Abstract
Werner syndrome is a premature aging disorder that is caused by defects in the Werner protein (WRN). WRN is a member of the RecQ helicase family and possesses helicase and exonuclease activities. It is involved in various aspects of DNA metabolism such as DNA repair, telomere maintenance, and replication. Poly(ADP-ribose) polymerase 1 (PARP1) is also involved in these processes by catalyzing the formation of the nucleic-acid-like biopolymer poly(ADP-ribose) (PAR). It was previously shown that WRN interacts with PARP1 and that WRN activity is inhibited by PARP1. Using several bioanalytical approaches, here we demonstrate that the enzymatic product of PARP1, i.e., PAR, directly interacts with WRN physically and functionally. First, WRN binds HPLC-size-fractionated short and long PAR in a non-covalent manner. Second, we identified and characterized a PAR-binding motif (PBM) within the WRN sequence and showed that several basic and hydrophobic aminoacids are of critical importance for mediating the PAR binding. Third, PAR-binding inhibits the DNA-binding, the helicase and the exonuclease activities of WRN in a concentration-dependent manner. Based on our results we propose that the transient nature of PAR produced by living cells would provide a versatile and swiftly reacting control system for WRN’s function. More generally, our work underscores the important role of non-covalent PAR-protein interactions as a regulatory mechanism of protein function.
INTRODUCTION
Poly(ADP-ribosyl)ation (PARylation) is a post-translational modification catalyzed by poly(ADP-ribose) polymerases (PARPs). PARP1 accounts for the bulk of the cellular PARylation capacity and is involved in various cellular processes, such as DNA repair, telomere regulation, transcription, and the regulation of cell death (1, 2). It is catalytically activated by DNA strand breaks and forms linear and branched chains of poly(ADP-ribose) (PAR) consisting of up to 200 ADP-ribose units, which are covalently bound to acceptor proteins, such as PARP1 itself (automodification) and histones (Figure 1) (1, 2). Poly(ADP-ribose) glycohydrolase (PARG), on the other hand, hydrolyzes PAR both in an endo- and exoglycosidic manner, which leads to rapid turnover of PAR in the cell and to the formation of free PAR. In addition to covalent modification, free PAR can bind to proteins in a non-covalent manner, which may alter their functions or subcellular localization. Although PAR and DNA are structurally related, non-covalent PAR-protein interactions are not mediated via common DNA binding domains, since general DNA binding enzymes such as polymerases, ligases, and DNase1 do not bind PAR (3, 4). Instead the PAR-protein interaction is mediated by at least three specific PAR binding motifs: (i) distinct macrodomains, (ii) a PAR-binding zinc finger motif, and (iii) a weakly conserved ~20 aminoacid (aa) PAR binding motif (PBM) (5–9). Whereas the first two binding motifs are present in a limited number of human proteins (<50), the 20 aa PBM has been identified in several hundred human protein sequences (6, 9). This motif consists of (i) a cluster rich in basic aa and (ii) a pattern of hydrophobic aa interspersed with basic residues (6, 9). Most of the PAR interaction partners identified so far are involved in genomic maintenance and cell-cycle control. For example, the recruitment of the base-excision repair (BER) protein XRCC1 to sites of DNA damage is completely dependent on efficient PAR formation (10). Moreover, the binding of PAR to the DEK oncoprotein promotes the formation of DEK multimers with potential impact on gene transcription and maintenance of genomic stability (11). Also, the ability of p53 to bind to DNA is decreased upon non-covalent interaction with PAR, suggesting a PAR-dependent regulation of transactivation functions of p53 (12).
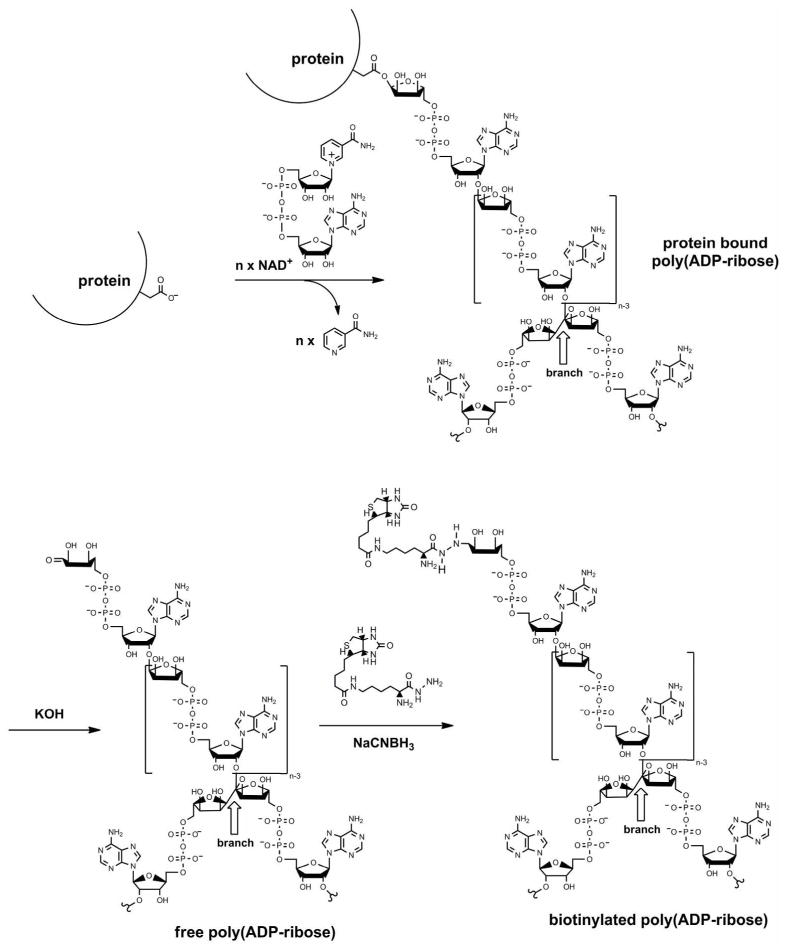
Figure 1. In vitro production and biotinylation of poly(ADP-ribose).
PARPs cleave the glycosidic bond of NAD+ between nicotinamide and ribose followed by the covalent modification of acceptor proteins with an ADP-ribosyl unit. PARPs also catalyze adduct elongation, giving rise to linear polymers with chain lengths of up to 200 ADP-ribosyl units, characterized by their unique ribose (1″→ 2′) ribose phosphate-phosphate backbone. Some of the PARP family members also catalyze a branching reaction by creating ribose (1‴→2″) ribose linkages. In vitro-synthesized poly(ADP-ribose) (PAR) was detached from proteins by KOH treatment, purified by phenol-chloroform extraction, and for some experiments HPLC-size-fractionated and labeled with biotin via a carbonyl-reactive biotin analog (biocytin hydrazide) for detection purposes.
