Abstract
Membrane phospholipid synthesis is a vital facet of bacterial physiology. Although the spectrum of phospholipid headgroup structures produced by bacteria is large, the key precursor to all of these molecules is phosphatidic acid (PtdOH). Glycerol-3-phosphate derived from the glycolysis via glycerol-phosphate synthase is the universal source for the glycerol backbone of PtdOH. There are two distinct families of enzymes responsible for the acylation of the 1-position of glycerol-3-phosphate. The PlsB acyltransferase was discovered in Escherichia coli, and homologs are present in many eukaryotes. This protein family primarily uses acyl-acyl carrier protein (ACP) endproducts of fatty acid synthesis as acyl donors, but may also use acyl-CoA derived from exogenous fatty acids. The second protein family, PlsY, is more widely distributed in bacteria and utilizes the unique acyl donor, acyl-phosphate, which is produced from acyl-ACP by the enzyme PlsX. The acylation of the 2-position is carried out by members of the PlsC protein family. All PlsCs use acyl-ACP as the acyl donor, although the PlsCs of the γ-proteobacteria also may use acyl-CoA. Phospholipid headgroups are precursors in the biosynthesis of other membrane-associated molecules and the diacylglycerol product of these reactions is converted to PtdOH by one of two distinct families of lipid kinases. The central importance of the de novo and recycling pathways to PtdOH in cell physiology suggest these enzymes are suitable targets for the development of antibacterial therapeutics in Gram-positive pathogens. This article is part of a Special Issue entitled Phospholipids and Phospholipid Metabolism.
Keywords: bacteria, acyltransferase, phosphatidic acid, glycerol-phosphate, acyl carrier protein, coenzyme A, diacylglycerol
1. Introduction
Bacteria produce a bewildering variety of phospholipids that play critical roles in the adaptation to the environment. Phosphatidic acid (PtdOH) (Fig. 1) is the universal precursor required for the production of these molecules. In contrast to the variety of enzymes involved in producing the broad spectrum of bacterial phospholipid structures, there are only a limited number of enzymes required for the formation of PtdOH, the key intermediate in their synthesis. This review covers the enzymes and pathways responsible for the de novo formation of PtdOH and the recycling enzymes that produce PtdOH from the diacylglycerol formed from the utilization of phospholipids in the biosynthesis of other molecules. In the early days of research on bacterial lipid metabolism, the field thought that understanding these pathways would provide important insight into how all cells construct their phospholipids. Indeed, the elucidation of the formation of sn-glycerol-3-phosphate (G3P) and the consecutive acylation of the 1-position followed by the 2-position of G3P in E. coli identified enzymes and genes that have homologous sequences and functions in mammalian systems. More recently, it has become apparent that a primary bacterial enzyme for the acylation of the 1-position of G3P in many human pathogens has no mammalian homologs and uses a different acyl donor that other acyltransferases in biology. This review covers the current knowledge and future directions for research on these two primary acyltransferase pathways to PtdOH and the kinases involved in PtdOH metabolism in bacteria.
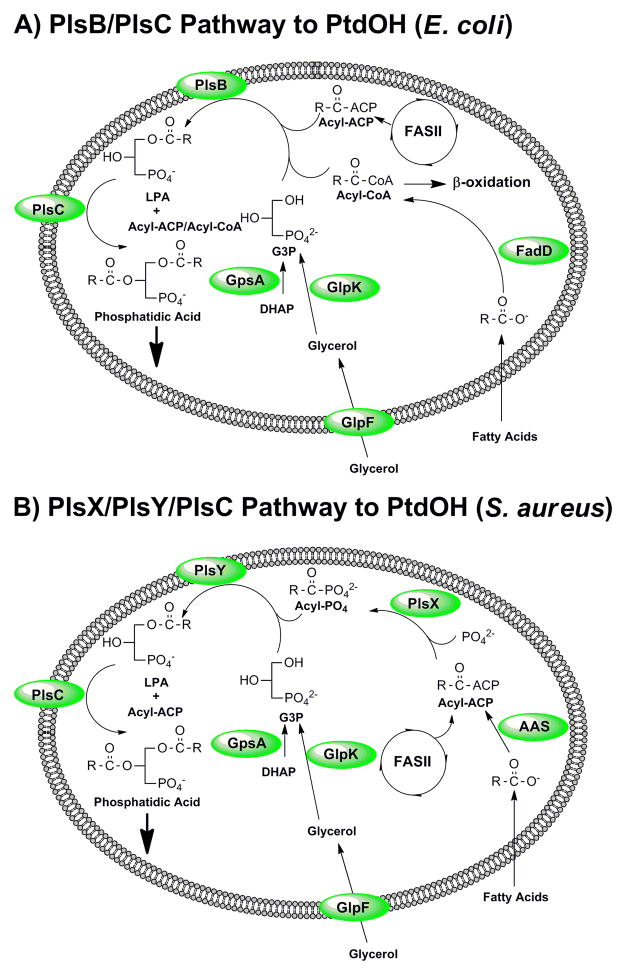
Fig. 1.
Pathways for the biosynthesis of PtdOH in bacteria. (A) PtdOH metabolism in E. coli is representative of the bacteria that utilize the PlsB/PlsC acyltransferase pathway to PtdOH. These acyltransferases use either acyl-ACP substrates produced by type II fatty acid synthesis (FASII) or acyl-CoA thioesters generated by the activation of exogenous fatty acids by acyl-CoA synthetase (FadD). The PlsB pathway is largely confined to the γ-proteobacteria. (B) PtdOH metabolism in S. aureus is representative of bacteria that utilize the PlsX/PlsY/PlsC acyltransferase pathway to PtdOH. Acyl-ACP generated by FASII is either used by PlsC to acylate the 2-position of LPA or is converted by PlsX to acyl-PO4 for incorporation into the 1-position by PlsY. This is the only pathway present in most Gram-positive pathogens. In both schemes, the G3P backbone is produced by GpsA, but may also be obtained from the environment by the GlpF/GlpK pathway. Many bacteria also have a transport system for G3P (not shown). Exogenous fatty acids are incorporated into phospholipid following their ligation to ACP by acyl-ACP synthetases (AAS). PlsB, PlsC, PlsY and GlpF are intrinsic membrane proteins. AAS, FadD and PlsX are soluble proteins that are thought to interact with the membrane interface.
2. Glycerol-3-Phosphate
PtdOH is the biosynthetic product of the esterification of two fatty acids onto the two hydroxyl groups of sn-glycerol-3-phosphate (G3P). The formation of G3P from the reduction of dihydroxyacetone phosphate by the G3P synthase (GpsA) is the only de novo pathway to G3P in bacteria [1–4] (Fig. 1). Dihydroxyacetone phosphate is diverted from the glycolytic pathway, so GpsA links intermediary and lipid metabolism. GpsA is different from GlpD, the aerobic G3P dehydrogenase, which breaks down G3P for energy production [5]. The inactivation of the gpsA genein E. coli and Stapholococcus aureus gives rise to glycerol or G3P auxotrophs illustrating that GpsA is required for the biosynthesis of G3P in vivo [1,6]. E. coli GpsA is a soluble enzyme that is strongly inhibited by its product, G3P. The stringent regulation of the production of G3P buffers the intracellular G3P concentration to ensure a steady supply of G3P for lipid biosynthesis. However, the experimental manipulation of the intracellular G3P concentration showed that it does not have a role in regulating phospholipid formation [7]. In contrast, Bacillus subtilis GpsA is not feedback inhibited by G3P, possibly due to Gram positive bacteria requiring more G3P units for cell wall biosynthesis than for phospholipid formation [8]. A major Gram-positive cell wall component lipotechoic acid contains 14–33 G3P units [9,10]. The increased metabolic demand for G3P correlates with the relaxed regulation of the GpsA in Gram-positive bacteria.
Bacteria also obtain G3P directly from the environment (GlpT) or through the uptake and phosphorylation of glycerol by GlpF and GlpK. E. coli can use G3P and glycerol as the sole carbon source through utilization of the genes encoded in the glp regulon, spread over 5 operons that allow the import and metabolism of G3P, glycerol, and glycerophosphodiesters [11,12]. Expression of the glp regulon is controlled at two levels. At the global level, the expression of the glp genes are suppressed when preferred carbon sources, such as glucose, are present through the regulation by the cAMP-CRP complex as a part of global catabolite repression [13]. At the local level, glp regulon expression is controlled by the glp represser, GlpR [14]. GlpR is a tetrameric protein that binds to the operators of the glp operons to prevent transcription [11,12]. G3P induces the expression of the regulon by binding to GlpR and decreasing the affinity of GlpR for the operators. The glp operons have differential sensitivity to the repressor, with glpFK operon approximately 3 times more sensitive to repression than the glpTQ operon [14]. The regulatory mechanism of glycerol and G3P metabolism from E. coli is not representative of all other bacteria. Bacillus subtilis glycerol and G3P utilizing genes are organized differently and expression of the glp regulon is controlled by the antiterminator protein GlpP [2,15]. Certain bacteria, such as Streptococcus pneumonia don’t have the genes for glycerol or G3P metabolism and therefore can’t metabolize exogenous G3P or glycerol at all. Most of the genes in the glp regulon are involved in breaking down G3P for energy, and therefore not the subject of this review. However, three gene products from the glp regulon, GlpT, GlpF, and GlpK, are involved in assimilating G3P from the environment and are important to understanding how gpsA mutants are used to study PtdOH metabolism.
