Abstract
As energy sources and structural components, sugars are the central regulators of plant growth and development. In addition to the abundant natural sugars in plants, more than 50 different kinds of rare sugars exist in nature, several of which show distinct roles in plant growth and development. Recently, one of the rare sugars, D-allose, an epimer of D-glucose at C3, is found to suppress plant hormone gibberellin (GA) signaling in rice. Scaffold protein RACK1A in the model plant Arabidopsis is implicated in the GA pathway as rack1a knockout mutants show insensitivity to GA in GA-induced seed germination. Using genetic knockout lines and a reporter gene, the functional role of RACK1A in the D-allose pathway was investigated. It was found that the rack1a knockout seeds showed hypersensitivity to D-allose-induced inhibition of seed germination, implicating a role for RACK1A in the D-allose mediated suppression of seed germination. On the other hand, a functional RACK1A in the background of the double knockout mutations in the other two RACK1 isoforms, rack1b/rack1c, showed significant resistance to the D-allose induced inhibition of seed germination. The collective results implicate the RACK1A in the D-allose mediated seed germination inhibition pathway. Elucidation of the rare sugar signaling mechanism will help to advance understanding of this less studied but important cellular signaling pathway.
Keywords: RACK1, rare sugar, D-allose, gibberellin, germination, Arabidopsis, scaffold protein, cell signaling
Text
Rare sugars – monosaccharides that rarely exist in nature – are known to regulate diverse physiological responses in both plants and animals. With the advent of the discovery of “Izumoring,” an in vitro enzymatic approach to the synthesis of all rare sugars,1,2 research in this area has implicated rare sugars under many physiological conditions. These include but are not limited to the immunosuppressive activity in liver transplantation,3 protection against liver ischemia reperfusion injury,4,5 protection from reactive oxygen species (ROS)6,7 and anticancer activity on different cancer cell lines.8-11 Although not abundant, rare sugars – D-allose, D-psicose, D-allitol, L-galactose, tagatose or their derivatives – have been found in the tissues of higher plants as well.12-18 It was found that D-psicose inhibits plant root growth via hexokinase-independent pathway; it also inhibits bacterial blight disease in rice.19,20 D-allose was found to inhibit the rice growth and prevent bacterial blight disease in rice as well.21
Recently, Akimitsu’s group, a pioneer in rare sugar research, through the use of powerful genetics study has shown that the rare sugar D-allose suppresses GA signaling pathway in rice.22 D-allose strongly inhibited GA mediated α-amylase induction in embryo-less rice half seeds,22 implicating a negative role of D-allose in the GA pathway. Earlier, we have shown that Arabidopsis scaffold protein RACK1A (Receptor for Activating C Kinase 1) positively regulates GA signaling pathway as rack1a knockout seeds showed insensitivity to GA-induced seed germination.23 Here we tested our hypothesis that the suppression of GA signaling pathway by D-allose would impact the RACK1A mediated positive regulation of GA signaling pathway.
