Abstract
Background
Controversy exists regarding CYP2D6 genotype and tamoxifen efficacy.
Methods
A matched case-control study was conducted utilizing the Austrian Breast and Colorectal Cancer Study Group Trial 8 that randomized post-menopausal women with estrogen receptor positive breast cancer to tamoxifen for 5 years (Arm A) or tamoxifen for 2 years followed by anastrozole for 3 years (Arm B). Cases had disease recurrence, contralateral breast cancer, second non-breast cancer, or died. For each case, controls were identified from the same treatment arm of similar age, surgery/radiation, and TNM stage. Genotyping was performed for alleles associated with no (PM; *3, *4, *6); reduced (IM; *10, and *41); and extensive (EM: absence of these alleles) CYP2D6 metabolism.
Findings
The common CYP2D6 *4 allele was in Hardy Weinberg Equilibrium. In Arm A during the first 5 years of therapy, women with 2 poor alleles (PM/PM: OR=2.45, 95% CI: 1.05–5.73, p=0.04) and women with one poor allele (PM/IM or PM/EM: OR=1.67, 95% CI: 0.95–2.93, p=0.07) had a higher likelihood of an event than women with two extensive alleles (EM/EM). In years 3–5 when patients remained on tamoxifen (Arm A) or switched to anastrozole (Arm B), PM/PM tended towards a higher likelihood of a disease event relative to EM/EM (OR= 2.40, 95% CI: 0.86–6.66, p=0.09) among women on Arm A but not among women on Arm B (OR= 0.28; 95% CI: 0.03–2.30).
Conclusion
In ABCSG8, the negative effects of reduced CYP2D6 metabolism were observed only during the period of tamoxifen administration, and not after switching to anastrozole.
Keywords: Tamoxifen, CYP2D6, metabolism, anastrozole, breast cancer, estrogen receptor
Background
In the adjuvant treatment of postmenopausal estrogen receptor (ER) positive breast cancer, 5 years of an aromatase inhibitor (AI) or a sequencing regimen of 2 years of tamoxifen followed by 3 years of an AI significantly prolongs disease free survival (DFS) compared to 5 years of tamoxifen (1). Based on these data, practice guidelines recommend either an AI for 5 years, or the sequence of tamoxifen followed by an AI (2).
Tamoxifen is a weak anti-estrogen with agonistic properties that is extensively metabolized into potent anti-estrogens, 4-hydroxy tamoxifen and endoxifen, which exhibit similar potency in terms of binding affinity to ERs (3), suppression of estradiol-stimulated cell proliferation (4) and gene expression (3). Endoxifen is formed by the CYP2D6-mediated oxidation of n-desmethyl tamoxifen (4, 5). Common genetic variations in CYP2D6 and/or drug-induced inhibition of CYP2D6 enzyme activity are associated with significant reductions in endoxifen concentrations in tamoxifen treated humans (6–8). These data led to the hypothesis that CYP2D6 variation may affect the clinical outcomes of women treated with tamoxifen but not anastrozole, since CYP3A and not CYP2D6 is the major P450 isoform involved in the metabolism of anastrozole (9).
There has been great heterogeneity with regard to the reported association between CYP2D6 metabolism and clinical outcomes. Retrospective data from a randomized clinical trial of women treated with tamoxifen monotherapy for early stage ER positive breast cancer (NCCTG 89-30-52) (10), a pooled analysis of the women from NCCTG 89-30-52 and a respective German cohort (11) and other reports (reviewed in (12)) have demonstrated an association between CYP2D6 genotype and disease-free survival (DFS). However, multiple other reports have not demonstrated an association (reviewed in (12)) including a recent analyses of a subset of patients enrolled in two prospective adjuvant clinical trials, ATAC and BIG 1-98 (13, 14). However, concern has been raised (15) given the observation of substantial departure from Hardy-Weinberg equilibrium (HWE) for the most important CYP2D6 allele, *4 in both studies (13,14) Moreover, a review evaluating published studies found large between-study variability in the classification of genotypes and choice of primary endpoint as well as substantial methodological shortcomings leading to inconsistencies in study conclusions (16). Therefore, we sought to obtain independent determination whether the odds of a disease event differs by CYP2D6 genotype in women with early stage ER positive breast cancer who received 5 years of tamoxifen as adjuvant therapy by performing a secondary analysis of a large prospective tamoxifen study. We also aimed to determine the relationship between CYP2D6 genotype and outcomes in women receiving 2 years of tamoxifen followed by 3 years of anastrozole.