The organophosphate:phosphate antiporter GlpT actively transports G3P into the cell using the energy from the efflux of phosphate [16–18]. E. coli GlpT is the best characterized family member and serves as a model for the GlpT from other bacteria [16]. E. coli GlpT consists of twelve transmembrane helices spanning the inner membrane. The crystal structure of E. coli GlpT has been solved to 3.3 Å [19,20]. GlpT operates via a single binding site mechanism. The antiporter alternates between two conformations: the Ci conformation where the active site is accessible from the cytosol and the Co conformation where the active site is accessible from the periplasm. Inorganic phosphate binding to the Ci conformation causes the transporter to adopt the Co conformation, transporting the phosphate across the membrane and allowing G3P to bind. G3P binding to the Co conformation causes conformation change back into the Ci conformation, transporting G3P into the cytosol and allowing another cycle of transport.
The aquaglyceroporin GlpF facilitates the passive diffusion of glycerol through the cell membrane [21,22], and the glycerol kinase, GlpK, phosphorylates glycerol to trap G3P inside the cell [23,24]. GlpF mediated influx is rapid, highly selective for glycerol and other polyols, and essentially nonsaturable [25,26]. The crystal structure of GlpF shows that each GlpF monomer of the associated tetramer forms a glycerol conducting channel with two half-membrane-spanning and six transmembrane α helices [21]. The helices form a narrow amphipathic selectivity channel that is wide enough to accommodate a single CH-OH group that force the glycerol hydroxyls to traverse the channel in single file. Intracellular glycerol is trapped by GlpK phosphorylation. E. coli GlpK is a soluble protein that associates as a homotetramer [24]. E. coli GlpK operates via an ordered mechanism where glycerol binds first to the enzyme followed by ATP. Fructose-1,6-bisphosphate allosterically inhibits E. coli GlpK, and the feedback inhibition prevents the overproduction of G3P when glucose is present in the media [27,28]. The growth of E. coli constitutively expressing a mutant GlpK that is refractory to fructose-1,6-bisphosphate regulation is inhibited by extracellular glycerol [29] illustrating that the overproduction of intracellular G3P causes growth stasis in E. coli.
3. Acyl Donors
3.1. Acyl-ACP (acyl-acyl carrier protein)
ACP is the predominant acyl group carrier in bacterial fatty acid synthesis [30] (Fig. 1). ACP is a 9 kDa protein with a 4′-phosphopantetheine prosthetic group that that carries the acyl chains as thioesters. In most bacteria, the dissociated type II fatty acid biosynthesis pathway (FASII) is the de novo pathway to fatty acids, and all of the intermediates are attached to ACP, which shuttles the acyl groups through the elongation process [31]. Long-chain acyl-ACP intermediates undergo either additional rounds of elongation or become substrates for the acyltransferase system depending on the length of the acyl group. Increasing acyl-ACP chain lengths become progressively poorer substrates for the elongation condensing enzymes of FASII, and at the same time become better substrates for the acyltransferases. Thus, the chain-length composition of the phospholipids is dependent upon the competition between the elongation activity of FASII and the rate of incorporation by the acyltransferase system. The combined effect of these substrate specificities means that most bacteria phospholipids are composed of 15–20 carbon fatty acids [32,33]. Increasing the relative rate of elongation compared to acyltransfer by either overexpressing the elongation condensing enzymes or decreasing acyltransferase activity increases the average chain lengths of the fatty acids incorporated into PtdOH [34,35]. The ratio of unsaturated:saturated fatty acids in PtdOH is determined by kinetic competition at the 10-carbon branch-point in FASII [31], and is not directly affected by acyltransferase activity. Many Gram-positive bacteria are capable of ligating exogenous fatty acids onto ACP. The resulting acyl-ACPs may either be elongated by FASII or utilized for phospholipid synthesis. Acyl-ACP synthetases are known [36]; however, the gene(s) responsible for the acyl-ACP synthetase activity in Gram-positive pathogens remains to be identified [37].
3.2. Acyl-PO4 (acyl-phosphate)
While the G3P acyltransferase responsible for PtdOH synthesis in E. coli (PlsB) uses acyl-ACP as the acyl donor (Fig. 1A), the most widely distributed bacterial G3P acyltransferase (PlsY) uses acyl-PO4 as the donor [38,39] (Fig. 1B). Acyl-PO4 is a phosphoric acid mixed anhydride generated from acyl-ACP and phosphate by the PlsX enzyme. Acyl-PO4 was synthesized by Lehninger in 1945 [40], but there is no known role for acyl-PO4 in mammalian fatty acid biosynthesis. It has poorer solubility and shorter half-life (12 hours) than acyl-ACP, but also exists at low concentrations and presumed to be a short-lived intermediate. PlsX is a soluble enzyme with solved crystal structures [41,42]. PlsX structure has homology to phosphotransacetylase, and is presumed to operate via an analogous mechanism, although no substrate binding or catalysis data is available. In Bacillus subtilis, PlsX associates with the membrane, but the basis for this association has not been explored [43]. E. coli has genes for both the plsB and plsX/plsY G3P acyltransferase systems; however, plsB is an essential gene, whereas neither plsX nor plsY are [44]. But this does not mean that PlsX has no role in E. coli metabolism because the plsX gene was first discovered as a second site gene mutation necessary to observe the G3P-dependent growth phenotype in strains expressing the plsB26 allele [45]. In Pseudomonas aeruginosa, plsB is not essential [46] suggesting that PlsX/Y pathway functions in this γ-proteobacter species. Neither PlsX nor PlsY homologs are found in mammalian genomes [38,40]. The role of PlsX and acyl-PO4 in organisms where the PlsX/PlsY pathway is not operational remains a major unanswered question in bacterial lipid metabolism. Perhaps acyl-PO4 has some regulatory role, as known for acyl-ACP and acyl-CoA (see below), but there is no evidence to support this interesting idea.
3.3. Acyl-CoA
Acyl-CoA functions as an alternate acyl donor in E. coli and presumably other γ-proteobacteria (Fig. 1). There is no mechanism for forming acyl-CoA from acyl-ACP in E. coli, thus acyl-CoAs are exclusively formed from exogenous fatty acids. Extracellular fatty acids traverse the outer membrane via the FadL porin and flip to the inner aspect of the cytoplasmic membrane where they are activated by acyl-CoA synthetase (FadD) (Fig. 1A) [47]. Acyl-CoA cannot be converted into acyl-ACP [48], which explains in part why supplementation with exogenous fatty acids cannot replace de novo biosynthesis. Acyl-CoAs of appropriate chain length are alternate substrates for PlsB and PlsC in E. coli. An alternate, and significant, fate for exogenous fatty acids is the degradation of acyl-CoAs via β-oxidation to generate energy [48]. In both Gram-positive and Gram-negative organisms, the FadD enzyme is co-regulated with the gene set encoding an inducible β-oxidation system. Many of these bacteria, like Bacillus subtilis, encode a fadD, but cannot use acyl-CoA for phospholipid synthesis [49,50]. This genetic connection between FadD and the β-oxidation genes suggests that the primary purpose of salvaging exogenous fatty acids is for energy generation via β-oxidation rather than phospholipid synthesis. However, the production of fatty acids is the most energy expensive process in phospholipid synthesis and the ability of γ-proteobacteria to utilize exogenous fatty acids for phospholipid synthesis is an adaptation that saves energy. Gram-positive bacteria, like Streptococcus pneumonia and Staphylococcus aureus, lack the genes for a β-oxidation system and acyl-CoAs have no role in phospholipid synthesis.