Scaffold protein RACK1 in metazoan plays a major role in coordinating different signal transduction pathways ranging from cell division to ion channel regulation by interacting with diverse proteins.24-28 RACK1 proteins with seven WD-40 repeats are highly conserved (70–80% at the protein level) in wide range of species, including plants, humans, rats, chickens, flies, nematodes, algae and yeasts. Although not recognized as such, the first RACK1 homolog in plants was identified in tobacco BY-2 suspension cells as a plant hormone auxin inducible gene29 and later in Arabidopsis, rice, rape and alfalfa.30-33 As opposed to a single gene in metazoan, the model plant Arabidopsis genome maintains three different RACK1 genes – termed as RACK1A, RACK1B and RACK1C.34 These Arabidopsis genes are found to regulate plant development with unequal genetic redundancy.34 The analysis of double and triple Arabidopsis RACK1 mutants revealed that the difference in gene expression level and the cross-regulation may determine the role played by the individual RACK1 gene in regulating plant development.34 In addition to regulating plant development, Arabidopsis RACK1 mediates multiple hormonal and stress responses.23,35,36
In order to elucidate the role of D-allose in Arabidopsis seed germination pathways, age-matched seeds of designated genotypes were grown in the presence of D-allose, GA3 and in both (Fig. 1). The germination of the WT seeds, as expected, has shown considerable inhibition by the D-allose application (Fig. 1D). Application of GA3 along with D-allose has slightly alleviated the D-allose induced inhibition of seed germination as the seeds showed enhanced germination growth compared with the seeds treated with D-allose alone. However, the degree of germination inhibition in the rack1a knockout seeds by D-allose was significantly higher compared with that in the WT seeds (Fig. 1E). The rack1a knockout seed coats remained almost intact even with the GA3 application (Fig. 1H) implicating RACK1A in the D-allose mediated suppression of GA3-induced seed germination pathway. As discussed above, Arabidopsis RACK1 genes act with unequal genetic redundancy due to the cross-regulation among the RACK1 genes and the differences in individual gene expression level.34 It is quite possible that the presence of RACK1B and RACK1C in the rack1a mutant background can potentially exert their own regulation on the pathway. While the RACK1B expression in the rack1a/rack1c and RACK1C expression in the rack1a/rack1c background were significantly downregulated compared with that in the WT seedlings, RACK1A expression was not significantly downregulated in the rack1b/rack1c mutant background.34 If RACK1B/RACK1C collectively exert negative regulation on RACK1A expression, it will be quite informative to see the response of rack1b/rack1c double mutant to D-allose induced seed germination inhibition. Quite interestingly, it was found that neither D-allose nor D-allose plus GA3 could significantly affect the seed germination rate and the early seedling development in the rack1b/rack1c double mutant background (Fig. 1F and I). On the other hand, all the seeds without any D-allose application did germinate and showed normal growth and development (Fig. 1A–C). As D-allose is an epimer of D-glucose, we used D-glucose as a control to make sure that the effect is very specific to D-allose. As can be seen from Figure 1J–L, all three genotypes of the tested seeds (WT, rack1a-1 knockout and rack1b/rack1c double knockout) did not show germination inhibition from D-glucose treatment, effectively eliminating the possibility that the D-allose acts as a simple carbon source. The results from the WT seedlings, where a functional RACK1A is present, can be explained by the presence of the two more isoforms of RACK1A – which can potentially cross-regulate the RACK1A function.34 It is reported that the knockout phenotype of rack1a is enhanced in either rack1b or rack1c knockout combination, implying the role of RACK1B or RACK1C in the RACK1A function.
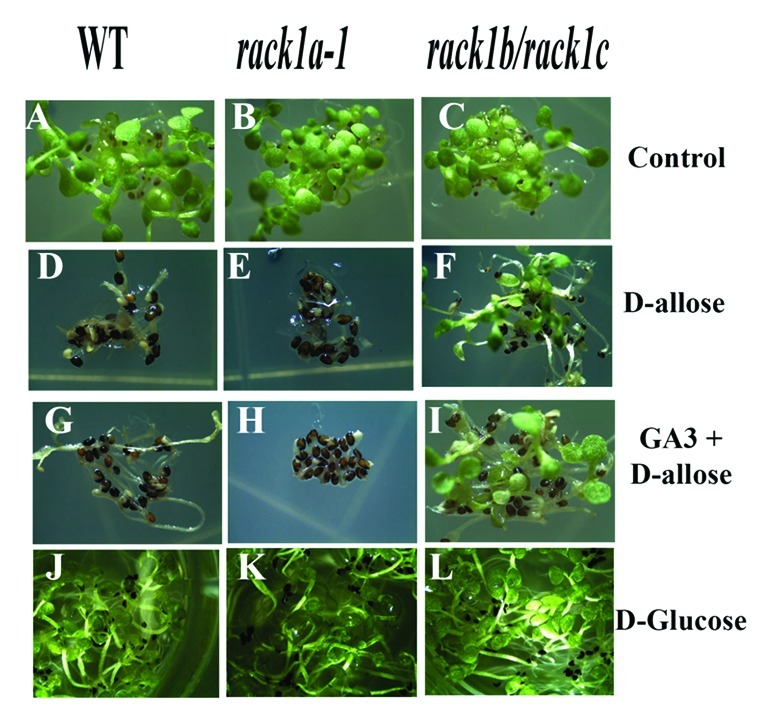
