Abstract
Study Design
Cells isolated from moderately and severely degenerated human intervertebral discs (IVDs) cultured in an alginate scaffold.
Objective
To compare the regenerative potential of moderately vs. severely degenerated cells using three pro-anabolic stimulants.
Summary of Background Data
Injection of soluble cell signaling factors has potential to slow the progression of IVD degeneration. While degenerative grade is thought to be an important factor in targeting therapeutic interventions it remains unknown whether cells in severely degenerated IVDs have impaired metabolic functions compared to lesser degenerative levels or if they are primarily influenced by the altered microenvironment.
Methods
NP cells were cultured in alginate for 21 days and treated with three different pro-anabolic stimulants: a growth factor/anti-inflammatory combination of TGFβ3+Dex, or matricellular proteins CTGF or Link-N. They were assayed for metabolic activity, DNA content, glycosaminoglycan (GAG), and qRT-PCR gene profiling.
Results
Moderately degenerated cells responded to stimulation with increased proliferation, decreased IL-1β, MMP9 and COL1A1 expression, and upregulated HAS1 as compared to severely degenerated cells. TGFβR1 (ALK5) receptors were expressed at greater levels in moderately than severely degenerated cells. TGFβ3+Dex had a notable stimulatory effect on moderately degenerated NP cells with increased anabolic gene expression, and decreased COL1A1 and ADAMTS5 gene expression. Link-N and CTGF had similar responses in all assays, and both treatments up-regulated IL-1β expression and had a more catabolic response than TGFβ3+Dex, particularly in the more severely degenerated group. All groups, including different degenerative grades, produced similar amounts of GAG.
Conclusion
Pro-anabolic stimulants alone had limited capacity to overcome the catabolic and pro-inflammatory cytokine expression of severely degenerated NP cells and likely require additional anti-inflammatory treatments. Moderately degenerated NP cells had greater TGFβ receptor 1 expression and better responded to anabolic stimulation.
Keywords: TGFβ, CTGF, Link-N, Degeneration, Nucleus Pulposus, Regenerative Therapy
Introduction
Current surgical treatments for IVD related low back pain such as fusion and disc replacement do not attempt to slow the degenerative process or promote repair. Stimulating native matrix repair in degenerated discs by soluble cell signaling factors is an attractive alternative that would be minimally invasive1. While degenerative grade is thought to be an important factor in targeting therapeutic interventions it remains unknown whether cells in severely degenerated IVDs have an impaired metabolic function compared to lesser degenerative levels or if these differences are associated with the IVD microenvironment. It was hypothesized that targeting an earlier degenerative grade would provide a more effective therapy since NP cells are likely to be more metabolically active and therefore more responsive to a biological stimulus.
Since it is unknown whether degenerative differences are dependent on a chosen stimulatory agent, this study focused on well established anabolic agents to evaluate potential differences in degenerative grades. Therefore a mitogenic/anti-inflammatory cocktail: Transforming Growth Factor Beta 3 (TGFβ3) + Dexamethasone (Dex), and matricellular proteins Connective Tissue Growth Factor (CTGF) and N-terminal peptide of Link Protein (Link-N) were chosen.2-6 Link-N functions to stabilize and protect aggrecan and hyaluronan aggregates7, 8 and stimulates synthesis of proteoglycans and collagens in NP cells in vitro.3, 4 It has also demonstrated potential as an IVD repair in vivo by partially restoring disc height in a rabbit puncture model.9 CTGF binds to aggrecan and enhances production and secretion of aggrecan by chondrocytes.2 NP cells transfected with CTGF increased synthesis of collagen type II and proteoglycans.6 Additionally, CTGF is secreted by notochordal cells10 and is speculated to be part of the limited reparative response of degenerated discs, regulated by TGFβ3.11 TGFβ3 is expressed during development of the IVD12 and enhances matrix development,13, 14 making it an important candidate in the IVD for promoting repair. Dexamethasone is an anti-inflammatory steroid that has been shown to reduce the effects of pro-inflammatory cytokines on human NP cells.15 TGFβ3 and Dex are components of chondrogenic media,16 and in combination have also been shown to stimulate mesenchymal stem cells to adopt an IVD phenotype.17 In addition, TGFβ3+Dex has been shown to stimulate degenerated human NP cells to proliferate, increase cell metabolism, and reduce catabolism.18
Degeneration results in a catabolic phenotype, an increase in cell death, and cellular senescence in the NP.19-21 A successful stimulation of degenerated NP cells therefore must promote genes consistent with a young rather than degenerated NP phenotype, increase cell numbers and metabolic activity, as well as stimulate glycosaminoglycan (GAG) production. To evaluate these outcome measures, a 3D alginate scaffold (to mimic the in situ morphology of the cell) was chosen to compare effects of the three pro-anabolic stimuli (Link-N, CTGF, and TGFβ3+Dex) on cells isolated from moderately and severely degenerated human IVDs.
Materials and Methods
Isolation and culture of Human NP cells
Cells were obtained from human NP tissue and cultured as described previously.18 Briefly, cells were enzymatically released (0.2% protease for 1 hour, 0.2% collagenase for 4 hours, Sigma-Aldrich, St. Louis, MO) from tissue obtained from patients undergoing anterior interbody fusions for low back pain secondary to degenerative disc disease, with institutional review board approval, and graded by the surgeon as either moderately (n=3) or severely (n=3) degenerated (Table 1). Cells were suspended in alginate beads at a density of 2×106cells/mL of 1.2% low viscosity alginate (Sigma-Aldrich). Human NP cells in alginate were cultured in 12 well plates at a density of 10 beads/well in Basal media which was defined as: low glucose DMEM, 1% penicillin/streptomycin, 0.05% fungizone, 2% NaCl/KCl, and 1% Insulin-Transferrin-Selenium (Gibco) containing either 10 ng/mL TGFβ3 (Gibco, recombinant human Cat#PHG9305) and 0.1 μM Dexamethasone18, 100 ng/mL of Link-N (Fisher, MW:1921.38, Cat#BP251001, Sequence: DHLSDNYTLDHDRAI)4, or 100 ng/ml10 of full-length recombinant human CTGF produced and purified as described22. Cells were maintained for 21 days in hypoxia (5% O2, 5% CO2, 37°C) with 2 media changes a week. Dependent variables included DNA content, metabolic activity, histology, GAG content, cell viability, and gene expression profiling.
Table 1.
