Abstract
Rationale
Hypertension (HTN) affects ~30% of adults in industrialized countries and is the major risk factor for cardiovascular disease.
Objective
We sought to study the genetic effect of coding and conserved non-coding variants in syndromic HTN genes on systolic (SBP) and diastolic (DBP) blood pressure to assess their overall impact on essential hypertension (EH).
Methods and Results
We resequenced 11 genes (AGT, CYP11B1, CYP17A1, HSD11B2, NR3C1, NR3C2, SCNN1A, SCNN1B, SCNN1G, WNK1 and WNK4) in 560 European (EA) and African (AA) ancestry GenNet participants with extreme SBP. We investigated genetic associations of 2,535 variants with BP in 19,997 EAs and 6,069 AAs in three types of analyses. First, we studied the combined effects of all variants in GenNet. Second, we studied 1000 Genomes imputed polymorphic variants in 9,747 EA and 3,207 AA ARIC subjects. Lastly, we genotyped 37 missense and common noncoding variants in 6,591 EAs and 6,521 individuals (3,659 EA/2,862 AA) from the CLUE and FBPP studies. None of the variants individually reached significant false-discovery rates (FDR≤0.05) for SBP and DBP. However, upon pooling all coding and non-coding variants we identified at least 5 loci (AGT, CYP11B1, NR3C2, SCNN1G and WNK1), with higher association at evolutionary conserved sites.
Conclusions
Both rare and common variants at these genes affect BP in the general population with modest effects sizes (<0.05 standard deviation units) and much larger sample sizes are required to assess the impact of individual genes. Collectively, conserved noncoding variants affect BP to a greater extent than missense mutations.
Keywords: essential hypertension, blood pressure, population genetics, sequencing, genotype
INTRODUCTION
Essential hypertension (EH), or hypertension (HTN) not ascribable to secondary causes, affects ~30% of adults in industrialized countries and is largely of unknown molecular etiology1. Although measured blood pressure (BP) is moderately heritable (heritability ~40–60%)2, it also varies with age, BMI, diet, stress level and sympathetic tone. The major physiological hypothesis for BP variation is Guyton’s thesis that variation in kidney fluid regulation, in turn depending on salt clearance3, leads to BP differences. Indeed, the identification of numerous Mendelian syndromic hypotension and HTN genes is proof of Guyton’s hypothesis since the encoded proteins regulate renal salt-water balance4.
Recently, there has been increasing effort to systematically study common polymorphisms in inter-individual variation in HTN risk. Two large genome-wide association studies (GWAS), from the CHARGE5 and Global BPGEN6 consortia, and a recent meta-analysis from the International Consortium for Blood Pressure GWAS (ICBP),7 have made progress in this direction with the identification of 29 loci explaining 1–2% of systolic (SBP) and diastolic BP (DBP) in over 200,000 subjects of European ancestry (EA). The identified variants in ICBP also showed effects in individuals of East Asian (N=29,719), South Asian (N=23,977) and African ancestries (N=19,775). Other analogous studies on individuals of Asian (22,900 subjects) and African (9,608 subjects) ancestries have additionally identified 5 SBP and 3 DBP loci8–12. Nevertheless, the precise genes at each mapped locus, their functions in BP regulation, and how functional variants in them lead to physiologic variation in BP all remain unknown and are major challenges ahead. We undertook the alternative approach of trying to assess how sequence variants in known Mendelian syndromic hypertension genes, affect BP variation in multiple large cohorts.
For the detection of gene variants, we chose as exemplars angiotensinogen (AGT), with a known effect on EH and renal tubular dysgenesis, and, 10 additional genes (CYP11B1, CYP17A1, HSD11B2, NR3C1, NR3C2, SCNN1A, SCNN1B, SCNN1G, WNK1, WNK4) (Online Table I) known to harbor loss-/gain-of-function or dominant negative mutations leading to a variety of autosomal dominant or recessive HTN syndromes (Online Table II). The disease-associated variants and mutations at these genes are all rare, except for those at AGT. Thus, we enquired whether any sequence variants within these genes in the general population were associated with EH, i.e., did these genes play a larger role in elevating BP in the general population and lead to EH?
We first determined the DNA sequences of these genes to identify genetic variants at coding sequences, intron-exon junctions and all highly conserved non-coding elements in the vicinity of each gene. Our sequenced sample included 560 individuals from the GenNet network of the Family Blood Pressure Program (FBPP)13, equally divided into 8 strata comprising European (EA) and African (AA) ancestry, males and females, and, the highest (top) and lowest (bottom) 70 individuals for each stratum, corresponding to ~15% sex-, age- and BMI-adjusted SBP thresholds. The 2,535 identified variants were studied in three ways. We first examined the effect of all identified variants, missense and non-coding variants, pooled as a class, in 280 EA and 280 AA of our original GenNet samples but by weighting them according to their evolutionary conservation or predicted deleterious effect. We then studied all polymorphisms (MAF≥1%) and, lastly, a selected group of coding variants predicted to be deleterious based on protein sequence conservation and the nature of the chemical substitution. These two classes of variants were studied for genetic association with SBP, DBP, by imputation in the population-based cohort ARIC (9,747 EAs and 3,207 AAs) using first visit measurements and by direct genotyping in the population-based CLUE (6,591 EA samples) cohort and the family-based FBPP (3,659 EA and 2,862 AA samples) study. Our general conclusion is that, despite not finding statistical significance at individual variants, these genes, in aggregate, do show statistically significant effects on BP. In addition, we conclude that rare coding variants have genetic effects of the same magnitude as that of common non-coding polymorphisms and that the contribution of non-coding variants is not negligible. Finally, assessing the contribution of individual genes will require much larger sample sizes.
