Abstract
Isopentenyl diphosphate isomerase (IDI) is a key enzyme of the isoprenoid pathway, catalyzing the interconversion of isopentenyl diphosphate and dimethylallyl diphosphate, the universal precursors of all isoprenoids. In plants, several subcellular compartments, including cytosol/ER, peroxisomes, mitochondria and plastids, are involved in isoprenoid biosynthesis. Here, we report on the unique triple targeting of two Catharanthus roseus IDI isoforms encoded by a single gene (CrIDI1). The triple localization of CrIDI1 in mitochondria, plastids and peroxisomes is explained by alternative transcription initiation of CrIDI1, by the specificity of a bifunctional N-terminal mitochondria/plastid transit peptide and by the presence of a C-terminal peroxisomal targeting signal. Moreover, bimolecular fluorescence complementation assays revealed self-interactions suggesting that the IDI likely acts as a multimer in vivo.
Keywords: isopentenyl diphosphate isomerase, isoprenoid, alkaloid, triple targeting, subcellular localization, Catharanthus roseus
All isoprenoids originate from the two fundamental building blocks: isopentenyl diphosphate (IPP) and its allylic isomer dimethylallyl diphosphate (DMAPP). Sequential condensations of these C5 units lead to the formation of increasing chain-length polyprenyl diphosphates, which are the precursors of a variety of isoprenoid end products.1 IPP and DMAPP may be synthesized via two different biosynthetic pathways. In fungi and mammals, the C5 units come from the mevalonic acid (MVA) pathway, while in most eubacteria they are produced through the methyl erythritol phosphate (MEP) pathway. By contrast, plants harbor both a cytosolic/peroxisomal MVA pathway2 and a plastidial MEP pathway.1
Interconversion of IPP and DMAPP is governed by IPP isomerase (IDI; EC 5.3.3.2). Based on cofactor requirement, two types of IDIs have been characterized.3 Type I IDIs correspond to zinc metalloproteins needing Mg2+ for activity and are widely distributed in different kingdoms of life such as in fungi, mammals and plants, whereas type II IDIs are restricted to archaea and some bacteria. While fungi possess a single Type I IDI gene copy, mammals and plant genomes typically contain two duplicated copies (IDI1 and IDI2).4,5 Both genes are differentially expressed and encode distinct protein isoforms displaying a complex subcellular distribution. In mammals, each gene produces a single isoform localized exclusively to peroxisomes in a tissue-dependant manner with a widely distributed IDI1 and a skeletal muscle-specific IDI2.5 The situation is more complex in plants. In Arabidopsis, both genes (AtIDI1, At5g16440 and AtIDI2, At3g02780) are expressed in all plant organs, both producing long and short transcripts encoding protein isoforms that differ in length at their N-terminal ends. The long AtIDI1 isoform (AtIDI1L) is targeted to plastids while the long AtIDI2 isoform (AtIDI2L) is sorted to mitochondria.6,7 Both short isoforms (AtIDI1S and AtIDI2S), devoid of a N-terminal transit peptide (TP), are targeted to peroxisomes demonstrating the occurrence of a dual targeting of both gene products (plastid/peroxisome and mitochondria/peroxisome).8
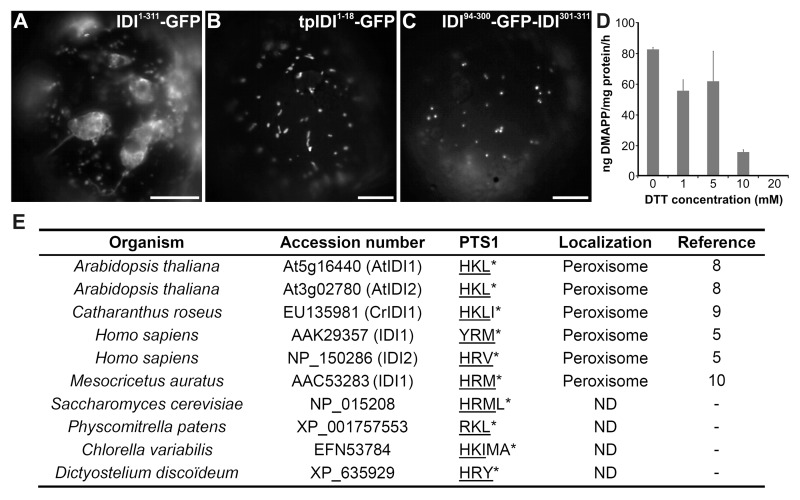
Our recent work9 showed that the situation is somehow different in Madagascar periwinkle (Catharanthus roseus), a medicinal plant widely used to study the architecture of the isoprenoid biosynthetic pathways leading to the formation of monoterpene indole alkaloids. Indeed, in C. roseus, CrIDI1 is highly expressed in all organs as long and short transcripts, while CrIDI2 is barely transcribed only in mature leaves.9 Furthermore, CrIDI1 gene products display an original triple targeting (Fig. 1A-C). Expression of green fluorescent protein fusions revealed that the CrIDI1 long isoform exhibits a N-terminal TP targeting the pseudo-mature protein to both mitochondria and plastids at an apparent similar efficiency (Fig. 1A). This dual targeting can be