Population of six patient samples used.
| Age | Sex | Diagnosis | Degenerative Grade | IVD level |
|---|---|---|---|---|
| 27 | F | Degenerative Disc Disease | Moderate | L5/S1 |
| 40 | F | Degenerative Disc Disease | Moderate | L2/L3 |
| 49 | M | Degenerative Disc Disease, L5 Spondylolysis | Moderate | L4/L5 |
| 44 | F | L5 Pars Defect | Severe | L5/S1 |
| 50 | M | Isthmic Spondylolistesis | Severe | L5/S1 |
| 53 | F | Degenerative Disc Disease | Severe | L5/S1 |
Dependent Variables
DNA content of samples was analyzed using the Picogreen dsDNA quantification Kit (Invitrogen) according to the manufacturer's instructions. Cells were dissociated with dissolving buffer (55mM Sodium citrate, 30mM EDTA, 0.15M NaCl), centrifuged, and lysed with 200 μL RLT buffer (Qiagen, Valencia, CA). Samples (10 μL, in duplicate) were incubated with Picogreen dsDNA in a 96-well plate and assessed on the spectrophotometer (extinction 485 nm, emission 528 nm) relative to DNA standards.
The 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT, Sigma Aldrich) assay was used to determine the cell's metabolic activity. Alginate beads were incubated in a 2 mg/mL solution of MTT 23 in DMEM for 4 hours. The cells were released by the dissolving buffer, the cell suspension was centrifuged, supernatant removed, and the cell pellet lysed with 200 μL of DMSO. The solution was read at an optical density of 570nm23 on a spectrophotometer in duplicate.
GAG content was assayed by measuring the amount of sulfated GAG accumulated during culture. The alginate constructs were first dissociated as described above, centrifuged, and 400 μL of a digestion buffer (100mM NaOAc, 10mM EDTA, 10mM L-cystine, 300 μg / mL Papain stock, in distilled water) was added to each pellet and supernatant and incubated for 20 hours at 60°C. The media collected over the culture period, as well as the digests were assessed using the Di-methyl methylene Blue (DMMB) assay with separate chondroitin sulfate standard curves corresponding to the digestion buffer and basal media to correct for background noise caused by the alginate. The results were summed for each group.
Histology was performed by washing the alginate beads 3 times for 30 seconds in HBSS, fixing them in 4 % paraformaldehyde for 30 min and rinsing them 3 times in HBSS for 10 minutes each. Next they were incubated in ½HBSS, ½Tissue-Tek Cryo-OCT Compound for two hours on a slow shaker and frozen at −20C in 100% Tissue-Tek Cryo-OCT Compound. Samples were thawed and polyvinyl media for frozen sections was removed from samples. They were placed in Histogel gel encapsulation media to maintain integrity and then placed in 10% Zinc formalin for 48 hours to fix. They were removed from fixative, rinsed and then dehydrated, and cleared using standard methods. After clearing, samples were infiltrated and polymerized with a hydrophobic acrylic resin. 4 μm thick sections were cut and mounted on silane-coated slides, and stained with either Safranin O24 or prepared for immunohistochemistry. For immunohistochemistry, sections were deplasticized and placed in a mild, non-heating antigen retrieval solution for 45 minutes. Sections were then rinsed with PBS and treated with a serum-free protein block for 10 minutes. After protein block treatment, TGFβRI primary antibody (1:100 dilution, ALK5, Cat#ab31013, Abcam, Cambridge, MA) was applied to sections and incubated overnight at 4°C. After primary antibody incubation, sections were rinsed with PBS and treated with a goat, anti-rabbit HRP conjugated secondary for 30 minutes. The secondary antibody was then rinsed off and DAB chromogen was applied to sections to visualize reaction. For a negative control non-immune rabbit serum was used. All sections were counter-stained with a dilute methylene blue.
Cell viability was performed with a Live/Dead kit (Invitrogen) where Calcein (excitation/ 494nm/ emission 517nm) stains the cytoplasm of live cells green and Ethidium (excitation 528nm/ emission 617nm) stains the DNA of dead cells red. The cells were dissociated from the alginate, re-suspended in 100μl of a 2μM Calcein AM/1μM Ethidium Homodimer-2 solution, and placed onto a microscope slide. Cells were incubated at 37°C in the dark for 30 minutes and visualized.
For assessing changes in gene expression, cells were released by dissociating the alginate in dissolving buffer for 10 minutes. They were washed with the dissolving buffer twice to remove all traces of alginate. Each pellet was lysed (300μl RLT buffer) and stored at −80C (1μL RNASE inhibitor). RNA was extracted, cDNA synthesized and custom RT2 Profiler™SYBR green PCR arrays (SABiosciences, Qiagen, CAPH-0817A) were run by the Vermont Cancer Centre DNA Analysis Facility18, 25. Relative gene expression was calculated using the comparative Ct method normalized to the gene expression of the cells before alginate culture (Day 0) and 3 housekeeping genes (18SrRNA, GAPDH, and ACTB). Genes of statistical significance and large effects were reported in figures, with full gene expression data included in Supplement 1. Since a high ratio of proteoglycan to collagen content is a known marker of a healthy NP phenotype,26 the Aggrecan/Collagen II ratio was also calculated as described previously,27 with a CT value of 40 substituted for the severely degenerated samples treated with Link-N and CTGF since they did not come up after 35 cycles.
Statistics
All data was assessed for normality using the Ryan–Joiner test. A two way ANOVA (factors: degenerative grade and media condition) was used for all dependent variables to determine if there was an effect of media condition or degenerative grade, and whether an interaction existed. For all dependent variables an n=3 for each degenerative grade was used. When there was an effect of media condition or an interaction a Fisher's least significant difference (LSD) test was used to evaluate which groups were significantly different (α=0.05). A one sample t-test or non-parametric sign test (for normally and non-normally distributed data, respectively) was also done on the ΔΔCT values to assess differences from day 0 for media conditions (ΔΔCT=0). All statistics were done with Minitab software (Version 16, State College, PA) and significance was always defined as p< 0.05.
Results
Degeneration related differences in proliferative and metabolic capabilities of human NP cells were determined by stimulating moderately and severely degenerated cells with three established stimulants (TGFβ3+Dex, CTGF, and Link-N). DNA content indicated that severely degenerated cells had a significant decrease in proliferation (Two-way ANOVA, main effect of degenerative grade p=0.004, Figure 1A) compared to moderately degenerated cells. There was also a trend of decreased metabolism (Figure 1B) in the severely degenerated cells treated with the matricellular proteins CTGF and Link-N, indicating distinct degeneration related differences in proliferation and metabolism.
