain, clear contrast was observed between gray matter (cerebral cortex) and white matter (medulla) (Figure 6A). In the gray matter, where neuronal cells (neurons) abundantly existed, cytoplasm was stained homogeneously, while nuclei were occasionally stained in white matter, which contained mainly glial cells.
Figure 6.
Immunohistochemical analysis of SH3GL1 in glioma cells. Immunohistochemical stain for SH3GL1 in whole normal brain, consisted of white matter and gray matter (A), and three representative results of normal white matter, low-grade glioma and high-grade glioma (B) were shown. Immunostaining for SH3GL1 was classified in five groups, and numbers of tissues in each group were scored (C).
It is known that glioma cells are commonly localized in white matter and progress along neural fibers [14]. Therefore, we compare the immunostaining levels between normal glial cells in white matter and glioma cells. In glioma tissues, strong positive staining of SH3GL1 was observed in the cytoplasms but not in the nucleus (Figure 6B). The levels of stain in white matter increased according to the malignancy of tumors; that is, high-grade glioma tissues were most heavily stained while normal glial cells were barely stained (Figures 6C). These results indicated that the protein levels of SH3GL1 were much higher in glioma cells than in normal glial cells in white matter.
Alteration of anti-SH3GL1 autoantibody level in rat glioma model
To confirm the changes in the serum anti-SH3GL1 autoantibody level, we used rat glioma models with C6 and 9 L cells which expressed its messenger RNA (data not shown). In the models, the brain tumors constantly became visible on MRI at 2-week after tumor inoculation and over 200 mm3 at 4-week (Figure 7A). All the tumor-bearing animals died within 5 weeks from the tumor inoculation. In the C6 glioma model, the serum levels of autoantibody to SH3GL1 significantly increased in the rats at 2-week after tumor inoculation compared with those at 3-day after the inoculation (p = 0.0028) (Figure 7B). In contrast, at the time of 4-week after the inoculation, the serum levels tended to decrease. In the other experiment using 9 L gliosarcoma cells, the result showed the same tendency without statistical significance (data not shown). These results show that the serum levels of autoantibody to SH3GL1 increased at the early stage of the animal models and turned to decrease at the late stage according to the increase of tumor volume as the time proceeded.
Figure 7.
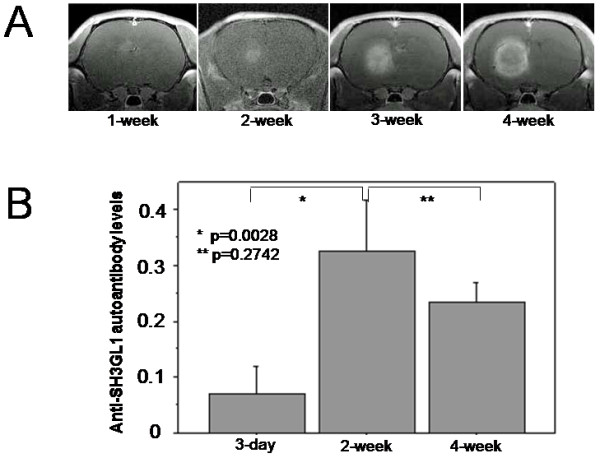
Changes in the serum autoantibody level to SH3GL1 in a rat brain tumor model using C6 rat glioblastoma cells which were confirmed to express SH3GL1 protein. MRI studies show a steady growth of tumor mass in the rat brain (A). The serum autoantibody levels were significantly increased at 2-week after tumor inoculation, and tended to decrease at 4-week after the inoculation (B).
Discussion
The molecular pathogenesis of glioblastoma has been well characterized and involves both gain and loss of a number of genes participating in proliferative or mitogenic signals. One of the most prevalent molecular changes consists of aberrant activation of EGFR, which occur in 50% of glioblastoma, but not seen in low-grade astrocytomas [12,15]. We have shown in this study that the SH3-domain of GRB2-like protein, which links the receptor tyrosine kinases activation to the ras pathway, had already overexpressed in low-grade gliomas and strongly induced a humoral immune response. In high-grade gliomas, the tissue expression of SH3GL1 was further increased, but the immune response was suppressed. Although there are few reports describing overexpression of this protein in human cancers, SH3GL1 protein is related to the activation of MLL proto-oncogene by chromosomal translocation [16]. Solitary SH3GL1 overexpression in NIH3T3 cells also reported to do some oncogenic behaviors in vivo [17,18]. It is not clear whether the overexpression is a result of amplification of receptor tyrosine kinases or not. However, the net result of these signaling complexes induces the shift of ras-GDP to its activated form ras-GTP, and may lead to activate the MAPK cascade and resultant alteration in gene expression concerning cell proliferation.