Methods
Patients
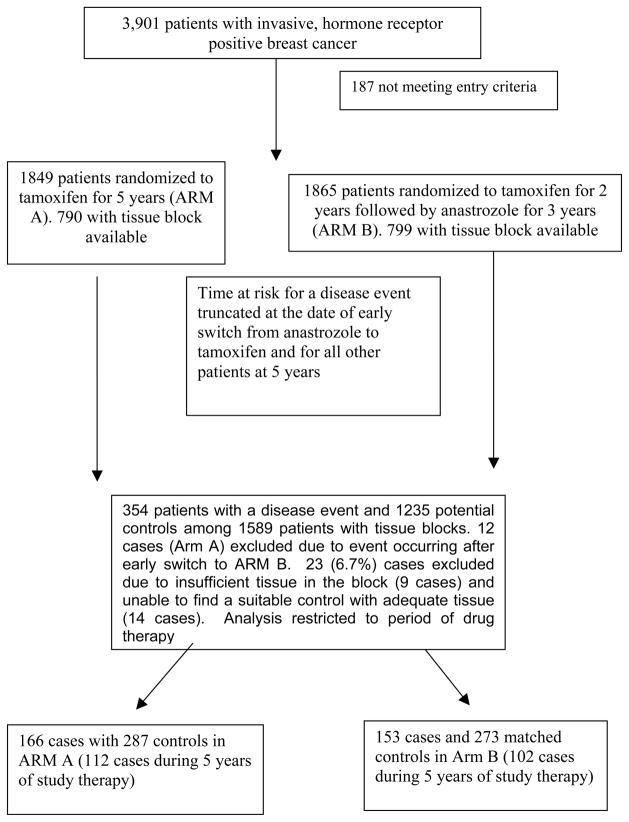
The source of patients was the Austrian Breast and Colorectal Study 8 (ABCSG trial 8, NCT00291759), a prospective, multicenter, randomized, open label trial that randomized 3,901 surgically resected early stage breast cancer patients within 6 weeks after surgery to either 5 years of tamoxifen (20 mg/daily) or to 2 years of tamoxifen (20 mg/daily) followed by a switch to anastrozole (1 mg/day) for 3 years (17). Eligible patients were postmenopausal women aged 80 years or younger with histologically verified ductal or lobular breast carcinoma that was invasive or minimally invasive, endocrine-responsive, and Nottingham grade 1 or grade 2. Neo-adjuvant chemotherapy (CT), hormone therapy (HT), or radiotherapy (RT) was not allowed. Patients underwent modified radical mastectomy or breast-conserving surgery with axillary lymph-node dissection or sentinel lymph-node biopsy (with or without subsequent RT). None of the patients received adjuvant CT. All patients provided written informed consent in accordance with the Declaration of Helsinki. The pharmacogenetics substudy was approved by the relevant ethics committees in Austria and the United States.
Sample Preparation
To overcome the potential problems related to somatic deletion of the CYP2D6 chromosomal locus on 22q13 (18, 19), three unmounted whole tissue sections (10-μm thick) derived from paraffin-embedded tissue blocks containing both normal and tumor tissue were prepared. One hematoxylin and eosin slide was also obtained to confirm tissue cellularity. From the unmounted whole sections, the tissue was deparaffinized, and DNA extracted using the modified method of Schroth et al (11).
Assay methods
DNA was assessed for the most common CYP2D6 single nucleotide polymorphisms (SNP) corresponding to alleles associated with null [*3 (2549 del A),*4(1846G>A), and *6(1707T>del)] reduced [*10 (100 C>T and 1846 G>A) and *41 (2988 G>A)] CYP2D6 enzyme activity as previously described (11) using the Applied Biosystems’ Taqman® Allelic Discrimination Assay (Foster City, CA) with the ABI Prism 7900HT Real Time System according to the manufacturer’s instructions. To ensure that non-specific amplification was not misinterpreted, the amplification plots were evaluated in addition to the end-point allelic discrimination plots. Two of the three triplicate reactions must concur and data derived from amplification beyond 45 cycles was not utilized. Samples from the Coriell Institute with known genotypes for each SNP and each possible allele combination (when available) were included on every plate and evaluated along with the unknown genotype samples. The presence of the alleles in the Coriell samples were confirmed by sequencing with validated methods that meet Hardy Weinberg equilibrium standards. The real-time methods were validated against the previously validated PCR and sequencing methods. A pooled DNA sample from ABI was also used in a standard curve to estimate the level of SNP detection for each run. The CYP2D6 *5 gene deletion allele and duplicated alleles could not be assessed because of DNA fragmentation which results from paraffin fixation.
CYP2D6 metabolism definition
CYP2D6 phenotype groups were defined as previously published (11) where ‘extensive’ metabolizers do not carry a null or reduced allele (EM/EM); those with one to two reduced alleles without a null allele (EM/IM, IM/IM); one null allele (PM/IM, PM/EM), and ‘poor’ metabolizers, those with 2 null alleles (PM/PM). Information regarding use of CYP2D6 inhibitors was unknown; however, the use of CYP2D6 inhibitors for the treatment of hot flashes was not recommended during the period of study enrollment on ABCSG 8.
