Abstract
Sonic hedgehog (Shh) signaling is critical for various developmental processes including specification of the midbrain dopamine (mDA) neurons in the ventral mesencephalon (vMes). While the timing of Shh and its response gene Gli1 segregates mDA neurons, their overall lineage contribution to mDA neurons heavily overlaps. Here, we demonstrate that the same set of mDA neuron progenitors sequentially respond to Shh signaling (Gli1 expression), induce Shh expression, and then turn off Shh responsiveness. Thus, at any given developmental stage, cells rarely co-express Shh and Gli1. Using ShhCre:GFP mice to delete the Smoothened receptor in the Shh pathway, we demonstrate that the loss of Shh signaling in Shh expressing cells results in a transient increase in proliferation and subsequent depletion of mDA neuron progenitors in the posterior vMes due to the facilitated cell cycle exit. Moreover, the change in duration of Shh signaling in vMes progenitors altered the timing of the contribution to the ventral tegmental area (VTA) and the substantia nigra pars compacta (SNc) mDA neurons. Taken together, our investigation on the relationship between the Shh-secreting and -responding cells revealed an intricate regulation of induction and cessation of Shh signaling that influences the distribution of mDA neurons in the VTA and SNc.
Keywords: Sonic hedgehog, Gli1, Midbrain dopamine neurons, Substantia nigra pars compacta, Ventral tegmental area, Genetic inducible fate mapping
INTRODUCTION
Sonic hedgehog (Shh) signaling plays essential roles in patterning and formation of many structures during development including the spinal cord, limb, and ventral mesencephalon (vMes) (Fuccillo et al., 2006; Ingham and Placzek, 2006; Chiang et al., 1996; Litingtung and Chiang, 2000; Zhang et al., 2001; Kraus et al., 2001; Bai et al., 2002; Wijgerde et al., 2002). Shh is a secreted molecule that diffuses away from the Shh-expressing cells. Upon Shh binding to the patched (Ptch1) receptor in Shh-responding cells, the Smoothened (Smo) receptor transduces intracellular signaling which converges on the Gli family of transcription factors (reviewed in Ingham and Placzek, (2006)). Among the Glis, Gli2 functions primarily as a transcriptional activator that induces expression of many target genes, such as Gli1, which is used as an accurate and sensitive read-out for active Shh signaling in Shh-responding cells (Bai et al., 2002; Ahn and Joyner, 2004; Ahn and Joyner, 2005).
Various tissues, including the neural tube and limbs, are properly patterned and their cell types specified through the dynamic temporal and spatial control of Shh expression and responsiveness during development. Interestingly, Shh-responsiveness is necessary and sufficient for induction of Shh ligand expression (Matise et al., 1998; Ye et al., 1998). Thus, the tight regulation of Shh responsiveness is controlled by Shh ligand expression and the ability of the receiving cells to transduce the Shh signal.
The vMes is an ideal model for studying the dynamic nature of Shh signaling because Shh expression and Shh-responsiveness (Gli1 expression) are temporally and spatially regulated during vMes development (Hayes et al., 2011; Zervas et al., 2004; Blaess et al., 2006; Joksimovic et al., 2009a). Dynamic changes in Shh and Gli1 expression in the vMes are translated into a distinct contribution pattern of midbrain dopamine (mDA) neurons (Hayes et al., 2011; Blaess et al., 2011; Joksimovic et al., 2009a), which are subdivided into the ventral tegmental area (VTA) and substantia nigra pars compacta (SNc) mDA neurons based on their anatomical location (Van den Heuvel and Pasterkamp, 2008). Interestingly, Gli1 expression is rapidly downregulated in Shh responding cells after induction of the Shh ligand (Hayes et al., 2011). This raises the possibility that the duration of Shh signaling in vMes progenitors may differ as some progenitors become refractory and lose their ability to respond to Shh signaling. In the developing limb and neural tube, changes in the duration of active Shh signaling determine digit identity (Zhu et al., 2008) and ventral neuronal cell types (Ribes et al., 2010), respectively. However, whether a similar mechanism contributes to mDA neuronal subtype development has not been addressed.
In this study, we manipulated the timing and duration of Shh signaling in the vMes and assessed the development of mDA neurons. Our comprehensive comparison of the expression pattern and short term genetic lineage analysis of Shh and Gli1 revealed a unique relationship in which Shh expression is induced in the Shh-responding cells. Furthermore, our genetic manipulations, which alter the timing and duration of Shh signaling by removing the Shh signaling receptor, Smo, in Shh-expressing cells, revealed a functional role for the tight temporal regulation of Shh signaling. Together, these studies demonstrate a functional requirement for dynamic Shh signaling in regulating the cell cycle status of mDA progenitors to ultimately influence their final distribution in the VTA and SNc.
MATERIALS AND METHODS
Animals
Mouse lines were maintained on an outbred Swiss Webster background. See Supplementary Table 1 for a description of each mouse allele. For breeding, male Shh>Cre:GFP/+ mice were crossed with Gli1nLacZ/+ females to generate ShhCre:GFP/+;Gli1nLacZ/+ embryos. Additionally, male Gli1CreER/+;R26tdTomato/tdTomato mice were crossed with wildtype or Gli1nLacZ/+ females to generate Gli1CreER/+;R26tdTomato/+ and Gli1CreER/nLacZ;R26tdTomato/+ embryos, respectively. For Smo loss of function experiments, male ShhCre:GFP/+;R26YFP/YFP mice were crossed with wildtype females to generate ShhCre:GFP/+;R26YFP/+ control embryos. For mutant embryos, male ShhCre:GFP/+;Smo+/− mice were crossed with SmoFlox/Flox;R26YFP/YFP females to generate ShhCre:GFP/+;SmoFlox/−;R26YFP/+ mutant embryos. All animals were housed and handled according to the National Institutes of Health Institutional Animal Care and Use Committee guidelines.
Tamoxifen, EdU, and BrdU injections
Briefly, between 9:00 and 10:00 a.m. on E7.5, E8.5, or E9.5, 2 mg of tamoxifen (TM) was delivered by oral gavage to the timed-pregnant dams using a disposable feeding needle (FST 9921) (Brown et al., 2009).
EdU (5-ethynyl-2'-deoxyuridine, Invitrogen, A10044) and BrdU (5-bromo-2'-deoxyuridine) were prepared as 2.5 μg/μl and 10 μg/μl stock solutions, respectively, in sterile PBS and stored at −20°C. EdU or BrdU was warmed to 37°C and delivered by intraperitoneal injection to the pregnant dams in the evening of E10.5, 11.5 or E13.5 at a dose of 20.8 mg/kg of body weight for EdU and 200 mg/kg of body weight for BrdU. Animals were sacrificed 1 hr after injection for proliferation analysis and at E13.5 for cell cycle exit study.
Tissue processing
The collection and processing of tissues were as described (Hayes et al., 2011). Briefly, tissue was fixed in 4% paraformaldehyde (PFA) overnight, rinsed in PBS, cryoprotected in a sucrose gradient, embedded in optimal cutting temperature (OCT), frozen in liquid nitrogen-chilled isopentane, and sectioned on the Leica Cryostat (CM3050S) (Brown et al., 2009). Sections were collected at 10 μm (E10.5), 12 μm (E11.5 and E13.5), and 14 μm (E16.5) and stored at −80°C.
RNA in situ hybridization
Shh and Gli1 probes were described previously (Platt et al., 1997). RNA in situ hybridization was performed essentially as described (Blaess et al., 2006). The hybridized RNA in situ probe was detected within 6 hours for Shh and 24 hours for Gli1. The developed sections were washed with PBS, fixed in 4% PFA, washed with PBS again, and coverslipped with Fluoromount-G (SouthernBiotech) mounting media.
Fluorescent immunohistochemistry and X-gal histochemistry
Immunohistochemistry was performed as described (Hayes et al., 2011; Wang et al., 2011). The antibodies used are listed in Supplemental Table 2. EdU detection was performed using the Click-iT EdU Imaging Kit (Invitrogen, C10340) according to the manufacture's guidelines. Briefly, after incubation in secondary antibodies and washing in 0.2 % TritonX-100/PBS (PBT), the sections were incubated in EdU staining solution (1× Click-iT reaction buffer, CuSO4, Alexa 647, 1× reaction buffer additive) for 30 minutes in a dark humid chamber, washed in PBT several times, counterstained in Hoescht (Invitrogen, H3569), washed in PBS, and coverslipped with Fluoromount-G mounting media. X-gal histochemistry to detect LacZ expression was performed as described (Hayes et al., 2011; Ahn and Joyner, 2004).
Microscopy
All fluorescent images were captured using a Leica DM6000 upright microscope equipped with a Hamamatsu ORCA-ER digital camera and the Volocity software (PerkinElmer) or Zeiss Axiovert 200M microscope with LSM510 Meta confocal system. Bright field images were captured with a MacroFire (Optronics) digital camera and PictureFrame (Optronics) software. Images were processed with Photoshop in Adobe Creative Suite 3 (San Jose, CA) for brightness and contrast levels.