Another factor involved in many aspects of DNA metabolism such as replication, DNA repair, and telomere maintenance is the Werner syndrome protein (WRN) (13). In the hereditary disease Werner syndrome (WS), the WRN gene is mutated and dysfunctional. Patients show premature aging starting after puberty, with age-associated characteristics such as osteoporosis, atherosclerosis and high cancer incidence, which is paralleled and probably caused by high susceptibility to genotoxic stress at a cellular level (14). WRN is a RecQ helicase with additional exonuclease and ATPase activities (13). WRN binds to various DNA substrates such as forked structures, DNA/RNA duplexes, and repair intermediates including Holliday junctions, bubbles, recessed ends, and telomeric substrates (15). Both WRN’s helicase and exonuclease activities work in 3′→5′ direction and can act independently of each other (16, 17). Enzymatic properties of WRN can be altered by post-translational modifications such as acetylation, which regulates WRN function in BER (18), and phosphorylation, which inhibits WRN’s enzymatic activities (19). A potential covalent PARylation of WRN by PARP1 has been reported (20). Furthermore, a physical and functional interaction between PARP1 and WRN has been discovered. First, cells derived from WS patients carrying a mutant form of WRN are severely deficient in their PARylation activity under conditions of genotoxic stress. Furthermore, WRN directly interacts with PARP1 physically in human cells (21, 22). The WRN-PARP1 interaction is mediated via three WRN regions, i.e., the N-terminus, the helicase domain and a C-terminal region containing the RecQ-conserved (RQC) domain; and via two PARP1 regions, i.e., the DNA-binding and BRCT domain (23). Interestingly, the WRN-PARP1 interaction seems to be dependent on the PARylation state of PARP1, because PARylated PARP1 binds WRN less efficiently than unmodified PARP1 (21). Also dependent on its PARylation state, PARP1 inhibits both exonuclease and helicase activities of WRN (23). Thus, unmodified PARP1 has a stronger inhibitory effect on WRN activity than PARylated PARP1 (23). In vivo, Parp1−/−/WrnΔhel/Δhel mice develop cancer at higher incidence and exhibit a shorter lifespan than corresponding single knock-outs. Furthermore, cells derived from those mice display major signs of genomic instability (24) as well as a pronounced dysregulation of gene transcription, including genes involved in apoptosis, cell cycle control and metabolism (25).
The present work highlights the role of non-covalent PAR-protein interactions as a regulatory mechanism of protein function. WRN is considered an important factor in genome maintenance, with implications in aging and cancer biology (13). The cellular functions of WRN and PARP1 are highly overlapping and the interplay between both factors leads to an inhibition of WRN’s catalytic activities (23). Here we are using a series of bioanalytical approaches, e.g., HPLC size-fractionation of PAR, PAR-protein interaction assays with biotin end-labeled PAR, and quantitative isotope dilution mass spectrometry to reveal that WRN directly and specifically interacts with free PAR in a non-covalent manner and that this interaction regulates WRN activity. We demonstrate that the PAR-protein interaction is mediated via at least one PBM located in WRN’s exonuclease domain. In addition, binding of PAR to WRN decreases WRN’s ability to bind to a physiologically relevant DNA substrate and also inhibits its exonuclease and helicase activities. We propose that the production of PAR provides a versatile, swiftly reacting and highly efficient system to control WRN’s function in a spatio-temporal manner.
RESULTS AND DISCUSSION
PAR binds to WRN in a non-covalent manner
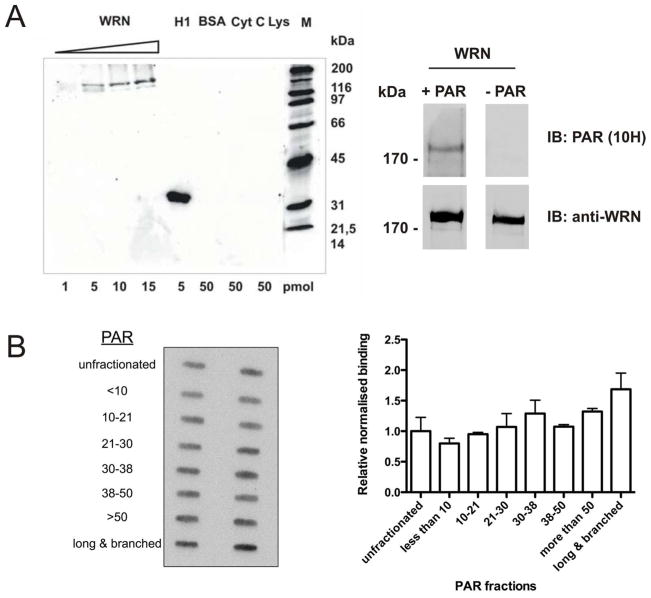
A direct physical and functional interaction of WRN with PARP1 was reported previously (21, 23). To test the hypothesis that WRN is capable of binding the enzymatic product of PARP1, i.e., PAR, we performed interaction studies using purified recombinant WRN. Increasing amounts of WRN were separated by SDS-PAGE, immobilized on a nitrocellulose membrane and incubated with in-vitro-synthesized, purified PAR (see Figure 1 for synthesis). After high-stringency salt washes, protein-bound PAR was detected using the PAR-specific antibody 10H (Figure 2A). As expected, no detectable levels of PAR bound to negative controls, such as bovine serum albumin, cytochrome C or lysozyme, whereas strong binding was observed to histone H1, which served as a positive control. Importantly, a PAR-specific signal was associated with WRN demonstrating that the latter is capable of specific non-covalent interaction with free PAR (Figure 2A).
Figure 2. PAR binds to WRN in a non-covalent manner.
A. PAR-overlay blots demonstrating WRN-PAR interaction. Proteins were separated by SDS-PAGE, immobilized on a nitrocellulose membrane and incubated with 0.2 μM (referring to ADP-ribosyl units) of unfractionated PAR. Protein-bound PAR was detected using the anti-PAR monoclonal antibody 10H. Left panel. Histone H1 served as a positive control; bovine serum albumin (BSA), cytochrome C (Cyt C), lysozyme (Lys) served as negative controls. Right panel. Specificity control of the 10H antibody. 5 pmol of WRN were loaded per lane. After blotting, the membrane was cut and incubated with or without 0.2 μM PAR (+/− PAR), respectively. Apart from this, both membranes were processed identically. After probing with the 10H antibody, membranes were stripped and incubated with an anti-WRN antibody as loading control. WRN-PAR binding was only observed for the PAR-incubated membrane, whereas no signal was detected for the control membrane. B. Left panel. PAR overlay slot-blot evaluating the dependency of WRN-PAR binding on PAR chain length. WRN (15 pmol/slot) was immobilized on a nitrocellulose membrane (in duplicates) and incubated with 0.2 μM PAR of chain length as indicated. Right panel. Densitometric quantification of PAR-overlay slot-blot, normalized to signals obtained from binding of unfractionated PAR.
The finding that WRN is potentially covalently modified with PAR on the one hand (20) and that it interacts with PAR in a non-covalent fashion on the other hand fits with data on other proteins such as histone H1 and DEK, where dual regulation by PAR has been shown (11, 26–28). This highlights the complex regulation of proteins by PAR on multiple levels.