Study Design and Analysis Plan
A matched case control study was conducted to examine whether the odds of a disease event differed with respect to CYP2D6 genotype. The definitions of a case and a control take into account the early release of the trial results in 2005 (17) as well as the opening of ABCSG 16 [Secondary Adjuvant Long-Term Study With Arimidex; (SALSA); NCT00295620]. That is, time at risk for a disease event was truncated at the date of switch to anastrozole for those women on Arm A who elected to switch from tamoxifen to anastrozole following the release the publication of the combined ABCSG-8/ARNO95 analysis (17). For other patients, events after 5 years of therapy were not eligible for analysis given that ABCSG 8 patients either enrolled onto ABCSG 16 or information regarding extended adjuvant hormonal therapy after 5 years was not collected. This analysis plan was consistent with the approach in the published parent ABCSG8 trial (20). Using the definition of invasive disease free survival by Hudis et al (21), a case was defined as a woman who had a documented local, regional, or distant recurrence of breast cancer; a contralateral breast cancer, or a 2nd non-breast primary cancer or died from any cause during her time at risk. For a given case, two controls were selected using an optimal matching (22, 23) from among women randomized to the same treatment arm whose age at randomization was within 5 years for the case; whose primary treatment (modified radical mastectomy +/− radiation therapy [RT] or breast conserving surgery [BCS] + RT versus BCS alone), tumor stage (I versus II/III), nodal status (positive versus negative) was the same as the case; and whose time at risk was longer than that of the case. In some situations, only one control was available for the case, either because matching criteria could not be met or adequate tissue was not available for genotyping of selected controls.
Conditional logistic regression modeling (CLRM) for matched triplets and pairs was used to examine whether the odds of a disease event differed with respect to CYP2D6 genotype in the following situations: 1) the first 5 years of treatment, for each arm separately 2) years 3–5 of treatment, for each arm separately, 3) in the first 2 years of treatment, (when all patients were assigned tamoxifen). Two-sided P-values < 0.05 were considered statistically significant. Analysis was performed using SAS (Version 9.2, SAS Institute Inc., Cary, NC). Assuming that 5% of the controls would be poor CYP2D6 metabolizers and the number of disease events would be 200, a two-sided alpha=0.05 chi-square test for the 2 to 1 matched odds ratio would have a 80% power for detecting an odds ratio of 2.5 or greater.
Results
Characteristics of the Patients
Of the 3,901 women enrolled in ABCSG 8, 1849 eligible patients were randomized to tamoxifen (Arm A) and 1865 eligible patients were randomized to the tamoxifen followed by anastrozole (Arm B) There were 790 patients from Arm A and 799 patients from Arm B with tissue blocks available. There were 354 patients with a disease event and 1235 potential controls among these 1589 patients. However, 12 cases from Arm A had an event after switching to anastrozole following early release of ABCSG 8 results (17) and as such were ineligible. Of the remaining 342 cases, 23 (6.7%) cases were excluded from the analysis due to insufficient tissue in the block (9 cases) and unable to find a suitable control with adequate tissue (14 cases) (Figure 1). Thus, 319 cases were matched to 557 matched controls (2 controls/case for 238 cases and 1 control/case for 81 cases). The number of cases eligible for analysis based on therapy period (years 1–2, 3–5) are given in Table 3. Cases occurring after the first 5 years of therapy were not eligible for analysis as outlined above. The breakdown of the type of disease events is given in Table 1 and the characteristics of the cases and controls overall and by treatment arm are given in Table 2.
Figure 1.
Consort diagram
Table 3.
Number of cases analyzed in specific follow-up time periods relative to drug therapy
| Within 2 Years | Years 3–5 | Within 5 Years | After Year 5* | Total | |
|---|---|---|---|---|---|
| Arm A | 28 | 84 | 112 | 54 | 166 |
| Arm B | 41 | 61 | 102 | 51 | 153 |
| Total | 69 | 145 | 214 | 105 | 319 |
Primary analysis excluded events occurring after 5 years (see methods).
Table 1.
First events among 319 cases.
| Event N (%) | |
|---|---|
| Local recurrence1 | 41 (12.9) |
| Distant recurrence1,2 | 90 (28.2) |
| Contralateral cancer | 28 (8.8) |
| Second primary2 | 91 (28.5) |
| Death | 78 (24.5) |
Seven patients experienced both local and distant recurrence as first event.
Two patients experienced both distant recurrence and second primary as first event.
Table 2.