Figure 1. Cells obtained from severely degenerated IVDs had a decreased proliferative capability and an impaired metabolism compared to cells obtained from moderately degenerated IVDs.
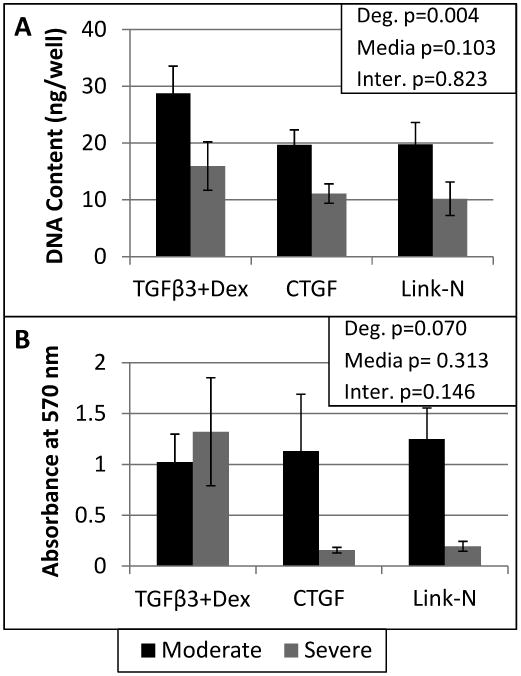
DNA content was determined by the Picogreen assay (A). The MTT assay was used to assess the metabolic activity of the human degenerated NP cells (B). A two way ANOVA was performed with degeneration and media condition as factors, with the results shown in the upper right corner for both assays. Error bars represent SEM.
Stimulating native matrix repair is the goal of an injectable stimulant, and all three stimulants were chosen based on their known anabolic potential. Histology of the alginate beads, showed similar amounts of proteoglycan staining for all groups (Figure 2A). DMMB supported the histology findings, showing substantial GAG produced, with no differences between groups (Figure 2B). However, comparison of gene expression values revealed that aggrecan and collagen II are only upregulated significantly in moderately degenerated cells treated with TGFβ3+Dex (Figure 2C). The Aggrecan/Collagen II ratio also demonstrated a notable trend of lower ratios with severe degeneration (Figure 2D). Therefore the high ratio of proteoglycan/collagen that is demonstrated by healthy NP cells, and would be the goal of an injectable stimulant, was not seen in severely degenerated cells in this study.
Figure 2. All groups created a GAG rich matrix, however an increase in ACAN and COL2A1 and high ACAN/COL2A1 ratios were only demonstrated in moderately degenerated cells.
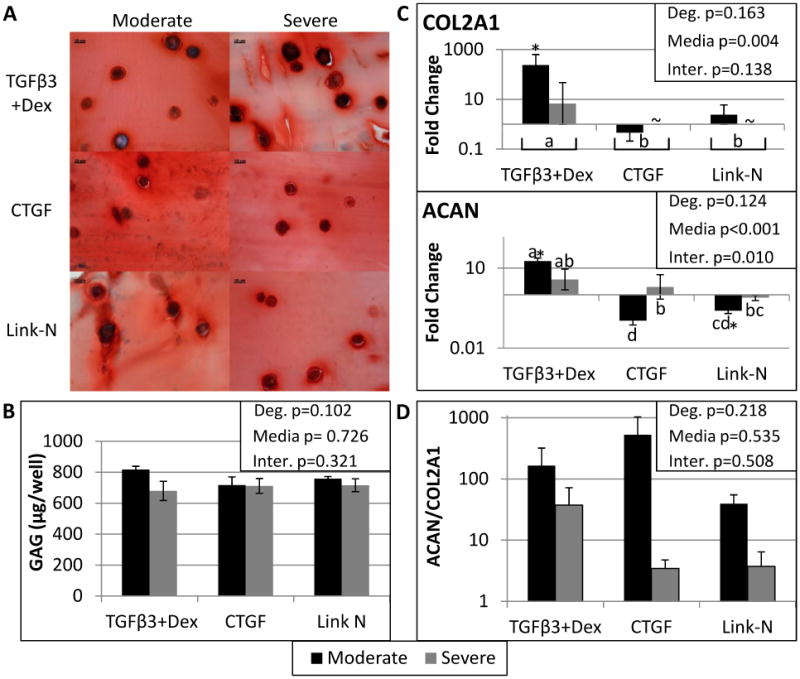
Histology of 10 micron frozen slices through the alginate bead constructs stained with Safranin O at 60X, where the scale bar is 10 microns (A). DMMB analysis was used to quantify the amount of GAG produced by the human degenerated NP cells (B). There were no significant differences in GAG production between media groups or with degree of degeneration. qRT-PCR results for Aggrecan (ACAN) and Collagen type II alpha 1 (COL2A1) (C) are also shown, with ACAN and COL2A1 only significantly upregulated in moderately degenerated cells stimulated with TGFβ3+Dex. Finally, the ratio of relative gene expression was calculated with the formula: 2−(ΔCt ACAN−ΔCt COL2A1). A CT value of 40 was substituted for COL2A1 for the severely degenerated samples treated with Link-N and CTGF since there was nothing detected after 35 cycles of qRT-PCR. A two way ANOVA was performed with degeneration and media condition as factors, with the results shown in the upper right corner for all assays. Treatment groups with different letters are significantly different (Fisher's post hoc test α=0.05 was run for each media condition with pooled degenerative grades or when there was a significant interaction between media and degeneration a post hoc test was run with all six groups). ∼ indicates the gene was not expressed by that group. Student's t-test indicated significant differences from day 0 represented by a *. Significance was always defined as p<0.05. Error bars represent SEM.