3.4. Metabolic and Genetic Regulation by Acyl Donors
The acyl donor pools play an important regulatory role in bacterial lipid metabolism. At the biochemical level, long-chain acyl-ACPs inhibit the initiation of fatty acid biosynthesis through feedback inhibition of FabH and acetyl-CoA carboxylase in E. coli [51,52]. Whether this biochemical regulatory paradigm can be extended to Gram-positive pathogens is an open question. Experiments with intact cells suggest that acyl-ACP may be a potent feedback regulator in S. pneumoniae, but not in S. aureus [37]. However, other ligands, like acyl-PO4 may be involved and candidate ligands need to be tested in vitro to determine their role in regulating the initiation of fatty acid synthesis in these organisms. Regulation at the genetic level is more diverse. In S. pneumoniae, the FASII enzymes are encoded together in the fab gene cluster, and transcriptionally repressed by the regulator FabT [53]. Long-chain acyl-ACP exerts feedback regulation of the lipid biosynthetic pathway by binding to FabT to increase FabT affinity for the promoter of the fab cluster, suppressing expression of the cluster [54]. Mutant strains with genetic knockouts of fabT have longer acyl chain lengths and increased ratio of saturated fatty acids in their lipids [53]. In contrast, the B. subtilis FASII transcription repressor is FapR, which represses the dispersed FASII genes when unbound to ligands [55]. Malonyl-CoA binding to FapR releases FapR from the DNA to allow transcription in a feed-forward mode of regulation. In E. coli, the ratio of saturated to unsaturated fatty acids is controlled by the FadR and FabR transcriptional regulators [48,56]. FadR is a repressor of β-oxidation and activator of fabA and fabB, which encode the two enzymes essential for unsaturated fatty acid biosynthesis [57,58]. Long-chain acyl-CoA binding to FadR dissociates it from DNA to reverse its activating effect. FabR monitors acyl-ACP pools to regulate the expression of fabA and fabB [58]. Unsaturated long chain acyl-ACP binding to FabR causes FabR to bind to the promoters of fabA and fabB to repress their expression. The binding of saturated acyl-ACP antagonizes this effect. Pseudomonas aeruginosa regulates the ratio of saturated to unsaturated fatty acids via the DesT transcription factor [59]. DesT binds to the promoter of the desCB operon, which encodes for an oxygen dependent desaturase that introduces a double bond into the fatty acid chain of saturated acyl-CoA [60]. Saturated acyl-CoA binding to DesT causes dissociation from the promoter to increase desCB expression, while unsaturated acyl-CoA binding to DesT triggers tighter promoter association and strong repression. DesT also regulates fabAB expression in P. aeruginosa to coordinate aerobic and anaerobic pathways for unsaturated fatty acid formation [61]. The DesT structures provide a striking example of how these transcriptional regulators sense the conformation of fatty acids to maintain membrane homeostasis [62]. A role for acyl-PO4 in regulating lipid metabolism appears attractive, but there is no evidence that this intermediate participates in either transcriptional or biochemical regulation of the pathway. Depletion of the PlsX enzyme in B. subtilis leads to cessation of both fatty acid and phospholipid biosynthesis, but depletion of PlsY and PlsC does not, supporting the regulatory role for acyl-ACP, but provides no evidence for pathway regulation by acyl-PO4 [43]. The acyl donor pools are involved in a multitude of biochemical and genetic regulatory processes that are sure to continue to grow in importance.
4. G3P Acyltransferases
4.1. PlsB
The first step in PtdOH formation is the acylation of the 1-position of G3P by the G3P acyltransferases [63]. The characterization of the first G3P acyltransferase identified in bacteria was facilitated by the isolation of mutants with a G3P auxotrophic phenotype that had a G3P Km defect in a membrane-associated G3P acyltransferase [64,65]. The acyltransferase, named PlsB, uses either acyl-ACP or acyl-CoA as the donor to acylate the 1-position of G3P [66–68] (Fig. 1A). The PlsB pathway is found in a subset of bacterial genomes, mostly from γ-proteobacteria. Four regions of high homology are shared between PlsBs from different species. Mutagenesis studies on these conserved residues have defined the roles of these conserved motifs in substrate binding and catalysis [69,70]. An HXSXXD motif (also known as the HX4D motif) constitutes homology block I (residues 306–311; E. coli numbering). The H306A and D311G mutants are inactive enzymes, while the S308A mutant has only modest decrease in activity [69,71]. His306 and Asp311 are suggested to form a charge relay system to abstract a proton similar to serine hydrolases [72]. Blocks II (GXXFIRR, residues 348–354) and III (FXEGXRXRXG, residues 383–393) participate in G3P binding, with mutations of the arginine residues in the blocks causing an increase in G3P Km. Block IV is a less conserved hydrophobic patch (ITLIPIYI, residues 417–425), with proline as the only invariant residue. The P421S mutant is completely inactive. It seems unlikely that the proline is directly involved in catalysis, and is thought to have an important, albeit undefined, structural role. The structure of a soluble G3P acyltransferase from squash has been solved [73]. The structure confirms that the HX4D motif forms a charge relay for catalysis similar to serine hydrolases. One nitrogen atom of the aromatic imidazole ring of histidine activates the 1-OH of G3P for nucleophilic attack on the acyl thioester of ACP, while the other nitrogen interacts with the aspartate to form the charge relay system. The two blocks of amino acids are rich in basic amino acids that are separated by a 42 residue spacer to form ionic bonds with the phosphate group of bound G3P. One block has the sequence of GGRXR, similar to the GXRXR motif in block III of E. coli PlsB. However, E. coli PlsB is a significantly larger protein than the squash G3P acyltransferase (817 aa vs 368 aa). The extra residues in E. coli PlsB are not likely involved in catalysis, but rather function in membrane binding and regulation.
PlsB is the key point of regulation in E. coli PtdOH biosynthesis [74], [75]. PlsB is biochemically and genetically regulated by guanosine tretraphosphate (ppGpp) and σE, respectively. The alarmone ppGpp accumulates following amino acid starvation and inhibits several major biosynthetic pathways including stable RNA, protein, and phospholipid biosynthesis [76,77]. PlsB is inhibited by ppGpp, with increasing ppGpp concentrations causing an accumulation of PlsB substrates in vivo [75]. The inhibition of PlsB by ppGpp allows coordination between the major biosynthetic pathways during nutrient limitation because PlsB catalyzes the first committed step in phospholipid biosynthesis. The σE factor is also induced following environmental stresses, but regulates gene expression instead enzyme activity [78]. Overexpression of σE increases the expression of a number of genes involved in pathogenesis, including the biosynthetic pathway involved in phospholipid biosynthesis via plsB induction. The plsB gene has two promoters: one for basal expression of the plsB gene and a distal promoter responsible for σE activation [74]. The prevailing model posits a key role for PlsB in the control of phospholipid and fatty acid synthesis. However, there are few firmly established mechanisms to support this regulatory role. The basis for the biochemical regulation of PlsB by ppGpp remains to determined, and a more comprehensive analysis of how other metabolites affect PlsB activity would help understand how membrane formation is integrated into metabolism. The role of membrane structure in PlsB activity is an unexplored area.
4.2. PlsY
PlsY is the most widely distributed G3P acyltransferase in Gram-positive bacteria, and is the sole G3P acyltransferase in medically important bacteria such as S. pneumonia and S. aureus [38] (Fig. 1B). Almost all bacteria only have a single plsY gene; however, there are a few bacteria, like Bacillus anthracis, that have multiple plsY genes. The function of these redundant genes is not understood. The PlsY protein is an acyl-PO4 dependent G3P acyltransferase. It is an integral membrane protein of five transmembrane segments, with three short nonconserved extracellular loops as well as three extensive and conserved cytoplasmic loops required for catalysis [39]. The first cytoplasmic loop composed of residues 35–46 (S. pneumoniae numbering) has the consensus sequence of GSGNXGXTNXXR. The glycine rich motif is similar to other known phosphate binding motifs, and mutation of the positively charged Arg46 into Ala causes a complete loss of activity confirming the motif as the substrate binding site for either negatively charged substrate of G3P or acyl-PO4. The second cytoplasmic loop composed of residues 100–107 has the consensus sequence of FXGGKXVA. The GGK motif is similar to the ATP binding loop in prototypical kinases [79]. The small size of glycine functions to permit the negatively charged substrate to interact with the lysine. The K104A mutation causes complete loss of PlsY activity, while adding side chains to Gly102 and Gly103 cause an over six-fold increase in the Km of G3P with minimal change in the acyl-PO4 Km. Therefore, loop 2 is deduced to be the G3P substrate binding site while loop 1 is the acyl-PO4 binding site. Loop 3 is the C-terminal cytoplasmic domain consisting of residues 185–197 with the consensus sequence of HX2NX8E. Mutants of the conserved His185 and Asn188 are catalytically defective. His185 is suggested to act as the general base to activate the 1-OH of G3P for nucleophilic attack on acyl-PO4, similar to the active site histidine of PlsB [39]. The E197A mutant fails to assemble into the membrane, suggesting a key role in protein folding. However, whether His185 alone is a sufficient general base or if other residues such as Glu197 participate to form a charge relay system such as in PlsB remains to be determined.
The role of PlsY in the regulation of fatty acid and phospholipid biosynthesis is unclear. S. pneumoniae PlsY is inhibited by long chain acyl-CoAs, but this form of regulation is not biologically relevant because acyl-CoAs have no role in S. pneumoniae lipid biosynthesis [80]. In a conditional plsY knockout of B. subtilis, fatty acid and acyl-PO4 biosynthesis continues despite inactivation of PlsY leading to an accumulation of nonesterified fatty acids [43]. In constrast, PlsX inactivation causes a complete arrest of fatty acid and phospholipid biosynthesis, suggesting that PlsX is the regulatory point coordinating fatty acid and phospholipid biosynthesis. At the genetic level, plsX and plsC, but not plsY, is regulated by the global lipid transcriptional regulator FapR in B. subtilis [55] However, neither plsX nor plsY are regulated by the global lipid transcriptional regulator FabT in S. pneumonia, but plsC and other fatty acid biosynthesis genes are [53]. The mechanism of coordinating fatty acid and phospholipid biosynthesis in bacteria utilizing the PlsX/Y pathway for PtdOH synthesis is basically unknown, and may or may not have elements in common with the E. coli paradigm with regard to feedback regulation of FabH and acetyl-CoA carboxylase.
The plsX/plsY genes are widely distributed in bacteria, with only Xanthomonadales in the γ-proteobacteria lacking the genes [38]. The plsX and plsY genes are essential in bacteria with no PlsB acyltransferase system [81]. PlsB is an essential gene and is the primary pathway for phospholipid biosynthesis in E. coli, although this organism has both the plsB and plsX/plsY genes. The role of PlsX and PlsY in bacteria where PlsB appears to be the only operable G3P acyltransferase is unknown. Archaea and eukarya exclusively use the plsB/plsC system to generate phospholipids, and do not contain plsX or plsY homologs.