explained by TP specificities. Indeed, the entire 77-residue long TP needed for plastid targeting contains a specific sequence composed of the first 18-residues that is absent in the Arabidopsis orthologs and sufficient to direct proteins to mitochondria (Fig. 1B). In addition, the short CrIDI1 isoform, devoid of the TP, is directed to the peroxisome by a Peroxisomal Targeting Signal 1 (PTS1) located at the C-terminal end of the protein (Fig. 1C). CrIDI1 displays the same PTS1 (HKL) as its Arabidopsis orthologs, but differs in the presence of an additional isoleucine residue that does not alter the peroxisomal import. It is interesting to notice that even if PTS1 are present on the C-terminus extremity of both long and short isoforms in Arabidopsis and C. roseus, the peroxisomal sorting is observed only for short isoforms. This highlights the importance of alternative transcription initiation events that probably allow countering competition between N-term located plastid and/or mitochondrion targeting sequence(s) and C-term located PTS1. Similar PTS1 are present in the IDI1 and IDI2 mammalian isoforms targeted to peroxisomes via the PTS1-dependent import pathway.5,10 Although it was a question of debate for many years, it is now clear that peroxisomes are key actors of isoprenoid metabolism in higher plants11 and mammals.12 This may be extended to other taxa on the basis of the wide distribution of putative PTS1-containing IDI in fungi, amoeba, mosses and alga (Fig. 1E).
Figure 1. Subcellular localization of IDIs. (A) Dual plastid and mitochondrial targeting of the long isoform of CrIDI1 (residues 1 to 311) fused to GFP (IDI1–311-GFP). (B) Mitochondrial targeting of the first 18 residues of the CrIDI1 transit peptide fused to GFP (tpIDI1-18-GFP). (C) Peroxisomal targeting of the CrIDI1 short isoform bearing an internally fused GFP (IDI94–300-GFP-IDI301–311). (D) Effect of DTT on CrIDI1 activity as judged by incubation of IPP with CrIDI1 at different DTT concentrations. IPP isomerized to DMAPP was detected as isoprene gas following its acidification with phosphoric acid as previously described.15 Error bars signify standard deviation of two replicates. (E) Sequences of PTS1 and putative PTS1 in IDIs from different organisms. The tripeptide of PTS1 is underlined. The asterisk indicates the end of the polypeptide chain. Peroxisomal localization is indicated when experimentally validated. Accession numbers are from GenPept and from Arabidopsis Genome Initiative (for AtIDI1 and AtIDI2). ND, not determined; Bars, 10 µm.
Another result of our study9 showed self-interactions of CrIDI1. Unlike previous results based on gel exclusion chromatography stating that IDIs are monomeric proteins,13,14 bimolecular fluorescence complementation approaches established that CrIDI1 isoforms are capable of self-interactions within plastids, mitochondria and peroxisomes. Self-interactions were also confirmed by migration of the recombinant enzyme on native PAGE. In gel, slow migrating bands corresponding to high molecular complexes dissociate in the presence of 10 mM of dithiothreitol (DTT). Although low DTT concentrations had no significant effect on CrIDI1 activity in vitro, we observed that concentrations of 10 mM DTT or higher strongly inhibited enzyme activity (Fig. 1D), suggesting that self-association of monomers might be important for CrIDI1 activity.
In conclusion, CrIDI1 gene products are targeted to plastids, mitochondria and peroxisomes to form self-interacting complexes that allow the isomerization of IPP and DMAPP synthesized from both the MEP and MVA pathways. Such a triple subcellular targeting of the products of a single gene seems to be a mechanism rarely described so far. Moreover, our results suggest the occurrence of distinct targeting mechanisms of isoprenoid biosynthetic enzymes in plants producing high amounts of specialized isoprenoids (e.g., C. roseus) and plants mainly producing housekeeping isoprenoid metabolites (e.g., A. thaliana).