Other gene expression differences between degenerative grades were evident. Day 0 gene expression levels of moderately degenerated cells were compared to the severely degenerated subjects, demonstrating four genes that were different at the beginning of culture (SOX9 and COL2A1 were significantly down-regulated and MMP1 and MMP2 were significantly up-regulated in severely degenerated cells compared to moderately degenerate cells). After 21 days of culture, degenerative grade was a significant factor (Two way ANOVA, main effect of degeneration p<0.05) for MMP3, MMP9, and IL1β (Figure 3). MMP3 was more up-regulated in moderately degenerated cells, while MMP9 and IL1β were more up-regulated in severely degenerated cells. Other genes that demonstrated significant differences between moderately and severely degenerated cells (Two way ANOVA, main effect of degeneration p<0.05) were: BGN, CASP3, HAS1, LAMB1, MMP14, MMP2, NGF, PDGFA, TAC4, TGFβR1, TGFβR2, and TIMP2 (Table 2). Specifically HAS1 was up-regulated in moderately degenerated cells, while BGN was more upregulated with severe degeneration (Figure 4). Of note for severely degenerated NP cells was the lack of detectable mRNA signals for 18 genes measured for both Link-N and CTGF groups (Table 2), including TGFβR1 (ALK5).
Figure 3. Catabolic and Pro-inflammatory cytokine related gene expression.
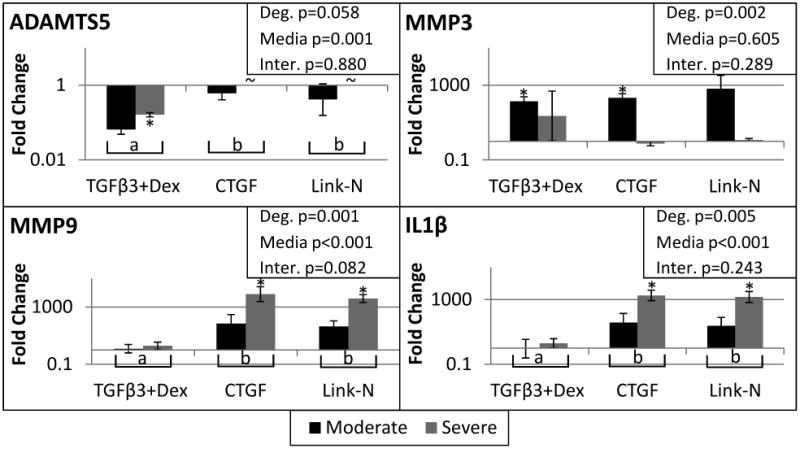
qRT-PCR results for A disintegrin and metalloproteinase with thrombospondin motifs 5 (ADAMTS5), matrix metalloproteinase 3 (MMP3), matrix metalloproteinase 9 (MMP9), and Interleukin 1 Beta (IL1β). Student's t-test indicated significant differences from day 0 represented by a *. A two way ANOVA was performed with degeneration and media condition as factors, with the results shown in the upper right corner for each gene. Treatment groups with different letters are significantly different (Fisher's post hoc test α=0.05 was run for each media condition with pooled degenerative grades). ∼ indicates the gene was not expressed by that group. Significance was always defined as p<0.05, error bars represent SEM.
Table 2.
Complete fold change gene expression data for moderately and severely degenerated cells stimulated with TGFβ3+Dex, CTGF, and Link-N. The gene expression was normalized to 18s, GAPDH and β-actin, and Day 0 cells. Red indicates a significant decrease in gene expression and blue indicates a significant increase in gene expression, p<0.05. ∼ indicates the gene was not expressed by that group. Of particular note was the large number of genes that were not expressed in the severely degenerated cells following stimulation by either of the matricellular proteins.
| Moderate Degeneration (n=3) | Severe Degeneration (n=3) | |||||
|---|---|---|---|---|---|---|
| TGF(β3+Dex | CTGF | Link-N | TGF(β3+Dex | CTGF | Link-N | |
| ADAMTS4 | 1.25 | 3.49 | ∼ | −1.21 | ∼ | ∼ |
| ADAMTS5 | −15.41 | −1.66 | −2.43 | −6.21 | ∼ | ∼ |
| ACAN | 18.48 | −9.34 | −3.99 | 3.78 | 1.98 | −1.28 |
| BDNF | 2.13 | 5.58 | 19.45 | 2.55 | ∼ | 4.15 |
| BGN | 9.63 | −1.15 | −2.52 | 10.75 | 8.22 | 6.48 |
| CASP3 | 1.96 | 4.39 | 9.56 | 1.71 | ∼ | ∼ |
| COL1A1 | −83.56 | −6.68 | −36.04 | −27.04 | 1.29 | −5.60 |
| COL10A1 | 26.64 | 114.70 | 59.94 | 13.95 | 382.38 | 413.02 |
| COL2A1 | 237.92 | −2.17 | 2.44 | 6.88 | ∼ | ∼ |
| COL3A1 | −1.65 | 1.00 | −1.24 | −2.01 | ∼ | −1.74 |
| CTGF | −2.92 | −10.23 | −10.53 | −4.26 | −1.10 | −9.50 |
| EGF | 8.80 | 38.86 | 17.20 | 6.90 | 218.02 | 32.84 |
| eln | 10.67 | 1.41 | −1.54 | 6.34 | ∼ | ∼ |
| FGF1 | −2.54 | −1.18 | 3.69 | −1.17 | 3.19 | ∼ |
| GPC1 | 2.34 | 2.82 | 2.78 | 1.39 | 1.58 | 1.28 |
| HAS1 | 9.31 | 13.42 | 67.22 | 3.77 | ∼ | ∼ |
| IGF1 | ∼ | 4.20 | 1.45 | 1.30 | ∼ | ∼ |
| IL1B | −1.08 | 37.85 | 23.89 | 2.01 | 1826.45 | 1467.56 |
| KRT19 | −5.28 | 1.07 | −1.13 | −2.92 | ∼ | ∼ |
| LAMB1 | 5.49 | 2.57 | 3.53 | 1.85 | −1.15 | −1.06 |
| MMP1 | 11.64 | 150.51 | 850.20 | 2.28 | 10.57 | 17.83 |
| MMP13 | 4.26 | 5.95 | 13.77 | 1.47 | 3.63 | ∼ |
| MMP14 | 2.05 | 1.65 | 5.48 | −1.89 | −1.52 | −2.23 |
| MMP2 | 2.44 | 6.80 | 5.02 | 1.54 | 1.59 | 1.42 |
| MMP3 | 138.53 | 209.03 | 653.21 | 22.82 | −1.32 | 1.19 |
| MMP9 | 1.23 | 70.46 | 44.54 | 1.96 | 7868.17 | 3831.99 |
| NGF | −2.02 | 3.79 | 8.25 | −3.72 | ∼ | 1.10 |
| BGLAP | 3.02 | 4.27 | 13.13 | 1.82 | 3.85 | 3.76 |
| PDGFA | 2.96 | 2.93 | 10.98 | 2.65 | ∼ | ∼ |
| PPARG | 5.78 | 3.81 | 16.53 | 4.74 | 14.24 | 4.19 |
| SOX9 | 8.31 | 4.33 | 19.88 | 15.51 | 75.65 | 28.61 |
| TAC4 | 14.29 | 16.69 | 18.59 | 14.27 | ∼ | ∼ |
| TGFB1 | 3.56 | 2.85 | 4.37 | 1.59 | 7.11 | 12.06 |
| TGFB2 | −4.30 | −1.70 | −1.45 | 1.40 | −1.80 | ∼ |
| TGFB3 | −3.49 | 2.50 | 1.93 | −2.51 | ∼ | 2.80 |
| TGFBR1 | 9.81 | 8.77 | 22.49 | 1.41 | ∼ | ∼ |
| TGFBR2 | 1.96 | 1.32 | 2.60 | 1.44 | ∼ | ∼ |
| TIMP1 | −1.18 | 2.30 | 4.37 | −1.07 | 4.69 | 6.43 |
| TIMP2 | 1.95 | 2.12 | 2.52 | 1.32 | 28.54 | 10.62 |
| TIMP3 | ∼ | ∼ | ∼ | ∼ | ∼ | ∼ |
| TNFα | 1.95 | 10.82 | 7.07 | 2.06 | ∼ | ∼ |
| WISP1 | 1.15 | 1.86 | 1.99 | 1.29 | −1.56 | ~ |
Figure 4. Hyaluronan synthase is upregulated more in moderately degenerated cells, while Biglycan is upregulated more in severely degenerated cells.