Characteristics of the cases and controls
| Arm A | Arm B | Overall | ||||
|---|---|---|---|---|---|---|
| Cases | Controls | Cases | Controls | Cases | Controls | |
| n = 166 | n = 287 | n = 153 | n = 270 | n = 319 | n = 557 | |
| Age, years, median (range) | 66 (49–80) | 66 (48–80) | 69 (49–80) | 67 (47–79) | 68 (49–80) | 66 (47–80) |
| Treatment, n (%) | ||||||
| BCS without RT | 29 (17.5) | 45 (15.7) | 25 (16.3) | 45 (16.7) | 54 (16.9) | 90 (16.2) |
| BCS with RT | 95 (57.2) | 186 (64.8) | 90 (58.8) | 171 (63.3) | 185 (58.0) | 357 (64.1) |
| Mastectomy without RT | 39 (23.5) | 52 (18.1) | 35 (22.9) | 47 (17.4) | 74 (23.2) | 99 (17.8) |
| Mastectomy with RT | 3 (1.8) | 4 (1.4) | 3 (2.0) | 7 (2.6) | 6 (1.9) | 11 (2.0) |
| Tumor stage, n (%) | ||||||
| I | 102 (61.4) | 181 (63.1) | 94 (61.4) | 165 (61.1) | 196 (61.4) | 346 (62.1) |
| II/III | 64 (38.6) | 106 (36.9) | 59 (38.6) | 105 (38.9) | 123 (38.6) | 211 (37.9) |
| Node status, n (%) | ||||||
| Positive | 66 (39.8) | 108 (37.6) | 56 (36.6) | 93 (34.4) | 122 (38.2) | 201 (36.1) |
| Negative | 100 (60.2) | 179 (62.4) | 97 (63.4) | 177 (65.6) | 197 (61.8) | 356 (63.9) |
| Grade, n (%) | ||||||
| I | 29 (18.7) | 64 (24.3) | 29 (20.9) | 65 (27.1) | 58 (19.7) | 129 (25.6) |
| II | 126 (81.3) | 199 (75.7) | 110 (79.1) | 175 (72.9) | 236 (80.3) | 374 (74.4) |
| Unknown | 11 | 24 | 14 | 30 | 25 | 54 |
| Her2 status, n (%) | ||||||
| Positive | 13 (8.1) | 16 (5.8) | 9 (6.1) | 16 (6.1) | 22 (7.2) | 32 (5.9) |
| Negative | 147 (91.9) | 262 (94.2) | 138 (93.9) | 248 (93.9) | 285 (92.8) | 510 (94.1) |
| Unknown | 6 | 9 | 6 | 6 | 12 | 15 |
Abbreviations: BCS – Breast conserving surgery, RT – Radiation therapy
Genotype and allele frequency
The five CYP2D6 SNPs associated with the CYP2D6 alleles: *4, *6, *10, and *41, and the CYP2D6 *3 SNP were genotyped with a success rate of greater than 98% and 84%, respectively. The number of samples for each observed genotype and the corresponding minor allelic frequencies are shown in Table 4. The variant allele frequencies were similar to published reports in a predominantly Caucasian population, the predominant ethnic group in Austria (PhamGKB.org). Tests for HWE demonstrated that *4 (p=0.07) *6 (p=1.0) and *10 (p=0.54) alleles were within HWE, with some deviation for the *41 (p=0.009) and rare *3 allele (p=0.003).
Table 4.
Number of observed genotypes and minor allele frequencies (q) for CYP2D6 *3, *4, *6, *10, and *41 among the cases and controls
| CYP2D6 allele | Number (out of 876 samples) | minor allele frequency |
|---|---|---|
|
| ||
| CYP2D6 *3 (2549 delA) | q=0.01 | |
| Wt/Wt | 717 | |
| Wt/*3 | 13 | |
| *3/*3 | 2 | |
| No Call | 144 | |
|
| ||
| CYP2D6 *4 (1846 G>A) | q=0.21 | |
| Wt/Wt | 558 | |
| Wt/*4 | 271 | |
| *4/*4 | 47 | |
| No Call | 0 | |
|
| ||
| CYP2D6 *6 (1707 delT) | q=0.01 | |
| Wt/Wt | 831 | |
| Wt/*6 | 23 | |
| *6/*6 | 0 | |
| No Call | 22 | |
|
| ||
| #CYP2D6 *10 (100 C>T and 1846 G>A) | q=0.03 | |
| Wt/Wt | 806 | |
| Wt/*10 | 49 | |
| *10/*10 | 1 | |
| No call | 20 | |
|
| ||
| CYP2D6 *41 (2988 A) | q=0.09 | |
| Wt/Wt | 735 | |
| Wt/*41 | 125 | |
| *41/*41 | 13 | |
| No Call | 2 | |
#CYP2D6 *10 defined as 100T in absence of 1846A
Association of CYP2D6 phenotype with the likelihood of a disease event by treatment arm during the 5 years of therapy
In Arm A during the 5 years of tamoxifen treatment, PM/PM (OR 2.45, 95% CI: 1.05–5.73; p=0.04) had a higher odds of a disease event relative to EM/EM (Table 5). There was also a trend for patients classified as IM/PM or EM/PM relative to EM/EM (OR 1.67, 95% CI: 0.95–2.93, p=0.07) but not in patients classified as EM/IM or IM/IM relative to EM/EM (OR 1.23, 95% CI: 0.58–2.61, p=0.60) to have a higher odds of a disease event. Conversely in Arm B, no significant association was found between CYP2D6 genotype and the likelihood of a disease event during the 5 years of treatment.
Table 5.