MATERIALS AND METHODS
An expanded Methods section is available in the Online Supplemental Materials
Cohorts and samples studied
FBPP
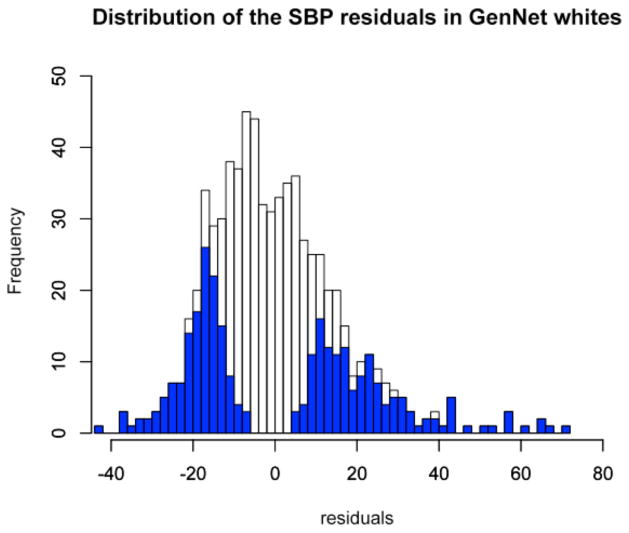
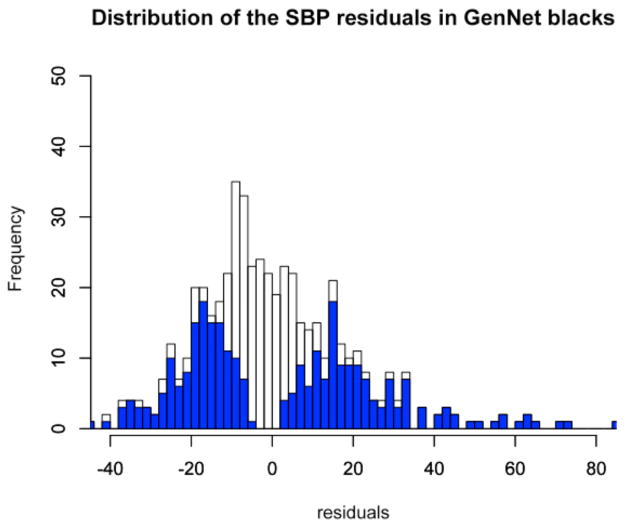
We chose 560 independent samples, equally divided between males/females and EA/AA ancestry, from the total of 705 EA and 521 AA unrelated GenNet subjects of FBPP13. We selected 70 individuals with the most extreme SBP residuals (top/bottom levels). Specifically, we selected 560 GenNet subjects, whose SBP residuals lay below the ~15th %tile (corresponding to residuals of −50.02 to −5.29 mmHg) or above the ~85th %tile (corresponding to residuals of 2.47 to 85.65 mmHg), see Figure 1.
Figure 1.
Distribution of systolic blood pressure (SBP) residuals in GenNet participants: (a) 705 unrelated European Americans (EA), and (b) 521 unrelated African Americans (AA). Residual SBP for the 280 EA and 280 AA individuals chosen for sequencing are highlighted in blue.
ARIC
For the polymorphic variant study, we used GWAS data from the Atherosclerosis Risk in Communities (ARIC)14 samples to study their association with BP in visit 1 of 9,747 EAs and 3,207 AAs (Online Table III.)
CLUE
For the rare putatively deleterious variants association studies, we had access and genotyped DNA samples from 7,065 Odyssey subjects in the CLUE study.15
DNA sequencing and genotyping methods
For each gene, we considered all exons±50nt flanking sequence, 2kb up-/down-stream of the gene, and mammalian-conserved non-coding elements within 5kb of the gene (>70% sequence identity across>100bp in human-mouse and human-rat alignments or LOD>50 from UCSC 17-species alignment) for Sanger resequencing (Online Methods). We designed primers for Sequenom pooled assay of 15 non-synonymous deleterious variants from RS&G (this study) and 26 replicated common variants from ICBP7 using the MassArray® Assay Design Software, Sequenom Inc.; we could develop successful assays for 11 and 26 variants, respectively, and a 27th variant was genotyped using Taqman. We followed the standard genotyping protocols for Sequenom16 and Taqman17.
Association analyses
For our analyses, we adjusted the BP measurements for potential medication effects by adding 15/10mmHg to SBP/DBP in individuals who were taking anti-hypertensive drugs at the time of ascertainment18. The residuals of SBP/DBP were adjusted on age, age2, gender and BMI, separately for the EAs and AAs.
The significance level was set to be FDR≤0.05. We analyzed the overall effects of our 2,535 variants by collapsing (pooling) them in two ways: weighting all variants by their conservation (phyloP) score and weighting only missense variants by their predicted deleterious effect (PolyPhen2 scores), for each gene in EAs and AAs separately, followed by pooling across all 11 genes, using the Madsen & Browning allele frequency weighted sum method in combination with conservation or deleterious effects as described by Price and collegues19.
We used BEAGLE20 (version 3.3) to impute RS&G and 1000 Genomes polymorphic variants (MAF≥1%) within 10kb boundaries of the 11 gene regions in ARIC, using 1000 Genomes EUR/AFR reference panels for EAs/AAs. Variants with imputation score r2≤0.3 were removed before association analysis with PLINK21 for SBP/DBP in visit 1. For genetic association analysis of individual rare putatively deleterious variants, we used MERLIN22 (-assoc) in CLUE and FBPP.
Power calculations
We assume independent sampling, additive genetic model and that BP continuous trait arose from a two-component normal mixture distribution, where one component corresponded to the variant allele whose genetic effect was shifted by s standard deviation units with respect to the other component corresponding to the reference allele. Their variances are assumed to be the same and equal to 1 and their mixing proportion reflect the allele frequencies q and p (=1−q), respectively. The power to detect the difference in the means of the two components is then:
where Φ is the standard normal cumulative distribution function of sample size n, is the quantile of the standard normal distribution at α significance level. The second summand is very small and can be ignored.
RESULTS
DNA sequencing and variant detection
For each individual, we obtained on average 150,448bp of DNA sequence of which 47,091bp (31%) was coding and 103,357bp (69%) was conserved non-coding. Across all 560 individuals this led to a dataset comprising ~85Mb with a variant distribution as shown in Table 1. A variable pattern of genetic variation is observed across the 11 genes. The data suggests modest variation in the numbers of SNVs across the 11 genes in comparison to the expectation based on the length of sequence scanned (P=0.044 in EAs; P=1.7×10−4 in AAs) but this is highly significant for the total number of coding and conserved non-coding variants (P=3.15×10−62 in EAs; P=7.22×10−57 in AAs). Some of this difference in statistical significance is likely due to the absolute smaller numbers of coding than total variants. Nevertheless, these results suggest significant variation in the evolutionary constraint on conserved non-coding and intronic sequences as well. Notable outliers among these genes are CYP11B1 and NR3C1 with significant excess and deficiency of coding variants, respectively. Among all variants, seven genes are outliers, with NR3C2, SCNN1B, SCNN1G and WNK1 supporting a significant increase and CYP17A1, NR3C1 and WNK4 supporting a significant decrease, in variation as compared to the length of the sequence scanned. A similar overall trend is observed with INDELs where the total numbers of variants is non-random across genes with respect to the length scanned (P=1.78×10−6 in EAs; P=1.99×10−8 in AAs). These data on variation suggest the great variability in the observed numbers and types of variants both across genes, and coding versus non-coding segments of each gene, regardless of the genes’ GC content. This implies that since the individuals sequenced harbor a mixture of functionally relevant and neutral variants, and consequently phenotypically relevant and irrelevant variants, the detection of genetic effects at a specific gene is dependent on factors beyond sample size. In other words, despite our extensive sequencing, we might not have sampled functional variants equally across each gene.
Table 1.