As we have shown previously, non-covalent binding of PAR to some proteins, e.g., DEK and XPA, depends on the chain length of the polymer (29). We therefore investigated the ability of WRN to bind to PAR of different, well-defined chain length. As is shown in Suppl. Figure 1, PAR of a chain length ranging from 5 to >65 units was size-fractionated using a previously established anion exchange HPLC protocol (29, 30). In a PAR-overlay assay equal amounts of membrane-immobilized WRN were incubated with equal amounts of PAR of defined chain length. Binding of PAR to WRN was observed for all PAR fractions, with a slight tendency for stronger binding of WRN to longer PAR chains (Figures 2B). Nonetheless, PAR chains smaller than 10 units could still bind strongly to WRN, with signal intensities similar to those of unfractionated PAR, indicating that WRN is able to bind even very short PAR chains with high affinity (Figures 2B). A control experiment using XPA showed a strong dependency of PAR-XPA binding on PAR chain length, which confirmed our previous results (Suppl. Figure 2) (29). The finding that some PAR-interacting proteins, such as XPA and DEK, exhibit a strong preference for long PAR chain lengths (11, 29), whereas others, such as WRN and H1, also efficiently bind to short polymer underscores the importance of PAR chain length and probably also branching complexity in the regulation of protein function.
Identification of a PAR-binding motif in the WRN exonuclease domain
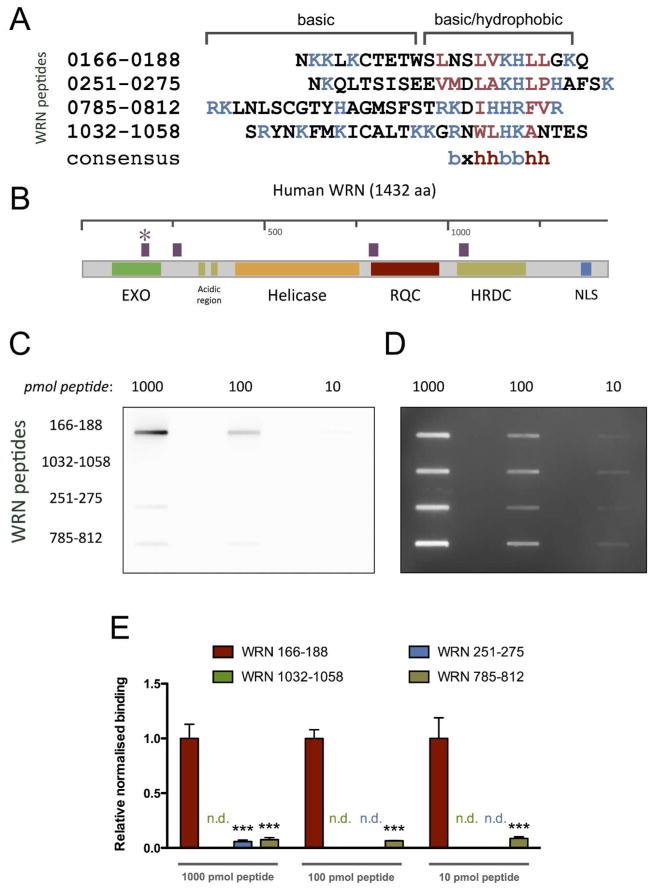
The most frequent feature of proteins that associate with PAR non-covalently is the presence of a weakly conserved PBM (6). This PBM consensus sequence consists of a stretch of basic aa that are separated by hydrophobic aa from a further cluster of N-terminally located basic aa (Figure 3 A). A homology search based on the PBM consensus sequence reported by Pleschke et al. revealed a total of four potential PBMs within the WRN protein (Figure 3A, B). The first PBM is located in the WRN exonuclease domain (aa 166-188), the second in between the exonuclease domain and the acidic region (aa 251-275), the third in the RecQ conserved (RQC) domain (aa 785-812), and the fourth in the helicase and RNase D C-terminal (HRDC) domain (aa 1032-1058) (Figure 3B). These sequences were synthesized as oligopeptides and tested for their ability to bind PAR using a PAR overlay assay. Increasing amounts of each peptide were immobilized on a nitrocellulose membrane and incubated with biotin end-labeled PAR (Figure 1) to detect PAR with high specificity and sensitivity (29). Biotinylated PAR was detected after high-stringency salt washes using streptavidin-coupled horse radish peroxidase (HRP). Peptide ‘166-188’ at the N-terminus of WRN revealed strong PAR-binding (indicated by an asterisk, Figure 3B), whereas weak PAR-binding was observed for peptides ‘251-275’ and ‘785-812’. No or very weak PAR-binding was detected for peptide ‘1032-1058’, further demonstrating the specificity of the PAR binding to peptide ‘166-188’ (Figure 3 C–E). The finding that peptide ‘166-188’ has been identified in an in silico search for putative PBMs in combination with the result that this peptide binds PAR with high affinity provides strong evidence for the existence of a PBM within aa 166-188 of the WRN protein.
Figure 3. WRN exhibits a PAR-binding motif in its exonuclease domain.
A. Alignment of the PAR-binding motif (PBM) consensus sequence with the WRN polypeptide sequence revealed four putative candidates. Basic and hydrophobic aa are depicted in blue and red, respectively. B. Scheme of the WRN protein highlighting functional domains as well as the location of putative PBMs (in purple). Asterisk indicates the PBM that was confirmed experimentally. C. Experimental confirmation of the PBM. WRN peptides ‘166–188’, ‘251–275’, ‘785–812’, and ‘1032–1058’ were tested for their ability to bind PAR, by using a PAR-overlay assay. Peptides were immobilized on a nitrocellulose membrane and incubated with 3 nM biotinylated PAR. PAR was detected using streptavidin-HRP. Peptide ‘166–188’ bound PAR strongly, whereas peptides ‘251–275’ and ‘785–812’ showed weak PAR-binding. No PAR-binding was detected with peptide ‘1032–1058’. D. Peptide loading control using SYPRO Ruby staining E. Densitometric quantification of C. Data represent means ± SEM from three independent experiments. Statistical analysis was performed using ANOVA and Dunnett’s post-test. *** p<0.001; n.d., not detectable; EXO, exonuclease domain; RQC, RecQ C-terminal domain; HRDC, helicase and RNase D C-terminal domain; NLS, nuclear localization signal.
Basic and hydrophobic amino acids in the PBM contribute to PAR binding
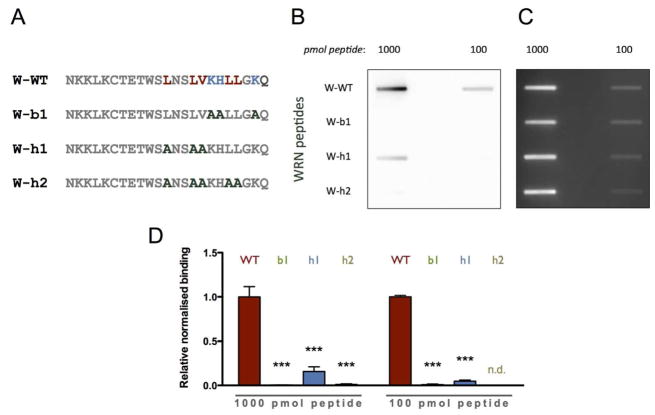
To determine which aa residues within the PBM mediate PAR-binding, peptide ‘166-188’ was subjected to an aa exchange analysis. Both hydrophobic and basic aa have been described to contribute to the specificity of PAR-binding within PBMs (6, 9). To this end, the PBM was mutated consecutively either by changing basic or hydrophobic aa in groups of two or three (Figure 4A). A PAR-overlay assay was performed to test the interaction of the four peptides with PAR (Figure 4). As expected, the wild type (WT) peptide displayed the strongest PAR-binding affinity. PAR-binding was significantly weaker with peptide W-h1, carrying exchanges of three hydrophobic aa to alanine. Peptide W-h2 with exchanges of five hydrophobic aa as well as peptide W-b1 with exchanges of three basic aa displayed no detectable PAR-binding.
