Abstract
Transcription factors require coactivators and corepressors to modulate transcription in mammalian cells. The vitamin D receptor (VDR) utilizes coactivators and corepressors to gain tight control over the activity of a diverse set of genes that can regulate calcium transport, slow proliferation and promote immune responses. We have recently established the VDR/RXR cistrome in human colon cancer cells and have linked these binding sites to the genes that are regulated by 1α,25-dihydroxyvitamin D3 (1,25(OH)2D3). In additional studies described herein, we demonstrate that the coactivators SRC1, CBP and MED1 are recruited to upregulated genes to facilitate transcription as expected. SRC1 was the most highly correlated to VDR/RXR binding (50%). However, we also found that corepressor molecules such as NCoR and SMRT were present along with SRC1, CBP or MED1 at these 1,25(OH)2D3 activated gene enhancers. Interestingly, genome-wide NCoR binding mimicked VDR binding by increasing its association with VDR binding in response to 1,25(OH)2D3 treatment. Overall, these data indicate a complex role for corepressor and coactivator complexes in the activation or active repression of 1,25(OH)2D3 responsive genes.
Keywords: VDR, ChIP-seq, corepressor, coactivator, coregulator, vitamin D
1. INTRODUCTION
The steroid hormone 1α,25-dihydroxyvitamin D3 (1,25(OH)2D3) tightly regulates calcium and phosphorus homeostasis [1]. 1,25(OH)2D3 acts to regulate genes through its receptor, the vitamin D receptor (VDR), and its heterodimeric partner the retinoid X receptor (RXR). These genes are responsible for retention of calcium in the kidney, absorption of dietary calcium in the intestine and direct remodeling processes across the bone through osteoblasts and bone-resorbing osteoclasts. For the regulation of these processes, 1,25(OH)2D3 induces genes such as TRPV6, PMCA1B, and S100G whose products are directly involved in the transepithelial transfer of calcium from the gut lumen and SLC34A1 and SLC34A3 that facilitate phosphate uptake [2, 3] and genes such as CYP3A4, CYP3A7, CYP2B6, and ABCB1 that function to detoxify lithocholic acid and other secondary bile acids or are involved in the transport and metabolism of foreign compounds [4–6]. It is also known that 1,25(OH)2D3 exerts activity and regulation on the VDR gene itself as well as its catabolic enzyme CYP24A1 [7, 8].
Coactivators are essential for transcription initiation and are classically believed to provide linkage from receptor complexes to the basal transcriptional machinery [9]. Indeed it is now known that coactivators have catalytic domains that are responsible for modification of the chromatin environment such as acetylation, methylation, phosphorylation and many others [10]. The modulation of these chromatin marks defines the epigenome that drives cell-type and tissue-type specificity. Currently, there are over 300 transcriptional coregulators that have been described in the literature [11]. Coactivators, those believed to facilitate transcription, and corepressors, those believed to inhibit transcription, have been interchangeably described in the activation and/or repression of genes, making characterization difficult. It has been demonstrated, for example, that for full activation of estrogen responsive genes, the corepressor SMRT is required in the activation complex [12]. The coactivators and corepressors are able to directly interact with nuclear receptors like the VDR through LXXLL protein motifs [13] and SMRT is directly involved with vitamin D-mediated transcription [14] as well as other coregulatory molecules [15]. It is believed that ligand activated receptors can change conformations and preference for coactivators based upon which ligands are receptor bound [16]. Furthermore, post translational modifications of coactivators further diversify the activities transferred during coactivation of genes.