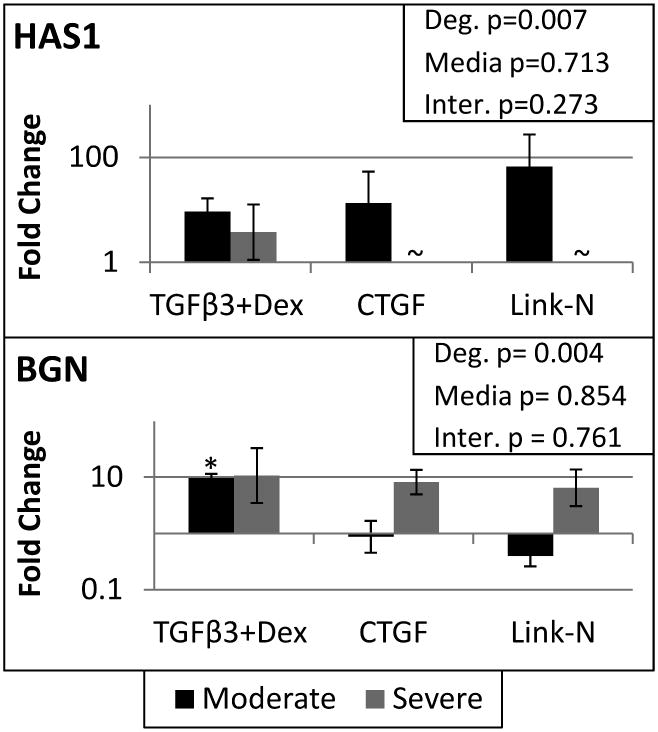
qRT-PCR results for hyaluronan synthase 1 (HAS1) and biglycan (BGN). Student's t-test indicated a significant difference from day 0 represented by a *. A two way ANOVA was performed with degeneration and media condition as factors, with the results shown in the upper right corner for each gene. Treatment groups with different letters are significantly different (Fisher's post hoc test α=0.05 was run for each media condition with pooled degenerative grades or when there was a significant interaction between media and degeneration a post hoc test was run with all six groups). ∼ indicates the gene was not expressed by that group. Significance was always defined as p<0.05, error bars represent SEM.
Since TGFβR1 was speculated to be related to the decreased expression of the other genes, the gene expression results were verified with protein immunohistochemistry of TGFβR1 (ALK5). All groups exhibited positive immunostaining for TGFβR1; however, there was reduced immunostaining for the severely as compared to the moderately degenerated groups, in a manner consistent with qRT-PCR findings (Figure 5). The reduction in immunostaining with degeneration was related to reduced intensity of TGFβR1 staining.
Figure 5. TGFβR1 is more upregulated in moderately degenerate cells compared to severely degenerate cells.
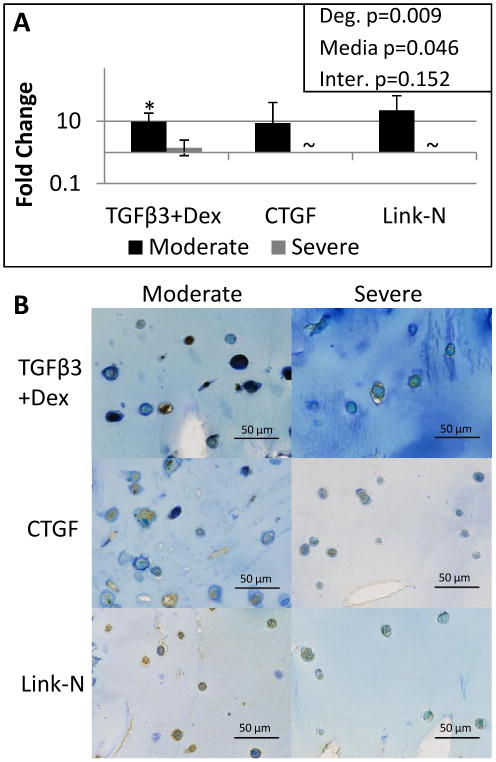
qRT-PCR results for TGFβR1. Student's t-test indicated a significant difference from day 0 represented by a *. A two way ANOVA was performed with degeneration and media condition as factors, with the results shown in the upper right corner. ∼ indicates the gene was not expressed by that group. Significance was always defined as p<0.05, error bars represent SEM. To confirm gene expression levels, histology of 10 micron frozen slices through the alginate bead constructs were stained with TGFβR1 and imaged at 40X, where the scale bar is 50 microns (B). All groups exhibited positive immunostaining for TGFβR1; however, there was reduced immunostaining for the severely degenerated groups, in a manner consistent with qRT-PCR findings.
