Abstract
The let-7 microRNA (miRNA) is highly conserved across animal phyla and generally regulates cellular differentiation and developmental timing pathways. In C. elegans, the mature let-7 miRNA starts to accumulate in the last stages of larval development where it directs cellular differentiation programs required for adult fates. Here we show that expression of the let-7 gene in C. elegans is under complex transcriptional control. The onset of let-7 transcription begins as early as the first larval stage in some tissues, and as late as the third larval stage in others, and is abrogated at the gravid adult stage. Transcription from two different start sites in the let-7 promoter oscillates during each larval stage. We show that transcription is regulated by two distinct cis-elements in the promoter of let-7, the previously described temporal regulatory element (TRE), and a novel element downstream of the TRE that we have named the let-7 transcription element (LTE). These elements play distinct and redundant roles in regulating let-7 expression in specific tissues. In the absence of the TRE and LTE, transcription of let-7 is undetectable and worms exhibit the lethal phenotype characteristic of let-7 null mutants. We also identify several genes that affect the transcription of let-7 generally and tissue-specifically. Overall, spatio-temporal regulation of let-7 transcription is orchestrated by multiple cis- and trans-acting factors to ensure appropriate expression of this essential miRNA during worm development.
Keywords: let-7, C. elegans, microRNA, molting, developmental timing
Introduction
MicroRNAs (miRNAs) are ~22 nucleotide (nt) non-coding RNAs that regulate gene expression by imperfectly base pairing with sequences in target mRNAs, resulting in translational repression or degradation (Huntzinger and Izaurralde, 2011; Pasquinelli, 2012). Misregulation of miRNA expression has been implicated in a wide range of diseases (Sayed and Abdellatif, 2011), highlighting the importance of understanding the control of miRNA biogenesis at the transcriptional and post-transcriptional levels. Abnormal expression of the let-7 miRNA in particular has been linked to numerous human afflictions, particularly cancer (Boyerinas and Sipkins, 2010). The sequence of let-7 miRNA is perfectly conserved across much of bilaterian phylogeny (Pasquinelli et al., 2000). The temporal expression of the mature let-7 miRNA is also highly conserved: mature let-7 is expressed in most differentiated cell types but is undetectable in healthy, undifferentiated stem cells (Mondol and Pasquinelli, 2012). In humans, let-7 miRNAs function as tumor suppressors by regulating cell cycle genes, and misregulation of let-7 has been implicated in breast, colon and lung cancer (Boyerinas and Sipkins, 2010).
In the general miRNA biogenesis pathway, RNA polymerase II transcribes primary miRNA (pri-miRNA) transcripts that are typically hundreds to thousands of nt long (Kim et al., 2009). Drosha, an RNAse III enzyme, excises the ~70 nt precursor miRNA (pre-miRNA) from the pri-miRNA nascent transcript. The next step of maturation involves Dicer, another RNAse III enzyme, which cuts a ~22 nt duplex from the hairpin; one strand will become the mature guide and the other will be discarded as the passenger strand. The single-stranded mature miRNA is then bound by an Argonaute protein, forming the miRNA Induced Silencing Complex (miRISC). Through imperfect binding, miRNAs serve as guide molecules for miRISC complexes to bind specific target mRNAs, usually in the 3’ UTR resulting in reduced target expression (Huntzinger and Izaurralde, 2011; Pasquinelli, 2012).
In C. elegans three primary let-7 transcripts are expressed from the let-7 locus: ~1730 nt transcripts from the A start site and ~890 nt transcripts from the B start site (B) and one or both of these species undergo trans-splicing to create the third isoform of ~750 nt (SL1) (Bracht et al., 2004). Although all three transcripts are detectable in each larval stage, the precursor and mature let-7 miRNA do not accumulate until the third larval stage (L3) (Van Wynsberghe et al., 2011). Early in development, the RNA binding protein, LIN-28, co-transcriptionally associates with pri-let-7 transcripts and prevents Drosha processing (Van Wynsberghe et al., 2011). Regulation of let-7 biogenesis by Lin28 also occurs in mammalian cells where Lin28 binds both primary and precursor let-7 and negatively regulates their processing or stability (Thornton and Gregory, 2012). Inhibition of let-7 biogenesis is relieved upon the expression of miRNAs that target Lin28 for down-regulation, which occurs as stem cells undergo differentiation and as worms progress to later larval stages.
Transcriptional regulation of the C. elegans let-7 gene has been previously studied by fusing the region upstream of the mature miRNA to green fluorescent protein (GFP) coding sequences. These reporters indicated that transcription might start as early as embryogenesis and continue into adulthood (Esquela-Kerscher et al., 2005; Johnson et al., 2003; Martinez et al., 2008b). These transcriptional reporters were expressed in most somatic cell types with the hypodermal seam and anchor cells showing temporally regulated expression starting in L3 (Esquela-Kerscher et al., 2005; Johnson et al., 2003). The temporal regulatory element (TRE) was identified as a promoter region important for hypodermal seam cell expression and for full rescue activity of the let-7 gene (Johnson et al., 2003). The transcription factors, HBL-1 and DAF-12, have been reported to directly regulate the expression of let-7. HBL-1 represses transcription of let-7 in the hypodermal seam and vulval precursor cells until the third larval stage (Roush and Slack, 2009). DAF-12 is a nuclear hormone receptor transcription factor that negatively or positively regulates the expression of let-7 depending upon its ligand bound status (Bethke et al., 2009; Hammell et al., 2009). Additionally, using yeast one-hybrid analysis of the promoter regions of C. elegans miRNAs, one group found that 14 different transcription factors bound to the promoter of let-7 (Martinez et al., 2008a). The importance of these factors and potentially additional promoter elements for the transcription of let-7 in vivo is yet to be determined.
To identify cis- and trans-acting factors required for the endogenous let-7 transcriptional expression pattern, we created reporters based on the mapped transcription start sites. We show that the let-7 primary transcripts are first detected at the end of the first larval stage and exhibit pulses of expression near the molt of each larval stage. Our reporters recapitulate this oscillating pattern, indicating that cycling of primary let-7 during each larval stage is largely regulated at the transcriptional level. Through promoter deletion analyses, we identified two cis-elements required for let-7 transcription. In agreement with Johnson et al., 2003, we found that the TRE is necessary for hypodermal seam cell expression (Johnson et al., 2003), and also that it regulates expression from the A start site. We identified a new element, the let-7 transcription element (LTE), which regulates expression from the B start site and is required for transcription in intestinal cells. The TRE and LTE appear to be redundant for promoting transcription in other cell types, such as neuronal and muscle, since removal of both but not either element alone completely abrogates transcription of let-7 and, hence, rescue activity of let-7 transgenes. Using RNAi against transcription factors in our transgenic GFP animals, we have found several positive and negative regulators of let-7 transcription. Some of these regulators appear to act globally on let-7 transcription, while others have tissue specific effects. Overall, our results indicate that multiple protein factors and two distinct promoter elements regulate transcription of let-7 in different tissues and at different times during development.
Materials and Methods
Nematode strains
C. elegans were grown at 25°C and synchronized using standard hypochlorite treatment. At time zero, starved L1 animals were plated on bacteria expressing OP50, and then collected at selected time points based on previously published time course analyses of worm development and molting at 25°C (Hirsh et al., 1976; Jeon et al., 1999; Zisoulis et al., 2012). Strains used in this study include the following: wild type (WT) Bristol N2, SP231 let-7(mn112) [mnDp1(X;V)/+ V; unc-3(e151) let-7(mn112) X]. Non-integrated transgenic strains: let-7A::GFP PQ490a [pha-1(e2123); apEX490a [pha-1(+); let-7A::GFP]], PQ490b [pha-1(e2123); apEX490b [pha-1(+); let-7A::GFP]], PQ490c [pha-1(e2123); apEX490c [pha-1(+); let-7A::GFP]], let-7B::GFP PQ491 [pha-1(e2123); apEX491 [pha-1(+); let-7A::GFP]], PQ491a [pha-1(e2123); apEX491a [pha-1(+); let-7B::GFP]], PQ491b [pha-1(e2123); apEX491b [pha-1(+); let-7B::GFP]], PQ491c [pha-1(e2123); apEX491c [pha-1(+); let-7B::GFP]], TRElet-7B::GFP PQ376 [pha-1(e2123); apEX376 [pha-1(+); TRElet-7B::GFP]], PQ377 [pha-1(e2123); apEX377 [pha-1(+); TRElet-7B::GFP]], PQ378 [pha-1(e2123); apEX378 [pha-1(+); TRElet-7B::GFP]], PQ379 [pha-1(e2123); apEX379 [pha-1(+); TRElet-7B::GFP]], LTElet-7B::GFP PQ417 [pha-1(e2123); apEX417 [pha-1(+); LTElet-7B::GFP]], PQ418 [pha-1(e2123); apEX418 [pha-1(+); LTElet-7B::GFP]], PQ419 [pha-1(e2123); apE419 [pha-1(+); TRElet-7B::GFP]], TRE LTElet-7B::GFP PQ413 [pha-1(e2123); apEX413 [pha-1(+); TRE LTElet -7B::GFP]], PQ414 [pha-1(e2123); apEX414 [pha-1(+); TRE LTElet -7B::GFP]], PQ415 [pha-1(e2123); apEX415 [pha-1(+); TRE LTElet -7B::GFP]], Integrated transgenic strains: iWT PQ488 [let-7(mn112) [mnDp1(X;V)/+ V; unc-3(e151) let-7(mn112) X]; apEX488 [iWTlet-7(II)]], ilet-7B::GFP PQ462 [let-7(mn112) [mnDp1(X;V)/+ V; unc-3(e151) let-7(mn112) X]; apEX462 [ilet-7B::GFP(II)]], i TRE PQ483 [let-7(mn112) [mnDp1(X;V)/+ V; unc-3(e151) let-7(mn112) X]; apEX483 [i TRElet-7(II)]], i LTE PQ477 [let-7(mn112) [mnDp1(X;V)/+ V; unc-3(e151) let-7(mn112) X]; apEX477 [i LTElet-7(II)]], i TRE LTE PQ478 [let-7(mn112) [mnDp1(X;V)/+ V; unc-3(e151) let-7(mn112) X]; apEX478 [i TRE LTElet-7(II)]], PQ481 [let-7(mn112) [mnDp1(X;V)/+ V; unc-3(e151) let-7(mn112) X]; apEX481 [i TRE LTElet-7(II)]], i TRE LTE PQ482 [let-7(mn112) [mnDp1(X;V)/+ V; unc-3(e151) let-7(mn112) X]; apEX482 [i TRE LTElet-7(II)]]. Strains expressing extra chromosomal transgenes were created by injecting pha-1(e2123) animals with let-7::GFP constructs at 50 ng/µl, the pha-1 rescue construct at 50 ng/µl, and luciferase, over digested by HindIII, at 50 ng/µl. Single copy let-7 rescue constructs were injected and integrated as previously described (Frokjaer-Jensen et al., 2008) and then crossed into SP231 let-7(mn112) animals and genotyped. At least 100 animals were scored for each construct. Staging of animals was done by gonad development.