Figure 1. Seed germination inhibition by D-allose is enhanced in rack1a knockout line. Age matched seeds from indicated genotypes were grown under continuous light (75 μmol/m2/sec) at 22°C in MS minus salt liquid media (Upper panel) containing either 10 mM D-allose or 10 mM D-allose and 10 μM of Gibberellins (GA3) or 10 mM of D-glucose for two weeks. Compared with the no treatment control, D-allose inhibited WT seed germination and inhibition is enhanced in the rack1a knockout line. rack1b/rack1c double mutant line partially overcomes the D-allose mediated inhibition of seed germination. D-glucose control treatment does not affect the seed germination in any of the genotypes (lower panel)
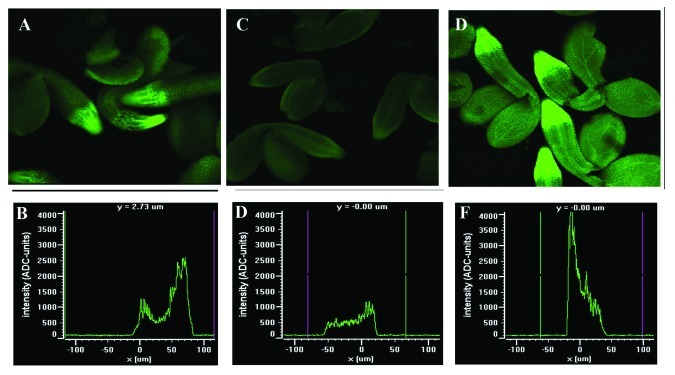
To understand the cellular mechanism of this signaling network, it is proposed that D-allose will not only negatively regulate GA3 induced seed germination and growth, it may potentially downregulate RACK1A expression to attenuate the positive regulation of GA3 signaling by RACK1A. We utilized a transgenic line expressing GFP fused to the RACK1A gene which is driven by the RACK1A native promoter. As can be seen in Figure 2, the application of D-allose to the transgenic seeds (Fig. 2C and D), compared with the no-treatment control (Fig. 2A and B) did significantly downregulated RACK1A expression in the root tip region of the developing embryo isolated from seeds treated for 72 h. Application of GA3 significantly enhanced the RACK1A expression in the same tissue regions (Fig. 2E and F). D-Glucose was used as a control, which, contrary to the D-allose, slightly increased RACK1A expression (data not shown), indicating that the D-allose specifically downregulates RACK1A expression in the embryonic root tip region. Although the gene expression was restricted to the root tip growth area, it remains to be seen whether the similar regulation takes place in the shoot apical meristems of young seedlings. Even though it is known that D-allose exert its inhibitory role on GA signaling through hexokinase-dependent pathway, it will be an intriguing study to investigate the precise role hexokinase plays in the currently elucidated pathway. Figure 3 presents a possible working model of the pathway. The simple explanation for the presented data are that D-allose negatively regulates GA-mediated seed germination and early seedling development through the inhibition of RACK1A expression. Though RACK1A is reported to positively regulate GA signaling pathway,23 in the absence of epistasis studies between rack1a and known GA signaling mutants, it is not quite possible to indicate a downstream or upstream regulatory position of RACK1A in the GA signaling pathway. However, the presumptive position of the RACK1A in the current model is partially consistent with the presented data but with future genetic studies the more concrete regulatory position in this pathway will be ascertained.
Figure 2. D-Allose inhibits RACK1A:GFP expression in the embryos while GA induces expression. Seeds from the pRACK1A::RACK1A:GFP expressing plants were incubated for 72h under the respective treatment regimens. The RACK1A expression is concentrated in the root tip region (A); RACK1A expression is downregulated with 10 mM of D-allose (C) and the expression is upregulated with 10 μM GA3 application (D). The GFP fluorescence was captured with a NIKON ECLIPSE TE2000-E fluorescence microscope with a Nikon C1 Confocal imaging system. To alleviate the concerns of image intensity on different samples, the image acquisition settings were set as same for all three panels. The gain setting was set at 6.65B; pixel dwell time was 61.44 μS and the 408 and 488 laser lines power were set at 29.2% and 38.4% respectively. A representative root tip from another run is used to quantify the expression level by calculating the intensity of the pixels along a line drawn horizontally across a single root tip (delimited zone is shown by the green and purple vertical lines). The pixel intensity is shown with an arbitrary intensity unit along the vertical axis with D-allose (D); with GA3 (E) and no treatment (B).