Quantification and statistical analyses
At E10.5, 11.5, and E13.5, the Lmx1a and EdU co-staining was quantified using the measurement function in Volocity (PerkinElmer). Lmx1a and EdU staining was quantified by measuring the area that contained pixels with intensity greater than 1 standard deviation from the peak of the pixel intensity distribution. Lmx1a and EdU are both nuclear stains, which allowed Volocity to measure the area of their co-expression. We then calculated the percentage of proliferating Lmx1a area by dividing EdU and Lmx1a double positive area by total Lmx1a area (Lmx1a+ EdU+/Total Lmx1a+). Coronal sections were matched for anterior/posterior level based on the distribution pattern of TH+ or Lmx1a+ cells.
At E16.5, stereological counting was performed on 14 μm thick samples collected in 8 sets. Using the StereoInvestigator System (MicroBright Field, Inc.), the SNc and VTA mDA neuron area was outlined, and 40 μm by 40 μm counting frames were systematically placed every 100 μm by 100 μm over the outlined area. The number of TH+ EdU+ cells and only TH+ cells were counted in each counting frame. The percentage of mDA neurons derived from progenitors that were proliferating at E11.5 or E13.5 were determined by the ratio of TH+ EdU+ co-expressing cells counted to total TH+ cells counted (TH+ EdU+/Total TH+) for each area (SNc, VTA, and total mDA neurons). Number of brains/embryos used is indicated as n in the Results section and the quantified data are presented as the mean value ± the standard error of means (s.e.m). The statistical analyses were performed using the Student's t-test and p ≤ 0.05 was considered statistically significant.
RESULTS
Dynamic expression of Shh and Gli1 in the developing vMes
Shh and Gli1 expression in the vMes is spatially and temporally dynamic. Previous studies demonstrated dynamic lateral expansion of the Shh and Gli1 expression domains (Hayes et al., 2011; Blaess et al., 2011; Joksimovic et al., 2009a). In order to test whether the progenitor cells migrate laterally as the tissue expands or the expression is being induced in naïve cells located laterally, we performed expression analyses using two methods: mRNA transcript analysis and short-term lineage tracing. We used ShhCre:GFP/+;Gli1nLacZ/+ embryos to label and analyze the Shh-secreting (GFP) and Shh-responding cells (Gli1-expressing, β-gal) across vMes development. Operationally, we use Shh (GFP) as a short-term lineage tracer and Shh (mRNA) to delineate the current Shh expression. Similarly, we use Gli1 (β-gal) as a short-term lineage tracer to track Shh-responding cells and Gli1 (mRNA) to identify the current Gli1 expression. The transcript analysis accurately reports all cells that express Shh or Gli1 at the time of analysis. The short-term lineage tracing, based on reporter protein expression, is temporally delayed due to the time required for new protein synthesis. Therefore, cells that recently induced Shh or Gli1 expression will be labeled by the transcript analysis but not by the short-term lineage tracer. On the other hand, cells which recently downregulated Shh or Gli1 will retain the short term lineage tracer, due to the longer half-life of reporter proteins, but not the transcript.
We first assessed the changes in gene expression patterns of both Shh and Gli1. Shh expression initiates in the node at E7.5 where it extends rostro-caudally along the ventral midline in the notochord, a non-neural tissue (Echelard et al., 1993). At the level of Otx2+ vMes, we observed Shh (GFP) and Shh (mRNA) expression in the notochord at E8.5 (6–8 somite stage) (Figure 1A, 1I, and 1M). The cells in the floorplate of the vMes responded to notochord-derived Shh as evident by the induced expression of both Gli1 (β-gal) and Gli1 (mRNA) at E8.5 (Figure 1E, 1I, 1M, and 1Q).
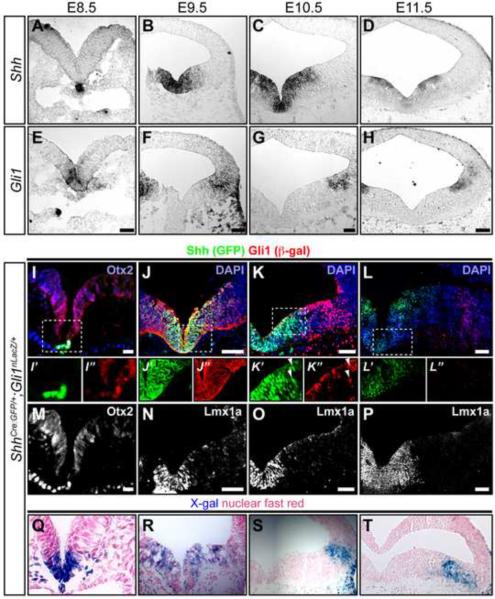
Figure 1. Shh and Gli1 expression in the vMes.
(A–D) Shh (mRNA) expression at E8.5 (A), E9.5 (B), E10.5 (C), and E11.5 (D). (E–H) Gli1 (mRNA) expression at E8.5 (E), E9.5 (F), E10.5 (G), and E11.5 (H). (I–L) Shh (GFP, green) and Gli1 (β-gal, red) expression in ShhCre:GFP;Gli1nLacZ embryos at E8.5 (I), E9.5 (J), E10.5 (K), and E11.5 (L). Boxed regions demarcate higher magnification images shown below (` for GFP, green and “ for β-gal, red). Arrowheads in K' and K” indicate Shh (GFP) and Gli1 (β-gal) double positive cells. (M–P) Otx2 expression at E8.5 (M) and Lmx1a expression in the vMes at E9.5 (N), E10.5 (O), and E11.5 (P) indicate the analyzed tissue as the midbrain. (Q–T) X-gal histochemistry was performed on adjacent sections I–L to confirm the β-gal immunofluorescent staining results. Scale bars are 32 μm in A,E and 60 μm in B–D, F–H, and I–P.
At E9.5, Shh expression was detected within the Lmx1a+ mDA neuron progenitor domain of the vMes by both Shh (GFP) and Shh (mRNA) (Figure 1B, 1J, and 1N). Shh responsiveness, reported by Gli1 (β-gal) and Gli1 (mRNA) expression, expanded from the Lmx1a+ ventromedial domain into more lateral Lmx1a- vMes (Figure 1F, 1J, 1N and 1R). Interestingly, while Gli1 (β-gal)+ cells were found in the most ventromedial vMes and co-expressed Shh and Lmx1a (Figure 1J and 1N), the Gli1 (mRNA) expression was already completely downregulated in the medial vMes by E9.5 (Figure 1F). Thus, our expression analysis suggests that the same vMes progenitors in the ventromedial domain co-express Shh and Gli1 only for a brief period before Gli1 gets downregulated medially.
At E10.5, Shh (GFP) and Shh (mRNA) expression expanded further into the lateral vMes domain where Lmx1a was not expressed (Figure 1C, 1K, and 1O). The perdurance of Gli1 (β-gal) protein revealed that a few cells located at the medial boundary of its expression domain co-expressed Lmx1a and Shh (GFP) (Figure 1K, arrowheads in K′ and K″, and 1O). However, as in the E9.5 analysis, Gli1 (mRNA)+ cells were found only in the lateral vMes outside the Lmx1a+ domain, again suggesting that co-expression of Shh, Gli1, and Lmx1a was very transient (Figure 1G, 1K, 1O, and 1S).
At E11.5, in contrast to previous stages, Shh (GFP) and Shh (mRNA) expression was slightly downregulated medially but still maintained in the lateral vMes just outside the Lmx1a+ domain (Figure 1D, 1L, and 1P). Gli1 (β-gal) and Gli1 (mRNA) expression further downregulated medially and was only detected in the lateral vMes outside the Shh+ and Lmx1a+ domains (Figure 1H, 1L, 1P, and 1T). Together, our analysis demonstrate that the very dynamic induction and cessation of Shh expression closely follows the Gli1 expression pattern from a day earlier, but the overlap between current Shh and Gli1 expression was transitory in the developing vMes.
Induction of Shh expression in the Gli1 lineage cells
We next performed lineage mapping experiments to determine the relative position of early marked Gli1 lineage cells to the later Shh and Gli1-expressing cells. This comparison of lineage versus current expression can distinguish whether the vMes progenitors continuously responded to Shh but migrated laterally or whether the vMes progenitors progressively induced Gli1 more laterally due to Shh being secreted from an expanding medial domain.
We used a combination of temporal and cumulative fate mapping approaches to follow the contribution of the early Gli1-expressing cells, in the medial vMes, to their final location, and then compared that with the current Gli1 expression domain. Specifically, we delivered a single dose of TM each morning to Gli1CreER/+;R26tdTomato/+ mice at the indicated embryonic stages shown in Figure 2. We analyzed the spatial distribution of the Gli1 lineage cells in the developing vMes 24, 48, or 72 hours after the first TM administration (Figure 2).