Plasmid Constructs
All GFP constructs were made using a GFP vector construct from pSEH33, which contains three NLS repeats, four GFP exons separated by SV40 introns, followed by ~430 nt of the let-858 3’ UTR. The let-7 promoter was amplified from genomic DNA using A632 forward with A607 or A608 reverse, to create plet-7A::GFP (Fig. 1A, Fig. 4 B6) and plet-7B::GFP (Fig. 1A), respectively. The PCR fragments and pSEH33 were digested with KpnI and HindIII, and then ligated. To create the TRE rescue construct (Fig. 7 A2), the let-7 rescue fragment was sequentially digested with NheI and BstEII, blunt ended with Klenow, and then self-ligated (Johnson et al., 2003). The TREplet-7B::GFP reporter (Fig. 4 B1) was made by amplifying the TRE rescue construct with primers A632 and A608. The PCR fragments and plet-7B::GFP construct were digested with KpnI and HindIII, and then ligated. LTEplet-7B::GFP (Fig. 4 B5) was made by amplifying genomic DNA with A2256 and A632 digesting the PCR product and plet-7B::GFP with SacII and HindIII, blunt ending with Klenow, and then self-ligating. LTE rescue construct (Fig. 7A) was made by amplifying LTEplet-7B::GFP with A1348 and A1338, then digesting the PCR product and let-7 rescue construct with XmaI and KpnI, and then ligating. The ΔTRE LTE plet-7B::GFP construct (Fig. 4 B9) was made by digesting LTEplet-7B::GFP with BstEII and NheI, blunt ended with Klenow, and then self-ligated. The ΔTRE LTE rescue construct (Fig. 7) was made by amplifying TRE LTEplet-7B::GFP with A1348 and A1338, then digesting the PCR product and let-7 rescue construct with XmaI and KpnI, and then ligating. Construct 2 (Fig. 4) was made by digesting plet-7B::GFP with NheI and HindII, blunt ending with Klenow, and then self-ligating. Construct 3 (Fig. 4) was made by digesting plet-7B::GFP with EcoRV and BstEII, blunt ending with Klenow, and then self-ligating. Construct 4 (Fig. 4) was created by digesting plet-7B::GFP with EcoRV and HindIII, blunt ending with Klenow, and then self-ligating. Construct 7 (Fig. 4) was made by amplifying genomic DNA with A2252 and A632 digesting the PCR product and plet-7B::GFP with SacII and HindIII, blunt ending with Klenow, and then self-ligating. To make the mos-SCI rescue constructs, each rescue construct was amplified with primers A1347 and A1650. The PCR product and mos-SCI vector CFJ151 were digested with SpeI and XhoI then ligated together.
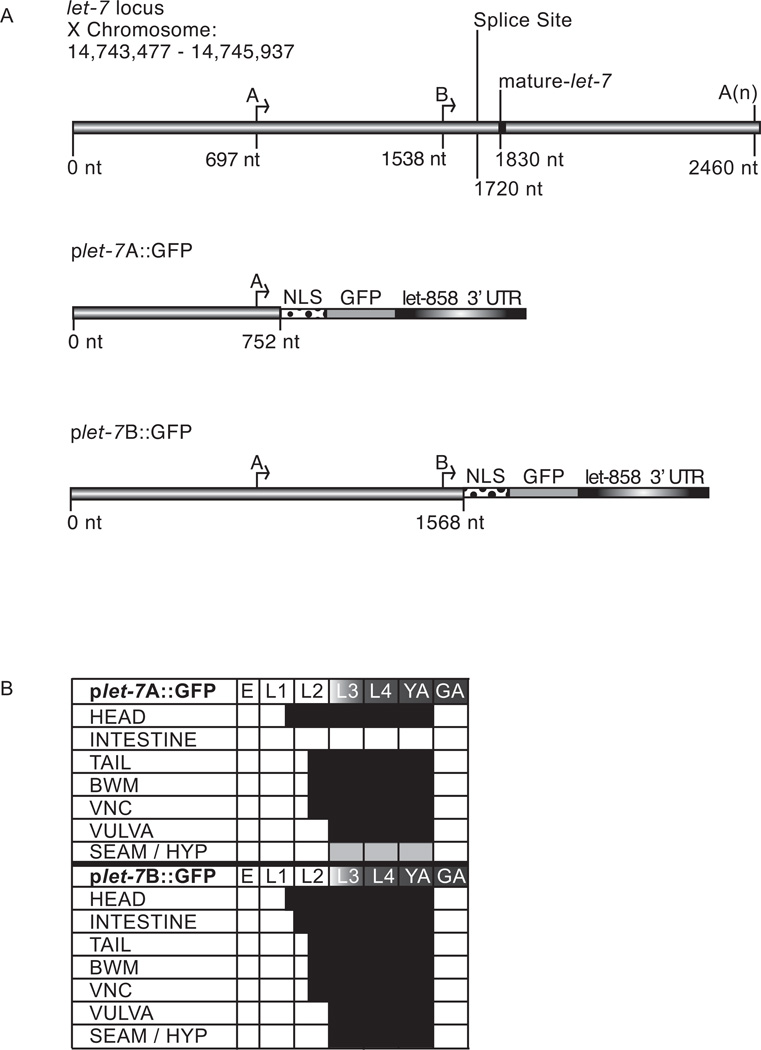
Fig. 1. Expression of plet-7::GFP reporters.
(A) Diagram of the endogenous let-7 gene and reporter constructs. The plet-7A::GFP and plet-7B::GFP constructs contain 752 and 1568 nt of endogenous let-7 promoter, respectively, fused to GFP with a nuclear localization signal (NLS) and the let-858 3’UTR. (B) The spatio-temporal expression pattern of plet-7::GFP in transgenic animals was analyzed in embryos (E), larval stages (L1-L4), young (YA) and gravid (GA) adults. Endogenous mature let-7 is present from L3 to GA (gradient shading). The black and light gray boxes represent stages with strong or weak GFP expression, respectively. Expression in the head and tail is seen in most neurons and muscle cells. Body wall muscle (BWM); ventral nerve cord (VNC); vulva includes precursor as well as mature cells; SEAM/ HYP, seam and hypodermal cells.
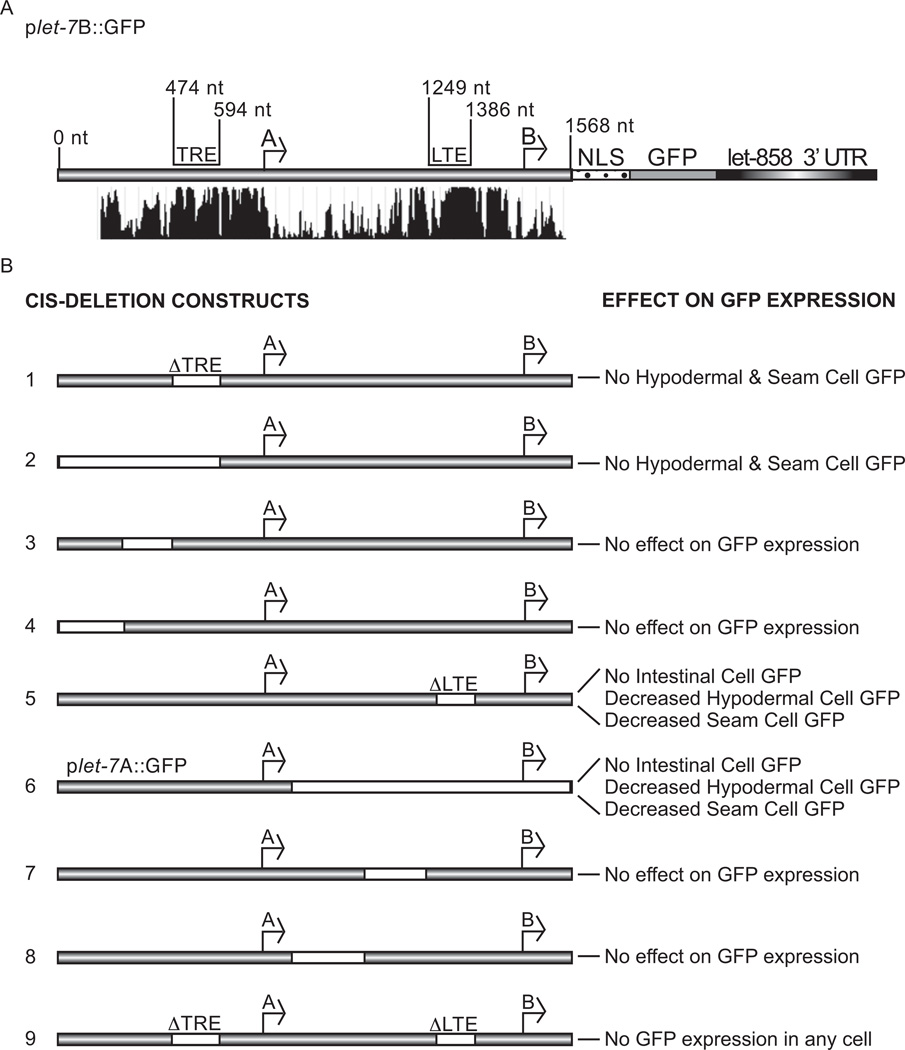
Fig. 4. Identification of cis-regulatory elements in the let-7 promoter.