Recent advances in transcription research have revealed an extensive array of instructional epigenetic marks that are inserted across the genome in a cell-type specific manner [17]. Some epigenetic marks are associated specifically with regulatory regions, indicating that cellular phenotype is a direct consequence of the establishment of cell-specific enhancers by early lineage-specific transcription factors [18, 19]. Functional binding sites for the VDR/RXR revealed transcription factor interactions as well as the coregulator recruitment processes which identified at least some of the consequences of these interactions at sites on the genome [20]. In the current studies, we expanded on our recent discovery of the VDR/RXR cistrome by analyzing the coactivators (SRC1, CBP, MED1) as well as corepressors (NCoR, SMRT) involved during 1,25(OH)2D3-mediated transcription. We found a high correlation between SRC1 and CBP occupancy and transcriptional activation; however, we also found that repressors such as NCoR and SMRT were present in the activation complexes. These studies highlight the complex nature of transcription and coregulatory molecules.
2. MATERIALS AND METHODS
2.1 Reagents
1,25(OH)2D3 was obtained from Tetrionics, Inc. (Madison, WI). Antibodies to SRC-1 (M-341, sc-8495), CBP (A-22, sc-3996), MED1 (M-255, sc-8998), NCoR (C-20, sc-1609) were purchased from Santa Cruz Biotechnology, Inc. (Santa Cruz, CA). SMRT (PA1-843) antibody was purchased from Affinity Bioreagents (Thermo Fisher, Rockford, IL).
2.2 Cell Culture
Human LS180 CRC cells were obtained from ATCC (Manassas, VA). LS180 cells were cultured in minimum Eagle’s medium supplemented with 10% non-heat-inactivated fetal bovine serum from Hyclone (Logan, UT), 1% non-essential amino acids, 1% sodium pyruvate, and 1% penicillin-streptomycin from Invitrogen as previously reported [21].
2.3 Chromatin Immunoprecipitation Sequencing (ChIP-seq)
LS180 cells were treated for 3 hrs with vehicle or 10−7M 1,25(OH)2D3 prior to Chromatin ImmunoPrecipitation which was performed as described previously [21, 22]. ChIP-DNA was prepared and amplified using the Illumina ChIP-seq DNA preparation kit (1003473, #11257047 RevA), clusters formed and sequenced on the Illumina GAIIx or HiSeq2000 sequencers by the University of Wisconsin - Madison DNA Sequencing Facility in the University of Wisconsin- Madison Biotechnology Center [22]. Samples were further processed by two methods QuEST and HOMER. Peaks were accepted if they passed criteria of both methods. The data were analyzed with QuEST 2.4 [23] and HOMER [18] as previously reported [22]. All data and tracks have been deposited in the Gene Expression Omnibus (GEO) at GSE39277.
3. RESULTS AND DISCUSSION
3.1 VDR/RXR interact with coactivators to modulate transcription
We have previously quantitated the number of DNA binding sites for the vitamin D receptor (VDR) and its heterodimer partner retinoid x receptor (RXR) across the LS180 (human colon adenocarcinoma) genome using ChIP-seq analysis (FDR < 0.001) [22]. Cells were treated with either ethanol vehicle or 1,25(OH)2D3 for 3 hr and then subjected to the above analyses using validated antibodies to either VDR or RXR. We determined the number of high confidence peaks for VDR and RXR overlap in the 1,25(OH)2D3 condition to be 638 sites. A mapping analysis of peaks to surrounding genes revealed that 98% of VDR/RXR binding sites were located within either intergenic or intronic regions, which is consistent with findings for most transcription factors [24–26].
