Abstract
Membrane curvature and lipid composition regulate important biological processes within a cell. Currently, several proteins have been reported to sense and/or induce membrane curvatures, e.g. Synaptotagmin-1 and Amphiphysin. However, the large protein scaffold of these curvature sensors limits their applications in complex biological systems. Our interest focuses on identifying and designing peptides that can sense membrane curvature based on established elements observed in natural curvature-sensing proteins. Membrane curvature remodeling also depends on their lipid composition, suggesting strategies to specifically target membrane shape and lipid components simultaneously. We have successfully identified a 25-mer peptide, MARCKS-ED, based on the effector domain sequence of the intracellular membrane protein myristoylated alanine-rich C-kinase substrate that can recognize PS with preferences for highly curved vesicles in a sequence specific manner. These studies further contribute to the understanding of how proteins and peptides sense membrane curvature, as well as provide potential probes for membrane shape and lipid composition.
INTRODUCTION
Membrane curvature plays a vital role in cell signaling, endo- and exocytosis, membrane fusion and protein trafficking (1). Naturally-occurring proteins with a Bin-Amphiphysin-Rvs domain or with the ArfGAP1 Lipid Packing Sensor (ALPS) motif are known to be able to sense membrane curvature (2, 3). However, these large proteins are not optimal for large-scale production, limiting their uses in biotechnology developments. Our primary goal is to identify peptides with curvature-sensing ability that can potentially be used for extracellular vesicle detection based on shape and lipid composition. Herein, we report a short peptide derived from the effector domain of myristoylated alanine-rich C-kinase substrate (MARCKS-ED) that selectively recognizes highly curved membrane surfaces. MARCKS-ED preferentially binds to highly curved surfaces of both synthetic lipid vesicles and isolated extracellular vesicles in rat blood plasma. Furthermore, we also observed that the MARCKS-ED peptide recognizes vesicle surfaces not only based on size but also on their lipid component, detecting the negatively charged phosphatidylserine (PS) exposed on the cell surface of a C. elegans animal model. These results demonstrated that MARCKS-ED recognizes PS lipid composition and membrane curvature, shedding insight into the understanding of protein-lipid interactions in curvature sensing.
MARCKS is an 87-kDa, intracellular protein whose functions involve sequestering phosphatidylinositol 4,5-bisphosphate (PIP2) and regulating Phospholipase C signaling (4). The MARCKS protein also recognizes PS, the negatively charged lipid enriched on the inner leaflet of the cytoplasmic membrane, using its ED (a.a. 151–175) region (5). This protein-membrane association can be regulated and reversed by its binding to Calmodulin (CaM) in the presence of Ca2+ (6).
We focused on the ED region of the MARCKS protein in our search for curvature-sensing peptides based on the following rationales: First, it has been established that the membrane binding by the MARCKS protein is driven by electrostatic interactions between the cationic residues (i.e. Lys, Arg) within the ED peptide region and PS, while the secondary structure is not important (7). This suggests that the unfolded, truncated MARCKS-ED peptide may still retain the membrane binding ability of the full-length protein. Secondly, curved membranes are known to expose more phospholipid packing defects at the membrane surface due to the asymmetric stretching of the bilayer (8). Previous studies have shown that the BAR domains sense curvature partially by detecting these lipid packing defects (2). The MARCKS protein was reported to insert the aromatic side chains of five Phe residues from the ED region into the lipid bilayers (9). We hypothesized that such insertion could in turn stabilize the membrane defects of highly curved vesicles. This hypothesis is consistent with the previous observation that MARCKS is localized in areas of the cell membrane with positive curvature (10).
RESULTS AND DISCUSSION
The MARCKS-ED peptide (Table 1) was synthesized using standard microwave-assisted solid phase Fmoc chemistry (11). To study the roles of particular amino acid residues, two mutant MARCKS peptides were prepared: (i) the five Phe residues that have been previously suggested to vertically insert into the bilayers were mutated to Ala to generate MARCKSmut1; and (ii) the positively charged residues (Lys, Arg) were mutated to Ala to generate MARCKSmut2. We also synthesized MARCKS-ED-scr with a scrambled sequence to investigate the sequence specificity of MARCKS-ED. Fluorescently-labeled MARCKS-ED, MARCKSmut1, MARCKSmut2, and MARCKS-ED-scr peptides were prepared by conjugating either NBD or Alexa Fluor 546™ to their N- termini via a flexible ε–aminohexanoic acid linker. To observe the secondary structure of MARCKS-ED, circular dichroism (CD) analysis was performed for both untreated and vesicle-treated peptide. In agreement with previous reports (12, 13), MARCKS-ED does not demonstrate any predominant secondary structures (Supplementary Figure 1) in aqueous solution. In the presence of lipid vesicles, its CD spectrum did not differ significantly, suggesting that MARCKS-ED remains primarily unstructured.
Table 1.
Sequences of the MARCKS peptides.a
 |
Cationic residues critical for the electrostatic interactions with PS are highlighted in red. The Phe residues speculated to stabilize the curvature defects are shown in blue.
We first prepared synthetic vesicles of various sizes (diameter Ø = 30, 100 and 400 nm) and with various lipid compositions (Supplementary Figure 2) that closely resemble the lipid composition of biological membranes (14). Dynamic light scattering (Supplementary Figure 3) and negative-stain transmission electron microscopy (TEM) (Supplementary Figure 4) were used to validate the lipid vesicle sizes with commercial polystyrene beads as the calibration standard. Furthermore, a time-course experiment was carried out to confirm the stability of the vesicles up to 16 hours (Supplementary Figure 3).