Distribution of identified sequence variants by mutation type. For each of the 11 genes, the size of the sequenced target (coding/non-coding) and the observed numbers of variants (substitutions (SNV)/indels (INDEL); coding/non-coding) in 280 EAs and 280 AAs are shown.
| Gene | Sequenced length (bp) | EA (N=280) | AA (N=280) | |||||||
|---|---|---|---|---|---|---|---|---|---|---|
|
| ||||||||||
| SNV | INDEL | SNV | INDEL | |||||||
|
| ||||||||||
| Coding | Non-coding | Coding | Non-coding | Coding | Non-coding | Coding | Non-coding | Coding | Non-coding | |
| AGT | 2,584 | 6,490 | 5 | 59 | - | 1 | 9 | 113 | - | 1 |
| CYP11B1 | 3,535 | 4,129 | 14 | 52 | - | - | 27 | 86 | - | 1 |
| CYP17A1 | 1,870 | 3,237 | 6 | 19 | - | - | 7 | 30 | - | 0 |
| HSD11B2 | 1,884 | 4,909 | 5 | 52 | - | 4 | 8 | 63 | - | 9 |
| NR3C1 | 6,614 | 24,717 | 6 | 91 | - | 5 | 11 | 163 | - | 11 |
| NR3C2 | 5,898 | 23,483 | 8 | 325 | - | 29 | 13 | 494 | - | 54 |
| SCNN1A | 3,200 | 5,788 | 7 | 64 | - | 1 | 16 | 136 | - | 2 |
| SCNN1B | 2,597 | 3,947 | 9 | 63 | - | 1 | 9 | 109 | - | 2 |
| SCNN1G | 3,499 | 4,116 | 11 | 131 | - | 8 | 9 | 120 | - | 12 |
| WNK1 | 11,232 | 12,170 | 18 | 216 | 1 | 20 | 41 | 281 | - | 34 |
| WNK4 | 4,178 | 10,371 | 10 | 34 | - | 2 | 25 | 72 | - | 4 |
|
| ||||||||||
| Totals | 47,091 | 103,357 | 99 | 1,106 | 1 | 71 | 175 | 1,667 | - | 130 |
|
| ||||||||||
| 150,448 | 1,205 | 72 | 1,842 | 130 | ||||||
|
| ||||||||||
| 1,277 | 1,972 | |||||||||
Global tests of BP effects
We first examined the distribution of variants by their allele frequency class and their residual SBP phenotype, as affected by their membership in only the top BP class or only the bottom class versus those present in both classes. This analysis constitutes a global test of association between SBP and the entire set of genetic variants we identified (Table 2). Overall, there is no enrichment of variants at either the higher or the lower SBP threshold (265 vs. 291, P=0.27 for EAs; 432 vs. 378, P=0.06 for AAs). Interestingly, only a borderline significance was observed at the protein sequence level where non-synonymous variants were slightly enriched at the extremes (P=0.049 for EAs; P=0.021 for AAs). When this pattern was tested across the 11 genes the results were highly significant across the three BP classes (P=5.0×10−8 for EAs; P=6.4×10−3 for AAs) but not for the bottom versus top comparisons (P=0.25 for EAs; P=0.07 for AAs), suggesting that either the effect is weak or the trend is owing to the differences in numbers of variants across genes, as demonstrated earlier. These results are not unexpected since the vast majority of variants we detected, even at bona fide BP genes, are not related to the BP phenotype. If there is an effect it must be relegated to only a few variants; the small over-representation for non-synonymous sites may arise from a larger fraction of these variants affecting BP.
Table 2.
Distribution of identified sequence variants by blood pressure (BP) threshold. The observed numbers of variants (total; frequency distribution; SNV/INDEL; coding/non-coding; synonymous/non-synonymous) classified by their occurrence at the top BP threshold or the bottom BP threshold or present in both classes, are shown.
| EA (N=280) | AA (N=280) | ||||||||
|---|---|---|---|---|---|---|---|---|---|
|
| |||||||||
| BP group | Bottom | Both | Top | Total | Bottom | Both | Top | Total | |
| All variants | 291 | 721 | 265 | 1,277 | 378 | 1,162 | 432 | 1,972 | |
|
| |||||||||
| Variant frequency | <1% | 284 | 168 | 261 | 713 | 376 | 301 | 423 | 1,100 |
| 1–5% | 6 | 182 | 4 | 192 | 2 | 400 | 9 | 411 | |
| ≥5% | 1 | 371 | 0 | 372 | 0 | 461 | 0 | 461 | |
|
| |||||||||
| SNV/INDEL | 272/19 | 677/44 | 256/9 | 1,205/72 | 353/25 | 1,086/76 | 403/29 | 1,842/130 | |
|
| |||||||||
| Coding/Non-coding | 28/263 | 49/671 | 23/242 | 100/1,176 | 37/341 | 90/1,072 | 48/384 | 175/1,797 | |
|
| |||||||||
| Synonymous/Non-Synonymous | 11/17 | 27/22 | 8/15 | 46/54 | 16/21 | 49/41 | 16/32 | 81/94 | |
To test this last hypothesis, we performed an alternate analysis of the association effect of all variants on BP by weighting each variant by its presumed functional effect. Since we sequenced both coding and conserved non-coding elements, our first analysis on all variants used conservation (phyloP) score as weights; our second analysis focused on non-synonymous variants only, which are generally more conserved, and used PolyPhen2 scores, an index of deleterious effect, as weights. All analyses were performed using the Madsen-Browning weighted sum method from the pooling test by Price and colleagues19 (Table 3). Interestingly, the class of all variants (1,205 in EAs and 1,842 in AAs) was significantly associated with SBP in both populations (P= 0.008 in EAs; P= 0.004 in AAs) but not with DBP in either (P= 0.073 in EAs; P= 0.189 in AAs). When the analyses were restricted to missense alleles, none of the associations were significant since they were based on only 39 and 70 variants in EAs and AAs, respectively. Individual genes showed considerable variation but, being based on few variants, few of these tests were significant. However, for the test of all variants in individual genes, 5 of the 11 genes were statistically significant (FDR≤0.05) for SBP: AGT (P=0.009), CYP11B1 (P=0.005), NR3C2 (P=4×10−5), SCNN1G (P=3.5×10−4) in EAs and WNK1 (P=0.004) in AAs. When we studied non-coding variants separately and also weighted them by conservation phyloP scores, the effect in individual gene locus is even more significant with 2 additional loci: CYP17A1 (P=0.026) and HSD11B2 (P=0.010) with FDR≤0.05 (Online Table IV.)
Table 3.