Figure 4. PAR-overlay blot with mutant WRN peptides derived from peptide ‘166-188’.
A. Peptide sequences used for PAR binding studies with aminoacid exchanges highlighted in bold. B. Peptides were immobilized on a nitrocellulose membrane in concentrations as indicated and incubated with 3 nM biotinylated PAR. PAR was detected using streptavidin-HRP. The WT PAR binding motif displays the strongest binding to PAR followed by the peptide W-h1, which possesses only minor PAR-binding ability. For both W-h2 and W-b1 binding is strongly reduced. C. Peptide loading control using SYPRO Ruby staining. D. Densitometric quantification of B. Data represent means ± SEM of three independent experiments. Statistical analysis was performed as described in Figure 3. *** p<0.001; n.d.: not detectable.
In summary, we have identified a PBM at aa position 166-188 of the WRN protein. Within this motif, both basic and hydrophobic aa contribute to efficient binding of the polymer, although basic aa appeared to have a slightly greater impact on PAR-binding than hydrophobic ones. Next, we tested three different functional end points of WRN activity, i.e., DNA binding properties as well as WRN’s enzymatic activities to analyze potential functional consequences of the WRN-PAR interaction.
PAR inhibits WRN-DNA interaction
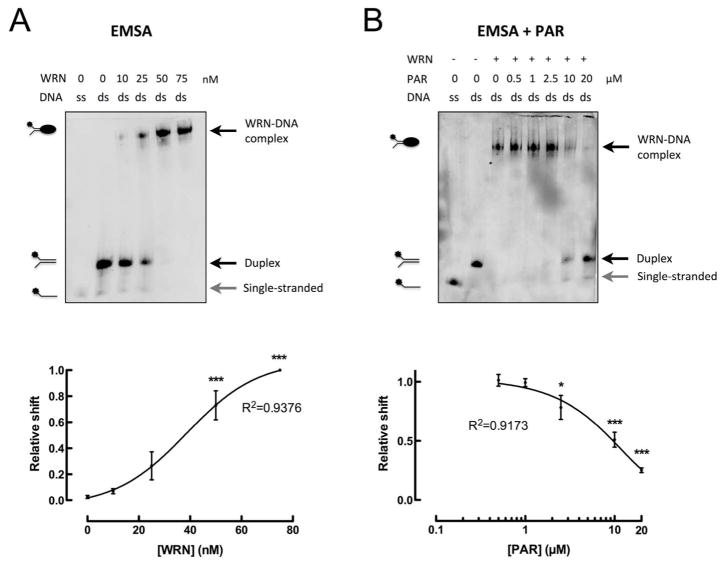
For WRN-DNA interaction studies, we chose a commonly used DNA oligoduplex substrate which comprises a forked structure carrying a 5′-biotin end label to allow detection via streptavidin-HRP. As shown previously such a DNA structure can be potentially formed during processes such as transcription, replication and at telomeric ends and serves as a good substrate for WRN, because it can be recognized as ssDNA, dsDNA, a DSB, or a forked structure (15). Binding of full-length WRN to this oligoduplex was analyzed using an electrophoretic mobility shift assay (EMSA). As it is evident from Figure 5A, WRN efficiently bound to the oligoduplex in a concentration-dependent manner as expected. An almost complete and highly significant shift was observed at a molar ratio of WRN:oligoduplex of 2.5:1, where even residual single-stranded DNA (from incompletely annealed oligoduplexes) is bound by the protein (Figure 5A). To analyze if the WRN-PAR interaction interferes with WRN-DNA binding, we pre-incubated WRN with increasing concentrations of PAR before adding the oligoduplex. As no major influence of PAR chain length on its WRN-binding affinity was observed, EMSA experiments were performed with unfractionated PAR representing a mixture of short and long chains ranging from 1 to over 200 units, with an estimated mean chain length of 100 ADP-ribosyl units (PAR100mer). Figure 5B demonstrates that with increasing concentrations of PAR fewer WRN-DNA complexes were formed. At a concentration of 10 μM PAR (PAR concentration refers to monomeric ADP-ribose if not stated otherwise) a highly significant reduction of the electrophoretic shift was observed to ~50% compared to control (2:1 molar ratio PAR100mer:WRN). Maximum reduction of the electrophoretic shift by ~75% was observed at a PAR concentration of 20 μM (4:1 molar ratio of PAR100mer:WRN) (Figure 5B).
Figure 5. WRN-PAR interaction inhibits binding of WRN to DNA.
A. Upper panel: Increasing concentrations of WRN were incubated with 200 fmol of biotin-labeled forked oligoduplex. A WRN-dose-dependent shift of the oligoduplex is indicative of WRN-DNA binding. Lower panel: Densitometric quantification of three independent experiments (means ± SEM). B. Upper panel: Effect of PAR on the WRN-DNA interaction. WRN (50 nM) was pre-incubated with increasing concentrations of PAR. 10 μM PAR correspond to a 2:1 molar ratio of PAR100mer:WRN. The presence of PAR inhibited the formation of the WRN-DNA complex in a dose-dependent manner. Lower panel: Densitometric quantification of three independent experiments (means ± SEM). Statistical analysis was performed using ANOVA and Dunnett’s multiple comparison test, *** p<0.001. Curves were fitted using a sigmoidal dose-response curve with variable slope. Single-stranded oligonucleotide (grey arrow), duplex oligonucleotide and WRN-DNA complex (black arrow) are indicated. ds, double-stranded biotinylated oligonucleotide; ss, single-stranded biotinylated oligonucleotide.
These results demonstrate a direct functional effect of free PAR by interfering with WRN’s DNA-binding ability. WRN harbors three DNA-binding domains with distinct substrate specificities (15). One of these DNA-binding domains is located N-terminally within the exonuclease domain, the other two C-terminally, i.e., within the RQC and helicase domains. Because all three of these DNA-binding domains bind to forked substrates, as used in this study, with high affinity, we speculate that steric and/or structural changes within the WRN protein are responsible for the reduced WRN-DNA interaction upon PAR-binding. Furthermore, PAR may compete with the DNA substrate for WRN-binding via WRN’s N-terminal DNA-binding domain which overlaps with the N-terminal PBM. Conversely, it is possible that DNA is able to dissociate PAR from WRN. In this respect, it was shown previously that PAR and DNA compete for PAR binding sites in histone H1 at a high molar excess of DNA (3, 6). Because WRN shows a weaker PAR binding compared to histone H1 (Figure 2A), it is therefore likely that at high concentrations DNA is able to dissociate PAR from WRN as well. If this is of any physiological relevance remains to be elucidated.