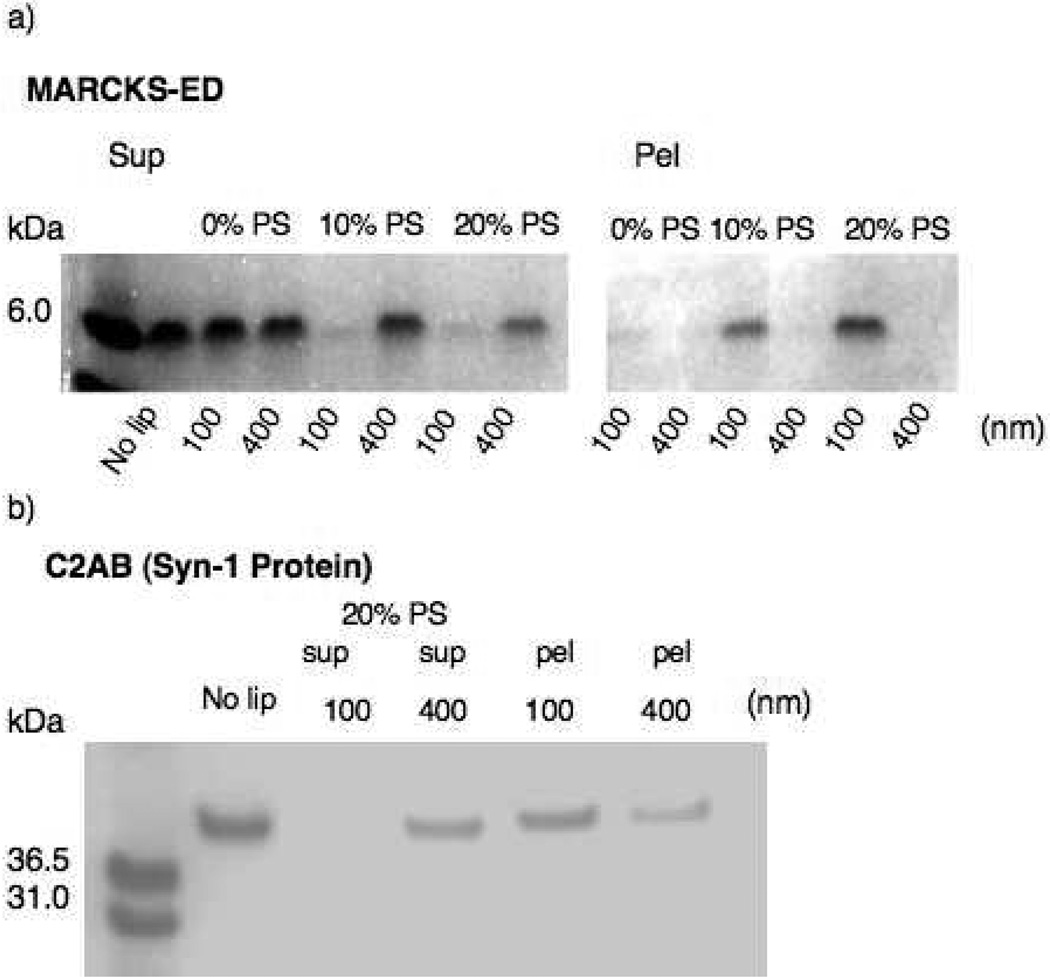
A previously established cosedimentation assay was performed to test the curvature sensing behavior of MARCKS-ED (15). Two vesicle pore sizes, 100 and 400 nm, were used in this experiment because vesicles smaller than 100 nm were difficult to pellet even at high centrifugation speed. We also used a previously reported curvature sensing protein, C2AB, as a positive control (16). Figure 1 shows the gel electrophoresis results of the supernatant and pellet samples collected after sedimentation with MARCKS-ED or C2AB, respectively. A reduction in band intensity indicates that more MARCKS-ED or C2AB was pulled down by the 100 nm PS-containing vesicles than by the larger 400 nm vesicles. Also, less MARCKS-ED was pulled down by the lipid vesicles without PS, suggesting that both the peptide-lipid binding and its curvature sensing depends on the presence of PS. Taken together, these results showed that, similarly to the C2AB protein, MARCKS-ED preferentially binds to highly curved vesicles containing PS.
Figure 1.
Cosedimentation of synthetic liposomes of different sizes with (a) MARCKS-ED ; or (b) the curvature-sensing C2AB domain of Synatptotagmin-1. Sup = supernatant. Pel = pellet. No lip = no lipid vesicles. [C2AB] = 1.5 µM. [Ca2+], for C2AB only = 1 mM. [MARCKS-ED] = 10 µM. [Lipid VesicleC2AB] = 300 µM. [Lipid VesicleMARCKS-ED] = 600 µM.
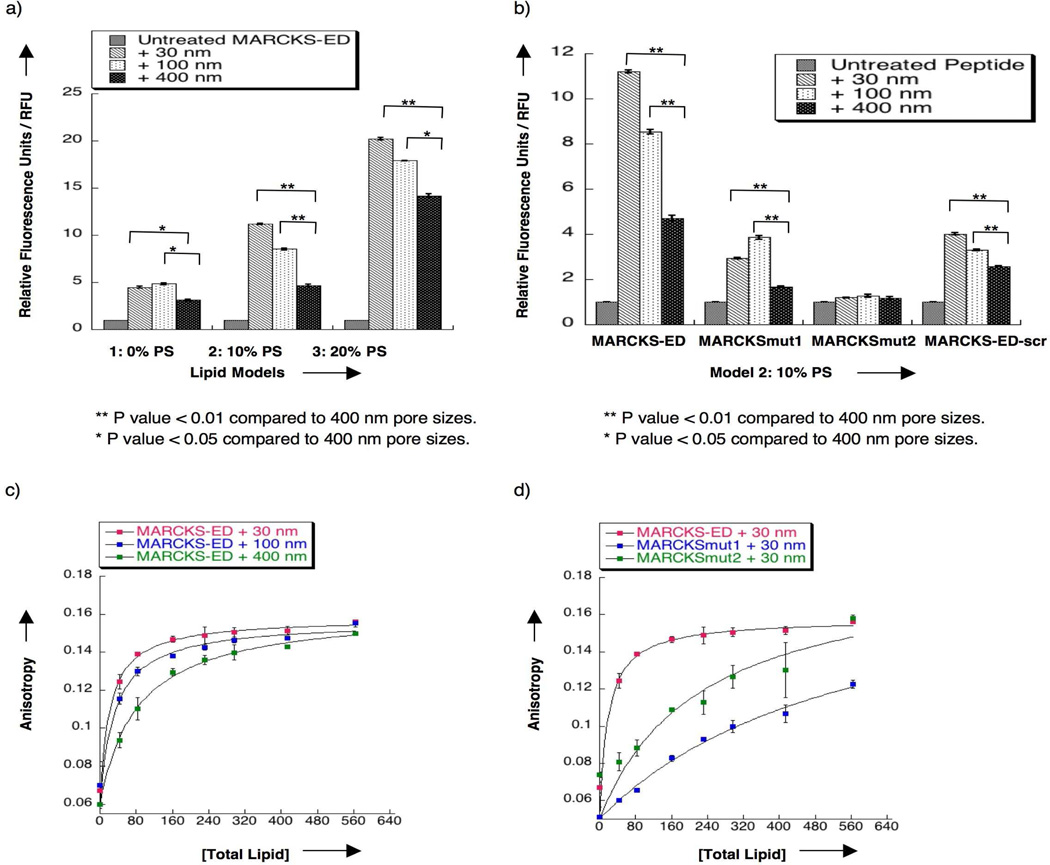
Next, we conducted a fluorescence enhancement assay to further validate the curvature-and PS-sensing behavior of MARCKS-ED. Upon binding to lipid vesicles of different sizes and lipid compositions, the fluorescence intensity of the NBD-labeled MARCKS-ED peptide increased due to the elevated hydrophobicity of the surrounding environment of the fluorophore (Supplementary Figure 5). Figure 2a shows that the fluorescence enhancement of MARCKS-ED treated with the 30 nm pore size lipid vesicles containing PS is significantly higher than the samples treated with both the 100 and 400 nm vesicles (statistic analysis was carried out using the ANOVA method, Supplementary Tables 2, 3 and 4). Appropriate controls were performed to confirm that the fluorophore alone has no effect on the observed fluorescence enhancement (Supplementary Figure 6a and 6b). Compared to the samples treated with the 400 nm vesicles, samples treated with the 30 nm vesicles showed a fluorescence intensity increase of approximately 1.5 fold, a change that is greater than the ones induced by the positive control protein, C2AB (Supplementary Figure 6c). MARCKS-ED-scr also showed a binding preference to highly curved vesicles containing 10% PS (Figure 2b) but not to vesicles containing 0% or 20% PS (Supplementary Figure 7). The fluorescence intensity enhancement by MARCKS-ED-scr, however, is remarkably lower at all these different PS concentrations. MARCKSmut1 and MARCKSmut2 both showed significantly reduced fluorescence enhancement and lacked the curvature sensing behavior observed in the wild type MARCKS-ED (Figure 2b, Supplementary Figure 8). Furthermore, the specificity of MARCKS-ED was confirmed by the observation that its binding can be partially reversed by the addition of CaM (Supplementary Figure 6d), which is in agreement with the behavior of the full-length MARCKS protein. Lastly, the fluorescence intensity differences were less significant with vesicles containing no PS. Taken together, we concluded that MARCKS-ED recognizes PS with a preference for highly curved vesicles in a sequence specific manner.