Genetic association results of pooled variants by gene, population and type. We assessed association by pooling (collapsing) either all variants, missense and non-coding variants within each gene, and across all genes, within EAs and AAs. In addition, missense variants were weighted by PolyPhen2 scores (*note that PolyPhen2 scores are not available on all missense variants) whereas phyloP scores weighted all and non-coding variants. The numbers of variants (k) involved are provided; P-values with FDR≤0.05 are highlighted in yellow.
| EA | AA | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
|
| ||||||||||||
| All variants | Missense | All variants | Missense | |||||||||
|
| ||||||||||||
| Gene | SBP | DBP | k | SBP | DBP | k* | SBP | DBP | k | SBP | DBP | k* |
| All | 0.008 | 0.073 | 1,205 | 0.157 | 0.460 | 39 | 0.004 | 0.189 | 1,842 | 0.124 | 0.259 | 70 |
| AGT | 0.009 | 0.183 | 64 | 0.035 | 0.501 | 3 | 0.698 | 0.763 | 122 | 0.462 | 0.775 | 6 |
| CYP11B1 | 0.005 | 0.047 | 66 | 0.181 | 0.115 | 4 | 0.388 | 0.434 | 113 | 0.520 | 0.624 | 9 |
| CYP17A1 | 0.401 | 0.350 | 25 | 0.996 | 0.943 | 4 | 0.115 | 0.598 | 37 | 0.567 | 0.616 | 4 |
| HSD11B2 | 0.072 | 0.113 | 57 | 0.972 | 0.938 | 2 | 0.111 | 0.766 | 71 | 0.168 | 0.221 | 3 |
| NR3C1 | 0.476 | 0.790 | 97 | 0.835 | 0.756 | 2 | 0.014 | 0.104 | 174 | 0.879 | 0.778 | 3 |
| NR3C2 | 4.0×10−5 | 0.024 | 333 | 0.006 | 0.096 | 3 | 0.013 | 0.629 | 507 | 0.742 | 0.790 | 8 |
| SCNN1A | 0.106 | 0.286 | 71 | 0.016 | 0.164 | 6 | 0.085 | 0.380 | 152 | 0.021 | 0.285 | 12 |
| SCNN1B | 0.071 | 0.078 | 72 | 0.290 | 0.165 | 5 | 0.110 | 0.560 | 118 | 0.033 | 0.017 | 7 |
| SCNN1G | 3.5×10−4 | 0.073 | 142 | 0.081 | 0.067 | 5 | 0.122 | 0.455 | 129 | 0.278 | 0.728 | 3 |
| WNK1 | 0.297 | 0.181 | 234 | 0.020 | 0.218 | 2 | 0.004 | 0.003 | 322 | 0.688 | 0.360 | 12 |
| WNK4 | 0.870 | 0.812 | 44 | 0.999 | 0.990 | 3 | 0.022 | 0.400 | 97 | 0.080 | 0.089 | 3 |
Association studies of common variants
To further elaborate the effects of individual common variants we tested genetic association in EA and AA subjects in two general population samples (ARIC: N=12,954; CLUE: N=6,591); Online Table III provides summaries of demographic and BP-related phenotypic data for these samples. Our sequencing screen identified 564 of 1,277 variants in EAs and 872 of 1,972 variants in AAs that were polymorphic (MAF≥1%). Although these variants could be directly tested for association, statistical power would be greater if we could additionally use imputed variants. We used data from the 1000 Genomes Project,23 together with our RS&G data and ARIC’s Affymetrix 6.0 marker data5, to perform imputation at the 11 loci (Online Table I) using the computer program BEAGLE20, onto 9,747 EAs and 3,207 AAs in ARIC. As a check on the utility of sequencing in study samples versus imputation from reference panels, we compared, for the sequenced targets only, the numbers of variants in RS&G only, in 1000 Genomes only and shared by both for each locus and in aggregate (Online Figure I). Despite locus-specific variation, the overall pattern is clear: there were more variants identified by RS&G and 1000 Genomes (1,605 variants found in RS&G only, 567 variants in 1000 Genomes only and 858 in both). Although there are numerous systematic technical differences between RS&G (Sanger technology, comprehensive coverage, alignment to sequenced portion only) and 1000 Genomes (next generation sequencing, low coverage, alignment to whole genome), and the combined sample size is larger, we believe that the use of BP enriched samples in RS&G (280 EAs & 280 AAs), as compared to the random samples in 1000 Genomes (379 EUR & 246 AFR), is one reason that led to a larger number of variants.
Two of the 11 genes, namely HSD11B2 and WNK4, have unusual minor allele frequency distributions in 1000 Genomes with many variants under 10%, few above 40% and none at intermediate frequencies. This suggests that the variation patterns in these two regions may result in improper imputation; therefore, they were not included in our association analyses. There were a remainder of 1,821 EUR variants and 2,534 AFR variants in the combined panel of RS&G and 1000 Genomes to be imputed into ARIC. After imputation, we excluded variants with imputation score r2<0.3, leaving 731 variants in EUR and 827 in AFR. We performed genetic association studies in ARIC for visit 1 SBP and DBP using 727 EA and 807 AA variants in 9 gene regions. Only 6 highly correlated variants in CYP17A1 reached statistical significance (FDR≤0.05) in EAs (N=9,747) and none in AAs (N=3,207) (Online Table V). All 6 positive variants have been previously identified in EA GWAS5,6 and were not unique to RS&G. Thus, despite the CYP17A1 association, common RS&G variants did not contribute to this finding.
The above results could be due to an absence of common causal coding variation in the 9 genes studied or low statistical power. To test this aspect directly, we performed a positive control experiment and genotyped 27 replicated common variants known to be associated with BP5–7 in available samples from highly selected families that are expected to be enriched for BP variants (GenNet and HyperGEN networks of FBPP: N=6,521) and in CLUE (N=6,591); see Methods and Online Table III. The results, taken together (Online Table VI), show significant associations at only ATP2B1 and FES in the EAs only. This clearly demonstrates, using true positive SNVs, the low statistical power (empirically, 2/27 or 7%) of these non-coding variants in ~6,500 EA subjects in CLUE and even lower power in the ~3,000 EA/AA subjects in FBPP. Admittedly, the average allelic effect of these 27 variants in the ICBP study is ~0.6mm Hg and ~0.4 mmHg for SBP and DBP, respectively.7 Given that the population phenotypic variance for SBP and DBP is ~16mmHg and ~10mmHg, respectively, these average effect sizes are ~0.04σ for both SBP and DBP, where the effect is measured in units of the phenotypic standard deviation (σ). Comparing these results to power calculations (Online Table VIII) at the CLUE and FBPP sample sizes and allele frequencies, and various assumed effect sizes, suggests that these non-coding polymorphisms have statistical power <33 and <10 in CLUE and FBPP subjects, respectively (Online Table VII).