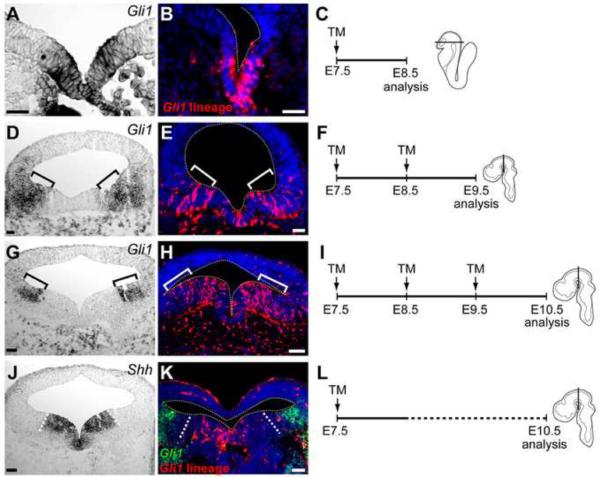
Figure 2. Gli1 expressing cells contribute to the Shh domain.
(A,D,G) Gli1 (mRNA) expression at the time of analysis, E8.5 (A), E9.5 (D), and E10.5 (G). (J) Shh (mRNA) expression at E10.5. (B,E,H) Gli1 lineage cells (red) marked with TM at E7.5 (B), E7.5–E8.5 (E), and E7.5–E9.5 (H). (K) Gli1 lineage cells marked at E7.5 (red) and Gli1 (β-gal, green) expression at E10.5. (C,F,I,L) Schematic of the experimental paradigms. The embryos received TM (down arrow) at E7.5, E8.5, or E9.5, and TM mediated recombination for 24–30 hours (solid line), and the marking was retained for the life of the animal (dashed line). Embryos were analyzed at E8.5 (A–C), E9.5 (D–F), or E10.5 (G–L). Embryo schematics indicating the sectioning plane for each stage. White dashed lines flank the Shh (mRNA) expression domain in J–K. White outlines demarcate the tissue in B, E, H, K. Brackets indicate the Gli1 (mRNA) domain in D, E, G, and H. Gli1 lineage (red) cells in the dorsal midbrain parenchyma and meninges were due to the autofluorescent background signals. Scale bars are 32 μm in A,B,D,E and 60 μm in G,H,J,K.
When we marked the earliest Gli1 lineage by delivering TM at E7.5 and determined their location at E8.5, we found that the Gli1 lineage cells in the most medial vMes were within the current Gli1 (mRNA)+ expression domain (Figure 2A–2C). Since Cre recombinase activity continues up to 36 hours following TM delivery, the lineage analysis after 24 hours indicated that the initial population being marked was the same as the cells currently expressing Gli1 at the time of analysis (Figure 2A and 2B).
Next, we cumulatively marked the Gli1 lineage by delivering TM at E7.5 and E8.5 and determined their location at E9.5 (Figure 2D–2F). Any cell that expressed Gli1 between E7.5 and E9.5 would be labeled with the reporter protein (tdTomato). Gli1 lineage cells located in the medial vMes no longer expressed Gli1 (mRNA) at E9.5 (Figure 2D), indicating that the medial Gli1 lineage cells were derived from the earlier Gli1 expressing cells that ceased to express Gli1 (Figure 2B). We also found Gli1 lineage cells located within the Gli1 (mRNA)+ domain at E9.5 (Figure 2D and 2E, brackets). By comparing the position of E7.5-marked Gli1 lineage cells in the medial domain with Gli1 lineage cells that overlap with the current Gli1 expression in the lateral domain, we could conclude that the lateral lineage cells were derived from progenitors marked at E8.5 (Figure 2D and 2E, brackets). Furthermore, Gli1 lineage cells were never observed outside the lateral boundary of the current Gli1 expression domain (Figure 2D and 2E), indicating there was a progressive shift of Shh-responsiveness to more lateral vMes progenitors.
Finally, we cumulatively marked the cells that expressed Gli1 between E7.5 and E10.5 by delivering TM at E7.5, E8.5, and E9.5 and determined their location at E10.5 (Figure 2G–2I). We again observed lineage cells in the medial domain where Gli1 (mRNA) was no longer expressed at the time of analysis, further supporting the idea that the progenitors in the medial vMes did not migrate laterally (Figure 2G and 2H). In addition, the Gli1 lineage cells were located throughout the vMes, including the Gli1 (mRNA) expression domain (Figure 2G and 2H). However, the Gli1 lineage cells were not located at the most lateral extent of the Gli1 (mRNA)+ domain, suggesting that these cells recently induced Gli1 expression but were unavailable to undergo CreER-mediated recombination to express the lineage marker (tdTomato) (Figure 2G and 2H, brackets).
We further confirmed that the medial Gli1 lineage cells originated from early, medial Gli1 expressing cells by comparing the E7.5-marked lineage cells to the current expression of Gli1 (β-gal)+ cells at E10.5 (Figure 2J–2L). We found early Gli1 expressing cells, marked at E7.5, that were only located in the very medial vMes, within the Shh (mRNA)+ domain and not in the lateral Gli1 (β-gal)+ domain at E10.5 (Figure 2J–2K). These results further support the idea that the progenitors in the medial vMes only responded to Shh for a brief period of time and new cells induced Gli1 or Shh expression in the lateral domain (Hayes et al., 2011; Blaess et al., 2011; Joksimovic et al., 2009a). Together, our data demonstrate that the same set of vMes progenitors express Gli1 first and then induce Shh under tight temporal regulation.
Loss of Shh signaling sustains proliferation of mDA neuron progenitors
We tested whether local Shh signaling (Shh signaling in Shh-expressing cells) was critical for vMes development by eliminating the Shh signaling receptor, Smo, in the Shh expressing cells. Specifically, we used the ShhCre:GFP/+ allele (Harfe et al., 2004) and SmoFlox/− (Long et al., 2001; Zhang et al., 2001) to conditionally remove Smo in the medial vMes beginning at E9 (Hayes et al., 2011). We used the Smo allele because it encodes the signal transducing protein for Shh and previous studies showed that conditional removal of Smo caused a severe reduction in mDA neurons (Blaess et al., 2006). In addition, we used the R26YFP/+ allele (Srinivas et al., 2001) to mark the mutant cells and follow their contribution to the vMes. Thus, we compared ShhCre:GFP/+;R26YFP/+ embryos (referred to as control) to ShhCre:GFP/+;SmoFlox/−;R26YFP/+ embryos (referred to as Smo conditional mutant).
First, we analyzed mutant and control embryos at E10.5 to determine if the mDA neuron progenitors were properly induced in the vMes. We compared the anterior and posterior vMes because our previous work indicated that Shh expression proceeded in an anterior to posterior order (Hayes et al., 2011). At E10.5, we observed no change in the establishment of the Lmx1a+ mDA neuron progenitors between the mutant and control embryos in the anterior or posterior vMes (Supplementary Figure 1A–1E). In addition, proliferation in the vMes at E10.5 was not changed at both the anterior and posterior levels in the mutant and control embryos (Supplementary Figure 1F–1J). ShhCre:GFP/+-mediated removal of Smo began at ~E9 in the Gli1-expressing cells that had recently induced Shh ligand expression (Figure 1A, 1B, 1I, and 1J). Therefore, by E10.5, only ~ 36 hours were allowed for Cre-mediated deletion of the Smo allele, degradation of the remaining Smo protein, and arrest of the downstream signaling. Thus, E10.5 may be too early to observe the effects of the loss of Smo on identity or survival of the mDA neuron progenitors.
Next, we investigated the vMes at E11.5, when mDA progenitors start to differentiate (Bayer et al., 1995). We labeled the progenitors in the S-phase of the cell cycle by acutely delivering EdU (5-ethynyl-2'-deoxyuridine), a thymidine analog, to pregnant dams at E11.5 one hour prior to analysis (Figure 3A). EdU incorporation allowed us to distinguish between the proliferating mDA progenitors (EdU+ Lmx1a+) located in the ventricular zone and the post-mitotic immature mDA neurons (EdULmx1a+) located in the mantle zone that were progressing towards differentiation (Figure 3A–3E). Finally, we quantified the area of the vMes that was Lmx1a+ and EdU+ Lmx1a+ to assess the proportion of the mDA progenitor domain that was proliferating at E11.5 (Figure 3F).