5. 1-Acyl-G3P Acyltransferases
The 1-acyl-G3P (LPA) acyltransferase, PlsC, transfers a fatty acid to the 2 position of LPA to complete the synthesis of PtdOH [82,83]. PlsC is an intergral membrane protein, and essential in all bacteria. PlsC has not been characterized in detail, and most of our understanding is derived through sequence comparison to PlsB. Like PlsB, PlsC contains a conserved HX4D motif believed to be the catalytic core. PlsC also contains two motifs with conserved arginine residues believed to interact with the phosphate group of LPA for substrate binding. The PlsC enzymes from Gram-positive bacteria such as B. subtilis [43] or S. pneumoniae [38] use only acyl-ACP as the acyl donor, while E. coli PlsC is able to also use acyl-CoA [82]. The residues responsible for ACP/CoA specificity are unknown.
Some bacteria have more than one plsC gene ortholog. Two LPA acyltransferases have been characterized in Neisseria meningitides, and both can complement a temperature-sensitive E. coli plsC mutant [84,85]. Three PlsC homologs are found in Pseudomonas fluorescenes [86]. Two homologs (hdtS and patB) both complement the E. coli plsC mutant. However, growth defects were observed when either gene is knocked out, with the most significant changes in membrane acyl chain composition occuring in the hdtS gene knockout consistent with it being the major LPA acyltransferase activity. Why two LPA acyltransferases are required by Pseudomonas fluorescences is poorly understood. The third ortholog (olsA) does not have detectable LPA acyltransferase activity, and is involved in the acyltransfer reaction in the final step of ornithine lipid biosynthesis [86]. The OlsA protein from Rhodobacter capsulatus is also a PlsC homolog, and has both LPA acyltransferase and ornithine lipid acyltransferase activity [87]. The core PlsC enzyme structure has evolved to perform acylation reactions on substrates other than LPA. A detailed biochemical and structural characterization would advance the mechanistic and structural understanding of the PlsC mechanism and specificity.
Acyl-chain positional asymmetry is observed in bacterial PtdOH. In E. coli, the 1-OH position of G3P is primarily occupied by 16:0 and 18:1, while the 2-OH position is primarily 16:1 and 18:1 [30]. In contrast, the 1-OH position in S. aureus is primarily anteiso17:0 and the 2-OH position is primarily anteiso15:0 [37]. The fatty acids incorporated into the 1 and 2 positions of G3P are dependent upon the relative selectivity of the PlsB/PlsX/PlsY enzymes versus the PlsC enzyme as well as the available pool of acyl-ACP. PlsB, PlsX/PlsY, and PlsC do not demonstrate absolute specificity for their preferred fatty acids. In S. aureus strain RN4220, the PlsX/PlsY acyltransferase system incorporates fatty acids other than anteiso17:0 at lower levels in the 1 position. While S. aureus PlsC is highly selective for anteiso15:0 under normal growth conditions, a S. aureus mutant that cannot initiate any fatty acid biosynthesis incorporates available exogenous fatty acids such as 18:1 into the 2 position [37]. An E. coli fabA mutant that can’t produce unsaturated fatty acids incorporates 16:0 into both positions [88]. Having correct acyl-composition is vital to bacterial cell survival [31], and is dependent on coordinating the FASII system to generate the appropriate length and unsaturation of acyl chains and the selectivity of the acyltransferase system. PlsC appears to be a fertile field for investigation. There is no information on the biochemical regulation of PlsC, and basic information regarding its topology and the key residues involved in catalysis are lacking. Furthermore, there remain many PlsC homologs with unknown functions waiting to be characterized.
6. Phospholipid Recycling
6.1. DgkA
PtdOH is also produced by the phosphorylation of diacylglycerol (DAG) by a DAG kinase (Dgk) (Fig. 2). Phospholipid headgroups are used as intermediates in other biosynthetic pathways, and the Dgk pathway to PtdOH provides a mechanism to re-introduce the DAG produced in these reactions into the phospholipid biosynthetic pathway. This process was first recognized in E. coli, where phosphatidylglycerol (PtdGro) functions as a sn-glycerol-1-phosphate donor for periplasmic membrane-derived oligosaccharides (MDO), which are required for osmotic homeostasis [89,90]. The DAG product in MDO biosynthesis flips to the cytoplasmic aspect of the inner membrane where it is converted to PtdOH by the membrane-bound kinase, DgkA [91]. Knockout of the dgkA gene results in accumulation of DAG and other neutral lipids when MDO synthesis is minimal, but in osmotically challenging environments that accelerate MDO formation, the absence of DgkA is lethal [90,92]. DgkAs are small (~150 amino acid) integral membrane proteins with three transmembrane helices. The structure of DgkA shows that it assembles into a trimer in the membrane and represents a distinct class of lipid kinases [93] (Fig. 2A). The DAG enters into the active site from the membrane via a gate formed by the third transmembrane domain of each subunit interacting with the first two transmembrane domains of the adjacent subunit. ATP approaches the active site from the cytosol, and the phosphorylation chemistry occurs near the water-membrane interface.
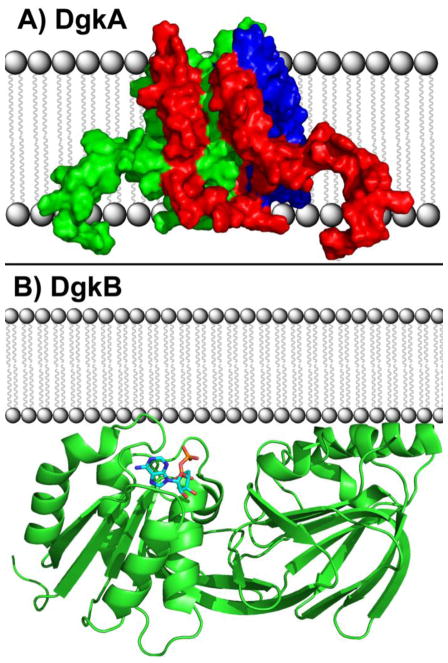
Fig. 2.
DAG kinases. (A) DgkA is the DAG kinase responsible for recycling DAG produced from the turnover of PtdGro in the synthesis of membrane-derived oligosaccharides in E. coli. DgkA is the prototypical DAG kinase of Gram-negative bacteria. It is an integral membrane protein that assembles as a trimer in the bilayer. The individual subunits of the homotrimer are colored red, green and blue. DAG enters the active site from the membrane via a gate formed by the third transmembrane domain of each subunit interacting with the first two transmembrane domains of the adjacent subunit. ATP approaches the active site from the cytosol. (B) DgkB is the DAG kinase responsible for recycling DAG produced from the synthesis of LTA in S. aureus, and is the prototypical DAG kinase of Gram-positive bacteria. DgkB is an interfacial enzyme that docks to the membrane via a positively-charged patch of amino acids located adjacent to the active site. A DgkB monomer is shown in green ribbon and ADP is shown in the structure as sticks.
The dgkA gene is located adjacent to and transcribed in the opposite direction of the plsB gene in E. coli, and the two genes are differentially regulated [74]. Expression of dgkA is increased and plsB decreased following activation of the stringent response, and dgkA transcription diminishes and plsB is activated by the stress response regulator, σE [94]. The opposing regulation reflects the opposing nature of the gene products. DgkA functions in recycling PtdOH from existing lipids while PlsB produces new PtdOH. Expression of the dgkA gene is also upregulated by the BasRS two-component regulator, which regulates the transcription of genes involved in LPS modification [95]. BasR directly binds to the promoter region of dgkA in vitro, and a mutant that constitutively increases signaling through this pathway has increased DgkA activity. Homologs of dgkA are widely distributed, but not ubiquitous, in eubacteria, and missing in all eukaryotes except for Viridiplantae and Rhizaria clades [96]. However, not all of the dgkA-related genes encode DAG kinases.
