Abstract
ARI12 belongs to a family of 16 potential E3 ligases in Arabidopsis and is strongly induced in leaves upon low and high fluence rates (HFR) of UV-B. We have shown that ARI12 is a downstream target of the UV-B receptor, UVR8, and the transcription factors HY5 and HYH under low fluence rates. However under HFR of broad band UV-B ARI12 expression was still downstream of HY5 and HYH but increased in uvr8 mutants. To determine if other photoreceptors are responsible for the induction of ARI12 we quantified its expression in double mutants of the UV-A and blue light receptors, CRY1/2 and PHOT1/2, and the red light receptors PHYA/B. While the expression of ARI12 was increased in cyr1/2 it was unaffected in phot1/2 and phyA/B. Therefore ARI12 expression is suppressed by UVR8 and cryptochromes, and independent of phototropins and phytochromes A and B upon HFR of broad band UV-B.
Keywords: UV-B, ARI12, phytochrome, cryptochrome, phototropin, UVR8
Following the depletion of the stratospheric ozone layer increasing solar UV (UV)-B (280–315 nm) radiation will reach the earth. The increased UV-B radiation will have significant effects on natural and agricultural ecosystems.1-3 While low doses of UV-B serve as signal to control growth and development, high doses inhibit growth and reduce yield.4 Moreover high UV-B radiation causes DNA damages but also induces the production of UV-B protecting flavonoids.4,5 While photoreceptors for UV-A, blue and red light have been known for decades the existence of an UV-B specific receptor has only recently been confirmed.6,7 UVR8 was identified in Arabidopsis because uvr8 mutants were hypersensitive to UV-B, exhibit reduced UV-B-induced flavonoid biosynthesis and CHS expression.8 Furthermore UVR8 mediates low fluence rates UV-B-dependent photomorphogenesis.9 UVR8 is constantly expressed and present as inactive dimer in the cytoplasm. Upon UV-B radiation, UVR8 monomerises due to the disruption of salt bridges between arginines in the proximity of two tryptophanes that serve as UV-B chromophores and interact in the nucleus with the ubiquitin E3 ligase and central light regulator COP1.7,10,11 Downstream of the UVR8-mediated signaling cascade are two transcription factors HY5 and HYH which have been proposed to regulate all UVR8 dependent genes.12
ARI12 is a member of a family of potential ubiquitin E3 ligases in Arabidopsis.13 Under white light conditions, ARI12 is expressed in roots and hypocotyls and hardly detectable in leaves.14 We have recently shown that ARI12 expression is strongly induced upon low and high fluence rates of UV-B radiation and under both conditions this expression depends on the transcription factors HY5 and its homolog HYH.15
While ARI12 expression depended on UVR8 at low fluence rates, ARI12 was higher expressed in uvr8 mutants upon broad band HFR conditions. To determine if other photoreceptors are responsible for the induction of ARI12 upon HFR we extended the expression analyses to mutants of the UV-A, blue light and the red light receptors. The receptors responsible for UV-A and blue light (320–500 nm) perception are cryptochromes and phototrophins.16,17 The genome of Arabidopsis codes for two redundantly acting cryptochromes (CRY1 and 2) and phototropins (PHOT1 and 2) and a family of five phytochromes (PHYA-E) that perceive red and far-red light (600–700 nm).18,19 PHYA and PHYB are the most prominent members20,21 and both act through the transcription factor HY5.22