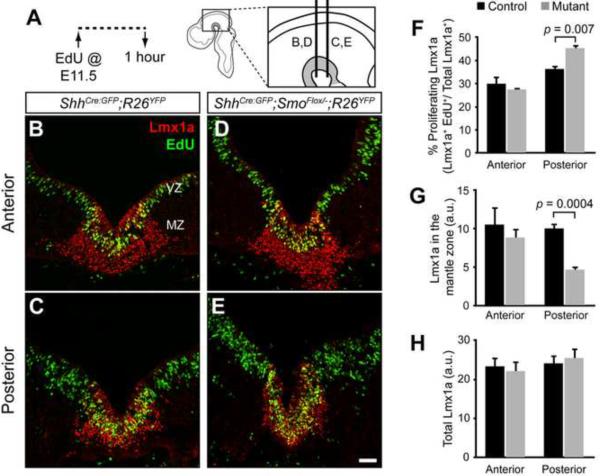
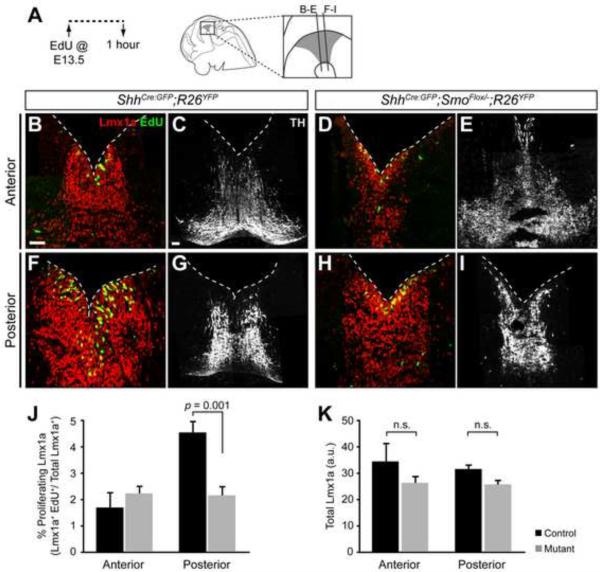
Figure 3. Loss of local Shh signaling affects cell proliferation in mDA neuron progenitors.
(A) Timeline of experiments shows that EdU was delivered at E11.5 and embryos were sacrificed and analyzed 1 hour later. Schematic indicates the anterior and posterior sectioning planes at E11.5. (B,C) Coronal sections of ShhCre:GFP/+;R26YFP/+ control embryos in the anterior (B) and posterior (C) vMes show the mDA neuron progenitors (Lmx1a, red) in the proliferative (EdU+, green) ventricular zone (VZ) and non-proliferative (EdU−) mantle zone (MZ). (D,E) Coronal sections of ShhCre:GFP/+;SmoFlox/−;R26YFP/+ mutant embryos show a decrease in the mDA neurons progenitors (Lmx1a, red) located in the MZ (EdU−) and an increase in the proliferative mDA neuron progenitors (Lmx1a+, EdU+) in the VZ of the posterior vMes (E) but not the anterior vMes (D). (F) Quantification of the percentage of the Lmx1a+ domain that incorporated EdU between the control (black) and the mutant (gray) shows that the posterior domain had significantly more proliferating Lmx1a in the mutant (p = 0.007). (G) Quantification of the Lmx1a expression in the MZ shows a reduction in the posterior vMes in mutants but not in the anterior vMes (p = 0.0004). (H) Quantification of total Lmx1a expression in the control (black) and the mutant (gray) shows no difference. a.u. = arbitrary units. Scale bars are 60 μm. n = 4 for control and mutant.
In the anterior vMes, both the control and Smo conditional mutants showed a comparable proportion of proliferating Lmx1a+ mDA progenitors in the ventricular zone(29.9 ± 3.1% in controls vs. 27.5 ± 0.88% in mutants, p = 0.47, n = 4 per genotype) as well as the extent of Lmx1a+ cells in the mantle zone (10.5 ± 2.2 a.u. in controls vs. 8.8 ± 1.2 a.u. in mutants, p = 0.70, n = 4 per genotype) (Figure 3B, 3D, 3F, and 3G). In contrast, the posterior vMes in the Smo contitional mutant embryos displayed an increase in the proportion of proliferating EdU+ Lmx1a+ cells in the ventricular zone(45.3 ± 1.6%) compared to control embryos (36.3 ± 1.6%) (p = 0.007, n = 4 per genotype) (Figure 3C, 3E, and 3F). Interestingly, the extent of the total Lmx1a+ domain in both anterior and posterior vMes was similar between the mutant and control embryos (Figure 3H). As a result, there was a considerable decrease in the Lmx1a+cells in the mantle zone of the Smo conditional mutants (p = 0.0004, Figure 3G). Notably, there was no significant difference in total EdU+ cells (data not shown) within the Lmx1a+ domain between controls and Smo conditional mutants, indicating that shortened Shh signaling did not result in a loss of proliferating cells. In agreement with our result, a previous study suggested that reduced availability of the Shh ligand also caused increased proliferation in the hindbrain and spinal cord floorplate (Joksimovic et al., 2009b). In conclusion, loss of Shh signaling in the Shh expressing cells resulted in more posterior mDA neuron progenitors that remained in a proliferative state and a reduction in post-mitotic Lmx1a+ cells in the posterior vMes. Together, our results suggest that local Shh signaling is required for cell cycle exit of the mDA neuron progenitors, specifically in the posterior vMes.
Late, local Shh signaling is required for proper allocation of SNc and VTA mDA neurons
We next tested if the changes in proliferation in the posterior vMes at E11.5 led to more mDA neurons in the ventral midbrain (vMb) at E16.5. First, we delivered EdU at E11.5 to label the progenitors in S-phase of the cell cycle and determined their contribution to the vMb at E16.5 (Figure 4A). We analyzed at E16.5 because the differentiation of mDA neurons is complete by this stage (Bayer et al., 1995). We qualitatively observed an increase in the non-DA cells that were born at E11.5 in ShhCre:GFP/+;SmoFlox/−;R26YFP/+ mutant embryos, and further analysis using the Shh-lineage marker (YFP) indicated that these EdU+ cells were also derived from Shh-expressing cells (Figure 4B–4G and data not shown). Accordingly, our quantification results showed a significant difference in the number of mDA neurons born at E11.5 (226 ± 18 cells in ShhCre:GFP/+;SmoFlox/−;R26YFP/+ mutants and 157 ± 16 cells in the control, n = 3 per genotype, p = 0.046) (Figure 4I). However, the subtle increase in the number of cells derived from E11.5 proliferating progenitors did not cause an increase in the total number of mDA neurons in mutant (790 ± 33) compared to the control (704 ± 65, p = 0.304) (Figure 4H), suggesting that there could be a reduced production of mDA neurons at other developmental stages in ShhCre:GFP/+;SmoFlox/−;R26YFP/+ mutants.
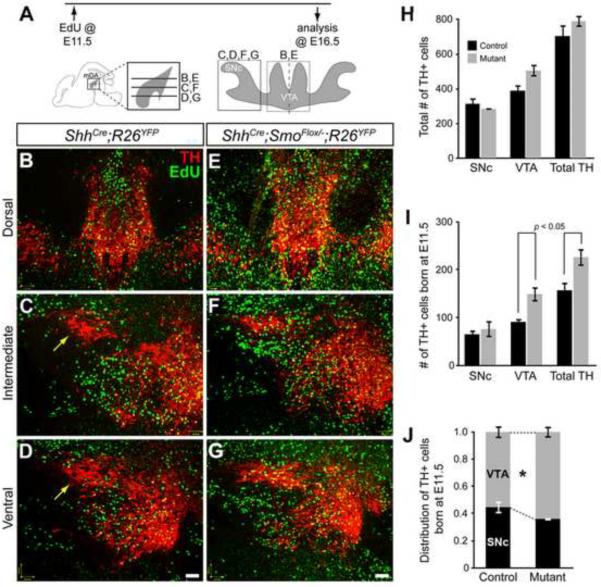
Figure 4. mDA neurons are generated but differentially allocated when local Shh signaling is lost.
(A) Timeline of experiments indicates that EdU was delivered at E11.5 and the location of the proliferative cells was observed at E16.5. Schematics indicate the horizontal sectioning plane at E16.5. Gray area indicates mDA neurons with the SNc and VTA boxed. (B–G) mDA neurons (red) labeled with EdU (green) at E11.5 in ShhCre:GFP/+;R26YFP/+ control (B–D) and ShhCre:GFP/+;SmoFlox/−;R26YFP/+ mutant (E–G) brains. Yellow arrows indicate anterolateral SNc where less EdU+ cells are found in the control compared to the mutants. There are more EdU+ cells found in the mutants compared to the control at all levels. The dorsal level sections show more VTA mDA neurons are derived from E11.5 EdU labeled cells in the mutant (E) than in the control (B). (H) The number of TH+ neurons in the SNc, VTA, and total TH was counted by a systematic random sampling of the total mDA neurons in the control and mutant and show no statistical differences. (I) Quantification of the number of TH+ neurons that incorporated EdU at E11.5 in the SNc, VTA, and total TH+ shows that there was a significant increase in mDA neurons born at E11.5 in the VTA and in total. (J) The distribution of E11.5-born mDA neurons show that there is a significant bias toward the VTA in ShhCre:GFP/+;SmoFlox/−;R26YFP/+ mutants. *, p = 0.029. Scale bar = 60 μm. n = 3 per genotype.