(A) Conservation of the let-7 promoter sequence among six nematode species, shown below the C. elegans let-7 gene with the positions of the TRE and LTE indicated (http://genome.ucsc.edu) (Kent et al., 2002). (B) White regions in let-7 promoter constructs represent deletions. The effect of each deletion construct on reporter GFP expression is indicated in the right column and construct numbers are in the left column.
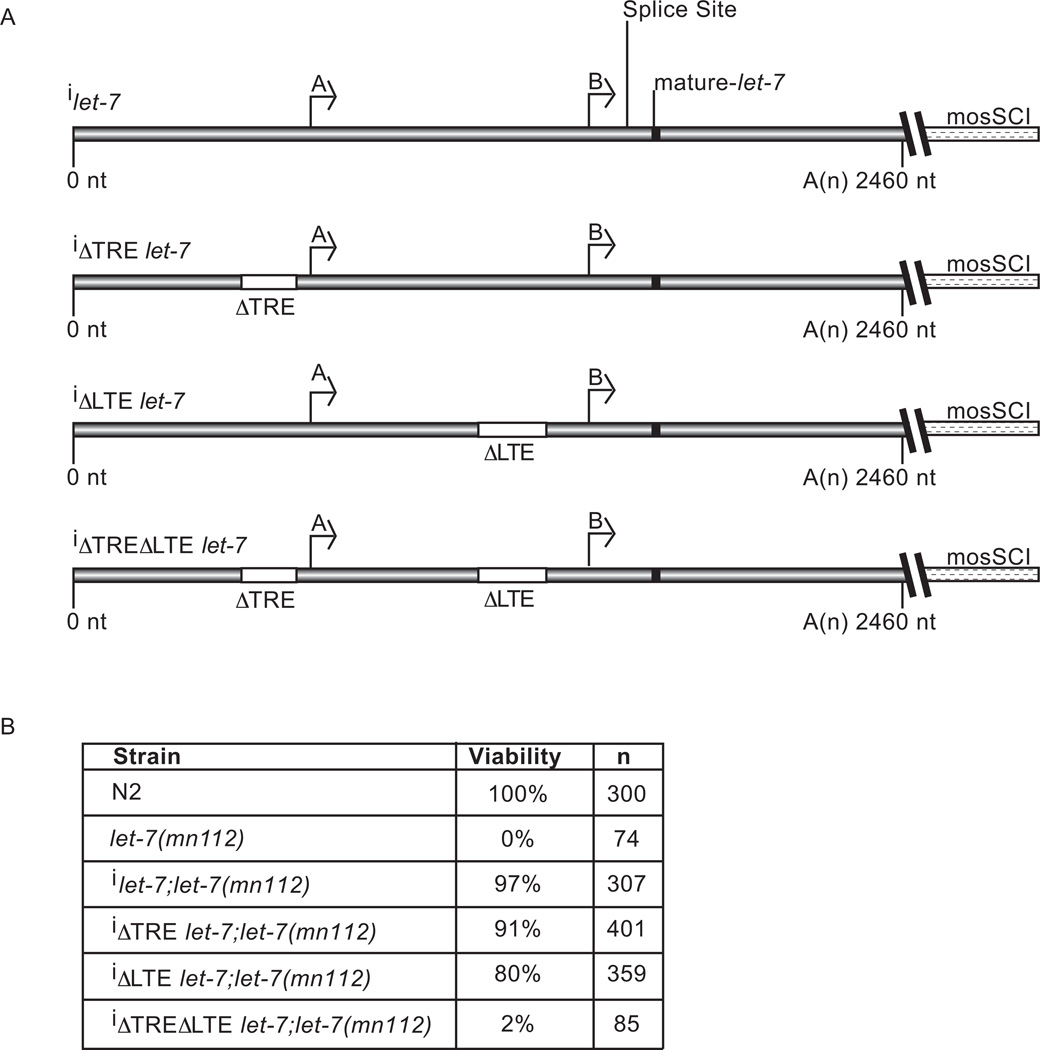
Fig. 7. The effect of promoter deletions on let-7 rescue activity.
(A) Constructs tested for let-7 rescue activity contained WT or the indicated deletions in the let-7 promoter sequences inserted into the mosSCI backbone to allow for single copy integration (i) into Chromosome II. (B) Rescue analysis was performed on integrated transgenic animals crossed to let-7(mn112) mutant animals. Viability was analyzed as the percent of animals alive at the early gravid adult stage. Shown is the average viability from three independent rescue experiments for each strain and n = the total number of animals analyzed.
RNAi Screen
For 1st generation RNAi, C. elegans were grown at 25°C and synchronized using standard hypochlorite treatment. At time zero, starved L1 animals were plated on bacteria expressing either gene specific RNAi or an empty vector, and then collected at different time points. For 2nd generation RNAi, animals were allowed to mature on RNAi plates until gravid. Then they were synchronized again and, at time zero, the starved L1s plated on bacteria expressing the same RNAi as their parents, and then collected at the desired time points.
RNA analyses
RNA was extracted from synchronized animals, using standard Trizol extraction methods (Bracht et al., 2004). Agarose Northern analysis was used for primary miRNA detection with probe templates shown in Supplementary Table 1. RNA ligase-mediated rapid amplification of cDNA ends (RACE) was completed with the GENERACER kit (Invitrogen) and primers listed in Supplementary Table 2. Total RNA was ligated to the kit 5′ linker and reverse-transcribed with Superscript III (Invitrogen) and a GFP primer specific to the first exon. PCR and nested PCR used 5′ linker and primers inside the first exon of GFP and the resulting PCR products were sequenced after TOPO cloning (Invitrogen).
RESULTS
Spatial and temporal expression regulated by the let-7 promoter
The let-7 miRNA is an important regulator of development across species (Mondol and Pasquinelli, 2012). Both the timing and level of let-7 miRNA expression are highly regulated by transcriptional and post-transcriptional mechanisms. While multiple factors have been found to control the processing and stability of the let-7 miRNA, less is known about transcriptional regulation of this important miRNA. Previous reports have detected let-7 expression from the embryonic to adult stages in most cell types, using transcriptional reporters in C. elegans (Esquela-Kerscher et al., 2005; Johnson et al., 2003; Martinez et al., 2008b). In some cases, the non-temporal expression pattern in particular tissues was considered unspecific background expression by the reporters (Johnson et al., 2003). To re-examine the spatio-temporal expression of let-7 transcription, we generated GFP reporters based on the mapped transcriptional start sites in the let-7 promoter. In C. elegans, two transcriptional start sites produce endogenous let-7 primary transcripts of ~1,731 nt, and ~890 nt, from the A and B start sites, respectively (Fig. 1A) (Bracht et al., 2004). Based on these mapped start sites, we fused 752 base pairs (bp) (plet-7A::GFP) or 1568 bp (plet-7B::GFP) of let-7 promoter sequence to a nuclear localized GFP gene and the 3’UTR of a constitutively expressed gene (let-858) (Kelly et al., 1997).
Each let-7 transcriptional reporter construct was co-injected with pha-1 rescue sequence into pha-1(e2123) worms. Four independent lines for plet-7A::GFP and three independent lines for plet-7B::GFP were isolated based on pha-1 rescue activity and examined for GFP expression. No differences in GFP expression patterns for the independent lines were observed for each of the transcriptional reporters. The spatio-temporal expression patterns driven by each reporter are summarized in Figure 1B and representative images are shown in Figure 2. Both of the plet-7::GFP constructs drove temporally regulated expression, initially observed at the end of the first larval stage (L1) (Fig. 1B), consistent with the initial detection of endogenous primary let-7 transcripts (Van Wynsberghe et al., 2011). Starting in late L1 (~10 h at 25°C), robust GFP was seen in muscle and neuronal cells in the head, including the nerve ring (Fig. 1B). Midway through L2, GFP accumulated in the neurons and muscle cells of the tail, body wall muscle (BWM), and ventral nerve cord (VNC) (Fig. 1B). Beginning in L3, all transgenic lines began to express GFP in vulva precursor cells (Fig. 1B). After the onset of GFP expression in a particular tissue, it was maintained until adults became gravid with embryos (~52 h at 25° C) (Fig. 1B). While GFP was not detected in the germ line, this could be attributable to silencing of multicopy extra-chromosomal transgenes in this tissue. Thus, we also inserted a single copy of the plet-7B::GFP transgene into Chromosome II using the Mos-transposon mediated integration system (Frokjaer-Jensen et al., 2008). These transgenics displayed the same expression pattern as the array-based strains, including undetectable GFP in the germ line (data not shown).