PAR inhibits WRN’s helicase and exonuclease activities
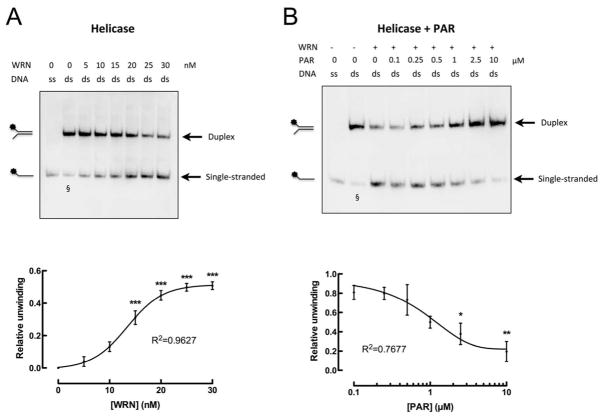
To studied if the WRN-PAR interaction affects WRN’s enzymatic functions, first a WRN helicase assay was developed based on a previously published protocol (31). Instead of using a radioactively end-labeled substrate, a non-radioactive biotin-labeled forked DNA oligoduplex was used. The oligoduplex was incubated with increasing concentrations of WRN, separated by native PAGE, immobilized on a nylon membrane, and detected via streptavidin-HRP. As expected, a dose-dependent unwinding of the substrate was observed (Figure 6A). At a molar ratio of WRN:oligoduplex of 0.75:1 significant formation of single-stranded DNA was observed that reached saturation at a molar ratio of WRN:oligoduplex of 1.25:1 (Figure 6A). When WRN was preincubated with increasing concentrations of PAR before adding the oligoduplex substrate, WRN’s unwinding activity was inhibited in a concentration-dependent manner (Figure 6B). PAR significantly inhibited DNA unwinding by 62% at a concentration of 2.5 μM (1:1 molar ratio of PAR100mer:WRN). A maximum inhibitory effect of 80% was observed at 10 μM PAR (4:1 molar ratio of PAR100mer:WRN) (Figure 6B).
Figure 6. WRN-PAR interaction inhibits WRN’s helicase activity.
A. WRN helicase assay. Upper panel: Unwinding of a forked oligoduplex in a WRN-concentration-dependent manner. Lower panel: Densitometric quantification from three independent experiments (means ± SEM). Reactions were performed at 37°C for 20 min. B. Impact of PAR binding on WRN’s helicase activity. Upper panel: Recombinant WRN (25 nM) was pre-incubated with increasing concentrations of PAR as indicated. Helicase reaction was started by addition of the biotin-labeled oligoduplex. The presence of PAR inhibited WRN’s helicase activity in a concentration-dependent manner. Lower panel: Densitometric quantification of three independent experiments (means ± SEM). 10 μM PAR correspond to a 4:1 molar ratio of PAR100mer:WRN. § indicates single-stranded DNA due to incomplete annealing of oligonucleotides. This signal was background-subtracted in quantitative analyses. Statistical analysis and curve fitting were performed as described in Figure 5. * p<0.05; ** p<0.01; *** p<0.001.
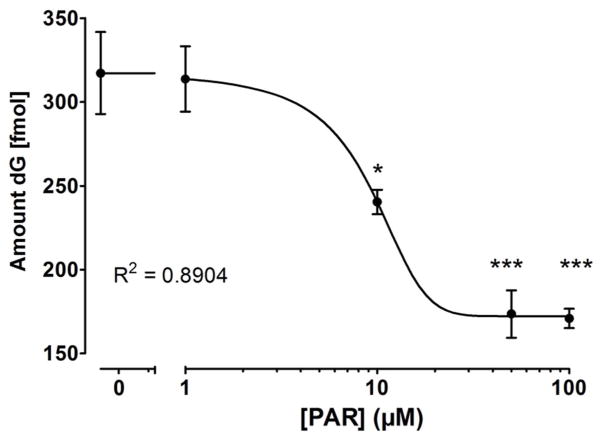
As the WRN-PBM is located in the exonuclease domain, we examined a potential impact of PAR on WRN’s exonuclease function using a recently developed method to quantify WRN’s exonuclease activity (32). In this method WRN’s exonuclease activity is analyzed by detecting the release of free deoxyguanosine (dG) via isotope dilution mass spectrometry (LC-MS/MS). A forked oligoduplex mimicking the telomeric repeat sequence was used as a substrate. A concentration-dependent reduction in WRN’s exonuclease activity was observed in the presence of PAR (Figure 7). A significant inhibition of exonuclease activity by ~25% was observed at a PAR concentration of 10 μM (2.5:1 molar ratio of PAR100mer:WRN). Maximum inhibition of exonuclease activity of ~45% was observed with PAR concentrations >50 μM. These results were confirmed using a classical WRN exonuclease activity assay by detection of a 5′-biotin-end-labeled oligonucleotide after electrophoretic separation under denaturing conditions (Suppl. Figure 3)
Figure 7. WRN-PAR interaction inhibits WRN’s exonuclease activity.
WRN (40 nM) was pre-incubated with increasing concentrations of PAR. Exonuclease reaction was started by addition 75 fmol of a forked oligoduplex. Exonuclease reaction was carried out at 37°C for 45 min, subsequently the mixture was placed on ice, 15N-labeled dG was added to samples as an internal standard to account for technical variability, and nucleotides were dephosphorylated by alkaline phosphatase. After removal of enzymes, free deoxguanosine (dG), which directly correlates with WRN’s exonuclease activity, was quantified via isotope dilution LC-MS/MS. 10 μM PAR correspond to a 2.5:1 molar ratio of PAR100mer:WRN. Statistical analysis and curve fitting were performed as described in Figure 5. * p<0.05; *** p<0.001.
In summary, our results demonstrate that WRN-PAR interaction significantly interferes with WRN’s helicase and exonuclease functions in vitro. The effect of PAR on WRN’s exonuclease and helicase activities may be induced by conformational changes upon PAR-binding leading to allosteric inhibition of the enzyme or by the reduced DNA-binding ability of WRN upon PAR-binding. The effect of PAR on WRN’s exonuclease activity is conclusive, considering that the PBM as identified in this study is located in this domain. Notably, molar ratios of WRN and PAR as used in this study are in agreement with physiological ratios. Previous studies estimated that cells contain ~60,000 WRN and ~45,000 PAR100mer molecules, respectively, under conditions of genotoxic stress (33, 34). Furthermore, PAR production in cells is highly controlled in a spatio-temporal manner, e.g. at sites of DNA damage, potentially resulting in very high local concentrations.
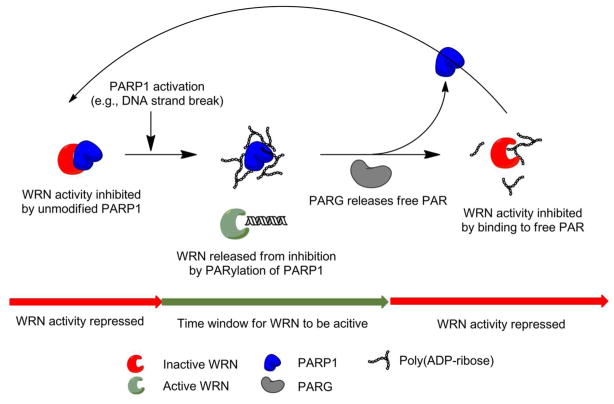
It is important to note that WRN activity needs to be tightly controlled in vivo, because uncontrolled WRN activity may cause genomic instability. Von Kobbe et al. showed that the unmodified PARP1 is able to inhibit both catalytic activities of WRN, while enzymatic activation of PARP1 released WRN from its enzymatic inhibition (23). Considering our finding that PAR itself controls WRN activity, it is tempting to speculate that the transient nature of PAR produced by living cells in response to DNA strand breakage and specific DNA structures like four-way junctions would provide a versatile and swiftly reacting control system for WRN’s function (Figure 8). In such a scenario, under physiological conditions unmodified PARP1 would inhibit WRN by ‘taming’ its exonuclease and helicase activities. Upon a genotoxic stimulus, e.g. a DNA strand break, PARP1 is PARylated and WRN is released from its repression, opening a time window during which WRN can take action on its DNA substrates. Shortly thereafter, PARG releases free PAR leading to a non-covalent WRN-PAR complex which shuts down WRN activity, as we have shown in the present work.