The mDA neuron progenitors that incorporated EdU at E11.5 contributed to both the SNc and VTA at E16.5. Interestingly, the anterior-lateral SNc showed numerous TH+ EdU+ cells in the mutant while the corresponding region in the control showed no TH+ EdU+ cells (Figure 4C–4D and 4F–4G, arrow). However, quantification of the SNc neurons born at E11.5 did not show a significant increase in the mutant (76 ± 17 cells, n = 3) compared to the control (65 ± 9 cells, p = 0.603, n = 3) (Figure 4I). In contrast, the VTA showed a significant increase in TH+ cells born at E11.5 in the mutant (149 ± 16) compared to the control (91 ± 6) (p = 0.027) (Figure 4I). In summary, loss of Shh signaling in the Shh expressing cells resulted in an increase in the amount of mDA neuron progenitors that were maintaining the proliferative state at E11.5 and then contributed to the VTA with a significant bias (Figure 4J, p = 0.029).
Loss of Shh signaling depletes mDA neuron progenitors by E13.5
Next, we investigated the vMes at E13.5, toward the end of mDA neuron neurogenesis (Bayer et al., 1995) to test whether the increased proliferation observed in the E11.5 Smo conditional mutant was transient or sustained. We again labeled the proliferating cells with EdU, allowed 1 hour for incorporation, and then analyzed the anterior and posterior vMes (Figure 5A). In the anterior vMes, there was a comparable amount of proliferating cells in the control and Smo conditional mutants (Figure 5B, 5D and 5J). Also, the TH+ cells in both the anterior and posterior vMes at E13.5 were similarly distributed in the control and Smo conditional mutants indicating that there was not a loss of mDA neurons (Figure 5C and 5E) or Lmx1a+ mDA progenitors (Figure 5K). Interestingly, many Lmx1a+ cells were still proliferating in the posterior vMes in the control (Figure 5F), but not in the Smo conditional mutant (Figure 5H and 5J). Together, these results indicate that the loss of local Shh signaling transiently increases mDA progenitors in active cell cycle at E11.5, but subsequently depletes proliferating progenitors by E13.5 through the increased cell cycle exit (Figures 3 and 5). Thus the timing and duration of Shh signaling may be important for the mDA neuron maturation.
Figure 5. Loss of local Shh signaling depletes proliferative mDA neuron progenitors at E13.5.
(A) Timeline indicates that EdU was delivered at E13.5 and the location of the labeled cells was analyzed 1hr later at the indicated section levels for the panels B–E (Anterior) and F–I (Posterior). (B–E) The proliferating cells (EdU+, green) at E13.5 show similar distribution between control (B) and ShhCre:GFP/+;SmoFlox/−;R26YFP/+ mutant (D) in the anterior midbrain based on TH expression pattern (C,E). (F–I) In contrast, the EdU+ cells were more abundant in the control (F) compared to the ShhCre:GFP/+;SmoFlox/−;R26YFP/+ mutant (H) in the posterior midbrain where the level was matched based on the distribution pattern of TH+ mDA neurons (G,I). (J) Quantification of the percentage of proliferating Lmx1a+ domain per section confirmed that the posterior midbrain contains significantly less proliferating cells in ShhCre:GFP/+;SmoFlox/−;R26YFP/+ mutant (p < 0.001, n = 3 per genotype). (K) Quantification of total Lmx1a expression indicates no change between control and mutant. Scale bars are 50 μm.
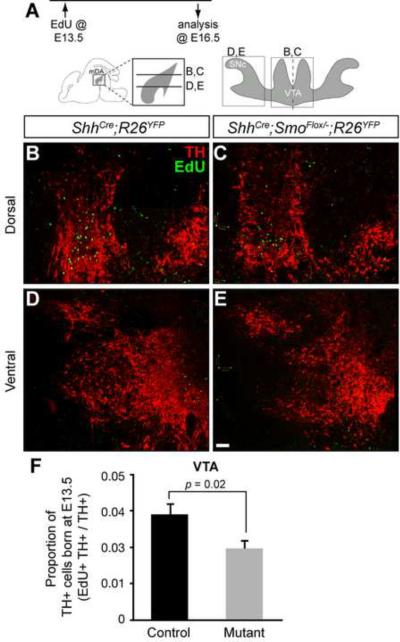
Finally, the cells labeled at E13.5 were followed to their final location within the E16.5 midbrain (Figure 6). Consistent with the reduced number of proliferating cells in the posterior vMes of ShhCre:GFP/+;SmoFlox/−;R26YFP/+ mutants at E13.5, there were fewer EdU+ cells found in the VTA of the mutant compared to the control (Figure 6B, 6C, and 6F). Furthermore, there were hardly any EdU+ cells found in the SNc, confirming that most of the mDA neurons in the SNc were derived from cells born earlier than E13.5 (Figure 6D and 6E) (Hayes et al., 2011). Together, the E13.5 data supports our idea that the reduced proliferation at E13.5 compensated for the transient increase in proliferation of mDA progenitors at E11.5 in ShhCre:GFP/+;SmoFlox/−;R26YFP/+ mutants to produce similar numbers of mDA neurons overall.
Figure 6. mDA neurons born at E13.5 in Smo conditonal mutants contribute less to VTA, and none to the SNc.
(A) Timeline shows that EdU was delivered at E13.5 and the location of the labeled cells observed at E16.5. Schematics indicate the horizontal sectioning planes at E16.5. Gray area indicates mDA neurons with the SNc and VTA boxed. (B,C) Dorsal VTA contains few mDA neurons (TH, red) derived from E13.5 labeling (EdU, green) in both control (B) and mutant (C), but mutants show less labeled cells reflecting the decrease in proliferation at E13.5. (D,E) The SNc has hardly any cells derived from E13.5 proliferative progenitors in both control and mutant. (F) Quantification of TH+ cells born at E13.5 shows a smaller proportion of VTA cells were derived from E13.5 progenitors in mutants compared to the control (n = 3 per genotype). Scale bar is 50 μm.
Loss of local Shh signaling promotes cell cycle exit of vMes progenitors
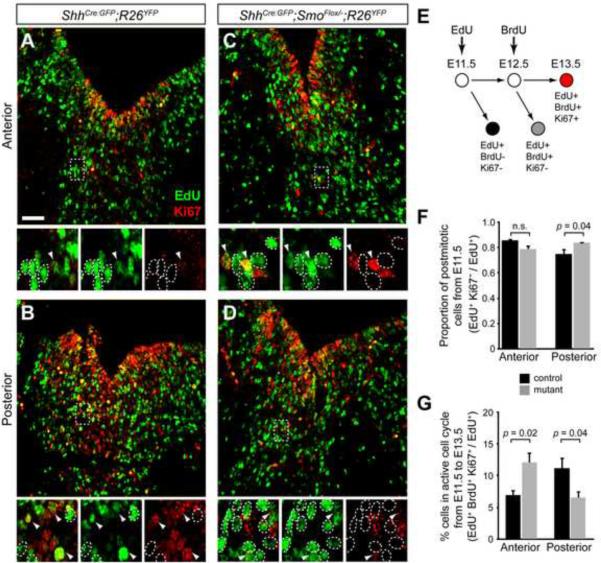
In order to account for the changes in proliferation of vMes progenitors between E11.5 and E13.5 in ShhCre:GFP/+;SmoFlox/− mutants, we performed a cell cycle exit analysis to determine the duration that progenitors spent in active cell cycle. We injected thymidine analogs, EdU and BrdU, at E11.5 and E12.5, respectively, to label the proliferating cells and analyzed their cell cycle status at E13.5 with Ki67 expression (Figure 7E) (Wang et al., 2011). The proportion of cells that are thymidine analog+ Ki67− among the total number of thymidine analog+ cells represents the proportion of cells that became postmitotic since the labeling (Figure 7E). In the anterior vMes, the majority of cells proliferating at E11.5 (EdU+) became postmitotic (Ki67−) in the control (85.6 ± 1.2 %, n = 3) as in the mutant (78.7 ± 3.1 %, n = 4, p = 0.086) (Figure 7A, 7C, and 7F). Surprisingly, in the anterior vMes, there was an increase in cells that maintained an active cell cycle from E11.5–E13.5 in the mutant compared to the control, which may correspond to the continued contribution to the anterior lateral SNc neurons observed in the mutant (Figure 4G and 7G). In contrast, a significantly greater proportion of proliferating cells at E11.5 (EdU+) became postmitotic in the posterior vMes in the Smo conditional mutants (74.8 ± 4.4 % and 83.7 ± 1.0 % in control and mutant, respectively. p = 0.043) (Figure 7F). As expected, there was a corresponding decrease in the number of cells that remained in active cell cycle from E11.5 to E13.5 in the posterior vMes, which resulted in the decreased contribution to the VTA (Figure 6F and 7G).
Figure 7. Loss of local Shh signaling promotes cell cycle exit of vMes progenitors.