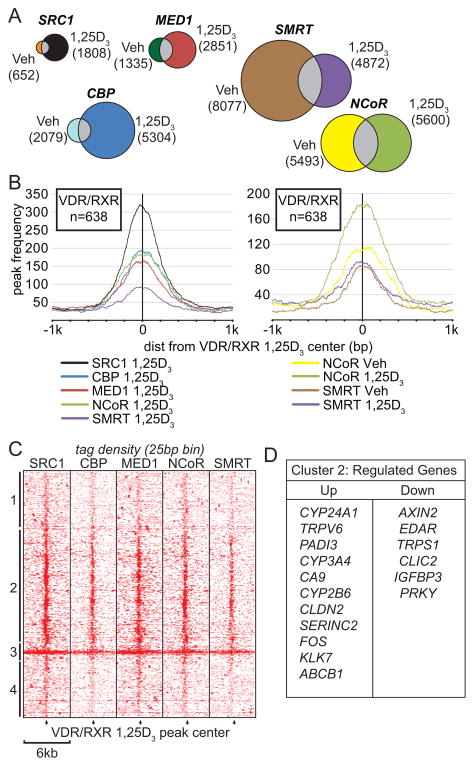
Transcription factor nuclear receptors, such as VDR and RXR, are known to recruit a variety of coregulatory complexes that are essential for altering gene expression [9]. To assess the ability of VDR to recruit these coregulatory complexes to target genes in a genome-wide manner, we conducted individual ChIP-seq analyses in LS180 cells using antibodies to the coactivators SRC1, MED1 and CBP, and to the corepressors NCoR and SMRT following treatment with either vehicle or 1,25(OH)2D3. Peaks were identified and quantitated as described in [22] using QuEST and HOMER [18, 23]. As can be seen in Figure 1a, while a number of sites for each of the coregulators was present under basal conditions, as might be anticipated for these general coregulators, an overlapping yet unique set of sites for each of these transcription factors was induced by 1,25(OH)2D3 as well. With the exception of those for SMRT, the total number of binding sites for all coregulators increased in the presence of 1,25(OH)2D3. The number of binding sites for VDR is greatly increased by 1,25(OH)2D3 treatment [22]. These data suggest that 1,25(OH)2D3 likely promotes the recruitment of not only coactivators but also corepressors such as NCoR or SMRT to gene targets as well. We next correlated these coregulatory sites to the peak centers of the 638 1,25(OH)2D3-induced VDR/RXR-bound sites. In this analysis, SRC1 peak frequency was most highly correlated with VDR/RXR peak frequency (50%) and the peaks were located very near VDR/RXR peak centers. Moreover, we also found that SRC1 tracked most closely with regulated genes of interest suggesting a strong functional relationship between SRC1 and VDR. Only the peak frequency of NCoR binding was reduced in the presence of 1,25(OH)2D3, suggesting that at some sites ligand activation of the VDR could result in corepressor displacement. Clearly, while corepressors may function at genes to mediate repression by 1,25(OH)2D3, they may also exert coactivator function as has been observed for the estrogen receptor (ER) [12]. These data suggest that coregulators may interact uniquely at VDR/RXR-bound sites and that these sites may display unique functional activities as well.
Figure 1.
VDR/RXR 1,25(OH)2D3 peaks interact with coregulators near actively regulated genes. A, Venn diagrams depict peak interactions from ChIP-seq data for SRC1, MED1, CBP, SMRT and NCoR in the Vehicle (Veh) and 1,25(OH)2D3 (1,25) conditions. Peak numbers are indicated next to condition for each cofactor. B, Peak interactions of coregulators and VDR/RXR NRABS peaks (peaks/n=638) are shown ±1kb near the VDR/RXR peak centers. The distance of maximal peak frequency in basepairs (bp) relative to VDR/RXR peak center is provided in the legend. C, The tag density (averaged in 25bp bins) from SRC1, CBP, MED1, NCoR, and SMRT ChIP-seq samples were correlated to ±3kb around each VDR/RXR peak center. Each row corresponds to a different peak (n=638). The heat map was clustered using hierarchical average linkage clustering into 4 major clusters numbered 1–4 on the left. VDR/RXR peak centers are indicated by black arrows. Red color denotes high density, white is zero. D, Cluster 2 was annotated to the nearest surrounding promoter and these promoters were correlated with gene expression analysis fold change (>95% confidence, >2-fold). The down-regulated genes and an abbreviated list of those that are up-regulated genes (>2-fold) and associated with Cluster 2 are displayed.