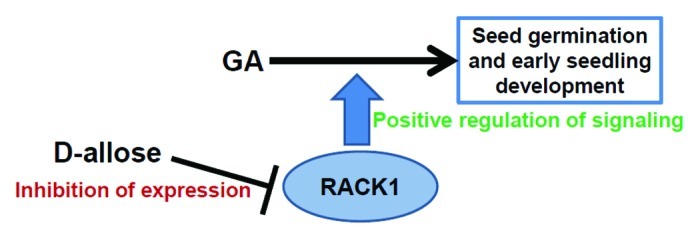
Figure 3. Working model of role of RACK1A in D-allose regulated GA induced seed germination and early seedling development signal transduction pathway. D-allose may negatively regulate GA-mediated seed germination and early seedling development through the inhibition of RACK1A expression. Molecular genetics studies indicate that RACK1A regulates gibberellin induced seed germination and early seedling development and that this process is inhibited by rare sugar D-allose. In the absence of epistasis data, the precise position of RACK1A in the GA3 signaling pathway cannot be ascertained. Based on the current data, a presumed position of RACK1A is indicated that is promoting GA3 induced seed germination and seedling development. For the lack of concrete data, role of other isoforms of RACK1 genes are omitted from the model.
Conclusion
D-allose has shown its promise in diverse physiological and biological applications in animal and plant studies. The current study fills up a gap in the understanding of the prominent role of D-allose in the seed germination signaling pathway in Arabidopsis. Considering the huge economic values in understanding the seed germination signaling pathway, the current study results will help in advancing the molecular elucidation of this pathway. In addition, the role of growth hormone GA induced seed germination process is an intensely studied physiological processes and the precise understanding of the signaling pathways will potentially contribute to the better elucidation of the pathway. Understanding the D-allose mediated cellular signaling mechanism during the seed germination process, can potentially contribute to wider applied use of this rather under-estimated important physiological regulator.
Acknowledgments
This work was supported by the Laboratory Directed Research and Development Program of Oak Ridge National Laboratory. Oak Ridge National Laboratory is managed by UT-Battelle, LLC, for the US. Department of Energy under Contract No. DE-AC05–00OR22725 and an NSF grant to HU (HRD-1036285).
Glossary
Abbreviations:
- GA
Gibberellin
- RACK1
Receptor for Activated C Kinase 1
- MS
Murashige and Skoog
Footnotes
Previously published online: www.landesbioscience.com/journals/psb/article/21995
References
- 1.Granström TB, Takata G, Tokuda M, Izumori K. Izumoring: a novel and complete strategy for bioproduction of rare sugars. J Biosci Bioeng. 2004;97:89–94. doi: 10.1263/jbb.97.89. [DOI] [PubMed] [Google Scholar]
- 2.Izumori K. Izumoring: a strategy for bioproduction of all hexoses. J Biotechnol. 2006;124:717–22. doi: 10.1016/j.jbiotec.2006.04.016. [DOI] [PubMed] [Google Scholar]
- 3.Hossain MA, Wakabayashi H, Goda F, Kobayashi S, Maeba T, Maeta H. Effect of the immunosuppressants FK506 and D-allose on allogenic orthotopic liver transplantation in rats. Transplant Proc. 2000;32:2021–3. doi: 10.1016/S0041-1345(00)01540-2. [DOI] [PubMed] [Google Scholar]
- 4.Hossain MA, Izuishi K, Maeta H. Protective effects of D-allose against ischemia reperfusion injury of the rat liver. J Hepatobiliary Pancreat Surg. 2003;10:218–25. doi: 10.1007/s00534-002-0785-8. [DOI] [PubMed] [Google Scholar]
- 5.Hossain MA, Wakabayashi H, Izuishi K, Okano K, Yachida S, Tokuda M, et al. Improved microcirculatory effect of D-allose on hepatic ischemia reperfusion following partial hepatectomy in cirrhotic rat liver. J Biosci Bioeng. 2006;101:369–71. doi: 10.1263/jbb.101.369. [DOI] [PubMed] [Google Scholar]
- 6.Murata A, Sekiya K, Watanabe Y, Yamaguchi F, Hatano N, Izumori K, et al. A novel inhibitory effect of D-allose on production of reactive oxygen species from neutrophils. J Biosci Bioeng. 2003;96:89–91. doi: 10.1016/s1389-1723(03)90104-6. [DOI] [PubMed] [Google Scholar]