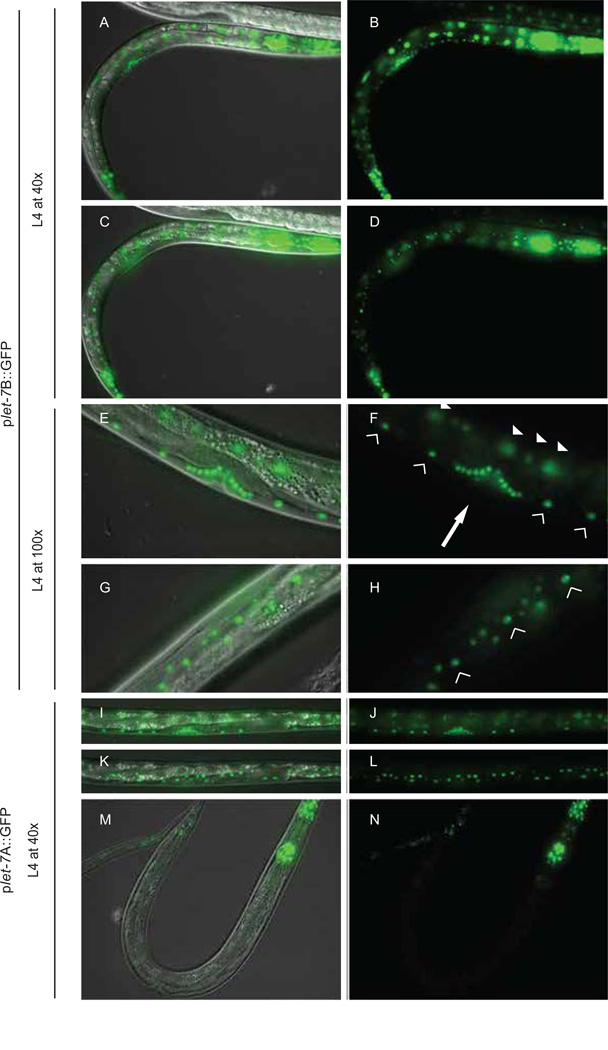
Fig. 2. Spatial expression of plet-7::GFP reporters.
Micrographs show GFP and DIC overlays (A, C, E, G, I, K, M) and GFP alone (B, D, F, H, J, L, N). (A–D) Expression of GFP from plet-7B::GFP in worms is detected in all major somatic tissues by the L4 stage. (E,F) L4 transgenic animals expressing plet-7B::GFP in the vulva cells (arrow), ventral cord neurons (hollow triangles) and intestine (solid triangles). (G,H) Transgenic animals expressing plet-7B::GFP in the hypodermal cells and seam cells (seam cells shown with hollow triangles). Transgenic L4 animals expressing plet-7A::GFP in vulval and ventral cord neurons (I, J) and hypodermal cells (K–L). (M–N) Transgenic plet-7A::GFP L4 animals display GFP in the head, tail and vulva, but not intestinal cells; hypodermal cell expression is generally weaker and not visible in this plane of view. Images are representative of more than 100 animals scored in at least 3 independent lines for each construct and time point.
Although plet-7A::GFP and plet-7B::GFP transgenics showed similar spatio-temporal expression patterns in most cell types, they were not entirely overlapping. In hypodermal and seam cells, the plet-7B::GFP reporter produced a robust GFP signal at the L3 stage, similar to the expression pattern previously reported for other let-7 transcriptional reporters (Fig. 1B) (Esquela-Kerscher et al., 2005; Johnson et al., 2003; Martinez et al., 2008b). However, plet-7A::GFP animals exhibited only weak, transient expression of GFP in the hypodermal cells beginning at the L3 stage (Fig. 1B). Early in the second stage of development, all intestinal cells showed high expression of GFP in plet-7B::GFP transgenic animals, while GFP was undetectable in the intestine of plet-7A::GFP transgenic animals (Fig. 1B). Taken together, these results suggest that there is at least one cis-element, downstream of the A start site, that enhances expression in hypodermal and seam cells and is essential for expression in intestinal cells.
Cycling transcription of let-7
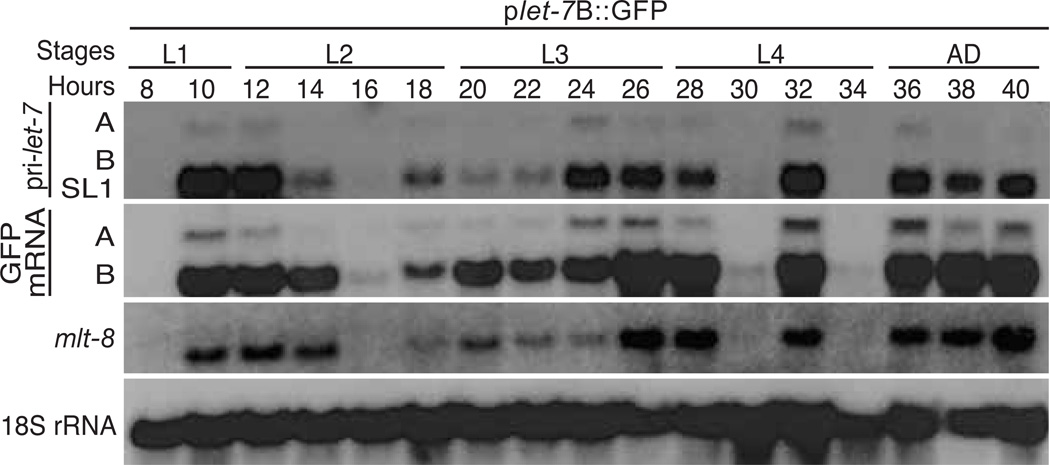
We previously showed that primary let-7 expression oscillates during each larval stage of development (Van Wynsberghe et al., 2011). To test if this pattern is transcriptionally regulated, we analyzed the expression of GFP mRNA from plet-7B::GFP transgenic worms every two hours from mid L1 to early adulthood. GFP transcripts from both the A and B start sites oscillated throughout larval development in a pattern very similar to that of endogenous primary let-7 transcripts (Fig. 3). These results show that the let-7 promoter is sufficient to produce the cycling pattern of primary let-7 expression during worm development.
Fig. 3. Cycling expression of let-7.
Total RNA was isolated at the indicated time points from synchronized transgenic worms expressing the plet-7B::GFP reporter construct. Levels of endogenous primary let-7 transcripts, transgenic GFP mRNA, mlt-8 mRNA, and 18S rRNA were analyzed by northern blotting. The similar sized B and SL1 transcripts often do not clearly resolve.
The pulses of pri-let-7 transcription typically preceded each larval molt (Fig. 3). This pattern is reminiscent of GFP expression driven by promoters from genes in the molting pathway, including mlt-8, mlt-9, mlt-10, and mlt-11 (Frand et al., 2005). By Northern blot analyses, we found that endogenous mlt-8 transcripts have the same dynamic cycling pattern as pri-let-7 (Fig. 3). These results suggest that let-7 and some genes in the molting pathway are under common transcriptional regulatory mechanisms.
Identification of cis-regulatory elements in the let-7 promoter
To identify cis-element(s) in the let-7 promoter that regulate the spatio-temporal expression pattern, we analyzed a series of deletions in the plet-7B::GFP reporter for effects on GFP expression. Comparison of the let-7 promoter region among six nematode species revealed several highly conserved stretches of sequence (Fig. 4A). One conserved region upstream of the A start site, the TRE, was previously shown to regulate the timing of hypodermal cell expression (Johnson et al., 2003). In agreement with that study, deletion of the TRE ( TRE) from our reporter abrogated GFP expression in the hypodermal and seam cells (Johnson et al., 2003) (Figs. 4B and 5). Likewise, removal of the TRE and all upstream sequence resulted in the same expression pattern (Fig. 4B). However, deletion of smaller segments upstream of the A start site that left the TRE intact had no effect on GFP expression.
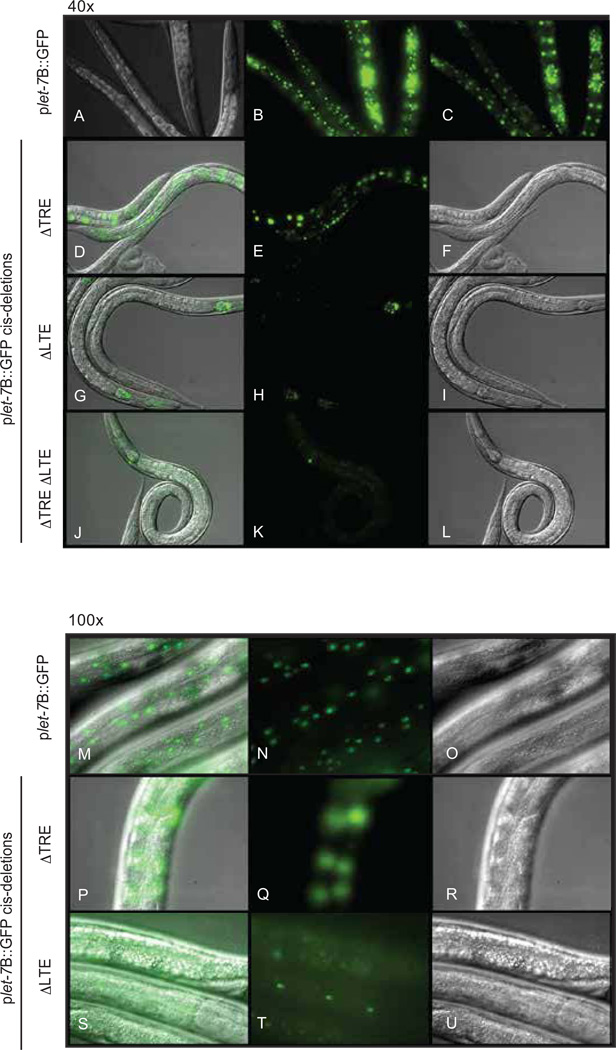
Fig. 5. Examples of GFP expression from transgenic animals carrying plet-7B::GFP or cis deleted plet-7B::GFP constructs.
(A–L) 40× magnification of L4 stage transgenic animals expressing plet-7B::GFP (A–C) at two different planes of focus to observe hypodermal (B) or intestinal (C) expression, TRE plet-7B::GFP (D–F) focused on intestinal cells, which have robust GFP expression, LTE plet-7B::GFP (G–I) focused on intestinal cells, which lack GFP expression, or TRE LTE plet-7B::GFP (J–L), which lack GFP expression in any plane of focus. (M–U) 100× magnification of L4 stage transgenic animals expressing plet-7B::GFP focused on hypodermal GFP expression (M–O), TRE plet-7B::GFP (P–R) focused on hypodermal cells, which have undetectable GFP expression, LTE plet-7B::GFP (S–U) focused on hypodermal cells, which have diminished GFP expression. Images are representative of more than 100 animals in at least two lines for each strain.
