SUMMARY
Polarization of mammalian neurons with a specified axon requires precise regulation of microtubule and actin dynamics in the developing neurites. Here we show that mammalian partition defective 3 (mPar3), a key component of the Par polarity complex that regulates the polarization of many cell types including neurons, directly regulates microtubule stability and organization. The N-terminal portion of mPar3 exhibits strong microtubule binding, bundling and stabilization activity, which can be suppressed by its C-terminal portion via an intra-molecular interaction. Interestingly, the inter-molecular oligomerization of mPar3 is able to relieve the intra-molecular interaction and thereby promote microtubule bundling and stabilization. Furthermore, disruption of this microtubule regulatory activity of mPar3 impairs its function in axon specification. Together, these results demonstrate a role for mPar3 in directly regulating microtubule organization that is crucial for neuronal polarization.
INTRODUCTION
Neurons are among the most polarized cells in living organisms. They develop two morphologically, molecularly and physiologically distinct subcellular compartments – axon and dendrite (Barnes and Polleux, 2009). This axon-dendrite polarity is the basis of unidirectional information flow in the nervous system – the axon is responsible for sending signals whereas the dendrites are responsible for receiving signals. Extensive studies have been carried out to characterize the neuronal polarization process, mostly using hippocampal neurons in culture that recapitulate many aspects of in vivo axon specification (Bradke and Dotti, 2000; Craig and Banker, 1994; Dotti et al., 1988). A crucial event during this process is the differential growth rate of the future axon versus dendrites. The polarization of neurons begins with one and only one of the cellular processes (i.e. neurites) growing faster than the remaining processes. As development proceeds, this fast-growing process becomes the axon and the remaining processes become the dendrites.
Neurite growth largely takes place at the tip, the growth cone, which harbors a peripheral domain enriched in actin and a central domain predominantly composed of microtubules (Forscher and Smith, 1988). As the growth cone advances, neurites extend and grow. Moreover, the dynamics of the actin and microtubule cytoskeleton control the motility of the growth cone, thereby determining the growth rate of the neurites. A fast-growing neurite often possesses a growth cone with a less condensed and more dynamic actin cytoskeleton and, at the same time, more stabilized microtubules (Forscher and Smith, 1988). Indeed, it has been demonstrated that actin and microtubule cytoskeleton dynamics are differentially regulated at the neurite growth cones of a polarizing neuron and this differential regulation is critical for axon specification and the establishment of neuronal polarity (Bradke and Dotti, 1999; Stiess and Bradke, 2011; Witte and Bradke, 2008; Witte et al., 2008). While these studies provide important insights into neuronal polarization, our knowledge of the molecular mechanisms underlying the differential regulation of the actin and microtubule cytoskeleton at the growth cones of the developing neurites in polarizing neurons remains limited. Recently, it was shown that the evolutionarily conserved partition defective (Par) protein complex plays an essential role in establishing the axon-dendrite polarity of mammalian neurons (Shi et al., 2003). The Par protein complex consists of Par3 and Par6, two PDZ domain-containing proteins, as well as atypical protein kinase C (aPKC) (Goldstein and Macara, 2007; Kemphues, 2000). During mammalian neuron polarization, the subcellular distribution of mammalian Par3 (mPar3) and mPar6 becomes polarized, resulting in their selective enrichment in the future axon. Disruption of this polarized distribution of mPar3 or mPar6 impairs axon specification and neuronal polarization (Shi et al., 2003). Since this initial report, mounting evidences point to a critical role for the mPar3/mPar6/aPKC protein complex in regulating axon specification and neuronal polarization (Higginbotham et al., 2006; Nishimura et al., 2004; Nishimura et al., 2005; Schwamborn et al., 2007a; Shi et al., 2004; Vohra et al., 2007; Yi et al., 2010).
Both Par3 and Par6 are considered to be scaffold proteins. Par6 forms a stable complex with aPKC and contains a semi-Cdc42/Rac interactive binding (CRIB) domain that specifically binds to the active GTP-bound form of the small GTPases, Cdc42 and Rac1 (Joberty et al., 2000). Par3, on the other hand, interacts with both Par6 and the T-lymphoma invasion and metastasis 1 (Tiam1) protein, a guanine nucleotide exchange factor for Rac (Kunda et al., 2001; Nishimura et al., 2005). Small GTPases are central regulators of actin cytoskeleton dynamics (Hall, 1998). Polarization of the mPar3/mPar6/aPKC complex facilitates local activation of small GTPases at the tip of the future axon (Schwamborn et al., 2007a; Schwamborn et al., 2007b; Schwamborn and Puschel, 2004). Further evidences for the role of mPar3 and mPar6 in the regulation of actin cytoskeleton came from studies showing that they control the morphogenesis of dendritic spines, cellular structures largely supported by filamentous actin (Zhang and Macara, 2006; Zhang and Macara, 2008). While these studies link the mPar3/mPar6/aPKC complex to local regulation of the actin cytoskeleton at the future axon tip, it is unclear whether the polarization of Par proteins mediates differential regulation of microtubule dynamics in the neurites of developing neurons for polarization and axon specification.
RESULTS
The N-terminal portion of mPar3 bundles and stabilizes microtubules
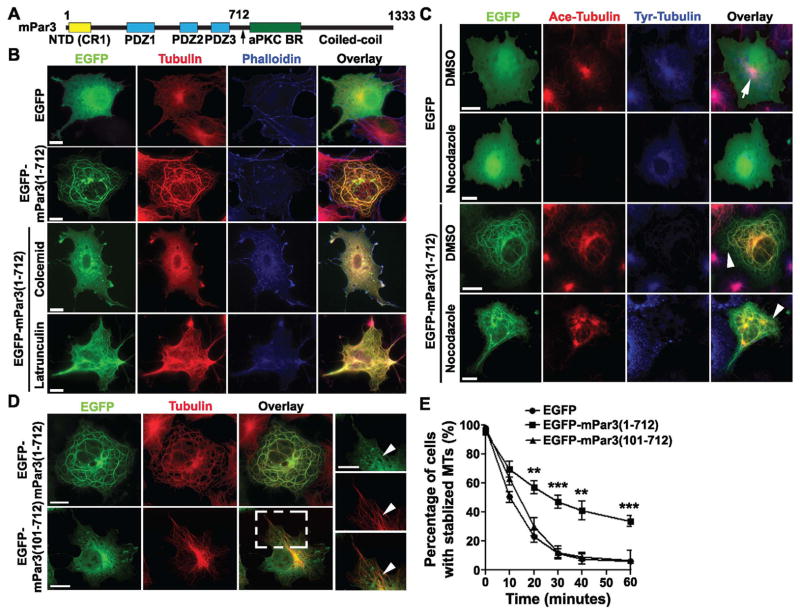
To investigate the role of mPar3 in regulating microtubule organization, we performed structure-function analyses of mPar3 (Figure 1A). We generated a series of full-length or fragments of mPar3 fused with enhanced green fluorescent protein (EGFP) at the N-terminus and examined their distribution in COS-7 cells. Remarkably, when we expressed the N-terminal portion of mPar3 lacking the C-terminal aPKC binding domain and coiled coil region (i.e. amino acids 1 to 712, mPar3(1-712)), it formed thick, rounded tubular structures in the cells (Figure 1B and Figure S1, green). Moreover, these tubular structures co-localized with the microtubule cytoskeleton revealed by tubulin immunostaining (Figure 1B, red), but not with the actin cytoskeleton labeled by phalloidin staining (Figure 1B, blue). Notably, while microtubules in EGFP-expressing cells or non-transfected cells appeared as a profuse array of thin filaments emanating from the centrosome (the major microtubule-organizing center), they became unusually thick, rounded bundles in EGFP-mPar3(1-712) expressing cells (Figure 1B). In addition, we generated mPar3(1-712) constructs with EGFP fused at the C-terminus, or fused with a small His tag, or expressed EGFP-mPar3(1-712) in HEK293 cells (Figure S1). Under all these conditions, mPar3(1-712) fusion proteins formed thick tubular structures and co-localized with microtubules, suggesting that this is an intrinsic property of mPar3(1-712).
Figure 1. The N-terminal portion of mPar3 bundles and stabilizes microtubules.
(A) Schematic illustration of the domain structure of mPar3. Numbers refer to amino acid positions. NTD, N-terminal oligomerization domain; CR1, conserved region 1; PDZ, PSD-95/Dlg/ZO-1; aPKC BR, aPKC binding region. (B) Images of COS7 cells expressing EGFP or EGFP-mPar3(1-712) (green) stained with the antibodies against tubulin (red) and phalloidin (blue). Scale bars: 20μm. (C) Images of COS7 cells expressing EGFP or EGFP-mPar3(1-712) (green) treated with DMSO or nocodazole (10 μM), a microtubule-depolymerizing agent, for 60 minutes and stained with the antibodies against acetylated tubulin (red) and tyrosinated tubulin (blue). Arrow and arrowheads indicate stable acetylated microtubules in cells expressing EGFP or EGFP-mPar3(1-712), respectively. Scale bars: 20 μm. (D) Images of COS7 cells expressing EGFP-mPar3(1-712) or EGFP-mPar3(101-712) (green) and stained with the antibody against tubulin (red). High magnification images of a region in EGFP-mPar3(101-712)-expressing cell (broken lines) are shown to the right. Arrowheads indicate co-localization of EGFP-mPar3(101-712) with microtubules. Scale bars: 20 μm and 10 μm. (E) Quantification of the fraction of cells with stabilized microtubules expressing EGFP, EGFP-mPar3(1-712), or EGFP-mPar3(101-712) at different time points after nocodazole (10 μM) treatment. Error bars indicate s.e.m. of independent sets of experiments (EGFP, n=6; EGFP-mPar3(1-712), n=14; EGFP-mPar3(101-712), n=5). **, p<0.005; ***, p<0.0001. (See also Figure S1 and S2)
To confirm that the thick, rounded tubular structures formed by mPar3(1-712) are indeed microtubule bundles, we treated the cells with colcemid, a microtubule-depolymerizing drug, or latrunculin, an actin filament destabilizing drug (Figure 1B). We found that, while the tubular structures formed by mPar3(1-712) remained largely intact after latrunculin treatment, they were completely eliminated by colcemid treatment. Taken together, these results strongly suggest that mPar3(1-712) not only localizes to microtubules, but also induces the formation of unusually thick, rounded microtubule bundles in cells.