Footnotes
Previously published online: www.landesbioscience.com/journals/psb/article/21892
References
- 1.Bouvier F, Rahier A, Camara B. Biogenesis, molecular regulation and function of plant isoprenoids. Prog Lipid Res. 2005;44:357–429. doi: 10.1016/j.plipres.2005.09.003. [DOI] [PubMed] [Google Scholar]
- 2.Simkin AJ, Guirimand G, Papon N, Courdavault V, Thabet I, Ginis O, et al. Peroxisomal localisation of the final steps of the mevalonic acid pathway in planta. Planta. 2011;234:903–14. doi: 10.1007/s00425-011-1444-6. [DOI] [PubMed] [Google Scholar]
- 3.Berthelot K, Estevez Y, Deffieux A, Peruch F. Isopentenyl diphosphate isomerase: A checkpoint to isoprenoid biosynthesis. Biochimie. 2012;94:1621–34. doi: 10.1016/j.biochi.2012.03.021. [DOI] [PubMed] [Google Scholar]
- 4.Cunningham FX, Jr., Gantt E. Identification of multi-gene families encoding isopentenyl diphosphate isomerase in plants by heterologous complementation in Escherichia coli. Plant Cell Physiol. 2000;41:119–23. doi: 10.1093/pcp/41.1.119. [DOI] [PubMed] [Google Scholar]
- 5.Clizbe DB, Owens ML, Masuda KR, Shackelford JE, Krisans SK. IDI2, a second isopentenyl diphosphate isomerase in mammals. J Biol Chem. 2007;282:6668–76. doi: 10.1074/jbc.M610922200. [DOI] [PubMed] [Google Scholar]
- 6.Phillips MA, D’Auria JC, Gershenzon J, Pichersky E. The Arabidopsis thaliana type I Isopentenyl Diphosphate Isomerases are targeted to multiple subcellular compartments and have overlapping functions in isoprenoid biosynthesis. Plant Cell. 2008;20:677–96. doi: 10.1105/tpc.107.053926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Okada K, Kasahara H, Yamaguchi S, Kawaide H, Kamiya Y, Nojiri H, et al. Genetic evidence for the role of isopentenyl diphosphate isomerases in the mevalonate pathway and plant development in Arabidopsis. Plant Cell Physiol. 2008;49:604–16. doi: 10.1093/pcp/pcn032. [DOI] [PubMed] [Google Scholar]
- 8.Sapir-Mir M, Mett A, Belausov E, Tal-Meshulam S, Frydman A, Gidoni D, et al. Peroxisomal localization of Arabidopsis isopentenyl diphosphate isomerases suggests that part of the plant isoprenoid mevalonic acid pathway is compartmentalized to peroxisomes. Plant Physiol. 2008;148:1219–28. doi: 10.1104/pp.108.127951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Guirimand G, Guihur A, Phillips MA, Oudin A, Glévarec G, Melin C, et al. A single gene encodes isopentenyl diphosphate isomerase isoforms targeted to plastids, mitochondria and peroxisomes in Catharanthus roseus. Plant Mol Biol. 2012;79:443–59. doi: 10.1007/s11103-012-9923-0. [DOI] [PubMed] [Google Scholar]
- 10.Paton VG, Shackelford JE, Krisans SK. Cloning and subcellular localization of hamster and rat isopentenyl diphosphate dimethylallyl diphosphate isomerase. A PTS1 motif targets the enzyme to peroxisomes. J Biol Chem. 1997;272:18945–50. doi: 10.1074/jbc.272.30.18945. [DOI] [PubMed] [Google Scholar]
- 11.Clastre M, Papon N, Courdavault V, Giglioli-Guivarc’h N, St-Pierre B, Simkin AJ. Subcellular evidence for the involvement of peroxisomes in plant isoprenoid biosynthesis. Plant Signal Behav. 2011;6:2044–6. doi: 10.4161/psb.6.12.18173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kovacs WJ, Tape KN, Shackelford JE, Duan X, Kasumov T, Kelleher JK, et al. Localization of the pre-squalene segment of the isoprenoid biosynthetic pathway in mammalian peroxisomes. Histochem Cell Biol. 2007;127:273–90. doi: 10.1007/s00418-006-0254-6. [DOI] [PubMed] [Google Scholar]
- 13.Dogbo O, Camara B. Purification of isopentenyl pyrophosphate isomerase and geranylgeranyl pyrophosphate synthase from Capsicum chromoplasts by affinity chromatography. Biochim Biophys Acta. 1987;920:140–8. doi: 10.1016/0005-2760(87)90253-0. [DOI] [Google Scholar]
- 14.Bruenger E, Chayet L, Rilling HC. Isopentenyl pyrophosphate isomerase:dimethylallyl pyrophosphate isomerase: isolation from Claviceps sp. SD 58 and comparison to the mammalian enzyme. Arch Biochem Biophys. 1986;248:620–5. doi: 10.1016/0003-9861(86)90516-3. [DOI] [PubMed] [Google Scholar]
- 15.Brüggemann N, Schnitzler JP. Relationship of isopentenyl diphosphate (IDP) isomerase activity to isoprene emission of oak leaves. Tree Physiol. 2002;22:1011–8. doi: 10.1093/treephys/22.14.1011. [DOI] [PubMed] [Google Scholar]