SH3GL1 is known to be predominantly localized in the nuclei of haematopoietic cells and fibroblasts in contrast to cytoplasmic localization in neurons and osteoblasts [19,20]. In the adult brain, SH3GL1 is highest in the neurons of the granular layer of the cerebral cortex [21] and known to be involved in the development of central nervous system [22,23]. These published data were compatible with our results of immunohistochemical staining with SH3GL1 antibody. In glioma tissues, strong positive staining of SH3GL1 was obtained in the cytoplasms but not in the nucleus, and the levels of staining in white matter increased according to the advance of its malignancy. These results suggested that the SH3GL1 overexpression might have some oncogenic roles in gliomas. However, the levels of serum autoantibodies to SH3GL1 in the patients with high-grade glioma were not increased in our study, while the levels in the patients with low-grade glioma were increased. It is believed that the abnormal cytoplasmic SH3GL1 overexpression in glioma cell has a potential to induce immune responses, but various mechanisms of immunosuppression prevent the reaction in high-grade glioma [24-27]. All the other candidate genes identified in this study showed the same low immunoreactivity in patients with high-grade gliomas. The suppression of the immunosurveillance mechanism in high-grade glioma would attenuate the recognition of SEREX-derived antigens in antigen presenting cells (APC). In fact, it has been known that various immunosuppressive molecules, such as TGF-β, IL-10, and prostaglandins, are highly expressed in cancers including high-grade glioma [24,25], and these molecules could inhibit the maturation of professional APCs. Such an evading immune destruction has now added to the hallmark of cancer [28].
The major cause of the lower level of anti-SH3GL1 autoantibody in high-grade glioma patients would be the non-specific immunosuppression caused by increased immunosuppressive cytokines [24,25]. However, the animal experiment provides an additional hypothesis that the depressed autoantibody levels could be partly due to the antigen-specific immune tolerance induced by the existence of large tumor and long-term antigen exposure. The early stage of the rat glioma models indicates a relatively small tumor and short-term antigen exposure, and the late stage indicates a large tumor and long-term antigen exposure to the immune system. The long-term antigen exposure from a large tumor could generally induce immune tolerance through development of immune resistant tumor variants and the tumor microenvironment inducing immune cell anergy or death [26,27]. It is usually accepted that gliomas often progress from low-grade tumors to higher-grade tumors as the time proceeds, although low-grade gliomas are not always in an early-stage of the disease and secondary glioblastoma is less frequent than de novo glioblastoma [12]. The possible contribution of antigen-specific immune tolerance to the depressed autoantibody levels in high-grade glioma patients remains to be elucidated.
SEREX is one of the most powerful tools to find novel tumor antigen for various cancers [5], and some autologous antigens to esophageal cancer have been identified in our groups [29-31]. Compared with other screening techniques such as transcriptomics or proteomics, SEREX offers a crucial advantage that subtle changes in the protein expressions can be detected through immunological reactions [32,33]. Several authors have already applied SEREX to glioma, and some antigens, including glioma-expressed antigen 2 (GLEA2) [7], PHD finger protein 3 (PHF3) [7,34], and SRY-box 6 (SOX6) [8] have been identified. It should be noted that we found autologous antibodies against SH3GL1 to be a low-grade glioma-specific marker with similar experimental systems to others. Our unique approach was the quantitative comparison of the levels of serum antibodies using the ELISA, while the approach of others was qualitative analysis. The application of ELISA in the validation step could lead to the discovery of a low-grade glioma-specific high titer of the autoantibody and the decrease in high-grade gliomas. Although some candidates of glioma biomarkers have been identified by various screening methods [6-8,34-37], no serum marker for early diagnosis has been found yet. Therefore, it is quite valuable to find a novel serum biomarker for its early diagnosis, prediction of the prognosis in each patient, and development of a new molecular target. Indeed,
The results of an overlap peptide array and ELISA using deletion mutants of SH3GL1 showed that 12 amino acids in the C-terminal portion, FPLSYVEVLVPL, were indicated as a major epitope site. By using a synthetic peptide corresponding to the epitope as an antigen, a more accurate screening for the patients with low-grade gliomas and a specific peptide vaccine therapy would be achieved in the future.
Competing interests
The authors declare that they have no competing interests.