Figure 2.
Fluorescence enhancement assay (a and b) with the NBD label attached to the N-terminus via a flexible ε-aminohexanoic acid linker to MARCKS-ED, MARCKSmut1, MARCKSmut2 and MARCKS-ED-scr. [Peptide] = 500 nM. [Total Lipid] = 500 µM. The y-axis is described in relative fluorescence units (RFU). Fluorescence was normalized to the untreated NBD-MARCKS-ED peptide samples (RFU=1). (a) Bar graphs showing greater fluorescence of MARCKS-ED treated with 30 nm lipid vesicles containing PS. (b) Bar graphs showing reduced binding and curvature-sensing behavior with mutant peptides, MARCKSmut1, MARCKSmut2 and MARCKS-ED-scr, with lipid vesicles containing 60% POPC: 15% cholesterol: 15% POPE: 10% POPS. ** P value < 0.01 compared to 400 nm pore sizes. * P value < 0.05 compared to 400 nm pore sizes. (c) Fluorescence anisotropy titration of MARCKS-ED with various sized lipid vesicles containing 60% POPC: 15% cholesterol: 15% POPE: 10% POPS. (d) Fluorescence anisotropy titration of various MARCKS peptides with 30 nm pore size lipid vesicles containing 60% POPC: 15% cholesterol: 15% POPE: 10% POPS. The plots were fitted using a standard one-site saturation equation (18). [Peptide] = 1 µM. Phosphate-buffered saline (PBS, pH= 7.4) buffer was used in all sample preparation.
Since fluorescence enhancement assays could not distinguish the contributions between different degrees of membrane penetration and binding affinity, we performed a fluorescence anisotropy assay to specifically measure the binding affinity of the MARCKS peptides. Lipid vesicles were titrated to NBD-labeled MARCKS-ED, MARCKSmut1, or MARCKSmut2 peptides. Since the peptide partitions between the hydrophobic lipid bilayer and the aqueous solvent, the molar partition coefficient (Kp) is often reported (17). By definition, the apparent dissociation constant (Kd), described as the lipid concentration where 50% of the peptide is bound, is the reciprocal of the molar partition coefficient (18). Our results indicated that MARCKS-ED binds more tightly to highly curved vesicles. As shown in Figure 2c and Supplementary Figure 9, MARCKS-ED was found to bind to 30, 100, and 400 nm pore size lipid vesicles containing 10% PS with Kd values of 24 ± 3, 42 ± 13, 86 ± 20 µM, respectively. As a comparison, C2AB showed a 1.9 fold increase in binding to smaller vesicles (105 nm) relative to the larger ones (252 nm) (16). Statistic analysis was performed using ANOVA single factor analysis (Supplementary Tables 5, 6). By contrast, MARCKSmut1 showed a reduced binding affinity (Figure 2d), confirming the critical roles of the Phe residues (10). Similarly, electrostatic interactions also contribute importantly to the binding affinity as MARCKSmut2 is observed to have a weaker binding affinity. These data suggest that both aromatic and cationic residues are required for MARCKS-ED, which is in agreement with our speculation that the interactions between MARCKS-ED and small, PS-enriched vesicles are driven by both electrostatic interactions and the inserted Phe residues that presumably stabilize defects of curved bilayers.