(A–D) Proliferative cells labeled at E11.5 (EdU+, green) were analyzed for their status in cell cycle at E13.5 (Ki67, red) in the anterior (A, C) and posterior (B, D) vMes of control (A–B) and mutant (C–D) embryos. White outlines demarcate cells proliferating at E11.5 that became postmitotic by E13.5 and white arrowheads point to cells that remained in active cell cycle from E11.5 to E13.5. (E) Schematic of cell cycle exit experiments shows that EdU and BrdU were injected at E11.5 and E12.5, respectively, and active cell cycle status was analyzed by Ki67 expression at E13.5. (F) Quantification shows the proportion of EdU+ cells that exit the cell cycle between E11.5 and E13.5 is significantly greater in the posterior vMes. (G) Quantification shows a greater anterior and smaller posterior proportion of mutant progenitors maintain anactive cell cycle between E11.5 and E13.5. Scale bar = 50 μm. n = 3 per genotype.
DISCUSSION
Our careful analysis of the dynamic expression and short-term lineage tracing of the Shh-secreting and Shh-responding cells revealed a unique relationship between these two cell populations. Using Gli1 as a sensitive readout of Shh signaling, we demonstrated that Shh signaling precedes the emergence of Shh expression in the developing vMes. In addition, we found that Shh and Gli1 expressing cells are basically the same population of cells that sequentially turn on Gli1 and then Shh expression. The progressive cumulative fate mapping results showed that the Gli1 lineage contributed to the entire vMes demonstrating that cells in the vMes at one point in their history responded to Shh signaling (Hayes et al., 2011). As a result, most, if not all, of the Shh-expressing cells must have been derived from the Gli1 lineage, indicating that the Shh and Gli1 lineages are not distinct cell populations, but are intricately linked by dynamic temporal regulation.
The importance of Gli1 expression preceding induction of Shh expression has been demonstrated in Gli2 null mice, which lacks the main activator of Shh signaling and fails to turn on Shh expression in the floorplate (Matise et al., 1998; Bai et al., 2002). Subsequently, Shh and Gli1 expression expands laterally through a progressive induction of Gli1 as the Shh expression is induced in the Gli1 expressing cells and Shh ligand is diffused to the neighboring lateral cells (see the Wave model, Figure 8A). When Shh signaling is removed very early across the vMes including both Shh-expressing and –responding domains, there is a substantial loss of mDA neurons, such as in En1-Cre;SmoFlox/− mutants (Blaess et al., 2006). However, in the En1-Cre;SmoFlox/− mutants, there is an initial burst of Shh signaling in the most medial vMes because En1 expression emerges after Gli1 but before Shh expression (Li et al., 2002; Blaess et al., 2006). As a result, Gli1 expression is induced in the medial vMes by Shh secreted from the notochord, but the progressive lateral induction of Shh signaling across the vMes is shut down leading to a drastic decrease in mDA neurons (Blaess et al., 2006). Thus, the few remaining mDA neurons in En1-Cre;SmoFlox/− mutants are perhaps due to the initial burst of Shh signaling in response to Shh from the notochord before the En1-Cre terminated Shh signaling (Blaess et al., 2006; Tran et al., 2010; Omodei et al., 2008; Trokovic et al., 2003; Li et al., 2002).
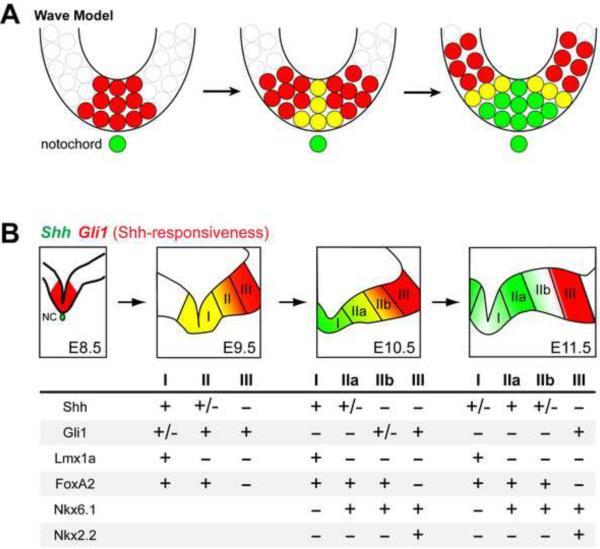
Figure 8. Summary of Shh and Gli1 lineage contribution to mDA neurons.
(A) In the wave model, the Gli1+ cells (red) in the medial vMes co-express Shh (yellow). The medial expansion of Shh (green) allows the ligand to diffuse more laterally to induce Gli1 expression in the lateral domain. Induction of Shh continues in the medial domain and the Gli1 is downregulated. Yellow indicates co-expression of Shh and Gli1. (B) Schematic of the developing vMes showing three distinct domains of Shh and Gli1 expression. Domain I expresses FoxA2 and Lmx1a and consists of cells that expressed Gli1 early (E7–E9) and Shh continuously (E8–E11). Domain I contributes to mDA neurons of both SNc and VTA. Domain II expresses FoxA2 and Nkx6.1 but not Lmx1a, and consists of cells that express Gli1 after its downregulation in the medial domain (E9–E10) and Shh in expansion (E9–E11). This domain II contributes primarily to the VTA mDA neurons. Domain II is divided into IIa and IIb to account for the expansion of Shh expression before it reaches the lateral limit of FoxA2 expression from E10.5 to E11.5. Gli1 expressing cells in the most lateral domain (E10–E11, Domain III) are outside the Lmx1a domain, express Nkx6.1 and Nkx2.2 and do not become mDA neurons. NC is notochord.
Even though our expression analysis showed little overlap between Shh expression and Shh-responsiveness, activation of Shh signaling within Shh-expressing cells had biological significance as evidenced by our Smo conditional mutant analysis. The loss of local Shh signaling primarily affected the posterior vMes at E11.5 and E13.5, which resulted in an altered contribution pattern to the VTA mDA neurons at E16.5. Specifically, we found that the loss of Shh local signaling resulted in a transient increase in proliferation of Lmx1a+ vMes progenitors at E11.5, which corresponded to an increased contribution to the VTA. Subsequent gradual depletion of proliferating Lmx1a+ vMes progenitors at E13.5 in the Smo conditional mutant was due to accelerated cell cycle exit and resulted in decreased contribution to the VTA. However, the combinatorial effects of the transient increase and subsequent depletion made the overall number of mDA neurons unchanged. In our previous study, we suggested that the VTA progenitors maintain their proliferative state longer based on the contribution of both early and late Gli1 lineage to the VTA (Hayes et al., 2011). In support of this idea, our findings demonstrate that premature cell cycle exit primarily affects the VTA mDA neurons in the Smo conditional mutants. This transient shift in the cell types (more VTA from E11.5 to the less VTA from E13.5 progenitors) in Smo conditional mutants indicates that the duration of Shh signaling could serve as another regulatory means of differentially allocating the mDA neuron subtypes.
Previous fate mapping studies demonstrated a correlation between the dynamic temporal and spatial changes in Shh expression and responsiveness in the vMes that contributed to distinct mDA neurons (Joksimovic et al., 2009a; Hayes et al., 2011; Blaess et al., 2011). The present study shows that Shh-expressing cells are derived from previous Gli1-expressing cells; therefore, the earliest Shh-responding cells (Gli1-expressing cells E7.5–E8.5) and Shh-expressing cells (E8.5–E9.5) located in the medial vMes (domain I) become primarily SNc mDA neurons and also VTA mDA neurons to a lesser extent (Joksimovic et al., 2009a; Hayes et al., 2011; Blaess et al., 2011). These early cells in domain I co-express FoxA2 and Lmx1A (Figure 1, 8B, and Supplementary Figure 2) (Joksimovic et al., 2009a; Hayes et al., 2011; Blaess et al., 2011; Ferri et al., 2007; Ono et al., 2007, Andersson et al., 2006). Next, the intermediate domain II includes Shh-responding cells between E8.5–E9.5 that express Shh between E9.5–E11.5. At E10.5 and E11.5, this domain can be subdivided into IIa and IIb due to their dynamic changes in Shh expression and Shh-responsiveness: Shh expression is first restricted to IIa and Foxa2 expression extends into IIb, and then Shh expression expands into IIb around E11 (Figure 1, 8B, and Supplementary Figure 2). Cells in the intermediate domain II express FoxA2 and Nkx6.1, but not Lmx1a, and contribute to some SNc but mostly VTA mDA neurons, as well as non-dopaminergic cells (Figure 8B and Supplementary Figure 2) (Joksimovic et al., 2009a; Hayes et al., 2011; Blaess et al., 2011; Ferri et al., 2007; Ono et al., 2007, Andersson et al., 2006). Finally, cells in the most lateral vMes domain III respond to Shh signaling, express Nkx2.2 and Nkx6.1, but never express Shh themselves and do not become mDA neurons (Ferri et al., 2007; Blaess et al., 2011; Hayes et al., 2011). Our comparative analysis of current expression and short term lineage mapping compliment the previous studies because we were able to demonstrate the dynamic nature of the developing vMes which allowed for a more definitive characterization of the expression domains (I–III).