Bundling of microtubules often stabilizes them (Takemura et al., 1992). To determine whether mPar3(1-712) bundles and thereby stabilizes microtubules, we treated the cells expressing EGFP or EGFP-mPar3(1-712) with a low concentration of nocodazole (10 μM), another microtubule-depolymerizing agent, and stained them with the antibodies against acetylated tubulin and tyrosinated tubulin, which mark stabilized and destabilized microtubules, respectively (Westermann and Weber, 2003) (Figure 1C and E). Upon nocodazole treatment, control cells expressing EGFP showed a progressive decrease in the level of acetylated tubulin and a concurrent increase in tyrosinated tubulin. Nearly all cells lost acetylated tubulin 60 minutes after treatment (Figure 1C and E). In contrast, a substantial fraction of cells expressing EGFP-mPar3(1-712) possessed high levels of acetylated tubulin and low levels of tyrosinated tubulin (Figure 1C, 1E, S2A and S2B). Usually stabilized microtubules marked by acetylated tubulin are found near the center of the cell, where the centrosome is located (Bulinski et al., 1988; Piperno et al., 1987; Schulze et al., 1987; Schulze and Kirschner, 1987) (Figure 1C, arrow). However, stabilized microtubules in cells expressing mPar3(1-712) extended to the cell periphery (Figure 1C, arrowheads), indicating that these stabilized microtubule bundles are not necessarily nucleated from the centrosome, similar to the long microtubule bundles in the neurites of developing neurons. These results suggest that mPar3(1-712) bundles and stabilizes microtubules, and affects the state of post-translational modification of tubulin. In addition, microtubule bundles stabilized by mPar3(1-712) can exist in the cytoplasm independently of the centrosome.
Microtubule association domain in mPar3(1-712)
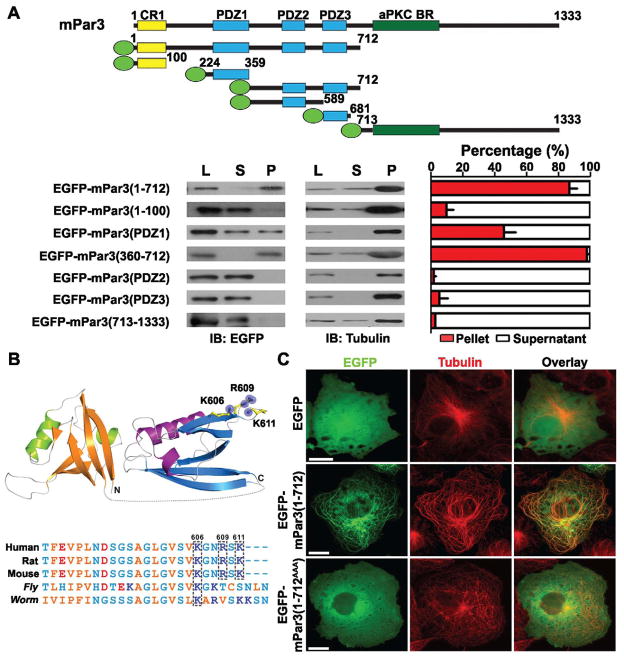
To identify the domain in mPar3(1-712) responsible for microtubule association, we carried out further structure-function analysis. We found that removal of the N-terminal conserved oligomerization domain (amino acids 1-100, NTD or CR1) completely prevented the protein, mPar3(101-712), from forming thick microtubule bundles (Figure 1D) and the stabilization of microtubules (Figures 1E). However, this domain was not required for microtubule association, as mPar3(101-712) remained localized to microtubule arrays (Figure 1D, arrowheads). In addition, point mutations in the NTD (V13D/D70K) that selectively disrupt the oligomerization activity without affecting protein folding (Feng et al., 2007) prevented the formation of thick microtubule bundles while retaining microtubule association (Figure S2C, arrowheads). These results suggest that the oligomerization domain is required for bundling microtubules, while the microtubule association domain lies within amino acids 101-712. To further narrow down this domain, we performed microtubule co-sedimentation assays using lysates of COS7 cells expressing various fragments of mPar3 fused to EGFP (Figure 2A, top) and found that mPar3(1-712) and mPar3(360-712) containing both PDZ domains 2 and 3 exhibited strong co-sedimentation with microtubules, whereas the N-terminal oligomerization domain (1-100), PDZ 2 (360-589), PDZ 3 (590-681), and the C-terminal portion (713-1333) did not (Figure 2A, bottom). Notably, PDZ1 (224-359), a region previously shown to indirectly associate with dynein motor proteins (Schmoranzer et al., 2009), exhibited partial co-sedimentation with microtubules.
Figure 2. Microtubule-association domain in mPar3(1-712).
(A) (top) Schematic illustrations of the domain structure of full-length and fragments of mPar3 fused to EGFP (green oval). (bottom) Co-sedimentation analysis of mPar3 fragments with microtubules. Sample western blot images are shown to the left and quantification of the percentage of proteins in the pellet (P, red bars) versus supernatant (S, white bars) is shown to the right. Error bars represent s.e.m. of four independent sets of experiments. (B) A conserved cluster of positively charged residues (K606, R609, and K611 in mouse Par3) revealed by sequence alignment (top) and structural modeling (bottom). (C) Images of COS7 cells expressing EGFP, EGFP-mPar3(1-712), or EGFP-mPar3(1-712AAA) in which K606, R609 and K611 were mutated to alanine (A) (green) and stained with the antibody against tubulin (red). Scale bars: 20 μm. (See also Figure S3)
To map out the critical residues for microtubule association in PDZ domains 2 and 3, we modeled the three-dimensional structure of this region (360-712) based on the existing protein structure database (Figure 2B, top). The binding of many microtubule-associated proteins (MAPs) to microtubules is thought to involve an ionic interaction between the basic tubulin-binding domain and the acidic C-terminus of tubulin (Littauer et al., 1986; Paschal et al., 1989; Serrano et al., 1984). Therefore, we searched for positively charged residues at the surface of the predicted structure and identified a cluster of basic residues including K606, R609 and K611 that are well conserved between humans and rodents, and partly conserved in C. elegans Par3 protein and to a lesser degree in Drosophila Bazooka protein (Figure 2B, bottom). Interestingly, mutations of these three residues to alanine (A) prevented the protein, mPar3(1-712AAA), from associating with and bundling microtubules (Figure 2C), suggesting that this cluster of positively charged residues is crucial for the association of mPar3(1-712) with microtubules. These three mutations in mPar3 did not interfere with the complex formation among mPar3, mPar6 and aPKC (Figure S3A) or the lipid binding of mPar3 PDZ domains (Wu et al., 2007) (Figure S3B), indicating that the loss of microtubule association is unlikely due to an overall protein mis-folding.
Direct microtubule association and bundling by mPar3(1-712)
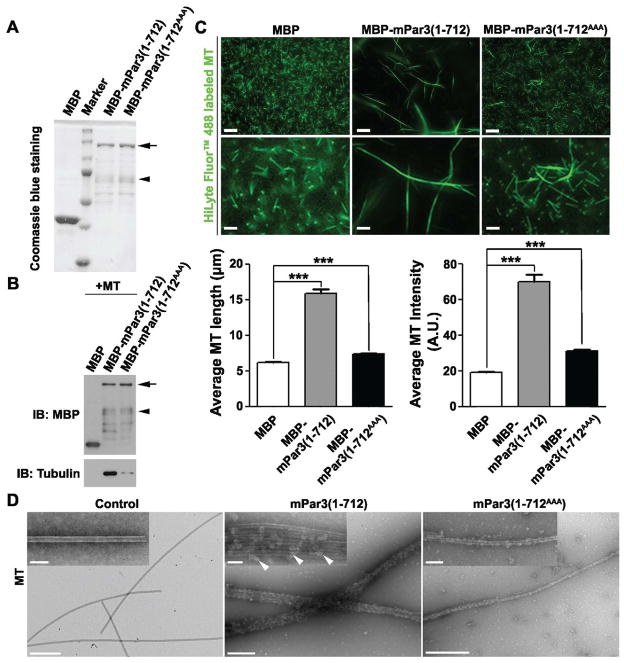
After identifying the microtubule association domain, we next evaluated whether the association between mPar3(1-712) and microtubules was direct. We purified mPar3(1-712) or mPar3(1-712AAA) fused to Maltose-Binding Protein (MBP), or MBP alone expressed in bacteria (Figure 3A), incubated them with in vitro assembled microtubules, and performed the immunoprecipitation (Figure 3B) and microtubule co-pelleting experiments (Figure S4A). While MBP did not bind microtubules, MBP-mPar3(1-712) bound microtubules robustly. Moreover, this direct association was largely abolished by mutating the three positively charged residues that we identified, as MBP-mPar3(1-712AAA) barely bound microtubules.
Figure 3. Direct microtubule association and bundling by mPar3(1-712).