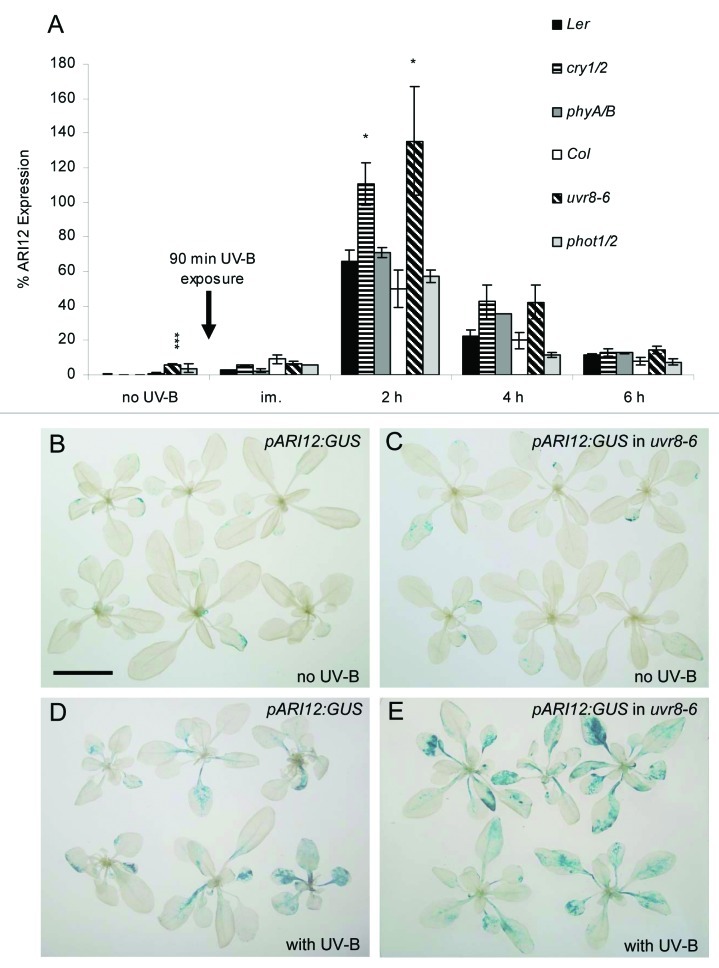
Similar to the previous analyses with uvr8 and hy5/hyh mutants, double mutants of the photoreceptors cry1/2, phot1/2 and phyA/B were cultivated under 140 µmol m−2 s−1 white light conditions and were exposed on day 25 for 90 min with 4.0 µmol·m−2·s−1 broad band UV-B. Leaves were harvested before and at different time points after UV-B exposure. ARI12 expression was quantified by qRT-PCR as reported by Lang-Mladek et al.15 While the expression of ARI12 is very low before UV-B exposure in wildtype plants (Fig. 1A) it is significantly higher in uvr8–6 mutants indicating that UVR8 might act as a suppressor of ARI12 expression in white light and UV-B. However histochemical analyses of the ARI12 promoter GUS reporter (pARI12:GUS) in uvr8–6 does not support the qPCR results of the white light conditions (Fig. 1B and C). Since the expression of ARI12 in the uvr8–6 background is very low, the difference might be due to the lower sensitivity of the reporter construct compared with the qPCR quantification. Consistently, the difference of the ARI12 expression between uvr8–6 and wildtype is apparent with the histochemical staining after UV-B exposure probably because of its at least one magnitude higher expression (Fig. 1D and E). Independent of the mutant background the RNA abundance of ARI12 peaked at about 2 h after UV-B exposure. The expression of ARI12 in phot1/2 and phyA/B was not significantly different from their wildtype backgrounds, indicating that these two photoreceptors are not involved in the UV-B specific induction of ARI12. That phototropins and phytochromes are not involved in ARI12 expression agrees with our survey of the public available microarrays that have been explored with the Bio-Array Resource and the Genevestigator tool.23,24 In these data sets ARI12 was not significantly induced by blue, red nor high or low light conditions nor differently regulated in phyA or phyB mutants.25-28
Figure 1.ARI12 expression in uvr8–6, cry1/2, phot1/2 and phyA/B single and double mutants upon high fluence rates of broad band UV-B radiation. (A) Time course of ARI12 expression before (no UV-B, 140 µmol m−2 s−1 white light), immediately (im) and at different times after a 90 min addition of 4 µmol m−2 s−1 of UV-B. qRT-PCR data were normalized to the expression of the reference gene TUB9. Data represent means and standard errors of at least three independent biological replicates. Significant difference were calculated with Student’s T-tests and * indicates p-values of ≤ 0.05, and *** of ≤ 0.001, respectively. (B-E) Histochemical staining of pARI12:GUS (B,D) and pARI12:GUS in uvr8–6 mutants (C,E) before (B,C) and 6 h after UV-B exposure (D,E). Pictures were taken with the same magnification and the size bar in B corresponds to 20 mm.
In contrast ARI12 was higher expressed in uvr8–6 and the double mutant cry1/2 at 2 h after UV-B exposure indicating that upon HFR of broad band UV-B radiation UVR8 and the CRYs are probably inhibiting ARI12 expression.
In summary we present evidences that UVR8 and CRY1/2 are required to avoid excess of ARI12 expression under HFR conditions. Thus ARI12 is the first gene that is positively regulated by HY5/HYH and negatively by UVR8 at HFR of UV-B. The functional significance of this specific regulation however has to be determined yet.
Disclosure of Potential Conflicts of Interest
There were no potential conflicts of interest to expose.