Since the let-7A promoter alone was insufficient for full expression in intestinal, hypodermal and seam cells (Figs. 1B, 2I-N and 4B), we predicted that at least one additional element between the A and B start sites contributes to let-7 transcription in those cell types. Through deletion analysis, we identified a highly conserved region of 138 nt that was required for intestinal expression (Figs. 4, 5 and Supplementary Fig. 1). Deletion of this element, which we have named the let-7 transcriptional element (LTE), eliminated intestinal GFP expression and diminished hypodermal and seam cell GFP expression (Fig. 5). This pattern was analogous to the expression from the plet-7A::GFP reporter (Fig. 2). Consistent with the comparable expression patterns of the plet-7A::GFP and TREplet-7B::GFP reporters, removal of other regions between the A start site and the LTE had no effect on GFP expression (Fig. 4B). Interestingly, when the LTE and TRE were both deleted, GFP expression was undetectable in any stage or tissue (Figs. 4B and 5).
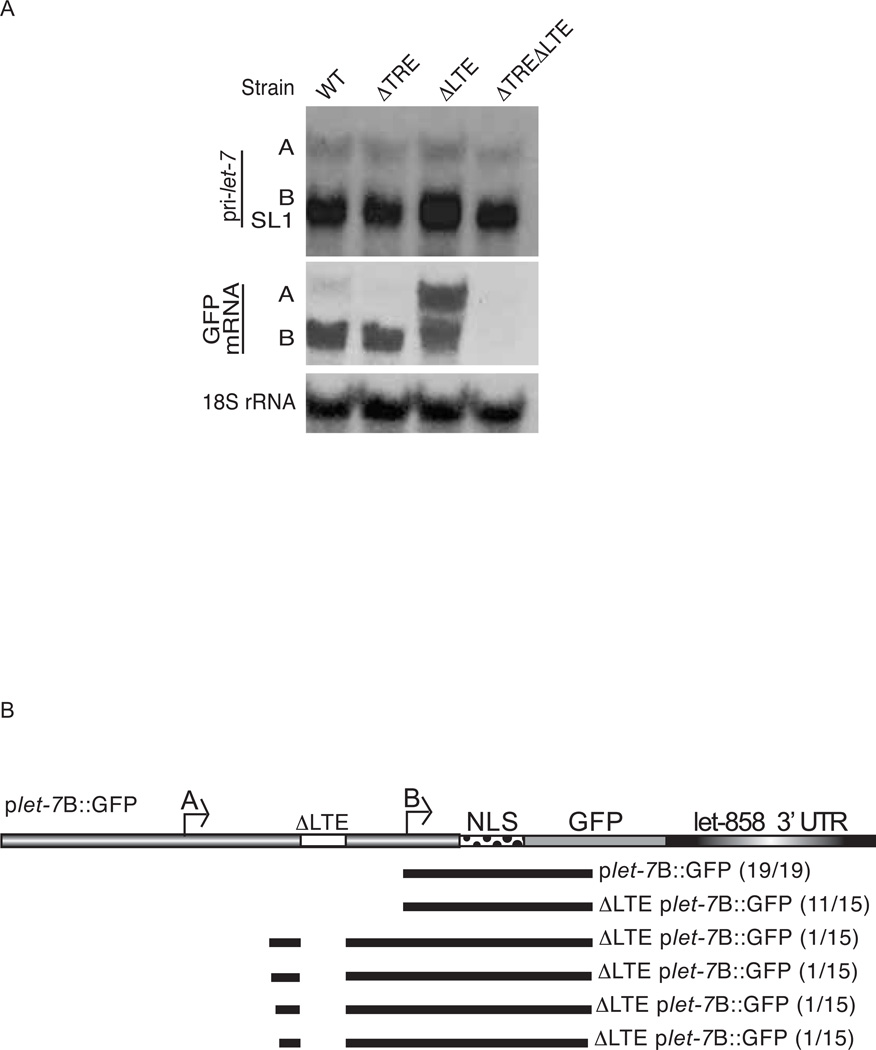
To test the effect of the promoter deletions on expression of transcripts from the A and B start sites, we performed Northern blotting to detect the GFP transcripts. Compared to the WT reporter, the ΔTREplet-7B::GFP strain produced similar levels of B but undetectable levels of A transcripts (Fig. 6A). These results suggest that the TRE is important for transcription from the A start site. In the ΔLTE strain, we detected elevated levels of A transcripts and heterogeneous transcripts around the same size and larger than the B form. To characterize the unexpected transcripts, we performed 5’ rapid amplification of cDNA ends (RACE) using RNA from WT and ΔLTEplet-7B::GFP animals. While all of the transcripts cloned from WT transgenics mapped to the previously identified B start site, several of the transcripts from ΔLTEplet-7B::GFP animals mapped ~176 nt upstream of the B start site (Fig. 6B). Removal of both the TRE and LTE resulted in virtually undetectable levels of GFP mRNA transcripts (Fig. 6A), consistent with the absence of GFP fluorescence in those transgenic animals (Fig. 5J–L). These results show that two distinct elements regulate expression from the A and B start sites and that loss of both elements eliminates transcription from the let-7 promoter.
Fig. 6. Effects of the let-7 promoter deletions on reporter mRNA expression.
(A) Total RNA was isolated from L4 stage transgenic animals and levels of endogenous primary let-7 transcripts, transgenic GFP mRNA, and 18S rRNA were analyzed by northern blotting. (B) The 5’ ends of GFP expressed from the WT and LTE constructs were mapped by 5’ RACE. The number of clones mapping to a particular position for WT plet-7B::GFP and LTE plet-7B::GFP are indicated.
To assess the importance of the TRE and LTE in vivo, we tested transgenic constructs for rescue of let-7(mn112) mutant animals, which produce no mature let-7 miRNA (Reinhart et al., 2000). We introduced integrated, single copy let-7 transgenes containing the WT and each of the promoter deletions into let-7(mn112) animals (Fig. 7A). Strains with the rescue constructs that deleted either the TRE or LTE grew to gravid adulthood and produced viable progeny in numbers comparable to that of the WT strain. However with respect to WT, overall development was slightly delayed in the individual ΔTRE and ΔLTE transgenics and the rescue activity of the ΔLTE construct was also reduced (Fig. 7B). Worms with the ΔTRE rescue construct had gapped (50%) and missing alae (22%), consistent with a role for this element in driving seam cell expression. Simultaneous deletion of both the TRE and LTE elements in the rescue construct resulted in lethality comparable to nontransgenic let-7(mn112) mutant animals (Fig. 7B). Taken together, the TRE and LTE largely compensate for each other in directing sufficient let-7 expression, but loss of both elements is incompatible with viability.
Regulation of let-7 transcription by trans-acting factors
So far, the nuclear hormone receptor DAF-12 and the hunchback-like 1 (HBL-1) transcription factors have been reported to regulate the expression of let-7 in C. elegans (Abrahante et al., 2003; Bethke et al., 2009; Hammell et al., 2009; Roush and Slack, 2009). To identify additional factors that might contribute to the dynamic spatial and temporal expression pattern of let-7, we used RNAi to screen for genes that disrupted transcription from the plet-7B::GFP reporter. Candidates for the RNAi screen included a list of predicted transcription factors previously reported to produce phenotypes consistent with altered let-7 expression when depleted by RNAi (Table 1 and Supplementary Table 2). Since the cycling pattern of primary let-7 expression resembles the transcription of genes in the molting pathway (Fig. 3), we also included molting-related genes in the list of candidates (Frand et al., 2005). Additionally, we screened transcription factors that have been reported to directly bind the let-7 promoter through yeast one hybrid or chromatin immunoprecipitation (ChIP) assays (Celniker et al., 2009; Martinez et al., 2008a). In these screens, we expected increased, precocious, or persistent GFP expression, or any combination of these, upon RNAi of a negative regulator and decreased or delayed GFP expression upon depletion of a positive regulator. Several genes positively or negatively affected global GFP expression when depleted by RNAi (Table 1). RNAi of many other candidates had tissue specific effects. In particular, intestinal expression appeared to be sensitive to RNAi of several genes. As expected, RNAi against hbl-1 resulted in precocious expression of GFP in the hypodermal and seam cells of plet-7B::GFP transgenic animals (Roush and Slack, 2009).
Table 1.
Transcriptional regulators of let-7. RNAi of the listed genes was performed on plet-7B::GFP animals and compared to empty vector controls.
| RNAi | Sequence | Name | Effect on GFP Expression |
|---|---|---|---|
| 1 | D1081.8* Y | phi-7 | No GFP expression |
| 1 | F38H4.9 | let-92 | No GFP expression |
| 2 | T24H7.1* | phb-2 | No GFP expression |
| 2 | W05E10.3* | ceh-32 | Decreased overall GFP |
| 2 | F58E6.10* | unc-42 | Decreased overall GFP |
| 2 | F21H11.3 * | tbx-2 | Decreased overall GFP |
| 1 | R06A4.9 | pfs-2 | No GFP expression in vulva |
| 1 | F10C1.5* Y | dmd-5 | No intestinal GFP; Decreased SEAM/ HYP GFP |
| 1 | W01F3.3 | mlt-11 | No intestinal GFP |
| 2 | M88.6 | pan-1 | No intestinal GFP |
| 1 | W08F4.6 | mlt-8 | No intestinal GFP, few with low GFP in head and tail |
| 1 | W09B6.1 | pod-2 | No intestinal GFP, few with low GFP in head and tail |
| 2 | Y48B6A.3 | xrn-2 | No intestinal GFP, few with low GFP in head and tail |
| 2 | C32F10.6* | nhr-2 | Decreased intestinal GFP |
| 2 | Y75B8A.2* | nob-1 | Decreased intestinal GFP |
| 2 | M142.4* | vab-7 | Decreased intestinal GFP |
| 2 | Y66A7A.8* | tbx-33 | Decreased intestinal GFP |
| 2 | R119.6* | taf-4 | Decreased intestinal GFP |
| 2 | R13A5.5* | ceh-13 | Decreased intestinal GFP |
| 2 | R07B1.1* | vab-15 | Decreased intestinal GFP |
| 2 | F55A8.1* | egl-18 | Decreased intestinal GFP |
| 2 | ZK430.8 | mlt-7 | Decreased intestinal, SEAM / HYP GFP |
| 1 | C17G1.6 | nas-37 | Decreased SEAM / HYP GFP |
| 2 | F08C6.1 | adt-2 | Decreased SEAM / HYP GFP |
| 2 | F11C1.6 | nhr-25 | No HYP GFP; Decreased GFP starting in L2 |
| 2 | C01H6.5 | nhr-23 | Decreased GFP starting in L2 |
| 2 | F31E3.1* | ceh-20 | Increased overall GFP |
| 2 | K10G6.1* | lin-31 | Increased overall GFP |
| 2 | F58A3.1* | ldb-1 | Increased overall GFP |
| 1 | C09G5.6 | bli-1 | Increased overall GFP |
| 2 | K04A8.6 | dre-1 | Increased overall GFP |
| 2 | R53.3* | egl-43 | Increased GFP in head |
| 2 | F26C11.2* | unc-4 | Increased GFP in head and tail |
| 1 | F13D11.2* | hbl-1 | Precocious SEAM/ HYP GFP |
| 2 | F57B9.2 | let-711 | Mosaic GFP in adults; Precocious GFP in vulva |
transcription factor;
yeast one hybrid;
bold, molting factor;
1 or 2, first or second generation RNAi, respectively.