Authors’ contributions
TM performed experiments, analyzed data and participated in writing; TH, MT, NS, and YI conceived the idea, designed and supervised the study; TO carried out immunohistochemistry; MK performed the overlap peptide array. All authors read and approved the final manuscript.
Author details
1Departments of Neurological Surgery, Chiba University, Graduate School of Medicine, 1-8-1, Inohana, Chuo-ku, Chiba 260-8670, Japan. 2Genetics and Biochemistry, Chiba University, Graduate School of Medicine, 1-8-1, Inohana, Chuo-ku, Chiba 260-8670, Japan. 3Diagnostic Pathology, Chiba University, Graduate School of Medicine, 1-8-1, Inohana, Chuo-ku, Chiba 260-8670, Japan. 4Department of Biochemistry, Graduate School of Life Science, Nagoya Women’s University, 3-40, Shioji-cho, Mizuho-ku, Nagoya 467-8610, Japan.
Contributor Information
Tomoo Matsutani, Email: tomoomastu@nifty.com.
Takaki Hiwasa, Email: hiwasa_takaki@faculty.chiba-u.jp.
Masaki Takiguchi, Email: mtak@faculty.chiba-u.jp.
Takashi Oide, Email: oide@faculty.chiba-u.jp.
Mitoshi Kunimatsu, Email: kunimatu@nagoya-wu.ac.jp.
Naokatsu Saeki, Email: nsaeki@faculty.chiba-u.jp.
Yasuo Iwadate, Email: iwadatey@faculty.chiba-u.jp.
References
- Ohgaki H, Kleihues P. Epidemiology and etiology of gliomas. Acta Neuropathol. 2005;109:93–108. doi: 10.1007/s00401-005-0991-y. [DOI] [PubMed] [Google Scholar]
- Anderson E, Grant R, Lewis SC, Whittle IR. Randomized Phase III controlled trials of therapy in malignant glioma: where are we after 40 years? Br J Neurosurg. 2008;22:339–349. doi: 10.1080/02688690701885603. [DOI] [PubMed] [Google Scholar]
- van den Bent MJ, Afra D, de Witte O, Ben Hassel M, Schraub S, Hoang-Xuan K, Malmstrom PO, Collette L, Pierart M, Mirimanoff R, Karim AB. Long-term efficacy of early versus delayed radiotherapy for low-grade astrocytoma and oligodendroglioma in adults: the EORTC 22845 randomised trial. Lancet. 2005;366:985–990. doi: 10.1016/S0140-6736(05)67070-5. [DOI] [PubMed] [Google Scholar]
- Sanai N, Berger MS. Glioma extent of resection and its impact on patient outcome. Neurosurgery. 2008;62:753–764. doi: 10.1227/01.neu.0000318159.21731.cf. [DOI] [PubMed] [Google Scholar]
- Sahin U, Tureci O, Schmitt H, Cochlovius B, Johannes T, Schmits R, Stenner F, Luo G, Schobert I, Pfreundschuh M. Human neoplasms elicit multiple specific immune responses in the autologous host. Proc Natl Acad Sci U S A. 1995;92:11810–11813. doi: 10.1073/pnas.92.25.11810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmits R, Cochlovius B, Treitz G, Regitz E, Ketter R, Preuss KD, Romeike BF, Pfreundschuh M. Analysis of the antibody repertoire of astrocytoma patients against antigens expressed by gliomas. Int J Cancer. 2002;98:73–77. doi: 10.1002/ijc.10170. [DOI] [PubMed] [Google Scholar]
- Pallasch CP, Struss AK, Munnia A, Konig J, Steudel WI, Fischer U, Meese E. Autoantibodies against GLEA2 and PHF3 in glioblastoma: tumor-associated autoantibodies correlated with prolonged survival. Int J Cancer. 2005;117:456–459. doi: 10.1002/ijc.20929. [DOI] [PubMed] [Google Scholar]
- Ueda R, Iizuka Y, Yoshida K, Kawase T, Kawakami Y, Toda M. Identification of a human glioma antigen, SOX6, recognized by patients’ sera. Oncogene. 2004;23:1420–1427. doi: 10.1038/sj.onc.1207252. [DOI] [PubMed] [Google Scholar]
- Behin A, Hoang-Xuan K, Carpentier AF, Delattre J-Y. Primary brain tumors in adults. Lancet. 2003;361:323–331. doi: 10.1016/S0140-6736(03)12328-8. [DOI] [PubMed] [Google Scholar]
- Rozakis-Adcock M, Fernley R, Wade J, Pawson T, Bowtell D. The SH2 and SH3 domains of mammalian Grb2 coupled the EGF receptor to the Ras activator mSos1. Nature. 1993;363:83–85. doi: 10.1038/363083a0. [DOI] [PubMed] [Google Scholar]