Acknowledgments
We thank Gareth Jenkins for cry1/2, phot1/2 and phyA/B double mutants, Roman Ulm for uvr8–6 and Christina Lang-Mladek for the pARI12:GUS line. The research was supported by the Austrian Science Fund (FWF) projects (P17888-B14, F3707) to M. -T. H. and the COST-FA0906 action UV4growth. LSX received a China scholarship council (CSC) fellowship.
Glossary
Abbreviations:
- ARIADNE12 (ARI12)
UV RESISTANCE LOCUS (UVR8), CRYPTOCHROME 1/2 (CRY1/2), PHOTOTROPIN 1/2 (PHOT1/2), PHYTOCHROME A/B (PHYA/B), beta-glucuronidase (GUS), CONSTITUTIVELY PHOTOMORPHOGENIC 1 (COP1), Arabidopsis thaliana (Arabidopsis), CHALCONE SYNTHASE (CHS), ELONGATED HYPOCOTYL 5 (HY5), HOMOLOG OF ELONGATED HYPOCOTYL (HYH), TUBULIN BETA-9 (TUB9), high fluence rates (HFR)
Footnotes
Previously published online: www.landesbioscience.com/journals/psb/article/22052
References
- 1.Searles PS, Flint SD, Caldwell MM. A meta-analysis of plant field studies simulating stratospheric ozone depletion. Oecologia. 2001;127:1–10. doi: 10.1007/s004420000592. [DOI] [PubMed] [Google Scholar]
- 2.Kakani VG, Reddy KR, Zhao D, Sailaja K. Field crop responses to ultraviolet-B radiation: a review. Agric For Meteorol. 2003;120:191–218. doi: 10.1016/j.agrformet.2003.08.015. [DOI] [Google Scholar]
- 3.Flint SD, Ryel RJ, Caldwell MM. Ecosystem UV-B experiments in terrestrial communities: a review of recent findings and methodologies. Agric For Meteorol. 2003;120:177–89. doi: 10.1016/j.agrformet.2003.08.014. [DOI] [Google Scholar]
- 4.Jenkins GI. Signal transduction in responses to UV-B radiation. Annu Rev Plant Biol. 2009;60:407–31. doi: 10.1146/annurev.arplant.59.032607.092953. [DOI] [PubMed] [Google Scholar]
- 5.Agati G, Tattini M. Multiple functional roles of flavonoids in photoprotection. New Phytol. 2010;186:786–93. doi: 10.1111/j.1469-8137.2010.03269.x. [DOI] [PubMed] [Google Scholar]
- 6.Brown BA, Cloix C, Jiang GH, Kaiserli E, Herzyk P, Kliebenstein DJ, et al. A UV-B-specific signaling component orchestrates plant UV protection. Proc Natl Acad Sci U S A. 2005;102:18225–30. doi: 10.1073/pnas.0507187102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rizzini L, Favory J-J, Cloix C, Faggionato D, O’Hara A, Kaiserli E, et al. Perception of UV-B by the Arabidopsis UVR8 protein. Science. 2011;332:103–6. doi: 10.1126/science.1200660. [DOI] [PubMed] [Google Scholar]
- 8.Kliebenstein DJ, Lim JE, Landry LG, Last RL. Arabidopsis UVR8 regulates ultraviolet-B signal transduction and tolerance and contains sequence similarity to human regulator of chromatin condensation 1. Plant Physiol. 2002;130:234–43. doi: 10.1104/pp.005041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Favory J-J, Stec A, Gruber H, Rizzini L, Oravecz A, Funk M, et al. Interaction of COP1 and UVR8 regulates UV-B-induced photomorphogenesis and stress acclimation in Arabidopsis. EMBO J. 2009;28:591–601. doi: 10.1038/emboj.2009.4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Christie JM, Arvai AS, Baxter KJ, Heilmann M, Pratt AJ, O’Hara A, et al. Plant UVR8 photoreceptor senses UV-B by tryptophan-mediated disruption of cross-dimer salt bridges. Science. 2012;335:1492–6. doi: 10.1126/science.1218091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wu D, Hu Q, Yan Z, Chen W, Yan C, Huang X, et al. Structural basis of ultraviolet-B perception by UVR8. Nature. 2012;484:214–9. doi: 10.1038/nature10931. [DOI] [PubMed] [Google Scholar]