Significant differences from day 0 demonstrated that the stimulants had some differential effects on the cells (Table 2). TGFβ3+Dex, in particular increased gene expression of SOX9 (moderate p<0.001 and severe p=0.033), COL2A1 (moderate p=0.030), ACAN (moderate p=0.008) and GPC1 (moderate p=0.019), and downregulated COL1A1 (moderate p=0.003 and severe p=0.019) and ADAMTS5 (severe p=0.005). CTGF and Link-N on the other hand upregulated MMP9 (CTGF p=0.017, Link-N p=0.006), IL1β (CTGF =0.010, Link-N p=0.011) and COL10A1 (CTGF p=0.019, Link-N p=0.017) in severely degenerated cells.
Discussion
This study evaluated the stimulatory potential of three pro-anabolic agents on moderately and severely degenerated human NP cells isolated from surgical tissue, to screen their potential use for treating IVD degeneration. The goal was to counter the degeneration related shift towards catabolism, reduced cellularity, and decreased metabolic rates, and to evaluate whether degree of degeneration affects the cellular response to stimulus. As hypothesized, moderately degenerated cells demonstrated greater stimulatory potential than severely degenerated cells by having an increased proliferative capacity, a limited pro-inflammatory response, and a gene expression profile more consistent with remodeling rather than degeneration associated matrix breakdown and proinflammatory cytokines (Table 3 for summary of results).
Table 3.
Summary of results.
| Characteristics of a healthy phenotype | Moderate | Severe | ||||
|---|---|---|---|---|---|---|
| TGFβ3+Dex | CTGF | Link-N | TGFβ3+Dex | CTGF | Link-N | |
| Proliferative capability | ++ | ++ | ++ | + | − | − |
| High metabolic rate | ++ | ++ | ++ | ++ | − | − |
| GAG rich matrix | ++ | ++ | ++ | ++ | ++ | ++ |
| High ACAN/COL2A1 ratio | ++ | ++ | ++ | + | − | − |
| Low MMP9 expression | − | + | + | − | ++ | ++ |
| Low Il1β expression | − | + | + | − | ++ | ++ |
| TGFβR1 expression | ++ | ++ | ++ | + | − | − |
Qualitative grading: ++ highest increase, + moderate increase, − negligible increase or smallest effect
Italics indicates no differences were observed between groups.
 indicates an assay where a negligible increase or small effect demonstrates an effective stimulus.
indicates an assay where a negligible increase or small effect demonstrates an effective stimulus.
Severely degenerated NP cells had impaired proliferative capacity, metabolic activity, and ratio of ACAN/COL2A1 as compared to moderately degenerated NP cells, indicating that cell differences are as important as microenvironment differences with degeneration. Proliferative capacity, metabolic activity and ratio of ACAN/COL2A1 are all important criteria for cellular responsiveness of an IVD relevant therapy,18, 27 since degeneration is marked by an increase in cell death and cellular senescence20, 21 and a healthy NP is characterized by a high ratio of proteoglycan to collagen.26 As expected from NP cells treated with anabolic stimuli, DMMB and safranin O staining demonstrated a GAG and proteoglycan rich matrix, respectively, for all groups. DMMB measures sulfated GAGs (not aggrecan), and with age there is an increase in keratan sulfated GAGs at the expense of chondroitin sulfated side chains,28 which could potentially account for the observed similarities for moderately and severely degenerated cells. Safranin O, likewise, does not discriminate between keratin sulfated and chondroitin sulfated side chains.29 The lower ratio of ACAN/COL2A1 for severely degenerated cells indicated that there could be altered ratios of matrix production that are dependent on the degenerative state of the cells.
Severely degenerated cells had several gene expression patterns that were distinct from moderately degenerated NP cells after 21 days in an alginate scaffold. MMP9, which is related to IVD damage30 and is present in degenerated IVDs,31 was up-regulated to a greater extent in severely degenerated cells. However, although degeneration is associated with MMP expression and activity;32 certain MMPs including MMP1, MMP2, MMP3, and MMP14, are also present during development in human fetal IVDs.33 Interestingly, in this study MMP2, MMP3, and MMP14 were more up-regulated in the moderately degenerated cells and may be representative of a remodeling response rather than catabolism. Biglycan gene expression was up-regulated with severe degeneration, while hyaluronan synthase and laminin gene expression were expressed at lower levels with severe degeneration. Unlike other proteoglycans of the IVD, biglycan gene expression is increased with advancing degeneration,34 and is also expressed more in adult NP cells compared to notochordal cells.35 Therefore, this study continues to support the finding that increased biglycan expression is a characteristic of degenerative changes in NP cellular responses.
Severely degenerated cells had a large number of genes that were not detectable with qRT-PCR under certain media conditions. In particular, both matricellular proteins did not stimulate TGFβR1 (ALK5) gene expression in severely degenerated cells. This was in contrast to the upregulation of these receptors in the moderately degenerated cells with particularly intense immunostaining. The decrease in ALK5 expression and reduced immunostaining in severely degenerated cells is consistent with the observed shift from the protective ALK5 toward the destructive ALK1 in cartilage with aging and during OA36, suggesting the comparison of ALK1 and ALK5 may be important in future work. Furthermore, IL-1β and MMP9 were up-regulated to a greater extent in severely degenerated cells when the receptor was not expressed. Taken together these results suggest that severely degenerated NP cells have an altered responsiveness to pro-anabolic stimulants with enhanced pro-inflammatory and catabolic phenotypes when expression of TGFβR1 (ALK5) is diminished.
TGFβ3+Dex, CTGF, and Link-N were chosen based on their anabolic potential.2, 4, 13 Of all the groups analyzed, TGFβ3+Dex had the greatest effect on moderately degenerated NP cells, increasing proliferation, up-regulating anabolic gene expression (ACAN, COL2A1, SOX9, BGN, and GPC1), and decreasing fibrotic COL1A1 and catabolic ADAMTS5 gene expression. The matricellular proteins Link-N and CTGF showed remarkably similar trends in gene expression and proliferation to each other, but increased IL1β expression relative to the nascent degenerative state and consequently exhibited a higher catabolic response than the TGFβ3+Dex group. The increased IL1β expression with Link-N and CTGF in severely degenerated NP cells suggests that matricellular protein treatments may require additional supplementation with an anti-inflammatory, such as Dex, to counter the increased NFκB signaling of severely degenerated NP cells.37.