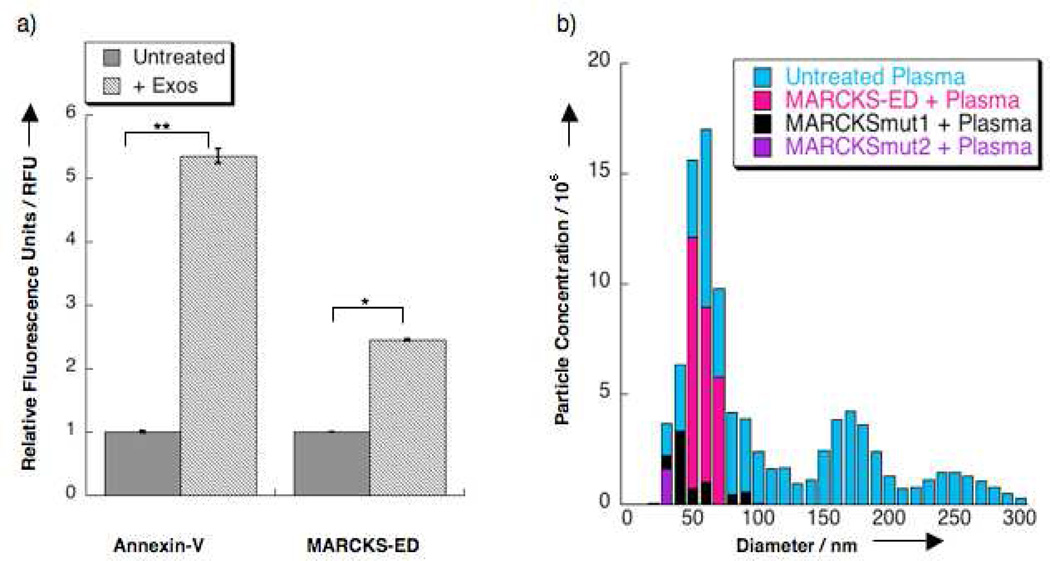
Having established the simultaneous curvature- and PS-sensing by MARCKS-ED in synthetic lipid vesicles, we investigated if MARCKS-ED could detect highly curved, PS-enriched extracellular particles (e.g. microvesicles and exosomes) in a complex biological system. Microvesicles (Ø = 0.1 – 1 1 µm) and exosomes (Ø = 0.03 – 0.1 1 µm) are highly curved lipid vesicles that are shed into bodily fluids (e.g. blood, urine, ascitic fluid) (19). They are released by stressed or cancerous cells in which lipid asymmetry is de-regulated, thus, resulting in the externalization and enrichment of PS on their outer leaflet (20). A direct correlation between the overexpression of these extracellular vesicles in the blood and cancer metastasis has been observed in B16 mouse melanoma cells (21). We investigated if MARCKS-ED could detect extracellular vesicles using plasma samples from a stressed rat model (22). The extracellular vesicles in these samples were characterized by TEM imaging (Supplementary Figure 10a) and immunoblot analysis of the signature CD63 protein exposed on the surface of exosomes and microvesicles (Supplementary Figure 10b) (22). These nano-sized, isolated vesicles were measured by nanoparticle tracking analysis (20) with an average size of Ø = 56 nm. Lastly, we also confirmed any exposed PS on their membrane surface with an established PS-sensing protein, Annexin-V. Previous reports showed that upon binding to PS-enriched synthetic lipid vesicles, the fluorescence from W187 of Annexin-V would increase (23). Indeed, fluorescence enhancement was observed for the Annexin-V protein incubated with the isolated extracellular vesicles from our tested animal models, indicating specific PS recognition (Figure 3a).
Figure 3.
Ex vivo fluorescence assays. a) Fluorescence enhancement of MARCKS-ED and Annexin-V after incubation with isolated rat extracellular vesicles. Fluorescence was normalized to the untreated NBD-MARCKS-ED peptide (0.5 µM) and Annexin-V (0.32 µM) samples. Fluorescence was normalized as 1.0 to the untreated protein or peptide samples in relative fluorescence units (RFU). ** P value <0.01 compared to untreated samples. *P value <0.05 compared to untreated samples. b) Nanoparticle tracking analysis (20) results showing extracellular vesicles in plasma from stressed rats treated with fluorescently-labeled MARCKS-ED at concentrations of 55 nM. The untreated plasma samples (blue) were detected using the scatter mode and treated samples were monitored by tracking the fluorescence of Alexa Fluor 546™ conjugated to the MARCKS-ED (pink), MARCKSmut1 (5) and MARCKSmut2 (purple) peptides.
Figure 3a shows fluorescence enhancement with MARCKS-ED incubated with the isolated extracellular vesicles, demonstrating that MARCKS-ED can indeed bind to these biological particles. We further quantified the diameter size and particle count of the isolated extracellular vesicles detected by MARKCKS-ED. Nanoparticle tracking analysis (NTA) uses a laser source under either scatter or fluorescence detection mode to track small particles by Brownian motion (20), providing a robust method for analyzing vesicle size distribution (24). We first sought to detect the particles of all sizes in whole plasma samples from our stressed rat model under the scatter mode (Supplementary Video 1). Under the fluorescence mode with the emission filter set for the Alexa Fluor 546™ label, the particles that bound to the MARCKS-ED (Supplementary Video 3), MARCKSmut1 (Supplementary Video 4), and MARCKSmut2 (Supplementary Video 5) peptides were observed. We found that MARCKS-ED selectively binds to exosomes (Ø = 0.03 – 0.1 1 µm) in whole plasma (Figure 3b). MARCKSmut1 also showed some preferential binding to smaller vesicles but with much weaker fluorescence signal. The MARCKSmut2 peptide showed only negligible binding. Furthermore, tests with blank samples were performed to rule out possible artifacts caused by background fluorescence from the peptides, the vesicles (Supplementary Video 2) or the unconjugated dye (Supplementary Figure 11). Taken together, these data demonstrate that MARCKS-ED can selectively detect biologically relevant extracellular vesicles with highly curved, PS-enriched surfaces in a complex rat plasma solution.