- 12.Brown BA, Jenkins GI. UV-B signaling pathways with different fluence-rate response profiles are distinguished in mature Arabidopsis leaf tissue by requirement for UVR8, HY5, and HYH. Plant Physiol. 2008;146:576–88. doi: 10.1104/pp.107.108456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Eisenhaber B, Chumak N, Eisenhaber F, Hauser MT. The ring between ring fingers (RBR) protein family. Genome Biol. 2007;8:209. doi: 10.1186/gb-2007-8-3-209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mladek C, Guger K, Hauser MT. Identification and characterization of the ARIADNE gene family in Arabidopsis. A group of putative E3 ligases. Plant Physiol. 2003;131:27–40. doi: 10.1104/pp.012781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lang-Mladek C, Xie L, Nigam N, Chumak N, Binkert M, Neubert S, et al. UV-B signaling pathways and fluence rate dependent transcriptional regulation of ARIADNE12. Physiol Plant. 2012;145:527–39. doi: 10.1111/j.1399-3054.2011.01561.x. [DOI] [PubMed] [Google Scholar]
- 16.Christie JM. Phototropin blue-light receptors. Annu Rev Plant Biol. 2007;58:21–45. doi: 10.1146/annurev.arplant.58.032806.103951. [DOI] [PubMed] [Google Scholar]
- 17.Jiao Y, Lau OS, Deng XW. Light-regulated transcriptional networks in higher plants. Nat Rev Genet. 2007;8:217–30. doi: 10.1038/nrg2049. [DOI] [PubMed] [Google Scholar]
- 18.Aihara Y, Tabata R, Suzuki T, Shimazaki K-i, Nagatani A. Molecular basis of the functional specificities of phototropin 1 and 2. Plant J. 2008;56:364–75. doi: 10.1111/j.1365-313X.2008.03605.x. [DOI] [PubMed] [Google Scholar]
- 19.Chen M, Chory J. Phytochrome signaling mechanisms and the control of plant development. Trends Cell Biol. 2011;21:664–71. doi: 10.1016/j.tcb.2011.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kami C, Lorrain S, Hornitschek P, Fankhauser C. Light-regulated plant growth and development. In: Marja CPT, ed. Current Topics in Developmental Biology: Academic Press, 2010:29-66. [DOI] [PubMed] [Google Scholar]
- 21.Franklin KA, Quail PH. Phytochrome functions in Arabidopsis development. J Exp Bot. 2010;61:11–24. doi: 10.1093/jxb/erp304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Saijo Y, Sullivan JA, Wang H, Yang J, Shen Y, Rubio V, et al. The COP1-SPA1 interaction defines a critical step in phytochrome A-mediated regulation of HY5 activity. Genes Dev. 2003;17:2642–7. doi: 10.1101/gad.1122903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Winter D, Vinegar B, Nahal H, Ammar R, Wilson GV, Provart NJ. An “Electronic Fluorescent Pictograph” browser for exploring and analyzing large-scale biological data sets. PLoS One. 2007;2:e718. doi: 10.1371/journal.pone.0000718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zimmermann P, Hennig L, Gruissem W. Gene-expression analysis and network discovery using Genevestigator. Trends Plant Sci. 2005;10:407–9. doi: 10.1016/j.tplants.2005.07.003. [DOI] [PubMed] [Google Scholar]
- 25.Kilian J, Whitehead D, Horak J, Wanke D, Weinl S, Batistic O, et al. The AtGenExpress global stress expression data set: protocols, evaluation and model data analysis of UV-B light, drought and cold stress responses. Plant J. 2007;50:347–63. doi: 10.1111/j.1365-313X.2007.03052.x. [DOI] [PubMed] [Google Scholar]
- 26.Caldana C, Degenkolbe T, Cuadros-Inostroza A, Klie S, Sulpice R, Leisse A, et al. High-density kinetic analysis of the metabolomic and transcriptomic response of Arabidopsis to eight environmental conditions. Plant J. 2011;67:869–84. doi: 10.1111/j.1365-313X.2011.04640.x. [DOI] [PubMed] [Google Scholar]
- 27.Foreman J, Johansson H, Hornitschek P, Josse E-M, Fankhauser C, Halliday KJ. Light receptor action is critical for maintaining plant biomass at warm ambient temperatures. Plant J. 2011;65:441–52. doi: 10.1111/j.1365-313X.2010.04434.x. [DOI] [PubMed] [Google Scholar]
- 28.Leivar P, Tepperman JM, Cohn MM, Monte E, Al-Sady B, Erickson E, et al. Dynamic antagonism between phytochromes and PIF family basic helix-loop-helix factors induces selective reciprocal responses to light and shade in a rapidly responsive transcriptional network in Arabidopsis. Plant Cell. 2012;24:1398–419. doi: 10.1105/tpc.112.095711. [DOI] [PMC free article] [PubMed] [Google Scholar]