Our findings further demonstrate the importance of temporal dynamics of Shh signaling in developing vMes progenitors for specifying mDA neuron subtypes. Much attention has been focused on production of mDA neurons in vitro for their potential in cell replacement therapy. While most in vitro protocols for mDA neurons include the treatment of neural progenitors with Shh, the final characterization and validation of mDA neurons mostly rely on the expression of TH and do not address the heterogeneity among mDA neurons. Our findings provide an additional potential regulatory means to control the subtypes of mDA neurons by changing the responsiveness to Shh signaling to produce more selective types of mDA neurons such as VTA versus SNc. Together, our study demonstrates that the duration of Shh signaling controls the proliferative state of mDA neuron progenitors, which may ultimately affect their mature identity and function.
CONCLUSION
In this study, we compared the temporal and spatial dynamics of Shh expression and Shh responsiveness during vMes development. Previous studies identified that early Shh signaling in the vMes is important for the generation of mDA neuron progenitors (Blaess et al., 2006; Chiang et al., 1996; Matise et al., 1998), but did not distinguish between Shh expressing cells (local Shh signaling) versus purely responsive cells (paracrine Shh signaling). We report here that the Shh expressing cells are responsive to the local Shh ligand and the signaling has a functional role in the medial vMes. We identified that the vMes progenitors initially respond to Shh signaling and then induce Shh expression (Gli1+, Shh+). Our conditional mutant analysis of ShhCre:GFP/+;SmoFlox/− indicates that the later local Shh signaling regulates the cell cycle status of mDA progenitors and may shape the distribution of the mDA neurons within the VTA and SNc. Thus, through the tight control of Shh signaling duration at both induction and cessation time points, Shh signaling can influence the neurogenesis and distribution of the mDA neurons in the ventral midbrain.
Supplementary Material
(A) Schematic of E10.5 embryo indicating the anterior and posterior sectioning planes. Gray shading indicates vMes (B,C) Coronal sections of ShhCre:GFP/+;R26YFP/+ control embryos in the anterior (B) and posterior (C) vMes showing the mDA progenitors (Lmx1a, red) from the Shh lineage (GFP, green). (D,E) Coronal sections of ShhCre:GFP/+;SmoFlox/−;R26YFP/+ mutant embryos in the anterior (D) and posterior (E) vMes showing mDA progenitors (Lmx1a, red) that have lost Shh signaling (GFP, green). Boxed regions in B-E indicate higher magnification images below panels marked i (GFP) and ii (Lmx1a). (F–I) EdU was delivered at E10.5 and embryos were sacrificed and analyzed 1 hour later. Coronal sections of ShhCre:GFP/+;R26YFP/+ control (F#x2013;G) and ShhCre:GFP/+;SmoFlox/−;R26YFP/+ mutant (H–I) embryos in the anterior (F, H) and posterior (G, I) vMes. (J) Quantification of the percentage of Lmx1a+ domain that incorporated EdU in the control (black) and mutant (gray) shows no difference in the anterior or posterior domain. Scale bars are 60 μm. n = 3 per genotype.
(A–C) Schematic of Shh (green) and Gli1 (red) expression in the vMes at E9.5, E10.5, and E11.5. (D–F) Lmx1a expression identifies the medial domain I. (G–I) FoxA2 expression defines the lateral extent of intermediate domain II and largely overlaps with Shh expression. (J,K) Nkx6.1 expression is mutually exclusive to Lmx1a+ domain I but extends into domain II with an additional lateral patch at the limit of Gli1 expression in domain III. (L,M) Nkx2.2 expression overlaps with Gli1 expression at E10.5 and E11.5 and defines the most lateral domain III, and also includes lateral patchy expression domain of Nkx6.1. The subdivision between IIa and IIb occurs at E9.5 because the lateral expansion of Shh expression has not reached the lateral limit of FoxA2 expression.
Highlights
-
■
Shh signaling is dynamic in the vMes.
-
■
Shh-expressing cells are derived from earlier Shh-responding cells.
-
■
Local Shh signaling regulates the cell cycle status in the vMes.
-
■
Duration of Shh signaling contributes to the distribution of mDA neurons in the VTA and SNc.
Acknowledgments
We would like to thank Drs. M. Zervas, J. Li and Y. Mukoyama for their critical comments throughout the course of this project. We also would like to thank Drs. M. German for Lmx1a antibody and S. Mackem for advice on the Smo allele. The confoncal microscopy was done at Microscopy Image Center, NICHD. This work was supported by the Intramural Research Program at NIH/NICHD.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- Ahn S, Joyner AL. Dynamic changes in the response of cells to positive hedgehog signaling during mouse limb patterning. Cell. 2004;118:505–516. doi: 10.1016/j.cell.2004.07.023. [DOI] [PubMed] [Google Scholar]
- Ahn S, Joyner AL. In vivo analysis of quiescent adult neural stem cells responding to Sonic hedgehog. Nature. 2005;437:894–897. doi: 10.1038/nature03994. [DOI] [PubMed] [Google Scholar]
- Andersson E, Tryggvason U, Deng Q, Friling S, Alekseenko Z, Robert B, Perlmann T, Ericson J. Identification of intrinsic determinants of midbrain dopamine neurons. Cell. 2006;124:393–405. doi: 10.1016/j.cell.2005.10.037. [DOI] [PubMed] [Google Scholar]
- Bai CB, Auerbach W, Lee JS, Stephen D, Joyner AL. Gli2, but not Gli1, is required for initial Shh signaling and ectopic activation of the Shh pathway. Development. 2002;129:4753–4761. doi: 10.1242/dev.129.20.4753. [DOI] [PubMed] [Google Scholar]
- Bayer SA, Wills KV, Triarhou LC, Ghetti B. Time of neuron origin and gradients of neurogenesis in midbrain dopaminergic neurons in the mouse. Exp Brain Res. 1995;105:191–199. doi: 10.1007/BF00240955. [DOI] [PubMed] [Google Scholar]
- Blaess S, Bodea GO, Kabanova A, Chanet S, Mugniery E, Derouiche A, Stephen D, Joyner AL. Temporal-spatial changes in Sonic Hedgehog expression and signaling reveal different potentials of ventral mesencephalic progenitors to populate distinct ventral midbrain nuclei. Neural Dev. 2011;6:29. doi: 10.1186/1749-8104-6-29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blaess S, Corrales JD, Joyner AL. Sonic hedgehog regulates Gli activator and repressor functions with spatial and temporal precision in the mid/hindbrain region. Development. 2006;133:1799–1809. doi: 10.1242/dev.02339. [DOI] [PubMed] [Google Scholar]
- Brown A, Brown S, Ellisor D, Hagan N, Normand E, Zervas M. A practical approach to genetic inducible fate mapping, a visual guide to mark and track cells in vivo. J Vis Exp. 2009;34:1687. doi: 10.3791/1687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiang C, Litingtung Y, Lee E, Young KE, Corden JL, Westphal H, Beachy PA. Cyclopia and defective axial patterning in mice lacking Sonic hedgehog gene function. Nature. 1996;383:407–413. doi: 10.1038/383407a0. [DOI] [PubMed] [Google Scholar]
- Echelard Y, Epstein DJ, St-Jacques B, Shen L, Mohler J, McMahon JA, McMahon AP. Sonic hedgehog, a member of a family of putative signaling molecules, is implicated in the regulation of CNS polarity. Cell. 1993;75:1417–1430. doi: 10.1016/0092-8674(93)90627-3. [DOI] [PubMed] [Google Scholar]
- Ferri AAM, Lin W, Mavromatakis YE, Wang JC, Sasaki H, Whitsett JA, Ang S-L. Foxa1 and Foxa2 regulate multiple phases of midbrain dopaminergic neuron development in a dosage-dependent manner. Development. 2007;134:2761–2769. doi: 10.1242/dev.000141. [DOI] [PubMed] [Google Scholar]
- Fuccillo M, Joyner AL, Fishell G. Morphogen to mitogen, the multiple roles of hedgehog signalling in vertebrate neural development. Nat Rev Neurosci. 2006;7:772–783. doi: 10.1038/nrn1990. [DOI] [PubMed] [Google Scholar]
- Harfe BD, Scherz PJ, Nissim S, Tian H, McMahon AP, Tabin CJ. Evidence for an expansion-based temporal Shh gradient in specifying vertebrate digit identities. Cell. 2004;118:517–528. doi: 10.1016/j.cell.2004.07.024. [DOI] [PubMed] [Google Scholar]