Of the eleven candidates previously found to bind the let-7 promoter through yeast one hybrid or ChIP assays, two scored as strong regulators of plet-7B::GFP (Table 1 and Supplementary Table 2). RNAi against phi-7 appeared to disrupt let-7 transcription in all cell types, resulting in undetectable GFP expression at all stages (Table 1 and Supplementary Figs. 2–3). RNAi of dmd-5 resulted in abrogated GFP expression in intestinal cells, with diminished hypodermal and seam cell GFP expression (Table 1 and Supplementary Figs. 2–3). To eliminate the possibility that RNAi of dmd-5 or phi-7 caused nonspecific silencing of the extrachromosomal transgene, single-copy, integrated plet-7B::GFP transgenic animals (iplet-7B::GFP) were also tested and found to be equally sensitive to depletion of these factors (Supplementary Figs. 2–3). Overall, our screen showed that multiple factors contribute to the complex temporal and spatial expression pattern of let-7 during C. elegans development.
Discussion
The basic mechanism of miRNA biogenesis has been revealed during the last decade (Kim et al., 2009). The distinct pattern of tissue and stage specific expression of individual miRNAs is regulated at both the transcriptional and post-transcriptional levels. Regulation of let-7 miRNA is of particular interest as changes in levels of this miRNA have been linked to human diseases, including cancer (Mondol and Pasquinelli, 2012). The let-7 miRNA is developmentally regulated across species with generally increasing levels as cells differentiate. This expression pattern is at least partly due to factors that both negatively, such as LIN-28 and hnRNP A1, and positively, such as KSRP, control processing of this miRNA (Choudhury and Michlewski, 2012; Thornton and Gregory, 2012). However, less is understood about transcriptional regulation of let-7. Here we show that multiple cis- and trans-acting factors contribute to the dynamic spatial and temporal expression of let-7 in C. elegans. We demonstrate that the let-7 promoter drives bursts of transcription during each larval stage, generally coinciding with the molts. We characterized two promoter elements that have redundant as well as non-overlapping roles in directing transcription in specific tissues. The spatio-temporal control of let-7 expression appears complex as multiple proteins were found to have distinct effects on expression of our transcriptional reporters. As let-7 is a key regulatory molecule in species as diverse as C. elegans and humans, elucidation of the elements controlling expression of this important miRNA may provide insights into how it becomes mis-regulated in disease tissues.
In most cases, the spatio-temporal expression from our reporters agreed with previous studies showing that let-7 is expressed in most somatic tissues during worm development (Esquela-Kerscher et al., 2005; Johnson et al., 2003; Martinez et al., 2008b). However, there were a few discrepancies. Martinez et al., 2008, observed expression starting in late embryos (Martinez et al., 2008b), while we did not detect expression from the plet-7::GFP reporters until late L1, which is consistent with the timing of endogenous primary let-7 accumulation (Van Wynsberghe et al., 2011). In agreement with Johnson et al., 2003, expression driven by our let-7 promoter reporters commenced at the L3 stage in the hypodermal seam cells (Johnson et al., 2003). Additionally, we were able to detect temporally punctuated expression in different tissues, initiating at late L1 in the head, followed by intestine, tail region, body wall muscle cells and the ventral nerve cord in L2 and finally the vulval and hypodermal cells during the L3 stage. Expression in all tissues terminated by the gravid adult stage, consistent with the reduction in mature let-7 noted during aging (Kato et al., 2011). Differences between our reporters and others designed to evaluate transcription of let-7 miRNA in vivo include incorporation of promoter sequence based on the transcriptional start sites, instead of location of the mature miRNA, and fusion of nuclear localization signals to the GFP to facilitate identification of tissue specific expression.
Multiple regulatory mechanisms contribute to the appropriate expression levels of let-7 miRNA in C. elegans. Here we show that the cycling pattern of primary let-7 expression is transcriptionally controlled. This pattern is reminiscent of several molting pathway genes (Frand et al., 2005). Inactivation of the nuclear hormone receptor nhr-23 by RNAi was previously shown to prevent expression of reporters driven by molting pathway genes, including mlt-8 (Frand et al., 2005). Similarly, we found that RNAi against nhr-23 or nhr-25, another molting pathway transcription factor, abrogated transcription of let-7 late in the second larval stage (Table 1). However, most of the animals subjected to nhr-23 or nhr-25 RNAi never matured past the L2 stage, though the few that did also showed reduced GFP expression from the plet-7B::GFP reporter. These results agree with previous findings that these factors are important for progression through the L2-L3 larval transition (Asahina et al., 2000; Frand et al., 2005; Gissendanner and Sluder, 2000; Kostrouchova et al., 2001). They also raise the caveat that it might be the arrested development of the animals that is affecting let-7 transcription, rather than direct regulation. Inactivation of several other genes involved in the molting pathway also perturbed plet-7B::GFP reporter expression, particularly in the intestine. These observations raise the possibility that feeding signals could be linked to the rhythmic expression of let-7 and other genes that cycle with each molt.
Systematic deletion of let-7 promoter elements revealed two regions important for transcription. The TRE, which is upstream of the A start site, was originally identified as an element required for expression in hypodermal seam cells at the L3 stage (Johnson et al., 2003). Consistent with Johnson et al., we found that the TRE is the only element upstream of the A start site that seems to be essential for positively regulating expression of reporters driven by the let-7 promoter. Reporters lacking the TRE failed to express GFP protein in the hypodermal seam cells and to produce GFP mRNA initiating from the A start site. These data suggest that the TRE regulates transcription from the A start site and that this activity is required for let-7 expression in certain tissues.
The absence of intestinal expression by the plet-7A::GFP reporter indicated that an element between the A and B start sites regulates expression in that tissue. We found that deletion of a single region upstream of the B start site disrupted intestinal GFP expression. This let-7 transcriptional element (LTE) covers a highly conserved region in the let-7 promoter that is not obviously related to the TRE in sequence, except for the shared heptamer TCACGCA identified by SCOPE analysis (Supplementary Fig. 1) (Carlson et al., 2007). Removal of the LTE also resulted in increased GFP mRNA expression from the A start site and the production of aberrant transcripts from a position ~176 nt upstream of the usual B start site. Thus, the LTE might not only promote intestinal GFP expression, but it might also antagonize expression from positions other than the B start site.
The TRE and LTE have redundant roles in positively regulating the expression of let-7 in most tissues. Only the hypodermal seam cells or the intestine were sensitive to individual deletions of the TRE or LTE, respectively. Removal of both elements was necessary to eliminate expression from the plet-7B::GFP reporter in all tissues. Furthermore, rescue activity was largely maintained in let-7 constructs lacking either the TRE or LTE alone. These results suggest that the redundancy of these elements is sufficient to produce adequate let-7 for viability. The TRE construct rescued the lethality of let-7(mn112) but these worms still displayed defects in alae formation. These results are consistent with the diminished transcription by TRE plet-7B::GFP in the hypodermal seam cells, which are responsible for producing alae at the adult stage. The delayed growth of the worms rescued with the LTE construct could reflect the possible absence of let-7 expression in the intestine, as feeding is linked to growth rate in developing larvae. Since individual deletion of the TRE, which is required for hypodermal seam cell expression, or the LTE, which is required for intestinal cell expression, did not abolish let-7 rescue activity, the lack of let-7 transcription in these tissues alone does not seem to contribute to the lethality of null let-7 worms.
We identified 35 different candidate genes as regulators of let-7 transcription. Most of these genes appear to be positive regulators of let-7 expression as their depletion resulted in decreased expression by the plet-7B::GFP reporter. Two of these candidates, phi-7 and dmd-5, may be direct regulators of let-7 transcription as they were previously reported to bind the let-7 promoter in yeast one hybrid assays (Martinez et al., 2008a). RNAi of phi-7 generally abrogated GFP transcription in the plet-7B::GFP transgenic animals. In C. elegans, RNAi of phi-7 (protein homeostasis interference) results in a wide range of phenotypes, including lethality and molting defects (Frand et al., 2005; Rual et al., 2004). The human ortholog of PHI-7, CDC5L, is a Myb family transcription factor and a core component in complexes that regulate RNA splicing, DNA replication and DNA damage response, as well as S-phase cell cycle and neuronal fate specification (Ajuh et al., 2000; Boudrez et al., 2000; Lu et al., 2008; Urano-Tashiro et al., 2010; Zhang et al., 2009). Depletion of dmd-5 (Doublesex/MAB-3 domain) eliminated expression from plet-7B::GFP in intestinal cells and diminished GFP intensity in hypodermal and seam cells. This expression pattern recapitulates that of LTE plet-7B::GFP transgenic animals, suggesting that DMD-5 could work through the LTE to promote transcription. The dmd-5 gene appears to be expressed in neuronal cells with intermittent intestinal expression from the mid-embryo stage throughout larval development (Reece-Hoyes et al., 2007). RNAi of dmd-5 causes phenotypes consistent with decreased expression of let-7, such as protruding and rupturing of the vulva, lethality and molting defects (Frand et al., 2005; Simmer et al., 2003). DMD-5 is related to the vertebrate DMRT (Doublesex- and mab-3-related transcription factor) family of Zinc-finger containing DNA binding proteins that have been shown to function in sexual development and neurogenesis (Hong et al., 2007; Yoshizawa et al., 2011; Zhu et al., 2000). Interestingly, DMD-5 also was found to bind lin-4 and the highly related mir-237 promoter sequences through yeast one hybrid experiments (Martinez et al., 2008a). The lin-4 miRNA functions upstream of let-7 in the developmental timing pathway and shares its broad tissue expression pattern (Esquela-Kerscher et al., 2005; Johnson et al., 2003; Martinez et al., 2008b), raising the possibility that DMD-5 has a role in directing the transcription of these heterochronic miRNAs during larval development.