Since all 638 VDR/RXR binding sites were annotated to specific genomic locations and to their nearest gene neighbors, we linked these gene loci to the gene expression data documented in our previous work [22]. We then analyzed individual ChIP-seq tag densities for SRC1, CBP, MED1, NCoR and SMRT (25 bp bins) across a 6kb interval with the VDR/RXR site at its center (± 3kb). As shown in Figure 1c, each row of tag density represents a different VDR/RXR peak (each peak correlates to nearest gene). Based upon tag density of the coregulators, the data were clustered using hierarchical clustering with centroid linkage and 4 clusters were identified. Data clusters 1 and 4 contained reduced tag densities for many of the coregulators. Indeed, only a few of the genes located adjacent to these VDR/RXR peaks were regulated by 1,25(OH)2D3. Cluster 2, on the other hand, contained very high tag densities that were associated with all the coregulators and centered specifically on the VDR/RXR peaks themselves. This cluster of sites was linked most closely to the genes highly regulated by 1,25(OH)2D3 such as those listed in Figure 1d. Cluster 3 represents a small number (eight) of anomalous VDR/RXR binding sites that appeared to be unrelated to gene loci. These data suggest that 1,25(OH)2D3 induces both VDR/RXR binding and coregulator recruitment at specific sites and that these sites are capable of regulating genes that are located nearby.
3.2 c-FOS is controlled by coordinated coregulator action
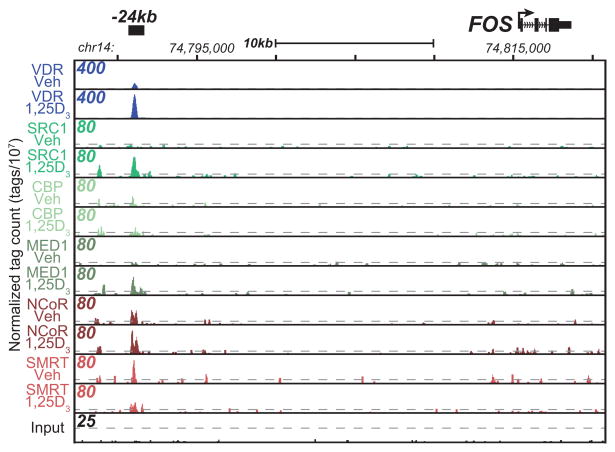
The results of the above studies confirm and extend mechanistic studies conducted over the past decade on several genes known to be regulated by 1,25(OH)2D3 in the intestine or colon, including CYP24A1, TRPV6, CYP2B6 and CYP3A4 [22]. c-FOS is a gene that is also known to be regulated by vitamin D and has the ability to function as a pro-proliferation or a pro-differentiation factor. As we previously discovered, c-FOS is upregulated by 1,25(OH)2D3 and there is a large amount of VDR/RXR binding at upstream of the c-FOS gene [22] (also seen in Figure 2). We found many, but not all, of the coregulators present at this site as well. SRC1 is not detectable before the addition of 1,25(OH)2D3 and afterwards, displays a sharp and strong peak of binding that correlates directly with the VDR/RXR binding. MED1 follows a similar pattern of recruitment and is not present before 1,25(OH)2D3 treatment. The corepressor NCoR is present at this site and is further increased with 1,25(OH)2D3 treatment (from 50 to 80 normalized reads). SMRT, on the other hand, is slightly decreased at this particular enhancer consistent with the mechanism of a corepressor being alleviated in response to coactivator recruitment. We believe that the accumulation of NCoR and/or SMRT in coordination with VDR/RXR binding leads to the conclusion that these factors are directly involved.
Figure 2.