Figure 8. Potential molecular mechanism for temporally controlled regulation of WRN activity by PAR and PARP1.
The model summarizes the results from the present study and Ref (23). For details see Text.
Thus, the WRN-PAR complex may represent an intermediate state to control WRN activity until unmodified PARP1 is fully reconstituted and able to take over the function of repressing WRN activity. Because PARP1 and WRN are both multifunctional proteins, additional factors are presumably involved in vivo.
The physiological relevance of a functional regulation of WRN has impressively been demonstrated in a recent study which characterized the PAR-associated proteome in response to alkylating DNA-damage-mediated PARP activation (35). This study identified WRN as factor that is strongly modified by PAR upon DNA damage. Whether this occurs in a covalent or non-covalent manner remains to be clarified.
In conclusion, by using an array of bioanalytical approaches we provide new insight into the regulation of the multifunctional WRN protein via non-covalent interaction with the nucleic-acid-like biopolymer PAR. Results from this study and other recent findings provide evidence that PARP1 and its enzymatic product PAR work cooperatively to modulate WRN activity in a spatio-temporal manner. This may have important implications in DNA metabolism and genomic maintenance, aging as well as cancer biology. Notably, inhibitors of PARP catalytic activity are currently being tested in cancer therapy either as radio- or chemosensitizers or as stand-alone drugs following the concept of synthetic lethality (36). Overall, this work exemplifies how the non-covalent interaction of a protein, e.g., WRN with a scaffold and signaling molecule like PAR can mediate efficient regulation of protein functionality.
MATERIAL AND METHODS
Sequence alignment of PAR-binding motifs
In silico alignment was performed using the PattInProt motif search tool (http://npsa-pbil.ibcp.fr/cgi-bin/npsa_automat.pl?page=npsa_pattinprot.html) with the algorithm [HKR]-X-[AVILFWP]-[AVILFWP]-[HKR]-[HKR]-[AVILFWP]-[AVILFWP] allowing one mismatch, according to Pleschke et al. (6).
Expression and purification of human His-WRN
Human His-WRN was overexpressed in Sf9 cells and purified as described (32). WRN oligopeptides were custom-synthesized by Genscript.
Synthesis and purification of PAR
Human PARP1 was expressed in Sf9 cells and purified as described (29, 37). Synthesis of PAR was performed as described (29). Briefly, 75 nM of recombinant PARP1 was incubated in a mixture containing Tris-HCl pH 7.8 (100 mM), MgCl2 (10 mM), DTT (1 mM), histone H1 (60 μg/ml), histone H2a (180 μg/ml), EcoRI linker [5′-GGAATTCC-3′] (50 μg/ml), βNAD+ (1 mM) for 20 min at 37°C. The reaction was stopped by adding ice-cold trichloroacetic acid (TCA) to a final concentration of 10% (w/v). After precipitation and centrifugation at 9000 × g for 10 min at 4°C, the pellet was washed twice with ice-cold ethanol. PAR was detached from proteins by incubation in 0.5 M KOH and 50 mM EDTA for 10 min at 37°C. After adjustment of pH to 7.5, DNA and proteins were digested using DNase I [200 μg/ml] and proteinase K [100 μg/ml]. PAR was finally purified by phenol-chloroform extraction and ethanol precipitation.
Biotinylation of PAR and HPLC fractionation
Biotinylation and preparative anion-exchange HPLC fractionation was performed as described (29). Briefly, in-vitro-synthesized and purified PAR was incubated with 4 mM biocytin hydrazide under reductive amination conditions in sodium acetate buffer pH 5.5 for 8 h at RT. Samples were dialyzed and PAR was precipitated using ethanol. Biotinylated PAR was size-fractionated using a Shimadzu LC-8A HPLC system with a semi-preparative DNA Pac PA100 column (Dionex). PAR fractions were eluted using a multistep NaCl gradient in 25 mM Tris-HCl pH 9.0 [modified from (30)]. PAR fractions were ethanol-precipitated, dissolved in water, concentration determined via absorption at 258 nm, and characterized on a silver-stained sequencing gel (GELCODE Color silver stain, Pierce).
Binding of immobilized proteins and peptides to PAR (PAR-overlay blot)
Proteins were separated by 8% SDS-PAGE and immobilized on a nitrocellulose membrane (GE Healthcare). Peptides were immobilized directly by slot-blotting. Subsequently, membranes were incubated with PAR as indicated in TBST buffer for 1 h at RT before unspecific binding was removed by high-stringency washes using 1 M NaCl in TBST (3 times for 5 min at RT). Blots were blocked in 5% (w/v) milk powder and bound PAR was detected using 10H antibody or streptavidin-HRP (GE Healthcare).
Oligonucleotides
Oligonucleotides were purchased from Sigma-Aldrich. For WRN helicase assays and EMSAs 200 fmol of 5′-biotin-(TTT)5-GAGTGTGGTGTACATGCACTAC-3′ oligonucleotide was annealed with 400 fmol of 5′-GTAGTGCATGTACACCACACTC-(TTT)5-3′ complementary oligonucleotide by heating to 95°C for 5 min in TE buffer supplemented with 50 mM NaCl followed by cooling to RT. For exonuclease assays, 5′-biotin-(TTT)5-(TTAGGG)4-CATGCACTAC-3′ oligonucleotide was annealed with 5′-GTAGTGCATG-(CCCTAA)4-(TTT)5-3′ complementary oligonucleotide. Annealing of the oligonucleotides was confirmed via 20% TBE-PAGE followed by semi-dry TBE-blotting. After immobilizing on nylon membranes (GE Healthcare) biotin-labeled oligonucleotides were detected via streptavidin-HRP.
Electophoretic mobility shift assay (EMSA)
WRN (in concentrations as indicated) was incubated with 200 fmol of the annealed oligoduplex duplex in EMSA buffer containing 40 mM Tris-HCl pH 8.0, 4 mM MgCl2 0.1 mg/ml BSA, 5 mM DTT and 0.1% (v/v) Nonidet P40 for 30 min at 4°C in a total reaction volume of 10 μl. To test the influence of PAR on WRN-DNA binding, PAR was added in concentrations as indicated and mixtures incubated for 20 min at RT prior to the addition of oligoduplex. After addition of 4 μl loading dye [40% (v/v) glycerol, 0.05% (w/v) Orange G and 0.05% (w/v) bromophenol blue], DNA and DNA-WRN complexes were separated via 4% TBE-PAGE and detected as described above.