Association studies of rare variants
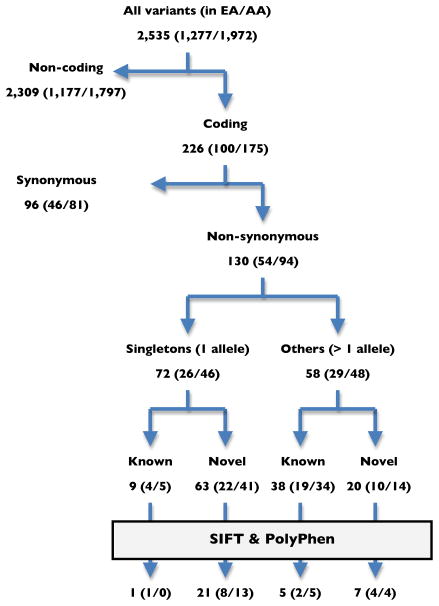
As demonstrated earlier, the totality of all variants identified by sequencing, of which the vast majority were rare (Table 2), showed significant association with SBP in both EAs and AAs (Table 3) but effects at individual genes were not well resolved. Consequently, for rare variant analysis, we first attempted to assess their functional impact since they engender low statistical power by virtue of their rarity; in other words, a higher probability of causality would decrease false positives. Assessing function is straightforward for coding non-synonymous variants where predictions of likely effect are based on protein conservation and the nature of specific substitutions; however, this is tenuous for non-coding variants whose functions are poorly understood. We gauge their impact using evolutionary conservation, although recent advances in the ENCODE project24 may lead to future improvements. Consequently, we restricted attention to 54 EA and 94 AA non-synonymous variants of which 26 and 46 variants were singletons and the remaining 29 and 48 variants were present in multiple copies, respectively (Figure 2). It is not surprising that the fraction of novel variants (not in dbSNP 129) variants are higher for singleton 88% (63/72) as compared to multiplex variants (34% or 20/58). We predicted whether the non-synonymous variants were deleterious or not using the computer programs SIFT25 and PolyPhen26. The fraction of these ‘functional candidate variants’, defined as those predicted to be deleterious by both algorithms, was 26% (34 of 130) overall, but higher for the singleton (31% or 22/72) than multiplex (21% or 12/58) variants owing to natural selection. From the total of 15 EA predicted deleterious non-synonymous variants we were able to genotype 11 in larger cohorts (Table 4). We genotyped these 11 variants in a sample of 6,591 unrelated EA subjects from CLUE and 6,521 related individuals (3,659 EAs and 2,862 AAs) from FBPP. The characteristics of the genotyped individuals are provided in Online Table III and summary genetic association results in Table 4, with detailed results in Online Table VII. None of the variants show statistical significance. Of these 11 variants, 7 are rare (MAF=0.01%–0.6%) in both EAs and AAs; the remaining 4 variants are polymorphic (MAF≥1%) in either the EAs or AAs. In other words, the infrequency of these variants suggest that they do not appear in phenotypically validated EH subjects and so cannot be a major determinant of risk.
Figure 2.
The classification of all 1,277 and 1,972 sequence variants observed, by functional category and relative abundance, in the 11 genes we examined.
Table 4.
Genetic association study of 11 rare predicted deleterious variants, and a blood pressure positive control (*), in CLUE and FBPP samples (variants with FDR≤0.05 are highlighted in yellow; nr = not relevant; standardized effect size of significant variant in brackets).
| rsID | Gene | Chr | Rare allele | Coding change | CLUE | FBPP EA | FBPP AA | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
|
| |||||||||||||
| MAF (%) | P-value
|
MAF (%) | P-value
|
MAF (%) | P-value
|
||||||||
| SBP | DBP | SBP | DBP | SBP | DBP | ||||||||
| rs72645625 | NR3C2 | 4 | G | c.C1651G:p.P551A | 0 | nr | nr | 0.4 | 0.099 | 0.305 | 0.03 | 0.704 | 0.871 |
| rs2286007 | WNK1 | 12 | T | c.C1994T:p.T665I | 7.8 | 0.814 | 0.79 | 7.2 | 0.354 | 0.488 | 0.9 | 0.83 | 0.712 |
| rs17755373 | WNK1 | 12 | T | c.C5468T:p.P1823L | 1.0 | 0.825 | 0.072 | 1.4 | 0.314 | 0.675 | 0.3 | 0.364 | 0.234 |
| rs72650764 | WNK1 | 12 | G | c.A5851G:p.T1951A | 0.03 | 0.3768 | 0.117 | 0.01 | 0.451 | 0.379 | 0.03 | 0.866 | 0.128 |
| rs72657550 | SCNN1A | 12 | G | c.G1559C:p.G520A | 0 | nr | nr | 0.04 | 0.717 | 0.859 | 0 | nr | nr |
| rs5742912 | SCNN1A | 12 | C | c.T1477C:p.W493R | 2.4 | 0.695 | 0.101 | 1.7 | 0.094 | 0.35 | 0.3 | 0.66 | 0.651 |
| rs72646501 | SCNN1G | 16 | A | c.C776A:p.T259N | 0.2 | 0.182 | 0.169 | 0.1 | 0.758 | 0.573 | 1.8 | 0.198 | 0.115 |
| rs72647528 | SCNN1G | 16 | A | c.G1458A:p.W486X | 0.1 | 0.53 | 0.842 | 0.01 | 0.43 | 0.342 | 0 | nr | nr |
| rs72647542 | SCNN1G | 16 | T | c.C1868T:p.P623L | 0 | nr | nr | 0.1 | 0.143 | 0.175 | 0 | nr | nr |
| rs72654338 | SCNN1B | 16 | A | c.G880A:p.G294S | 0 | nr | nr | 0.6 | 0.321 | 0.141 | 0.05 | 0.937 | 0.057 |
| rs56030257 | WNK4 | 17 | C | c.T1888C:p.S630P | 0.1 | 0.825 | 0.819 | 0.1 | 0.636 | 0.975 | 0 | nr | nr |
| rs2681472* | ATP2B1 | 12 | C | nr | 17.1 | 5.18×10−4 (−0.09) | 1.96×10−4 (−0.09) | 16 | 0.023 | 0.006 | 10.1 | 0.182 | 0.175 |
The outcome of our analyses is the result of either an absence of a true effect or a small effect at individual SNVs that we do not have the power to detect. This distinction is important since the expectation is that rare variants (MAF≤5%) should have much larger effect sizes than common variants and, moreover, we have already demonstrated a cumulative effect of all SNVs in GenNet (Table 3). We suggest instead that the majority of the effect of rare variants, or any variant for that matter, is small. Thus, the power to detect associations is low unless the variant frequency is well above 5% or the allelic effect is >0.25σ (Online Table VIII). For polymorphic variants at 1% frequency, the calculated statistical power is 19% and 55% at sample sizes of 3,000 and 7,000, respectively, and an allelic effect is >0.25σ. The paucity of positive results from this study suggests that the true effect size is considerably smaller and probably of the same order of magnitude as those for non-coding polymorphisms (0.05σ). Published data shows that the allelic effect of the positive control rs2681472 in ATP2B1 is ~0.06–0.07σ5,6, which at an allele frequency of between 15–20% and a sample size between 3,000–7,000 has a power between 13–40% but in 10,000 samples has power >60%. Consequently, rs2681472 is highly significant in ARIC but much less so in CLUE and FBPP.
DISCUSSION
By studying the role of syndromic HTN genes in BP regulation and EH, in non-syndromic subjects from the general population at the extremes of the corrected SBP residual distribution, we found meager, but not an absence of, evidence of effects at individual rare or common variants at known HTN genes. However, upon pooling all variants we obtained statistically significant association of these same genes to SBP; the much smaller collection of missense variants was non-significant. The statistical significance of all elements (coding and conserved non-coding), suggests both the low statistical power of testing effects of coding alleles with the sample sizes at hand and strongly implicates the importance of conserved non-coding variants to inter-individual BP variation.