Aiming to test our hypothesis that MARCKS-ED simultaneously detects shape and PS lipid composition, we further examined its ability to selectively bind to PS in live animals. We carried out fluorescence staining assays in an established C. elegans model with PS-exposing cell membranes (25). PS is normally kept in the inner leaflet of plasma membranes in living cells and is exposed on the cell surface only under certain cellular events, i.e., when cells undergo apoptosis or lose the ability to maintain the PS asymmetry with a tat-1 gene mutation (26). Mutations of the tat-1 gene in C. elegans, which encodes for a phospholipid translocase that maintains PS plasma membrane asymmetry (26), resulted in PS externalization to the outer plasma membrane leaflet. Knock-out of the ced-7 gene greatly reduced the macrophage clearance of apoptotic cell corpses with PS-exposed on their cell surface (26). The NBD-labeled MARCKS-ED peptide was used to stain the dissected gonads of wild type (N2) animals, tat-1(tm3117) mutant animals, and engulfment-deficient ced-7 (n2094) mutant animals. As shown in Figure 4, MARCKS-ED can recognize PS exposed on the surface of all germ cells in the tat-1(tm3117) mutant and un-engulfed apoptotic germ cell corpses in the ced-7(n2094) mutant without staining the wild type controls. By contrast, MARCKSmut1 did not appear to detect PS-exposing membranes in either tat-1 or ced-7 mutant animals, confirming that MARCKS-ED detects PS in a sequence specific manner. Further, Annexin-V, a known PS-sensor, was also used to stain the gonads of tat-1 and ced-7 mutant animals. These staining results with MARCKS-ED were comparable to the staining results observed with Annexin-V (Supplementary Figure 12), indicating that MARCKS-ED may serve as a small peptide alternative to Annexin V and detect PS exposed on the membrane surface in live animals.
Figure 4.
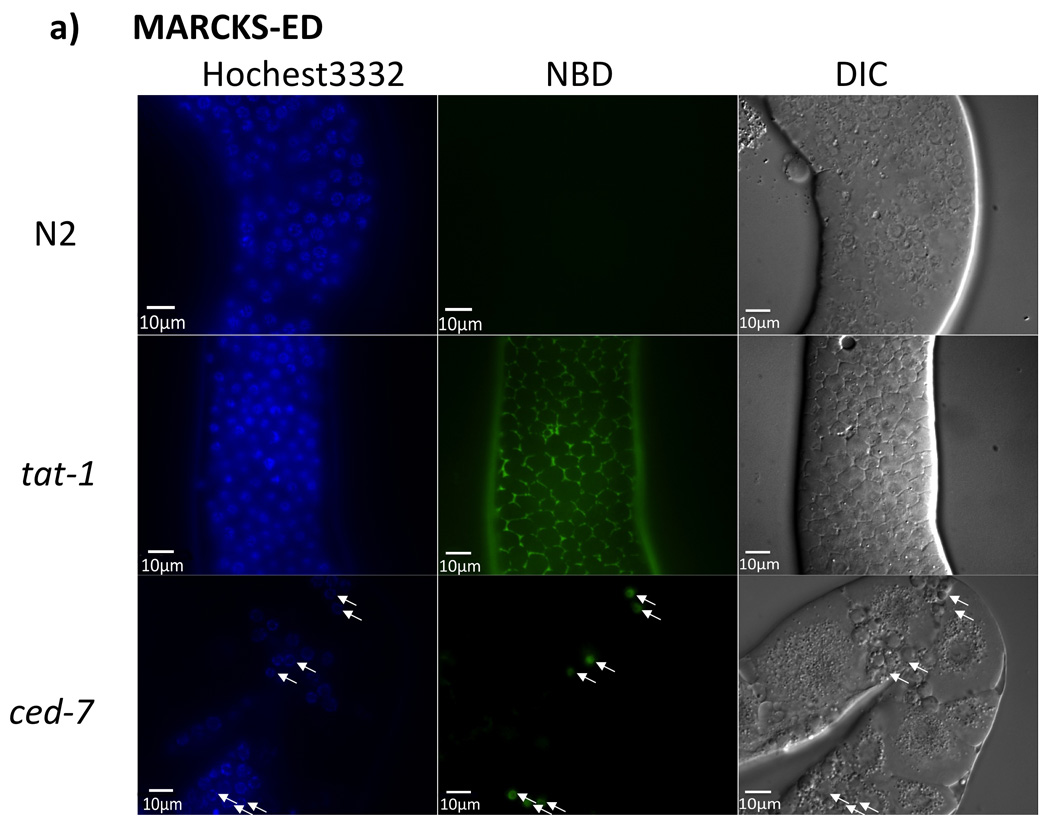
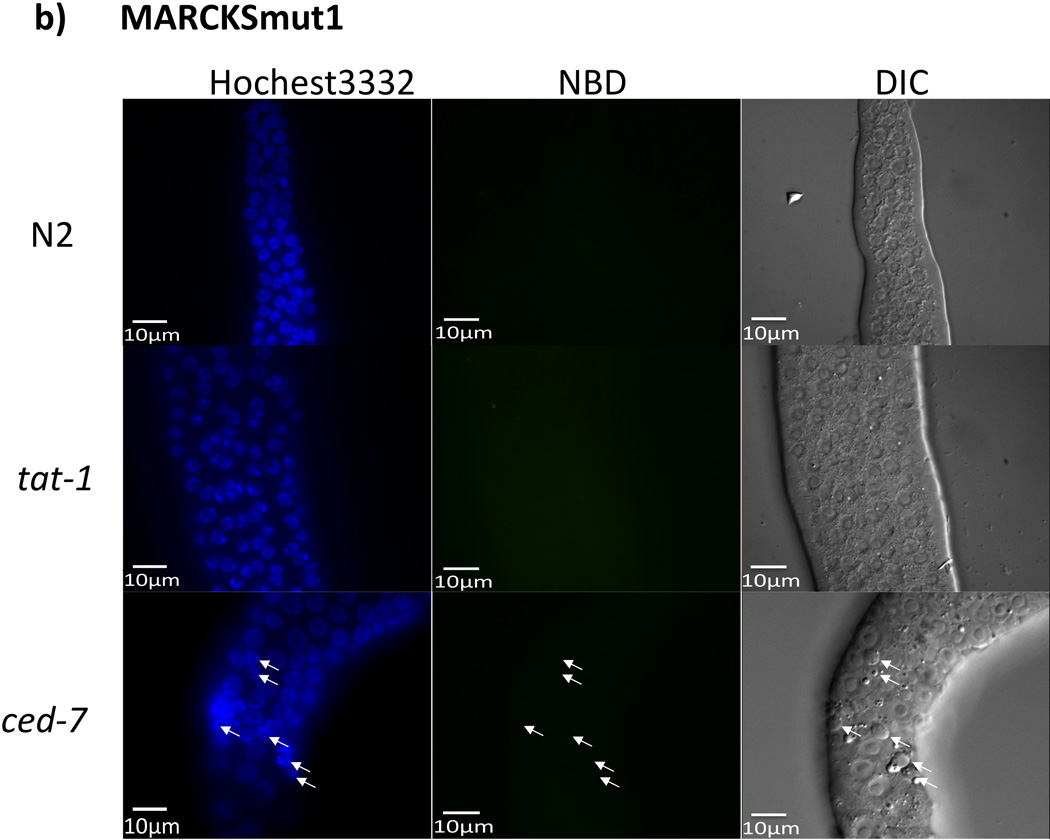
In vivo C. elegans fluorescence assay. The exposed gonads of a wild type N2 hermaphrodite animal (top row), a tat-1(tm3117) mutant animal (middle row), and a ced-7(n2094) mutant animal (bottom row), were stained with NDB-labeled a) MARCKS-ED or b) MARCKSmut1. Images of Hoechst 33342 staining (cell nucleus), MARCKS peptide staining, and differential interference contrast (DIC) microscopy (22) are shown. Arrowheads indicate apoptotic cell corpses. Scale bar = 10 µm.