(A) Purification of MBP, MBP-mPar3(1-712) or MBP-mPar3(1-712AAA) recombinant proteins from E. coli. The arrow indicates full-length MBP-mPar3(1-712) and MBP-mPar3(1-712AAA), and the arrowhead indicates some degradation products. (B) Purified MBP, MBP-mPar3(1-712), or MBP-mPar3(101-712) proteins were mixed with in vitro assembled microtubules. Immunoprecipitation experiments were then performed using the amylose beads to assess microtubules bound to the purified proteins. The arrow indicates full-length MBP-mPar3(1-712) and MBP-mPar3(1-712AAA), and the arrowhead indicates some degradation products. (C) Purified MBP, MBP-mPar3(1-712), or MBP-mPar3(1-712AAA) proteins were mixed with in vitro assembled HyLyte 488 labeled microtubules. The morphology of microtubules was then analyzed by fluorescence microscopy. (top) Images of HyLyte 488 labeled microtubules assembled in vitro in the presence of purified recombinant protein MBP (left), MBP-mPar3(1-712) (middle) or MBP-mPar3(1-712AAA) (right). High magnification images are shown at the bottom. Scale bars: 20 μm and 5 μm. (bottom) Quantification of the average length (left, MBP, n=405; MBP-mPar3(1-712), n=377; MBP-mPar3(1-712AAA), n=726; from three independent sets of experiments) and fluorescence intensity (right, MBP, n=329; MBP-mPar3(1-712), n=295; MBP-mPar3(1-712AAA), n=369; from three independent sets of experiments) of microtubules. A.U., arbitrary unit. Error bars indicate s.e.m.; ***, p<0.0001. (D) Purified MBP, MBP-mPar3(1-712), or MBP-mPar3(1-712AAA) proteins were mixed with in vitro assembled microtubules. The morphology of microtubules was then analyzed by transmission electron microscopy (TEM). TEM images of in vitro assembled microtubules in the presence of MBP (left), MBP-mPar3(1-712) (middle) or MBP-mPar3(1-712AAA) (right). High magnification images are shown as insets. Scale bars: 500 nm and 100 nm. (See also Figure S4)
To test whether this direct association between mPar3(1-712) and microtubules leads to microtubule bundling, we incubated purified recombinant proteins with in vitro assembled fluorescence-labeled microtubules (Figure 3C and Figure S4B). We found that in the presence of MBP, microtubules existed as dense, short and thin filaments. In contrast, in the presence of MBP-mPar3(1-712), microtubules formed long and thick filaments at a low density, which were decorated by MBP-mPar3(1-712) recombinant proteins, suggesting that the short and thin microtubule filaments are effectively bundled into long and thick filaments by mPar3(1-712). As a result, the average microtubule filament length was substantially longer and the average microtubule filament fluorescence intensity was significantly higher in the presence of MBP-mPar3(712) than that of MBP (Figure 3C). Furthermore, this microtubule bundling activity of mPar3(1-712) critically depends on the positively charged residue cluster in the microtubule association domain, as mutating them to alanine strongly inhibited the formation of long and thick microtubule bundles (MBP-mPar3(1-712AAA), Figure 3C and Figure S4B).
To further reveal the nature of mPar3(1-712) microtubule bundling activity, we examined microtubule configuration by transmission electron microscopy (TEM) (Figure 3D). We found that while microtubules in the control condition existed mostly as single filaments (Figure 3D, left), they became thick bundles containing on average four to six individual filaments in the presence of mPar3(1-712), which decorated the length of the thick microtubule bundles (Figure 3D, middle, arrowheads). Again, this microtubule bundling activity was largely abolished when the positively charged residues in the microtubule association domain were mutated (Figure 3D, right). Taken together, these results clearly demonstrated that the N-terminal portion of mPar3 acts as a microtubule-associating protein that directly binds and bundles microtubules.
Suppression of microtubule bundling activity by the C-terminal portion
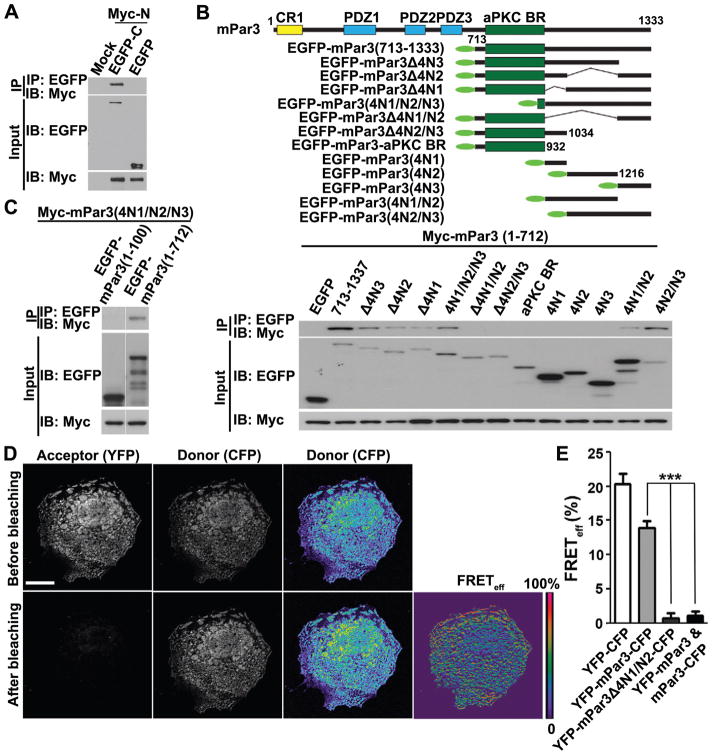
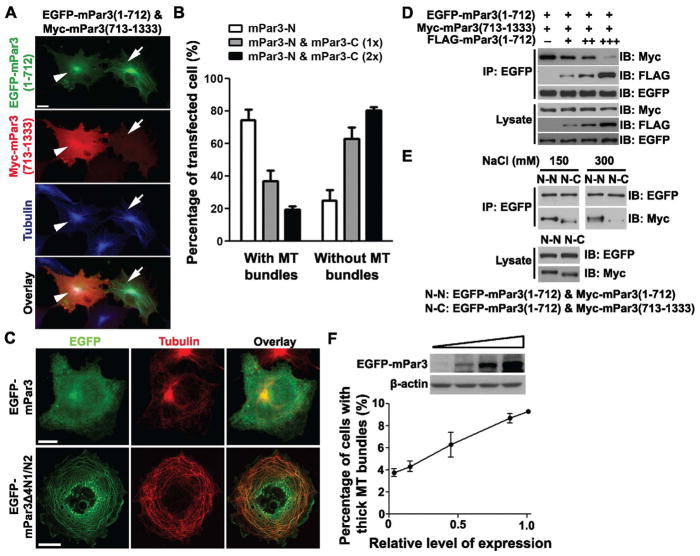
Having established that the N-terminal portion of mPar3 binds, bundles and stabilizes microtubules, we went on to explore this activity in full-length mPar3. In our initial structure-function analysis, we noted that full-length mPar3 only occasionally localized to microtubules, raising the possibility that the microtubule binding and bundling activity was mostly masked in full-length mPar3. We postulated that the C-terminal portion of mPar3 might interact with its N-terminal portion and thereby suppress its microtubule binding and bundling activity. To test this, we first examined the potential interaction between the N- and C-terminal portions of mPar3. We found that a Myc-tagged N-terminal portion of mPar3, Myc-mPar3(1-712), co-precipitated with an EGFP-tagged C-terminal portion, EGFP-mPar3(713-1333) (Figure 4A), suggesting an intra-molecular interaction between the N-terminal and C-terminal portions of mPar3. Furthermore, we found that co-expression of the C-terminal portion of mPar3 suppressed the N-terminal portion’s microtubule binding and bundling activity in a dose-dependent manner (Figure 5A and 5B). The higher the level of mPar3 C-terminus expression, the less frequent the cells with thick microtubule bundles were observed.
Figure 4. Intra-molecular interaction between the N- and C-terminal portions of mPar3.
(A) COS7 cells were transfected with plasmids carrying Myc-mPar3(1-712) and EGFP-mPar3(713-1333). Cell lysates were harvested 48 hours post-transfection and immunoprecipitated with anti-EGFP antibody and agarose beads. Input lysate and bound proteins were separated by SDS-PAGE and immunoblotted using antibodies against EGFP and Myc. (B) (top) Schematic illustration of the C-terminal fragments fused to EGFP (green oval), used to map the domain interacting with the N-terminal portion. (bottom) Immunoprecipitation of the C-terminal portion fragments with the N-terminal portion. (C) The C-terminal coiled-coil region is sufficient to bind the N-terminal portion of mPar3, but does not bind the N-terminal oligomerization domain. (D) Images of COS7 cells expressing YFP-mPar3-CFP before and after photobleaching the acceptor YFP. Pseudocolored images of the donor CFP and FRET efficiency (FRETeff (%) = ((CFPpost-CFPpre)/CFPpost) are shown to the right. Scale bars: 20 μm. (E) Quantification of FRET efficiency for YFP-CFP (n=30), YFP-mPar3-CFP (n=43), YFP-mPar3Δ4 N1/N2-CFP (n=28), and YFP-mPar3/mPar3-CFP (n=31). ror bars indicate s.e.m.Er ***, p<0.0001. (See also Figure S5)
Figure 5. Intra-molecular interaction suppresses, while oligomerization of mPar3 promotes microtubule binding and bundling.
(A) Images of COS7 cells expressing EGFP-mPar3(1-712) (green) and Myc-mPar3(713-1333) (red), and stained for tubulin (blue). Scale bars: 20 μm. (B) Quantification of the percentage of cells expressing the N-terminal portion of mPar3 with microtubule bundles. (C) Images of COS7 cells expressing full-length (left) or Δ4N1/N2 (right) mPar3 fused to EGFP (green) and stained for tubulin (red). Scale bars: 20 μm. (D) Immunoprecipitation of EGFP-mPar3(1-712) with Myc-mPar3(713-1333) in the presence of different levels of FLAG-mPar3(1-712). (E) Immunoprecipitation efficiency between the N-terminal portions or the N- and C-terminal portions at different salt concentrations. (F) (top) Expression levels of EGFP-mPar3 revealed by western blot. (bottom) Quantification of the percentage of cells expressing EGFP-mPar3 with thick microtubule bundles. Error bars indicate s.e.m. ***, p<0.0001.