Some limitations impact the interpretation of the findings in this study. We chose to use day 0 as the most relevant baseline control to compare alterations in mRNA expression relative to the degenerated state, as we were comparing moderate vs severe degeneration and not concerned with a comparison between media groups. Several significant effects of grade of degeneration were detected, yet with 3 human subjects per degenerative level there is limited statistical power to conclude lack of degeneration differences. The data suggests that the TGFβ3+Dex cocktail had the greatest effect, particularly on moderately degenerated cells which may be due to combined anabolic and anti-inflammatory effects of TGFβ3+Dex. Therefore future studies investigating synergistic effects of these pro-anabolic matricellular proteins with Dex or other inhibitors of NFκB, and growth factors, are necessary to demonstrate further potential of matricellular proteins as an augmenting treatment for severely degenerated IVDs.
We conclude that severely degenerated NP cells had impaired gene expression patterns, proliferative capacity and metabolic rates as compared with moderately degenerated NP cells indicating degenerative grade is an important factor when considering cellular responsiveness to pro-anabolic stimulants. Furthermore, it was demonstrated that severely degenerated NP cells had impaired function and diminished TGFβ responsiveness irrespective of microenvironmental changes due to matrix and nutritional changes the cells experience in vivo. When an anabolic stimulant was combined with an anti-inflammatory agent (Dex), cells were able to overcome pro-inflammatory and catabolic phenotypes, suggesting anabolic stimulants should be paired with anti-inflammatory supplements for increased responsiveness.
Acknowledgments
We gratefully acknowledge Ilana Stock for invaluable technical assistance with histological imaging.
Source of Funding: This work supported by grants from the NIH (R21AR056037 & R01AR051146) and by the AO Research Fund (project F-09-10I) of the AO Foundation.
Footnotes
Conflicts of Interest: The authors do not have any conflicts of interest to disclose.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Masuda K, An HS. Prevention of disc degeneration with growth factors. Eur Spine J. 2006;15(3):S422–32. doi: 10.1007/s00586-006-0149-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Aoyama E, Hattori T, Hoshijima M, et al. N-terminal domains of CCN family 2/connective tissue growth factor bind to aggrecan. Biochem J. 2009;420:413–20. doi: 10.1042/BJ20081991. [DOI] [PubMed] [Google Scholar]
- 3.Petit A, Yao G, Rowas SA, et al. Effect of Synthetic Link N Peptide on the Expression of Type I and Type II Collagens in Human Intervertebral Disc Cells. Tissue Eng Part A. 2010 doi: 10.1089/ten.TEA.2010.0494. [DOI] [PubMed] [Google Scholar]
- 4.Mwale F, Demers CN, Petit A, et al. A synthetic peptide of link protein stimulates the biosynthesis of collagens II, IX and proteoglycan by cells of the intervertebral disc. J Cell Biochem. 2003;88:1202–13. doi: 10.1002/jcb.10479. [DOI] [PubMed] [Google Scholar]
- 5.Liu H, McKenna LA, Dean MF. An N-terminal peptide from link protein can stimulate biosynthesis of collagen by human articular cartilage. Arch Biochem Biophys. 2000;378:116–22. doi: 10.1006/abbi.2000.1758. [DOI] [PubMed] [Google Scholar]
- 6.Liu Y, Kong J, Chen BH, Hu YG. Combined expression of CTGF and tissue inhibitor of metalloprotease-1 promotes synthesis of proteoglycan and collagen type II in rhesus monkey lumbar intervertebral disc cells in vitro. Chin Med J (Engl) 2010;123:2082–7. [PubMed] [Google Scholar]
- 7.Neame PJ, Barry FP. The link proteins. Experientia. 1993;49:393–402. doi: 10.1007/BF01923584. [DOI] [PubMed] [Google Scholar]
- 8.Rodriguez E, Roughley P. Link protein can retard the degradation of hyaluronan in proteoglycan aggregates. Osteoarthritis Cartilage. 2006;14:823–9. doi: 10.1016/j.joca.2006.02.008. [DOI] [PubMed] [Google Scholar]
- 9.Mwale F, Masuda K, Pichika R, et al. The efficacy of Link N as a mediator of repair in a rabbit model of intervertebral disc degeneration. Arthritis Res Ther. 2011;13:R120. doi: 10.1186/ar3423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Erwin WM, Ashman K, O'Donnel P, Inman RD. Nucleus pulposus notochord cells secrete connective tissue growth factor and up-regulate proteoglycan expression by intervertebral disc chondrocytes. Arthritis Rheum. 2006;54:3859–67. doi: 10.1002/art.22258. [DOI] [PubMed] [Google Scholar]
- 11.Tran CM, Markova D, Smith HE, et al. Regulation of CCN2/connective tissue growth factor expression in the nucleus pulposus of the intervertebral disc: role of Smad and activator protein 1 signaling. Arthritis Rheum. 2010;62:1983–92. doi: 10.1002/art.27445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pelton RW, Dickinson ME, Moses HL, Hogan BL. In situ hybridization analysis of TGF beta 3 RNA expression during mouse development: comparative studies with TGF beta 1 and beta 2. Development. 1990;110:609–20. doi: 10.1242/dev.110.2.609. [DOI] [PubMed] [Google Scholar]
- 13.Hiyama A, Gogate SS, Gajghate S, Mochida J, Shapiro IM, Risbud MV. BMP-2 and TGF-beta stimulate expression of beta1,3-glucuronosyl transferase 1 (GlcAT-1) in nucleus pulposus cells through AP1, TonEBP, and Sp1: role of MAPKs. J Bone Miner Res. 2010;25:1179–90. doi: 10.1359/jbmr.091202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Reza AT, Nicoll SB. Serum-free, chemically defined medium with TGF-beta(3) enhances functional properties of nucleus pulposus cell-laden carboxymethylcellulose hydrogel constructs. Biotechnology and bioengineering. 2010;105:384–95. doi: 10.1002/bit.22545. [DOI] [PubMed] [Google Scholar]
- 15.Jee BK, Surendran S, Park KM, et al. Role of tumor necrosis factor-alpha, interleukin-8, and dexamethasone in the focal adhesion kinase expression by human nucleus pulposus cells. Spine (Phila Pa 1976) 2007;32:30–5. doi: 10.1097/01.brs.0000250997.24617.a4. [DOI] [PubMed] [Google Scholar]