Association between CYP2D6 phenotype and disease event during the 5 years of drug therapy in arms A and B
| Tamoxifen only (Arm A) | Tamoxifen followed by anastrozole (Arm B) | |||
|---|---|---|---|---|
| Odds Ratio (95% CI) | p-value | Odds Ratio (95% CI) | p-value | |
| PM/PM relative to EM/EM | 2.45 (1.05, 5.73) | 0.04 | 0.60 (0.15, 2.37) | 0.47 |
| EM/PM and PM/IM relative to EM/EM | 1.67 (0.95, 2.93) | 0.07 | 0.76 (0.43, 1.31) | 0.32 |
| EM/IM and IM/IM relative to EM/EM | 1.23 (0.58, 2.61) | 0.60 | 1.02 (0.52, 2.01) | 0.96 |
Association of CYP2D6 phenotype with the likelihood of a disease event in the first 2 years and last 3 years of treatment
Having observed marked differences between Arms A and B in terms of the association CYP2D6 genotype and the likelihood of a disease event over the 5 entire years of treatment, we performed a secondary analysis to evaluate the nature of this association in the first 2 years of treatment when all patients received tamoxifen and in years 3–5 when patients on Arm A continued tamoxifen and patients on Arm B switched to anastrozole. There were a limited number of events during years 1–2 (Table 3); however, a similar non-significant higher odds of a disease event was observed for PM/PM relative to EM/EM in the first 2 years of tamoxifen for both arms: Arm A: OR = 2.54, p = 0.25 and Arm B: OR = 2.60, p = 0.46.
During years 3–5, for women on Arm A who remained event free during the first 2 years of tamoxifen therapy, PM/PM (OR 2.40, 95% CI: 0.86 to 6.66, p = 0.09) and IM/PM or EM/PM (OR 1.70, 95% CI: 0.91–3.17, p=0.09) had a trend towards an increased odds of a disease event relative to EM/EM. In contrast, women on Arm B who remained event-free during the first 2 years of tamoxifen, the odds of disease event for PM/PM (OR 0.28, p = 0.23) and the IM/PM or EM/PM group (OR 0.63, p = 0.22) were found to be non-significantly decreased during years 3–5 relative to EM/EM.
Discussion
Among the post-menopausal women with ER positive breast cancer enrolled on ABCSG 8 who were randomized to 5 years of tamoxifen (Arm A), there was a significantly higher odds of a disease event for those with CYP2D6 PM/PM phenotype relative to those with the CYP2D6 EM/EM phenotype, but this was not observed in patients treated with anastrozole following tamoxifen (Arm B). Moreover, in Arm A, there was a strong trend towards a higher likelihood of a disease event (relative to EM/EM phenotype) in those that carried at least one poor allele (PM/IM and PM/EM), but not for patients without poor alleles (IM/IM and EM/IM). Because of the small number of patients with the IM/IM phenotype, no conclusions can be drawn regarding this group. The overall findings are consistent with pharmacokinetic data demonstrating a stepwise reduction in endoxifen concentrations based on number of PM alleles (8).
For ABCSG 8 patients randomized to 2 years of tamoxifen followed by 3 years of anastrozole (Arm B), CYP2D6 genotype was not associated with the odds of a disease event. However, breaking the treatment period into the tamoxifen phase and the anastrozole phase, we found non-significant higher odds of a disease event among PM/PM relative to EM/EM in the first 2 years of tamoxifen similar to Arm A but no evidence of significantly increased odds of a disease event during anastrozole treatment in years 3–5 for PM/PM phenotype relative to EM/EM phenotype. These data suggest that AI use following tamoxifen negates or even reverses the higher likelihood of disease recurrence observed in patients with reduced CYP2D6 metabolism. The observation that reduced metabolism is detrimental only during the period of tamoxifen administration may explain some of the controversy surrounding CYP2D6 genotype and tamoxifen efficacy, given that switching from tamoxifen to an AI is recommended (24). While early studies (prior to the routine use of AIs following tamoxifen) demonstrated an association between CYP2D6 and tamoxifen efficacy (11, 25) multiple recent studies have been negative (26–28). However, a major concern of the negative studies is the lack of assurance that the confounding impact of a switch to an AI was adequately considered.
Simon et al have proposed a refined system for biomarker studies that incorporates a hierarchal level of evidence scale for tumor marker studies using archived specimens (29). This “prospective-retrospective” design stipulates that the clinical and pathologic characteristics of patients in the biomarker study be representative of patients in the parent trial and that a sufficient number of patients with archived tissue be included for adequate statistical power. Notably the ATAC CYP2D6 analysis included 588 (18.9%) of the 3116 women randomized to tamoxifen and the clinical characteristics of genotyped patients differed significantly (p<0.005) in multiple important clinical characteristics (e.g., chemotherapy, radiation therapy, hormone receptor status), both compared with non-genotyped UK patients and rest of the world patients. Furthermore, only 89 (17%) of the 535 distant recurrences were included in the tamoxifen CYP2D6 analysis. Given the variants genotyped in ATAC, Schroth et al estimated that more than 1200 patients would be required to detect a hazard ratio of 1.85 between CYP2D6 poor (PM) and extensive metabolizer (EM) with 90% power (11). Therefore, the ATAC clinical analysis of CYP2D6 was neither representative of the entire population nor adequately powered.