To identify the region(s) in the C-terminal portion of mPar3 that interacts with the N-terminal portion, we expressed various fragments of the C-terminal portion together with the N-terminal portion and performed co-precipitation experiments (Figure 4B). We found that the C-terminal coiled-coil region was critical for the intra-molecular interaction with the N-terminal portion, as deletion of any two of the three previously characterized fragments (4N1, 4N2 and 4N3) in this region (Nishimura et al., 2005) impaired the interaction (Figure 4B). Moreover, the coiled-coil region alone was sufficient for the interaction with the N-terminal portion (Figure 4C). Should the coiled-coil region at the C-terminal portion of mPar3 mediate the intra-molecular interaction that suppresses the microtubule binding and bundling activity of the N-terminal portion, we expect that removal of this region would obviate the intra-molecular interaction and thereby promote microtubule binding and bundling activity. Consistent with this, we found that a deletion in the coiled-coil region substantially enhanced the microtubule binding and bundling activity of mPar3 (Figure 5C).
To further confirm the intra-molecular interaction between the N- and C-terminal portions of mPar3, we fused yellow fluorescence protein (YFP) and Cerulean (a cyan fluorescence protein, CFP, variant) to the N-terminus and C-terminus of mPar3, respectively, and performed Förster Resonance Energy Transfer (FRET) experiments (Figures 4D, 4E and S5). We found that upon bleaching the acceptor fluorophore YFP, the fluorescence intensity of the donor fluorophore Cerulean was markedly increased (Figure 4D and S5), indicating that FRET occurs between the two fluorophores and thereby implying the proximity between the N- and C-termini of mPar3. As expected, a direct fusion of Cerulean and YFP led to strong FRET (Figures 4E and S5). However, when YFP and Cerulean were fused to the N-terminus or the C-terminus of mPar3 separately, no obvious FRET was detected when the two proteins were co-expressed (Figures 4E and S5). Furthermore, we found that a deletion in the coiled-coil region that disrupted the intra-molecular interaction between the N- and C-terminal portions of mPar3 eliminated FRET (Figures 4E and S5). Taken together, these results further strongly support the notion that the N-terminal portion of mPar3 interacts with its C-terminal portion.
Oligomerization of mPar3 promotes microtubule binding and bundling
Previous studies have shown that the conserved N-terminal oligomerization domain mediates strong inter-molecular interaction and is critical for Par3 function (Benton and St Johnston, 2003a; Benton and St Johnston, 2003b; Feng et al., 2007; Mizuno et al., 2003). We found that while this oligomerization domain did not bind microtubules, it was required for microtubule bundling (Figure 1D and 1E). Given that an intra-molecular interaction between the N- and C- terminal portions of mPar3 suppresses microtubule binding and bundling, we hypothesized that the inter-molecular interaction of mPar3 through the oligomerization domain could disrupt the intra-molecular interaction and thereby expose the microtubule binding region at the N-terminal portion. To test this possibility, we expressed EGFP-tagged mPar3(1-712) and Myc-tagged mPar3(713-1333) in COS7 cells and tested whether co-expression of FLAG-tagged mPar3(1-712) was able to compete with Myc-tagged mPar3(713-1333) in interacting with EGFP-tagged mPar3(1-712). Indeed, we found that the interaction between the N- and C-terminal portions of mPar3 was effectively competed off by the oligomerization interaction between the N-terminal portions of mPar3 (Figure 5D).
Consistent with the notion that the inter-molecular oligomerization of mPar3 is able to compete off the intra-molecular interaction, we found that the inter-molecular interaction between the N-terminal portions of mPar3 was more stable under high salt conditions than the intra-molecular interaction between the N- and C-terminal portions (Figure 5E), suggesting that the intermolecular oligomerizing interaction is stronger than the intra-molecular interaction. Furthermore, we found that increasing the level of mPar3 expression facilitated its microtubule binding and bundling activity (Figure 5F).
Microtubule regulation in polarizing neurons by mPar3
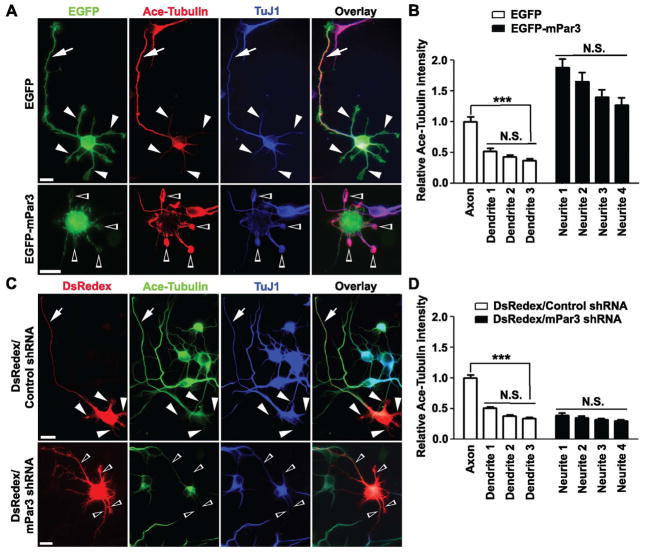
Mammalian Par3 plays critical roles in neuronal polarization (Figure S6) (Higginbotham et al., 2006; Nishimura et al., 2004; Nishimura et al., 2005; Schwamborn et al., 2007a; Shi et al., 2004; Shi et al., 2003; Vohra et al., 2007; Yi et al., 2010). Having found that mPar3 acts as a microtubule-associated protein that bundles and stabilizes microtubules, we next wanted to know whether mPar3 does indeed regulate microtubule dynamics and organization in polarizing neurons. We first examined the distribution of endogenous mPar3 in developing hippocampal neurons and found that mPar3 decorated stabilized microtubule filaments in the neurites (Figure S7). Previous studies have shown that during neuronal polarization, microtubules are preferentially stabilized in the designated future axon and this polarized microtubule stability is both necessary and sufficient to drive axon specification (Witte and Bradke, 2008; Witte et al., 2008). Consistent with this, we found that in polarizing hippocampal neurons expressing EGFP, the level of acetylated tubulin was significantly higher in the emerging axon than in the other neurites (Figure 6A, top and 6B). Interestingly, in neurons expressing EGFP-mPar3, high levels of acetylated tubulin were found in multiple neurites (Figure 6A, bottom and 6B). Consequently, neurons failed to polarize (Figure 6A, bottom), as shown previously (Shi et al., 2004; Shi et al., 2003). On the other hand, in neurons expressing mPar3 shRNA that specifically knocks down endogenous mPar3 (Bultje et al., 2009), the level of acetylated tubulin was low in all neurites, and neurons also failed to polarize (Figure 6C, bottom and 6D). Taken together, these results clearly suggest that mPar3 in developing neurons not only stabilizes microtubules, but also controls the spatial polarization of stabilized microtubules, resulting in their selective enrichment in the emerging axon. Given that mPar3 becomes enriched in the future axon, this polarization of stabilized microtubules is likely a consequence of the polarization of mPar3 proteins.
Figure 6. Microtubule regulation in polarizing neurons by mPar3.
(A) Images of neurons expressing EGFP (top) or EGFP-mPar3 (bottom) (green) stained with the antibodies against acetylated tubulin (red), a stabilized microtubule marker, and TuJ1 (blue), an immature neuron marker. Scale bars: 10 μm. (B) Quantification of the relative intensity of acetylated tubulin in different neurites compared to that in the axon of control neurons (EGFP, n=16; EGFP-mPar3, n=16). Error bars indicate s.e.m. ***, p<0.0001. N.S., not significant. (C) Images of neurons expressing DsRedex/Control shRNA (top) or DsRedex/mPar3 shRNA (bottom) (red) stained with the antibodies against acetylated tubulin (green) and TuJ1 (blue). Note that in control neurons expressing DsRedex/Control shRNA, acetylated tubulin was selectively enriched in the emerging axon (arrows), but not in the remaining neurites (arrowheads). In contrast, in neurons expressing DsRedex/mPar3 shRNA, the level of acetylated microtubules was low in all neurites (open arrowheads) and neurons failed to polarize. Scale bars: 10 μm. (D) Quantification of the relative intensity of acetylated tubulin in different neurites compared to that in the axon of control neurons (Control shRNA, n=16; mPar3 shRNA, n=16). Error bars indicate s.e.m. ***, p<0.0001; N.S., not significant. (See also Figure S6 and S7)
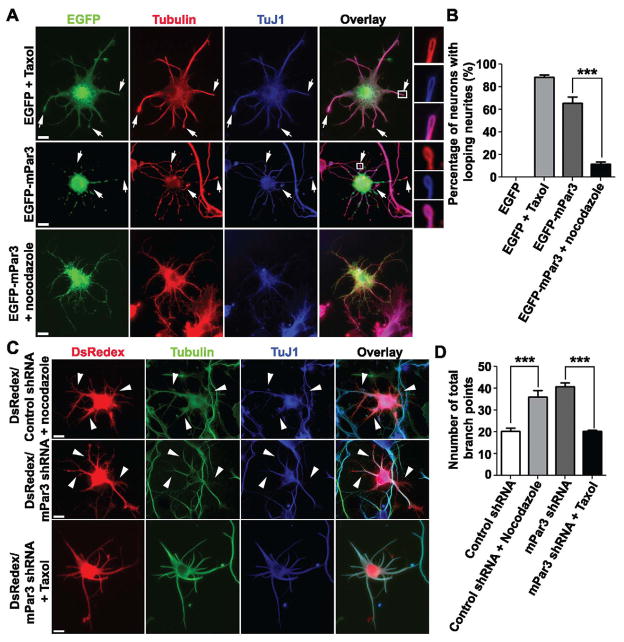
To further confirm that mPar3 regulates microtubule stability in developing neurons, we examined the functional link between overexpression or depletion of mPar3 and stabilization or destabilization of microtubules. As shown previously, treatment of neurons with 1 μM Taxol, which globally stabilizes microtubules, impaired axonal outgrowth and neuron polarization (Figure 7A, top) (Chuckowree and Vickers, 2003; Dehmelt et al., 2003; Letourneau and Ressler, 1984; Witte et al., 2008); moreover, it caused extensive microtubule looping at the tip of neurites (Figure S7A top and S7B), characteristic of excessive microtubule assembly and stabilization that rarely occurs under normal conditions. Interestingly, expression of EGFP-mPar3 resulted in similar defects in neuronal polarization and microtubule looping (Figure 7A middle and 7B). Furthermore, the microtubule looping defect caused by mPar3 over-expression was suppressed when neurons were treated with nocadozole (1.6 μM), which destabilizes microtubules (Figure 7A bottom and 7B). It was expected that global treatment with nocodazole would not rescue the polarization defect, as it inhibits axonal outgrowth and neuronal polarization (Witte et al., 2008). Notably, nocodazole treatment also smoothed out the punctate distribution of EGFP-mPar3 (Figure 7A bottom), further indicating a direct link between mPar3 and microtubules.