In summary, the MARCKS-ED peptide was found to differentiate lipid vesicle sizes in both synthetic phospholipid models and extracellular vesicles generated in a rat animal model. Our data suggest that electrostatic interactions and aromatic Phe residues play a critical role in curvature sensing. MARCKS-ED may recognize PS-enriched, curved membranes by filling in the defects in asymmetrically stretched bilayers with its Phe residues, supported by the observation that its binding was greatly reduced with both MARCKSmut1 and MARCKSmut2 peptides. In vivo C. elegans studies confirmed specific detection of PS exposed on the membrane surface by the MARCKS-ED peptide. These results implies that MARCKS-ED could become a prototype for a new generation of peptide sensors that can simultaneously detect both PS and curvature and facilitate investigations of critical biological events, such as extracellular vesicles shedding and apoptosis.
METHODS
Solid phase peptide synthesis
Peptides were synthesized using a CEM Liberty microwave-assisted peptide synthesizer following the standard solid phase Fmoc chemistry (11). For fluorophore labeling, 4-chloro-7-nitrobenzo-2-oxa-1,3-diazole (NBD) or Alexa Fluor 546™ was conjugated to the N-terminus of the peptide via a flexible linker, e-aminohexanoic acid, using the previously reported coupling method (11). A Kaiser test was performed to confirm the efficiency of the fluorophore labeling (27). Following purification, peptides were lyophilized to produce a TFA salt powder. The prepared peptides were characterized by matrix-assisted laser-desorption ionization time-of-flight (MALDI) to confirm their identity.
Synthetic lipid vesicle preparation
To produce various lipid vesicles, we followed a previously established protocol (28). The phospholipids 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphoethanolamine (POPE), 1-palmitoyl-2-oleoyl-sn-glycero-3-phospho-L-serine (POPS), 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine (POPC) and cholesterol (Avanti Lipids) were mixed at the appropriate amounts to produce stock solutions for all synthetic lipid models, using lipids present in physiological membranes (14). For Annexin-V fluorescence enhancement studies, sphingomyelin and phosphatidylinositol were added to mimic the C. elegans outer leaflet membrane (25).
Circular dichroism (CD) spectroscopy
The peptide solutions were prepared at 10 µM in 10 mM phosphate buffer (pH= 7.40) in the presence of 500 µM lipid vesicles (30 nm pore size with 20% PS). Circular dichroism spectra were recorded using the Chirascan CD spectrometer (Applied Photophysics) with a 1 mm path length quartz cuvette at 20°C using phosphate buffer as a blank. The reading was then converted to molar residue ellipticity (θ). Five scans from 190 to 260 nm with data points taken every 1.0 nm were obtained and averaged for each sample.
Cosedimentation assay
MARCKS-ED (10 µM) was incubated with 600 µM synthetic vesicles of sizes 100 nm and 400 nm. The positive control was the intact C2A–C2B cytoplasmic domains (C2AB) of rat Synaptotamin-1 (a.a. 96–421). C2AB (1.5 µM) was treated with CaCl2 (1 mM) and incubated with 300 µM synthetic vesicles of sizes 100 nm and 400 nm, based on the vesicle concentrations previously reported (16). C2AB treated with vesicles was incubated at room temperature for 30 minutes, followed by centrifugation of 65,000 rpm for 45 minutes at 20°C. MARCKS-ED treated with vesicles was incubated at room temperature for 2 hours, followed by 75,000 rpm for 45 minutes at 20°C. The supernatant for each sample was collected as well as the pellets from the MARCKS-ED-treated samples and assayed on a pre-casted 12–15% Tris-Bis gel (Invitrogen).
Fluorescence enhancement assay
The emission spectra of all NBD-labeled peptides were recorded using a Fluorolog-3 fluorometer (Horiba Jobin Yvon) with λex = 480 nm. The peptides and protein were tested at a concentration of 500 nM in PBS (pH 7.40) treated with 500 µM synthetic vesicles of different vesicles sizes. Fluorescence was observed with an emission range of 500–650 nm. Monitored at λex = 295 nm, Annexin-V (0.32 µM) was treated with CaCl2 (3 mM) in TES buffer and with 500 µM lipid vesicles of varying PS content. An emission spectrum of 300–450 nm was recorded. The positive control C2AB (200 nM) from the rat Synaptotagmin-1 (Syn-1) protein (G374, residues 96–421) was treated with CaCl2 (2.5 mM) and observed with λex of 275 nm and emission range of 300–450 nm. The untreated peptide and Ca2+-C2AB samples were corrected by the PBS (pH=7.40) blank solution for the peptides and PBS with Ca2+ for the Ca2+-C2AB sample. All samples were prepared and incubated overnight in 4°C.
Fluorescence anisotropy assay
The NBD-labeled peptides (1 µM) were titrated by synthetic liposomes of various sizes (30, 100, and 400 nm). Fluorescence anisotropy was recorded using a Fluorolog-3 fluorometer. The mixture was allowed to equilibrate for 2 minutes prior to the next titration. The excitation wavelength was set to λex = 480 nm whereas the emission filter was set to λem = 545 nm. The voltage was set to 250 V throughout the experiment. Blank PBS (pH= 7.40) was titrated to NBD-labeled peptides as a negative control, where negligible anisotropy change was observed.