6.2. DgkB
PtdGro is the sn-glycerol-1-phosphate donor for the synthesis of the lipoteichoic acids (LTA) in Gram-positive bacteria such as B. subtilis. As in MDO biosynthesis, DAG is a product of the reaction [9]; however unlike MDO, LTA is a major cell wall constituent and its synthesis is constitutive. Each LTA contains polyphosphoglycerol chains of 14–33 units, meaning that there is a constant, significant turnover of PtdGro to DAG in Gram-positive bacteria [9,10]. However, the dgkA gene homolog in B. subtilis is not essential [81], and it was subsequently shown that the B. subtilis dgkA gene is actually an undecaprenol kinase with no DAG kinase activity [97]. Rather, these Gram-positive bacteria express a soluble DAG kinase (DgkB) that belongs to the eukaryotic DAG kinase superfamily (Pfam00781) that is essential for recycling DAG [97]. Inactivation of dgkB leads to cessation of LTA biosynthesis, the accumulation of DAG and eventual loss of viability [97]. The crystal structure of the homodimeric S. aureus DgkB shows that each monomer is composed two domains with the catalytic site located in a cleft between the two domains [98] (Fig. 2B). The key active site residues and the components of the structural Asp-water-Mg2+ network are conserved in the catalytic cores of the mammalian signaling DAG kinases, indicating that these enzymes use the same mechanism and have similar structures as the bacterial DgkBs. The DgkB surface is overall electronegative, except for an electropositive patch consisting of Lys15, Arg20, and Lys165 adjacent to the active site entrance. DgkB binds to anionic phospholipid vesicles with high affinity, and analysis of the K15A, R20A, and K165A mutant proteins confirm the role of these residues in interfacial docking [99]. Binding of DgkB to anionic phospholipids lowers the Km for ATP from 3.7 mM to 32 μM illustrating that interfacial binding triggers a conformational change that activates the kinase. Thus, interfacial binding is a prerequisite for the DgkB activity because it is needed to locate the DAG substrate and promote a conformational change that permits ATP binding. As with dgkA, not all of the dgkB homologs are DAG kinases, although their substrates are largely unknown. Of the 3 dgkB homologs in B. subtilis, only one has DAG kinase activity. The other two family members have similar ATP binding domains and are presumed to be lipid kinases. However, this is a very loose prediction and much more work is needed to determine the substrates for the multiple dgkB homologs found in Gram-positive bacteria.
7. Targeting PtdOH Synthesis for Antibacterial Drug Discovery
The central importance of PtdOH to bacterial physiology suggest that targeting PtdOH synthesis may offer opportunities to develop novel antibacterial therapeutics. Targeting GpsA has been generally discarded due to the numerous ways that bacterial cells can obtain glycerol from the environment and the availability of these nutrients in the animal host. The acyltransferases are essential enzymes and cannot be circumvented by acquisition of phospholipids from the host. Phenethyl alcohol is a bacteriostatic agent that causes membrane perturbations in Gram-negative [100–102] and Gram-positive bacteria [103]. Sublethal concentrations of the drug selectively inhibit incorporation of acetate into fatty acids [104], although this effect is secondary to that on phospholipid synthesis [105]. Phenethyl alcohol inhibits sn-glycerol-3-phosphate acyltransferase (PlsB) in vitro [106]. However, there have been no attempts to develop this scaffold further. One reason the acyltransferases have not been seriously targeted is that they are integral membrane enzymes presenting a barrier to preparing sufficient amounts of enzyme to support high-throughput screens. Also, there are close homologs of PlsB and PlsC found in mammals that are thought to carry out important acyltransferase reactions in intermediary metabolism [70] opening the potential for candidate compounds to affect host lipid metabolism. This is not a concern in the case of PlsY, which does not have mammalian homologs [38] and is essential for PtdOH synthesis in Gram-positive pathogens. The approach that has been taken with PlsY is to design isosteric, non-hydrolyzable acyl-PO4 analogs as substrate competitive inhibitors of PlsY, and potentially product inhibitors of PlsX [80]. The latest generation of these mimics, the acylsulfamates, exhibit on-target activity against S. aureus, but the hydrophobic nature of these inhibitors presents a challenge to their use as drugs [107]. Structural biology is where progress needs to be made in this area. Determining the structure of acyl-PO4 bound to either PlsY or PlsX would greatly facilitate the design of the next generation inhibitors. There have been no efforts to target the DAG kinases. DgkA is only essential in osmotically challenging environments, reducing the enthusiasm for targeting this enzyme. On the other hand, DgkB is essential to deal with the products of Gram-positive bacterial LTA synthesis and inhibitors of this soluble enzyme would be effective agents against many pathogens. Clearly, chemical targeting of the bacterial PtdOH synthesis is in its infancy and it remains to be seen whether the significant challenges in protein biochemistry and compound design can be overcome to target these essential enzymes.
Highlights.
Phosphatidic acid is the key intermediate in bacterial phospholipid synthesis
Acyltransferases control the positional distribution of fatty acyl chains in the phospholipids.
Acyl-ACP and acyl-CoA are important biochemical and transcriptional regulators.
Acknowledgments
This work was supported by National Institutes of Health grant GM034496, Cancer Center (CORE) Support Grant CA21765 and the American Lebanese Syrian Associated Charities.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Cronan JE, Bell RM. Mutants of Escherichia coli defective in membrane phospholipid synthesis: mapping of the structural gene for L-glycerol 3-phosphate dehydrogenase. J Bacteriol. 1974;118:598–605. doi: 10.1128/jb.118.2.598-605.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Beijer L, Nilsson RP, Holmberg C, Rutberg L. The glpP and glpF genes of the glycerol regulon in Bacillus subtilis. J Gen Microbiol. 1993;139:349–359. doi: 10.1099/00221287-139-2-349. [DOI] [PubMed] [Google Scholar]
- 3.Kito M, Pizer LI. Purification and regulatory properties of the biosynthetic L-glycerol-3-phophate dehydrogenase from Escherichia coli. J Biol Chem. 1969;244:3316–3323. [PubMed] [Google Scholar]
- 4.Ray TK, Cronan JE., Jr Acylation of glycerol 3-phosphate is the sole pathway of de novo phospholipid synthesis in Escherichia coli. J Bacteriol. 1987;169:2896–2898. doi: 10.1128/jb.169.6.2896-2898.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hayashi Si, Koch JP, Lin ECC. Active transport of L-glycerophosphate in Escherichia coli. J Biol Chem. 1964;239:3098–3105. [PubMed] [Google Scholar]
- 6.Ray PH, White DC. Effect of glycerol deprivation on the phospholipid metabolism of a glycerol auxotroph of Staphylococcus aureus. J Bacteriol. 1972;109:668–677. doi: 10.1128/jb.109.2.668-677.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Goelz SE, Cronan JE., Jr The positional distribution of fatty acids in Escherichia coli phospholipids is not regulated by sn-glycerol 3-phosphate levels. J Bacteriol. 1980;144:462–464. doi: 10.1128/jb.144.1.462-464.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Morbidoni HR, de Mendoza D, Cronan JE. Synthesis of sn-glycerol 3-phosphate, a key precursor of membrane lipids, in Bacillus subtilis. J Bacteriol. 1995;177:5899–5905. doi: 10.1128/jb.177.20.5899-5905.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Koch HU, Haas R, Fischer W. The role of lipoteichoic acid biosynthesis in membrane lipid metabolism of growing Staphylococcus aureus. Eur J Biochem. 1984;138:357–363. doi: 10.1111/j.1432-1033.1984.tb07923.x. [DOI] [PubMed] [Google Scholar]
- 10.Taron DJ, Childs WC, III, Neuhaus FC. Biosynthesis of D-alanyl-lipoteichoic acid: role of diglyceride kinase in the synthesis of phosphatidylglycerol for chain elongation. J Bacteriol. 1983;154:1110–1116. doi: 10.1128/jb.154.3.1110-1116.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Larson TJ, Ye SZ, Weissenborn DL, Hoffmann HJ, Schweizer H. Purification and characterization of the repressor for the sn-glycerol 3-phosphate regulon of Escherichia coli K12. J Biol Chem. 1987;262:15869–15874. [PubMed] [Google Scholar]