Figure 7. Functional link between mPar3 and microtubule stability in polarizing neurons.
(A) Images of neurons expressing EGFP (green) treated with Taxol (1 μM) (top) or EGFP-mPar3 (green) (middle) or EGFP-mPar3 (green) in the presence of nocodazole (1.6 μM) (bottom) stained with the antibodies against tubulin (red) and TuJ1 (blue). Arrows indicate looping neurites. High magnification images of looping neurites (boxed region) are shown to the right. Scale bars: 10 μm. (B) Quantification of the percentage of neurons with looping neurites (EGFP, n=13; EGFP/Taxol, n=14; EGFP-mPar3, n=16; EGFP-mPar3/nocodazole, n=14). Error bars indicate s.e.m. ***, p<0.0001. (C) Images of neurons expressing DsRedex/Control shRNA (red) treated with nocodazole (1.6 μM) (top) or DsRedex/mPar3 shRNA (red) (middle) or DsRedex/mPar3 shRNA (red) in the presence of Taxol (1 μM) (bottom) stained with the antibodies against tubulin (green) and TuJ1 (blue). Arrowheads indicate short branches. Scale bars: 10 μm. (D) Quantification of the number of total branch points in individual neurons (DsRedex/Control shRNA, n=17; DsRedex/Control shRNA/nocodazole, n=15; DsRedex/mPar3 shRNA, n=27; DsRedex/mPar3 shRNA/Taxol, n=16). Error bars indicate s.e.m. ***, p<0.0001. (See also Figure S6 and S7)
Conversely, we found that neurons expressing mPar3 shRNA exhibited defects similar to those treated with nocodazole (1.6 μM); they failed to polarize and grew numerous short protrusions (Figure 7C and D). Again, the abnormal growth of short protrusions was rescued by Taxol (1 μM) treatment (Figure 7C and D). Collectively, these results strongly suggest that mPar3 stabilizes microtubules in developing neurons.
Microtubule binding and bundling activity of mPar3 is required for neuronal polarization
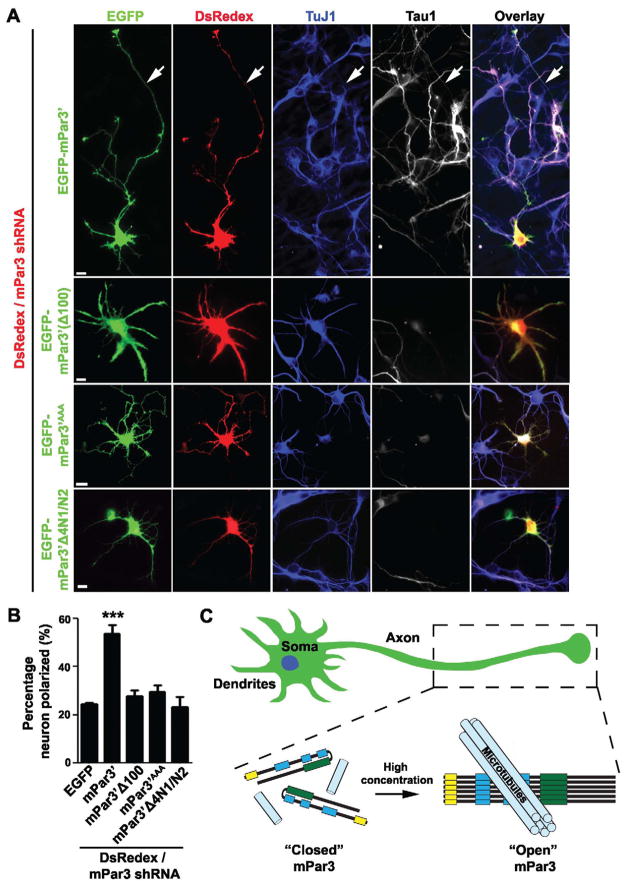
Given that mPar3 regulates the stability and spatial organization of microtubules in developing neurons, we proceeded to ask whether this microtubule regulatory activity is required for mPar3 control of axon specification and neuron polarization. Over-expression or depletion of mPar3 impairs neuron polarization (Nishimura et al., 2005; Schwamborn et al., 2007a; Shi et al., 2003), which precludes direct functional analysis of mPar3 mutants that fail to bind and/or bundle microtubules. To overcome this, we established a rescue experiment paradigm. We knocked down endogenous mPar3 using shRNA (Bultje et al., 2009) in developing hippocampal neurons and at the same time expressed either wild type or various mutant forms of mPar3 resistant to shRNA, and examined their effect on neuron polarization (Figure 8).
Figure 8. Microtubule binding and bundling activity of mPar3 is required for neuronal polarization.
(A) Images of neurons expressing DsRedex/mPar3 shRNA (red) and one of four forms of shRNA-resistant mPar3 fused with EGFP (green): wild type (EGFP-mPar3′), with a deletion of the oligomerization domain (EGFP-mPar3′Δ100), with mutations in the positively charged residue cluster (EGFP-mPar3′AAA), or with a deletion in the coiled-coil region (EGFP-mPar3′Δ4N1/N2), and stained with the antibodies against TuJ1 (blue) and Tau1 (white), an axonal marker. Arrows indicate the axon. Scale bars: 10 μm. (B) Quantification of the percentage of neurons that are polarized with a single axon (EGFP, n=8; EGFP-mPar3′, n=13; EGFP-mPar33100, n=8; EGFP-mPar3′Δ4N1/N2, n=6; EGFP-mPar3AAA, n=6). Error bars indicate s.e.m. ***, p<0.0001. (C) A model illustrating the conformation dependent regulation of microtubule bundling and stabilization by mPar3 in specifying neuronal polarity. In the closed conformation, the intra-molecular interaction between the N- and C-terminal portions suppresses mPar3 microtubule regulatory activity. As mPar3 accumulates, inter-molecular oligomerization promotes an open conformation that directly binds, bundles and stabilizes microtubules through the N-terminal portion. This microtubule regulatory activity of mPar3 is crucial for axon specification and polarization of mammalian neurons. (See also Figure S8)
We found that while expression of wild type mPar3 rescued the neuronal polarization defect caused by mPar3 shRNA, expression of mPar3 carrying the mutations in the microtubule binding domain, mPar3AAA, or a deletion of the oligomerization domain required for microtubule bundling, mPar3(Δ100), or a deletion in the coiled-coil region critical for the intra-molecular interaction at a similar level failed to rescue this defect (Figure 8A and B). These results demonstrated that the microtubule binding and bundling activity of mPar3 is required for its function in axon specification and neuronal polarization. It is worth noting that similar results were obtained in another well-characterized assay for mPar3 function – junction assembly of Madin-Darby canine kidney (MDCK) cells in a Ca2+-switch assay (Figure S8), suggesting that the microtubule regulatory activity is likely crucial for mPar3 function in other cellular contexts.
DISCUSSION
Previous studies established that microtubule stabilization can specify axon formation (Witte and Bradke, 2008; Witte et al., 2008); however, it remained largely unclear how microtubules are preferentially stabilized in the emerging axons of developing neurons. Our results provide evidences that mPar3 acts as a microtubule-associated protein that binds and bundles microtubules likely in a conformation-dependent manner. In the closed conformation, the intra-molecular interaction between the N- and C-terminal portions suppresses mPar3 microtubule regulatory activity. As mPar3 accumulates, inter-molecular oligomerization promotes an open conformation that directly binds, bundles and stabilizes microtubules (Figure 8C). Previous studies have shown that mPar3 accumulates in the emerging axon of polarizing neurons (Nishimura et al., 2004; Schwamborn et al., 2007a; Schwamborn and Puschel, 2004; Shi et al., 2004; Shi et al., 2003). Taken together, these results suggest that selective accumulation of mPar3 stabilizes microtubules preferentially in the emerging axon. It is interesting to note that polarization of mPar3 to the emerging axon relies on microtubule-based transportation (Nishimura et al., 2004; Shi et al., 2004). This raises an intriguing possibility that the selective stabilization of microtubules facilitates the further accumulation of mPar3 in the emerging axon and this positive feedback loop drives neuronal polarization.