WRN helicase assay
WRN was incubated (at concentrations as indicated) in helicase reaction buffer containing 30 mM HEPES-KOH pH 7.4, 5% (v/v) glycerol, 40 mM KCl, 0.1 mg/ml BSA, 1 mM MgCl2, and 1 mM ATP in a total volume of 10 μl. To test the influence of PAR on the WRN helicase activity, PAR was added as indicated and mixtures were incubated for 20 min at RT prior to the addition of oligoduplex. Helicase reaction was carried out for 20 min at 37°C and was stopped by the addition of 4 μl stop dye [1% (w/v) SDS, 40% (v/v) glycerol, 50 mM EDTA, 0.05% (w/v) Orange G, 0.05% (w/v) bromophenol blue and 0.05% (w/v) xylene cyanol]. Afterwards oligonucleotides were separated by 12% TBE-PAGE and detected as described above.
WRN exonuclease assay
The assay was carried out as described recently (32). Briefly, 40 nM WRN was incubated in a reaction buffer containing 40 mM Tris-HCl pH 8.0, 4 mM MgCl2, 0.1 mg/ml BSA and 4 mM DTT together with 75 fmol oligonucleotide substrate and concentrations of PAR as indicated for 45 min at 37°C in a total volume of 10 μl. After completion of the reaction, the mixture was immediately transferred on ice, and 90 μl of 30 mM sodium acetate (pH 7.8) and 2 pmol of 15N-labeled-dG (internal standard) were added to the mixture before filtering through a 10-kDa cut-off spin column filter (Pall). Subsequently, nucleotides were dephosphorylated by incubation with 34 U alkaline phosphatase (Sigma-Aldrich) at 37°C overnight. Samples were filtered through a 10-kDa cut-off spin column filter and subjected to LC-MS/MS analysis using a Waters LC-MS/MS system consisting of an HPLC 2695 Separations Module and a Micromass Quattro Micro mass spectrometer equipped with an electrospray ionization source. Samples were resolved using a Hypersil Gold column (150 mm × 2.1 mm; 3 μm particle size; Thermo Fisher) under isocratic conditions with a solvent composition of 97% of 0.1% acetic acid in water and 3% of 0.1% acetic acid in acetonitrile at a flow rate of 0.2 ml/min and a column temperature of 25°C. The MS was operated in positive ion mode. Multiple reaction monitoring (MRM) mode was used for data acquisition. Response ratios were obtained by plotting the MRM area ratios between the labeled and unlabeled dG against their corresponding concentration ratios.
Supplementary Material
Acknowledgments
We thank M. Malanga for her initial help with in silico analysis. Furthermore we thank M. Miwa (Nagahama, Japan) and T. Sugimura (Tokyo, Japan) for the kind gift of 10H antibody and A. Marx (University of Konstanz) for sharing scientific equipment. This work was supported by the Deutsche Forschungsgemeinschaft (FOR434, Konstanz Research School Chemical Biology [KoRS-CB], and Research Training Group [RTG] 1331) and by the Intramural Program of the National Institute on Aging, National Institutes of Health. O. Popp and S. Veith were supported by fellowships of the KoRS-CB and the RTG 1331, respectively.
Footnotes
ASSOCIATED CONTENT
This material is available free of charge via the Internet at http://pubs.acs.org.
References
- 1.Hottiger MO, Hassa PO, Luscher B, Schuler H, Koch-Nolte F. Toward a unified nomenclature for mammalian ADP-ribosyltransferases. Trends Biochem Sci. 2010;35:208–219. doi: 10.1016/j.tibs.2009.12.003. [DOI] [PubMed] [Google Scholar]
- 2.Rouleau M, Patel A, Hendzel MJ, Kaufmann SH, Poirier GG. PARP inhibition: PARP1 and beyond. Nat Rev Cancer. 2010 doi: 10.1038/nrc2812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Malanga M, Atorino L, Tramontano F, Farina B, Quesada P. Poly(ADP-ribose) binding properties of histone H1 variants. Biochim Biophys Acta. 1998;1399:154–160. doi: 10.1016/s0167-4781(98)00110-9. [DOI] [PubMed] [Google Scholar]
- 4.Althaus FR, Bachmann S, Hofferer L, Kleczkowska HE, Malanga M, Panzeter PL, Realini C, Zweifel B. Interactions of poly(ADP-ribose) with nuclear proteins. Biochimie. 1995;77:423–432. doi: 10.1016/0300-9084(96)88155-7. [DOI] [PubMed] [Google Scholar]
- 5.Gagne JP, Hunter JM, Labrecque B, Chabot B, Poirier GG. A proteomic approach to the identification of heterogeneous nuclear ribonucleoproteins as a new family of poly(ADP-ribose)-binding proteins. Biochem J. 2003;371:331–340. doi: 10.1042/BJ20021675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Pleschke JM, Kleczkowska HE, Strohm M, Althaus FR. Poly(ADP-ribose) binds to specific domains in DNA damage checkpoint proteins. J Biol Chem. 2000;275:40974–40980. doi: 10.1074/jbc.M006520200. [DOI] [PubMed] [Google Scholar]
- 7.Ahel I, Ahel D, Matsusaka T, Clark AJ, Pines J, Boulton SJ, West SC. Poly(ADP-ribose)-binding zinc finger motifs in DNA repair/checkpoint proteins. Nature. 2008;451:81–85. doi: 10.1038/nature06420. [DOI] [PubMed] [Google Scholar]
- 8.Timinszky G, Till S, Hassa PO, Hothorn M, Kustatscher G, Nijmeijer B, Colombelli J, Altmeyer M, Stelzer EH, Scheffzek K, Hottiger MO, Ladurner AG. A macrodomain-containing histone rearranges chromatin upon sensing PARP1 activation. Nat Struct Mol Biol. 2009;16:923–929. doi: 10.1038/nsmb.1664. [DOI] [PubMed] [Google Scholar]
- 9.Gagne JP, Isabelle M, Lo KS, Bourassa S, Hendzel MJ, Dawson VL, Dawson TM, Poirier GG. Proteome-wide identification of poly(ADP-ribose) binding proteins and poly(ADP-ribose)-associated protein complexes. Nucleic Acids Res. 2008;36:6959–6976. doi: 10.1093/nar/gkn771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.El-Khamisy SF, Masutani M, Suzuki H, Caldecott KW. A requirement for PARP-1 for the assembly or stability of XRCC1 nuclear foci at sites of oxidative DNA damage. Nucleic Acids Research. 2003;31:5526. doi: 10.1093/nar/gkg761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fahrer J, Popp O, Malanga M, Beneke S, Markovitz DM, Ferrando-May E, Bürkle A, Kappes F. High-affinity interaction of poly(ADP-ribose) and the human DEK oncoprotein depends upon chain length. Biochemistry. 2010;49:7119–7130. doi: 10.1021/bi1004365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Malanga M, Pleschke JM, Kleczkowska HE, Althaus FR. Poly(ADP-ribose) binds to specific domains of p53 and alters its DNA binding functions. J Biol Chem. 1998;273:11839–11843. doi: 10.1074/jbc.273.19.11839. [DOI] [PubMed] [Google Scholar]