Genetic association studies are important but remain difficult due to the lack of statistical power to definitively identify associations. Statistical power of such studies depend on both the sample size and the population variance explained, the latter being a function of allelic effect size and its frequency. Thus, low power can stem from: (1) the use of inadequate sample sizes given the numerous variants tested, and (2) the small genetic effects of these variants. In this study, we started with genes that are known to impact BP physiology, and examined both common and predicted deleterious rare variants within these genes, to focus the analyses on variants that are expected to have higher impact on BP. Moreover, enrichment of variants by sequencing BP extremes should have also enriched for causal variants. Additionally, we used a large sample size given the few genes we examined: 19,997 EAs (3,659 in FBPP, 6,591 in CLUE and 9,747 in ARIC), along side 6,069 AAs (2,862 in FBPP and 3,207 in ARIC). Nevertheless, we did not detect pervasive associations that survived multiple testing correction, although common and rare variants were studied in two different populations.
The fundamental question in blood pressure genetics is: what is the expected genetic effect of any functional or causal allele? To understand our results, consider the average allelic effect from other BP studies. We estimated these effects, by calculating the median allelic BP effect of each genetic variant identified, from five groups of alleles: (a) 91 disease causing mutations (DMs) in 111 syndromic patients across the 11 genes from the Human Gene Mutation Database (HGMD) with BP values as cited in the published literature; (b) 12 DMs from HGMD in 69 GenNet individuals (in 7 of 11 genes, Online Table IX); (c) 2,379 SNVs in all 560 GenNet individuals (in all 11 genes); (d) 11 predicted deleterious mutations we studied in ~13,000 CLUE and FBPP subjects; and, (e) 27 validated ICBP variants we studied in ~13,000 CLUE and FBPP subjects (Online Table X). This classification attempts to produce an allelic series from an expected largest to smallest effect: we estimated the median effects to be 3.57, 0.73, 0.55, 0.11 and 0.01, respectively, with corresponding standard errors 0.34, 0.42, 0.02, 0.15 and 0.003. This suggests that although the genes we selected for study do have rare mutations of very large effect (class a: 3.57σ), these mutations are not observed in the GenNet individuals we sequenced. However, of the previously identified HGMD mutations we did detect in GenNet (class b), their effect size is considerably (5X) smaller at 0.73σ. These mutations are enriched since the background effect of all variants (class c) we identified in GenNet, individuals already selected for SBP extremes, was smaller still at 0.55σ. Despite predictions of deleterious effects at the 11 variants (class d) we genotyped in a much larger sample of ~13,000 subjects in FBPP and CLUE, these variants have a smaller effect yet at 0.11σ. As a comparator, the replicated GWAS variants (class e) in ~13,000 subjects in FBPP and CLUE had an effect size of 0.01σ, smaller than the original ICBP study and likely demonstrates the “winners curse”. None of the variants we identified or examined from HGMD had average population frequencies ≥5%, when found in the EVS database27, had frequencies and were usually much smaller. These results make it clear that for a homeostatically controlled trait like BP, allelic effects in the general population are unlikely to be larger than 0.11σ even at recognized BP genes. Even more broadly, if all BP allelic effects are <0.25σ then for alleles at 1%, 5% and 10%, statistical power is never >80% unless the numbers of individuals studied are >110,000, >22,000 and >11,000, respectively; the detection is even more difficult for DBP.
Although analyses of individual rare and common variants did not yield significant associations with SBP or DBP, we identified 5 loci that are significantly associated with SBP by pooling all variants in each gene and across all genes. Additionally, 12 variants in 7 genes, from the total of 2,535 originally identified, were also present in HGMD as disease causing mutations (Table 3). Three of these 7 genes (AGT, CYP11B1, and SCNN1G) are also statistically associated with SBP by pooling of all variant. Two additional genes (CYP17A1 and HSD11B2) are found to be statistically associated with SBP by pooling only non-coding elements (Online Table IV). Furthermore, two additional loci (NR3C2 and WNK1), not noted in HGMD, are significantly associated with SBP in the pooled variant test. Hence, our results show that at least 5 of the 11 syndromic HTN loci also contribute to BP and EH in the general population and that conservation (phyloP score) provided greater statistical significance than classifying missense variants by their deleterious effect (PolyPhen2 score). This implies that conservation analysis, based on numerous genome sequences, may be more informative than restricting to only missense variants and their prediction of deleterious effect, at least for complex traits like BP. The underlying reasons for this are that our ability to predict the deleterious effect may be poor for the numerous missense mutations we identified except for the severest alleles and that variation at non-coding elements is a very significant contributor to complex diseases. Indeed, the recent study by Yang and colleagues, who demonstrate the existence of numerous variants at both coding and non-coding elements proximal to genes, is consistent with this view28. Consequently, studies of both the exome and the conserved genomic segments in the human genome need to be comprehensively examined in very large samples for fully elaborating the contributions to BP physiology and EH.
Supplementary Material
NOVELTY & SIGNIFICANCE.
Part I
-
What is known?
Essential hypertension (EH), or high blood pressure (BP) without secondary causes, affects ~30% of adults in industrialized countries but whose molecular etiology is largely unknown.
Despite its moderate heritability (~40–60%), BP varies with age, BMI, diet, stress level and sympathetic tone.
Rare variants in numerous renal genes have been identified in many rare Mendelian hypo-/hypertension (HTN) syndromes with deleterious alleles that have a large impact on BP and also lead to electrolyte abnormalities. In addition, common variants in >60 loci have been discovered to impact BP variation using genome-wide association studies (GWAS) explaining ~1–2% of systolic (SBP) and diastolic BP (DBP) variation
-
What new information does this article contribute?
Rare variants in the same hypo-/hypertension (HTN) syndromic genes do not have large effects in the general population.
Conserved non-coding sequences, at these same genes, although lacking precise functional information, contribute significantly to BP variation.
If all genetic effects are small, genetic studies of association are probably not meaningful unless a minimum of 50,000 subjects are included.
Part II
We resequenced the coding and conserved noncoding regions of 10 syndromic hypertension genes and angiotensinogen, genes known to impact BP in some families, in the general population by focusing on individuals at the extremes of SBP. Analyses of common and rare variants at these 11 genes, individually, did not yield significant association with SBP or DBP. However, by pooling coding and conserved noncoding elements, and weighting their genetic contribution by allele frequency and nucleotide conservation, we showed a strong association with BP’s in at least 5 loci. Our study leads us to believe that both common and rare variants have very small effects (~0.05 standard deviation unit) on BP and EH. For the first time, our results reveal the significant contribution of conserved noncoding elements in syndromic HTN genes to BP traits in the general population. Consequently, both the exome and the conserved genomic segments in the human genome need to be comprehensively examined in very large samples to allow full elucidation of the genetic contributions to BP physiology and EH.
Acknowledgments
We appreciate the work of and technical help from Dr. Mark Rieder (University of Washington, Department of Genome Sciences, Seattle, WA) and Dr. Samuel Levy (J. Craig Venter Institute, Rockville, MD) for DNA sequencing in the RS&G. We thank the personnel of the CLUE and FBPP studies for sample selection and gratefully acknowledge the efforts of Gina Hilton and Joan Ritho for genotyping in the CLUE cohort. The authors thank the staff and participants of the ARIC study for their important contributions. We used the HGMD database under a license to the Aravinda Chakravarti laboratory at the Johns Hopkins University School of Medicine.