- Hayes L, Zhang Z, Albert P, Zervas M, Ahn S. Timing of Sonic hedgehog and Gli1 expression segregates midbrain dopamine neurons. J Comp Neurol. 2011;519:3001–3018. doi: 10.1002/cne.22711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ingham PW, Placzek M. Orchestrating ontogenesis, variations on a theme by sonic hedgehog. Nat Rev Genet. 2006;7:841–850. doi: 10.1038/nrg1969. [DOI] [PubMed] [Google Scholar]
- Joksimovic M, Anderegg A, Roy A, Campochiaro L, Yun B, Kittappa R, McKay R, Awatramani R. Spatiotemporally separable Shh domains in the midbrain define distinct dopaminergic progenitor pools. Proc Natl Acad Sci U S A. 2009a;106:19185–19190. doi: 10.1073/pnas.0904285106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joksimovic M, Yun BA, Kittappa R, Anderegg AM, Chang WW, Taketo MM, McKay RD, Awatramani RB. Wnt antagonism of Shh facilitates midbrain floor plate neurogenesis. Nat Neurosci. 2009b;12:125–131. doi: 10.1038/nn.2243. [DOI] [PubMed] [Google Scholar]
- Kraus P, Fraidenraich D, Loomis CA. Some distal limb structures develop in mice lacking Sonic hedgehog signaling. Mech Dev. 2001;100:45–58. doi: 10.1016/s0925-4773(00)00492-5. [DOI] [PubMed] [Google Scholar]
- Li JY, Lao Z, Joyner AL. Changing requirements for Gbx2 in development of the cerebellum and maintenance of the mid/hindbrain organizer. Neuron. 2002;36:31–43. doi: 10.1016/s0896-6273(02)00935-2. [DOI] [PubMed] [Google Scholar]
- Litingtung Y, Chiang C. Specification of ventral neuron types is mediated by an antagonistic interaction between Shh and Gli3. Nat Neurosci. 2000;3:979–985. doi: 10.1038/79916. [DOI] [PubMed] [Google Scholar]
- Long F, Zhang XM, Karp S, Yang Y, McMahon AP. Genetic manipulation of hedgehog signaling in the endochondral skeleton reveals a direct role in the regulation of chondrocyte proliferation. Development. 2001;128:5099–5108. doi: 10.1242/dev.128.24.5099. [DOI] [PubMed] [Google Scholar]
- Matise MP, Epstein DJ, Park HL, Platt KA, Joyner AL. Gli2 is required for induction of floor plate and adjacent cells, but not most ventral neurons in the mouse central nervous system. Development. 1998;125:2759–2770. doi: 10.1242/dev.125.15.2759. [DOI] [PubMed] [Google Scholar]
- Omodei D, Acampora D, Mancuso P, Prakash N, Di Giovannantonio LG, Wurst W, Simeone A. Anterior-posterior graded response to Otx2 controls proliferation and differentiation of dopaminergic progenitors in the ventral mesencephalon. Development. 2008;135:3459–470. doi: 10.1242/dev.027003. [DOI] [PubMed] [Google Scholar]
- Ono Y, Nakatani T, Sakamoto Y, Mizuhara E, Minaki Y, Kumai M, Hamagushi A, Nishimura M, Inoue Y, Hayashi H, Takahashi J, Imai T. Differences in neurogenic potential in floor plate cells along an anteroposterior location: midbrain dopaminergic neurons originate from mesencephalic floor plate cells. 2007;134:3213–3225. doi: 10.1242/dev.02879. [DOI] [PubMed] [Google Scholar]
- Perez-Balaguer A, Puelles E, Wurst W, Martinez S. Shh dependent and independent maintenance of basal midbrain. Mech Dev. 2009;126:301–313. doi: 10.1016/j.mod.2009.03.001. [DOI] [PubMed] [Google Scholar]
- Platt KA, Michaud J, Joyner AL. Expression of the mouse Gli and Ptc genes is adjacent to embryonic sources of hedgehog signals suggesting a conservation of pathways between flies and mice. Mech Dev. 1997;62:121–135. doi: 10.1016/s0925-4773(96)00648-x. [DOI] [PubMed] [Google Scholar]
- Ribes V, Balaskas N, Sasai N, Cruz C, Dessaud E, Cayuso J, Tozer S, Yang LL, Novitch B, Marti E, Briscoe J. Distinct Sonic Hedgehog signaling dynamics specify floor plate and ventral neuronal progenitors in the vertebrate neural tube. Genes Dev. 2010;24:1186–1200. doi: 10.1101/gad.559910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srinivas S, Watanabe T, Lin CS, William CM, Tanabe Y, Jessell TM, Costantini F. Cre reporter strains produced by targeted insertion of EYFP and ECFP into the ROSA26 locus. BMC Dev Biol. 2001;1:4. doi: 10.1186/1471-213X-1-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tran TH, Jarrell A, Zentner GE, Welsh A, Brownell I, Scacheri PC, Atit R. Role of canonical Wnt signaling/β-catenin via Dermo1 in cranial dermal cell development. Development. 2010;137:3973–984. doi: 10.1242/dev.056473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trokovic R, Trokovic N, Hernesniemi S, Pirvola U, Vogt Weisenhorn DM, Rossant J, McMahon AP, Wurst W, Partanen J. FGFR1 is independently required in both developing mid- and hindbrain for sustained response to isthmic signals. EMBO J. 2003;22:1811–823. doi: 10.1093/emboj/cdg169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van den Heuvel DM, Pasterkamp RJ. Getting connected in the dopamine system. Prog Neurobiol. 2008;85:75–93. doi: 10.1016/j.pneurobio.2008.01.003. [DOI] [PubMed] [Google Scholar]
- Wang H, Ge G, Uchida Y, Luu B, Ahn S. Gli3 is required for maintenance and fate specification of cortical progenitors. J Neurosci. 2011;31:6440–6448. doi: 10.1523/JNEUROSCI.4892-10.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wijgerde M, McMahon JA, Rule M, McMahon AP. A direct requirement for Hedgehog signaling for normal specification of all ventral progenitor domains in the presumptive mammalian spinal cord. Genes Dev. 2002;16:2849–2864. doi: 10.1101/gad.1025702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye W, Shimamura K, Rubenstein JL, Hynes MA, Rosenthal A. FGF and Shh signals control dopaminergic and serotonergic cell fate in the anterior neural plate. Cell. 1998;93:755–766. doi: 10.1016/s0092-8674(00)81437-3. [DOI] [PubMed] [Google Scholar]
- Zervas M, Millet S, Ahn S, Joyner AL. Cell behaviors and genetic lineages of the mesencephalon and rhombomere 1. Neuron. 2004;43:345–357. doi: 10.1016/j.neuron.2004.07.010. [DOI] [PubMed] [Google Scholar]
- Zhang XM, Ramalho-Santos M, McMahon AP. Smoothened mutants reveal redundant roles for Shh and Ihh signaling including regulation of L/R asymmetry by the mouse node. Cell. 2001;105:781–792. [PubMed] [Google Scholar]
- Zhu J, Nakamura E, Nguyen MT, Bao X, Akiyama H, Mackem S. Uncoupling Sonic hedgehog control of pattern and expansion of the developing limb bud. Dev Cell. 2008;14:624–632. doi: 10.1016/j.devcel.2008.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(A) Schematic of E10.5 embryo indicating the anterior and posterior sectioning planes. Gray shading indicates vMes (B,C) Coronal sections of ShhCre:GFP/+;R26YFP/+ control embryos in the anterior (B) and posterior (C) vMes showing the mDA progenitors (Lmx1a, red) from the Shh lineage (GFP, green). (D,E) Coronal sections of ShhCre:GFP/+;SmoFlox/−;R26YFP/+ mutant embryos in the anterior (D) and posterior (E) vMes showing mDA progenitors (Lmx1a, red) that have lost Shh signaling (GFP, green). Boxed regions in B-E indicate higher magnification images below panels marked i (GFP) and ii (Lmx1a). (F–I) EdU was delivered at E10.5 and embryos were sacrificed and analyzed 1 hour later. Coronal sections of ShhCre:GFP/+;R26YFP/+ control (F#x2013;G) and ShhCre:GFP/+;SmoFlox/−;R26YFP/+ mutant (H–I) embryos in the anterior (F, H) and posterior (G, I) vMes. (J) Quantification of the percentage of Lmx1a+ domain that incorporated EdU in the control (black) and mutant (gray) shows no difference in the anterior or posterior domain. Scale bars are 60 μm. n = 3 per genotype.
(A–C) Schematic of Shh (green) and Gli1 (red) expression in the vMes at E9.5, E10.5, and E11.5. (D–F) Lmx1a expression identifies the medial domain I. (G–I) FoxA2 expression defines the lateral extent of intermediate domain II and largely overlaps with Shh expression. (J,K) Nkx6.1 expression is mutually exclusive to Lmx1a+ domain I but extends into domain II with an additional lateral patch at the limit of Gli1 expression in domain III. (L,M) Nkx2.2 expression overlaps with Gli1 expression at E10.5 and E11.5 and defines the most lateral domain III, and also includes lateral patchy expression domain of Nkx6.1. The subdivision between IIa and IIb occurs at E9.5 because the lateral expansion of Shh expression has not reached the lateral limit of FoxA2 expression.