- Woods SA, Marmor E, Feldkamp M, Lau N, Apicelli AJ, Boss G, Gutmann DH, Guha A. Aberrant G protein signaling in nervous system tumors. J Neurosurg. 2002;97:627–642. doi: 10.3171/jns.2002.97.3.0627. [DOI] [PubMed] [Google Scholar]
- Ohgaki H, Dessen P, Jourde B, Horstmann S, Nishikawa T, Di Patre PL, Burkhard C, Schüler D, Probst-Hensch NM, Maiorka PC, Baeza N, Pisani P, Yonekawa Y, Yasargil MG, Lütolf UM, Kleihues P. Genetic pathways to glioblastoma: a population-based study. Cancer Res. 2004;64:6892–6899. doi: 10.1158/0008-5472.CAN-04-1337. [DOI] [PubMed] [Google Scholar]
- Kato R, Kaga C, Kunimatsu M, Kobayashi T, Honda H. Peptide array-based interaction assay of solid-bound peptides and anchorage-dependent cells and its effectiveness in cell-adhesive peptide design. J Biosci Bioeng. 2006;101:485–495. doi: 10.1263/jbb.101.485. [DOI] [PubMed] [Google Scholar]
- Giese A, Westphal M. Glioma invasion in the central nervous system. Neurosurgery. 1996;39:235–252. doi: 10.1097/00006123-199608000-00001. [DOI] [PubMed] [Google Scholar]
- Verhaak RG, Hoadley KA, Purdom E. et al. Integrated genomic analysis identifies clinically relevant subtypes of glioblastoma characterized by a abnormalities in PDGFRA, IDH1, EGFR, and NF1. Cancer Cell. 2010;17:98–110. doi: 10.1016/j.ccr.2009.12.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- So CW, Caldas C, Liu MM, Chen SJ, Huang QH, Gu LJ, Sham MH, Wiedemann LM, Chan LC. EEN encodes for a member of a new family of proteins containing an Src homology 3 domain and is the third gene located on chromosome 19p13 that fuses to MLL in human leukemia. Proc Natl Acad Sci U S A. 1997;94:2563–2568. doi: 10.1073/pnas.94.6.2563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yam JW, Jin DY, So CW, Chan LC. Identification and characterization of EBP, a novel EEN binding protein that inhibits Ras signaling and is recruited into the nucleus by the MLL-EEN fusion protein. Blood. 2004;103:1445–1453. doi: 10.1182/blood-2003-07-2452. [DOI] [PubMed] [Google Scholar]
- Ma LH, Liu H, Xiong H, Chen B, Zhang XW, Wang YY, Le HY, Huang QH, Zhang QH, Li BL, Chen Z, Chen SJ. Aberrant transcriptional regulation of the MLL fusion partner EEN by AML1-ETO and its implication in leukemogenesis. Blood. 2007;109:769–777. doi: 10.1182/blood-2006-02-003517. [DOI] [PubMed] [Google Scholar]
- Cheung N, So CW, Yam JW, So CK, Poon RY, Jin DY, Chan LC. Subcellular localization of EEN/endophilin A2, a fusion partner gene in leukaemia. Biochem J. 2004;383:27–35. doi: 10.1042/BJ20040041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giachino C, Lantelme E, Lanzetti L, Saccone S, Bella Valle G, Migone N. A novel SH3-containing human gene family preferentially expressed in the central nervous system. Genomics. 1997;41:427–434. doi: 10.1006/geno.1997.4645. [DOI] [PubMed] [Google Scholar]
- So CW, Sham MH, Chew SL, Cheung N, So CK, Chung SK, Caldas C, Wiedemann LM, Chan LC. Expression and protein-binding studies of the EEN gene family, new interacting partners for dynamin, synaptojanin and huntingtin proteins. Biochem J. 2000;348(Pt 2):447–458. [PMC free article] [PubMed] [Google Scholar]
- Ringstad N, Nemoto Y, De Camilli P. The SH3p4/Sh3p8/SH3p13 protein family: binding partners for synaptojanin and dynamin via a Grb2-like Src homology 3 domain. Proc Natl Acad Sci U S A. 1997;94:8569–8574. doi: 10.1073/pnas.94.16.8569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ringstad N, Nemoto Y, De Camilli P. Differential expression of endophilin 1 and 2 dimers at central nervous system synapses. J Biol Chem. 2001;276:40424–40430. doi: 10.1074/jbc.M106338200. [DOI] [PubMed] [Google Scholar]