- 12.Zeng G, Ye S, Larson TJ. Repressor for the sn-glycerol 3-phosphate regulon of Escherichia coli K-12: primary structure and identification of the DNA-binding domain. J Bacteriol. 1996;178:7080–7089. doi: 10.1128/jb.178.24.7080-7089.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hogema BM, Arents JC, Inada T, Aiba H, Van Dam K, Postma PW. Catabolite repression by glucose 6-phosphate, gluconate and lactose in Escherichia coli. Mol Microbiol. 1997;24:857–867. doi: 10.1046/j.1365-2958.1997.3991761.x. [DOI] [PubMed] [Google Scholar]
- 14.Zhao N, Oh W, Trybul D, Thrasher KS, Kingsbury TJ, Larson TJ. Characterization of the interaction of the glp repressor of Escherichia coli K-12 with single and tandem glp operator variants. J Bacteriol. 1994;176:2393–2397. doi: 10.1128/jb.176.8.2393-2397.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Glatz E, Persson M, Rutberg B. Antiterminator protein GlpP of Bacillus subtilis binds to glpD leader mRNA. Microbiology. 1998;144:449–456. doi: 10.1099/00221287-144-2-449. [DOI] [PubMed] [Google Scholar]
- 16.Lemieux MJ, Huang Y, Wang DN. Glycerol-3-phosphate transporter of Escherichia coli: Structure, function and regulation. Res Microbiol. 2004;155:623–629. doi: 10.1016/j.resmic.2004.05.016. [DOI] [PubMed] [Google Scholar]
- 17.Saier MH. Families of transmembrane sugar transport proteins. Mol Microbiol. 2000;35:699–710. doi: 10.1046/j.1365-2958.2000.01759.x. [DOI] [PubMed] [Google Scholar]
- 18.Elvin CM, Hardy CM, Rosenberg H. Pi exchange mediated by the GlpT-dependent sn-glycerol-3-phosphate transport system in Escherichia coli. J Bacteriol. 1985;161:1054–1058. doi: 10.1128/jb.161.3.1054-1058.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lemieux MJ, Huang Y, Wang DN. Crystal structure and mechanism of GlpT, the glycerol-3-phosphate transporter from E. coli. J Electron Microsc. 2005;54:i43–i46. doi: 10.1093/jmicro/54.suppl_1.i43. [DOI] [PubMed] [Google Scholar]
- 20.Huang Y, Lemieux MJ, Song J, Auer M, Wang DN. Structure and mechanism of the glycerol-3-phosphate transporter from Escherichia coli. Science. 2003;301:616–620. doi: 10.1126/science.1087619. [DOI] [PubMed] [Google Scholar]
- 21.Fu D, Libson A, Miercke LJW, Weitzman C, Nollert P, Krucinski J, Stroud RM. Structure of a glycerol-conducting channel and the basis for its selectivity. Science. 2000;290:481–486. doi: 10.1126/science.290.5491.481. [DOI] [PubMed] [Google Scholar]
- 22.Stroud RM, Miercke LJ, O’Connell J, Khademi S, Lee JK, Remis J, Harries W, Robles Y, Akhavan D. Glycerol facilitator GlpF and the associated aquaporin family of channels. Curr Opin Struct Biol. 2003;13:424–431. doi: 10.1016/s0959-440x(03)00114-3. [DOI] [PubMed] [Google Scholar]
- 23.Weissenborn DL, Wittekindt N, Larson TJ. Structure and regulation of the glpFK operon encoding glycerol diffusion facilitator and glycerol kinase of Escherichia coli K-12. J Biol Chem. 1992;267:6122–6131. [PubMed] [Google Scholar]
- 24.Thorner JW, Paulus H. Catalytic and allosteric properties of glycerol kinase from Escherichia coli. J Biol Chem. 1973;248:3922–3932. [PubMed] [Google Scholar]
- 25.Hénin J, Tajkhorshid E, Schulten K, Chipot C. Diffusion of Glycerol through Escherichia coli Aquaglyceroporin GlpF. Biophys J. 2008;94:832–839. doi: 10.1529/biophysj.107.115105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Heller KB, Lin EC, Wilson TH. Substrate specificity and transport properties of the glycerol facilitator of Escherichia coli. J Bacteriol. 1980;144:274–278. doi: 10.1128/jb.144.1.274-278.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zwaig N, Lin ECC. Feedback inhibition of glycerol kinase, a catabolic enzyme in Escherichia coli. Science. 1966;153:755–757. doi: 10.1126/science.153.3737.755. [DOI] [PubMed] [Google Scholar]
- 28.Zwaig N, Kistler WS, Lin ECC. Glycerol kinase, the pacemaker for the dissimilation of glycerol in Escherichia coli. J Bacteriol. 1970;102:753–759. doi: 10.1128/jb.102.3.753-759.1970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cozzarelli NR, Koch JP, Hayashi S, Lin ECC. Growth stasis by accumulated L-glycerophosphate in Escherichia coli. J Bacteriol. 1965;90:1325–1329. doi: 10.1128/jb.90.5.1325-1329.1965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rock CO, Jackowski S. Forty years of fatty acid biosynthesis. Biochem Biophys Res Commun. 2002;292:1155–1166. doi: 10.1006/bbrc.2001.2022. [DOI] [PubMed] [Google Scholar]
- 31.Zhang Y-M, Rock CO. Membrane lipid homeostasis in bacteria. Nat Rev Microbiol. 2008;6:222–233. doi: 10.1038/nrmicro1839. [DOI] [PubMed] [Google Scholar]
- 32.Cronan JE, Jr, Rock CO. Biosynthesis of Membrane LipidsEscherichia coli and Salmonella typhimurium. In: Neidhardt FC, Ingraham JL, Low KB, Magasanik B, Schaechter M, Umbarger HE, editors. Cellular and Molecular Biology. Amer Soc Microbiol; Washington, DC: 1987. pp. 474–497. [Google Scholar]
- 33.Rock CO, Goelz SE, Cronan JE., Jr Phospholipid synthesis in Escherichia coli. Characteristics of fatty acid transfer from acyl-acyl carrier protein to sn-glycerol-3-phosphate. J Biol Chem. 1981;256:736–742. [PubMed] [Google Scholar]
- 34.Cronan JE, Jr, Weisberg LJ, Allen RG. Regulation of membrane lipid synthesis in Escherichia coli. Accumulation of free fatty acids of abnormal length during inhibition of phospholipid synthesis. J Biol Chem. 1975;250:5835–5840. [PubMed] [Google Scholar]
- 35.Garwin JL, Klages AL, Cronan JE., Jr β-Ketoacyl-acyl carrier protein synthase II of Escherichia coli. Evidence for function in the thermal regulation of fatty acid synthesis. J Biol Chem. 1980;255:3263–3265. [PubMed] [Google Scholar]
- 36.Jiang Y, Chan CH, Cronan JE. The soluble acyl-acyl carrier protein synthetase of Vibrio harveyi B392 is a member of the medium chain acyl-CoA synthetase family. Biochemistry. 2006;45:10008–10019. doi: 10.1021/bi060842w. [DOI] [PubMed] [Google Scholar]
- 37.Parsons JB, Frank MW, Subramanian C, Saenkham P, Rock CO. Metabolic basis for the differential susceptibility of Gram-positive pathogens to fatty acid synthesis inhibitors. Proc Natl Acad Sci U S A. 2011;108:15378–15383. doi: 10.1073/pnas.1109208108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lu Y-J, Zhang Y-M, Grimes KD, Qi J, Lee RE, Rock CO. Acyl-phosphates initiate membrane phospholipid synthesis in gram-positive pathogens. Molec Cell. 2006;23:765–772. doi: 10.1016/j.molcel.2006.06.030. [DOI] [PubMed] [Google Scholar]
- 39.Lu Y-J, Zhang F, Grimes KD, Lee RE, Rock CO. Topology and active site of PlsY: the bacterial acylphosphate:glycerol-3-phosphate acyltransferase. J Biol Chem. 2007;282:11339–11346. doi: 10.1074/jbc.M700374200. [DOI] [PubMed] [Google Scholar]
- 40.Lehninger AL. Synthesis and properties of the acyl phosphates of some higher fatty acids. J Biol Chem. 1945;162:333–342. [PubMed] [Google Scholar]
- 41.Kim Y, Li H, Binkowski TA, Holzle D, Joachimiak A. Crystal structure of fatty acid/phospholipid synthesis protein PlsX from Enterococcus faecalis. J Struct Funct Genomics. 2008 doi: 10.1007/s10969-008-9052-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Badger J, Sauder JM, Adams JM, Antonysamy S, Bain K, Bergseid MG, Buchanan SG, Buchanan MD, Batiyenko Y, Christopher JA, Emtage S, Eroshkina A, Feil I, Furlong EB, Gajiwala KS, Gao X, He D, Hendle J, Huber A, Hoda K, Kearins P, Kissinger C, Laubert B, Lewis HA, Lin J, Loomis K, Lorimer D, Louie G, Maletic M, Marsh CD, Miller I, Molinari J, Muller-Dieckmann HJ, Newman JM, Noland BW, Pagarigan B, Park F, Peat TS, Post KW, Radojicic S, Ramos A, Romero R, Rutter ME, Sanderson WE, Schwinn KD, Tresser J, Winhoven J, Wright TA, Wu L, Xu J, Harris TJR. Structural analysis of a set of proteins resulting from a bacterial genomics project, Proteins: Structure. Function, and Bioinformatics. 2005;60:787–796. doi: 10.1002/prot.20541. [DOI] [PubMed] [Google Scholar]