The c-FOS locus is regulated by novel VDR/RXR binding. c-FOS genomic locus (chr14:74,787,306–74,820,798). ChIP-seq tag density profiles for VDR-Veh, VDR-1,25D3, SRC1-1,25D3, CBP-1,25D3, MED1-1,25D3, NCoR-1,25D3, SMRT-1,25D3, and input in the presence of 1,25(OH)2D3 are displayed centered around the c-FOS locus with gene transcriptional direction indicated by the arrow. The ChIP-seq tag densities have been normalized to 1×107 tags with the tag maximum for the data depicted on the top left of each track. FDR threshold of 0.001 is represented as a dashed line for each tag density track.
As we examine the coactivator activities throughout the genome, we find different combinations of coactivators being required for transcription. We believe that there is a direct association of the coactivators with the VDR/RXR and these activation complexes are diverse in the protein partners that are present. Since these ChIP assays are population assays with cells at different stages of activation, there is the possibility of the corepressors and coactivators to be in different complexes, yet appear to be co-bound by the ChIP-seq analysis. Extensive ChIP, re-ChIP assays might be able to tease apart these relationships further, however complexes may include transcription factors like VDR/RXR as well as NCoR and SRC1 or VDR/RXR with SMRT and CBP. Since there are over 300 coactivators known to interact in the genome, there are likely innumerable combinations of coactivation and/or corepression complexes [11]. Our studies represent the first step of identifying the coregulatory sites in the presence of 1,25(OH)2D3 activation. Many of these activities will need to be examined further through siRNA and other deletions, which is outside the scope of the studies herein. Through these experiments, the role of coactivators in 1,25(OH)2D3-mediated VDR/RXR transcription throughout the genome will be further defined which will help in therapeutic strategies that target coactivator activities.
Highlights.
We performed genome-wide ChIP-seq analysis on coactivators and corepressors during 1,25(OH)2D3 treatments.
The coactivator SRC-1 was most highly correlated with VDR/RXR during 1,25(OH)2D3 activation.
Corepressors NCoR and SMRT were also recruited to activation complexes near genes stimulated by 1,25(OH)2D3.
NCoR showed an increased association with VDR only after 1,25(OH)2D3 treatment, as exemplified by the gene c-FOS.
Acknowledgments
This work was supported by National Institutes of Health grant DK-073995 to J.W.P.
We thank members of the Pike Lab for their helpful discussions and contributions to this manuscript. We also acknowledge the University of Wisconsin DNA Sequencing Facility supported by Marie Adams, Josh Hyman and Eric Cabot. Linux server was maintained by Mindy Preston and Rebecca Hudson.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Plum LA, DeLuca HF. Vitamin D, disease and therapeutic opportunities. Nat Rev Drug Discov. 2010;9(12):941–955. doi: 10.1038/nrd3318. [DOI] [PubMed] [Google Scholar]
- 2.Bouillon R, Van Cromphaut S, Carmeliet G. Intestinal calcium absorption: Molecular vitamin D mediated mechanisms. J Cell Biochem. 2003;88(2):332–339. doi: 10.1002/jcb.10360. [DOI] [PubMed] [Google Scholar]
- 3.Nijenhuis T, Hoenderop JG, Nilius B, Bindels RJ. (Patho)physiological implications of the novel epithelial Ca2+ channels TRPV5 and TRPV6. Pflugers Arch. 2003;446(4):401–409. doi: 10.1007/s00424-003-1038-7. [DOI] [PubMed] [Google Scholar]