In contrast to ATAC, the ABCSG 8 analysis of CYP2D6 included 52% (214/408) of all first events occurring during the first 5 years (20). Our analysis plan was to match cases to two controls from a pool of 1235 ABCSG patients who did not have a disease event based on known prognostic factors and exposure time in order to increase the power to detect a smaller odds ratio. Because our study design focused on early events occurring during the period of drug administration (years 1–5), an unanswered question remains whether alterations in CYP2D6 metabolism affect the risk of late recurrences (after tamoxifen discontinuation).
Simon et al additionally propose that for a “retrospective-prospective” design, the biomarker assay must be analytically and pre-analytically validated for use with archived tissues (29). CYP2D6 enzyme activity (and therefore endoxifen concentrations) results from both CYP2D6 germ-line variation, and the potency and duration of CYP2D6 inhibitors co-administered with tamoxifen. A limitation of this and other prospective clinical trials evaluating tamoxifen is that germ-line DNA was never collected, and information regarding concomitant medications is unknown. Because paraffin embedded tumors blocks which contain normal tissue are often collected, germline DNA may be extracted and used for genotyping. However, loss of heterozygosity (LOH) involving chromosome 22q13.1, the location of the CYP2D6 gene (18, 19) has been noted in ER positive tumors. Therefore, it is critical to assess for HWE, which states that both allele and genotype frequencies in a population remain constant assuming no new mutations, no selection, and random mating. Substantial departure from HWE may point to genotyping error or other biases. Notably, the BIG1-98 utilized tumor cores for CYP2D6 genotyping and demonstrated marked deviation from HWE for nearly all CYP2D6 alleles, including the most important CYP2D6 variant (*4) (HWE: p=10−91).
Within the ABCSG 8 cohort, in order to obtain sufficient numbers of normal epithelial cells for the detection of germline genotypes, we extracted DNA from tissue sections that contained both normal and tumor tissue, and the *4, *6, and *10 alleles were within HWE. However, moderate deviation was observed for the rare *3 allele, as well as reduced metabolism alleles (Table 4). The latter observation may relate to somatic deletion of the CYP2D6 chromosomal locus (18, 19) or the presence of germline deletion of the entire CYP2D6 gene (CYP2D6 *5) (30), leading to a deficit of observed heterozygotes. It should be noted, however, that the measured allele frequencies in this study were similar to previously published data from other European Caucasian populations (PharmGKB http://www.pharmgkb.org/index.jsp).
Prior studies evaluating the importance of adherence have indicated higher rates of non-adherence in patients without close follow-up(31) as well as in younger patients (< age 45) (32). While adherence was not formally monitored, ABCSG 8 enrolled postmenopausal women with a median age of and regular follow-up visits were required.
In summary, in the ABCSG 8 clinical trial, the CYP2D6 PM/PM phenotype was associated with a higher likelihood of an early disease event in women treated with 5 years of tamoxifen, but not in patients treated with sequential tamoxifen followed by anastrozole. Prospective studies are needed to determine whether altering the dose, the duration, or choice of adjuvant hormonal therapy based on CYP2D6 genotype or the pharmacological monitoring of endoxifen levels will improve the clinical outcomes of postmenopausal women with early stage ER positive breast cancer.
Translational Relevance.
There is controversy whether the efficacy of tamoxifen is altered in women with genetic or drug-induced alterations in CYP2D6, the rate limiting enzyme responsible for the metabolism of tamoxifen to its active metabolite, endoxifen. Whereas many negative studies have evaluated CYP2D6 polymorphisms in patients with a history of tamoxifen use, data from ABCSG 8 demonstrate that variation in CYP2D6 metabolism is associated with a higher risk of recurrence only during the period of tamoxifen administration, and not after switching to anastrozole. These data suggest that future studies should prospectively evaluate novel strategies to overcome the limitations of CYP2D6 metabolism, including the direct administration of endoxifen
Acknowledgments
Supported in part by 1R01CA133049-01 (Goetz, Suman, Weinshilboum, Ames)
Footnotes
Financial Disclosures: none
Dr. Goetz reports that he has been a consultant for Gtx (not reimbursed).
Dr. Gnant reports receiving research support from and serving as a consultant for AstraZeneca, Novartis, and Pfizer, and receiving lecture fees and honoraria for participation on advisory boards from AstraZeneca, Novartis, Sanofi-Aventis, Roche, Schering, Amgen, and Pfizer.