- 43.Paoletti L, Lu Y-J, Schujman GE, de Mendoza D, Rock CO. Coupling of fatty acid and phospholipid synthesis in Bacillus subtilis. J Bacteriol. 2007;189:5816–5824. doi: 10.1128/JB.00602-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Yoshimura M, Oshima T, Ogasawara N. Involvement of the YneS/YgiH and PlsX proteins in phospholipid biosynthesis in both Bacillus subtilis and Escherichia coli. BMC Microbiol. 2007;7:69. doi: 10.1186/1471-2180-7-69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Larson TJ, Ludtke DN, Bell RM. sn-Glycerol-3-phosphate auxotrophy of plsB strains of Escherichia coli: Evidence that a second mutation, plsX, is required. J Bacteriol. 1984;160:711–717. doi: 10.1128/jb.160.2.711-717.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Jacobs MA, Alwood A, Thaipisuttikul I, Spencer D, Haugen E, Ernst S, Will O, Kaul R, Raymond C, Levy R, Chun-Rong L, Guenthner D, Bovee D, Olson MV, Manoil C. Comprehensive transposon mutant library of Pseudomonas aeruginosa. Proc Natl Acad Sci U S A. 2003;100:14339–14344. doi: 10.1073/pnas.2036282100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Black PN, DiRusso CC. Transmembrane movement of exogenous long-chain fatty acids: proteins, enzymes, and vectorial esterification. Microbiol Mol Biol Rev. 2003;67:454–472. doi: 10.1128/MMBR.67.3.454-472.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Cronan JE, Jr, Subrahmanyam S. FadR, transcriptional co-ordination of metabolic expediency. Mol Microbiol. 1998;29:937–943. doi: 10.1046/j.1365-2958.1998.00917.x. [DOI] [PubMed] [Google Scholar]
- 49.DiRusso CC, Black PN, Weimar JD. Molecular inroads into the regulation and metabolism of fatty acids, lessons from bacteria. Prog Lipid Res. 1999;38:129–197. doi: 10.1016/s0163-7827(98)00022-8. [DOI] [PubMed] [Google Scholar]
- 50.Matsuoka H, Hirooka K, Fujita Y. Organization and function of the YsiA regulon of Bacillus subtilis involved in fatty acid degradation. J Biol Chem. 2007;282:5180–5194. doi: 10.1074/jbc.M606831200. [DOI] [PubMed] [Google Scholar]
- 51.Davis MS, Cronan JE., Jr Inhibition of Eschericia coli acetyl coenzyme A carboxylase by acyl-acyl carrier protein. J Bacteriol. 2001;183:1499–1503. doi: 10.1128/JB.183.4.1499-1503.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Heath RJ, Rock CO. Inhibition of β-ketoacyl-acyl carrier protein synthase III (FabH) by acyl-acyl carrier protein in Escherichia coli. J Biol Chem. 1996;271:10996–11000. doi: 10.1074/jbc.271.18.10996. [DOI] [PubMed] [Google Scholar]
- 53.Lu Y-J, Rock CO. Transcriptional regulation of fatty acid biosynthesis in Streptococcus pneumoniae. Mol Microbiol. 2006;59:551–566. doi: 10.1111/j.1365-2958.2005.04951.x. [DOI] [PubMed] [Google Scholar]
- 54.Jerga A, Rock CO. Acyl-acyl carrier protein regulates transcription of fatty acid biosynthetic genes via the FabT repressor in Streptococcus pneumoniae. J Biol Chem. 2009;284:15364–15368. doi: 10.1074/jbc.C109.002410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Schujman GE, Paoletti L, Grossman AD, de Mendoza D. FapR, a bacterial transcription factor involved in global regulation of membrane lipid biosynthesis. Dev Cell. 2003;4:663–672. doi: 10.1016/s1534-5807(03)00123-0. [DOI] [PubMed] [Google Scholar]
- 56.Zhu K, Zhang YM, Rock CO. Transcriptional regulation of membrane lipid homeostasis in Escherichia coli. J Biol Chem. 2009;284:34880–34888. doi: 10.1074/jbc.M109.068239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Kass LR, Bloch K. On the enzymatic synthesis of unsaturated fatty acids in Escherichia coli. Proc Natl Acad Sci U S A. 1967;58:1168–1173. doi: 10.1073/pnas.58.3.1168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Feng Y, Cronan JE. Escherichia coli unsaturated fatty acid synthesis. J Biol Chem. 2009;284:29526–29535. doi: 10.1074/jbc.M109.023440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Zhang Y-M, Zhu K, Frank MW, Rock CO. A Pseudomonas aeruginosa transcription factor that senses fatty acid structure. Mol Microbiol. 2007;66:622–632. doi: 10.1111/j.1365-2958.2007.05934.x. [DOI] [PubMed] [Google Scholar]
- 60.Zhu K, Choi K-H, Schweizer HP, Rock CO, Zhang Y-M. Two aerobic pathways for the formation of unsaturated fatty acids in Pseudomonas aeruginosa. Mol Microbiol. 2006;60:260–273. doi: 10.1111/j.1365-2958.2006.05088.x. [DOI] [PubMed] [Google Scholar]
- 61.Subramanian C, Zhang Y-M, Rock CO. DesT coordinates the expression of anaerobic and aerobic pathways for unsaturated fatty acid biosynthesis in Pseudomonas aeruginosa. J Bacteriol. 2010;192:280–285. doi: 10.1128/JB.00404-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Miller DJ, Zhang Y-M, Subramainian C, Rock CO, White SW. Transcriptional regulation of membrane lipid homeostasis. Nat Struct Mol Biol. 2010;17:971–975. doi: 10.1038/nsmb.1847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Zhang Y-M, Rock CO. Acyltransferases in bacterial glycerophospholipid synthesis. J Lipid Res. 2008;49:1867–1874. doi: 10.1194/jlr.R800005-JLR200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Bell RM, Cronan JE., Jr Mutants of Escherichia coli defective in membrane phospholipid synthesis. Phenotypic suppression of sn-glycerol-3-phosphate acyltransferase Km mutants by loss of feedback inhibition of the biosynthetic sn-glycerol-3-phosphate dehydrogenase. J Biol Chem. 1975;250:7153–7158. [PubMed] [Google Scholar]
- 65.Bell RM. Mutants of Escherichia coli defective in membrane phospholipid synthesis: properties of wild type and Km defective sn-glycerol-3-phosphate acyltransfersae activities. J Biol Chem. 1975;250:7147–7152. [PubMed] [Google Scholar]
- 66.Lightner VA, Bell RM, Modrich P. The DNA sequences encoding plsB and dgk loci of Escherichia coli. J Biol Chem. 1983;258:10856–10861. [PubMed] [Google Scholar]
- 67.Lightner VA, Larson TJ, Tailleur P, Kantor GD, Raetz CRH, Bell RM, Modrich P. Membrane phospholipid synthesis in Escherichia coli: Cloning of a structural gene (plsB) of the sn-glycerol-3-phosphate acyltransferase. J Biol Chem. 1980;255:9413–9420. [PubMed] [Google Scholar]
- 68.Green PR, Merrill AH, Jr, Bell RM. Membrane phospholipid synthesis in Escherichia coli: Purification, reconstitution, and characterization of sn-glycerol-3-phosphate acyltransferase. J Biol Chem. 1981;256:11151–11159. [PubMed] [Google Scholar]
- 69.Lewin TM, Wang P, Coleman RA. Analysis of amino acid motifs diagnostic for the sn-glycerol-3-phosphate acyltransferase reaction. Biochemistry. 1999;38:5764–5771. doi: 10.1021/bi982805d. [DOI] [PubMed] [Google Scholar]
- 70.Coleman RA, Lee DP. Enzymes of triacylglycerol synthesis and their regulation. Prog Lipid Res. 2004;43:134–176. doi: 10.1016/s0163-7827(03)00051-1. [DOI] [PubMed] [Google Scholar]
- 71.Heath RJ, Rock CO. A conserved histidine is essential for glycerolipid acyltransferase catalysis. J Bacteriol. 1998;180:1425–1430. doi: 10.1128/jb.180.6.1425-1430.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Polgár L. The catalytic triad of serine peptidases. Cell Mol Life Sci. 2005;62:2161–2172. doi: 10.1007/s00018-005-5160-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Turnbull AP, Rafferty JB, Sedelnikova SE, Slabas AR, Schierer TP, Kroon JTM, Simon JW, Fawcett T, Nishida I, Murata N, Rice DW. Analysis of the structure, substrate specificity, and mechanism of squash glycerol-3-phosphate (1)-acyltransferase. Structure. 2001;9:347–353. doi: 10.1016/s0969-2126(01)00595-0. [DOI] [PubMed] [Google Scholar]
- 74.Wahl A, My L, Dumoulin R, Sturgis JN, Bouveret E. Antagonistic regulation of dgkA and plsB genes of phospholipid synthesis by multiple stress responses in Escherichia coli. Mol Microbiol. 2011;80:1260–1275. doi: 10.1111/j.1365-2958.2011.07641.x. [DOI] [PubMed] [Google Scholar]
- 75.Heath RJ, Jackowski S, Rock CO. Guanosine tetraphosphate inhibition of fatty acid and phospholipid synthesis in Escherichia coli is relieved by overexpression of glycerol-3-phosphate acyltransferase (plsB) J Biol Chem. 1994;269:26584–26590. [PubMed] [Google Scholar]
- 76.Svitil AL, Cashel M, Zyskind JW. Guanosine tetraphosphate inhibits protein synthesis in vivo. J Biol Chem. 1993;268:2307–2311. [PubMed] [Google Scholar]
- 77.Cashel M, Gentry DR, Hernadez VJ, Vinella D. The Stringent ResponseEscherichia coli and Salmonella typhimurium. In: Neidhardt FC, Curtis R, Lin ECC, Ingraham JL, Low BL, Magasanik B, Reznikoff WS, Riley M, Schaechter M, Umbarger HE, editors. Cellular and Molecular Biology. Vol. 2. American Society for Microbiology; Washington, D.C: 1996. pp. 1458–1496. [Google Scholar]