- 16.Boeuf S, Richter W. Chondrogenesis of mesenchymal stem cells: role of tissue source and inducing factors. Stem Cell Res Ther. 2010;1:31. doi: 10.1186/scrt31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Steck E, Bertram H, Abel R, Chen B, Winter A, Richter W. Induction of intervertebral disc-like cells from adult mesenchymal stem cells. Stem Cells. 2005;23:403–11. doi: 10.1634/stemcells.2004-0107. [DOI] [PubMed] [Google Scholar]
- 18.Abbott RD, Purmessur D, Monsey RD, Iatridis JC. Regenerative potential of TGFbeta3 + Dex and notochordal cell conditioned media on degenerated human intervertebral disc cells. J Orthop Res. 2011 doi: 10.1002/jor.21534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Le Maitre CL, Freemont AJ, Hoyland JA. Localization of degradative enzymes and their inhibitors in the degenerate human intervertebral disc. J Pathol. 2004;204:47–54. doi: 10.1002/path.1608. [DOI] [PubMed] [Google Scholar]
- 20.Gruber HE, Ingram JA, Norton HJ, Hanley EN., Jr Senescence in cells of the aging and degenerating intervertebral disc: immunolocalization of senescence-associated beta-galactosidase in human and sand rat discs. Spine (Phila Pa 1976) 2007;32:321–7. doi: 10.1097/01.brs.0000253960.57051.de. [DOI] [PubMed] [Google Scholar]
- 21.Le Maitre CL, Freemont AJ, Hoyland JA. Accelerated cellular senescence in degenerate intervertebral discs: a possible role in the pathogenesis of intervertebral disc degeneration. Arthritis Res Ther. 2007;9:R45. doi: 10.1186/ar2198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ball DK, Moussad EE, Rageh MA, Kemper SA, Brigstock DR. Establishment of a recombinant expression system for connective tissue growth factor (CTGF) that models CTGF processing in utero. Reproduction. 2003;125:271–84. doi: 10.1530/rep.0.1250271. [DOI] [PubMed] [Google Scholar]
- 23.Chen J, Yan W, Setton LA. Static compression induces zonal-specific changes in gene expression for extracellular matrix and cytoskeletal proteins in intervertebral disc cells in vitro. Matrix Biol. 2004;22:573–83. doi: 10.1016/j.matbio.2003.11.008. [DOI] [PubMed] [Google Scholar]
- 24.Lillie RD. Histopathologic Technic and Practical Histochemistry. New York, NY: McGraw-Hill Inc.; 1976. [Google Scholar]
- 25.Purmessur D, Schek RM, Abbott RD, Ballif BA, Godburn KE, Iatridis JC. Notochordal conditioned media from tissue increases proteoglycan accumulation and promotes a healthy nucleus pulposus phenotype in human mesenchymal stem cells. Arthritis Res Ther. 2011;13:R81. doi: 10.1186/ar3344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mwale F, Roughley P, Antoniou J. Distinction between the extracellular matrix of the nucleus pulposus and hyaline cartilage: a requisite for tissue engineering of intervertebral disc. Eur Cell Mater. 2004;8:58–63. doi: 10.22203/ecm.v008a06. discussion -4. [DOI] [PubMed] [Google Scholar]
- 27.Gantenbein-Ritter B, Benneker LM, Alini M, Grad S. Differential response of human bone marrow stromal cells to either TGF-beta(1) or rhGDF-5. Eur Spine J. 2011;20:962–71. doi: 10.1007/s00586-010-1619-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Scott JE, Bosworth TR, Cribb AM, Taylor JR. The chemical morphology of age-related changes in human intervertebral disc glycosaminoglycans from cervical, thoracic and lumbar nucleus pulposus and annulus fibrosus. Journal of anatomy. 1994;184(Pt 1):73–82. [PMC free article] [PubMed] [Google Scholar]
- 29.Camplejohn KL, Allard SA. Limitations of safranin ‘O’ staining in proteoglycan-depleted cartilage demonstrated with monoclonal antibodies. Histochemistry. 1988;89:185–8. doi: 10.1007/BF00489922. [DOI] [PubMed] [Google Scholar]
- 30.Loreto C, Musumeci G, Castorina A, Martinez G. Degenerative disc disease of herniated intervertebral discs is associated with extracellular matrix remodeling, vimentin-positive cells and cell death. Ann Anat. 2011;193:156–62. doi: 10.1016/j.aanat.2010.12.001. [DOI] [PubMed] [Google Scholar]
- 31.Roberts S, Caterson B, Menage J, Evans EH, Jaffray DC, Eisenstein SM. Matrix metalloproteinases and aggrecanase: their role in disorders of the human intervertebral disc. Spine (Phila Pa 1976) 2000;25:3005–13. doi: 10.1097/00007632-200012010-00007. [DOI] [PubMed] [Google Scholar]
- 32.Le Maitre CL, Pockert A, Buttle DJ, Freemont AJ, Hoyland JA. Matrix synthesis and degradation in human intervertebral disc degeneration. Biochem Soc Trans. 2007;35:652–5. doi: 10.1042/BST0350652. [DOI] [PubMed] [Google Scholar]
- 33.Rutges JP, Nikkels PG, Oner FC, et al. The presence of extracellular matrix degrading metalloproteinases during fetal development of the intervertebral disc. Eur Spine J. 2010;19:1340–6. doi: 10.1007/s00586-010-1378-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Cs-Szabo G, Ragasa-San Juan D, Turumella V, Masuda K, Thonar EJ, An HS. Changes in mRNA and protein levels of proteoglycans of the anulus fibrosus and nucleus pulposus during intervertebral disc degeneration. Spine (Phila Pa 1976) 2002;27:2212–9. doi: 10.1097/00007632-200210150-00006. [DOI] [PubMed] [Google Scholar]
- 35.Chen J, Yan W, Setton LA. Molecular phenotypes of notochordal cells purified from immature nucleus pulposus. Eur Spine J. 2006;15(3):S303–11. doi: 10.1007/s00586-006-0088-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Blaney Davidson EN, Remst DF, Vitters EL, et al. Increase in ALK1/ALK5 ratio as a cause for elevated MMP-13 expression in osteoarthritis in humans and mice. J Immunol. 2009;182:7937–45. doi: 10.4049/jimmunol.0803991. [DOI] [PubMed] [Google Scholar]
- 37.Wuertz K, Vo N, Kletsas D, Boos N. Inflammatory and catabolic signalling in intervertebral discs: the roles of NF-kappaB and MAP kinases. Eur Cell Mater. 2012;23:103–19. doi: 10.22203/ecm.v023a08. discussion 19-20. [DOI] [PubMed] [Google Scholar]