References
- 1.Dowsett M, Cuzick J, Ingle J, Coates A, Forbes J, Bliss J, et al. Meta-analysis of breast cancer outcomes in adjuvant trials of aromatase inhibitors versus tamoxifen. Journal of clinical oncology: official journal of the American Society of Clinical Oncology. 2010;28:509–18. doi: 10.1200/JCO.2009.23.1274. [DOI] [PubMed] [Google Scholar]
- 2.Burstein HJ, Prestrud AA, Seidenfeld J, Anderson H, Buchholz TA, Davidson NE, et al. American Society of Clinical Oncology clinical practice guideline: update on adjuvant endocrine therapy for women with hormone receptor-positive breast cancer. Journal of clinical oncology: official journal of the American Society of Clinical Oncology. 28:3784–96. doi: 10.1200/JCO.2009.26.3756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Johnson MD, Zuo H, Lee KH, Trebley JP, Rae JM, Weatherman RV, et al. Pharmacological characterization of 4-hydroxy- N -desmethyl tamoxifen, a novel active metabolite of tamoxifen. Breast cancer research and treatment. 2004;85:151–9. doi: 10.1023/B:BREA.0000025406.31193.e8. [DOI] [PubMed] [Google Scholar]
- 4.Stearns V, Johnson MD, Rae JM, Morocho A, Novielli A, Bhargava P, et al. Active tamoxifen metabolite plasma concentrations after coadministration of tamoxifen and the selective serotonin reuptake inhibitor paroxetine. J Natl Cancer Inst. 2003;95:1758–64. doi: 10.1093/jnci/djg108. [DOI] [PubMed] [Google Scholar]
- 5.Desta Z, Ward BA, Soukhova NV, Flockhart DA. Comprehensive evaluation of tamoxifen sequential biotransformation by the human cytochrome P450 system in vitro: prominent roles for CYP3A and CYP2D6. J Pharmacol Exp Ther. 2004;310:1062–75. doi: 10.1124/jpet.104.065607. [DOI] [PubMed] [Google Scholar]
- 6.Jin Y, Desta Z, Stearns V, Ward B, Ho H, Lee KH, et al. CYP2D6 genotype, antidepressant use, and tamoxifen metabolism during adjuvant breast cancer treatment. J Natl Cancer Inst. 2005;97:30–9. doi: 10.1093/jnci/dji005. [DOI] [PubMed] [Google Scholar]
- 7.Madlensky L, Natarajan L, Tchu S, Pu M, Mortimer J, Flatt SW, et al. Tamoxifen metabolite concentrations, CYP2D6 genotype, and breast cancer outcomes. Clin Pharmacol Ther. 2011;89:718–25. doi: 10.1038/clpt.2011.32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Murdter TE, Schroth W, Bacchus-Gerybadze L, Winter S, Heinkele G, Simon W, et al. Activity levels of tamoxifen metabolites at the estrogen receptor and the impact of genetic polymorphisms of phase I and II enzymes on their concentration levels in plasma. Clin Pharmacol Ther. 2011;89:708–17. doi: 10.1038/clpt.2011.27. [DOI] [PubMed] [Google Scholar]
- 9.Kamdem LK, Liu Y, Stearns V, Kadlubar SA, Ramirez J, Jeter S, et al. In vitro and in vivo oxidative metabolism and glucuronidation of anastrozole. British journal of clinical pharmacology. 2010;70:854–69. doi: 10.1111/j.1365-2125.2010.03791.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Goetz MP, Rae JM, Suman VJ, Safgren SL, Ames MM, Visscher DW, et al. Pharmacogenetics of tamoxifen biotransformation is associated with clinical outcomes of efficacy and hot flashes. Journal of clinical oncology: official journal of the American Society of Clinical Oncology. 2005;23:9312–8. doi: 10.1200/JCO.2005.03.3266. [DOI] [PubMed] [Google Scholar]
- 11.Schroth W, Goetz MP, Hamann U, Fasching PA, Schmidt M, Winter S, et al. Association between CYP2D6 polymorphisms and outcomes among women with early stage breast cancer treated with tamoxifen. Jama. 2009;302:1429–36. doi: 10.1001/jama.2009.1420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sideras K, Ingle JN, Ames MM, Loprinzi CL, Mrazek DP, Black JL, et al. Coprescription of tamoxifen and medications that inhibit CYP2D6. J Clin Oncol. 2010;28:2768–76. doi: 10.1200/JCO.2009.23.8931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rae JM, Drury S, Hayes DF, Stearns V, Thibert JN, Haynes BP, et al. CYP2D6 and UGT2B7 Genotype and Risk of Recurrence in Tamoxifen-Treated Breast Cancer Patients. Journal of the National Cancer Institute. 2012;104:452–60. doi: 10.1093/jnci/djs126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Regan MM, Leyland-Jones B, Bouzyk M, Pagani O, Tang W, Kammler R, et al. CYP2D6 Genotype and Tamoxifen Response in Postmenopausal Women with Endocrine-Responsive Breast Cancer: The Breast International Group 1-98 Trial. Journal of the National Cancer Institute. 2012;104:441–51. doi: 10.1093/jnci/djs125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Brauch HB, Schroth W, Ingle JN, Goetz MP. CYP2D6 and Tamoxifen: Awaiting the Denouement. J Clin Oncol. 2011;29:4589–90. doi: 10.1200/JCO.2011.38.8611. [DOI] [PubMed] [Google Scholar]