SOURCES OF FUNDING
This work was supported by grant HL086694 from the National Heart, Lung, and Blood Institute (NHLBI) to A.C. and by The Resequencing and Genotyping Service, funded by the NHLBI, who performed sequencing at the University of Washington, Department of Genome Sciences, Seattle, WA and the J. Craig Venter Institute, Rockville, MD [N01-HV-48194 and N01-HV-48196 to A.C.]. The Atherosclerosis Risk in Communities Study was carried out as a collaborative study supported by NHLBI contracts (HHSN268201100005C, HHSN268201100006C, HHSN268201100007C, HHSN268201100008C, HHSN268201100009C, HHSN268201100010C, HHSN268201100011C, and HHSN268201100012C), R01HL087641, R01HL59367 and R01HL086694; National Human Genome Research Institute contract U01HG004402; and National Institutes of Health contract HHSN268200625226C. Infrastructure was partly supported by Grant Number UL1RR025005, a component of the National Institutes of Health and NIH Roadmap for Medical Research.
Non-standard Abbreviation and Acronyms
- 1000 Genomes
Thousand Genomes Project
- AA
African American
- AFR
African and African American individuals in the 1000 Genomes Project
- AsA
Asian American
- BP
blood pressure
- DBP
diastolic blood pressure
- EA
European American
- EH
essential hypertension
- EUR
European individuals in the 1000 Genomes Project
- FBPP
Family Blood Pressure Program
- HA
Hispanic American
- HTN
hypertension
- INDEL
insertion/deletion
- LTA
long-term average
- MAF
minor allele frequency
- RS&G
Resequencing and Genotyping Services
- SBP
systolic blood pressure
- SD
standard deviation
- SNV
single nucleotide variant
Footnotes
Author Contributions:
Study conception and design of experiments: GBE, SKG, AC; molecular genetic analysis: KHN; data analysis: KHN, VP, GBE, AK, AC; contribution of samples and editing of manuscript: RC, SCH, BIF, JC, WHLK, ACM, EB; drafting, editing and completing manuscript: KHN, VP, GBE, SKG, AC.
Conflict of Interest Disclosure: None
References
- 1.Pickering G. Hyperpiesis: high blood-pressure without evident cause: essential hypertension. Brit Med J. 1965;2:1021–1026. doi: 10.1136/bmj.2.5469.1021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Oldham PD, Pickering G, Roberts JA, Sowry GS. The nature of essential hypertension. Lancet. 1960;1:1085–1093. doi: 10.1016/s0140-6736(60)90982-x. [DOI] [PubMed] [Google Scholar]
- 3.Guyton AC. Blood pressure control–special role of the kidneys and body fluids. Science. 1991;252:1813–1816. doi: 10.1126/science.2063193. [DOI] [PubMed] [Google Scholar]
- 4.Lifton RP, Gharavi AG, Geller DS. Molecular mechanisms of human hypertension. Cell Rev. 2001;104:545–556. doi: 10.1016/s0092-8674(01)00241-0. [DOI] [PubMed] [Google Scholar]
- 5.Levy D, Ehret GB, Rice K, Verwoert GC, Launer LJ, Dehghan A, Glazer NL, Morrison AC, Johnson AD, Aspelund T, Aulchenko Y, Lumley T, Kottgen A, Vasan RS, Rivadeneira F, Eiriksdottir G, Guo X, Arking DE, Mitchell GF, Mattace-Raso FUS, Smith AV, Taylor K, Scharpf RB, Hwang S, Sijbrands EJG, Bis J, Harris TB, Ganesh SK, O’Donnell CJ, Hofman A, Rotter JI, Coresh J, Benjamin EJ, Uitterlinden AG, Heiss G, Fox CS, Witteman JCM, Boerwinkle E, Wang TJ, Gudnason V, Larson MG, Chakravarti A, Psaty BM, van Duijn CM. Genome-wide association study of blood pressure and hypertension. Nat Genet. 2009;41:677–687. doi: 10.1038/ng.384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Newton-Cheh C, Johnson T, Gateva V, Tobin MD, Bochud M, Coin L, Najjar SS, Zhao JH, Heath SC, Eyheramendy S, Papadakis K, Voight BF, Scott LJ, Zhang F, Farrall M, Tanaka T, Wallace C, Chambers JC, Khaw K, Nilsson P, van der Harst P, Polidoro S, Grobbee DE, Onland-Moret NC, Bots ML, Wain LV, Elliott KS, Teumer A, Luan J, Lucas G, Kuusisto J, Burton PR, Hadley D, McArdle WL, Brown M, Dominiczak A, Newhouse SJ, Samani NJ, Webster J, Zeggini E, Beckmann JS, Bergmann S, Lim N, Song K, Vollenweider P, Waeber G, Waterworth DM, Yuan X, Groop L, Orho-Melander M, Allione A, Di Gregorio A, Guarrera S, Panico S, Ricceri F, Romanazzi V, Sacerdote C, Vineis P, Barroso I, Sandhu MS, Luben RN, Crawford GJ, Jousilahti P, Perola M, Boehnke M, Bonnycastle LL, Collins FS, Jackson AU, Mohlke KL, Stringham HM, Valle TT, Willer CJ, Bergman RN, Morken MA, Doring A, Gieger C, Illig T, Meitinger T, Org E, Pfeufer A, Wichmann HE, Kathiresan S, Marrugat J, O’Donnell CJ, Schwartz SM, Siscovick DS, Subirana I, Freimer NB, Hartikainen A, McCarthy MI, O’Reilly PF, Peltonen L, Pouta A, de Jong PE, Snieder H, van Gilst WH, Clarke R, Goel A, Hamsten A, Peden JF, Seedorf U, Syvanen A, Tognoni G, Lakatta EG, Sanna S, Scheet P, Schlessinger D, Scuteri A, Dorr M, Ernst F, Felix SB, Homuth G, Lorbeer R, Reffelmann T, Rettig R, Volker U, Galan P, Gut IG, Hercberg S, Lathrop GM, Zelenika D, Deloukas P, Soranzo N, Williams FM, Zhai G, Salomaa V, Laakso M, Elosua R, Forouhi NG, Volzke H, Uiterwaal CS, van der Schouw YT, Numans ME, Matullo G, Navis G, Berglund G, Bingham SA, Kooner JS, Connell JM, Bandinelli S, Ferrucci L, Watkins H, Spector TD, Tuomilehto J, Altshuler D, Strachan DP, Laan M, Meneton P, Wareham NJ, Uda M, Jarvelin M, Mooser V, Melander O, Loos RJF, Elliott P, Abecasis GR, Caulfield M, Munroe PB Wellcome Trust Case Control Consortium. Genome-wide association study identifies eight loci associated with blood pressure. Nat Genet. 2009;41:666–676. doi: 10.1038/ng.361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.The International Consortium for Blood Pressure genome-wide association studies. Genetic variants in novel pathways influence blood pressure and cardiovascular disease risk. Nature. 2011;478:103–109. doi: 10.1038/nature10405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kato N, Miyata T, Tabara Y, Katsuya T, Yanai K, Hanada H, Kamide K, Najura J, Kohara K, Takeuchi F, Mano H, Yasunami H, Kimura A, Kita Y, Ueshima H, Nakayama T, Soma M, Hata A, Fujioka A, Kawano Y, Nakao K, Sekine A, Yoshida T, Nakamura Y, Saruta T, Ogihara T, Sugano S, Miki T, Tomoike H. High-density association study and nomination of susceptibility genes for hypertension in the Japanese National Project. Hum Mol Genet. 2008;17:617–627. doi: 10.1093/hmg/ddm335. [DOI] [PubMed] [Google Scholar]