Our finding that mPar3 is a microtubule-associated protein directly regulating microtubule stability and organization is both interesting and surprising, although upon reflection, not entirely unexpected. Since its discovery, the function of Par3 has been extensively studied (Goldstein and Macara, 2007; Kemphues, 2000). While it plays essential roles in many cellular contexts that involve microtubule regulation, such as cell polarization and spindle orientation during asymmetric cell division, Par3 is largely considered to be a scaffolding protein that localizes other proteins including effector molecules like protein kinases and small GTPases. Nonetheless, a role for Par3 in microtubule anchoring and/or stabilization had been hypothesized since initial observations of defects in spindle orientation and positioning in Par3 mutant C. elegans embryos (Cheng et al., 1995; Kemphues and Strome, 1997). Consistent with this postulation, it has been shown that microtubules are more stable at the anterior than the posterior cortex in C. elegans embryos and this gradient in microtubule stability is dependent on the anterior localization of Par3 (Labbe et al., 2003). A similar anterior to posterior microtubule gradient has been revealed in the Drosophila oocyte (Becalska and Gavis, 2010; Doerflinger et al., 2010; Parton et al., 2011). Interestingly, this microtubule organization also depends on Bazooka, the Drosophila homolog of Par3 (Becalska and Gavis, 2010; Doerflinger et al., 2010). Despite these experimental evidences, the direct link between Par3 and microtubule regulation remained obscure. Our data provides important new insights into this link. It will be interesting to examine the function of Par3 as a microtubule-associated protein that directly regulates microtubule stability and organization in different cellular contexts including spindle orientation and centrosome behavior across species (Bultje et al., 2009; Grill et al., 2001; Januschke and Gonzalez, 2010; Knoblich, 2008; Siegrist and Doe, 2005; Siller and Doe, 2009; Wang et al., 2009).
Our results showed that the PDZ domains 2 and 3 of mPar3 bind microtubules, while the N-terminal oligomerization domain facilitates their bundling and stabilization. Previous studies showed that the evolutionarily conserved oligomerization domain is critical for Par3 localization and function (Benton and St Johnston, 2003a; Benton and St Johnston, 2003b; Mizuno et al., 2003). The oligomerization domain of mPar3 has recently been shown to form a higher order helical filament that contains six units in one complete helix (Feng et al., 2007). Interestingly, in our TEM configuration analysis of microtubule bundles in the presence of the purified N-terminal portion of mPar3, we observed up to six microtubule filaments in individual microtubule bundles, raising the possibility that the helical filament structure formed by the oligomerization domains provides the wrapping force for the bundling of microtubules.
Unlike conventional microtubule-associated proteins, such as Tau, mPar3 appears to bind and bundle microtubules in a conformation-dependent manner. Both the microtubule association domain and the oligomerization domain are located within the N-terminal portion of mPar3. Interestingly, the C-terminal portion of mPar3 interacts with the N-terminal region and masks its microtubule regulatory activity. Therefore, in such a closed conformation, mPar3 is not an effective microtubule-associated protein. Remarkably, we found that this closed conformation can be switched to an open conformation when mPar3 accumulates, due to a stronger intermolecular oligomerization activity that competes off the intra-molecular interaction. This conformational switch provides a platform for dynamic and intricate regulation of microtubules by mPar3. This is particularly interesting because an increasing number of proteins have been shown to interact with and sometimes even modify Par3 (Benton and St Johnston, 2003b; Feng et al., 2008; Laprise and Tepass, 2011; Morais-de-Sa et al., 2010; Nishimura et al., 2004; Nishimura et al., 2005; Schmoranzer et al., 2009; von Stein et al., 2005; Wu et al., 2007). We expect that these interactions and modifications can influence the conformational change in mPar3 and thus its microtubule regulatory activity. For example, phosphorylation of the conserved serine residue at position 151 (S151) in Drosophila Bazooka by Par1 recruits 14-3-3 and interferes with Bazooka oligomerization (Benton and St Johnston, 2003b), which is critical for microtubule bundling and stabilization. Future efforts to dissect the precise regulation of the conformational switch in mPar3 will likely provide crucial insights into mPar3 function.
As a regulated microtubule-associated protein, mPar3 is ideal for providing highly localized regulation of microtubule stability, which is central to neuron polarization (Stiess and Bradke, 2011; Witte and Bradke, 2008; Witte et al., 2008). Selective stabilization of microtubules in one and only one of the neurites in a polarizing neuron is both necessary and sufficient for axon specification. Our results showed that mPar3 functions as a key microtubule-associated protein that stabilizes microtubules selectively in the emerging axon and drives neuronal polarization. It has been shown that mPar3 accumulates in the neurite that becomes the axon (Nishimura et al., 2004; Schwamborn and Puschel, 2004; Shi et al., 2004; Shi et al., 2003). It is conceivable that once it accumulates in the emerging axon, mPar3 switches to an open conformation and actively stabilizes microtubules, which may facilitate further transportation of mPar3 to the emerging axon. While our results suggest that an increase in the concentration of mPar3 triggers the conformation switch, it is likely that the conformation switch is also regulated by additional signaling events, especially in vivo (Arimura and Kaibuchi, 2007; Barnes and Polleux, 2009; Insolera et al., 2011). It is worth noting that microtubule elongation in axons is promoted by CRMP2, which binds to tubulin dimers and transports them to the growing plus-end (Fukata et al., 2002; Inagaki et al., 2001). Therefore, multiple microtubule regulatory proteins need to function in concert to control neuron polarization.
In summary, we have demonstrated direct regulation of microtubule stability and organization by mPar3, which is essential for axon specification and neuron polarization. The strong conservation of both expression of Par3 and its tight link to microtubule regulation indicates that this activity of Par3 as a conformation-dependent microtubule-associated protein is likely fundamental to its function in many cellular contexts across species.
EXPERIMENTAL PROCEDURES
Cell culture, transfection and immunofluorescence microscopy
COS7 cells were plated at a density of 1×106 cells in either 100 mm plates for biochemistry experiments or 2×104 cells on 12 mm glass coverslips for immunofluorescence analysis, and grown overnight to ~70% confluence for transfection using FuGene 6 (Roche) according to the manufacturer’s instructions. Mouse hippocampal dissociated neurons were prepared as described previously (Shi et al., 2003) and transfected 2-3 hours after plating using Lipofectamine 2000 (Invitrogen) according to the manufacturer’s instructions. All procedures for animal handling and usage were approved by the institutional research animal resource center (RARC). Neuronal polarity was analyzed as described previously (Shi et al., 2003).
For immunofluorescence analysis, cells were fixed with 4% paraformaldehyde/4% sucrose in phosphate-buffered saline (PBS) at 4°C for 30 minutes, washed three times with PBS, and permeabilized by incubation in PBS with 0.3% Triton X-100 for 10 minutes. After blocking in PBS with 10% goat serum and 0.3% Triton X-100 for 1 hour at room temperature, cells were incubated with the primary antibodies at 4°C overnight, followed by incubation with the secondary antibodies at room temperature for 2 hours. Coverslips were mounted onto glass slides with anti-fade mounting medium (Vector Laboratories) and visualized with an inverted microscope equipped with epifluorescence illumination and a cooled CCD camera (Axio Observer, Zeiss). Images were taken with AxioVision (Zeiss) and analyzed with AxioVision (Zeiss), MetaMorph (Molecular Devices) and Adobe Photoshop (Adobe). Data were expressed as mean±s.e.m. and statistical differences were determined using nonparametric Mann-Whitney-Wilcoxon and Kruskal-Wallis tests.
Immunoprecipitation and microtubule co-sedimentation assays
At 48 hours post-transfection, cells were harvested and homogenized in a lysis buffer containing 50 mM Tris (pH 7.5), 150 mM NaCl, 10% glycerol, 1% Nonidet P-40, 2 mM EDTA, 5 mM NaVO4 and 1X protease inhibitor cocktail (Roche). The homogenate was incubated on ice for 15 minutes before centrifugation at 12,000 g for 10 minutes at 4°C. Cell lysates were immunoprecipitated with the anti-GFP mouse monoclonal antibody (Roche) followed by adsorption to protein A/G plus-Agarose beads (Santa Cruz). After three final washes with the lysis buffer, proteins were analyzed by sodium dodecyl sulfate polyacrylamide gel electrophoresis(SDS-PAGE) and immunoblotted with the antibodies against EGFP, Myc, FLAG (Sigma, 1:2000), β-actin (Sigma, 1:2000), and HA (Roche, 1:2000). The microtubule co-sedimentation assay was performed as described previously (Hergovich et al., 2003).
FRET analysis
At 48 hours post-transfection, COS7 cells grown on glass coverslips were mounted for acquiring FRET images using confocal laser scanning microscopy (SP2, Leica). To photobleach YFP, the cells were illuminated until 20% of the original fluorescence intensity was detected. Under these conditions, direct bleaching of CFP was kept to a minimum. The increase in CFP emission under CFP excitation after photobleaching YFP indicated FRET. The FRET efficiency, FRETeff (%), of individual cells was measured as (CFPpost-CFPpre) / CFPpost.
In vitro microtubule binding and bundling assay
MBP, MBP-mPar3(1-712) and MBP-mPar3(1-712AAA) recombinant proteins with or without C-terminal fusion of mCherry were expressed in E. coli and affinity purified. Polymerized microtubules were prepared as described above. After the incubation of purified MBP, MBP-mPar3(1-712) or MBP-mPar3(1-712AAA) proteins with in vitro assembled microtubules, amylose beads were used to absorb MBP or MBP fusion proteins. Bound proteins were separated by SDS-PAGE and immunoblotted using the antibodies against MBP and tubulin.