- 13.Rossi ML, Ghosh AK, Bohr VA. Roles of Werner syndrome protein in protection of genome integrity. DNA Repair (Amst) 2010;9:331–344. doi: 10.1016/j.dnarep.2009.12.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Muftuoglu M, Oshima J, von Kobbe C, Cheng WH, Leistritz DF, Bohr VA. The clinical characteristics of Werner syndrome: molecular and biochemical diagnosis. Hum Genet. 2008;124:369–377. doi: 10.1007/s00439-008-0562-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.von Kobbe C, Thoma NH, Czyzewski BK, Pavletich NP, Bohr VA. Werner syndrome protein contains three structure-specific DNA binding domains. J Biol Chem. 2003;278:52997–53006. doi: 10.1074/jbc.M308338200. [DOI] [PubMed] [Google Scholar]
- 16.Huang S, Beresten S, Li B, Oshima J, Ellis NA, Campisi J. Characterization of the human and mouse WRN 3′-->5′ exonuclease. Nucleic Acids Res. 2000;28:2396–2405. doi: 10.1093/nar/28.12.2396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ahn B, Bohr VA. DNA secondary structure of the released strand stimulates WRN helicase action on forked duplexes without coordinate action of WRN exonuclease. Biochem Biophys Res Commun. 2011;411:684–689. doi: 10.1016/j.bbrc.2011.06.184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Muftuoglu M, Kusumoto R, Speina E, Beck G, Cheng WH, Bohr VA. Acetylation regulates WRN catalytic activities and affects base excision DNA repair. PLoS ONE. 2008;3:e1918. doi: 10.1371/journal.pone.0001918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cheng WH, von Kobbe C, Opresko PL, Fields KM, Ren J, Kufe D, Bohr VA. Werner syndrome protein phosphorylation by abl tyrosine kinase regulates its activity and distribution. Mol Cell Biol. 2003;23:6385–6395. doi: 10.1128/MCB.23.18.6385-6395.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Adelfalk C. Physical and functional interaction of the Werner syndrome protein with poly-ADP ribosyl transferase. FEBS Letters. 2003;554:55–58. doi: 10.1016/s0014-5793(03)01088-3. [DOI] [PubMed] [Google Scholar]
- 21.von Kobbe C, Harrigan JA, May A, Opresko PL, Dawut L, Cheng WH, Bohr VA. Central role for the Werner syndrome protein/poly(ADP-ribose) polymerase 1 complex in the poly(ADP-ribosyl)ation pathway after DNA damage. Mol Cell Biol. 2003;23:8601–8613. doi: 10.1128/MCB.23.23.8601-8613.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lachapelle S, Gagne JP, Garand C, Desbiens M, Coulombe Y, Bohr VA, Hendzel MJ, Masson JY, Poirier GG, Lebel M. Proteome-wide identification of WRN-interacting proteins in untreated and nuclease-treated samples. J Proteome Res. 2011;10:1216–1227. doi: 10.1021/pr100990s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.von Kobbe C, Harrigan JA, Schreiber V, Stiegler P, Piotrowski J, Dawut L, Bohr VA. Poly(ADP-ribose) polymerase 1 regulates both the exonuclease and helicase activities of the Werner syndrome protein. Nucleic Acids Res. 2004;32:4003–4014. doi: 10.1093/nar/gkh721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lebel M, Lavoie J, Gaudreault I, Bronsard M, Drouin R. Genetic cooperation between the Werner syndrome protein and poly(ADP-ribose) polymerase-1 in preventing chromatid breaks, complex chromosomal rearrangements, and cancer in mice. Am J Pathol. 2003;162:1559–1569. doi: 10.1016/S0002-9440(10)64290-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Deschenes F, Massip L, Garand C, Lebel M. In vivo misregulation of genes involved in apoptosis, development, and oxidative stress in mice lacking both functional Werner Syndrome protein and poly(ADP-ribose) polymerase-1. Hum Mol Genet. 2005 doi: 10.1093/hmg/ddi362. [DOI] [PubMed] [Google Scholar]
- 26.Ogata N, Ueda K, Kagamiyama H, Hayaishi O. ADP-ribosylation of histone H1. Identification of glutamic acid residues 2, 14, and the COOH-terminal lysine residue as modification sites. J Biol Chem. 1980;255:7616–7620. [PubMed] [Google Scholar]
- 27.Panzeter PL, Realini CA, Althaus FR. Noncovalent interactions of poly(adenosine diphosphate ribose) with histones. Biochemistry. 1992;31:1379–1385. doi: 10.1021/bi00120a014. [DOI] [PubMed] [Google Scholar]
- 28.Kappes F, Fahrer J, Khodadoust MS, Tabbert A, Strasser C, Mor-Vaknin N, Moreno-Villanueva M, Bürkle A, Markovitz DM, Ferrando-May E. DEK is a poly(ADP-ribose) acceptor in apoptosis and mediates resistance to genotoxic stress. Molecular and cellular biology. 2008;28:3245–3257. doi: 10.1128/MCB.01921-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Fahrer J, Kranaster R, Altmeyer M, Marx A, Bürkle A. Quantitative analysis of the binding affinity of poly(ADP-ribose) to specific binding proteins as a function of chain length. Nucl Acids Res. 2007;35(21):e143. doi: 10.1093/nar/gkm944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kiehlbauch CC, Aboul-Ela N, Jacobson EL, Ringer DP, Jacobson MK. High resolution fractionation and characterization of ADP-ribose polymers. Anal Biochem. 1993;208:26–34. doi: 10.1006/abio.1993.1004. [DOI] [PubMed] [Google Scholar]
- 31.Brosh RM, Jr, Opresko PL, Bohr VA. Enzymatic mechanism of the WRN helicase/nuclease. Methods Enzymol. 2006;409:52–85. doi: 10.1016/S0076-6879(05)09004-X. [DOI] [PubMed] [Google Scholar]
- 32.Mangerich A, Veith S, Popp O, Fahrer J, Martello R, Bohr VA, Bürkle A. Quantitative Analysis of WRN Exonuclease Activity by Isotope Dilution Mass Spectrometry. Mech Ageing Dev. 2012 doi: 10.1016/j.mad.2012.06.005. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Moser MJ, Kamath-Loeb AS, Jacob JE, Bennett SE, Oshima J, Monnat RJ., Jr WRN helicase expression in Werner syndrome cell lines. Nucleic Acids Res. 2000;28:648–654. doi: 10.1093/nar/28.2.648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Juarez-Salinas H, Sims JL, Jacobson MK. Poly(ADP-ribose) levels in carcinogen-treated cells. Nature. 1979;282:740–741. doi: 10.1038/282740a0. [DOI] [PubMed] [Google Scholar]
- 35.Gagne JP, Pic E, Isabelle M, Krietsch J, Ethier C, Paquet E, Kelly I, Boutin M, Moon KM, Foster LJ, Poirier GG. Quantitative proteomics profiling of the poly(ADP-ribose)-related response to genotoxic stress. Nucleic Acids Res. 2012 doi: 10.1093/nar/gks486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mangerich A, Bürkle A. How to kill tumor cells with inhibitors of poly(ADP-ribosyl)ation. Int J Cancer. 2011;128:251–265. doi: 10.1002/ijc.25683. [DOI] [PubMed] [Google Scholar]
- 37.Beneke S, Alvarez-Gonzalez R, Bürkle A. Comparative characterisation of poly(ADP-ribose) polymerase-1 from two mammalian species with different life span. Exp Gerontol. 2000;35:989–1002. doi: 10.1016/s0531-5565(00)00134-0. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.