- 9.Yang H, Liang Y, Wu Y, Chung C, Chiang K, Ho H, Ting C, Lin T, Sheu S, Tsai W, Chen J, Leu H, Yin W, Chiu T, Chen C, Fann CSJ, Wu J, Lin T, Lin S, Chen Y, Chen J, Pan W. Genome-wide association study of young-onset hypertension in the Han Chinese population of Taiwan. Plos ONE. 2009;4:e5459. doi: 10.1371/journal.pone.0005459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cho YS, Go MJ, Kim YJ, Heo JY, Oh JH, Ban H, Yoon D, Lee MH, Kim D, Park M, Cha S, Kim J, Han B, Min H, Ahn Y, Park MS, Han HR, Jang H, Cho EY, Lee J, Cho NH, Shin C, Park T, Park JW, Lee J, Cardon L, Clarke G, McCarthy MI, Lee J, Lee J, Oh B, Kim H. A large-scale genome-wide association study of Asian populations uncovers genetic factors influencing eight quantitative traits. Nat Genet. 2009;41:527–534. doi: 10.1038/ng.357. [DOI] [PubMed] [Google Scholar]
- 11.Adeyemo A, Gerry N, Chen G, Herbert A, Doumatey A, Huang H, Zhou J, Lashley K, Chen Y, Christman M, Rotimi C. A Genome-Wide Association Study of Hypertension and Blood Pressure in African Americans. PLoS Genet. 2009;5:e1000564. doi: 10.1371/journal.pgen.1000564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fox ER, Young JH, Li Y, Dreisbach W, Keating BJ, Musani SL, Liu K, Morrison AC, Ganesh S, Kutlar A, Ramachandran VS, Polak JF, Fabsitz RR, Dries DL, Farlow DN, Redline S, Adeyemo A, Hirschorn JN, Sun YV, Wyatt SB, Penman AD, Palmas W, Rotter JI, Townsend RR, Doumatey AP, Tayo BO, Mosley TH, Lyon HN, Kang SJ, Rotimi SN, Cooper RS, Franceschini N, Curb JD, Martin LW, Eaton CB, Kardia SLR, Taylor HA, Caulfield MJ, Ehret GB, Johnson T, Chakravarti A, Zhu X, Levy D. Association of genetic variation with systolic and diastolic blood pressure among African Americans: the Candidate Gene Association Resource study. Hum Mol Genet. 2011;20:2273–2284. doi: 10.1093/hmg/ddr092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.FBPP: Multi-center genetic study of hypertension: the Family Blood Pressure Program (FBPP) Hypertension. 2002;39:3–9. doi: 10.1161/hy1201.100415. [DOI] [PubMed] [Google Scholar]
- 14.The ARIC Steering Committee. The atherosclerosis risk in communities (ARIC) study: design and objective. Am J Epidemiol. 1989;129:687–702. [PubMed] [Google Scholar]
- 15.Gallicchio L, Chang HH, Christo DK, Thuita L, Huang HY, Strickland P, Ruczinski I, Clipp S, Helzlsouer KJ. Single nucleotide polymorphisms in obesity-related genes and all-caused and cause-specific mortality: a prospective cohort study. BMC Med Genet. 2009;10:103. doi: 10.1186/1471-2350-10-103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gabriel S, Ziaugra L, Tabbaa D. SNP genotyping using the Sequenom MassARRAY iPLEX platform. Curr Prot in Hum Genet. 2009;60:2.12.1–2.12.18. doi: 10.1002/0471142905.hg0212s60. [DOI] [PubMed] [Google Scholar]
- 17.Hui L, DelMonte T, Ranade K. Genotyping using the TaqMan assay. Curr Prot in Hum Genet. 2008;56:2.10.1–2.10.8. doi: 10.1002/0471142905.hg0210s56. [DOI] [PubMed] [Google Scholar]
- 18.Tobin MD, Sheehan NA, Scurrah KJ, Burton PR. Adjusting for treatment effects in studies of quantitative traits: antihypertensive therapy and systolic blood pressure. Statist Med. 2005;24:2911–2935. doi: 10.1002/sim.2165. [DOI] [PubMed] [Google Scholar]
- 19.Price AL, Kryukov GV, Bakker PIW, Purcell SM, Staples J, Wei L, Sunyaev SR. Pooled association tests for rare variants in exon-resequencing studies. Am J Hum Genet. 2010;86:831–838. doi: 10.1016/j.ajhg.2010.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Browning SR, Browning BL. Rapid and accurate haplotype phasing and missing-data inference for whole-genome association studies by use of localized haplotype clustering. Am J Hum Genet. 2007;81:1084–1097. doi: 10.1086/521987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Purcell S, Neale B, Todd-Brown K, Thomas L, Ferreira MAR, Bender D, Maller J, Sklar P, de Bakker PIW, Daly MJ, Sham PC. PLINK: a toolset for whole-genome association and population-based linkage analysis. Am J Hum Genet. 2007;81:559–575. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Abecasis GR, Cherny SS, Cookson WO, Cardon LR. Merlin—rapid analysis of dense genetic maps using sparse gene flow trees. Nat Genet. 2002;30:97–101. doi: 10.1038/ng786. [DOI] [PubMed] [Google Scholar]
- 23.The 1000 Genomes Project Consortium A map of human genome variation from population-scale sequencing. Nature. 2010;467:1061–1073. doi: 10.1038/nature09534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.The ENCODE Project Consortium. Identification and Analysis of Functional Elements in 1% of the Human Genome by the ENCODE Pilot Project. Nature. 2007;447:799–816. doi: 10.1038/nature05874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ng PC, Henikoff S. SIFT: predicting amino acid changes that affect protein function. Nucl Acids Res. 2003;31:3812–3814. doi: 10.1093/nar/gkg509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ramensky V, Bork P, Sunyaev S. Human non-synonymous SNPs: server and survey. Nucl Acids Res. 2002;30:3894–3900. doi: 10.1093/nar/gkf493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Exome Variant Server. NHLBI GO Exome Sequencing Project (ESP) Seattle, WA: Aug, 2012. (URL: http://evs.gs.washington.edu/EVS/) [Google Scholar]
- 28.Yang J, Lee SH, Goddard ME, Visscher PM. GCTA: A Tool for Genome-wide Complex Trait Analysis. Am J Hum Genet. 2011;88:76–82. doi: 10.1016/j.ajhg.2010.11.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.