Microvesicle preparation
Adult male Fisher 344 rats (Harlan, 8–9 weeks old) weighing approximately 250–275 grams were used in all experiments. Animals were singly housed in clear Nalgene plastic cages (48×27×20 cm) and allowed access to food and water ad libitum. Temperature and humidity remained constant and animals were maintained on a 12-hour: 12-hour light-dark cycle (lights on at 7:00 AM). Animals were allowed to acclimate to these housing conditions for 1 week prior to any experimental manipulations and were handled each day. The care and treatment of the animals were in accordance with protocols approved by the University of Colorado Institutional Animal Care and Use Committee. An in-house exosome assay based on a reported ELISA method (22) was used for two common markers of exosomes, the tetraspanin CD63 and the membrane transport protein Rab5b. Particles expressing both proteins were captured by antibodies and quantified in a colorimetric endpoint assay.
Nanoparticle tracking analysis
Nanoparticle tracking analyses were performed using the NanoSight LM10-HS instrument equipped with a 532 nm laser and observed at scatter and fluorescence modes (filter = 550 nm). The instrument was calibrated using commercially available 50 nm polystyrene beads (Polysciences). Parallel experiments were done on isolated exosomes and plasma that were not treated with MARCKS-ED conjugated with Alexa Fluor 546™ to confirm that the suspended particles do not autofluoresce. Alexa Fluor 546™-labeled mouse anti-rat CD63 (AbD Serotec) and unconjugated Alexa Fluor 546™ were used as positive and negative controls, respectively. The profiles of MARCKS-ED-Alexa Fluor 546™ and Alexa Fluor 546™ in the absence of microvesicles were also investigated to confirm that these show negligible scatter and fluorescence background signals under identical NTA conditions.
In vivo staining of C. elegans gonads
Gonads of wild type (N2), tat-1(tm3117) or ced-7(n2094) C. elegans animals were stained as previously described (25). Briefly, 36 hour old hermaphrodite adult animals were collected and gently dissected by cutting their heads in a depression slide with a gonad dissection buffer (60 mM NaCl, 32 mM KCl, 3 mM Na2HPO4, 2 mM MgCl2, 20 mM HEPES, 50 µg mL−1 penicillin, 50 µg mL−1 streptomycin, 100 µg mL−1 neomycin, 10 mM glucose, 33% fetal calf serum, and 2 mM CaCl2) to expose the gonads. The exposed gonads were then washed once in the dissection buffer and transferred to a dissection buffer containing 4 µM of Hoechst 33342 and 20 µM of NBD labeled MARCKS-ED peptide, 20 µM NBD labeled MARCKSmut1 peptide, or 10 nM Alexa Fluor 488-conjugated Annexin-V for 45 minutes. Gonads were washed one more time in the dissection buffer, placed on a 5% agarose pad, and visualized using a Nomarski microscope equipped with an epifluorescence detector.
Supplementary Material
Acknowledgements
This work was supported by the Howard Hughes Medical Institute (HHMI) Collaborative Innovation Award to H. Yin and National Institutes of Health (GM59083 to D. Xue and MH061876 to E. Chapman). M. Fleshner thanks the National Science Foundation (NSF IOS 1022451) for financial support. E. Chapman is an HHMI Investigator. L. Morton is supported by a Ruth L. Kirschstein National Research Service Award (CA165349). We would like to thank M. Stowell (CU Boulder), D. Rees, and R. Phillips (Caltech) for their invaluable comments.
Footnotes
Supporting Information.
Supporting Information Available: This material is available free of charge via the Internet. Supplementary Videos, Tables, and Figures. Details about peptide preparation, dynamic light scattering, negative-stain TEM, fluorescence assays, nanoparticle tracking analyses, rat blood plasma exosomes, and C. elegans biological assays.
REFERENCES
- 1.Zimmerberg J, Kozlov MM. How proteins produce cellular membrane curvature. Nat. Rev. Mol. Cell Biol. 2006;7:9–19. doi: 10.1038/nrm1784. [DOI] [PubMed] [Google Scholar]
- 2.Bhatia VK, Madsen KL, Bolinger PY, Kunding A, Hedegard P, Gether U, Stamou D. Amphipathic motifs in BAR domains are essential for membrane curvature sensing. EMBO J. 2009;28:3303–3314. doi: 10.1038/emboj.2009.261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Mesmin B, Drin G, Levi S, Rawet M, Cassel D, Bigay J, Antonny B. Two lipid-packing sensor motifs contribute to the sensitivity of ArfGAP1 to membrane curvature. Biochemistry. 2007;46:1779–1790. doi: 10.1021/bi062288w. [DOI] [PubMed] [Google Scholar]
- 4.Ouimet CC, Wang JK, Walaas SI, Albert KA, Greengard P. Localization of the MARCKS (87 kDa) protein, a major specific substrate for protein kinase C, in rat brain. J. Neurosci. 1990;10:1683–1698. doi: 10.1523/JNEUROSCI.10-05-01683.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kim J, Blackshear PJ, Johnson JD, McLaughlin S. Phosphorylation reverses the membrane association of peptides that correspond to the basic domains of MARCKS and neuromodulin. Biophys. J. 1994;67:227–237. doi: 10.1016/S0006-3495(94)80473-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kim J, Shishido T, Jiang X, Aderem A, McLaughlin S. Phosphorylation, high ionic strength, and calmodulin reverse the binding of MARCKS to phospholipid vesicles. J. Biol. Chem. 1994;269:28214–28219. [PubMed] [Google Scholar]
- 7.Wang J, Gambhir A, Hangyas-Mihalyne G, Murray D, Golebiewska U, McLaughlin S. Lateral sequestration of phosphatidylinositol 4,5-bisphosphate by the basic effector domain of myristoylated alanine-rich C kinase substrate is due to nonspecific electrostatic interactions. J. Biol. Chem. 2002;277:34401–34412. doi: 10.1074/jbc.M203954200. [DOI] [PubMed] [Google Scholar]