- 7.Hirooka K, Miyamoto O, Jinming P, Du Y, Itano T, Baba T, et al. Neuroprotective effects of D-allose against retinal ischemia-reperfusion injury. Invest Ophthalmol Vis Sci. 2006;47:1653–7. doi: 10.1167/iovs.05-1018. [DOI] [PubMed] [Google Scholar]
- 8.Sui L, Dong Y, Watanabe Y, Yamaguchi F, Hatano N, Izumori K, et al. Growth inhibitory effect of D-allose on human ovarian carcinoma cells in vitro. Anticancer Res. 2005;25:2639–44. [PubMed] [Google Scholar]
- 9.Sui L, Dong Y, Watanabe Y, Yamaguchi F, Hatano N, Tsukamoto I, et al. The inhibitory effect and possible mechanisms of D-allose on cancer cell proliferation. Int J Oncol. 2005;27:907–12. [PubMed] [Google Scholar]
- 10.Yamaguchi F, Takata M, Kamitori K, Nonaka M, Dong Y, Sui L, et al. Rare sugar D-allose induces specific up-regulation of TXNIP and subsequent G1 cell cycle arrest in hepatocellular carcinoma cells by stabilization of p27kip1. Int J Oncol. 2008;32:377–85. [PubMed] [Google Scholar]
- 11.Hirata Y, Saito M, Tsukamoto I, Yamaguchi F, Sui L, Kamitori K, et al. Analysis of the inhibitory mechanism of D-allose on MOLT-4F leukemia cell proliferation. J Biosci Bioeng. 2009;107:562–8. doi: 10.1016/j.jbiosc.2008.12.021. [DOI] [PubMed] [Google Scholar]
- 12.Hough L, Stacey BE. The occurrence of D-ribohexulose in Itea ilicifolia Itea virginica, and Itea yunnanensis. Phytochemistry. 1963;2:315–20. doi: 10.1016/S0031-9422(00)84854-2. [DOI] [Google Scholar]
- 13.Hough L, Stacey BE. Variation in the allitol content of Itea plants during photosynthesis. Phytochemistry. 1966;5:171–5. doi: 10.1016/S0031-9422(00)85095-5. [DOI] [Google Scholar]
- 14.Beylis P, Howard AS, Perold GW. The occurrence of D-(+)-allose in nature. J Chem Soc. 1971;D:597a. doi: 10.1039/p19730000643. [DOI] [PubMed] [Google Scholar]
- 15.Perold GW, Beylis P, Howard AS. Metabolites of proteaceae. 8. The occurrence of (+)-D-allose in nature: rubropilosin and pilorubrosin from Protea rubropilosa Beard. J Chem Soc Perkin 1. 1973;6:643–9. doi: 10.1039/p19730000643. [DOI] [PubMed] [Google Scholar]
- 16.Wheeler GL, Jones MA, Smirnoff N. The biosynthetic pathway of vitamin C in higher plants. Nature. 1998;393:365–9. doi: 10.1038/30728. [DOI] [PubMed] [Google Scholar]
- 17.Chari VM, Grayer-Barkmeijer RJ, Harborne JB, Osterdahl B-G. An acylated allose-containing 8-hydroxyflavone glycoside from Veronica filiformis. Phytochemistry. 1981;20:1977–9. doi: 10.1016/0031-9422(81)84048-4. [DOI] [Google Scholar]
- 18.Weckwerth W, Loureiro ME, Wenzel K, Fiehn O. Differential metabolic networks unravel the effects of silent plant phenotypes. Proc Natl Acad Sci U S A. 2004;101:7809–14. doi: 10.1073/pnas.0303415101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kato-Noguchi H, Takaoka T, Izumori K. Psicose inhibits lettuce root growth via a hexokinase-independent pathway. Physiol Plant. 2005;125:293–8. doi: 10.1111/j.1399-3054.2005.00565.x. [DOI] [Google Scholar]
- 20.Kano A, Hosotani K, Gomi K, Yamasaki-Kokudo Y, Shirakawa C, Fukumoto T, et al. D-Psicose induces upregulation of defense-related genes and resistance in rice against bacterial blight. J Plant Physiol. 2011;168:1852–7. doi: 10.1016/j.jplph.2011.04.003. [DOI] [PubMed] [Google Scholar]
- 21.Kano A, Gomi K, Yamasaki-Kokudo Y, Satoh M, Fukumoto T, Ohtani K, et al. A rare sugar, d-allose, confers resistance to rice bacterial blight with upregulation of defense-related genes in Oryza sativa. Phytopathology. 2010;100:85–90. doi: 10.1094/PHYTO-100-1-0085. [DOI] [PubMed] [Google Scholar]
- 22.Fukumoto T, Kano A, Ohtani K, Yamasaki-Kokudo Y, Kim BG, Hosotani K, et al. Rare sugar D-allose suppresses gibberellin signaling through hexokinase-dependent pathway in Oryza sativa L. Planta. 2011;234:1083–95. doi: 10.1007/s00425-011-1463-3. [DOI] [PubMed] [Google Scholar]