- 16.Dahabreh I, Terasawa T, Castaldi P, Trikalinos TA. CYP2D6 testing to predict response to tamoxifen in women with breast cancer: Pharmacogenomic. PLoS Curr. 2010;2:RRN1176. doi: 10.1371/currents.RRN1176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jakesz R, Jonat W, Gnant M, Mittlboeck M, Greil R, Tausch C, et al. Switching of postmenopausal women with endocrine-responsive early breast cancer to anastrozole after 2 years’ adjuvant tamoxifen: combined results of ABCSG trial 8 and ARNO 95 trial. Lancet. 2005;366:455–62. doi: 10.1016/S0140-6736(05)67059-6. [DOI] [PubMed] [Google Scholar]
- 18.Castells A, Gusella JF, Ramesh V, Rustgi AK. A region of deletion on chromosome 22q13 is common to human breast and colorectal cancers. Cancer Res. 2000;60:2836–9. [PubMed] [Google Scholar]
- 19.Hirano A, Emi M, Tsuneizumi M, Utada Y, Yoshimoto M, Kasumi F, et al. Allelic losses of loci at 3p25.1, 8p22, 13q12, 17p13.3, and 22q13 correlate with postoperative recurrence in breast cancer. Clin Cancer Res. 2001;7:876–82. [PubMed] [Google Scholar]
- 20.Dubsky PC, Jakesz R, Mlineritsch B, Postlberger S, Samonigg H, Kwasny W, et al. Tamoxifen and anastrozole as a sequencing strategy: a randomized controlled trial in postmenopausal patients with endocrine-responsive early breast cancer from the austrian breast and colorectal cancer study group. J Clin Oncol. 2012;30:722–8. doi: 10.1200/JCO.2011.36.8993. [DOI] [PubMed] [Google Scholar]
- 21.Hudis CA, Barlow WE, Costantino JP, Gray RJ, Pritchard KI, Chapman JA, et al. Proposal for standardized definitions for efficacy end points in adjuvant breast cancer trials: the STEEP system. J Clin Oncol. 2007;25:2127–32. doi: 10.1200/JCO.2006.10.3523. [DOI] [PubMed] [Google Scholar]
- 22.Bergstralh E, Kosanke J. Computerized matching of cases to controls. Rochester: Department of Health Science Research, Mayo Clinic; 1995. [Google Scholar]
- 23.Rosenbaum PR. Optimal matching for observational studies. JASA. 1989;84:1024–32. [Google Scholar]
- 24.Burstein HJ, Griggs JJ, Prestrud AA, Temin S. American society of clinical oncology clinical practice guideline update on adjuvant endocrine therapy for women with hormone receptor-positive breast cancer. Journal of oncology practice/American Society of Clinical Oncology. 2010;6:243–6. doi: 10.1200/JOP.000082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Goetz MP, Rae JM, Suman VJ, Safgren SL, Ames MM, Visscher DW, et al. Pharmacogenetics of tamoxifen biotransformation is associated with clinical outcomes of efficacy and hot flashes. J Clin Oncol. 2005;23:9312–8. doi: 10.1200/JCO.2005.03.3266. [DOI] [PubMed] [Google Scholar]
- 26.Abraham JE, Maranian MJ, Driver KE, Platte R, Kalmyrzaev B, Baynes C, et al. CYP2D6 gene variants: association with breast cancer specific survival in a cohort of breast cancer patients from the United Kingdom treated with adjuvant tamoxifen. Breast Cancer Res. 12:R64. doi: 10.1186/bcr2629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Park IH, Ro J, Park S, Lim HS, Lee KS, Kang HS, et al. Lack of any association between functionally significant CYP2D6 polymorphisms and clinical outcomes in early breast cancer patients receiving adjuvant tamoxifen treatment. Breast Cancer Res Treat. 2012;131:455–61. doi: 10.1007/s10549-011-1425-2. [DOI] [PubMed] [Google Scholar]
- 28.Lash TL, Cronin-Fenton D, Ahern TP, Rosenberg CL, Lunetta KL, Silliman RA, et al. CYP2D6 inhibition and breast cancer recurrence in a population-based study in Denmark. Journal of the National Cancer Institute. 2011;103:489–500. doi: 10.1093/jnci/djr010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Simon RM, Paik S, Hayes DF. Use of archived specimens in evaluation of prognostic and predictive biomarkers. Journal of the National Cancer Institute. 2009;101:1446–52. doi: 10.1093/jnci/djp335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gaedigk A, Blum M, Gaedigk R, Eichelbaum M, Meyer UA. Deletion of the entire cytochrome P450 CYP2D6 gene as a cause of impaired drug metabolism in poor metabolizers of the debrisoquine/sparteine polymorphism. Am J Hum Genet. 1991;48:943–50. [PMC free article] [PubMed] [Google Scholar]
- 31.Osterberg L, Blaschke T. Adherence to medication. N Engl J Med. 2005;353:487–97. doi: 10.1056/NEJMra050100. [DOI] [PubMed] [Google Scholar]
- 32.Barron TI, Connolly R, Bennett K, Feely J, Kennedy MJ. Early discontinuation of tamoxifen: a lesson for oncologists. Cancer. 2007;109:832–9. doi: 10.1002/cncr.22485. [DOI] [PubMed] [Google Scholar]