A subset of genes from the RNAi screen emerged as potential negative regulators of let-7 expression. One of these genes, hbl-1 (hunchback-like), was previously reported to inhibit expression of let-7 in the seam cells, hypodermis and vulval precursor cells (VPCs) until the third larval stage (Roush and Slack, 2009). Expression of hbl-1 is, in turn, repressed by let-7 and its sister miRNAs, which then relieves hbl-1 inhibition of let-7 transcription by the L3 stage (Abbott et al., 2005; Abrahante et al., 2003; Lin et al., 2003). The HBL-1 responsive element was predicted to reside downstream of the TRE before the A start site (Roush and Slack, 2009). This sequence contains an A-rich element similar to the binding site defined for Hunchback in other organisms (Stanojevic et al., 1989; Treisman and Desplan, 1989).
About half of the genes identified in the RNAi screen are not predicted to encode transcription factors and, thus, may regulate the expression of let-7 indirectly. One of the negative regulators, dre-1 (daf-12 redundant function), encodes a highly conserved F box protein that functions in the molting and heterochronic pathways (Fielenbach et al., 2007; Frand et al., 2005). Depletion of dre-1 activity by RNAi or genetic lesions results in precocious seam cell fusion (Fielenbach et al., 2007). This phenotype is consistent with overexpression of let-7 (Reinhart et al., 2000). The precocious phenotype of dre-1 mutants was only partly rescued in let-7(n2853) mutants, suggesting that dre-1 also may work downstream or in parallel to let-7 (Fielenbach et al., 2007). DRE-1 is predicted to act in an E3 ubiquitin ligase complex but presumed proteolysis targets are yet to be determined. Considering the cycling expression of let-7, it is tempting to speculate that dre-1 could contribute to this pattern by targeting the destruction of positively acting regulators of let-7 transcription.
In summary, we have shown that transcription of let-7 miRNA oscillates during each larval stage. This pattern was unexpected given that levels of mature let-7 miRNA do not accumulate before the third larval stage and then steadily increase until adulthood (Van Wynsberghe et al., 2011). While the burst of let-7 transcription at each larval stage might enable rapid production of this miRNA and deter overexpression by limiting the amount of primary transcripts, it also necessitates post-transcriptional regulation by LIN-28 to prevent premature accumulation of mature miRNA (Lehrbach et al., 2009; Van Wynsberghe et al., 2011). Two transcriptional elements, the TRE and LTE, work redundantly and independently in specific cell types to coordinate spatio-temporal expression of let-7. The presence of at least one of these sites is required for let-7 expression and worm viability. We identified thirty-five genes with roles for positively or negatively regulating let-7 expression. Two of the candidates, dmd-5 and phi-7, are potential direct regulators of let-7 transcription, given their ability to associate with let-7 promoter sequences (Martinez et al., 2008a). The apparent overlapping functions of many trans-acting factors affecting let-7 transcription, combined with the redundancy, cooperation and independent regulation of transcription by two different cis-elements points to a complex network of regulatory molecules and signals dictating let-7 expression during worm development.
Supplementary Material
Highlights.
Transcription of let-7 from two start sites oscillates during each larval stage
2 promoter elements, the TRE and LTE, have redundant and non-overlapping roles
The novel let-7 transcription element regulates intestinal and hypodermal expression
An intact TRE or LTE is essential for let-7 transcription and, hence, viability
Multiple trans-acting factors regulate the spatio-temporal transcription of let-7
ACKNOWLEDGEMENTS
We thank members of the Pasquinelli lab for critical reading of the manuscript. Funding was provided by NIH CMG T32 GM007240 and NIH/NCI T32 CA009523 Training Grants (Z.S.K) and the US National Institutes of Health (GM071654), Keck, and Peter Gruber Foundations (A.E.P.).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- Abbott AL, Alvarez-Saavedra E, Miska EA, Lau NC, Bartel DP, Horvitz HR, Ambros V. The let-7 MicroRNA family members mir-48, mir-84, and mir-241 function together to regulate developmental timing in Caenorhabditis elegans. Dev Cell. 2005;9:403–414. doi: 10.1016/j.devcel.2005.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abrahante JE, Daul AL, Li M, Volk ML, Tennessen JM, Miller EA, Rougvie AE. The Caenorhabditis elegans hunchback-like gene lin-57/hbl-1 controls developmental time and is regulated by microRNAs. Developmental Cell. 2003;4:625–637. doi: 10.1016/s1534-5807(03)00127-8. [DOI] [PubMed] [Google Scholar]
- Ajuh P, Kuster B, Panov K, Zomerdijk JC, Mann M, Lamond AI. Functional analysis of the human CDC5L complex and identification of its components by mass spectrometry. Embo J. 2000;19:6569–6581. doi: 10.1093/emboj/19.23.6569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asahina M, Ishihara T, Jindra M, Kohara Y, Katsura I, Hirose S. The conserved nuclear receptor Ftz-F1 is required for embryogenesis, moulting and reproduction in Caenorhabditis elegans. Genes Cells. 2000;5:711–723. doi: 10.1046/j.1365-2443.2000.00361.x. [DOI] [PubMed] [Google Scholar]
- Bethke A, Fielenbach N, Wang Z, Mangelsdorf DJ, Antebi A. Nuclear hormone receptor regulation of microRNAs controls developmental progression. Science. 2009;324:95–98. doi: 10.1126/science.1164899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boudrez A, Beullens M, Groenen P, Van Eynde A, Vulsteke V, Jagiello I, Murray M, Krainer AR, Stalmans W, Bollen M. NIPP1-mediated interaction of protein phosphatase-1 with CDC5L, a regulator of pre-mRNA splicing and mitotic entry. J Biol Chem. 2000;275:25411–25417. doi: 10.1074/jbc.M001676200. [DOI] [PubMed] [Google Scholar]
- Boyerinas B, Sipkins DA. HSPCs in the balance: The vascular niche. Cell Stem Cell. 2010;7:645–646. doi: 10.1016/j.stem.2010.11.021. [DOI] [PubMed] [Google Scholar]
- Bracht J, Hunter S, Eachus R, Weeks P, Pasquinelli AE. Trans-splicing and polyadenylation of let-7 microRNA primary transcripts. RNA. 2004;10:1586–1594. doi: 10.1261/rna.7122604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carlson JM, Chakravarty A, DeZiel CE, Gross RH. SCOPE: a web server for practical de novo motif discovery. Nucleic Acids Res. 2007;35:W259–W264. doi: 10.1093/nar/gkm310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Celniker SE, Dillon LA, Gerstein MB, Gunsalus KC, Henikoff S, Karpen GH, Kellis M, Lai EC, Lieb JD, MacAlpine DM, Micklem G, Piano F, Snyder M, Stein L, White KP, Waterston RH. Unlocking the secrets of the genome. Nature. 2009;459:927–930. doi: 10.1038/459927a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choudhury NR, Michlewski G. Terminal loop-mediated control of microRNA biogenesis. Biochem Soc Trans. 2012;40:789–793. doi: 10.1042/BST20120053. [DOI] [PubMed] [Google Scholar]
- Esquela-Kerscher A, Johnson SM, Bai L, Saito K, Partridge J, Reinert KL, Slack FJ. Post-embryonic expression of C. elegans microRNAs belonging to the lin-4 and let-7 families in the hypodermis and the reproductive system. Dev Dyn. 2005;234:868–877. doi: 10.1002/dvdy.20572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fielenbach N, Guardavaccaro D, Neubert K, Chan T, Li D, Feng Q, Hutter H, Pagano M, Antebi A. DRE-1: an evolutionarily conserved F box protein that regulates C. elegans developmental age. Dev Cell. 2007;12:443–455. doi: 10.1016/j.devcel.2007.01.018. [DOI] [PubMed] [Google Scholar]
- Frand AR, Russel S, Ruvkun G. Functional genomic analysis of C. elegans molting. PLoS biology. 2005;3:e312. doi: 10.1371/journal.pbio.0030312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frokjaer-Jensen C, Davis MW, Hopkins CE, Newman BJ, Thummel JM, Olesen SP, Grunnet M, Jorgensen EM. Single-copy insertion of transgenes in Caenorhabditis elegans. Nature Genetics. 2008;40:1375–1383. doi: 10.1038/ng.248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gissendanner CR, Sluder AE. nhr-25, the Caenorhabditis elegans ortholog of ftz-f1, is required for epidermal and somatic gonad development. Developmental Biology. 2000;221:259–272. doi: 10.1006/dbio.2000.9679. [DOI] [PubMed] [Google Scholar]