- 78.Rhodius VA, Suh WC, Nonaka G, West J, Gross CA. Conserved and variable functions of the σE stress response in related genomes. PLoS Biol. 2005;4:e2. doi: 10.1371/journal.pbio.0040002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Saraste M, Sibbald PR, Wittinghofer A. The P-loop--a common motif in ATP- and GTP-binding proteins. Trends Biochem Sci. 1990;15:430–434. doi: 10.1016/0968-0004(90)90281-f. [DOI] [PubMed] [Google Scholar]
- 80.Grimes K, Lu YJ, Zhang YM, Luna V, Hurdle J, Carson E, Qi J, Kudrimoti S, Rock C, Lee R. Novel acyl phosphate mimics that target PlsY, an essential acyltransferase in gram-positive bacteria. ChemMedChem. 2008;3:1936–1945. doi: 10.1002/cmdc.200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Kobayashi K, Ehrlich SD, Albertini A, Amati G, Andersen KK, Arnaud M, Asai K, Ashikaga S, Aymerich S, Bessieres P, Boland F, Brignell SC, Bron S, Bunai K, Chapuis J, Christiansen LC, Danchin A, Debarbouille M, Dervyn E, Deuerling E, Devine K, Devine SK, Dreesen O, Errington J, Fillinger S, Foster SJ, Fujita Y, Galizzi A, Gardan R, Eschevins C, Fukushima T, Haga K, Harwood CR, Hecker M, Hosoya D, Hullo MF, Kakeshita H, Karamata D, Kasahara Y, Kawamura F, Koga K, Koski P, Kuwana R, Imamura D, Ishimaru M, Ishikawa S, Ishio I, Le Coq D, Masson A, Mauel C, Meima R, Mellado RP, Moir A, Moriya S, Nagakawa E, Nanamiya H, Nakai S, Nygaard P, Ogura M, Ohanan T, O’Reilly M, O’Rourke M, Pragai Z, Pooley HM, Rapoport G, Rawlins JP, Rivas LA, Rivolta C, Sadaie A, Sadaie Y, Sarvas M, Sato T, Saxild HH, Scanlan E, Schumann W, Seegers JFML, Sekiguchi J, Sekowska A, Seror SJ, Simon M, Stragier P, Studer R, Takamatsu H, Tanaka T, Takeuchi M, Thomaides HB, Vagner V, van Dijl JM, Watabe K, Wipat A, Yamamoto H, Yamamoto M, Yamamoto Y, Yamane K, Yata K, Yoshida K, Yoshikawa H, Zuber U, Ogasawara N. Essential Bacillus subtilis genes. Proc Natl Acad Sci U S A. 2003;100:4678–4683. doi: 10.1073/pnas.0730515100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Coleman J. Characterization of Escherichia coli cells deficient in 1-acyl-sn-glycerol-3-phosphate acyltransferase activity. J Biol Chem. 1990;265:17215–17221. [PubMed] [Google Scholar]
- 83.Coleman J. Characterization of the Escherichia coli gene for 1-acyl-sn-glycerol-3-phosphate acyltransferase (plsC) Mol Gen Genet. 1992;232:295–303. doi: 10.1007/BF00280009. [DOI] [PubMed] [Google Scholar]
- 84.Shih GC, Kahler CM, Swartley JS, Rahman MM, Coleman J, Carlson RW, Stephens DS. Multiple lysophosphatidic acid acyltransferases in Neisseria meningitidis. Mol Microbiol. 1999;32:942–952. doi: 10.1046/j.1365-2958.1999.01404.x. [DOI] [PubMed] [Google Scholar]
- 85.Swartley JS, Balthazar JT, Coleman J, Shafer WM, Stephens DS. Membrane glycerophospholipid biosynthesis in Neisseria meningitidis and Neisseria gonorrhoeae: identification, characterization, and mutagenesis of a lysophosphatidic acid acyltransferase. Mol Microbiol. 1995;18:401–412. doi: 10.1111/j.1365-2958.1995.mmi_18030401.x. [DOI] [PubMed] [Google Scholar]
- 86.Cullinane M, Baysse C, Morrissey JP, O’Gara F. Identification of two lysophosphatidic acid acyltransferase genes with overlapping function in Pseudomonas fluorescens. Microbiology. 2005;151:3071–3080. doi: 10.1099/mic.0.27958-0. [DOI] [PubMed] [Google Scholar]
- 87.Agun-Sunar S, Bilaloglu R, Goldfine H, Daldal F. Rhodobacter capsulatus OlsA is a bifunctional enzyme active in both ornithine lipid and phosphatidic acid biosynthesis. J Bacteriol. 2007;189:8564–8574. doi: 10.1128/JB.01121-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Jackson MB, Cronan JE., Jr An estimate of the minimum amount of fluid lipid required for the growth of Escherichia coli. Biochim Biophys Acta. 1978;512:472–479. doi: 10.1016/0005-2736(78)90157-8. [DOI] [PubMed] [Google Scholar]
- 89.Schulman H, Kennedy EP. Relation of turnover of membrane phospholipids to synthesis of membrane-derived oligosaccharides of Escherichia coli. J Biol Chem. 1977;252:4250–4255. [PubMed] [Google Scholar]
- 90.Kennedy EP. Osmotic regulation and biosynthesis of membrane-derived oligosaccharides in Escherichia coli. Proc Natl Acad Sci U S A. 1982;79:1092–1095. doi: 10.1073/pnas.79.4.1092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Raetz CRH, Newman KF. Diglyceride kinase mutants of Escherichia coli: inner membrane association of 1,2-diglyceride and its relation to synthesis of membrane-derived oligosaccharides. J Bacteriol. 1979;137:860–868. doi: 10.1128/jb.137.2.860-868.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Raetz CRH, Newman KF. Neutral lipid accumulation in the membranes of Escherichia coli mutants lacking diglyceride kinase. J Biol Chem. 1978;253:3882–3887. [PubMed] [Google Scholar]
- 93.Van Horn WD, Kim HJ, Ellis CD, Hadziselimovic A, Sulistijo ES, Karra MD, Tian C, Sönnichsen FD, Sanders CR. Solution nuclear magnetic resonance structure of membrane-integral diacylglycerol kinase. Science. 2009;324:1726–1729. doi: 10.1126/science.1171716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Durfee T, Hansen AM, Zhi H, Blattner FR, Jin DJ. Transcription profiling of the stringent response in Escherichia coli. J Bacteriol. 2008;190:1084–1096. doi: 10.1128/JB.01092-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Hagiwara D, Yamashino T, Mizuno T. A genome-wide view of the Escherichia coli BasS-BasR two-component system implicated in iron-responses. Biosci Biotechnol Biochem. 2004;68:1758–1767. doi: 10.1271/bbb.68.1758. [DOI] [PubMed] [Google Scholar]
- 96.Van Horn WD, Sanders CR. Prokaryotic diacylglycerol kinase and undecaprenol kinase. Annu Rev Biophys. 2011;41:81–101. doi: 10.1146/annurev-biophys-050511-102330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Jerga A, Lu Y-J, Schujman GE, de Mendoza D, Rock CO. Identification of a soluble diacylglycerol kinase required for lipoteichoic acid production in Bacillus subtilis. J Biol Chem. 2007;282:21738–21745. doi: 10.1074/jbc.M703536200. [DOI] [PubMed] [Google Scholar]
- 98.Miller DJ, Jerga A, Rock CO, White SW. Analysis of the Staphylococcus aureus DgkB structure reveals a common catalytic mechanism for the soluble diacylglycerol kinases. Structure. 2008;16:1036–1046. doi: 10.1016/j.str.2008.03.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Jerga A, Miller DJ, White SW, Rock CO. Molecular determinants for interfacial binding and conformational change in a soluble diacylglycerol kinase. J Biol Chem. 2009;284:7246–7254. doi: 10.1074/jbc.M805962200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Lucchini JJ, Corre J, Cremieux A. Antibacterial activity of phenolic compounds and aromatic alcohols. Res Microbiol. 1990;141:499–510. doi: 10.1016/0923-2508(90)90075-2. [DOI] [PubMed] [Google Scholar]
- 101.Corre J, Lucchini JJ, Mercier GM, Cremieux A. Antibacterial activity of phenethyl alcohol and resulting membrane alterations. Res Microbiol. 1990;141:483–497. doi: 10.1016/0923-2508(90)90074-z. [DOI] [PubMed] [Google Scholar]
- 102.Killian JA, Fabrie CH, Baart W, Morein S, de Kruijff B. Effects of temperature variation and phenethyl alcohol addition on acyl chain order and lipid organization in Escherichia coli derived membrane systems. A 2H- and 31P-NMR study. Biochim Biophys Acta. 1992;1105:253–262. doi: 10.1016/0005-2736(92)90202-w. [DOI] [PubMed] [Google Scholar]
- 103.Silva MT, Sousa JC, Macedo MA, Polonia J, Parente AM. Effects of phenethyl alcohol on Bacillus and Streptococcus. J Bacteriol. 1976;127:1359–1369. doi: 10.1128/jb.127.3.1359-1369.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Nunn WD, Tropp BE. Effects of phenethyl alcohol on phospholipid metabolism in Escherichia coli. J Bacteriol. 1972;109:162–168. doi: 10.1128/jb.109.1.162-168.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Nunn WD. Fatty acid synthesis in Escherichia coli is indirectly inhibited by phenethyl alcohol. Biochemistry. 1977;16:1077–1081. doi: 10.1021/bi00625a008. [DOI] [PubMed] [Google Scholar]
- 106.Nunn WD, Cheng PJ, Deutsch R, Tang CT, Tropp BE. Phenethyl alcohol inhibition of sn-glycerol 3-phosphate acylation in Escherichia coli. J Bacteriol. 1977;130:620–628. doi: 10.1128/jb.130.2.620-628.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Cherian P, Yao J, Leonardi R, Luna VA, Rock CO, Lee RE. Acyl-sulfamates target the essential glycerol-phosphate acyltransferase (PlsY) in Gram-positive bacteria. Bioorg Med Chem. 2012;20:4985–4994. doi: 10.1016/j.bmc.2012.06.029. [DOI] [PMC free article] [PubMed] [Google Scholar]