- 4.Jurutka PW, Thompson PD, Whitfield GK, Eichhorst KR, Hall N, Dominguez CE, Hsieh JC, Haussler CA, Haussler MR. Molecular and functional comparison of 1,25-dihydroxyvitamin D(3) and the novel vitamin D receptor ligand, lithocholic acid, in activating transcription of cytochrome P450 3A4. J Cell Biochem. 2005;94(5):917–943. doi: 10.1002/jcb.20359. [DOI] [PubMed] [Google Scholar]
- 5.Thummel KE, Brimer C, Yasuda K, Thottassery J, Senn T, Lin Y, Ishizuka H, Kharasch E, Schuetz J, Schuetz E. Transcriptional control of intestinal cytochrome P-4503A by 1a,25-dihydroxy vitamin D3. Mol Pharmacol. 2001;60(6):1399–1406. doi: 10.1124/mol.60.6.1399. [DOI] [PubMed] [Google Scholar]
- 6.Tachibana S, Yoshinari K, Chikada T, Toriyabe T, Nagata K, Yamazoe Y. Involvement of Vitamin D receptor in the intestinal induction of human ABCB1. Drug Metab Dispos. 2009;37(8):1604–1610. doi: 10.1124/dmd.109.027219. [DOI] [PubMed] [Google Scholar]
- 7.Meyer M, Goetsch P, Pike J. A downstream intergenic cluster of regulatory enhancers contributes to the induction of CYP24A1 Expression by 1alpha,25-dihydroxyvitamin D3. J Biol Chem. 2010;285(20):15599–15610. doi: 10.1074/jbc.M110.119958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zella L, Kim S, Shevde N, Pike J. Enhancers located within two introns of the vitamin D receptor gene mediate transcriptional autoregulation by 1,25-dihydroxyvitamin D3. Mol Endocrinol. 2006;20(6):1231–1247. doi: 10.1210/me.2006-0015. [DOI] [PubMed] [Google Scholar]
- 9.Klein-Hitpass L, Tsai SY, Weigel NL, Allan GF, Riley D, Rodriguez R, Schrader WT, Tsai MJ, O’Malley BW. The progesterone receptor stimulates cell-free transcription by enhancing the formation of a stable preinitiation complex. Cell. 1990;60(2):247–257. doi: 10.1016/0092-8674(90)90740-6. [DOI] [PubMed] [Google Scholar]
- 10.Johnson AB, Barton MC. Hypoxia-induced and stress-specific changes in chromatin structure and function. Mutat Res. 2007;618(1–2):149–162. doi: 10.1016/j.mrfmmm.2006.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lonard DM, O’Malley BW. Nuclear receptor coregulators: modulators of pathology and therapeutic targets. Nat Rev Endocrinol. 2012 doi: 10.1038/nrendo.2012.100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Peterson T, Karmakar S, Pace M, Gao T, Smith C. The silencing mediator of retinoic acid and thyroid hormone receptor (SMRT) corepressor is required for full estrogen receptor alpha transcriptional activity. Mol Cell Biol. 2007;27(17):5933–5948. doi: 10.1128/MCB.00237-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zella LA, Chang CY, McDonnell DP, Wesley Pike J. The vitamin D receptor interacts preferentially with DRIP205-like LxxLL motifs. Arch Biochem Biophys. 2007;460(2):206–212. doi: 10.1016/j.abb.2006.12.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kim JY, Son YL, Lee YC. Involvement of SMRT corepressor in transcriptional repression by the vitamin D receptor. Mol Endocrinol. 2009;23(2):251–264. doi: 10.1210/me.2008-0426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Oda Y, Sihlbom C, Chalkley RJ, Huang L, Rachez C, Chang CP, Burlingame AL, Freedman LP, Bikle DD. Two distinct coactivators, DRIP/mediator and SRC/p160, are differentially involved in vitamin D receptor transactivation during keratinocyte differentiation. Mol Endocrinol. 2003;17(11):2329–2339. doi: 10.1210/me.2003-0063. [DOI] [PubMed] [Google Scholar]
- 16.Bourguet W, Germain P, Gronemeyer H. Nuclear receptor ligand-binding domains: three-dimensional structures, molecular interactions and pharmacological implications. Trends Pharmacol Sci. 2000;21(10):381–388. doi: 10.1016/s0165-6147(00)01548-0. [DOI] [PubMed] [Google Scholar]