- Hammell CM, Karp X, Ambros V. A feedback circuit involving let-7-family miRNAs and DAF-12 integrates environmental signals and developmental timing in Caenorhabditis elegans. Proc Natl Acad Sci U S A. 2009;106:18668–18673. doi: 10.1073/pnas.0908131106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirsh D, Oppenheim D, Klass M. Development of the reproductive system of Caenorhabditis elegans. Developmental Biology. 1976;49:200–219. doi: 10.1016/0012-1606(76)90267-0. [DOI] [PubMed] [Google Scholar]
- Hong CS, Park BY, Saint-Jeannet JP. The function of Dmrt genes in vertebrate development: it is not just about sex. Developmental Biology. 2007;310:1–9. doi: 10.1016/j.ydbio.2007.07.035. [DOI] [PubMed] [Google Scholar]
- Huntzinger E, Izaurralde E. Gene silencing by microRNAs: contributions of translational repression and mRNA decay. Nat Rev Genet. 2011;12:99–110. doi: 10.1038/nrg2936. [DOI] [PubMed] [Google Scholar]
- Jeon M, Gardner HF, Miller EA, Deshler J, Rougvie AE. Similarity of the C. elegans developmental timing protein LIN-42 to circadian rhythm proteins. Science. 1999;286:1141–1146. doi: 10.1126/science.286.5442.1141. [DOI] [PubMed] [Google Scholar]
- Johnson SM, Lin SY, Slack FJ. The time of appearance of the C. elegans let-7 microRNA is transcriptionally controlled utilizing a temporal regulatory element in its promoter. Developmental Biology. 2003;259:364–379. doi: 10.1016/s0012-1606(03)00202-1. [DOI] [PubMed] [Google Scholar]
- Kato M, Chen X, Inukai S, Zhao H, Slack FJ. Age-associated changes in expression of small, noncoding RNAs, including microRNAs, in C. elegans. Rna. 2011;17:1804–1820. doi: 10.1261/rna.2714411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelly WG, Xu S, Montgomery MK, Fire A. Distinct requirements for somatic and germline expression of a generally expressed Caernorhabditis elegans gene. Genetics. 1997;146:227–238. doi: 10.1093/genetics/146.1.227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kent WJ, Sugnet CW, Furey TS, Roskin KM, Pringle TH, Zahler AM, Haussler D. The human genome browser at UCSC. Genome research. 2002;12:996–1006. doi: 10.1101/gr.229102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim VN, Han J, Siomi MC. Biogenesis of small RNAs in animals. Nat Rev Mol Cell Biol. 2009;10:126–139. doi: 10.1038/nrm2632. [DOI] [PubMed] [Google Scholar]
- Kostrouchova M, Krause M, Kostrouch Z, Rall JE. Nuclear hormone receptor CHR3 is a critical regulator of all four larval molts of the nematode Caenorhabditis elegans. Proceedings of the National Academy of Sciences of the United States of America. 2001;98:7360–7365. doi: 10.1073/pnas.131171898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lehrbach NJ, Armisen J, Lightfoot HL, Murfitt KJ, Bugaut A, Balasubramanian S, Miska EA. LIN-28 and the poly(U) polymerase PUP-2 regulate let-7 microRNA processing in Caenorhabditis elegans. Nat Struct Mol Biol. 2009;16:1016–1020. doi: 10.1038/nsmb.1675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin SY, Johnson SM, Abraham M, Vella MC, Pasquinelli A, Gamberi C, Gottlieb E, Slack FJ. The C elegans hunchback homolog, hbl-1, controls temporal patterning and is a probable microRNA target. Developmental Cell. 2003;4:639–650. doi: 10.1016/s1534-5807(03)00124-2. [DOI] [PubMed] [Google Scholar]
- Lu XY, Lu Y, Zhao YJ, Jaeweon K, Kang J, Xiao-Nan L, Ge G, Meyer R, Perlaky L, Hicks J, Chintagumpala M, Cai WW, Ladanyi M, Gorlick R, Lau CC, Pati D, Sheldon M, Rao PH. Cell cycle regulator gene CDC5L, a potential target for 6p12-p21 amplicon in osteosarcoma. Molecular cancer research : MCR. 2008;6:937–946. doi: 10.1158/1541-7786.MCR-07-2115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez NJ, Ow MC, Barrasa MI, Hammell M, Sequerra R, Doucette-Stamm L, Roth FP, Ambros VR, Walhout AJ. A C. elegans genome-scale microRNA network contains composite feedback motifs with high flux capacity. Genes & development. 2008a;22:2535–2549. doi: 10.1101/gad.1678608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez NJ, Ow MC, Reece-Hoyes JS, Barrasa MI, Ambros VR, Walhout AJ. Genome-scale spatiotemporal analysis of Caenorhabditis elegans microRNA promoter activity. Genome research. 2008b;18:2005–2015. doi: 10.1101/gr.083055.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mondol V, Pasquinelli AE. Let's make it happen: the role of let-7 microRNA in development. Curr Top Dev Biol. 2012;99:1–30. doi: 10.1016/B978-0-12-387038-4.00001-X. [DOI] [PubMed] [Google Scholar]
- Pasquinelli AE. MicroRNAs and their targets: recognition, regulation and an emerging reciprocal relationship. Nat Rev Genet. 2012;13:271–282. doi: 10.1038/nrg3162. [DOI] [PubMed] [Google Scholar]
- Pasquinelli AE, Reinhart BJ, Slack F, Martindale MQ, Kuroda MI, Maller B, Hayward DC, Ball EE, Degnan B, Muller P, Spring J, Srinivasan A, Fishman M, Finnerty J, Corbo J, Levine M, Leahy P, Davidson E, Ruvkun G. Conservation of the sequence and temporal expression of let-7 heterochronic regulatory RNA. Nature. 2000;408:86–89. doi: 10.1038/35040556. [DOI] [PubMed] [Google Scholar]
- Reece-Hoyes JS, Shingles J, Dupuy D, Grove CA, Walhout AJ, Vidal M, Hope IA. Insight into transcription factor gene duplication from Caenorhabditis elegans Promoterome-driven expression patterns. BMC Genomics. 2007;8:27. doi: 10.1186/1471-2164-8-27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reinhart BJ, Slack FJ, Basson M, Pasquinelli AE, Bettinger JC, Rougvie AE, Horvitz HR, Ruvkun G. The 21-nucleotide let-7 RNA regulates developmental timing in Caenorhabditis elegans. Nature. 2000;403:901–906. doi: 10.1038/35002607. [DOI] [PubMed] [Google Scholar]
- Roush SF, Slack FJ. Transcription of the C. elegans let-7 microRNA is temporally regulated by one of its targets, hbl-1. Developmental Biology. 2009;334:523–534. doi: 10.1016/j.ydbio.2009.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rual JF, Ceron J, Koreth J, Hao T, Nicot AS, Hirozane-Kishikawa T, Vandenhaute J, Orkin SH, Hill DE, van den Heuvel S, Vidal M. Toward improving Caenorhabditis elegans phenome mapping with an ORFeome-based RNAi library. Genome research. 2004;14:2162–2168. doi: 10.1101/gr.2505604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sayed D, Abdellatif M. MicroRNAs in development and disease. Physiol Rev. 2011;91:827–887. doi: 10.1152/physrev.00006.2010. [DOI] [PubMed] [Google Scholar]
- Simmer F, Moorman C, van der Linden AM, Kuijk E, van den Berghe PV, Kamath RS, Fraser AG, Ahringer J, Plasterk RH. Genome-wide RNAi of C. elegans using the hypersensitive rrf-3 strain reveals novel gene functions. PLoS Biol. 2003;1:E12. doi: 10.1371/journal.pbio.0000012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stanojevic D, Hoey T, Levine M. Sequence-specific DNA-binding activities of the gap proteins encoded by hunchback and Kruppel in Drosophila. Nature. 1989;341:331–335. doi: 10.1038/341331a0. [DOI] [PubMed] [Google Scholar]
- Thornton JE, Gregory RI. How does Lin28 let-7 control development and disease? Trends Cell Biol. 2012;22:474–482. doi: 10.1016/j.tcb.2012.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Treisman J, Desplan C. The products of the Drosophila gap genes hunchback and Kruppel bind to the hunchback promoters. Nature. 1989;341:335–337. doi: 10.1038/341335a0. [DOI] [PubMed] [Google Scholar]
- Urano-Tashiro Y, Sasaki H, Sugawara-Kawasaki M, Yamada T, Sugiyama A, Akiyama H, Kawasaki Y, Tashiro F. Implication of Akt-dependent Prp19 alpha/14-3-3beta/Cdc5L complex formation in neuronal differentiation. Journal of neuroscience research. 2010;88:2787–2797. doi: 10.1002/jnr.22455. [DOI] [PubMed] [Google Scholar]
- Van Wynsberghe PM, Kai ZS, Massirer KB, Burton VH, Yeo GW, Pasquinelli AE. LIN-28 co-transcriptionally binds primary let-7 to regulate miRNA maturation in Caenorhabditis elegans. Nat Struct Mol Biol. 2011;18:302–308. doi: 10.1038/nsmb.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshizawa A, Nakahara Y, Izawa T, Ishitani T, Tsutsumi M, Kuroiwa A, Itoh M, Kikuchi Y. Zebrafish Dmrta2 regulates neurogenesis in the telencephalon. Genes Cells. 2011;16:1097–1109. doi: 10.1111/j.1365-2443.2011.01555.x. [DOI] [PubMed] [Google Scholar]
- Zhang N, Kaur R, Akhter S, Legerski RJ. Cdc5L interacts with ATR and is required for the S-phase cell-cycle checkpoint. EMBO Rep. 2009;10:1029–1035. doi: 10.1038/embor.2009.122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu L, Wilken J, Phillips NB, Narendra U, Chan G, Stratton SM, Kent SB, Weiss MA. Sexual dimorphism in diverse metazoans is regulated by a novel class of intertwined zinc fingers. Genes & development. 2000;14:1750–1764. [PMC free article] [PubMed] [Google Scholar]
- Zisoulis DG, Kai ZS, Chang RK, Pasquinelli AE. Autoregulation of microRNA biogenesis by let-7 and Argonaute. Nature. 2012;486:541–544. doi: 10.1038/nature11134. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.