For the microtubule bundling assay, HiLyte Fluor 488 labeled tubulin (20 μg, Cytoskeleton Inc.) was resuspended in 5 μL G-PEM buffer (pH 6.9) containing 100 mM PIPES, 1 mM MgCl2, 1 mM EGTA and 1mM GTP, and incubated at 37°C for 20 minutes. About 0.7 μL Taxol (200 μM, Invitrogen) in DMSO was then added. After incubation at 37°C for 5 min, the solution was wrapped in foil as polymerized microtubule stock solution. One μL polymerized microtubule stock solution was added to 200 μL G-PEM buffer with 20 μM Taxol to reconstitute polymerized microtubule working solution. After immediately mixing 5 μL microtubule working solution with an equal volume of 0.5 μM recombinant protein solution on ice, samples were transferred to a glass-bottomed dish and covered with a cover glass. Images were taken using a pre-warmed inverted microscope (Axio Observer, Zeiss) equipped with epifluorescence illumination and a cooled CCD camera (ORCA, Hamamatsu).
Transmission Electron Microscopy (TEM)
Recombinant mPar3(1-712) or mPar3(1-712AAA) proteins expressed in E. coli were affinity purified and mixed with polymerized microtubules prepared in vitro as described above. The sample was deposited onto 200 square mesh copper grids with carbon film coating (Electron Microscopy Sciences) for 10 minutes prior to staining with 2% uranyl acetate. The TEM images were obtained at a magnification of 10,000× and 50,000× using the JOEL JEM 100cx transmission electron microscope.
Supplementary Material
HIGHLIGHTS.
The N-terminal portion of mPar3 directly binds, bundles and stabilizes MTs.
The intra-molecular interaction of mPar3 suppresses the MT regulatory activity.
Mammalian Par3 regulates MT organization in developing hippocampal neurons.
The MT regulatory activity of mPar3 is required for neuronal polarization.
Acknowledgments
We thank Dr. Alan Hall, Dr. Alexandra L. Joyner, and Kirsten M. Hively for comments on the manuscript, Drs. Yuh-Nung Jan and Lily Y. Jan for their support during the initial phase of this project, and the members of our laboratories for valuable input. We thank Dr. Kunihiro Uryu and the Electron Microscopy Resource Center of the Rockefeller University, and Dr. Sho Fujisawa and the Molecular Cytology Core Facility of Memorial Sloan-Kettering Cancer Center for technical support. This work was supported by grants from the National Institutes of Health (R01DA024681 and PO1NS048120), the McKnight Foundation, and the March of Dimes Foundation (to S.-H.S), and grants from RGC of Hong Kong to M.Z. (663610, 663811, HKUST6/CRF/10, RGC T13-607/12R and AoE/M-04/04). M.Z. is a Senior Fellow of IAS at HKUST.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Arimura N, Kaibuchi K. Neuronal polarity: from extracellular signals to intracellular mechanisms. Nat Rev Neurosci. 2007;8:194–205. doi: 10.1038/nrn2056. [DOI] [PubMed] [Google Scholar]
- Barnes AP, Polleux F. Establishment of axon-dendrite polarity in developing neurons. Annu Rev Neurosci. 2009;32:347–381. doi: 10.1146/annurev.neuro.31.060407.125536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Becalska AN, Gavis ER. Bazooka regulates microtubule organization and spatial restriction of germ plasm assembly in the Drosophila oocyte. Dev Biol. 2010;340:528–538. doi: 10.1016/j.ydbio.2010.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benton R, St Johnston D. A conserved oligomerization domain in drosophila Bazooka/PAR-3 is important for apical localization and epithelial polarity. Curr Biol. 2003a;13:1330–1334. doi: 10.1016/s0960-9822(03)00508-6. [DOI] [PubMed] [Google Scholar]
- Benton R, St Johnston D. Drosophila PAR-1 and 14-3-3 inhibit Bazooka/PAR-3 to establish complementary cortical domains in polarized cells. Cell. 2003b;115:691–704. doi: 10.1016/s0092-8674(03)00938-3. [DOI] [PubMed] [Google Scholar]
- Bradke F, Dotti CG. The role of local actin instability in axon formation. Science. 1999;283:1931–1934. doi: 10.1126/science.283.5409.1931. [DOI] [PubMed] [Google Scholar]
- Bradke F, Dotti CG. Establishment of neuronal polarity: lessons from cultured hippocampal neurons. Curr Opin Neurobiol. 2000;10:574–581. doi: 10.1016/s0959-4388(00)00124-0. [DOI] [PubMed] [Google Scholar]
- Bulinski JC, Richards JE, Piperno G. Posttranslational modifications of alpha tubulin: detyrosination and acetylation differentiate populations of interphase microtubules in cultured cells. J Cell Biol. 1988;106:1213–1220. doi: 10.1083/jcb.106.4.1213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bultje RS, Castaneda-Castellanos DR, Jan LY, Jan YN, Kriegstein AR, Shi SH. Mammalian Par3 regulates progenitor cell asymmetric division via notch signaling in the developing neocortex. Neuron. 2009;63:189–202. doi: 10.1016/j.neuron.2009.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng NN, Kirby CM, Kemphues KJ. Control of cleavage spindle orientation in Caenorhabditis elegans: the role of the genes par-2 and par-3. Genetics. 1995;139:549–559. doi: 10.1093/genetics/139.2.549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chuckowree JA, Vickers JC. Cytoskeletal and morphological alterations underlying axonal sprouting after localized transection of cortical neuron axons in vitro. J Neurosci. 2003;23:3715–3725. doi: 10.1523/JNEUROSCI.23-09-03715.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Craig AM, Banker G. Neuronal polarity. Annu Rev Neurosci. 1994;17:267–310. doi: 10.1146/annurev.ne.17.030194.001411. [DOI] [PubMed] [Google Scholar]
- Dehmelt L, Smart FM, Ozer RS, Halpain S. The role of microtubule-associated protein 2c in the reorganization of microtubules and lamellipodia during neurite initiation. J Neurosci. 2003;23:9479–9490. doi: 10.1523/JNEUROSCI.23-29-09479.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doerflinger H, Vogt N, Torres IL, Mirouse V, Koch I, Nusslein-Volhard C, St Johnston D. Bazooka is required for polarisation of the Drosophila anterior-posterior axis. Development. 2010;137:1765–1773. doi: 10.1242/dev.045807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dotti CG, Sullivan CA, Banker GA. The establishment of polarity by hippocampal neurons in culture. J Neurosci. 1988;8:1454–1468. doi: 10.1523/JNEUROSCI.08-04-01454.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng W, Wu H, Chan LN, Zhang M. The Par-3 NTD adopts a PB1-like structure required for Par-3 oligomerization and membrane localization. Embo J. 2007;26:2786–2796. doi: 10.1038/sj.emboj.7601702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng W, Wu H, Chan LN, Zhang M. Par-3-mediated junctional localization of the lipid phosphatase PTEN is required for cell polarity establishment. J Biol Chem. 2008;283:23440–23449. doi: 10.1074/jbc.M802482200. [DOI] [PubMed] [Google Scholar]
- Forscher P, Smith SJ. Actions of cytochalasins on the organization of actin filaments and microtubules in a neuronal growth cone. J Cell Biol. 1988;107:1505–1516. doi: 10.1083/jcb.107.4.1505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukata Y, Itoh TJ, Kimura T, Menager C, Nishimura T, Shiromizu T, Watanabe H, Inagaki N, Iwamatsu A, Hotani H, Kaibuchi K. CRMP-2 binds to tubulin heterodimers to promote microtubule assembly. Nat Cell Biol. 2002;4:583–591. doi: 10.1038/ncb825. [DOI] [PubMed] [Google Scholar]
- Goldstein B, Macara IG. The PAR proteins: fundamental players in animal cell polarization. Dev Cell. 2007;13:609–622. doi: 10.1016/j.devcel.2007.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grill SW, Gonczy P, Stelzer EH, Hyman AA. Polarity controls forces governing asymmetric spindle positioning in the Caenorhabditis elegans embryo. Nature. 2001;409:630–633. doi: 10.1038/35054572. [DOI] [PubMed] [Google Scholar]
- Hall A. Rho GTPases and the actin cytoskeleton. Science. 1998;279:509–514. doi: 10.1126/science.279.5350.509. [DOI] [PubMed] [Google Scholar]
- Hergovich A, Lisztwan J, Barry R, Ballschmieter P, Krek W. Regulation of microtubule stability by the von Hippel-Lindau tumour suppressor protein pVHL. Nat Cell Biol. 2003;5:64–70. doi: 10.1038/ncb899. [DOI] [PubMed] [Google Scholar]
- Higginbotham H, Tanaka T, Brinkman BC, Gleeson JG. GSK3beta and PKCzeta function in centrosome localization and process stabilization during Slit-mediated neuronal repolarization. Mol Cell Neurosci. 2006;32:118–132. doi: 10.1016/j.mcn.2006.03.003. [DOI] [PubMed] [Google Scholar]
- Inagaki N, Chihara K, Arimura N, Menager C, Kawano Y, Matsuo N, Nishimura T, Amano M, Kaibuchi K. CRMP-2 induces axons in cultured hippocampal neurons. Nat Neurosci. 2001;4:781–782. doi: 10.1038/90476. [DOI] [PubMed] [Google Scholar]
- Insolera R, Chen S, Shi SH. Par proteins and neuronal polarity. Dev Neurobiol. 2011;71:483–494. doi: 10.1002/dneu.20867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Januschke J, Gonzalez C. The interphase microtubule aster is a determinant of asymmetric division orientation in Drosophila neuroblasts. J Cell Biol. 2010;188:693–706. doi: 10.1083/jcb.200905024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joberty G, Petersen C, Gao L, Macara IG. The cell-polarity protein Par6 links Par3 and atypical protein kinase C to Cdc42. Nat Cell Biol. 2000;2:531–539. doi: 10.1038/35019573. [DOI] [PubMed] [Google Scholar]