- Zou JP, Morford LA, Chougnet C, Dix AR, Brooks AG, Torres N, Shuman JD, Coligan JE, Brooks WH, Roszman TL, Shearer GM. Human glioma-induced immunosuppression involves soluble factor(s) that alters monocyte cytokine profile and surface markers. J Immunol. 1999;162:4882–4892. [PubMed] [Google Scholar]
- Gomez GG, Kruse CA. Mechanisms of malignant glioma immune resistance and sources of immunosuppression. Gene Ther Mol Biol. 2006;10:133–146. [PMC free article] [PubMed] [Google Scholar]
- Mapara MY, Sykes M. Tolerance and cancer: mechanisms of tumor evasion and strategies for breaking tolerance. J Clin Oncol. 2004;22:1136–1151. doi: 10.1200/JCO.2004.10.041. [DOI] [PubMed] [Google Scholar]
- Wei J, Barr J, Kong L-Y, Wang Y, Wu A, Sharma AK, Gumin J, Henry V, Colman H, Sawaya R, Lang FF, Heimberger AB. Glioma-associated cancer initiating cells induce immunosuppression. Clin Cancer Res. 2010;16:461–473. doi: 10.1158/1078-0432.CCR-09-1983. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Hanahan D, Weinberg RA. Hallmarks of cancer: the next generations. Cell. 2011;144:646–674. doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- Nakashima K, Shimada H, Ochiai T, Kuboshima M, Kuroiwa N, Okazumi S, Matsubara H, Nomura F, Takiguchi M, Hiwasa T. Serological identification of TROP2 by recombinant cDNA expression cloning using sera of patients with esophageal squamous cell carcinoma. Int J Cancer. 2004;112:1029–1035. doi: 10.1002/ijc.20517. [DOI] [PubMed] [Google Scholar]
- Kuboshima M, Shimada H, Liu TL, Nakashima K, Nomura F, Takiguchi M, Hiwasa T, Ochiai T. Identification of a novel SEREX antigen, SLC2A1/GLUT1, in esophageal squamous cell carcinoma. Int J Oncol. 2006;28:463–468. [PubMed] [Google Scholar]
- Shimada H, Kuboshima M, Shiratori T, Nabeya Y, Takeuchi A, Takagi H, Nomura F, Takiguchi M, Ochiai T, Hiwasa T. Serum anti-myomegalin antibodies in patients with esophageal squamous cell carcinoma. Int J Oncol. 2007;30:97–103. [PubMed] [Google Scholar]
- Iwadate Y. Clinical proteomics in cancer research-promises and limitations of currenttwo-dimensional gel electrophoresis. Curr Med Chem. 2008;15:2393–2400. doi: 10.2174/092986708785909102. [DOI] [PubMed] [Google Scholar]
- Khalil AA. Biomarker discovery: a proteomic approach for brain cancer profiling. Cancer Sci. 2007;98:201–213. doi: 10.1111/j.1349-7006.2007.00374.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Struss AK, Romeike BF, Munnia A, Nastainczyk W, Steudel WI, Konig J, Ohgaki H, Feiden W, Fischer U, Meese E. PHF3-specific antibody responses in over 60% of patients with glioblastoma multiforme. Oncogene. 2001;20:4107–4114. doi: 10.1038/sj.onc.1204552. [DOI] [PubMed] [Google Scholar]
- Tanwar MK, Gilbert MR, Holland EC. Gene expression microarray analysis reveals YKL-40 to be a potential serum marker for malignant character in human glioma. Cancer Res. 2002;62:4364–4368. [PubMed] [Google Scholar]
- Fukuda ME, Iwadate Y, Machida T, Hiwasa T, Nimura Y, Nagai Y, Takiguchi M, Tanzawa H, Yamaura A, Seki N. Cathepsin D is a potential serum marker for poor prognosis in glioma patients. Cancer Res. 2005;65:5190–5194. doi: 10.1158/0008-5472.CAN-04-4134. [DOI] [PubMed] [Google Scholar]
- Iwadate Y, Hayama M, Adachi A, Matsutani T, Nagai Y, Hiwasa T, Saeki N. High serum level of plasminogen activator inhibitor-1 predicts histological grade of intracerebral gliomas. Anticancer Res. 2008;28:415–418. [PubMed] [Google Scholar]