- 23.Chen JG, Ullah H, Temple B, Liang J, Guo J, Alonso JM, et al. RACK1 mediates multiple hormone responsiveness and developmental processes in Arabidopsis. J Exp Bot. 2006;57:2697–708. doi: 10.1093/jxb/erl035. [DOI] [PubMed] [Google Scholar]
- 24.McCahill A, Warwicker J, Bolger GB, Houslay MD, Yarwood SJ. The RACK1 scaffold protein: a dynamic cog in cell response mechanisms. Mol Pharmacol. 2002;62:1261–73. doi: 10.1124/mol.62.6.1261. [DOI] [PubMed] [Google Scholar]
- 25.Alfarano C, Andrade CE, Anthony K, Bahroos N, Bajec M, Bantoft K, et al. The Biomolecular Interaction Network Database and related tools 2005 update. Nucleic Acids Res. 2005;33(Database issue):D418–24. doi: 10.1093/nar/gki051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sklan EH, Podoly E, Soreq H. RACK1 has the nerve to act: structure meets function in the nervous system. Prog Neurobiol. 2006;78:117–34. doi: 10.1016/j.pneurobio.2005.12.002. [DOI] [PubMed] [Google Scholar]
- 27.Chen S, Lin F, Shin ME, Wang F, Shen L, Hamm HE. RACK1 regulates directional cell migration by acting on G betagamma at the interface with its effectors PLC beta and PI3K gamma. Mol Biol Cell. 2008;19:3909–22. doi: 10.1091/mbc.E08-04-0433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Guo J, Wang S, Valerius O, Hall H, Zeng Q, Li JF, et al. Involvement of Arabidopsis RACK1 in protein translation and its regulation by abscisic acid. Plant Physiol. 2011;155:370–83. doi: 10.1104/pp.110.160663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ishida S, Takahashi Y, Nagata T. Isolation of cDNA of an auxin-regulated gene encoding a G protein beta subunit-like protein from tobacco BY-2 cells. Proc Natl Acad Sci U S A. 1993;90:11152–6. doi: 10.1073/pnas.90.23.11152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Iwasaki Y, Komano M, Ishikawa A, Sasaki T, Asahi T. Molecular cloning and characterization of cDNA for a rice protein that contains seven repetitive segments of the Trp-Asp forty-amino-acid repeat (WD-40 repeat) Plant Cell Physiol. 1995;36:505–10. doi: 10.1093/oxfordjournals.pcp.a078786. [DOI] [PubMed] [Google Scholar]
- 31.McKhann HI, Frugier F, Petrovics G, de la Peña TC, Jurkevitch E, Brown S, et al. Cloning of a WD-repeat-containing gene from alfalfa (Medicago sativa): a role in hormone-mediated cell division? Plant Mol Biol. 1997;34:771–80. doi: 10.1023/A:1005899410389. [DOI] [PubMed] [Google Scholar]
- 32.Vahlkamp L, Palme K. AtArcA. Accession No. U77381, the Arabidopsis thaliana homolog of the tobacco ArcA gene (PGR97-145) Plant Physiol. 1997;115:863. [Google Scholar]
- 33.Kwak JM, Kim SA, Lee SK, Oh SA, Byoun CH, Han JK, et al. Insulin-induced maturation of Xenopus oocytes is inhibited by microinjection of a Brassica napus cDNA clone with high similarity to a mammalian receptor for activated protein kinase C. Planta. 1997;201:245–51. doi: 10.1007/s004250050063. [DOI] [PubMed] [Google Scholar]
- 34.Guo J, Chen JG. RACK1 genes regulate plant development with unequal genetic redundancy in Arabidopsis. BMC Plant Biol. 2008;8:108–18. doi: 10.1186/1471-2229-8-108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ullah H, Scappini EL, Moon AF, Williams LV, Armstrong DL, Pedersen LC. Structure of a signal transduction regulator, RACK1, from Arabidopsis thaliana. Protein Sci. 2008;17:1771–80. doi: 10.1110/ps.035121.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Guo J, Wang J, Xi L, Huang W-D, Liang J, Chen JG. RACK1 is a negative regulator of ABA responses in Arabidopsis. J Exp Bot. 2009;60:3819–33. doi: 10.1093/jxb/erp221. [DOI] [PMC free article] [PubMed] [Google Scholar]