- 17.Zhou VW, Goren A, Bernstein BE. Charting histone modifications and the functional organization of mammalian genomes. Nat Rev Genet. 2011;12(1):7–18. doi: 10.1038/nrg2905. [DOI] [PubMed] [Google Scholar]
- 18.Heinz S, Benner C, Spann N, Bertolino E, Lin Y, Laslo P, Cheng J, Murre C, Singh H, Glass C. Simple combinations of lineage-determining transcription factors prime cis-regulatory elements required for macrophage and B cell identities. Mol Cell. 2010;38(4):576–589. doi: 10.1016/j.molcel.2010.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lin Y, Jhunjhunwala S, Benner C, Heinz S, Welinder E, Mansson R, Sigvardsson M, Hagman J, Espinoza C, Dutkowski J, Ideker T, Glass C, Murre C. A global network of transcription factors, involving E2A, EBF1 and Foxo1, that orchestrates B cell fate. Nat Immunol. 2010;11(7):635–643. doi: 10.1038/ni.1891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Raney BJ, Cline MS, Rosenbloom KR, Dreszer TR, Learned K, Barber GP, Meyer LR, Sloan CA, Malladi VS, Roskin KM, Suh BB, Hinrichs AS, Clawson H, Zweig AS, Kirkup V, Fujita PA, Rhead B, Smith KE, Pohl A, Kuhn RM, Karolchik D, Haussler D, Kent WJ. ENCODE whole-genome data in the UCSC genome browser (2011 update) Nucleic Acids Res. 2011;39(Database issue):D871–875. doi: 10.1093/nar/gkq1017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Meyer MB, Watanuki M, Kim S, Shevde NK, Pike JW. The human transient receptor potential vanilloid type 6 distal promoter contains multiple vitamin D receptor binding sites that mediate activation by 1,25-dihydroxyvitamin D3 in intestinal cells. Mol Endocrinol. 2006;20(6):1447–1461. doi: 10.1210/me.2006-0031. [DOI] [PubMed] [Google Scholar]
- 22.Meyer MB, Goetsch PD, Pike JW. VDR/RXR and TCF4/β-catenin cistromes in colonic cells of colorectal tumor origin: impact on c-FOS and c-MYC gene expression. Mol Endocrinol. 2012;26(1):37–51. doi: 10.1210/me.2011-1109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Valouev A, Johnson D, Sundquist A, Medina C, Anton E, Batzoglou S, Myers R, Sidow A. Genome-wide analysis of transcription factor binding sites based on ChIP-Seq data. Nat Methods. 2008;5(9):829–834. doi: 10.1038/nmeth.1246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Carroll JS, Liu XS, Brodsky AS, Li W, Meyer CA, Szary AJ, Eeckhoute J, Shao W, Hestermann EV, Geistlinger TR, Fox EA, Silver PA, Brown M. Chromosome-wide mapping of estrogen receptor binding reveals long-range regulation requiring the forkhead protein FoxA1. Cell. 2005;122(1):33–43. doi: 10.1016/j.cell.2005.05.008. [DOI] [PubMed] [Google Scholar]
- 25.Hatzis P, van der Flier L, van Driel M, Guryev V, Nielsen F, Denissov S, Nijman I, Koster J, Santo E, Welboren W, Versteeg R, Cuppen E, van de Wetering M, Clevers H, Stunnenberg H. Genome-wide pattern of TCF7L2/TCF4 chromatin occupancy in colorectal cancer cells. Mol Cell Biol. 2008;28(8):2732–2744. doi: 10.1128/MCB.02175-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nielsen R, Pedersen TA, Hagenbeek D, Moulos P, Siersbaek R, Megens E, Denissov S, Borgesen M, Francoijs KJ, Mandrup S, Stunnenberg HG. Genome-wide profiling of PPARgamma:RXR and RNA polymerase II occupancy reveals temporal activation of distinct metabolic pathways and changes in RXR dimer composition during adipogenesis. Genes Dev. 2008;22(21):2953–2967. doi: 10.1101/gad.501108. [DOI] [PMC free article] [PubMed] [Google Scholar]