- 8.Madsen KL, Bhatia VK, Gether U, Stamou D. BAR domains, amphipathic helices and membrane-anchored proteins use the same mechanism to sense membrane curvature. FEBS Lett. 2010;584:1848–1855. doi: 10.1016/j.febslet.2010.01.053. [DOI] [PubMed] [Google Scholar]
- 9.Ellena JF, Burnitz MC, Cafiso DS. Location of the myristoylated alanine-rich C-kinase substrate (MARCKS) effector domain in negatively charged phospholipid bicelles. Biophys. J. 2003;85:2442–2448. doi: 10.1016/s0006-3495(03)74667-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhang W, Crocker E, McLaughlin S, Smith SO. Binding of peptides with basic and aromatic residues to bilayer membranes: phenylalanine in the myristoylated alanine-rich C kinase substrate effector domain penetrates into the hydrophobic core of the bilayer. J. Biol. Chem. 2003;278:21459–21466. doi: 10.1074/jbc.M301652200. [DOI] [PubMed] [Google Scholar]
- 11.Merrifield RB. Merrifield Solid-Phase Peptide Synthesis. J. Am. Chem. Soc. 1963;85:2149. [Google Scholar]
- 12.Arbuzova A, Schmitz AA, Vergeres G. Cross-talk unfolded: MARCKS proteins. Biochem. J. 2002;362:1–12. doi: 10.1042/0264-6021:3620001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Matsubara M, Yamauchi E, Hayashi N, Taniguchi H. MARCKS, a major protein kinase C substrate, assumes non-helical conformations both in solution and in complex with Ca2+-calmodulin. FEBS Lett. 1998;421:203–207. doi: 10.1016/s0014-5793(97)01557-3. [DOI] [PubMed] [Google Scholar]
- 14.Rothman JE, Lenard J. Membrane asymmetry. Science. 1977;195:743–753. doi: 10.1126/science.402030. [DOI] [PubMed] [Google Scholar]
- 15.Peter BJ, Kent HM, Mills IG, Vallis Y, Butler PJ, Evans PR, McMahon HT. BAR domains as sensors of membrane curvature: the amphiphysin BAR structure. Science. 2004;303:495–499. doi: 10.1126/science.1092586. [DOI] [PubMed] [Google Scholar]
- 16.Hui E, Johnson CP, Yao J, Dunning FM, Chapman ER. Synaptotagmin-mediated bending of the target membrane is a critical step in Ca(2+)-regulated fusion. Cell. 2009;138:709–721. doi: 10.1016/j.cell.2009.05.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Arbuzova A, Wang L, Wang J, Hangyas-Mihalyne G, Murray D, Honig B, McLaughlin S. Membrane binding of peptides containing both basic and aromatic residues. Experimental studies with peptides corresponding to the scaffolding region of caveolin and the effector region of MARCKS. Biochemistry. 2000;39:10330–10339. doi: 10.1021/bi001039j. [DOI] [PubMed] [Google Scholar]
- 18.Rusu L, Gambhir A, McLaughlin S, Radler J. Fluorescence correlation spectroscopy studies of Peptide and protein binding to phospholipid vesicles. Biophys. J. 2004;87:1044–1053. doi: 10.1529/biophysj.104.039958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Graves LE, Ariztia EV, Navari JR, Matzel HJ, Stack MS, Fishman DA. Proinvasive properties of ovarian cancer ascites-derived membrane vesicles. Cancer Res. 2004;64:7045–7049. doi: 10.1158/0008-5472.CAN-04-1800. [DOI] [PubMed] [Google Scholar]
- 20.Camussi G, Deregibus MC, Bruno S, Cantaluppi V, Biancone L. Exosomes/microvesicles as a mechanism of cell-to-cell communication. Kidney Int. 2010;78:838–848. doi: 10.1038/ki.2010.278. [DOI] [PubMed] [Google Scholar]
- 21.Muralidharan-Chari V, Clancy JW, Sedgwick A, D'Souza-Schorey C. Microvesicles: mediators of extracellular communication during cancer progression. J. Cell Sci. 123:1603–1611. doi: 10.1242/jcs.064386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Logozzi M, De Milito A, Lugini L, Borghi M, Calabro L, Spada M, Perdicchio M, Marino ML, Federici C, Iessi E, Brambilla D, Venturi G, Lozupone F, Santinami M, Huber V, Maio M, Rivoltini L, Fais S. High levels of exosomes expressing CD63 and caveolin-1 in plasma of melanoma patients. PLoS One. 2009;4:e5219. doi: 10.1371/journal.pone.0005219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Meers P, Mealy T. Calcium-dependent annexin V binding to phospholipids: stoichiometry, specificity, and the role of negative charge. Biochemistry. 1993;32:11711–11721. doi: 10.1021/bi00094a030. [DOI] [PubMed] [Google Scholar]
- 24.Dragovic RA, Gardiner C, Brooks AS, Tannetta DS, Ferguson DJ, Hole P, Carr B, Redman CW, Harris AL, Dobson PJ, Harrison P, Sargent IL. Sizing and phenotyping of cellular vesicles using Nanoparticle Tracking Analysis. Nanomedicine. 2011;7:780–788. doi: 10.1016/j.nano.2011.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wang X, Wang J, Gengyo-Ando K, Gu L, Sun CL, Yang C, Shi Y, Kobayashi T, Mitani S, Xie XS, Xue D. C. elegans mitochondrial factor WAH-1 promotes phosphatidylserine externalization in apoptotic cells through phospholipid scramblase SCRM-1. Nat. Cell Biol. 2007;9:541–549. doi: 10.1038/ncb1574. [DOI] [PubMed] [Google Scholar]
- 26.Darland-Ransom M, Wang X, Sun CL, Mapes J, Gengyo-Ando K, Mitani S, Xue D. Role of C. elegans TAT-1 protein in maintaining plasma membrane phosphatidylserine asymmetry. Science. 2008;320:528–531. doi: 10.1126/science.1155847. [DOI] [PubMed] [Google Scholar]
- 27.Kaiser E, Colescott RL, Bossinger CD, Cook PI. Color test for detection of free terminal amino groups in the solid-phase synthesis of peptides. Anal. Biochem. 1970;34:595–598. doi: 10.1016/0003-2697(70)90146-6. [DOI] [PubMed] [Google Scholar]
- 28.Morton LA, Saludes JP, Yin H. Constant Pressure-controlled Extrusion Method for the Preparation of Nano-sized Lipid Vesicles. J. Vis. Exp. 2012;64:e4151. doi: 10.3791/4151. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.