- Kemphues K. PARsing embryonic polarity. Cell. 2000;101:345–348. doi: 10.1016/s0092-8674(00)80844-2. [DOI] [PubMed] [Google Scholar]
- Kemphues KJ, Strome S. Fertilization and Establishment of Polarity in the Embryo. In: Riddle DL, et al., editors. C. elegans II. 2. Chapter 13. Cold Spring Harbor Laboratory Press; 1997. [PubMed] [Google Scholar]
- Knoblich JA. Mechanisms of asymmetric stem cell division. Cell. 2008;132:583–597. doi: 10.1016/j.cell.2008.02.007. [DOI] [PubMed] [Google Scholar]
- Kunda P, Paglini G, Quiroga S, Kosik K, Caceres A. Evidence for the involvement of Tiam1 in axon formation. J Neurosci. 2001;21:2361–2372. doi: 10.1523/JNEUROSCI.21-07-02361.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Labbe JC, Maddox PS, Salmon ED, Goldstein B. PAR proteins regulate microtubule dynamics at the cell cortex in C. elegans. Curr Biol. 2003;13:707–714. doi: 10.1016/s0960-9822(03)00251-3. [DOI] [PubMed] [Google Scholar]
- Laprise P, Tepass U. Novel insights into epithelial polarity proteins in Drosophila. Trends Cell Biol. 2011;21:401–408. doi: 10.1016/j.tcb.2011.03.005. [DOI] [PubMed] [Google Scholar]
- Letourneau PC, Ressler AH. Inhibition of neurite initiation and growth by taxol. J Cell Biol. 1984;98:1355–1362. doi: 10.1083/jcb.98.4.1355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Littauer UZ, Giveon D, Thierauf M, Ginzburg I, Ponstingl H. Common and distinct tubulin binding sites for microtubule-associated proteins. Proc Natl Acad Sci U S A. 1986;83:7162–7166. doi: 10.1073/pnas.83.19.7162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizuno K, Suzuki A, Hirose T, Kitamura K, Kutsuzawa K, Futaki M, Amano Y, Ohno S. Self-association of PAR-3-mediated by the conserved N-terminal domain contributes to the development of epithelial tight junctions. J Biol Chem. 2003;278:31240–31250. doi: 10.1074/jbc.M303593200. [DOI] [PubMed] [Google Scholar]
- Morais-de-Sa E, Mirouse V, St Johnston D. aPKC phosphorylation of Bazooka defines the apical/lateral border in Drosophila epithelial cells. Cell. 2010;141:509–523. doi: 10.1016/j.cell.2010.02.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishimura T, Kato K, Yamaguchi T, Fukata Y, Ohno S, Kaibuchi K. Role of the PAR-3-KIF3 complex in the establishment of neuronal polarity. Nat Cell Biol. 2004;6:328–334. doi: 10.1038/ncb1118. [DOI] [PubMed] [Google Scholar]
- Nishimura T, Yamaguchi T, Kato K, Yoshizawa M, Nabeshima Y, Ohno S, Hoshino M, Kaibuchi K. PAR-6-PAR-3 mediates Cdc42-induced Rac activation through the Rac GEFs STEF/Tiam1. Nat Cell Biol. 2005;7:270–277. doi: 10.1038/ncb1227. [DOI] [PubMed] [Google Scholar]
- Parton RM, Hamilton RS, Ball G, Yang L, Cullen CF, Lu W, Ohkura H, Davis I. A PAR-1-dependent orientation gradient of dynamic microtubules directs posterior cargo transport in the Drosophila oocyte. J Cell Biol. 2011;194:121–135. doi: 10.1083/jcb.201103160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paschal BM, Obar RA, Vallee RB. Interaction of brain cytoplasmic dynein and MAP2 with a common sequence at the C terminus of tubulin. Nature. 1989;342:569–572. doi: 10.1038/342569a0. [DOI] [PubMed] [Google Scholar]
- Piperno G, LeDizet M, Chang XJ. Microtubules containing acetylated alpha-tubulin in mammalian cells in culture. J Cell Biol. 1987;104:289–302. doi: 10.1083/jcb.104.2.289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmoranzer J, Fawcett JP, Segura M, Tan S, Vallee RB, Pawson T, Gundersen GG. Par3 and dynein associate to regulate local microtubule dynamics and centrosome orientation during migration. Curr Biol. 2009;19:1065–1074. doi: 10.1016/j.cub.2009.05.065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schulze E, Asai DJ, Bulinski JC, Kirschner M. Posttranslational modification and microtubule stability. J Cell Biol. 1987;105:2167–2177. doi: 10.1083/jcb.105.5.2167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schulze E, Kirschner M. Dynamic and stable populations of microtubules in cells. J Cell Biol. 1987;104:277–288. doi: 10.1083/jcb.104.2.277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwamborn JC, Khazaei MR, Puschel AW. The interaction of mPar3 with the ubiquitin ligase Smurf2 is required for the establishment of neuronal polarity. J Biol Chem. 2007a;282:35259–35268. doi: 10.1074/jbc.M703438200. [DOI] [PubMed] [Google Scholar]
- Schwamborn JC, Muller M, Becker AH, Puschel AW. Ubiquitination of the GTPase Rap1B by the ubiquitin ligase Smurf2 is required for the establishment of neuronal polarity. EMBO J. 2007b;26:1410–1422. doi: 10.1038/sj.emboj.7601580. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Schwamborn JC, Puschel AW. The sequential activity of the GTPases Rap1B and Cdc42 determines neuronal polarity. Nat Neurosci. 2004;7:923–929. doi: 10.1038/nn1295. [DOI] [PubMed] [Google Scholar]
- Serrano L, de la Torre J, Maccioni RB, Avila J. Involvement of the carboxyl-terminal domain of tubulin in the regulation of its assembly. Proc Natl Acad Sci U S A. 1984;81:5989–5993. doi: 10.1073/pnas.81.19.5989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi SH, Cheng T, Jan LY, Jan YN. APC and GSK-3beta are involved in mPar3 targeting to the nascent axon and establishment of neuronal polarity. Curr Biol. 2004;14:2025–2032. doi: 10.1016/j.cub.2004.11.009. [DOI] [PubMed] [Google Scholar]
- Shi SH, Jan LY, Jan YN. Hippocampal neuronal polarity specified by spatially localized mPar3/mPar6 and PI 3-kinase activity. Cell. 2003;112:63–75. doi: 10.1016/s0092-8674(02)01249-7. [DOI] [PubMed] [Google Scholar]
- Siegrist SE, Doe CQ. Microtubule-induced Pins/Galphai cortical polarity in Drosophila neuroblasts. Cell. 2005;123:1323–1335. doi: 10.1016/j.cell.2005.09.043. [DOI] [PubMed] [Google Scholar]
- Siller KH, Doe CQ. Spindle orientation during asymmetric cell division. Nat Cell Biol. 2009;11:365–374. doi: 10.1038/ncb0409-365. [DOI] [PubMed] [Google Scholar]
- Stiess M, Bradke F. Neuronal polarization: the cytoskeleton leads the way. Dev Neurobiol. 2011;71:430–444. doi: 10.1002/dneu.20849. [DOI] [PubMed] [Google Scholar]
- Takemura R, Okabe S, Umeyama T, Kanai Y, Cowan NJ, Hirokawa N. Increased microtubule stability and alpha tubulin acetylation in cells transfected with microtubule-associated proteins MAP1B, MAP2 or tau. J Cell Sci. 1992;103(Pt 4):953–964. doi: 10.1242/jcs.103.4.953. [DOI] [PubMed] [Google Scholar]
- Vohra BP, Fu M, Heuckeroth RO. Protein kinase Czeta and glycogen synthase kinase-3beta control neuronal polarity in developing rodent enteric neurons, whereas SMAD specific E3 ubiquitin protein ligase 1 promotes neurite growth but does not influence polarity. J Neurosci. 2007;27:9458–9468. doi: 10.1523/JNEUROSCI.0870-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Stein W, Ramrath A, Grimm A, Muller-Borg M, Wodarz A. Direct association of Bazooka/PAR-3 with the lipid phosphatase PTEN reveals a link between the PAR/aPKC complex and phosphoinositide signaling. Development. 2005;132:1675–1686. doi: 10.1242/dev.01720. [DOI] [PubMed] [Google Scholar]
- Wang X, Tsai JW, Imai JH, Lian WN, Vallee RB, Shi SH. Asymmetric centrosome inheritance maintains neural progenitors in the neocortex. Nature. 2009;461:947–955. doi: 10.1038/nature08435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westermann S, Weber K. Post-translational modifications regulate microtubule function. Nat Rev Mol Cell Biol. 2003;4:938–947. doi: 10.1038/nrm1260. [DOI] [PubMed] [Google Scholar]
- Witte H, Bradke F. The role of the cytoskeleton during neuronal polarization. Curr Opin Neurobiol. 2008;18:479–487. doi: 10.1016/j.conb.2008.09.019. [DOI] [PubMed] [Google Scholar]
- Witte H, Neukirchen D, Bradke F. Microtubule stabilization specifies initial neuronal polarization. J Cell Biol. 2008;180:619–632. doi: 10.1083/jcb.200707042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu H, Feng W, Chen J, Chan LN, Huang S, Zhang M. PDZ domains of Par-3 as potential phosphoinositide signaling integrators. Mol Cell. 2007;28:886–898. doi: 10.1016/j.molcel.2007.10.028. [DOI] [PubMed] [Google Scholar]
- Yi JJ, Barnes AP, Hand R, Polleux F, Ehlers MD. TGF-beta signaling specifies axons during brain development. Cell. 2010;142:144–157. doi: 10.1016/j.cell.2010.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H, Macara IG. The polarity protein PAR-3 and TIAM1 cooperate in dendritic spine morphogenesis. Nat Cell Biol. 2006;8:227–237. doi: 10.1038/ncb1368. [DOI] [PubMed] [Google Scholar]
- Zhang H, Macara IG. The PAR-6 polarity protein regulates dendritic spine morphogenesis through p190 RhoGAP and the Rho GTPase. Dev Cell. 2008;14:216–226. doi: 10.1016/j.devcel.2007.11.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.